ABSTRACT
Current seasonal influenza vaccines are efficacious when vaccine strains are matched with circulating strains. However, they do not protect antigenic variants and newly emerging pandemic and outbreak strains. Thus, there is a critical need for developing so-called “universal” vaccines that protect against all influenza viruses. In the present study, we developed a bivalent heterologous DNA virus-like particle prime-boost vaccine strategy. We show that mice immunized with this vaccine were broadly protected against lethal challenge from group 1 (H1, H5, and H9) and group 2 (H3 and H7) viruses, with 94% aggregate survival. To determine the immune correlates of protection, we performed passive immunizations and in vitro assays. We show that this vaccine elicited antibody responses that bound HA from group 1 (H1, H2, H5, H6, H8, H9, H11, and H12) and group 2 (H3, H4, H7, H10, H14, and H15) and neutralized homologous and intrasubtypic H5 and H7 and heterosubtypic H1 viruses and hemagglutinin-specific CD4 and CD8 T cell responses. As a result, passive immunization with immune sera fully protected mice against H5, H7, and H1 challenge, whereas with both immune sera and T cells the mice survived heterosubtypic H3 and H9 challenge. Thus, it appears that (i) neutralizing antibodies alone fully protect against homologous and intrasubtypic H5 and H7 and (ii) neutralizing and binding antibodies are sufficient to protect against heterosubtypic H1, (iii) but against heterosubtypic H3 and H9, binding antibodies and T cells are required for complete survival. We believe that this vaccine regimen could potentially be a candidate for a “universal” influenza vaccine.
IMPORTANCE Influenza virus infection is global health problem. Current seasonal influenza vaccines are efficacious only when vaccine strains are matched with circulating strains. However, these vaccines do not protect antigenic variants and newly emerging pandemic and outbreak strains. Because of this, there is an urgent need to develop so-called “universal” influenza vaccines that can protect against both current and future influenza strains. In the present study, we developed a bivalent heterologous prime-boost vaccine strategy. We show that a bivalent vaccine regimen elicited broad binding and neutralizing antibody and T cell responses that conferred broad protection against diverse challenge viruses in mice, suggesting that this bivalent prime-boost strategy could practically be a candidate for a “universal” influenza vaccine.
KEYWORDS: influenza vaccine
INTRODUCTION
Current seasonal influenza vaccines are efficacious when vaccine strains are matched with circulating strains. However, these vaccines need to be reformulated frequently to elicit protective antibody responses against variants that arise via antigenic drift. They also do not protect humans from pandemics and outbreaks of newly emerging strains via antigenic shift, such as the emergence of the pandemic H1N1 influenza virus in 2009 and the avian H5N1, H5N6, H7N9, and H10N8 viruses (1, 2). Thus, the holy grail of influenza vaccine research is to develop “universal” vaccines that protect against both current and future influenza strains.
Influenza viruses are enveloped, negative-sense, single-strand RNA viruses with segmented genomes. Hemagglutinin (HA), neuraminidase (NA), and matrix 2 (M2) are three proteins on the virion surface. HA forms a trimer of covalently linked HA1/HA2 heterodimers. HA1 binds to sialic acid receptors, and HA2 mediates viral and endosomal membrane fusion. HA is a major target to host immune responses. Antigenically, HA in influenza A viruses comprises 18 subtypes, which are divided into two phylogenetically distinct groups (3–6). Group 1 comprises of H1, H2, H5, H6, H8, H9, H11, H12, H13, H16, H17, and H18, and group 2 consists of H3, H4, H7, H10, H14, and H15. HA subtypes within a group or between two groups have 40 or 60% sequence diversity, respectively.
The discovery of conserved epitopes in the HA stem region has spurred great efforts on development of stem-based “universal” vaccines (7–22). Basically, there are two approaches. One approach used sequential infection with different influenza strains (7) or sequential prime-boosts with head/stem chimera (cHA), in which heads from different HA subtypes were fused with a common stem (8–10). Sequential prime-boosts with cHA containing a common H1 stem were able to cross-protect mice from lethal challenge of H5N1 and H6N1 viruses (group 1) and to cross-neutralize H2N2 virus (group 1) (9), whereas sequential prime-boosts with cHA containing a common H3 stem were able to cross-protect mice from lethal challenge of H7N1 and H7N9 (group 2) (10). Another approach was to develop stem-only (or headless) HA (12–21). For example, two groups recently reported the construction of correctly folded H1 stem trimer (20) and self-assembled H1 stem-containing nanoparticle (21). Mice immunized with these immunogens had binding antibodies against HA from both group 1 and group 2 and neutralizing antibodies against H1 and H5 strains and were completely protected from H5 challenge (20, 21). Although the successful design of correctly folded stem is an important advancement toward “universal” influenza vaccine, the safety and the breadth of in vivo protection by stem-based “universal” vaccines remain to be improved.
In addition to HA stem-based “universal” vaccines, multivalent “universal” vaccines are also being developed. For example, Schwartzman et al. recently showed that an intranasal (i.n.) immunization with a cocktail of virus-like particle (VLP)-expressing group 1 (H1 and H5) and group 2 (H3 and H7) HA not only protects mice from homologous and intrasubtypic H1, H5 and H7 virus challenge but also partially protects mice from heterosubtypic H2, H6, H10, and H11 virus challenge, with a total of 94% aggregate survival (23).
Previously, we showed that priming mice twice with DNA plasmid encoding H5 HA and boosting once with VLP derived from A/Thailand/1(KAN)-1/2004 (TH04) strain (notated as TH DDV) induced antibody responses that cross-neutralize all clades and subclades of H5 viruses (24). To further improve the breadth and potency of antibody responses and immune protection, we developed a bivalent TH/NE DDV plus in vivo electroporation (EP) vaccine strategy (TH/NE DDV+EP), in which DNA plasmids and VLP encoding H5 (group 1) and H7 (group 2) HA were derived from TH04 and A/Netherlands/219/2003 (NE03) strains, respectively. It has been shown that immune sera elicited by live-attenuated NE03 strain cross-neutralizes diverse H7 strains from both Eurasian and American lineages (25). We first compared antibody responses against a panel of 12 influenza strains from subtypes H1, H3, H4, H5, H7, H9, and H10 between TH DDV and TH DDV+EP sera, between NE DDV and NE DDV+EP sera, and between a mixture of TH DDV+EP and NE DDV+EP sera and bivalent TH/NE DDV+EP sera. We then tested protective efficacy against diverse strains from subtypes H1, H3, H5, H7, and H9 by active and passive TH/NE DDV+EP immunizations. Finally, we systematically tested binding and neutralizing antibody and HA-specific T cell responses in immunized animals.
RESULTS
Enhancement of antibody responses by in vivo EP during DNA priming.
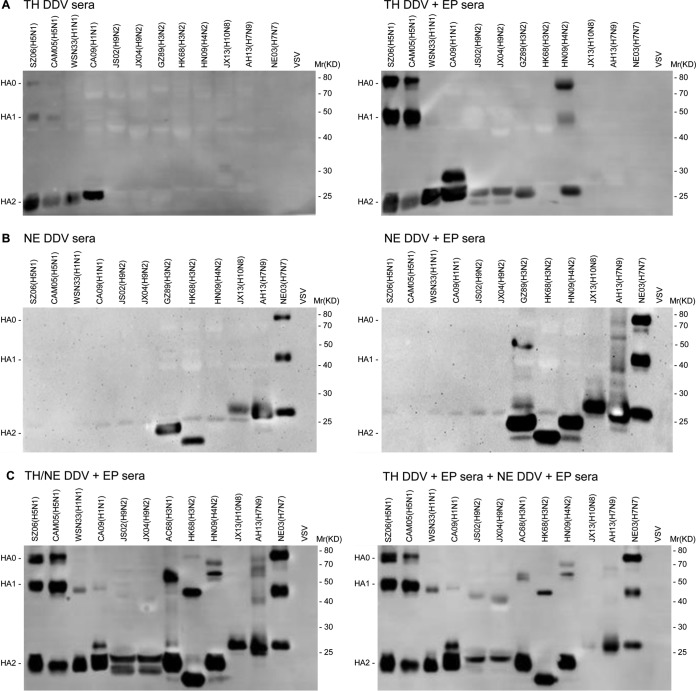
To test the effect of in vivo electroporation (EP) during DNA priming on antibody responses, we compared TH DDV and TH DDV+EP sera, as well as NE DDV and NE DDV+EP sera, in their ability to bind to HA from 12 influenza strains of both group 1 (H1, H5, and H9) and group 2 (H3, H4, H7, and H10) (see Table 1) and a control vesicular stomatitis virus (VSV). Figure 1A shows that TH DDV sera only bound both HA1 and HA2 of H5 HA (SZ06 and CAM05) and HA2 of H1 HA (WSN33 and CA09), but not HA from other eight strains of H9, H3, H4, H7, and H10 subtypes and the VSV control (the left panel). In contrast, TH DDV+EP sera not only increased the binding to both HA1 and HA2 of H5 HA and HA2 of H1 HA but also bound HA2 of H9 HA (JS02 and JX04) and H3 HA (GZ89), as well as HA1 and HA2 of H4 HA (HN09), but not HA from H3 HA (HK68), H10 HA, and H7 HA of group 2 and the VSV control (the right panel). Thus, we conclude that in vivo EP not only significantly enhances binding antibody responses of TH DDV to H5 and H1 HA but also broadens the responses to both group 1 (H9) and group 2 (H3 and H4) HA.
TABLE 1.
Influenza strains used in challenge experiments, plaque reduction assay, and Western blot analysis
| Strain | Abbreviation | Subtype | Analysis resulta |
||
|---|---|---|---|---|---|
| MLD50/50 μl | EID50/100 μl | PFU/ml | |||
| A/WSN/1933 | WSN33 | H1N1 | 102.3 | ND | 1.38 × 105 |
| A/California/07/2009 | CA09 | H1N1 | 104.5 | ND | 1.23 × 107 |
| A/Hong Kong/1/1968 | HK68 | H3N2 | 103.3 | ND | 5.25 × 105 |
| A/Aichi/1/1968 | AC68 | H3N1 | 105.75 | ND | 8.4 × 107 |
| A/Guizhou/54/1989 | GZ89 | H3N2 | ND | 107.5 | ND |
| A/duck/Hunan/8-19/2009 | HN09 | H4N2 | ND | 107.25 | ND |
| A/Cambodia/P0322095/2005 | CAM05 | H5N1 | 104.5 | ND | 3.0 × 104 |
| A/Shenzhen/406H/2006 | SZ06 | H5N1 | 106.75 | ND | 8.75 × 107 |
| A/Netherlands/219/2003 | NE03 | H7N7 | 102.75 | ND | 4.5 × 105 |
| A/Anhui/1/2013 | AH13 | H7N9 | ND | 106.5 | ND |
| A/swine/Jiangxi/1/2004 | JX04 | H9N2 | 102.75 | ND | 1.21 × 106 |
| A/chicken/Jiangsu/7/2002 | JS02 | H9N2 | 104.5 | ND | 5.63 × 107 |
| A/Jiangxi-Donghu/346/2013 | JX13 | H10N8 | ND | 107.75 | ND |
ND, not done.
FIG 1.
Comparison of binding antibodies between TH DDV and TH DDV+EP sera, between NE DDV and NE DDV+EP sera, and between TH/NE DDV+EP sera and combined TH DDV+EP sera and NE DDV+EP sera against HA from a panel of 12 influenza strains of seven HA subtypes and VSV control by Western blotting. (A) HA0, HA1, and HA2 bands revealed by TH DDV sera (left panel); HA0, HA1, and HA2 bands revealed by TH DDV+EP sera (right panel). (B) HA0, HA1, and HA2 bands revealed by NE DDV sera (left panel); HA0, HA1, and HA2 bands revealed by NE DDV+EP sera (right panel). (C) HA0, HA1, and HA2 bands revealed by TH/NE DDV+EP sera (the left panel); HA0, HA1, and HA2 bands revealed by combined TH DDV+EP sera and NE DDV+EP (right panel).
Similarly, NE DDV sera only bound both HA1 and HA2 of H7 HA (NE03) and HA2 of H7 HA (AH13), H3 HA (HK68 and GZ89) and H10 (JX13), but not HA from other eight strains of H4, H1, H5, and H9 HA and VSV control (the left panel in Fig. 1B). In contrast, NE DDV+EP sera not only increased the binding to HA1 and HA2 of H7 HA (NE03) and HA2 of H7 (AH13), H3 HA and H10 but also bound to HA2 of H4 HA (HN09) and HA1 of H7 (AH13) and H3 (GZ89), but not HA from group 1 viruses (the right panel in Fig. 1B). Thus, we conclude that in vivo EP also quantitatively and qualitatively enhances binding antibody responses of NE DDV. However, the enhancement is confined within group 2 viruses.
We next tested a bivalent TH/NE DDV+EP immunization. Figure 1C shows that TH/NE DDV+EP sera bound HA2 from all 12 strains of subtypes H1, H5, H9, H3, H4, H7, and H10 and bound HA1 from 11 strains except for strain JX13, subtype H10 (the left panel). The binding pattern was very similar to that detected by combined TH DDV+EP and NE DDV+EP sera (the right panel), indicating that in the bivalent TH/NE DDV+EP immunization, H5 and H7 HA did not interfere with each other in their immunogenicity. Therefore, we subsequently focused on TH/NE DDV+EP immunization.
Protection by active TH/NE DDV+EP immunization.
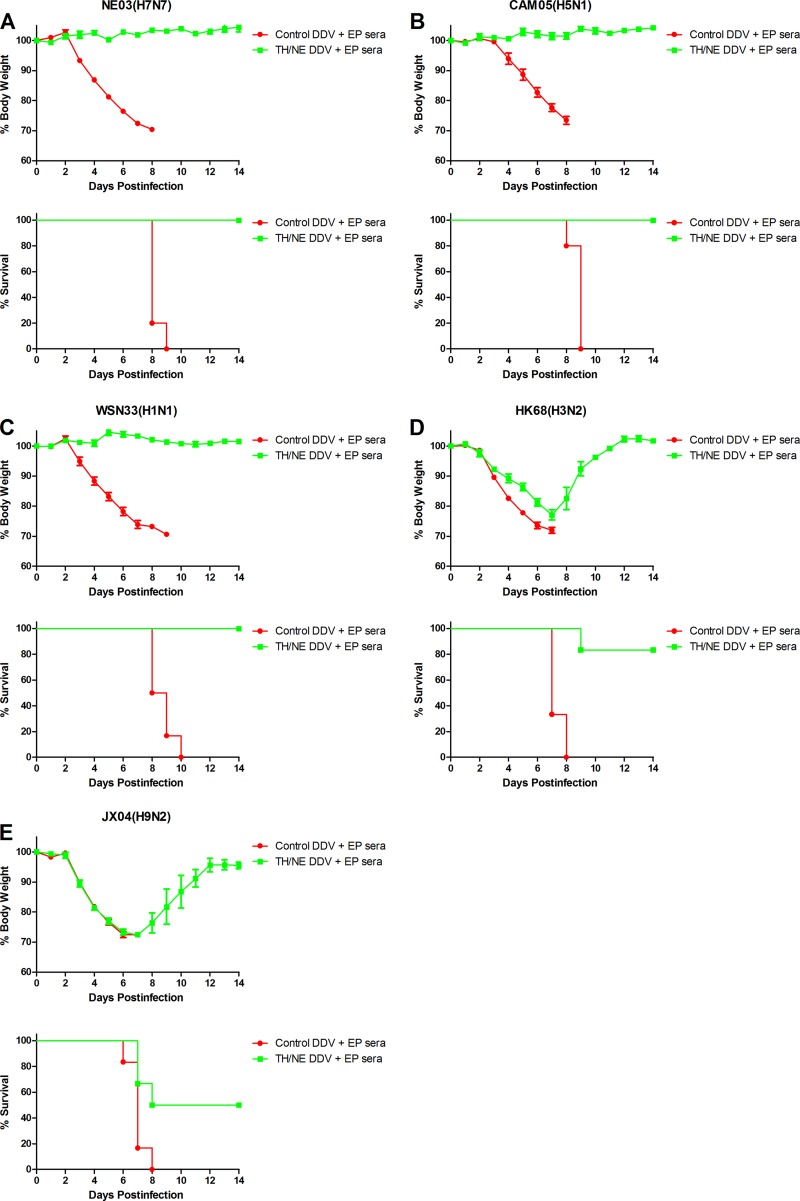
To test immune protection, TH/NE DDV+EP- and control DDV+EP-immunized mice were i.n. challenged with five 50% mouse lethal doses (MLD50) of NE03, CAM05, SZ06, AH13, WSN33, CA09, JS02, JX04, HK68, or AC68 strain (see Table 1). Figure 2A to I show that control DDV+EP-immunized mice, after challenge with the NE03, CAM05, SZ06, WSN33, CA09, JS02, JX04, HK68, or AC68 strain, died. In contrast, TH/NE DDV+EP-immunized mice, after challenge with the NE03, CAM05, SZ06, or WSN33 strain, did not have any sign of illness and weight loss, and all survived; after challenge with the CA09, JX04, or HK68 strain, the mice experienced a weight loss nadir of <20%, and all survived. When challenged with the JS02 or AC68 strain, the mice experienced a weight loss nadir of >20% and 5 of 6 mice or 4 of 6 mice, respectively, survived. In aggregate, 94.12% (48/51 animals) of TH/NE DDV+EP-immunized mice survived challenge compared to 0% (0/51 animals) of control DDV+EP-immunized mice (P < 0.001).
FIG 2.
In vivo efficacy of TH/NE DDV+EP immunization. (A to I) Time course of weight change (upper panels) and survival rate (lower panels), followed by homologous NE03 H7N7 (A), intrasubtypic CAM05 H5N1 (B), intrasubtypic SZ06 H5N1 (C), or heterosubtypic WSN33 H1N1 (D), CA09 H1N1 (E), HK68 H3N2 (F), AC68 H3N1 (G), JX04 H9N2 (H), and JS02 H9N2 (I) challenges. The survival rate was calculated as the percent survival within each experimental group (n = 5 or 6 mice per group). (J) Comparison of viral loads in lung tissues isolated at day 3 postchallenge of intrasubtypic AH13 H7N9 virus between TH/NE DDV+EP- versus control DDV+EP-immunized mice (three mice per group).
Figure 2J shows that after challenge with AH13 H7N9 the lung tissues in control DDV+EP-immunized mice exhibited significantly higher virus titers (average titer, 352 50% egg infectious doses [EID50]), whereas the lung tissues in TH/NE DDV+EP-immunized mice did not show evidence of any viruses. Thus, we conclude that TH/NE DDV+EP immunization not only completely protects against homologous and intrasubtypic H5 and H7 virus challenge but also completely or partially protects against heterosubtypic H1, H3, and H9 virus challenge.
Protection by passive immunization with immune sera alone or with immune sera and T cells.
To determine whether protection was mediated by immune sera, female BALB/c mice were i.p. injected with 700 μl of pooled TH/NE DDV+EP or control DDV+EP sera. At 24 h after passive immunization, mice were i.n. challenged with 5 MLD50 of the NE03, CAM05, WSN33, JX04, or HK68. Figure 3A to E show that, after the challenge, mice injected with control DDV+EP sera died within 10 days. In contrast, mice receiving TH/NE DDV+EP sera after challenge with the NE03, CAM05, or WSN33 strain experienced no weight loss, and all survived. After challenge with HK68 or JX04, the mice experienced a weight loss nadir of 20 or 25% and 5 of 6 mice or 3 of 6 mice, respectively, survived. In aggregate, 85.71% (24/28) of mice receiving TH/NE DDV+EP sera survived challenge compared to 0% (0/28 animals) of mice receiving control DDV+EP sera (P < 0.001).
FIG 3.
In vivo efficacy of passive immunization with immune sera or with immune sera and/or T cells. (A to E) Time course of weight change (upper panel) and survival rate (lower panel), followed by homologous NE03 H7N7 (A), intrasubtypic CAM05 H5N1 (B), or heterosubtypic WSN33 H1N1 (C), HK68 H3N2 (D), or JX04 H9N2 (E) challenge. The survival rate was calculated as the percent survival within each experimental group (n = 5 or 6 mice per group). (F and G) Time course of weight change (upper panel) and survival rate (lower panel) in mice receiving immune sera and/or T cells versus control sera, followed by HK68 H3N2 (F) or JX04 H9N2 (G) challenge. The survival rate was calculated as the percent survival within each experimental group (n = 6 mice per group).
Since protection against HK68 or JX04 by passive immunization with TH/NE DDV+EP sera (Fig. 3D and E) was less than that by active TH/NE DDV+EP immunization (Fig. 2F and H), we sought to determine whether T cells from TH/NE DDV+EP-immunized mice could also provide certain heterosubtypic protection. To test this, pooled immune sera or T cells either alone or combined were passively administered to BALB/c mice, followed by HK68 or JX04 challenge. Figure 3F shows that after challenge with HK68, all mice receiving both immune sera and T cells survived, whereas 5 of 6 mice receiving immune sera alone survived, and 2 of 6 mice receiving T cells alone survived. Figure 3G shows that, after challenge with JX04, mice receiving both immune sera and T cells experienced significantly less weight loss than mice receiving immune sera or T cells alone, and all survived, whereas 3 of 6 mice receiving immune sera alone survived, and 1 of 6 mice receiving T cells alone survived. Thus, we conclude that passive immunization with immune sera alone fully protects against homologous H7, intrasubtypic H5, and heterosubtypic H1 challenges, whereas mice treated with both immune sera and T cells completely survived heterosubtypic H3 and H9 challenges.
Broad binding antibody responses elicited by TH/NE DDV+EP immunization.
To investigate immune responses, we first measured total IgG, IgG1, and IgG2a antibody responses in TH/NE DDV+EP sera against H1, H3, H5, H7, and H9 HA by enzyme-linked immunosorbent assay (ELISA; gray shadow in Fig. 4A). Figure 4B shows that the 50% effective concentrations (EC50s) of total IgG, IgG1, and IgG2a against homologous H7 HA were 192,518, 70,636, and 110,552, respectively. Compared to the EC50s against homologous H7 HA, there were 4- to 10-fold decreases against intrasubtypic H5 HA, 20- to 25-fold decreases against H1 HA, 20- to 40-fold decreases against H3 HA, and 60- to 110-fold decreases against H9 HA. Thus, there is a hierarchy in HA-specific binding antibody responses in the immune sera against H7, H5, H1, H3, and H9 HA, which correlates the antigenic distance between the HA of a given subtype and the H5 or H7 HA immunogen (red color in Fig. 4A).
FIG 4.
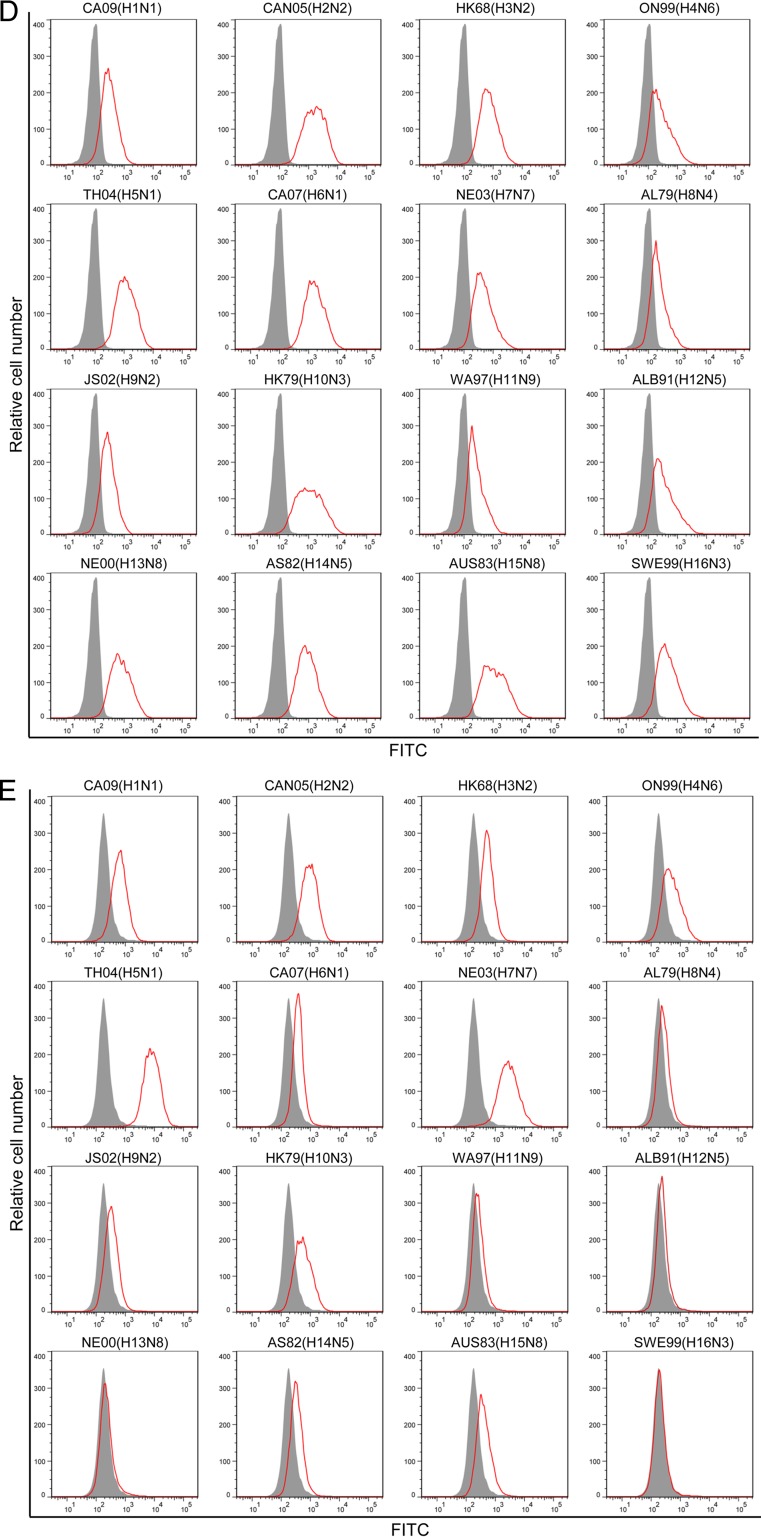
Binding antibody responses elicited by TH/NE DDV+EP immunization. (A) Phylogenetic tree of various HA used in binding assays. Red, homologous HA to immunogens (red); gray shading, HA used in ELISA; black, HA used to generate GPI-HA detected by FACS analysis. (B) Summary of EC50s and relative binding activity (in parentheses) of total IgG, IgG1, and IgG2a antibodies that bound to homologous H7 HA, intrasubtypic H5 HA, and heterosubtypic H1, H3, and H9 HA. The relative binding activity was calculated by dividing the EC50 against a given HA by the EC50 against homologous H7 HA. (C) Schematic diagram of the third-generation self-inactivating lentiviral vector pRRL-HA/hinge/His tag/foldon/DAF. HA, the ectodomain of HA from 16 HA subtypes (H1 to H16); hinge, a human IgG3 hinge region; His tag, a 6-histidine residue tag; foldon, a 27-residue trimerization domain at the C-terminal bacteriophage T4 fibritin; DAF, the C-terminal 34 amino acid residues of the decay-accelerating factor. The fusion gene was driven by the PGK promoter. (D) FACS analysis of cell surface expression of GPI-HA from 16 HA subtypes (H1 to H16) detected by anti-His tag antibody. Gray shading, mock-transduced CEM.NKR cells; red line, GPI-HA-transduced CEM.NKR cells. (E) FACS analysis of cell surface expression of GPI-HA from 16 HA subtypes (H1 to H16) detected by TH/NE DDV+EP sera, which had been repeatedly absorbed by CEM.NKR cells (see Materials and Methods). Gray shading, mock-transduced CEM.NKR cells; red line, GPI-HA-transduced CEM.NKR cells.
To further investigate the binding antibody responses, we generated a panel of transduced CEM.NKR cells expressing His-tagged glycosylphosphatidylinositol-anchored ectodomain of HA (GPI-HA) (Fig. 4C) from 16 HA subtypes (black color in Fig. 4A). Figure 4D shows that the cell surface expression of GPI-HA from all 16 subtypes were easily detected by anti-His tag antibody, although the levels of expression varied to some degrees among transduced cells. When mock- or GPI-HA-transduced CEM.NKR cells were stained with TH/NE DDV+EP sera, the GPI-HA from 14 of 16 subtypes was stained positively (Fig. 4E). TH/NE DDV+EP sera stained H5 and H7 HA extremely well, followed by H1, H2, H3, H4, and H10 HA and then by H6, H8, H9, H14, and H15 HA. Staining was low but measurable by H11 and H12 HA. Thus, like the EC50s shown in Fig. 4B, there is also a hierarchy in binding GPI-HA by TH/NE DDV+EP sera, which correlates antigenic distance between HA of a given subtype and H5 or H7 HA immunogen (red color in Fig. 4A).
Neutralizing antibody responses elicited by TH/NE DDV+EP immunization.
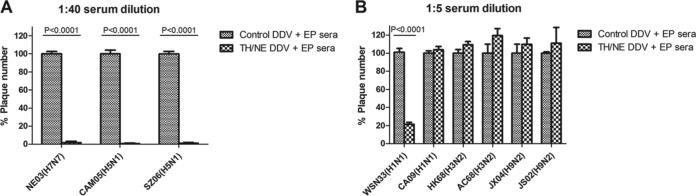
Figure 5A shows that TH/NE DDV+EP sera at 1:40 dilution completely inhibit plaque formation of NE03, CAM05, and SZ06 strains. Figure 5B shows that TH/NE DDV+EP sera at a 1:5 dilution significantly inhibit the plaque formation of WSN33 strain but not that of the other heterosubtypic strains. Thus, we conclude that TH/NE DDV+EP sera neutralize homologous and intrasubtypic H5 and H7 strains well but only moderately neutralize the limited heterosubtypic H1 strain.
FIG 5.
Neutralization measured by plaque reduction assay. (A) Comparison of percentages of plaques between TH/NE DDV+EP sera versus control DDV+EP sera at a 1:40 dilution against homologous NE03 H7N7 and intrasubtypic CAM05 H5N1 and SZ06 H5N1 strains. (B) Comparison of percentages of plaques between TH/NE DDV+EP sera versus control DDV+EP sera at 1:5 dilution against heterologous WSN33 H1N1, CA09 H1N1, HK68 H3N2, AC68 H3N1, JX04 H9N2, and JS02 H9N2 strains.
HA-specific T cell responses elicited by TH/NE DDV+EP immunization.
Finally, to test HA-peptide specific T cell responses, splenocytes from TH/NE DDV+EP- and control DDV+EP-immunized mice were assayed against a mixture of five HA peptides using intracellular cytokine staining assay. Figure 6A shows the gating strategy of single or double cytokine-secreted CD4 T cells upon peptide stimulation. On average, 0.28, 0.32, 0.24, 0.17, 0.22, 0.18, and 0.13% of CD4 T cells from TH/NE DDV+EP mice versus 0.05, 0.07, 0.06, 0.01, 0.03, 0.03, and 0.01% of CD4 T cells from control DDV+EP mice secreted gamma interferon (IFN-γ), interleukin-2 (IL-2), tumor necrosis factor alpha (TNF-α), IFN-γ + IL-2, IL-2 + TNF-α, IFN-γ + TNF-α, or IFN-γ + IL-2 + TNF-α, respectively. Statistical analyses between two groups yielded results that were all significantly different (P < 0.05) (Fig. 6B to H).
FIG 6.
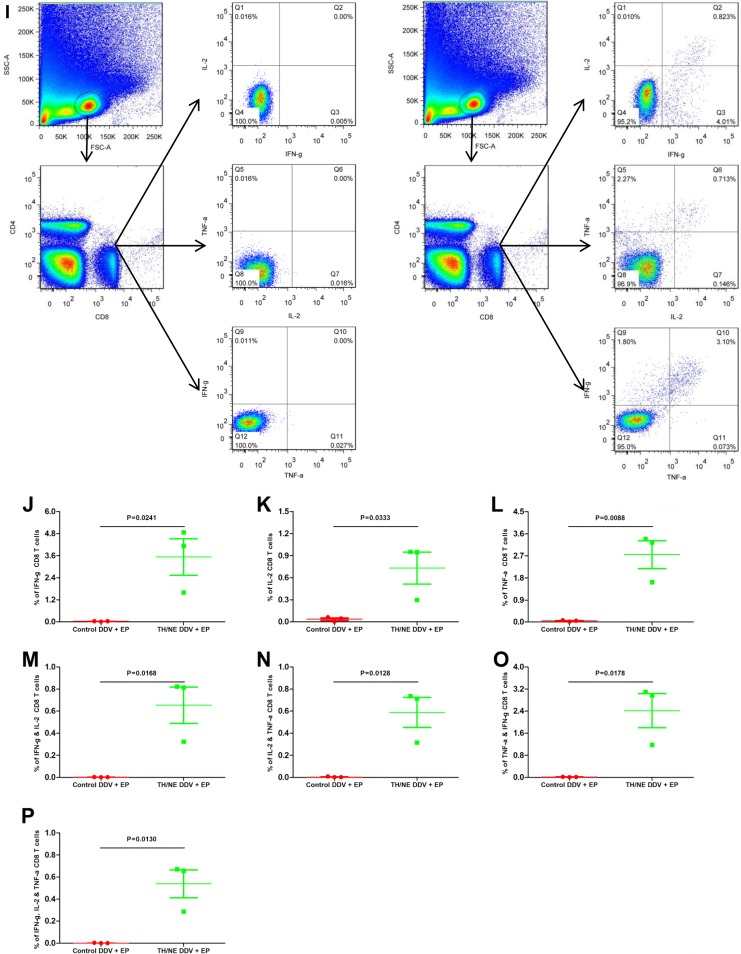
Intracellular cytokine staining analysis of CD4 and CD8 T cell responses against a panel of five HA peptides for TH/NE DDV+EP- and control DDV+EP-immunized mice. (A) Gating strategies of percentages of CD4 T cells that secreted IFN-γ, IL-2, and/or TNF-α. (B to D) The percentages of CD4 T cells that secreted IFN-γ, IL-2, TNF-α. (E to H) Percentages of CD4 T cells that secreted either both IFN-γ and IL-2 (IFN-γ/IL-2), IL-2/TNF-α, IFN-γ/TNF-α, or IFN-γ/IL-2/TNF-α. (I) Gating strategies of percentages of CD8 T cells that secreted IFN-γ, IL-2, and/or TNF-α. (J to L) Percentages of CD8 T cells that secreted IFN-γ, IL-2, and TNF-α. (M to P) Percentages of CD8 T cells that secreted either IFN-γ/IL-2, IL-2/TNF-α, IFN-γ/TNF-α, or IFN-γ/IL-2/TNF-α.
Figure 6I shows the gating strategy of single or double cytokine-secreted CD8 T cells upon peptide stimulation. On average, 3.53, 0.73, 2.75, 0.65, 0.59, 2.43, and 0.54% of CD8 T cells from TH/NE DDV+EP-immunized mice versus 0.03, 0.04, 0.05, 0.001, 0.002, 0.01, and 0.001% of CD8 T cells from control DDV+EP-immunized mice secreted IFN-γ, IL-2, TNF-α, IFN-γ plus IL-2 (IFN-γ/IL-2), IL-2/TNF-α, IFN-γ/TNF-α, or IFN-γ/IL-2/TNF-α, respectively. Statistical analyses between two groups were all significant different (P < 0.05) (Fig. 6J to P). Thus, we conclude that TH/NE DDV+EP immunization elicits both HA-peptide-specific CD4 and CD8 T cell responses.
DISCUSSION
In the present study, we developed a bivalent TH/NE DDV+EP vaccine strategy. We show that in vivo EP during DNA priming significantly enhances binding antibody responses quantitatively and qualitatively (Fig. 1A and B). As a result, immune sera elicited by bivalent TH/NE DDV+EP immunization bind HA2 (stem) from all 12 strains of both group 1 (H1, H5, and H9) and group 2 (H3, H4, H7, and H10) tested as well as HA1 (head) from 11 strains of subtypes H1, H3, H4, H5, H7, and H9 (Fig. 1C). Moreover, using fluorescence-activated cell sorting (FACS) analysis against transduced CEM.NKR cells expressing GPI-HA from 16 HA subtypes, we show that TH/NE DDV+EP sera bind 14 of 16 HA subtypes hierarchically (Fig. 4). Thus, this bivalent vaccine elicits very broader and hierarchical binding antibody responses against both the stems and the heads of diverse HA from both group 1 and 2 viruses, which correlates antigenic distance between a given HA subtype and an H5 or H7 immunogen.
In the present study, we also show that active TH/NE DDV+EP immunization confers (i) complete protection (no weight loss and all survival) against a lethal challenge of homologous H7 and intrasubtypic H5, and heterosubtypic H1 (WSN33), (ii) nearly complete protection (20% or less nadir weight loss and 100% survival) against heterosubtypic H1 (CA09), H3 (HK68), and H9 (JX04), and (iii) partial protection (25% or less nadir weight loss and 66 or 84% survival) against H3 (AC68) or H9 (JS02) (Fig. 2), resulting in >94% aggregate survival. Recently, Schwartzman et al. showed that an i.n. immunization with a cocktail of VLP-expressing group 1 (H1 and H5) and group 2 (H3 and H7) HA not only completely or near completely protects mice from homologous and intrasubtypic H1, H5, and H7 virus challenge but also partially protects mice from heterosubtypic H2, H6, H10, and H11 virus challenge, with a total of 94% aggregate survival (23). Thus, it appears that our bivalent (H5 and H7 HA) vaccine and their quadruple (H1, H3, H5, and H7 HA) VLP cocktail achieve similar levels of protection against homologous, intrasubtypic, and heterosubtypic strains even though the vaccine composition and administrations associated with these two regimens are quite different.
To correlate immune protection, we performed passive immunization, as well as in vitro neutralizing and binding antibody and HA-specific T cell responses. We demonstrated that passive immunization with immune sera alone completely protects mice against homologous H7, intrasubtypic H5, and heterosubtypic H1 challenge and that mice passively immunized with both immune sera and T cells completely survived heterosubtypic H3 or H9 challenge (Fig. 3). We also show that immune sera at 1:40 dilution completely inhibit the plaque formation of homologous H7 and intrasubtypic H5 viruses and, at a 1:5 dilution, significantly inhibit the plaque formation of heterosubtypic H1 (WSN33) virus, but not H1 (CA09), H3, and H9 viruses (Fig. 5). The EC50s of HA-specific total IgG, IgG1, and IgG2a in immune sera against homologous H7 are 4- to 10-fold higher than those against intrasubtypic H5, 20- to 25-fold higher against heterosubtypic H1, 20- to 40-fold higher against H3, and 60- to 110-fold higher against H9 (Fig. 4). Finally, we show that the vaccine elicits both HA-peptide-specific CD4 and CD8 T cell responses (Fig. 6). Thus, it appears that although this bivalent vaccine elicits HA-specific neutralizing and binding antibody and CD4 and CD8 T cell responses, neutralizing antibody responses alone fully protect against homologous and intrasubtypic H5 and H7 challenge, and neutralizing and binding antibody responses fully protect against heterosubtypic H1 challenge; however, both binding antibody and T cell responses are required for complete survival following heterosubtypic H3 and H9 challenge. However, more studies are needed to further correlate immune protections, such as mapping protective epitopes from diverse HA subtypes, as well as the role of CD4 versus CD8 T cells in immune protection.
Finally, DNA vaccines have been shown to be safe and well tolerated in various human trials against various pathogens (26–29). DNA plasmid has been shown to be a good priming regimen in animals and in humans against influenza viruses (30, 31). In addition, influenza VLPs have been shown to be safe and immunogenic in human trials (32, 33). Thus, the demonstration that this bivalent TH/NE DDV+EP vaccine elicits broad HA-specific antibody and T cell responses that confer broad protection against diverse influenza strains of subtypes H1, H3, H5, H7, and H9 in mice in the present study further supports the development of this vaccine in ferrets and in humans.
MATERIALS AND METHODS
Ethics statement.
The experimental protocol (CULATR-3064-13) was approved by the Animal Use Committee and the Safety Committee on BSL-3 Facility and Infectious Agents Li Ka-Shing Faculty of Medicine, the University of Hong Kong. All infection experiments were conducted at the biosafety level 3 facilities complying with the Ethics Committee regulations of University of Hong Kong, Hong Kong Special Administrative Region in accordance with EC directive 86/609/CEE. Embryonated chicken eggs were purchased from the Poultry Institute, Shandong Academy of Agricultural Science; the eggs were inoculated with viruses at day 9 of embryonation, and allantoic fluid was harvested at day 12 of embryonation.
Animals.
Female BALB/c mice at the age of 8 weeks were purchased from Charles River Laboratories (l'Arbresle, France) and housed in microisolator cages ventilated under negative pressure with HEPA-filtered air and a 12/12-h light/dark cycle. Before each inoculation or euthanasia procedure, the mice were anesthetized by an intraperitoneal (i.p.) injection of pentobarbital sodium (65 mg/kg; Sigma) to minimize suffering.
Cell lines.
The human embryonic kidney 293T cells were purchased from Invitrogen Life Technologies (Waltham, MA) and maintained in complete Dulbecco modified Eagle medium (high-glucose DMEM supplemented with 10% fetal bovine serum [FBS], 2 mM l-glutamine, 1 mM sodium pyruvate, penicillin [100 U/ml], and streptomycin [100 μg/ml]; Corning). The Madin-Darby canine kidney (MDCK) cell line was purchased from American Type Culture Collection and maintained in complete DMEM. The CEM.NKR CCR5+ Luc+ (CEM.NKR) cell line (34, 35) was obtained from the NIH AIDS Research and Reference Reagent Program (Germantown, MD) and maintained in complete RPMI 1640 medium (RPMI 1640 medium supplemented with 10% FBS, 2 mM l-glutamine, 1 mM sodium pyruvate, penicillin [100 U/ml], and streptomycin [100 μg/ml]).
Viruses.
Influenza viruses A/Cambodia/P0322095/2005 (CAM05) H5N1, A/Shenzhen/406H/2006 (SZ06) H5N1, A/WSN/1933 (WSN33) H1N1, A/California/07/2009 (CA09) H1N1, A/swine/Jiangxi/1/2004 (JX04) H9N2, A/chicken/Jiangsu/7/2002 (JS02) H9N2, A/Hong Kong/1/1968 (HK68) H3N2, A/Guizhou/54/1989 (GZ89) H3N2, and A/duck/Hunan/8-19/2009 (HN09) H4N2 were listed in Table 1. The reassortants A/Netherlands/219/2003 (NE03, RG 6+2) H7N7, A/Anhui/1/2013 (AH13, RG 6+2) H7N9, A/Jiangxi-Donghu/346/2013 (JX13, RG 6+2) H10N8, and A/Aichi/2/1968 (AC68, RG 7+1) H3N1 were generated by cotransfecting the 293T and the MDCK cell mixture with gene segments encoding HA and NA or HA protein derived from corresponding wild-type strains and the remaining six or seven gene segments encoding NA, PB2, PB1, PA, NP, M, and NS from A/PR8/1934 or A/WSN/1933 strain as described by Hoffmann et al. (36). Wild-type viruses and reassortants were propagated on MDCK cells or 9-day-old embryonated chicken eggs using standard viral culturing techniques. The PFU count was measured on MDCK cells, and the 50% mouse lethal doses (MLD50) of the viruses and reassortants were determined in BALB/c mice and calculated according to the method of Reed and Muench (37).
DNA plasmids.
Mammalian expression vector pCMV/R-HA and pCMV/R-NA from TH04 H5N1 and NE03 H7N7, as well as pCMV/R-HIV-1 Gag, were described previously (38).
To construct fusion genes encoding various GPI-anchored ectodomain HA, codon-optimized sequences encoding HA ectodomain from subtypes H1 to H16, the IgG3 hinge region, foldon (a 27-residue trimerization domain at the C-terminal bacteriophage T4 fibritin), and a histidine tag were genetically linked to the sequence encoding a GPI attachment signal (the C-terminal 34 amino acid residues of the decay-accelerating factor). The fusion genes were inserted into a third-generation lentiviral transfer vector pRRLsin-18.PPT.hPGK.Wpre (39). The resulting transfer constructs were designated pRRL-GPI-HAs.
VLP production.
TH and NE VLP expressing HA and NA from TH04 H5N1 and NE03 H7N7 strains and control VLP expressing HIV gag alone were generated as described before (24, 40). Briefly, to generate TH or NE VLP, 4.5 × 106 293T cells were cotransfected with 14 μg of pCMVΔ8.2, 2 μg of pCMV/R-H5 HA or pCMV/R-H7 HA, and 0.5 μg of pCMV/R-N1 NA or N7 NA using a calcium phosphate precipitation method. After overnight incubation, the cells were washed once with phosphate-buffered saline (PBS) and cultured in 10 ml of complete DMEM. The VLP-containing supernatants were harvested in 16 to 20 h, filtered through a 0.45-μm-pore-size filter, loaded onto 20% sucrose cushion, and ultracentrifuged at 25,000 rpm for 2 h at 4°C in an SW28 rotor (Beckman Coulter, Fullerton, CA). The pellets were resuspended in PBS and stored at −80°C in aliquots until use. The amount of HA on the surfaces of concentrated VLPs was measured with a standard hemagglutination assay.
Generation of stably transduced CEM.NKR cells expressing GPI-anchored ectodomain of HA from all 16 HA subtypes.
Recombinant lentiviruses were generated as described previously (41). Briefly, 4 × 106 293T cells were seeded onto a P-100 dish in 10 ml of complete DMEM. After overnight culture, the cells were cotransfected with 14 μg of one of sixteen pRRL-GPI-HA transfer constructs (see above), 14 μg of packaging construct encoding HIV-1 Gag/Pol (CMVRΔ8.2), and 2 μg of plasmids encoding the VSV-G protein envelope (pLP/VSV-G), using a calcium phosphate precipitation method. After 16 h, the culture supernatants were removed and replaced with fresh complete DMEM plus 1 mM sodium butyrate (Sigma). After an additional 8 h, later, the supernatants were again removed and replaced with fresh DMEM plus 4% FBS. After another 20 h, the culture supernatants were harvested and concentrated by ultracentrifugation as described previously (41). The vector pellets were resuspended in a small volume of DMEM and stored in aliquots in a −80°C freezer. Vector titers were determined as previously described (41).
To transduce CEM.NKR cells, 105 cells per well were seeded onto a 24-well plate. Cells were transduced with lentiviral vectors expressing GPI-HA at a multiplicity of infection of 20 in the presence of 8 μg of Polybrene/ml. After 24 h, the cells were washed with fresh complete RPMI 1640 and cultured in complete RPMI 1640. The stability of GPI-HA transgene expression in transduced CEM.NKR cells was checked periodically using anti-His tag antibody, followed by FACS analysis (see below).
Immunizations and immunization and challenge experiments.
To generate immune sera and splenocytes, 30 female BALB/c mice at the age of 8 weeks were randomly divided into five groups (6 mice per group). Mice in groups 1 and 2 were primed twice intramuscularly (i.m.) with 50 μg of pCMV/R-HA from TH04 or NE03 strain at week 0 and 3 and boosted once i.p. with 640 hemagglutinin units (HAU) of VLP from TH04 or NE03 strain at week 6 (designated the TH DDV and NE DDV groups, respectively). Mice in groups 3 and 4 were primed and boosted in the same way as in groups 1 and 2 except that during the priming DNA were delivered by in vivo EP, which consisted of six pulses with a 50-ms pulse length and a 1-s rest between adjacent pulses at a constant 60 V, using an EPT-I delivery device (TERESA, Shanghai, China) (designated the TH DDV+EP and NE DDV+EP groups, respectively). Mice in group 5 were i.m. primed twice with pCMV/R-HA from both TH04 and NE03 strains (50 μg each) at weeks 0 and 3, plus in vivo EP and i.p. boosted once with VLP from both the TH04 and NE03 strains (640 HAU each) at week 6 (designated the TH/NE DDV+EP group). At 7 days before the first prime and 14 days after the boost, serum samples were collected by retro-orbital plexus puncture, heat inactivated, and stored in aliquots at −20°C. The spleens were harvested 14 days after the boost, and splenocytes were isolated for intracellular cytokine staining (see below).
For the active immunization and challenge experiments, a total of 112 8-week-old female BALB/c mice were randomly divided into two groups (56 mice per group). Mice in the control group were i.m. primed twice with pCMV/R empty vector plus EP at weeks 0 and 3 and then i.p. boosted once with Gag-alone VLP at week 6. Mice in the immunization group were i.m. primed twice with pCMV/R-HA from both TH04 and NE03 strains (50 μg each) at weeks 0 and 3, plus in vivo EP, and i.p. boosted once with VLP from both the TH04 and the NE03 strains (640 HAU each) at week 6. At 2 weeks postboost, mice in both control and immunization groups were randomly divided into subgroups (five or six mice per subgroup) and challenged i.n. with 5 MLD50 of CAM05 H5N1, SZ06 H5N1, NE03 H7N7, WSN33 H1N1, CA09 H1N1, HK68 H3N2, AC68 H3N1, JX04 H9N2, or JS02 H9N2 strain or with 105 EID50 (50% egg infectious doses) of AH13 H7N9 virus. After the challenge, mice were monitored for the signs of illness for 14 days. Mice losing 30% of their initial weight were euthanized and scored as dead. For mice challenged with AH13 H7N9 virus, lung tissues (three mice per subgroup) were harvested and homogenized at day 3 postchallenge. EID50 in lung tissues were titrated on 9-day-old embryonated chicken eggs as described previously (42).
In passive immunization and challenge experiments, control and immune serum samples and splenocytes were collected from a total of 160 mice (80 in the control group and 80 in the TH/NE DDV+EP group) and combined at day 14 postimmunization. The serum samples were heat inactivated at 56°C for 30 min, and T cells were isolated by negative selection using mouse pan-T-cell isolation kit (Miltenyi Biotec) according to the manufacturer's instructions. Then, 700 μl of control or immune sera was i.p. transferred and/or 2 × 107 T cells (in 200 μl of PBS) were intravenously transferred into each naive recipient mouse. At 24 h after transfer, the mice were i.n. challenged with 5 MLD50 of the CAM05 H5N1, NE03 H7N7, WSN33 H1N1, HK68 H3N2, or JX04 H9N2 strain as described above. After the challenge, the survival and body weights were monitored daily for 14 days.
Plaque reduction assay.
To evaluate the neutralizing activity of TH/NE DDV+EP sera, MDCK cells (5 × 105 cells per well) in complete DMEM were seeded into six-well plates at 48 h before use. The medium was removed, the cells were washed with PBS, and 700 μl of serum-free DMEM was added to each well. The viruses (50 PFU per well) were incubated with 4-fold-diluted (starting at 1:40 against CAM05 H5N1, SZ06 H5N1, or NE03 H7N7 viruses) or 2-fold-diluted (starting at 1:5 against WSN33 H1N1, CA09 H1N1, AC68 H3N1, HK68 H3N2, JS02 H9N2, or JX04 H9N2 viruses) pooled TH/NE DDV+EP sera, with a final volume of 100 μl at 37°C for 1 h. The virus-serum mixture was then added onto the MDCK monolayer, followed by incubation at 37°C for 1 h. After the incubation, 0.8% (wt/vol) low-melting point agarose (Sigma) in MEM (3 ml per well) was added. At 72 h postinfection, the agarose overlay was discarded, and the cells were stained with 0.5% (wt/vol) crystal violet (1 ml per well) at room temperature for 1 h. Crystal violet was then removed, the cells were washed with H2O, and the number of plaques was counted as described previously (42). The assay was performed in triplicate.
Western blot analysis.
To determine the binding specificity of TH DDV, NE DDV, TH DDV+EP, NE DDV+EP, and TH/NE DDV+EP sera, 12 influenza viruses and a VSV control were produced and concentrated by ultracentrifugation. The concentrated virus samples were heated at 95°C for 10 min in the loading buffer containing 10% sodium dodecyl sulfate (SDS) and 5% β-mercaptoethanol and then loaded onto and separated by SDS–12% PAGE. The gels were transferred onto polyvinylidene difluoride membranes. The membranes were blocked with Tris-buffered saline containing 0.05% Tween 20 (TBST) and 5% nonfat dry milk overnight at 4°C. The membranes were subsequently incubated with 5 ml of 1:250-diluted pooled TH DDV, NE DDV, TH DDV+EP, NE DDV+EP, TH/NE DDV+EP, or combined TH DDV+EP and NE DDV+EP sera for 2 h at room temperature. After three washes with TBST, the membranes were incubated with 5 ml of 1:2,500-diluted horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG antibody for 1 h at room temperature. After an additional three washes with TBST, the membranes were probed with an EZ-ECL enhanced chemiluminescence detection kit for HRP (Biological Industries) according to the manufacturer's instructions.
ELISA.
To measure the total IgG, IgG1, and IgG2a antibody responses in TH/NE DDV+EP serum samples against divergent influenza HA, 96-well EIA/RIA flat-bottom plates (Corning) were coated with 100 ng of soluble ectodomain of HA proteins from WSN33 H1N1, HK68 H3N2, A/Anhui/1/2005 (AH05) H5N1, NE03 H7N7, or A/Hong Kong/1073/1999 (HK99) H9N2 (Sino Biological, Inc.) per well at 4°C overnight. The plates were then washed six times with PBS containing 0.05% Tween 20 (PBST) and blocked with PBS containing 5% FBS at 37°C for 1 h, followed by incubation with 4-fold-diluted pooled TH/NE DDV+EP serum samples (starting at a 1:40 dilution) at 37°C for 2 h. The plates were washed and incubated with HRP-conjugated goat anti-mouse IgG (1:5,000), goat anti-mouse IgG1 (1:500), or goat anti-mouse IgG2a (1:500) antibodies (Southern Biotech), respectively, at 37°C for 1 h. Then, the plates were washed six times with PBST buffer. Colorimetric analysis was performed using a TMB substrate kit (Thermo Scientific), and the absorbance was read at 450 nm by a spectrophotometer (Thermo Scientific). Titration curves were generated using sigmoid dose-response of nonlinear fit from GraphPad, and the EC50s were determined as the dilutions of immune serum that generated 50% of maximal optical densities at 450 nm.
FACS analysis.
To analyze the cell surface expression of HA from 16 HA subtypes (H1 to H16), CEM.NKR cells stably transduced with GPI-HA from CA09 H1N1, CAN05 H2N2, HK68 H3N2, ON99 H4N6, TH04 H5N1, CA07 H6N1, NE03 H7N7, AL79 H8N4, JS02 H9N2, HK79 H10N3, WA97 H11N9, ALB91 H12N5, NE00 H13N8, AS82 H14N5, AUS83 H15N8, or SWE99 H16N3 virus were incubated with a mouse anti-His tag antibody (Sigma), followed by fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG antibody (H&L; Life Technologies). The cells were washed twice with FACS buffer (PBS containing 1% BSA and 0.02% NaN3) and fixed with 2% formaldehyde in 0.3 ml of FACS buffer. FACS analysis was performed on an LSRII apparatus (Becton Dickinson, Mountain View, CA).
To evaluate the binding activity of TH/NE DDV+EP sera against various GPI-HA, CEM.NKR cells stably transduced with GPI-HA from the 16 HA subtypes described above were incubated with a 1:40-diluted pooled TH/NE DDV sera for 45 min on ice. Prior to the binding assay, pooled TH/NE DDV sera had been absorbed six times with CEM.NKR cells to remove the nonspecific binding activity. After the incubation, the cells were washed twice with FACS buffer and stained with FITC-conjugated goat anti-mouse IgG antibody (dilution of 1:500) for another 45 min on ice. The cells then were washed twice with FACS buffer and fixed with 2% formaldehyde in 0.5 ml of FACS buffer. FACS analysis was performed using an LSRII flow cytometer.
Intracellular cytokine staining assay.
To evaluate HA peptide-specific CD4 and CD8 T cell responses, an intracellular cytokine staining assay was performed as described previously (43). Briefly, spleens were harvested from control DDV+EP and TH/NE DDV+EP group mice at 14 days postboost and gently homogenized into a single-cell suspension. After erythrocyte lysis with ammonium-chloride-potassium (ACK), the splenocytes (2 × 106 cells/well) in complete RPMI 1640 medium were incubated with a mixture of five HA peptides—IYSTVASSL, LYEKVRLQL, HFEKIQIIPKS, KSSFFRNVVWLIKKN, and TIKRSYNNTNQE (5 μg/well for each peptide) (44, 45)—along with anti-mouse CD28 (2 μg/well) and anti-mouse CD49d (2 μg/well) antibodies in 24-well tissue culture plates at 37°C. After 2 h, Golgi plug (2 μl/well) was added. After a 6-h incubation, the cells were rested overnight at 4°C. The following morning, the cells were incubated with anti-mouse CD16/32 (Fc block) antibody, followed by surface staining with PerCP-conjugated anti-mouse CD4 and allophycocyanin-conjugated anti-mouse CD8 antibodies on ice for 45 min. The cells were then permeabilized with Cytofix/Cytoperm and stained with FITC-conjugated anti-mouse IFN-γ, phycoerythrin (PE)-conjugated anti-mouse IL-2, and PE-Cy7-conjugated anti-mouse TNF-α antibodies on ice for 45 min (all staining reagents were purchased from BD Biosciences). A total of 106 cells per sample were acquired on an LSRII flow cytometer. FACS data were analyzed using FlowJo software.
Statistical analysis.
Analyses were performed using GraphPad Prism v5.0. An unpaired Student t test was used to compare the two data sets, and differences were considered significant at a P of <0.05.
ACKNOWLEDGMENTS
We thank L. Naldini at the Turin University Medical School, Turin, Italy, for lentiviral transfer vector and other members of the Unit of Anti-Viral Immunity and Genetic Therapy, Institut Pasteur of Shanghai, Chinese Academy of Sciences, for helpful discussions during the course of this study. CEM.NKR CCR5+ Luc+ cells were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Germantown, MD. The cell line was originally developed and contributed by John Moore and Catherine Spenlehauer.
This study was supported by the 12-5 Mega Project (2013ZX10004003003003) and the 863 Project (2012AA02A404) from the Ministry of Science and Technology in China and by a grant from the Li Ka-Shing Foundation in Hong Kong.
REFERENCES
- 1.World Health Organization. 2016. Influenza at the human-animal interface. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.World Health Organization. 2014. Weekly epidemiological record. World Health Organization, Geneva, Switzerland: http://www.who.int/wer/2014/wer8911.pdf?ua=1. [Google Scholar]
- 3.Air GM. 1981. Sequence relationships among the hemagglutinin genes of 12 subtypes of influenza A virus. Proc Natl Acad Sci U S A 78:7639–7643. doi: 10.1073/pnas.78.12.7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fouchier RA, Munster V, Wallensten A, Bestebroer TM, Herfst S, Smith D, Rimmelzwaan GF, Olsen B, Osterhaus AD. 2005. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J Virol 79:2814–2822. doi: 10.1128/JVI.79.5.2814-2822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu X, Yu W, McBride R, Li Y, Chen LM, Donis RO, Tong S, Paulson JC, Wilson IA. 2013. Hemagglutinin homologue from H17N10 bat influenza virus exhibits divergent receptor-binding and pH-dependent fusion activities. Proc Natl Acad Sci U S A 110:1458–1463. doi: 10.1073/pnas.1218509110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO. 2013. New World bats harbor diverse influenza A viruses. PLoS Pathog 9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krammer F, Pica N, Hai R, Tan GS, Palese P. 2012. Hemagglutinin stalk-reactive antibodies are boosted following sequential infection with seasonal and pandemic H1N1 influenza virus in mice. J Virol 86:10302–10307. doi: 10.1128/JVI.01336-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hai R, Krammer F, Tan GS, Pica N, Eggink D, Maamary J, Margine I, Albrecht RA, Palese P. 2012. Influenza viruses expressing chimeric hemagglutinins: globular head and stalk domains derived from different subtypes. J Virol 86:5774–5781. doi: 10.1128/JVI.00137-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krammer F, Pica N, Hai R, Margine I, Palese P. 2013. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J Virol 87:6542–6550. doi: 10.1128/JVI.00641-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Margine I, Krammer F, Hai R, Heaton NS, Tan GS, Andrews SA, Runstadler JA, Wilson PC, Albrecht RA, Garcia-Sastre A, Palese P. 2013. Hemagglutinin stalk-based universal vaccine constructs protect against group 2 influenza A viruses. J Virol 87:10435–10446. doi: 10.1128/JVI.01715-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tran EE, Podolsky KA, Bartesaghi A, Kuybeda O, Grandinetti G, Wohlbold TJ, Tan GS, Nachbagauer R, Palese P, Krammer F, Subramaniam S. 2016. Cryo-electron microscopy structures of chimeric hemagglutinin displayed on a universal influenza vaccine candidate. mBio 7:e00257. doi: 10.1128/mBio.00257-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graves PN, Schulman JL, Young JF, Palese P. 1983. Preparation of influenza virus subviral particles lacking the HA1 subunit of hemagglutinin: unmasking of cross-reactive HA2 determinants. Virology 126:106–116. doi: 10.1016/0042-6822(83)90465-8. [DOI] [PubMed] [Google Scholar]
- 13.Sagawa H, Ohshima A, Kato I, Okuno Y, Isegawa Y. 1996. The immunological activity of a deletion mutant of influenza virus haemagglutinin lacking the globular region. J Gen Virol 77(Pt 7):1483–1487. doi: 10.1099/0022-1317-77-7-1483. [DOI] [PubMed] [Google Scholar]
- 14.Steel J, Lowen AC, Wang TT, Yondola M, Gao QS, Haye K, Garcia-Sastre A, Palese P. 2010. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. mBio 1:e00018-10. doi: 10.1128/mBio.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bommakanti G, Lu X, Citron MP, Najar TA, Heidecker GJ, ter Meulen J, Varadarajan R, Liang X. 2012. Design of Escherichia coli-expressed stalk domain immunogens of H1N1 hemagglutinin that protect mice from lethal challenge. J Virol 86:13434–13444. doi: 10.1128/JVI.01429-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mallajosyula VV, Citron M, Ferrara F, Lu X, Callahan C, Heidecker GJ, Sarma SP, Flynn JA, Temperton NJ, Liang X, Varadarajan R. 2014. Influenza hemagglutinin stem-fragment immunogen elicits broadly neutralizing antibodies and confers heterologous protection. Proc Natl Acad Sci U S A 111:E2514–E2523. doi: 10.1073/pnas.1402766111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valkenburg SA, Mallajosyula VV, Li OT, Chin AW, Carnell G, Temperton N, Varadarajan R, Poon LL. 2016. Stalking influenza by vaccination with pre-fusion headless HA mini-stem. Sci Rep 6:22666. doi: 10.1038/srep22666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu Y, Welsh JP, Swartz JR. 2014. Production and stabilization of the trimeric influenza hemagglutinin stem domain for potentially broadly protective influenza vaccines. Proc Natl Acad Sci U S A 111:125–130. doi: 10.1073/pnas.1308701110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wohlbold TJ, Nachbagauer R, Margine I, Tan GS, Hirsh A, Krammer F. 2015. Vaccination with soluble headless hemagglutinin protects mice from challenge with divergent influenza viruses. Vaccine 33:3314–3321. doi: 10.1016/j.vaccine.2015.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Impagliazzo A, Milder F, Kuipers H, Wagner MV, Zhu X, Hoffman RM, van Meersbergen R, Huizingh J, Wanningen P, Verspuij J, de Man M, Ding Z, Apetri A, Kukrer B, Sneekes-Vriese E, Tomkiewicz D, Laursen NS, Lee PS, Zakrzewska A, Dekking L, Tolboom J, Tettero L, van Meerten S, Yu W, Koudstaal W, Goudsmit J, Ward AB, Meijberg W, Wilson IA, Radosevic K. 2015. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 349:1301–1306. doi: 10.1126/science.aac7263. [DOI] [PubMed] [Google Scholar]
- 21.Yassine HM, Boyington JC, McTamney PM, Wei CJ, Kanekiyo M, Kong WP, Gallagher JR, Wang L, Zhang Y, Joyce MG, Lingwood D, Moin SM, Andersen H, Okuno Y, Rao SS, Harris AK, Kwong PD, Mascola JR, Nabel GJ, Graham BS. 2015. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med 21:1065–1070. doi: 10.1038/nm.3927. [DOI] [PubMed] [Google Scholar]
- 22.Fan X, Hashem AM, Chen Z, Li C, Doyle T, Zhang Y, Yi Y, Farnsworth A, Xu K, Li Z, He R, Li X, Wang J. 2015. Targeting the HA2 subunit of influenza A virus hemagglutinin via CD40L provides universal protection against diverse subtypes. Mucosal Immunol 8:211–220. doi: 10.1038/mi.2014.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwartzman LM, Cathcart AL, Pujanauski LM, Qi L, Kash JC, Taubenberger JK. 2015. An intranasal virus-like particle vaccine broadly protects mice from multiple subtypes of influenza A virus. mBio 6:e01044. doi: 10.1128/mBio.01044-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang G, Zhou F, Buchy P, Zuo T, Hu H, Liu J, Song Y, Ding H, Tsai C, Chen Z, Zhang L, Deubel V, Zhou P. 2014. DNA prime and virus-like particle boost from a single H5N1 strain elicits broadly neutralizing antibody responses against head region of H5 hemagglutinin. J Infect Dis 209:676–685. doi: 10.1093/infdis/jit414. [DOI] [PubMed] [Google Scholar]
- 25.Joseph T, McAuliffe J, Lu B, Jin H, Kemble G, Subbarao K. 2007. Evaluation of replication and pathogenicity of avian influenza a H7 subtype viruses in a mouse model. J Virol 81:10558–10566. doi: 10.1128/JVI.00970-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin JE, Sullivan NJ, Enama ME, Gordon IJ, Roederer M, Koup RA, Bailer RT, Chakrabarti BK, Bailey MA, Gomez PL, Andrews CA, Moodie Z, Gu L, Stein JA, Nabel GJ, Graham BS. 2006. A DNA vaccine for Ebola virus is safe and immunogenic in a phase I clinical trial. Clin Vaccine Immunol 13:1267–1277. doi: 10.1128/CVI.00162-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin JE, Louder MK, Holman LA, Gordon IJ, Enama ME, Larkin BD, Andrews CA, Vogel L, Koup RA, Roederer M, Bailer RT, Gomez PL, Nason M, Mascola JR, Nabel GJ, Graham BS, Team VS. 2008. A SARS DNA vaccine induces neutralizing antibody and cellular immune responses in healthy adults in a phase I clinical trial. Vaccine 26:6338–6343. doi: 10.1016/j.vaccine.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kibuuka H, Kimutai R, Maboko L, Sawe F, Schunk MS, Kroidl A, Shaffer D, Eller LA, Kibaya R, Eller MA, Schindler KB, Schuetz A, Millard M, Kroll J, Dally L, Hoelscher M, Bailer R, Cox JH, Marovich M, Birx DL, Graham BS, Michael NL, de Souza MS, Robb ML. 2010. A phase 1/2 study of a multiclade HIV-1 DNA plasmid prime and recombinant adenovirus serotype 5 boost vaccine in HIV-uninfected East Africans (RV 172). J Infect Dis 201:600–607. doi: 10.1086/650299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ledgerwood JE, Pierson TC, Hubka SA, Desai N, Rucker S, Gordon IJ, Enama ME, Nelson S, Nason M, Gu WJ, Bundrant N, Koup RA, Bailer RT, Mascola JR, Nabel GJ, Graham BS, Team VS. 2011. A West Nile Virus DNA vaccine utilizing a modified promoter induces neutralizing antibody in younger and older healthy adults in a phase I clinical trial. J Infect Dis 203:1396–1404. doi: 10.1093/infdis/jir054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei CJ, Boyington JC, McTamney PM, Kong WP, Pearce MB, Xu L, Andersen H, Rao S, Tumpey TM, Yang ZY, Nabel GJ. 2010. Induction of broadly neutralizing H1N1 influenza antibodies by vaccination. Science 329:1060–1064. doi: 10.1126/science.1192517. [DOI] [PubMed] [Google Scholar]
- 31.Ledgerwood JE, Wei CJ, Hu ZH, Gordon IJ, Enama ME, Hendel CS, McTamney PM, Pearce MB, Yassine HM, Boyington JC, Bailer R, Tumpey TM, Koup RA, Mascola JR, Nabel GJ, Graham BS, Team VS. 2011. DNA priming and influenza vaccine immunogenicity: two phase 1 open label randomised clinical trials. Lancet Infect Dis 11:916–924. doi: 10.1016/S1473-3099(11)70240-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khurana S, Wu J, Verma N, Verma S, Raghunandan R, Manischewitz J, King LR, Kpamegan E, Pincus S, Smith G, Glenn G, Golding H. 2011. H5N1 virus-like particle vaccine elicits cross-reactive neutralizing antibodies that preferentially bind to the oligomeric form of influenza virus hemagglutinin in humans. J Virol 85:10945–10954. doi: 10.1128/JVI.05406-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez-Macias C, Ferat-Osorio E, Tenorio-Calvo A, Isibasi A, Talavera J, Arteaga-Ruiz O, Arriaga-Pizano L, Hickman SP, Allende M, Lenhard K, Pincus S, Connolly K, Raghunandan R, Smith G, Glenn G. 2011. Safety and immunogenicity of a virus-like particle pandemic influenza A (H1N1) 2009 vaccine in a blinded, randomized, placebo-controlled trial of adults in Mexico. Vaccine 29:7826–7834. doi: 10.1016/j.vaccine.2011.07.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trkola A, Matthews J, Gordon C, Ketas T, Moore JP. 1999. A cell line-based neutralization assay for primary human immunodeficiency virus type 1 isolates that use either the CCR5 or the CXCR4 coreceptor. J Virol 73:8966–8974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spenlehauer C, Gordon CA, Trkola A, Moore JP. 2001. A luciferase-reporter gene-expressing T-cell line facilitates neutralization and drug-sensitivity assays that use either R5 or X4 strains of human immunodeficiency virus type 1. Virology 280:292–300. doi: 10.1006/viro.2000.0780. [DOI] [PubMed] [Google Scholar]
- 36.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A 97:6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am J Epidemiol 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 38.Tsai C, Caillet C, Hu H, Zhou F, Ding H, Zhang G, Zhou B, Wang S, Lu S, Buchy P, Deubel V, Vogel FR, Zhou P. 2009. Measurement of neutralizing antibody responses against H5N1 clades in immunized mice and ferrets using pseudotypes expressing influenza hemagglutinin and neuraminidase. Vaccine 27:6777–6790. doi: 10.1016/j.vaccine.2009.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Follenzi A, Ailles LE, Bakovic S, Geuna M, Naldini L. 2000. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat Genet 25:217–222. doi: 10.1038/76095. [DOI] [PubMed] [Google Scholar]
- 40.Ding H, Tsai CG, Gutierrez RA, Zhou F, Buchy P, Deubel V, Zhou P. 2011. Superior neutralizing antibody response and protection in mice vaccinated with heterologous DNA prime and virus like particle boost against HPAI H5N1 virus. PLoS One 6:e16563. doi: 10.1371/journal.pone.0016563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wen M, Arora R, Wang H, Liu L, Kimata JT, Zhou P. 2010. GPI-anchored single chain Fv: an effective way to capture transiently exposed neutralization epitopes on HIV-1 envelope spike. Retrovirology 7:79. doi: 10.1186/1742-4690-7-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ren HH, Wang GQ, Wang SS, Chen HL, Chen ZW, Hu HX, Cheng GH, Zhou P. 2016. Cross-protection of newly emerging HPAI H5 viruses by neutralizing human monoclonal antibodies: a viable alternative to oseltamivir. MAbs 8:1156–1166. doi: 10.1080/19420862.2016.1183083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou F, Wang G, Buchy P, Cai Z, Chen H, Chen Z, Cheng G, Wan XF, Deubel V, Zhou P. 2012. A triclade DNA vaccine designed on the basis of a comprehensive serologic study elicits neutralizing antibody responses against all clades and subclades of highly pathogenic avian influenza H5N1 viruses. J Virol 86:6970–6978. doi: 10.1128/JVI.06930-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng Y, Yewdell JW, Eisenlohr LC, Bennink JR. 1997. MHC affinity, peptide liberation, T cell repertoire, and immunodominance all contribute to the paucity of MHC class I-restricted peptides recognized by antiviral CTL. J Immunol 158:1507–1515. [PubMed] [Google Scholar]
- 45.Valkenburg SA, Li OT, Mak PW, Mok CK, Nicholls JM, Guan Y, Waldmann TA, Peiris JS, Perera LP, Poon LL. 2014. IL-15 adjuvanted multivalent vaccinia-based universal influenza vaccine requires CD4+ T cells for heterosubtypic protection. Proc Natl Acad Sci U S A 111:5676–5681. doi: 10.1073/pnas.1403684111. [DOI] [PMC free article] [PubMed] [Google Scholar]