ABSTRACT
The human roseoloviruses human herpesvirus 6A (HHV-6A), HHV-6B, and HHV-7 comprise the Roseolovirus genus of the human Betaherpesvirinae subfamily. Infections with these viruses have been implicated in many diseases; however, it has been challenging to establish infections with roseoloviruses as direct drivers of pathology, because they are nearly ubiquitous and display species-specific tropism. Furthermore, controlled study of infection has been hampered by the lack of experimental models, and until now, a mouse roseolovirus has not been identified. Herein we describe a virus that causes severe thymic necrosis in neonatal mice, characterized by a loss of CD4+ T cells. These phenotypes resemble those caused by the previously described mouse thymic virus (MTV), a putative herpesvirus that has not been molecularly characterized. By next-generation sequencing of infected tissue homogenates, we assembled a contiguous 174-kb genome sequence containing 128 unique predicted open reading frames (ORFs), many of which were most closely related to herpesvirus genes. Moreover, the structure of the virus genome and phylogenetic analysis of multiple genes strongly suggested that this virus is a betaherpesvirus more closely related to the roseoloviruses, HHV-6A, HHV-6B, and HHV-7, than to another murine betaherpesvirus, mouse cytomegalovirus (MCMV). As such, we have named this virus murine roseolovirus (MRV) because these data strongly suggest that MRV is a mouse homolog of HHV-6A, HHV-6B, and HHV-7.
IMPORTANCE Herein we describe the complete genome sequence of a novel murine herpesvirus. By sequence and phylogenetic analyses, we show that it is a betaherpesvirus most closely related to the roseoloviruses, human herpesviruses 6A, 6B, and 7. These data combined with physiological similarities with human roseoloviruses collectively suggest that this virus is a murine roseolovirus (MRV), the first definitively described rodent roseolovirus, to our knowledge. Many biological and clinical ramifications of roseolovirus infection in humans have been hypothesized, but studies showing definitive causative relationships between infection and disease susceptibility are lacking. Here we show that MRV infects the thymus and causes T-cell depletion, suggesting that other roseoloviruses may have similar properties.
KEYWORDS: herpesviruses, roseolovirus
INTRODUCTION
The study of herpesviruses has been fruitful, not only revealing mechanisms that drive disease but also providing insight into diverse processes, including oncogenesis and immune regulation (1). The Herpesviridae family includes several highly prevalent human pathogens (1–3). While they are related, these viruses have diverse genomes, particularly in their regions flanking conserved central core regions, and display distinct pathophysiologies. Many human herpesviruses belonging to the Alphaherpesvirinae (herpes simplex virus [HSV]), Betaherpesvirinae (human cytomegalovirus [HCMV]), and Gammaherpesvirinae (Epstein-Barr virus [EBV], Kaposi's sarcoma-associated virus [KSHV]) subfamilies have been scrupulously characterized. However, human herpesvirus 6A (HHV-6A), HHV-6B, and HHV-7 (HHV-6A/6B/7) are highly related members of the less-well-characterized Roseolovirus genus of the human Betaherpesvirinae subfamily. HHV-6B is among the most prevalent human herpesviruses, but little is known about its pathogenesis, disease sequelae, or the host immune response (4).
Ninety-five percent of humans are seropositive for either HHV-6A or HHV-6B by 2 years of age (5), and roseoloviruses are associated with many conditions, including encephalitis (6, 7), seizures (8, 9), dermatologic disease (10, 11), and multiple sclerosis (12–15). Epidemiological studies in humans linking HHV-6A/6B/7 to disease are limited because of the high prevalence of infection, leaving very few uninfected individuals for comparison. Furthermore, although many studies have identified correlations between diseases and HHV-6A/6B viremia, causality cannot be definitively proven, as the alternative interpretation that viremia and disease are caused by a common trigger remains. Thus, better experimental systems that allow for controlled study of roseolovirus biology may yield improved insight into a number of human diseases.
Tractable tools that allow for the study of herpesviruses in vitro and in animal models have been essential to understanding their mechanisms of pathogenesis. Many herpesviruses display species-specific tropism, which prevents direct study of important human pathogens, such as HCMV, EBV, KSHV, HHV-6A, HHV-6B, and HHV-7. However, in the cases of HCMV and EBV/KSHV, there exist the related rodent viruses murine cytomegalovirus (MCMV) and murine gammaherpesvirus 68 (γHV68), respectively, which have been useful in broadening insight into the diseases caused by these viruses. MCMV and γHV68 are clearly related to their human counterparts by sequence homology. Moreover, they share common mechanisms of pathogenesis and their homologous genes (3, 16, 17) exhibit similar functions. For instance, MCMV infection recapitulates most aspects of HCMV disease, with the exception of congenital infection due to the lack of placental infection (18). Similarly, a better understanding of HHV-6A/6B/7 pathophysiology would be aided by mouse homologs, but none have been previously identified.
Here we describe genome sequencing of a murine herpesvirus with properties that resemble those of murine thymic virus (MTV, murid herpesvirus 3), a previously identified virus (19). Genome analysis showed that it is a mouse betaherpesvirus most closely related to human herpesviruses 6A, 6B, and 7, indicating that the virus is a murine roseolovirus. Despite similarities between this roseolovirus and MTV, we were unable to verify that our virus stocks and MTV were derived from a common source. Furthermore, the genome of MTV was never published, preventing genetic comparisons. As such, we have named this agent murine roseolovirus (MRV).
RESULTS
MRV is a murine herpesvirus related to human roseoloviruses.
Infection with our virus stocks showed thymic necrosis and depletion of T cells in the thymus and spleen in BALB/c neonates (Fig. 1A and B). Depletion was evident in the CD4+ single positive (CD4 SP) subsets in the thymus, as previously reported for MTV (20). We observed reductions in double positive (CD4+ CD8+, DP) thymocytes, and both major T-cell subsets (CD4+ and CD8+) in the spleen. Transmission electron micrographic (TEM) images of cells from infected thymi revealed virus-like particles in most cells (Fig. 1C). Similar to the findings in prior publications on MTV (19), these hyperdense virions measured approximately 150 nm and were often found in concentric circles surrounding double-membrane cytoplasmic vesicles. In contrast, images obtained from age-matched control mice had no visible virus-like particles. While these phenotypes were also observed after MTV infection, the absence of freely available sequence information for MTV and our inability to directly link our virus stock to MTV prevented us from concluding that this virus and MTV are the same.
FIG 1.
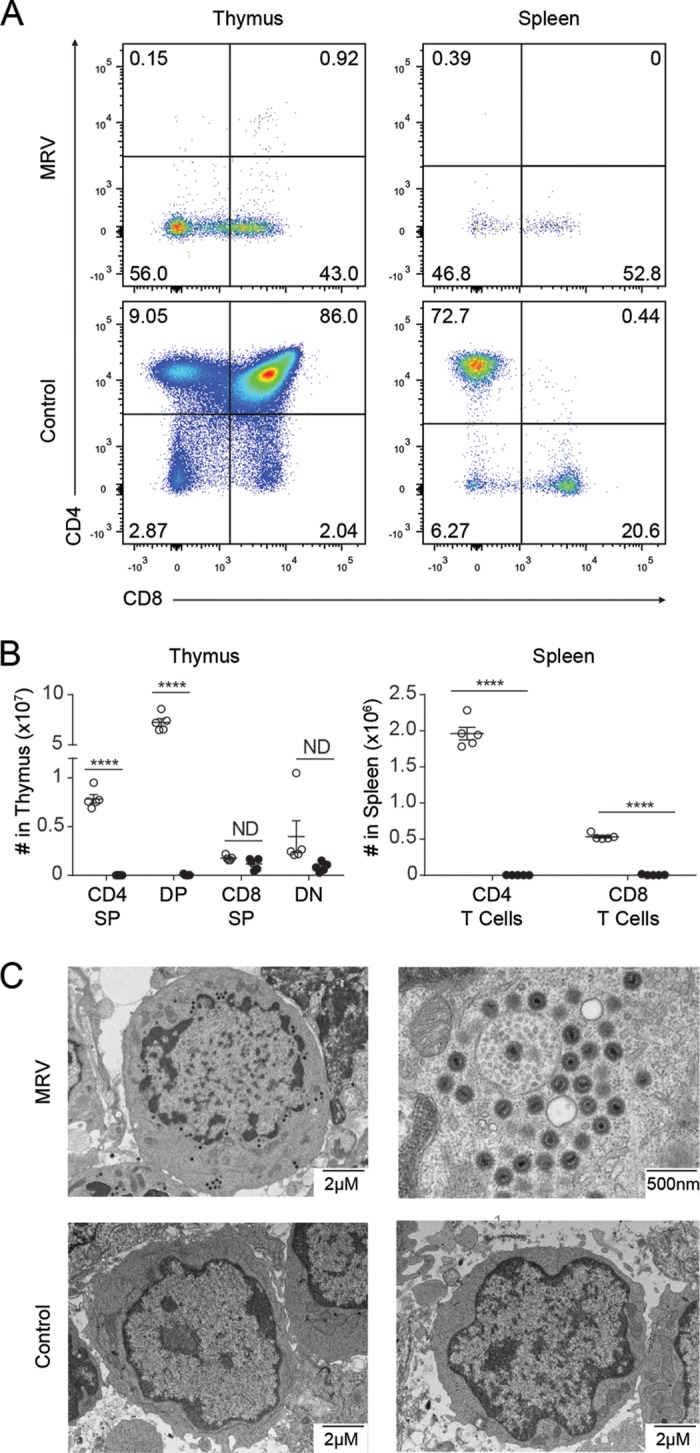
MRV infection depletes thymic and peripheral CD4+ cells, and virus particles are observable in thymi of infected mice. (A and B) Flow cytometry on thymus and spleen 10 days postinfection with MRV. Thymic cells were gated on live lymphocytes, and spleen cells were gated on live, CD3+ lymphocytes, with dot plots (A) and absolute quantification (B) with each point representing a single mouse. Open circles represent uninfected control mice; filled circles represent infected mice. Results for thymocyte subsets, CD4 SP (single positive CD4+), DP (CD4+ CD8+), CD8 SP (single positive CD8+), DN (CD4− CD8−), and splenic subsets are shown. (C) Transmission electron microscopy (TEM) of infected and uninfected thymocytes. Two different cells from an MRV-infected mouse are shown at two different resolutions (with the higher resolution of a cell with many virions) along with two different thymocytes from an uninfected control mouse. Data are representative of three or more independent experiments. ****, P < 0.0001 (unpaired t test).
We determined the primary genome sequence of this virus to assess its phylogeny. Since our virus, like MTV, has not been grown in tissue culture and a cloned virus is not available, DNA was harvested from the thymus of a BALB/c neonate at 7 days postinfection. We generated libraries using Pacific Biosciences (PacBio) RS II sequencing and Illumina MiSeq sequencing in tandem to obtain both long, low-fidelity reads and short, high-fidelity reads, respectively. After excluding reads that mapped to the reference mouse genome, we performed assembly using PacBio sequencing data and Illumina sequencing data separately. We then used the long contigs obtained from PacBio sequencing data assembly as a scaffold to anchor contigs assembled from high-fidelity Illumina sequencing data. The analysis resulted in an initial unambiguous single contiguous 156-kb contig (Fig. 2). We extended the sequence through anchoring of our known ends and searching through the PacBio and Illumina sequencing for reads and contigs extending beyond our assembled genome and were able to ultimately extend the genome to 174 kb, reaching the terminal direct repeats (DRs) on both ends of the genome. For single-nucleotide differences between the PacBio contig and the Illumina contig, we adopted the Illumina sequence because of the known propensity for errors to arise in PacBio data. There were 10 regions that were covered only by PacBio contigs and one region where Illumina data disagreed with PacBio data. We performed PCR amplification and Sanger sequencing of those regions and used Sanger sequencing data in the final genome. We analyzed the variability of the virus genome by remapping the high-fidelity Illumina reads onto the consensus reference genome. Although we suspected that the genome from an infected thymus would contain heterogeneity because the virus had not been previously cloned, we found that the genome had very little heterogeneity (Fig. 3), allowing for the establishment of the consensus sequence. Most differences were located near genome termini, with some positions having a variant frequency exceeding 0.5, which we attribute to multiple alleles contributing less than 50% of the reads mapped to those sites. In these cases, we assigned the nucleotide most frequently found to that site in the consensus sequence which may be refined once an MRV clone is sequenced.
FIG 2.
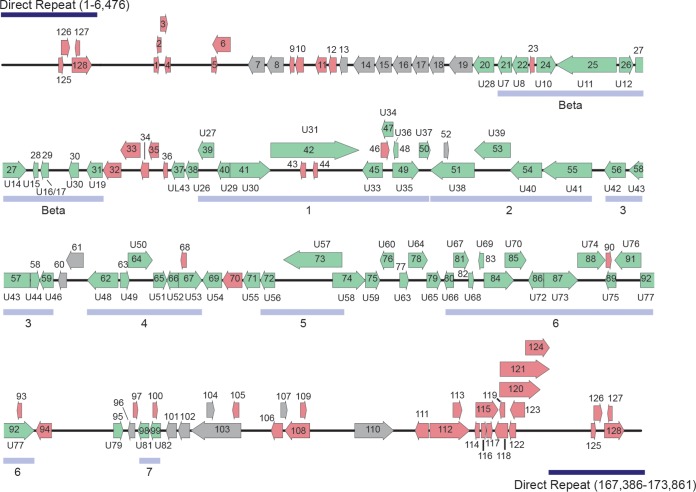
Linearized schematic diagram of MRV genome. The MRV genome is represented as a black line, and predicted ORFs are represented by arrows indicating the direction of transcription. ORFs predicted to be closely related to HHV-6/7 are green, ORFs related to other known proteins are gray, and ORFs with no similarity to NCBI nonredundant database proteins are shown in red. For MRV ORFs related to HHV-6/7, the corresponding HHV-6/7 homolog is listed adjacent to the MRV ORF.
FIG 3.
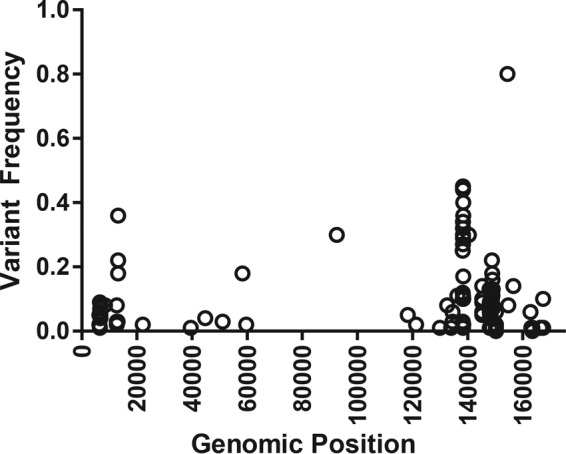
Linearized schematic of the MRV genome plotted against variant frequency. All Illumina sequencing data were remapped onto the MRV genome by using LoFreq.
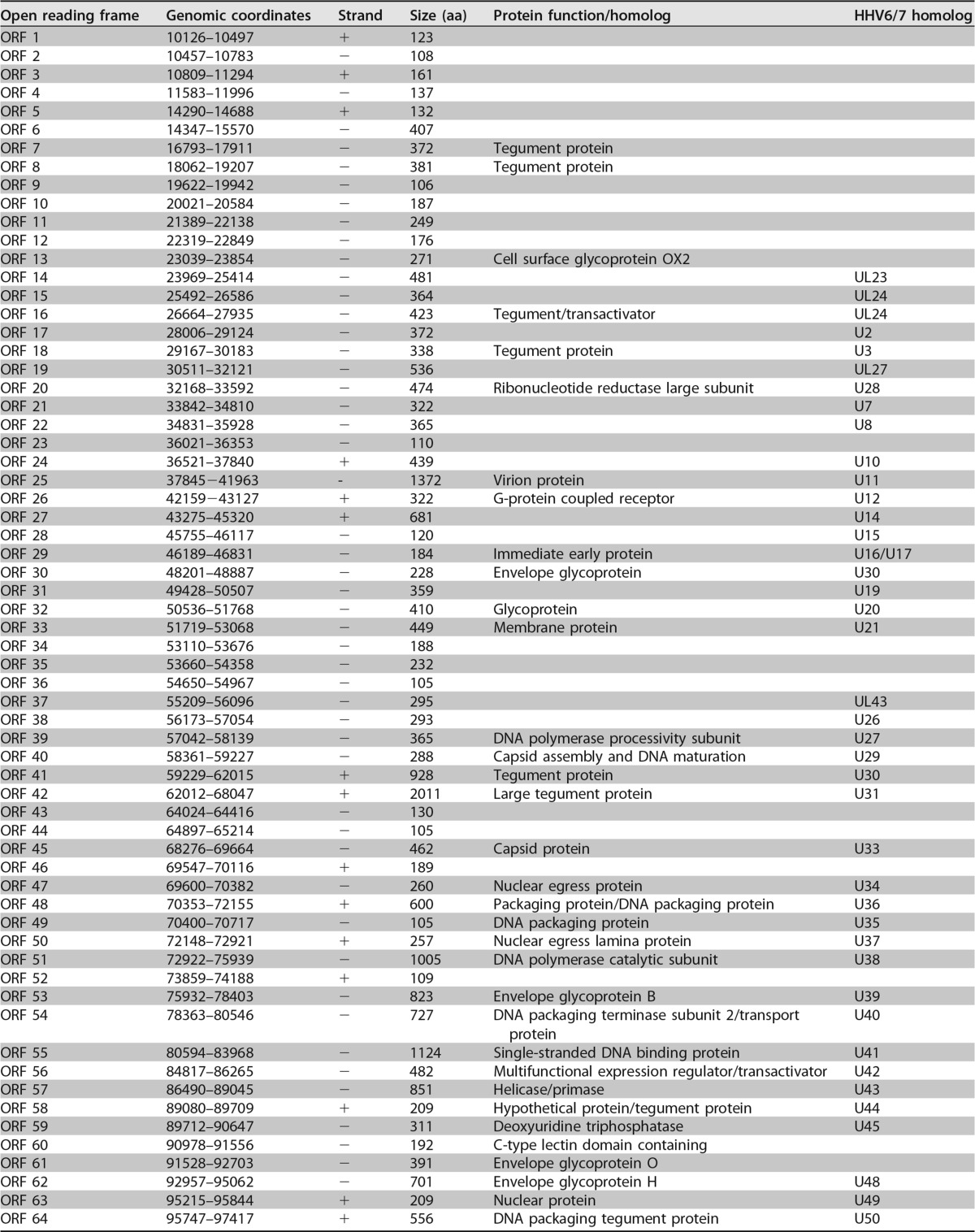
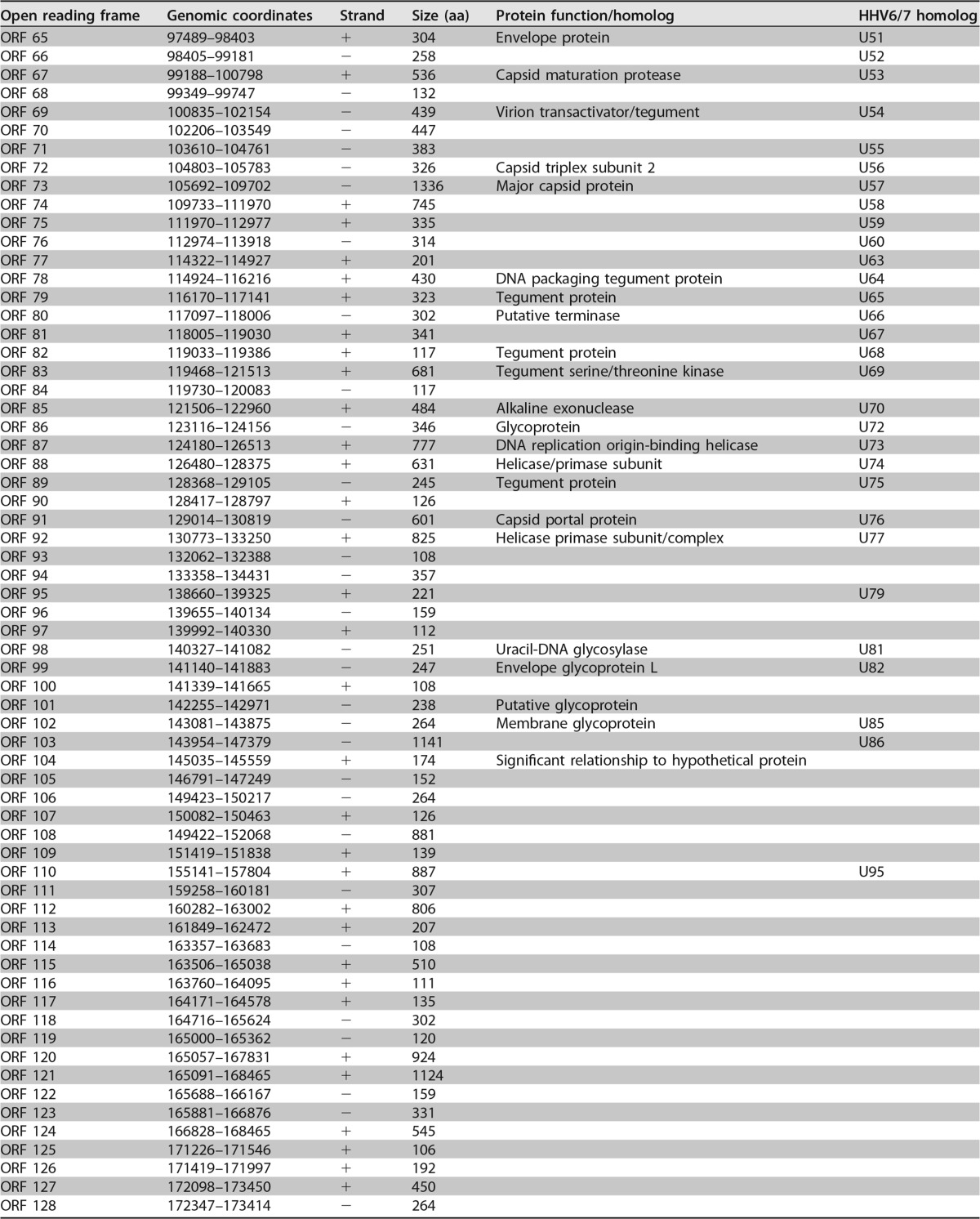
A total of 128 unique open reading frames (ORFs) were identified in silico as defined by the presence of a methionine start codon followed by at least 100 amino acids before a stop codon. Each translated ORF is listed with its length, genomic coordinates, and its most closely related protein in Table 1. Protein homologs were identified by BLASTP analysis of the deduced polypeptide encoded by each individual ORF against the nonredundant database of protein sequences curated by the National Center for Biotechnology Information (NCBI) (Table 1). ORFs predicted in the direct repeat regions of the genome are numbered based on their 3′ positions.
TABLE 1.
Predicted open reading frames of MRVa
ORFs are listed by genome coordinates, strand, size, and predicted function. The ORF nomenclature is based on the corresponding HHV6/7 homolog by BLASTP analysis.
The virus genome resembled a herpesvirus genome in structure. Polypeptide translations of 97 ORFs are related to known proteins, and 65 are closest in sequence similarity to deduced HHV-6A, HHV-6B, or HHV-7 proteins, suggesting that they are homologs of these roseolovirus molecules. Many of the remaining ORFs showed closest similarity to a different herpesvirus species, but they were also significantly similar to deduced HHV-6A/HHV-6B or HHV-7 proteins. As such, we have noted the corresponding HHV-6A/6B/7 homologs (2) for the ORFs in our virus genome (Fig. 2; Table 1).
The virus genome has orthologs of genes in every category of the core herpesvirus genes that encode proteins involved in gene regulation, nucleotide metabolism, DNA replication, and virion assembly and function (Table 1). We have annotated herpesvirus gene blocks 1 through 7 and the betaherpesvirus-specific gene block (1). The genome contains almost all betaherpesvirus signature genes (HHV-6 U27, U29, U31, U34 to U41, U43, U44, U46, U48 to U50, U53, U56, U57, U64 to U67, U70, U72, U74, U76, U77, U82 homologs) (2). While U48A, U80.5, and U99 homologs are missing in the virus genome, these data strongly support classification of this virus as a betaherpesvirus. Long stretches of the genome mimic the corresponding regions of the genomes of HHV-6A/6B/7 with the same gene order. Thus, the genomic architecture of this virus resembles that of betaherpesviruses.
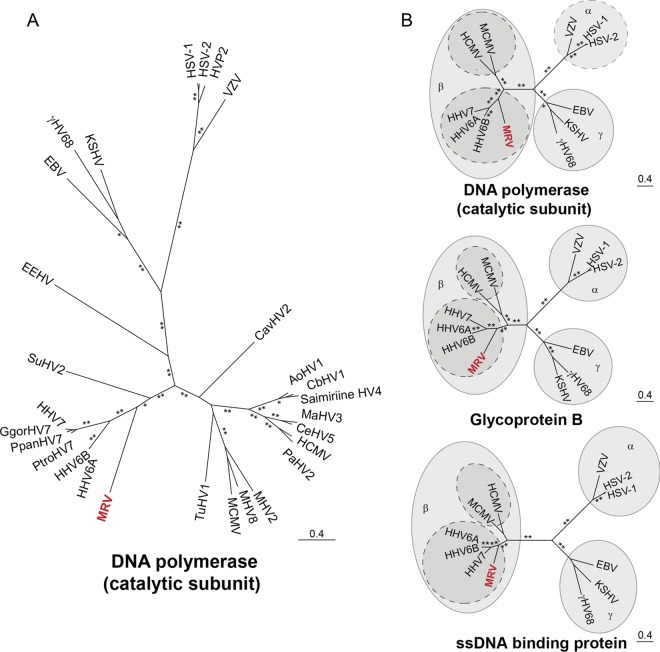
We also performed phylogenetic analyses comparing the catalytic domain of DNA polymerase, glycoprotein B, and viral single-stranded DNA (ssDNA) binding proteins. Maximum likelihood analysis of the catalytic subunit of DNA polymerase genes from all ICTV-annotated betaherpesviruses, NCBI-annotated roseoloviruses, and select mouse and human herpesviruses places the virus into a distinct cluster with betaherpesviruses, particularly with HHV-6A/6B/7 (Fig. 4A). This virus is more distantly related to another murine betaherpesvirus, mouse cytomegalovirus (MCMV). More focused examinations comparing the catalytic domain of DNA polymerase, glycoprotein B, and viral ssDNA binding proteins from the virus genome with corresponding proteins of representative mouse and human herpesviruses from each of the three subfamilies, Alphaherpesvirinae (HSV, varicella-zoster virus [VZV]), Betaherpesvirinae (MCMV, HCMV, HHV-6A, HHV-6B, HHV-7), and Gammaherpesvirinae (γHV68, EBV, KSHV), confirm this classification (Fig. 4B). Collectively, these data indicate that this virus is a murine roseolovirus, and therefore we named it murine roseolovirus (MRV).
FIG 4.
MRV is most closely related to human herpesviruses 6A, 6B, and 7. (A) Phylogeny based on amino acid sequences corresponding to the DNA polymerase catalytic subunit of ICTV-recognized herpesviruses and NCBI-classified roseoloviruses. (B) DNA polymerase catalytic subunit, glycoprotein B, and ssDNA binding protein are compared to those of known select mouse and human herpesviruses. Only bootstrap values above 90 are shown; values above 90 are indicated by a single asterisk, and values of 100 are indicated by two asterisks. Relative genetic distance scale bars are shown above short lines. Abbreviations: HHV-6A, human herpesvirus 6A; HHV-6B, human herpesvirus 6B; HHV-7, human herpesvirus 7; MCMV, murine cytomegalovirus, murid herpesvirus 1; HCMV, human cytomegalovirus, human herpesvirus 5; VZV, varicella-zoster virus, human herpesvirus 3; HSV-1, herpes simplex virus 1, human herpesvirus 1; HSV-2, herpes simplex virus 2, human herpesvirus 2; EBV, Epstein-Barr virus, human herpesvirus 4; KSHV, Kaposi's sarcoma-associated herpesvirus, human herpesvirus 8; γHV68, murine gammaherpesvirus 68, murid herpesvirus 4; AoHV1, aotine herpesvirus 1; CbHV1, cebine herpesvirus 1; CeHV5, cercopithecine herpesvirus 5; MaHV3, macacine herpesvirus 3; PaHV2, panine herpesvirus 2; HVP2, papiine herpesvirus 2; saimiriine HV4, saimiriine herpesvirus 4; MHV2, murid herpesvirus 2, rat cytomegalovirus; MHV8, murid herpesvirus 8; EEHV, elephantid herpesvirus 1; CavHV2, caviid herpesvirus 2; SuHV2, suid herpesvirus 2; TuHV1, tupaiid herpesvirus 1; GgorHV7, Gorilla gorilla herpesvirus 7; PpanHV7, Pan paniscus herpesvirus 7; PtroHV7, Pan troglodytes herpesvirus 7.
MRV contains novel ORFs, including potential immune evasion molecules.
Closely related herpesviruses often display distinct species tropism; for example, the related viruses HCMV and MCMV are capable of replicating only in human or mouse cells, respectively (1, 3). While their genomes are highly related, the terminal ends display the most variability because they encode proteins that interact specifically with molecules in their respective hosts. We conducted a prediction of deduced MRV ORF protein structure and function by Phyre2 and I-TASSER analysis to supplement the primary sequence homology-based BLASTP analysis.
Two MRV ORF proteins appear to be related to major histocompatibility complex (MHC) class I-like proteins. Phyre2 analyses showed that predicted proteins from ORF 32 and ORF 94 likely have immunoglobulin-like folds, and I-TASSER predicted that they were MHC class I-like proteins. Inasmuch as MHC class I-like molecules have diverse roles in herpesvirus biology, serving as immunomodulatory proteins in immune evasion, we predict similar functions for the MRV deduced proteins (21, 22).
ORF 60 is predicted to encode a protein that structurally resembles a C-type lectin protein and has 30% sequence identity to mouse C-type lectin domain family 2, member e (CLEC2e). Although there is no known function for CLEC2e, the CLEC family of proteins consists of Ca2+-dependent carbohydrate-binding proteins with a broad array of functions, including modulation of cell adhesion, signaling, and immune regulation (23, 24). Other related lectin-like molecules are functionally important natural killer cell receptors which can also recognize lectin-like ligands (25, 26). Thus, ORF 60 may modulate the function of these mammalian receptors or ligands.
Given the phenotype observed during MRV infection, we hypothesized that various viral proteins may specifically disrupt T-cell biology. ORF 69 is predicted to encode a homolog to HHV-6/7 protein U54. In HHV-6B, U54 inhibits expression of interleukin 2 (IL-2), an important growth factor for T-cell homeostasis (27), by preventing the dephosphorylation of NFAT (nuclear factor of activated T cells), thereby blocking its downstream transcriptional activation of the IL-2 gene (28). HHV-6A U24 is a protein that downregulates CD3, which signals downstream of the T-cell receptor (29). The MRV genome does not appear to have a U24 homolog at the corresponding position. However, the genomic interval between the U19 homolog and the U26 homolog contains six predicted ORFs, five of which encode novel proteins. Some of these ORFs may contain genes providing a similar function in MRV.
Herpesviruses encode various proteins modulating cell death pathways, including apoptosis and necroptosis (30–32). HHV-6B expresses U19, a protein known to impair p53-mediated apoptosis. MRV ORF 31 is predicted to encode a homolog of U19.
Innate immune sensors recognize viral nucleic acids and activate pattern recognition receptors, leading to transcription of type I interferons (IFNs) which initiate major pathways for antiviral immunity (33). HHV-6A and HHV-6B both encode proteins that disrupt type I IFN signaling (34), namely, the IE1 proteins which prevent IRF3 dimerization, which is required for initiation of interferon-dependent transcriptional programs. ORF 29 and ORF 31 encode MRV homologs of roseoloviruses IE1 and IE2, respectively, suggesting that they may dampen type I IFN signaling in mice.
As we expected from a previously uncharacterized genome, we identified many potential ORFs with no sequence similarity to those encoding any published proteins, such as ORF 120, which is predicted by both Phyre2 (100% confidence) and I-TASSER (Znorm = 4.8) to encode a protein resembling the unstructured alpha-helical structure of collagen 1 alpha 1. By I-TASSER analysis, ORF 123 is predicted to encode a protein similar to the head component of the yeast spliceosome with a normalized Z-score of 4.64, and ORF 124 resembles components of secretory IgA with a normalized Z-score of 4.14. The role of these ORFs in virus pathogenesis remains unclear.
MRV infects various tissues in vivo.
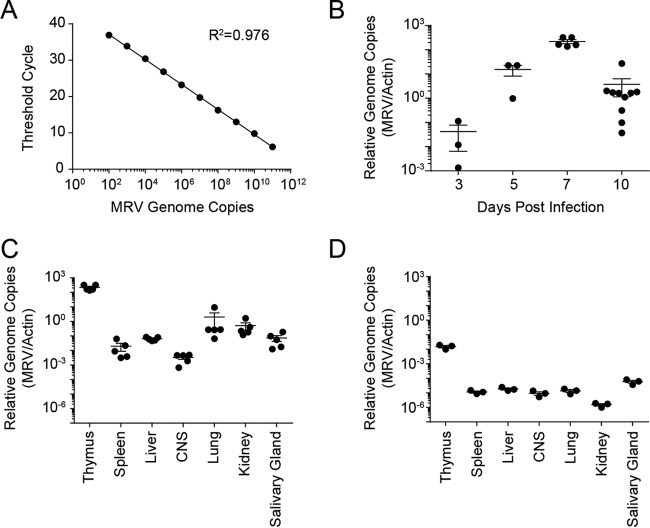
We have not been able to grow MRV in vitro, and an in vitro growth system for the related MTV has not been described. As such, traditional means of establishing virus burden, such as limiting dilution assays and plaque assays, are not possible. However, the MRV genome sequence allowed us to develop a sensitive quantitative PCR (qPCR) assay to detect genome copies of MRV as a measure of in vivo viral replication (Fig. 5A). Virus genome copies in the thymus peaked at 7 days postinfection (Fig. 5B). We assayed various organs known to be infected by herpesviruses (spleen, liver, kidney, salivary gland) or organs in which human roseoloviruses are known to be present (central nervous system). During acute infection, MRV was predominately localized to the thymi of infected mice, with low levels of virus detectable in other organs (Fig. 5C). We also measured MRV genome copy numbers in mice 7 weeks postinfection and again observed that MRV had a predilection for the thymus, albeit at significantly lower levels than during acute infection (Fig. 5D). These virus genome copies could represent latent infection in the thymus or low-level productive viral replication.
FIG 5.
MRV preferentially replicates in the thymus. (A) Standard curve of MRV genome copy numbers. The threshold cycle is plotted as a function of the absolute quantity of a plasmid containing the MRV genomic target. (B) Time course examining MRV genome copy numbers in thymi of neonatally infected BALB/c mice as a ratio to mouse actin genome copy numbers. (C and D) MRV genome copy numbers expressed as a ratio to mouse actin genome copy numbers at 1 week postinfection (C) and 7 weeks postinfection (D). Data are representative of three or more experiments. Each point represents a single mouse.
DISCUSSION
We performed ultrastructural microscopy and genome sequencing of MRV, which collectively indicate that it is a herpesvirus. The genomic architecture of MRV along with sequence similarity of each predicted ORF and phylogenetic analysis of three predicted ORFs strongly suggest that MRV is more closely related to the human roseoloviruses than to MCMV, a murine betaherpesvirus. Taken together, the genomic and sequence data strongly suggest that MRV belongs to the Roseolovirus genus of the Betaherpesvirinae subfamily in the family Herpesviridae.
MRV is likely related to the previously described MTV, a naturally occurring mouse herpesvirus (3, 35). MTV was originally identified by assaying tissue homogenates for viruses that drive mammary tumorigenesis (19). Blind passage of pooled organs from adult mice into neonatal mice by intraperitoneal injection reproducibly led to thymic “necrosis” in infected mice. Subsequent reports indicate that MTV infection of neonates leads to depletion of CD4+ T cells in the thymus of infected animals, whereas adult infections appear to be subclinical, with elevation of anti-MTV antibody titers but no overt thymic pathology (20, 36, 37). Neonatal depletion of CD4+ T cells is transient, as by 6 weeks postinfection, mice appear to have recovered a normal complement of CD4+ T cells in the spleen and thymus (38). Electron microscopy studies suggest that MTV has the morphological properties of a herpesvirus (19, 39). Similar to MCMV, MTV is shed through the salivary glands and can be horizontally transmitted (40, 41), but MTV is serologically distinct from MCMV (35). MTV has been mentioned to have a DNA polymerase sequence that is related to herpesviruses, but the sequence has not been reported (42). While MTV appears to be a herpesvirus, definitive assessment is not yet available, and detailed analysis, including that of its genome, is notably absent from the literature. While we have documented that MRV has many properties similar to those of MTV, the absence of these data and a direct link between our MRV stock and previous isolates of MTV prevented us from making determinations about the precise relationship of MTV to MRV and to other mouse and human herpesviruses. Without this certainty, we named the virus studied here MRV, as suggested by a reviewer of the manuscript.
Our report also shows the presence of virus genome copies in various organs. Herein we showed that MRV inoculation in the peritoneum results in MRV expansion in the thymus, as determined by our genome copy number assay. We found depletion of thymocyte subsets, suggesting lytic infection of T cells (20). Our study shows that MRV is also detectable at lower levels in peripheral organs of the mouse after acute infection, though this could be related to infected T-cell spread to these organs or viremia rather than direct infection of these organs.
In addition to the genome sequence similarities, there are other physiological and functional correlates between MRV and HHV-6. T-cell tropism has been described for HHV-6, which can replicate and cause lysis of infected T cells in vitro (43, 44). While we have not studied direct infection of T cells in vitro, our data strongly suggest that MRV has similar T-cell tropism. Furthermore, infection of humanized mice with HHV-6A showed severe loss of human CD4+ cells in the thymus (45–47), and ex vivo infection of human lymphoid tissues decreased the CD4+ T-cell number (48). The tissue tropism of human roseoloviruses may be very broad, involving a diverse array of organs and cell types (1). Taken together, the sequence and functional analyses indicate that MRV is a murine roseolovirus, related to HHV-6 and HHV-7.
While our data strongly suggest that MRV is a homolog of human roseoloviruses, the current analyses do not allow finer discrimination to determine if it is more closely related to HHV-6A, HHV-6B, or HHV-7. The genomes of HHV-6A/6B and HHV-7 can be distinguished by the presence of several HHV-6-specific ORFs. Our data indicate that MRV does not possess homologs of HHV-6 U6, U9, U22, U83, U78, or U94 signature genes (49). Nonetheless, it is unclear if evolutionarily divergent ORFs have compensated for these ORFs or whether the biology of MRV more closely mimics HHV-7.
Despite the correlates discussed above, there are some notable differences between MRV and human roseoloviruses, as one might expect from species-specific herpesviruses. MRV does possess many signature betaherpesvirus ORFs, but some genes thought to be roseolovirus specific are absent from the genome (2). However, these findings were not based on a comparison with nonhuman roseolovirus; our MRV genome sequence should shed additional light on roseolovirus-specific genes. HHV-6 has been reported to integrate in the telomeres of human chromosomes, while HHV-7 has not been observed to have this property (50–52). HHV-6 integration appears to be dependent on DR sequences at both ends of its genome, mirroring the hexameric TTAGGG sequence in human telomeres (53). Our genome does not show corresponding hexameric repeats in the MRV genome, but these data do not preclude the possibility that MRV could integrate into the mouse genome by unique mechanisms.
Furthermore, there has been no report of large-scale depletion of T cells during human roseolovirus infection, but to our knowledge, such studies may not have been systematically conducted, and T-cell depletion may occur at a different time with respect to the appearance of roseola rash, the usual presenting symptom of primary HHV-6 or HHV-7 infection. We hypothesize that similar CD4+ cell loss may occur during primary infection or reactivation of human roseoloviruses and that the thymus may be an important site for replication, especially in children. Alterations in T-cell number, diversity, or function may be especially important in the context of childhood vaccinations.
Models of HHV-6A and HHV-6B infection have been developed in marmosets and transgenic mice (46, 47, 54, 55). Additionally, identification of naturally occurring herpesviruses related to human roseoloviruses has been reported by various groups, notably in pig-tailed macaques (56, 57). Our report extends these studies by identifying a murine model for roseolovirus infection. The mouse is a particularly useful model organism for the study of human herpesviruses, and decades of intensive research have provided countless tools to study host-pathogen interactions.
The pathophysiological etiologies of many of the disorders associated with roseolovirus infection are unknown, but our studies suggest that they could also be related to the loss or altered function of CD4+ T cells, in addition to direct infection of specific organs. Furthermore, since thymus loss due to MRV occurs primarily in neonates, not adults, it is possible that there may be age-dependent effects due to HHV-6 or HHV-7 infection. Therefore, we posit that the study of MRV infections will provide insight into roseolovirus biology that may be more broadly applicable to human infections.
MATERIALS AND METHODS
Virus isolation and infection.
Our virus stocks were derived from a vial generously provided by Robert Livingston at the University of Missouri, Columbia, MO. Given the incomplete passage history and source of the virus stocks, we were unable to verify that this virus was directly linked to the originally described MTV (19). Virus stocks were produced by in vivo passaging as previously described (20, 38). A single virus stock was used for all experiments. Infected mice were intraperitoneally inoculated with 100 μl of a 1:10 dilution of thawed virus stock.
Flow cytometry.
Single-cell lymphocyte suspensions were prepared from thymi or spleens from infected animals and processed by manual disruption through a 70-μm filter. A total of 106 to 107 cells were stained with fixable viability dye, before incubation in 2.4G2 supernatant (anti-FcRII/III). Cells were stained with surface antibodies (Affymetrix, Santa Clara, CA): anti-CD3ε (145-2C11), anti-CD4 (RM4-5), and anti-CD8 (53-6.7). Samples were run by flow cytometry using a FACSCanto (BD Biosciences, San Jose, CA) and analyzed using FloJo X (TreeStar, Ashland, OR).
Transmission electron microscopy.
For ultrastructural analysis, cells were fixed in 2% paraformaldehyde–2.5% glutaraldehyde in 100 mM phosphate buffer, pH 7.2, for 1 h at room temperature. Samples were washed in sodium cacodylate buffer and postfixed in 1% osmium tetroxide (Polysciences, Inc.) for 1 h. Samples were then rinsed extensively in distilled H2O (dH2O) prior to en bloc staining with 1% aqueous uranyl acetate (Ted Pella, Inc., Redding, CA) for 1 h. Following several rinses in dH2O, samples were dehydrated in a graded series of ethanol and embedded in Eponate 12 resin (Ted Pella, Inc.). Sections of 95 nm were cut with a Leica Ultracut UCT ultramicrotome (Leica Microsystems, Inc., Bannockburn, IL), stained with uranyl acetate and lead citrate, and viewed on a JEOL 1200 EX transmission electron microscope (JEOL USA, Inc., Peabody, MA) equipped with an AMT 8-megapixel digital camera and AMT Image Capture Engine V602 software (Advanced Microscopy Techniques, Woburn, MA).
MRV genome sequencing.
Virus genomic DNA was extracted from the thymus of an infected neonatal mouse 7 days postinfection. Libraries were generated for sequencing using Pacific Biosciences RS sequencing (Pacific Biosciences, Menlo Park, CA) and Illumina MiSeq 2X250 sequencing (Illumina, San Diego, CA), both in accordance with the manufacturer's protocol. A total of 22,716 reads with a read length of approximately 113 to 2,201 bp (average, 681 bp) (short reads) and a total of 71,193 reads with a read length of 114 to 8,506 bp (average, 1,678 bp) (long reads) were obtained by Pacific Bioscience sequencing at the University of Michigan DNA Sequencing Core. A total of 62,966,095 reads were obtained by Illumina sequencing at the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine. CCS (circular consensus sequence) reads from Pacific Biosciences RS sequencing were analyzed using VirusHunter (58) to detect virus sequences and were used for de novo assembly to generate contigs. Illumina MiSeq data were analyzed using the VirusSeeker program (G. Zhao, unpublished). Contigs generated from both platforms were compared. Contig sequences generated using Pacific Biosciences RS sequencing data were used as a scaffold to orient contigs generated using Illumina data. Conflicts between the two assemblers were resolved by targeted PCR and cloning of the amplicon, followed by Sanger sequencing of cloned fragments. Single-nucleotide polymorphism (SNP) analysis was conducted using LoFreq (MIT, Boston, MA) (59).
Genome analysis.
Predicted ORFs were identified using Snapgene (GSL Biotech LLC, Chicago, IL), with only ORFs of greater than 100 amino acids in translated length analyzed. Analysis intended to identify spliced genes was not conducted. Each ORF was queried against the GenBank nonredundant protein database using the BLASTP algorithm. Novel ORFs were subjected to Phyre2 and I-TASSER analysis for protein structure modeling (59).
Phylogenetic analysis.
DNA polymerase catalytic subunit (1,005 amino acids [aa]), glycoprotein B (823 aa), and ssDNA binding protein (1,124 aa) genes were translated from MRV. These sequences were MUSCLE (60) aligned using SeaView (61). ModelGenerator (59) identified LG + G as the most appropriate amino acid substitution model. Maximum likelihood methods were implemented in RAxML (62) software. Phylogenetic trees were visualized using FigTree and stylistically finished using Adobe Illustrator (Adobe Systems, San Jose, CA). The viruses compared with MRV and their GenBank accession numbers are as follows: human herpesvirus 6A, NC_001664.2; human herpesvirus 6B, NC_000898.1; human herpesvirus 7, NC_001716.2; murid herpesvirus 1, NC_004065.1; human herpesvirus 5, FJ616285.1; human herpesvirus 1, X14112.1; human herpesvirus 2, NC_001798.2; human herpesvirus 3, NC_001348.1; human herpesvirus 8, NC_009333.1; human herpesvirus 4, NC_007605.1; murid herpesvirus 4, NC_001826; aotine herpesvirus 1, YP_004940081.1; cebine herpesvirus 1, AEW46233; cercopithecine herpesvirus 5, YP_004936030.1; macacine herpesvirus 3, AAC05256.1; panine herpesvirus 2, NP_612698.1; papiine herpesvirus 2, AHM96371.1; saimiriine herpesvirus 4, YP_004940227.1; murid herpesvirus 2, ADB44898.1; murid herpesvirus 8, YP_007016461.1; elephantid herpesvirus 1, AAG41999.1; caviid herpesvirus 2, YP_007417829.1; suid herpesvirus 2, ADV78258.1; tupaiid herpesvirus 1, AAD08667.1; Gorilla gorilla herpesvirus 7, AIN81109.1; Pan paniscus herpesvirus 7, AIN81107.1; and Pan troglodytes herpesvirus 7, AIN81106.1.
Virus genome copy quantification.
Genomic DNA was prepared from mouse tissues using a Puregene extraction kit (Qiagen, Valencia, CA) in accordance with the manufacturer's recommendations. Each qPCR utilized a TaqMan Universal PCR master mix, No AmpErase UNG (Life Technologies, Grand Island, NY), per the manufacturer's recommendations. MRV-specific primers amplifying the region from bp 69992 to bp 70144 are as follows: F, 5′ AAACTCCGTTACATGACGGTT 3′; R, 5′ GCCATACCGTTTCTTTGTCTTG 3′, TaqMan probe, 5′ TCACCCATACAGACAGATAAACTCTCA 3′. Actin-specific primers are as follows: F, 5′ AGCTCATTGTAGAAGGTGTGG 3′; R, 5′ GGTGGGAATGGGTCAGAAG 3′; TaqMan probe, 5′ TTCAGGGTCAGGATACCTCTCTTGCT 3′. Independent PCRs were run with MRV-specific primer/probes and actin-specific primer/probes. Amplification was performed on the Applied Biosystems StepOne Plus qPCR instrument (Life Technologies, Grand Island, NY). Each reaction was run in triplicate, and the absolute copy number was quantified using a standard curve.
Accession number(s).
The GenBank accession number for murine roseolovirus (MRV) is KY355735.
ACKNOWLEDGMENTS
We thank Robert Livingston for providing a virus inoculum, Yokoyama laboratory members for excellent discussions, University of Michigan DNA sequencing core for Pacific Biosciences sequencing, the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for Illumina sequencing, the Protein and Nucleic Acid Chemistry Laboratory for Sanger sequencing, and the Pathogen Discovery Facility in the Department of Pathology and Immunology for support generating Illumina libraries.
W. M. Yokoyama is an investigator of the Howard Hughes Medical Institute. S. J. Patel is supported by NIH grant T32GM007200, E. Park is supported by NIH grant F30DK108472, and E. J. Lauron is supported by a Robert D. Watkins Graduate Research Fellowship from the American Society for Microbiology.
REFERENCES
- 1.Knipe DM, Howley PM. 2013. Fields virology, 6th ed Wolters Kluwer/Lippincott Williams & Wilkins Health, Philadelphia, PA. [Google Scholar]
- 2.Arvin AM. 2007. Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed] [Google Scholar]
- 3.Fox JG. 2007. The mouse in biomedical research, 2nd ed Academic Press, Boston, MA. [Google Scholar]
- 4.Caserta MT, Krug LT, Pellett PE. 2014. Roseoloviruses: unmet needs and research priorities. Curr Opin Virol 9:167–169. doi: 10.1016/j.coviro.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Braun DK, Dominguez G, Pellett PE. 1997. Human herpesvirus 6. Clin Microbiol Rev 10:521–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blazsek A, Sillo P, Ishii N, Gergely P Jr, Poor G, Preisz K, Hashimoto T, Medvecz M, Karpati S. 2008. Searching for foreign antigens as possible triggering factors of autoimmunity: Torque Teno virus DNA prevalence is elevated in sera of patients with bullous pemphigoid. Exp Dermatol 17:446–454. doi: 10.1111/j.1600-0625.2007.00663.x. [DOI] [PubMed] [Google Scholar]
- 7.Shimazu Y, Kondo T, Ishikawa T, Yamashita K, Takaori-Kondo A. 2013. Human herpesvirus-6 encephalitis during hematopoietic stem cell transplantation leads to poor prognosis. Transpl Infect Dis 15:195–201. doi: 10.1111/tid.12049. [DOI] [PubMed] [Google Scholar]
- 8.Yamashita N, Morishima T. 2005. HHV-6 and seizures. Herpes 12:46–49. [PubMed] [Google Scholar]
- 9.Kondo K, Nagafuji H, Hata A, Tomomori C, Yamanishi K. 1993. Association of human herpesvirus 6 infection of the central nervous system with recurrence of febrile convulsions. J Infect Dis 167:1197–1200. doi: 10.1093/infdis/167.5.1197. [DOI] [PubMed] [Google Scholar]
- 10.Zerr DM, Meier AS, Selke SS, Frenkel LM, Huang ML, Wald A, Rhoads MP, Nguy L, Bornemann R, Morrow RA, Corey L. 2005. A population-based study of primary human herpesvirus 6 infection. N Engl J Med 352:768–776. doi: 10.1056/NEJMoa042207. [DOI] [PubMed] [Google Scholar]
- 11.Yamanishi K, Okuno T, Shiraki K, Takahashi M, Kondo T, Asano Y, Kurata T. 1988. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet i:1065–1067. [DOI] [PubMed] [Google Scholar]
- 12.Ablashi DV, Lapps W, Kaplan M, Whitman JE, Richert JR, Pearson GR. 1998. Human herpesvirus-6 (HHV-6) infection in multiple sclerosis: a preliminary report. Mult Scler 4:490–496. doi: 10.1177/135245859800400606. [DOI] [PubMed] [Google Scholar]
- 13.Challoner PB, Smith KT, Parker JD, MacLeod DL, Coulter SN, Rose TM, Schultz ER, Bennett JL, Garber RL, Chang M, Schad PA, Stewart PM, Nowinski RC, Brown JP, Burmer GC. 1995. Plaque-associated expression of human herpesvirus 6 in multiple sclerosis. Proc Natl Acad Sci U S A 92:7440–7444. doi: 10.1073/pnas.92.16.7440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chapenko S, Millers A, Nora Z, Logina I, Kukaine R, Murovska M. 2003. Correlation between HHV-6 reactivation and multiple sclerosis disease activity. J Med Virol 69:111–117. doi: 10.1002/jmv.10258. [DOI] [PubMed] [Google Scholar]
- 15.Goodman AD, Mock DJ, Powers JM, Baker JV, Blumberg BM. 2003. Human herpesvirus 6 genome and antigen in acute multiple sclerosis lesions. J Infect Dis 187:1365–1376. doi: 10.1086/368172. [DOI] [PubMed] [Google Scholar]
- 16.Flaño E, Woodland DL, Blackman MA. 2002. A mouse model for infectious mononucleosis. Immunol Res 25:201–217. doi: 10.1385/IR:25:3:201. [DOI] [PubMed] [Google Scholar]
- 17.Blackman MA. 2002. Persistent gamma-herpesvirus infections: what can we learn from an experimental mouse model? J Exp Med 195:F29–F32. doi: 10.1084/jem.20020243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price P, Olver SD. 1996. Syndromes induced by cytomegalovirus infection. Clin Immunol Immunopathol 80:215–224. doi: 10.1006/clin.1996.0117. [DOI] [PubMed] [Google Scholar]
- 19.Rowe WP, Capps WI. 1961. A new mouse virus causing necrosis of the thymus in newborn mice. J Exp Med 113:831–844. doi: 10.1084/jem.113.5.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morse SS, Valinsky JE. 1989. Mouse thymic virus (MTLV). A mammalian herpesvirus cytolytic for CD4+ (L3T4+) T lymphocytes. J Exp Med 169:591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith HR, Heusel JW, Mehta IK, Kim S, Dorner BG, Naidenko OV, Iizuka K, Furukawa H, Beckman DL, Pingel JT, Scalzo AA, Fremont DH, Yokoyama WM. 2002. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc Natl Acad Sci U S A 99:8826–8831. doi: 10.1073/pnas.092258599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mans J, Zhi L, Revilleza MJ, Smith L, Redwood A, Natarajan K, Margulies DH. 2009. Structure and function of murine cytomegalovirus MHC-I-like molecules: how the virus turned the host defense to its advantage. Immunol Res 43:264–279. doi: 10.1007/s12026-008-8081-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weis WI, Taylor ME, Drickamer K. 1998. The C-type lectin superfamily in the immune system. Immunol Rev 163:19–34. doi: 10.1111/j.1600-065X.1998.tb01185.x. [DOI] [PubMed] [Google Scholar]
- 24.Zelensky AN, Gready JE. 2005. The C-type lectin-like domain superfamily. FEBS J 272:6179–6217. doi: 10.1111/j.1742-4658.2005.05031.x. [DOI] [PubMed] [Google Scholar]
- 25.Yokoyama WM, Plougastel BF. 2003. Immune functions encoded by the natural killer gene complex. Nat Rev Immunol 3:304–316. doi: 10.1038/nri1055. [DOI] [PubMed] [Google Scholar]
- 26.Iizuka K, Naidenko OV, Plougastel BF, Fremont DH, Yokoyama WM. 2003. Genetically linked C-type lectin-related ligands for the NKRP1 family of natural killer cell receptors. Nat Immunol 4:801–807. doi: 10.1038/ni954. [DOI] [PubMed] [Google Scholar]
- 27.Boyman O, Sprent J. 2012. The role of interleukin-2 during homeostasis and activation of the immune system. Nat Rev Immunol 12:180–190. doi: 10.1038/nri3156. [DOI] [PubMed] [Google Scholar]
- 28.Iampietro M, Morissette G, Gravel A, Flamand L. 2014. Inhibition of interleukin-2 gene expression by human herpesvirus 6B U54 tegument protein. J Virol 88:12452–12463. doi: 10.1128/JVI.02030-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sullivan BM, Coscoy L. 2008. Downregulation of the T-cell receptor complex and impairment of T-cell activation by human herpesvirus 6 u24 protein. J Virol 82:602–608. doi: 10.1128/JVI.01571-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kofod-Olsen E, Moller JM, Schleimann MH, Bundgaard B, Bak RO, Oster B, Mikkelsen JG, Hupp T, Hollsberg P. 2013. Inhibition of p53-dependent, but not p53-independent, cell death by U19 protein from human herpesvirus 6B. PLoS One 8:e59223. doi: 10.1371/journal.pone.0059223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kofod-Olsen E, Ross-Hansen K, Schleimann MH, Jensen DK, Moller JM, Bundgaard B, Mikkelsen JG, Hollsberg P. 2012. U20 is responsible for human herpesvirus 6B inhibition of tumor necrosis factor receptor-dependent signaling and apoptosis. J Virol 86:11483–11492. doi: 10.1128/JVI.00847-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo H, Kaiser WJ, Mocarski ES. 2015. Manipulation of apoptosis and necroptosis signaling by herpesviruses. Med Microbiol Immunol 204:439–448. doi: 10.1007/s00430-015-0410-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McNab F, Mayer-Barber K, Sher A, Wack A, O'Garra A. 2015. Type I interferons in infectious disease. Nat Rev Immunol 15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaworska J, Gravel A, Fink K, Grandvaux N, Flamand L. 2007. Inhibition of transcription of the beta interferon gene by the human herpesvirus 6 immediate-early 1 protein. J Virol 81:5737–5748. doi: 10.1128/JVI.02443-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cross SS, Parker JC, Rowe WP, Robbins ML. 1979. Biology of mouse thymic virus, a herpesvirus of mice, and the antigenic relationship to mouse cytomegalovirus. Infect Immun 26:1186–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lussier G, Guenette D, Shek WR, Descoteaux JP. 1988. Evaluation of mouse thymic virus antibody detection techniques. Lab Anim Sci 38:577–579. [PubMed] [Google Scholar]
- 37.Lussier G, Guenette D, Shek WR, Descoteaux JP. 1988. Detection of antibodies to mouse thymic virus by enzyme-linked immunosorbent assay. Can J Vet Res 52:236–238. [PMC free article] [PubMed] [Google Scholar]
- 38.Morse SS, Sakaguchi N, Sakaguchi S. 1999. Virus and autoimmunity: induction of autoimmune disease in mice by mouse T lymphotropic virus (MTLV) destroying CD4+ T cells. J Immunol 162:5309–5316. [PubMed] [Google Scholar]
- 39.Parker JC, Vernon ML, Cross SS. 1973. Classification of mouse thymic virus as a herpesvirus. Infect Immun 7:305–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morse SS. 1989. Thymic necrosis following oral inoculation of mouse thymic virus. Lab Anim Sci 39:571–574. [PubMed] [Google Scholar]
- 41.St-Pierre Y, Potworowski EF, Lussier G. 1987. Transmission of mouse thymic virus. J Gen Virol 68:1173–1176. doi: 10.1099/0022-1317-68-4-1173. [DOI] [PubMed] [Google Scholar]
- 42.Ehlers B, Kuchler J, Yasmum N, Dural G, Voigt S, Schmidt-Chanasit J, Jakel T, Matuschka FR, Richter D, Essbauer S, Hughes DJ, Summers C, Bennett M, Stewart JP, Ulrich RG. 2007. Identification of novel rodent herpesviruses, including the first gammaherpesvirus of Mus musculus. J Virol 81:8091–8100. doi: 10.1128/JVI.00255-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lusso P, Malnati M, De Maria A, Balotta C, DeRocco SE, Markham PD, Gallo RC. 1991. Productive infection of CD4+ and CD8+ mature human T cell populations and clones by human herpesvirus 6. Transcriptional down-regulation of CD3. J Immunol 147:685–691. [PubMed] [Google Scholar]
- 44.Lusso P, Markham PD, Tschachler E, di Marzo Veronese F, Salahuddin SZ, Ablashi DV, Pahwa S, Krohn K, Gallo RC. 1988. In vitro cellular tropism of human B-lymphotropic virus (human herpesvirus-6). J Exp Med 167:1659–1670. doi: 10.1084/jem.167.5.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horvat B, Berges BK, Lusso P. 2014. Recent developments in animal models for human herpesvirus 6A and 6B. Curr Opin Virol 9:97–103. doi: 10.1016/j.coviro.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gobbi A, Stoddart CA, Malnati MS, Locatelli G, Santoro F, Abbey NW, Bare C, Linquist-Stepps V, Moreno MB, Herndier BG, Lusso P, McCune JM. 1999. Human herpesvirus 6 (HHV-6) causes severe thymocyte depletion in SCID-hu Thy/Liv mice. J Exp Med 189:1953–1960. doi: 10.1084/jem.189.12.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanner A, Carlson SA, Nukui M, Murphy EA, Berges BK. 2013. Human herpesvirus 6A infection and immunopathogenesis in humanized Rag2−/− γc−/− mice. J Virol 87:12020–12028. doi: 10.1128/JVI.01556-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grivel JC, Santoro F, Chen S, Faga G, Malnati MS, Ito Y, Margolis L, Lusso P. 2003. Pathogenic effects of human herpesvirus 6 in human lymphoid tissue ex vivo. J Virol 77:8280–8289. doi: 10.1128/JVI.77.15.8280-8289.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krug LT, Pellett PE. 2014. Roseolovirus molecular biology: recent advances. Curr Opin Virol 9:170–177. doi: 10.1016/j.coviro.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaufer BB, Flamand L. 2014. Chromosomally integrated HHV-6: impact on virus, cell and organismal biology. Curr Opin Virol 9:111–118. doi: 10.1016/j.coviro.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 51.Arbuckle JH, Medveczky MM, Luka J, Hadley SH, Luegmayr A, Ablashi D, Lund TC, Tolar J, De Meirleir K, Montoya JG, Komaroff AL, Ambros PF, Medveczky PG. 2010. The latent human herpesvirus-6A genome specifically integrates in telomeres of human chromosomes in vivo and in vitro. Proc Natl Acad Sci U S A 107:5563–5568. doi: 10.1073/pnas.0913586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luppi M, Marasca R, Barozzi P, Ferrari S, Ceccherini-Nelli L, Batoni G, Merelli E, Torelli G. 1993. Three cases of human herpesvirus-6 latent infection: integration of viral genome in peripheral blood mononuclear cell DNA. J Med Virol 40:44–52. doi: 10.1002/jmv.1890400110. [DOI] [PubMed] [Google Scholar]
- 53.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. 1988. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci U S A 85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leibovitch E, Wohler JE, Cummings Macri SM, Motanic K, Harberts E, Gaitan MI, Maggi P, Ellis M, Westmoreland S, Silva A, Reich DS, Jacobson S. 2013. Novel marmoset (Callithrix jacchus) model of human herpesvirus 6A and 6B infections: immunologic, virologic and radiologic characterization. PLoS Pathog 9:e1003138. doi: 10.1371/journal.ppat.1003138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reynaud JM, Jegou JF, Welsch JC, Horvat B. 2014. Human herpesvirus 6A infection in CD46 transgenic mice: viral persistence in the brain and increased production of proinflammatory chemokines via Toll-like receptor 9. J Virol 88:5421–5436. doi: 10.1128/JVI.03763-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Staheli JP, Dyen MR, Deutsch GH, Basom RS, Fitzgibbon MP, Lewis P, Barcy S. 2016. Complete unique genome sequence, expression profile, and salivary gland tissue tropism of the herpesvirus 7 homolog in pigtailed macaques. J Virol 90:6657–6674. doi: 10.1128/JVI.00651-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Staheli JP, Dyen MR, Lewis P, Barcy S. 2014. Discovery and biological characterization of two novel pig-tailed macaque homologs of HHV-6 and HHV-7. Virology 471-473:126–140. doi: 10.1016/j.virol.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao G, Krishnamurthy S, Cai Z, Popov VL, Travassos da Rosa AP, Guzman H, Cao S, Virgin HW, Tesh RB, Wang D. 2013. Identification of novel viruses using VirusHunter—an automated data analysis pipeline. PLoS One 8:e78470. doi: 10.1371/journal.pone.0078470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilm A, Aw PP, Bertrand D, Yeo GH, Ong SH, Wong CH, Khor CC, Petric R, Hibberd ML, Nagarajan N. 2012. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res 40:11189–11201. doi: 10.1093/nar/gks918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 62.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]