Abstract.
Optical intrinsic signal (OIS) imaging has been a powerful tool for capturing functional brain hemodynamics in rodents. Recent wide field-of-view implementations of OIS have provided efficient maps of functional connectivity from spontaneous brain activity in mice. However, OIS requires scalp retraction and is limited to superficial cortical tissues. Diffuse optical tomography (DOT) techniques provide noninvasive imaging, but previous DOT systems for rodent neuroimaging have been limited either by sparse spatial sampling or by slow speed. Here, we develop a DOT system with asymmetric source–detector sampling that combines the high-density spatial sampling (0.4 mm) detection of a scientific complementary metal-oxide-semiconductor camera with the rapid (2 Hz) imaging of a few () structured illumination (SI) patterns. Analysis techniques are developed to take advantage of the system’s flexibility and optimize trade-offs among spatial sampling, imaging speed, and signal-to-noise ratio. An effective source–detector separation for the SI patterns was developed and compared with light intensity for a quantitative assessment of data quality. The light fall-off versus effective distance was also used for in situ empirical optimization of our light model. We demonstrated the feasibility of this technique by noninvasively mapping the functional response in the somatosensory cortex of the mouse following electrical stimulation of the forepaw.
Keywords: diffuse optical tomography, functional neuroimaging, noninvasive imaging, optical imaging, structured illumination
1. Introduction
As advances in functional magnetic resonance imaging (fMRI) have transformed the study of human brain function, they have also widened the divide between standard research techniques used in humans and those used in mouse models. Although both task-based evoked responses1,2 and resting state networks3,4 have recently been observed in mice using fMRI, high signal-to-noise ratio (SNR) and resolution remain challenging due to the small volume of the mouse brain, and the logistics of fMRI hinder widespread application to high-throughput mouse studies. Therefore, a need exists for a fast benchtop modality for studying brain networks in mice. Optical imaging techniques, such as optical intrinsic signal imaging5 (OIS), have been developed and widely applied to task-based evoked responses.6,7 Most recently, OIS has been applied with wide field-of-view (FOV) to monitor functional connectivity in cases of disease8,9 and development10 in the mouse brain. However, traditional OIS methods are limited to planar imaging, providing only a two-dimensional view of cortical activity. In addition, OIS requires, at the least, a minimally invasive procedure of scalp reflection, making longitudinal imaging difficult or even impossible in some populations, such as infant mice.
In contrast to OIS, diffuse optical tomography (DOT) provides noninvasive volumetric imaging at depths extending to multiple centimeters, which in principle solves some of the limitations of OIS. In addition to DOT instrumentation, algorithms have been developed for handling arbitrary tissue geometries that can be matched to anatomy using numerical finite element modeling (FEM) of light transport.11 Although most papers have focused on humans,12,13 there have been some reports of the application of DOT to rodents.14,15 However, thus far, most animal DOT systems have either been fiber based, which are limited by sparse spatial sampling,14 or charge-coupled device based, which are limited by slow frame rates (0.1 Hz) that preclude imaging functional brain hemodynamics.15
Here, we present an imaging system that combines structured illumination (SI) with traditional DOT techniques (SI-DOT) to image a wide FOV () at high speed (). Successful implementation of SI-DOT for mouse functional neuroimaging requires optimizing for SI pattern sequences that preferentially sample deeper tissue. We introduce an analysis of the SNR for these patterns that quantifies the average light intensity as a function of an effective source-detector distance. This provides a light intensity versus distance measure analogous to methods used in traditional point illumination DOT. Following optimization, we validate SI-DOT for noninvasive imaging in mice by observing cortical responses to peripheral stimulation through the intact scalp.
2. Results
2.1. Structured Illumination-Diffuse Optical Tomography Imaging System
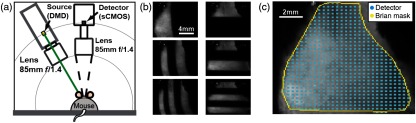
The goal of the SI-DOT instrument is to provide noninvasive functional neuroimaging of cortical hemodynamics (through both the scalp and the skull) at a speed of with an FOV of to cover the dorsal convexity of the mouse brain. The system leverages fast, low noise detection provided by a single scientific complementary metal-oxide-semiconductor (sCMOS) camera (Zyla 5.5, Andor Technology Ltd., South Windsor, Connecticut). For multicolor SI, we used a single digital micromirror device (DMD) projector (Lightcrafter 4500, Texas Instruments, Dallas, Texas). Within the projector, multicolored LEDs were reflected off a array of micromirrors to display arbitrarily complex two-dimensional illumination patterns. The sizes of the DMD chip, the mouse head, and the sCMOS sensor were all similar, which allowed for symmetric imaging optics. In particular, 85-mm lenses were used to maximize the FOV while allowing sufficient working distance, so that the projector could illuminate the mouse head from above [Fig. 1(a)]. To prevent specular reflection off the scalp from saturating the sensor, a polarizer ( 72 mm XS-Pro Kaesemann, Schneider Optics, Van Nuys, California) was placed on the projector with its polarization axis 90 deg relative to a second polarizer in front of the camera lens. A sequence of predefined illumination patterns were created in MATLAB (Mathworks, Natick, Massachusetts) and uploaded onto the projector’s on-board memory. Individual patterns were triggered one at a time and synchronized with each camera frame [Fig. 1(b)]. Several illumination sequences were explored, typically containing 40 different structured patterns. Each pattern ranged in spatial frequency from 0.08 to with two phases (180-deg phase shifts) and two orientations included per frequency. Images were collected at a camera frame rate of 80 Hz, providing a full DOT frame rate of 2 Hz. Each detection frame spanned a FOV using the central of the sensor. The data were binned to prior to reconstruction to improve SNR, yielding a pixel size of . The binned data had a dynamic range of , with the typical maximum values of and background standard deviations of approximately counts. The illumination patterns combined with detectors over the scalp provided total measurements [Fig. 1(c)].
Fig. 1.
SI-DOT system. (a) System schematic showing the relative positions and orientations of the DMD source projector and the sCMOS camera. Internal LEDs and optics illuminate the DMD with the desired wavelength, and illumination patterns are stored in the on-board projector memory and triggered consecutively to illuminate the head with the desired spatial frequencies. (b) Six example structured light patterns illuminating the intact mouse scalp, as collected by the sCMOS. (c) The planar-frame illumination of a mouse head as measured by the sCMOS camera, showing the positions of detectors over the intact scalp, after off-camera binning. A hand-drawn brain mask removes measurements that lie outside the exposed scalp for each mouse.
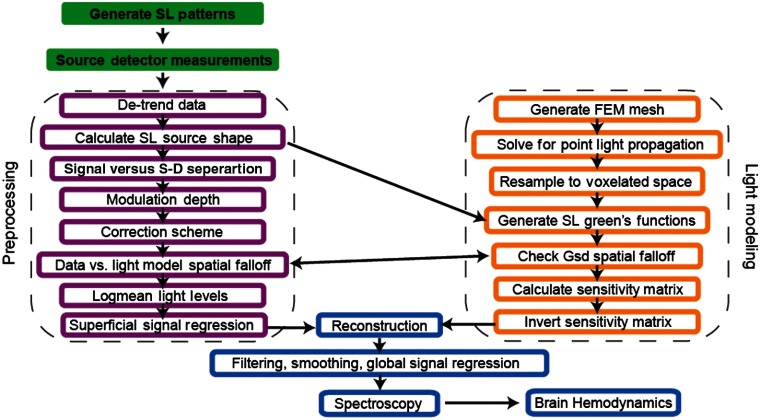
Light modeling and data analysis were conducted along parallel processing pipelines, adapted from our previous methods for high-density DOT (Fig. 2, see methods).12 Briefly, a finite element mesh was generated to model the mouse brain volume, and the fluence distributions for individual point light sources (Green’s functions) were calculated using NIRFAST.11 The mouse head was then resampled to a rectilinear grid (voxels). To generate Green’s functions specific to an SI pattern, the Green’s functions for all illuminated pixels within each SI pattern were summed (see Appendix A.1). To map measurements to voxels, a sensitivity matrix was calculated using a Jacobian approach and a linear Rytov forward model.12 Data were reconstructed using direct inversion and a regularized pseudoinverse of the forward sensitivity matrix following methods used commonly for human DOT data.11,12,16 Multiplying the inverted sensitivity matrix by the log-ratio of the light measurements produced a volumetric image sequence of absorption perturbations over time. Changes in absorption were converted to changes in chromophore concentration using the extinction coefficients of each hemoglobin species.17
Fig. 2.
SI-DOT processing stream. Raw data preprocessing (left, magenta boxes): raw measurement data collected by the sCMOS camera are subject to a series of quality assessment checks and prepared for reconstruction. Light modeling (right, orange boxes): a finite element mesh is created in NIRFAST and used for the forward model and calculation of the sensitivity matrix. Feedback between the model and the analysis of the raw data informs the optimization of the light model used in calculating the sensitivity matrix for each mouse. Once a satisfactory sensitivity matrix is generated, the data are reconstructed and spectroscopically unmixed to observe cortical hemodynamics (bottom, blue boxes).
2.2. Optimizing Measurements Using Effective Source–Detector Separation
A common technique for assessing measurement quality in DOT is to examine the measured light-level intensity as a function of source–detector separation. For SI-DOT, the source–detector separation is not as conceptually obvious as the Euclidean distance between the point-like sources and detectors in fiber-based DOT systems. However, a similar analysis would still be useful for evaluating measurement noise. Here, we defined an effective source–detector separation between each dark detector pixel and the source pattern for every measurement [Figs. 3(a) and 3(b)]. If one assumes a logarithmic fall-off of light fluence as a function of distance (e.g., following the Beer–Lambert law) and an effective attenuation coefficient of (a sensible value for the wavelengths in use), then a reasonable definition for an effective source–detector separation between a detector (pixel) and an illumination pattern is the exponentially weighted average of distances to each illuminated pixel. Briefly, for a dark detector pixel , source pixel , and source pattern , the effective distance is defined as
| (1) |
where and are the locations of dark detector pixel and source pixel , respectively, is defined to be , and is the set of bright pixels contained in the source pattern . Each pair of vertical bars denotes the two-norm of the enclosed expression, so is the ordinary Euclidean distance between and .
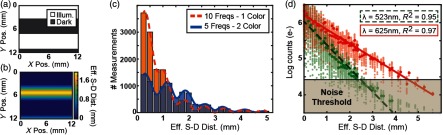
Fig. 3.
Measured light intensity versus effective source–detector separation. (a) Empirically determined edges of illuminated regions for one example pattern. (b) The effective source–detector separation is calculated for each detector in the dark region of each source pattern, shown here for one example pattern. (c) Total number of measurements as a function of distance, binned into 0.25 mm groups, for an illumination sequence consisting of 10 spatial frequencies and one color (orange) and a sequence of five frequencies and two colors (blue). (d) Signal intensity versus effective source–detector separation for each individual measurement, with averages at each unique distance emphasized and linearly fit. Data below the noise threshold ( background standard deviation) were cropped prior to reconstruction.
In principle, the set of possible patterns and pattern sequences is quite extensive as the problem expands combinatorially. We chose to evaluate binary square wave patterns that provide greater dynamic range and can be triggered faster using a DMD in comparison with sine waves, which are typically used in modulated light imaging18 or SI microscopy.19 We first evaluated ten spatial frequencies with two phases (180 deg offsets) and two orientations (vertical and horizontal) each. For this first pattern set, we found that over 75% of the measurements had an effective source–detector separation of 1 mm or less [Fig. 3(c), orange/dotted]. In a second pattern set, by sacrificing the five highest spatial frequencies, a dense set of measurements with short separation distances is preserved, whereas the relative sampling at larger distances is increased, improving the sensitivity to deeper tissue [Fig. 3(C), blue/solid]. With fewer spatial frequencies, the pattern sequence was expanded to include two wavelengths for spectroscopic reconstructions of multiple chromophores (i.e., oxy-, deoxy-, and total hemoglobin) while keeping the full frame rate at 2 Hz.
The average signal among all measurements at each unique effective source–detector separation showed an expected log-linear fall-off. In addition to guiding optimization of the illumination patterns, this technique helped us quantitatively identify noisy measurements. For example, the signals from green () and red () illumination patterns began to approach the noise floor of the camera at effective source–detector separations greater than 3 and 4.5 mm, respectively [Fig. 3(d)]. We empirically determined a threshold for cropping bad measurements by examining the standard deviation of light intensity in the background frames. Measurements greater than 20 times this value (e.g., electrons for a representative measurement) were used for all reconstructions. This noise threshold removed of all measurements, the majority of which fell outside the visually identified brain region (see Appendix A.2).
2.3. In Situ Estimation of Baseline Optical Properties
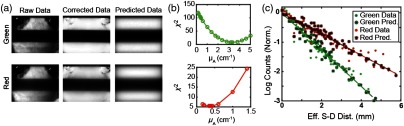
The sensitivity matrix used for DOT image reconstruction requires an estimate of the baseline tissue optical properties. Often, DOT algorithms use assumed attenuation coefficients from extant literature rather than in situ measurements. The data acquired in SI-DOT have the potential to permit in situ estimation of baseline optical properties by comparing the raw collected data with a light model. Although the continuous wave data used here cannot separate absorption and scattering, the data can be used to fit for the effective attenuation coefficient. In turn, in situ absorption coefficients can be deduced using an assumed reduced scattering coefficient of . Nonuniformities in illumination and skin pigment, however, can distort the interpretation of raw SI-DOT measurements. The spatial inhomogeneities in the reflected light intensity of each frame were corrected by subtracting a dark frame from each raw image and normalized by a frame of uniform illumination. Reflected light intensity in each corrected frame was then compared with a light model [Fig. 4(a)].
Fig. 4.
Correction scheme and light model quality assessment. (a) Raw data, corrected data, and predicted data for both green and red illumination. (b) The signal versus effective source–detector separation is calculated for the raw data, corrected data, and predicted data for a range of different absorption coefficients. Optimal values are shown as minima in the chi-squared plots between the corrected data and the predicted data and were empirically determined to be (top) and (bottom) for 523 and 625 nm wavelengths, respectively. (c) The signal versus effective source–detector separation for the corrected data fit with optimal values for red and green wavelengths.
For in situ estimation of optical properties, we compared the light fluence fall-off as a function of effective distance for both the SI-DOT measurements and the model predicted data, given by the source–detector Green’s functions (Gsd, see methods, Sec. 5.3). The chi-squared error between data and model was a continuous function of , with the best fit values of at 3.33 and for 523- and 625-nm wavelengths, respectively [Fig. 4(b)]. Fits using the optimal values showed excellent agreement to the data [Fig. 4(c)].
These fit lines also provided a means for refining our initial data quality assessment. Specifically, measurements with 40% or greater disagreement with the optimal predicted light fall-off were cropped from the data set prior to reconstruction. This model-deviation threshold typically removed of the total measurements (see Appendix A.2). Finally, superficial measurements (defined to be those with an effective source–detector separation of ) were averaged and regressed from all remaining measurements prior to reconstruction. This is analogous to superficial signal regression performed in human brain DOT and reduces the contribution of hemodynamics outside the brain (e.g., in the scalp) to the measured cortical signals.12,13,16
2.4. Noninvasive Evoked Responses In Vivo
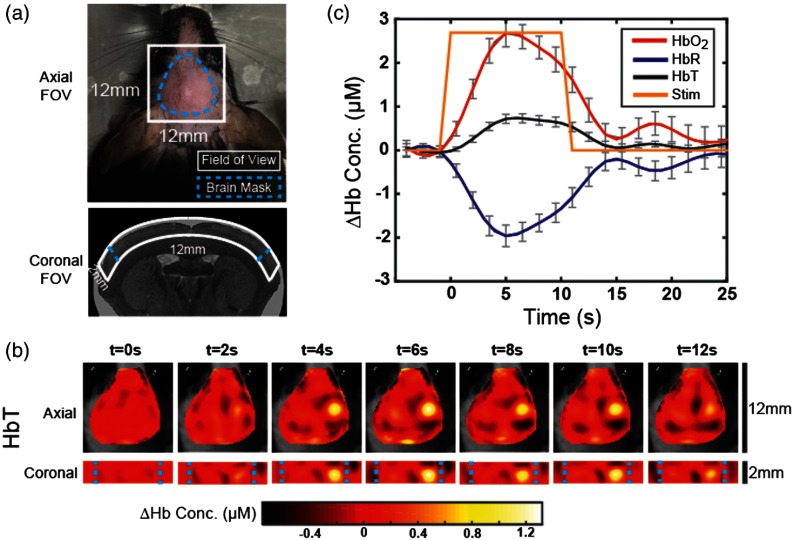
To validate SI-DOT for noninvasive functional mouse neuroimaging, we imaged anesthetized mice during peripheral stimulation of the left forepaw in a block design. Each block contained 10 s of , 0.5 mA pulses delivered at 3 Hz followed by 50 s of rest. A two-wavelength SI sequence allowed reconstruction of voxelwise changes in oxy-, deoxy-, and total hemoglobin (, HbR, and HbT, respectively). The reconstruction geometry (, with voxel dimensions of , ) covered an FOV consisting of scalp, skull, and upper cortical layers [Fig. 5(a)]. Five mice were imaged for 35 min each. Of this data set, 120 min of data passed measurement quality thresholds and were used for block averaging. Total hemoglobin block averaged images show focal activity in the contralateral (right) forepaw region [Fig. 5(b)]. Specifically, evoked hemodynamic responses demonstrate an increase in and HbT and a decrease in HbR [Fig. 5(c)] and were repeatable across mice (see Appendix A.3). Group averaged peak concentration changes of in , in HbR, and in HbT were observed at a depth of 1.7 mm beneath the scalp surface [Fig. 5(b)].
Fig. 5.
Noninvasive imaging of evoked cortical responses in the mouse. (a) Dotted lines show the FOVs for axial and coronal slices. The curvature of the cortex, as evident in the coronal FOV, was approximated by a slab geometry. (b) Following electrical stimulation of the left forepaw, we observe an increase in total hemoglobin concentration in the right hemisphere. An average axial slice through the maximum layer and its two neighboring axial slices is masked by a manually determined brain mask and overlaid on a grayscale image of the mouse head. Coronal slices through the peak voxel in the axial slice show the evoked response up to 2 mm beneath the surface of the scalp. (c) The average temporal response of voxels of the peak response were used to calculate time courses for , HbR, and HbT. Magnetic resonance image of the mouse brain is courtesy of the Duke Center for In Vivo Microscopy,20 accessed via the International Neuroinformatics Coordinating Facility Scalable Brain Atlas.21
3. Discussion
In this work, we developed a SI approach to DOT for noninvasive imaging of brain function in mice. The SI-DOT system combined the dense spatial sampling of camera-based DOT systems, finite element light modeling (common with fiber-based DOT), and the rapid scanning afforded by SI. To manage data quality, we developed several quality assessments to optimize the system and capitalize on its flexibility while also improving model accuracy by empirically calculating estimates of baseline optical properties. Feasibility was established by observing functional activations in vivo noninvasively and by reconstructing their depth as well as their lateral position. Although there are extensive reports of optical imaging of functional activations in mice with removal of either the scalp or both the scalp and the skull, noninvasive optical mapping of functional responses in mice has not yet been widely reported or explored.
A primary strength of the SI-DOT system is its great flexibility with regard to the source and detector grid density. However, the potential complexity of SI-DOT data sets presents challenges in evaluating data quality. SNR is of central importance in DOT system design and data quality optimization prior to image reconstruction.12,16 More specifically, the relationship between light level and source–detector distance guides many decisions throughout the construction of the instrument, including optics, sensor locations and sizes, and exposure times or frame rates. As a result, the ability to assess measurement quality before image reconstruction is important to instrument development and optimization. Thus, we developed a method for evaluating SI-DOT measurements using an effective distance between a detector pixel and an arbitrarily complex SI pattern. We have found that this metric enables the optimization of SI patterns and aids in identification and removal of bad or noisy measurements. Together, these techniques allow for efficient optimization and balancing of imaging speed, resolution, and FOV. In this paper, we optimized for a single view of the dorsal surface of the mouse head, consistent with our previous planar imaging systems,5 and imaged an FOV of using two wavelengths and 40 illumination patterns.
3.1. In Vivo Noninvasive Activations
Our DOT imaging techniques allow us to reconstruct hemodynamic activity up to 2 mm beneath the scalp surface. In response to electrical stimulation of the forepaw, we observe peak activity between 1.3 and 1.7 mm beneath the surface of the scalp, a finding consistent with the expected depth of activity given structural mouse studies examining the thickness of the scalp, skull, and cortical layers.22–24 The reconstructed images of , HbR, and HbT have magnitude, temporal response, and axial location that are consistent with previous functional imaging studies.25,26 As a function of depth, the SI-DOT images show an increase in and decrease in HbR corresponding to somatosensory layers 2 to 4. This depth-dependent hemodynamic activity is reasonably consistent with observations from invasive thinned-skull preparations in rats using multispectral imaging,27 laminar optical tomography,28 and noninvasive functional MRI studies in mice.29,30
The penetration depth and resolution of SI-DOT can be estimated from the system’s sensitivity matrix. Using the sensitivity matrix, we generated simulated measurements for a point activation. These data were then reconstructed using an inverted sensitivity matrix with regularization that reflects the operational SNR found in our experimental data. Taking the FWHM to be the point-spread function, this yields an axial resolution of 1.13 mm and a lateral resolution of 1.11 mm at a depth of 1.3 mm. Similarly, we quantified the depth sensitivity using a flat field imaging test by reconstructing a uniform perturbation throughout the volume.31 The sensitivity drops to 50% at a depth of 2.5 mm, setting a practical limit on the penetration depth of SI-DOT as constructed.
3.2. Context to Literature
SI addresses a critical issue that has limited either the speed or the spatial sampling of previous DOT techniques. A similar strategy has been applied in microscopy, where the use of SI has improved the speed and resolution of fluorescence microscopy beyond what was previously possible.19,32,33 In related mesoscopic imaging work, spatial frequency domain imaging (SFDI) utilizes SI patterns of varying spatial frequencies to probe different depths and provide three-dimensional (3-D) information.18,34–36 However, thus far, the SI literature has been limited to either applications outside brain imaging or brain imaging that is invasive and/or without tomographic reconstructions. Further, the relationship between source–detector distance and measurement SNR, which is central to assessing image quality in traditional point illumination DOT, has not been addressed using SFDI.
The primary application of wide-field SI has been for clinical noninvasive and depth-resolved assessment of skin and breast tissue in humans.37 Although the technique can provide 3-D tomographic reconstructions of absorption heterogeneities,35,36 thus far the implementations in rodent neuroimaging, for example to monitor stroke,38 cortical spreading depressions,39 or Alzheimer’s disease,40,41 have required invasive surgeries prior to imaging or have not been used in applications with fast dynamics (). Additionally, although SFDI is commonly used to assess baseline optical properties,42–44 our technique compares the light fall-offs as a function of effective distance in the optical measurements versus a finite element forward model, which will be easily translatable to calculating optical properties in complex geometries.
3.3. Limitations and Potential Improvements
In this work, there are three assumptions in the forward model that potentially limit the accuracy of reconstructions: homogeneous optical properties, geometrical boundaries of the mouse head, and the use of the diffusion approximation. Even though the absorption and scattering properties of different types of biological tissues are in fact relatively homogeneous,45 the accuracy of the forward model could be increased by modeling the scalp, skull, and brain separately. Additionally, although the scattering coefficient in tissue is relatively constant,46 implementing a wavelength-dependent power law47 to uncouple absorption and scattering would further improve the accuracy of the forward model. Further, an iterative procedure using determined optical properties to calculate new fits of signal versus effective source–detector separation, instead of assuming a of , might allow for better empirical optimization of both optical properties.
The localization accuracy of the functional response depth could be increased using a cylindrical head model, instead of the currently used slab geometry, or still further improved using an anatomical head model derived from MRI with coregistration and boundary identification, as is done in human fiber-based DOT imaging.12 The SI-DOT system could achieve boundary identification in an automated and data-driven way by including a pattern sequence with point illuminations for surface profiling. Accurate anatomical head modeling could enable transformations of the DOT data to a common atlas space for group comparisons of functional networks, as has been done previously for DOT studies in humans,12,48,49 while also accurately modeling both the boundary and the different layers of the mouse head.
Additionally, there is a slight model mismatch due to using a slab geometry to model the mouse head, which is curved. Because we constrained imaging to the top central portion of the skull, we estimate that there is approximately a displacement across the 10 mm FOV. Given the curvature of the mouse head [Fig. 5(a)], our calculations of illuminated pixel locations and distances near the edge of the FOV may be inaccurate up to due to our use of a slab instead of a cylindrical geometry, according to simple trigonometry. Due to the exponential weighting of large distances in the effective source–detector separation calculation, the subsequent maximum error in effective distance is . While small, if this error were used in baseline measurements,50 it would introduce significant image errors. However, our application is the imaging of differential brain activity, and these model errors are largely divided out through the use of the Rytov approximation.51,52 Anatomical head modeling will allow for more precise calculations of both effective source–detector separation and point illumination locations in the forward model.
In this study, we have used the diffusion approximation, whereas the radiative transport equation, as solved with Monte Carlo methods for example, would in principle be more accurate. Indeed a number of groups have shown similar reconstruction methods using Monte Carlo-generated Green’s functions.53,54 Although model errors are generally minimized using ratiometric data (in this study, the log-ratio data for temporal responses), the incorporation of Monte Carlo-derived forward models may provide better fits to the light intensity fall-off curves and better image quality.
Further exploration of parameter space might reveal ways to take further advantage of the system flexibility and optimize the trade-offs among imaging speed, resolution, and FOV. For example, shrinking the FOV would increase the camera speed and therefore increase the overall DOT frame rate or accommodate the addition of more source patterns while maintaining the same frame rate. Additionally, expanding the measurement set to consist of more spatially overlapping measurements and to include near infrared (NIR) wavelengths for deeper penetration has been shown to improve resolution and overall system performance in previous DOT systems.55 This could be done in SI-DOT by utilizing multiple views, which would allow for an FOV covering the entire mouse brain. This combined with NIR wavelengths would take advantage of greater source–detector separation to probe deeper into the brain, possibly with a resolution of 56 or slightly better at shallower depths.28
4. Summary/Conclusion
SI for DOT, with asymmetry between sources (few) and detectors (many), addresses a data rate limitation of previous DOT systems for rodents. SI-DOT improves upon current planar methods, which lack depth profiling and require surgical removal of the scalp, to provide noninvasive 3-D information of the brain. The SI-DOT system reported is built on an infrastructure with great flexibility. The techniques developed for assessing the quality of SI-DOT data sets before image reconstruction enable efficient optimization of the system to balance imaging speed, resolution, FOV, and computation time. Further, these techniques permit data quality analysis and an assessment of light model accuracy. The noninvasive 3-D imaging of the mouse cortex provided by SI-DOT has the potential to yield new insights into the functional architecture of the mouse brain and provide new avenues for studying healthy brain development, aging, disease, and therapies.
5. Methods
5.1. Animal Preparation and Imaging
All animal studies were approved by the Washington University School of Medicine Animal Studies Committee (protocol #20160217) under guidelines and regulations consistent with the guide for the Care and Use of Laboratory Animals, Public Health Service Policy on Humane Care and Use of Laboratory Animals, the Animal Welfare Act and Animal Welfare Regulations, and ARRIVE guidelines. Male C57/BL6 mice (, 5 to 7 weeks, 26 to 32 g, Jackson Laboratories, Bar Harbor, Maine) were used for imaging. At least 1 day before imaging, mice were anesthetized with 2% isoflurane for hair removal using Veet. For imaging, mice were anesthetized with a ketamine–xylazine mixture ( ketamine, xylazine) at with body temperature maintained at 37°C using an electric heating pad (mTCII, Cell Microcontrols, Norfolk, Virginia). Mice were secured using a Dazai imaging sled (Dazai Research Instruments, Toronto, Canada), with Velcro straps behind the ears and over the nose to prevent motion artifacts due to breathing. Each mouse was imaged for 35 min. Forepaw electrical stimulation was performed using A-M Systems Model 2100 Isolated Pulse Stimulator (A-M Systems, Sequim, Washington), with trigger sequences syncing stimulation, camera frames, and DMD patterns created in MATLAB (Mathworks, Natick, Massachusetts) and generated using a National Instruments Analog Output DAQ (National Instruments, Austin, Texas).
5.2. Light Modeling/Diffuse Optical Tomography Forward and Inverse Problem
A finite element mesh of an optically homogeneous tissue slab (right rectangular prism geometry) is generated using NIRFAST.11 For a set of chosen optical properties, we solve for the point light propagation from each mesh surface normal using a linearized Rytov forward model. The FEM mesh is then resampled to the desired voxelated space. The source patterns are estimated in situ to infer which locations on the tissue surface were illuminated by each pattern and to account for the curvature of the mouse head. Illuminated points are defined to be those of the maximum value detected in each frame for each illumination pattern. The corresponding point Green’s functions are summed to create the fluence distribution for each SI pattern. This is valid as only first-order interactions between the electromagnetic field and the tissue need to be considered; any nonlinear interactions, such as two-photon absorption, are negligible. This assumption is commonly invoked in DOT to model realistic point illumination shapes (i.e., exponential decay as a function of depth and finite beam areas).57 Additionally, linear approximations to the diffusion equation are used extensively in the DOT literature,58 namely the Rytov,59 Born,60 and normalized Born approximations.61 These linear assumptions are particularly well suited for detecting brain activity where the perturbations in absorption are small relative to baseline. To confirm that these linear assumptions are valid for SI-DOT measurements, we collected data with both extended illumination structures and as a series of separate images with point illuminations. Both approaches agree within experimental SNR (see Appendix A.1).
The predicted fluence at the tissue surface for each source–detector pair (Gsd, the unperturbed fluence) is converted into predicted detector counts for the corresponding measurement and compared with the raw data. Inaccuracies are addressed by iterating through the forward model’s generation of Green’s functions using different optical properties until the fit between raw data and modeled data is optimized. The products of the source and detector Green’s function values at each voxel are normalized by Gsd for the corresponding source–detector pair and scaled by the voxel volume. This quantity for each source–detector pair forms a row of the sensitivity matrix, whose columns correspond to voxels. A regularized pseudoinverse of the sensitivity matrix is then calculated using spatially variant Tikhonov regularization (with and , following the notation of Dehghani et al.31), and this regularized pseudoinverse is applied to the experimental measurements to reconstruct images (i.e., solve the inverse problem). These regularization and reconstruction procedures have been described in our previous work.12 This entire process is repeated to construct a separate sensitivity matrix and independently reconstruct images for each illumination wavelength.
5.3. Baseline Optical Property Estimation
Optimal optical properties are estimated by first averaging the raw data across all time points for each illumination pattern. Each frame has the background frame subtracted and is normalized by the planar frame. The average signal versus effective source–detector separation (see Sec. 2.2) is calculated for each wavelength at each unique distance. Gsd is calculated for a range of values of (see Sec. 5.2), which provides a predicted fit. The optimal is chosen to be the value that minimizes the chi-squared error between fit lines of corrected data and predicted data [Fig. 4(b)].
5.4. Data Analysis
Differential raw data are collected at covering a FOV and binned down to off camera for increased SNR and a more computationally efficient inverse problem. To account for systematic drift over the course of an imaging session, binned data are temporally detrended by subtracting the fit of a fifth-order polynomial to each pixel’s time trace. The shape of each individual structured light pattern is then empirically determined to generate the source Green’s functions. The signal versus effective source–detector separation is calculated for each measurement, and noisy measurements are identified and removed by setting a threshold at the typical background standard deviation [see Fig. 3(d)]. The denoised data are transformed by normalizing each measurement’s time trace by its mean and taking the logarithm to permit reconstruction of images. The average time trace of superficial measurements (those with effective source–detector separation , which mostly probe shallow, noncortical depths) is then regressed from the data set to remove systemic scalp signals. Analogous superficial regression procedures have been employed for similar purposes in previous DOT work.12,13,16 After reconstruction, data are temporally band-pass filtered between 0.009 and 0.25 Hz and spatially smoothed by convolution with a 3-D Gaussian kernel. Data from both wavelengths are reconstructed, with spectroscopic inversion using the LED spectra and known extinction coefficients from Prahl, adapted from our previous methods.5,17
Each individual 5-min run is block averaged. To include a run in group averages, data are required to have a visually identifiable evoked response, peaking temporally during or very near the stimulus period and with a magnitude at least twice as large as peak fluctuations outside the activation region. All runs that fit these criteria are averaged to generate temporal and spatial maps of , HbR, and HbT hemodynamics for each mouse. Time traces are plotted by determining the time and depth of the peak activation, taking the mean of that layer and the layers immediately above and below, and then averaging the time traces from all pixels that reach magnitude of the peak activation in the maximum frame. Group averaged maps are generated by 2-D translating the maps of each individual mouse in parallel axial planes such that the peak activations all align with their collective center of mass.
Acknowledgments
This study was supported by the National Institutes of Health [R01NS078223, P01NS080675, R01NS090874, R01NS084028 (JPC) and K25NS083754 (AQB)] and a fellowship from the Cognitive, Computational, and Systems Neuroscience Pathway at Washington University in St. Louis (ZEM). The authors would like to thank Mark A. Anastasio, Adam T. Eggebrecht, Patrick W. Wright, Jonathan R. Bumstead, Karla M. Bergonzi, Grant A. Baxter, and Andrew W. Kraft for helpful discussions, support, and feedback.
Biographies
Matthew D. Reisman received his BA degree in physics at the University of Colorado in 2012 and his MA degree in physics from Washington University in St. Louis in 2014. He is a PhD candidate in physics at Washington University in St. Louis. His research in the lab of Dr. Joseph Culver focuses on the development of optical neuroimaging systems and techniques.
Zachary E. Markow received his BA degree in physics and mathematics at Washington University in St. Louis in 2014. He is a PhD candidate in biomedical engineering at Washington University in St. Louis. His research in the labs of Drs. Joseph Culver and Mark Anastasio focuses on computational image reconstruction and optimization methods for human and small animal neuroimaging.
Adam Q. Bauer received his BS degree in physics at Truman State University in 2001 and his PhD in physics from Washington University in St. Louis in 2009 under Dr. James G. Miller. He is an assistant professor of radiology at Washington University in St. Louis. His research focuses on developing optical imaging methods and custom software algorithms to understand network neuroscience in the healthy brain and following neurological disease.
Joseph P. Culver received his BS degree in physics at the University of Washington in 1988 and his PhD in physics from the University of Pennsylvania in 1997 under Drs. Arjun G. Yodh and Robin M. Hochstrasser. He is a professor of radiology, physics, and biomedical engineering at Washington University in St. Louis. His lab explores ways of exploiting noninvasive optical measurements for both functional and molecular biological imaging.
Appendix.
A.1. Linearity of Diffuse Optical Tomography Measurements
We verified the use of summed “point” source Green’s functions to approximate patterned illumination experimentally. First [Fig. 6(a)], we generated a grid of equal-sized small square illuminations that cover the same area as a single broad illumination pattern [Fig. 6(b)]. We compared the sum of the small square illumination frames with the full structured pattern frame [Fig. 6(c)]. Within the illuminated region, the percent error is [Fig. 6(d)]. In the dark regions, experimental noise dominates the errors. Thus, our forward model, which treats the structured light Green’s functions as sums of point Green’s functions, is appropriate.
Fig. 6.
Linearity of SI-DOT measurements. (a) An grid of small square “points” is scanned on a phantom. One example point illumination frame is shown, before binning or background subtraction, with the grid indicating the locations of the remaining points. (b) The sum of the full grid of illumination points, after binning and background subtraction, averaged over 19 cycles. (c) An equivalent single-SI pattern, after binning, background subtraction, and averaging over the 19 cycles. (d) The % error between the SI frame and the summed point frame, showing uniform agreement between the two, within the expected variance due to greater noise (due to multiple readouts) in the point-summed frame.
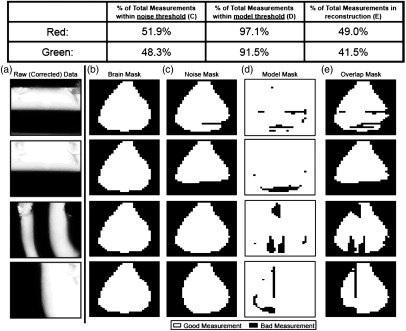
A.2. Measurement Removal
A series of masks are applied to reduce the measurement set to only include those of sufficient quality in the reconstruction. Upon data collection, 40 source patterns and the grid of binned detectors combine to provide 40,960 measurements. A visually identified and manually constructed brain mask excludes measurements in the FOV that do not lie over the scalp [Fig. 7(b)]. This mask typically identifies 40% to 45% of the total measurements. The noise mask discussed in Sec. 2.2 typically identifies 50% of the measurements, those that fall within the value of the standard deviation of the background. Nearly all of these measurements overlap those removed by the brain mask [Fig. 7(c)]. A model mask determines which measurements deviate from expectation, based on the optimized model (as discussed in Sec. 2.3). This mask typically removes of the measurements with green illumination and of the measurements with red illumination [Fig. 7(d)]. Finally, the overlap of these three masks is the complete set of measurements to be used in reconstructions [Fig. 7(e)].
Fig. 7.
Measurement removal. Table 1 shows the percentage of measurements from an entire data set within each threshold. (a) Raw corrected data for four different green illumination patterns, a high and low spatial frequency for each orientation. (b) The brain mask removes measurements from pixels that do not approximately align over the scalp and brain. This mask is identical for all illumination patterns for a given mouse. (c) The noise mask shows which measurements were below the noise threshold for the example pattern. (d) The model mask shows which measurements differed by more than 50% from the model’s prediction. (e) Only measurements surviving all three mask procedures are kept. This procedure is repeated for all illumination patterns and wavelengths to determine the full SI-DOT measurement set used for reconstruction.
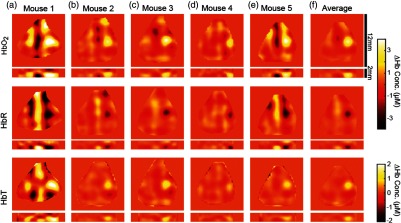
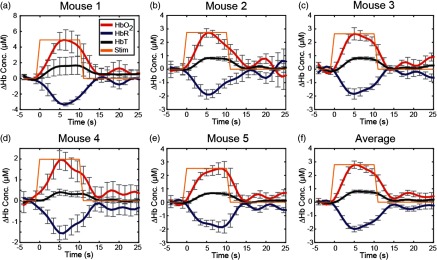
A.3. Repeatability
The data shown in Fig. 5 are averages of runs across five mice. Averaging data only across each individual mouse instead of across the entire group shows good repeatability. Maps of the peak frame show an ability to resolve the expected increase in and HbT, along with the decrease in HbT, at the individual mouse level (Fig. 8). Evoked responses also follow the expected temporal behavior at the individual mouse level (Fig. 9).
Fig. 8.
Maps of evoked responses in individual mice. Runs with visually identified evoked responses are block averaged (a–e) and then (f) averaged across all mice. For the five mice in this study, 15, 25, 30, 35, and 15 min of data were included for each mouse, respectively, out of 35 min total. The group average is calculated after a two-dimensional translation of each mouse’s data to spatially align the maximum value of each activation across mice. Reconstructions of , HbR, and HbT show the expected peak responses with good repeatability across mice, including the depths of the activations in coronal slices. DOT reconstruction artifacts, seen here mostly as vertical stripes, show where reconstruction algorithms could be further optimized to increase data quality.
Fig. 9.
Evoked response time courses. (a–e) Time traces of , HbR, and HbT for each mouse (mouse 1 to 5) are calculated by averaging the time traces of all pixels of the maximum response in Fig. 8 and for the average across all mice (f). The relative increases in and HbT and decrease in HbR follow expected time courses and show good repeatability across mice. Error bars show standard error across runs.
Disclosures
The authors declare no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Ahrens E. T., Dubowitz D. J., “Peripheral somatosensory fMRI in mouse at 11.7 T,” NMR Biomed. 14, 318–324 (2001). 10.1002/(ISSN)1099-1492 [DOI] [PubMed] [Google Scholar]
- 2.Mueggler T., et al. , “Bicuculline-induced brain activation in mice detected by functional magnetic resonance imaging,” Magn. Reson. Med. 46, 292–298 (2001). 10.1002/(ISSN)1522-2594 [DOI] [PubMed] [Google Scholar]
- 3.Nasrallah F. A., Tay H.-C., Chuang K.-H., “Detection of functional connectivity in the resting mouse brain,” NeuroImage 86, 417–424 (2014). 10.1016/j.neuroimage.2013.10.025 [DOI] [PubMed] [Google Scholar]
- 4.Mechling A. E., et al. , “Fine-grained mapping of mouse brain functional connectivity with resting-state fMRI,” NeuroImage 96, 203–215 (2014). 10.1016/j.neuroimage.2014.03.078 [DOI] [PubMed] [Google Scholar]
- 5.White B. R., et al. , “Imaging of functional connectivity in the mouse brain,” PLoS One 6, e16322 (2011). 10.1371/journal.pone.0016322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grinvald A., et al. , “Functional architecture of cortex revealed by optical imaging of intrinsic signals,” Nature 324, 361–364 (1986). 10.1038/324361a0 [DOI] [PubMed] [Google Scholar]
- 7.Arieli A., et al. , “Dynamics of ongoing activity: explanation of the large variability in evoked cortical responses,” Science 273, 1868–1871 (1996). 10.1126/science.273.5283.1868 [DOI] [PubMed] [Google Scholar]
- 8.Bero A. W., et al. , “Bidirectional relationship between functional connectivity and amyloid- deposition in mouse brain,” J. Neurosci. 32, 4334–4340 (2012). 10.1523/JNEUROSCI.5845-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bauer A. Q., et al. , “Optical imaging of disrupted functional connectivity following ischemic stroke in mice,” NeuroImage 99, 388–401 (2014). 10.1016/j.neuroimage.2014.05.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kozberg M. G., et al. , “Rapid postnatal expansion of neural networks occurs in an environment of altered neurovascular and neurometabolic coupling,” J. Neurosci. 36, 6704–6717 (2016). 10.1523/JNEUROSCI.2363-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dehghani H., et al. , “Near infrared optical tomography using NIRFAST: algorithm for numerical model and image reconstruction,” Commun. Numer. Methods Eng. 25, 711–732 (2009). 10.1002/cnm.v25:6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eggebrecht A. T., et al. , “Mapping distributed brain function and networks with diffuse optical tomography,” Nat. Photonics 8, 448–454 (2014). 10.1038/nphoton.2014.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White B. R., et al. , “Resting-state functional connectivity in the human brain revealed with diffuse optical tomography,” NeuroImage 47, 148–156 (2009). 10.1016/j.neuroimage.2009.03.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Culver J. P., et al. , “Volumetric diffuse optical tomography of brain activity,” Opt. Lett. 28, 2061 (2003). 10.1364/OL.28.002061 [DOI] [PubMed] [Google Scholar]
- 15.Patwardhan S., et al. , “Time-dependent whole-body fluorescence tomography of probe bio-distributions in mice,” Opt. Express 13, 2564–2577 (2005). 10.1364/OPEX.13.002564 [DOI] [PubMed] [Google Scholar]
- 16.Zeff B. W., et al. , “Retinotopic mapping of adult human visual cortex with high-density diffuse optical tomography,” Proc. Natl. Acad. Sci. U.S.A. 104, 12169–12174 (2007). 10.1073/pnas.0611266104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prahl S., “Optical absorption of hemoglobin,” Oregon Medical Laser Center, 2002, http://omlc.org/spectra/ (26 September 2016). [Google Scholar]
- 18.Cuccia D. J., et al. , “Modulated imaging: quantitative analysis and tomography of turbid media in the spatial-frequency domain,” Opt. Lett. 30, 1354–1356 (2005). 10.1364/OL.30.001354 [DOI] [PubMed] [Google Scholar]
- 19.Gustafsson M. G. L., “Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution,” Proc. Natl. Acad. Sci. U.S.A. 102, 13081–13086 (2005). 10.1073/pnas.0406877102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson G. A., et al. , “Waxholm space: an image-based reference for coordinating mouse brain research,” NeuroImage 53, 365–372 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bakker R., Tiesinga P., Kötter R., “The Scalable Brain Atlas: instant web-based access to public brain atlases and related content,” Neuroinformatics 13, 353–366 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Azzi L., et al. , “Gender differences in mouse skin morphology and specific effects of sex steroids and dehydroepiandrosterone,” J. Invest. Dermatol. 124, 22–27 (2005). 10.1111/j.0022-202X.2004.23545.x [DOI] [PubMed] [Google Scholar]
- 23.Hong G., et al. , “Through-skull fluorescence imaging of the brain in a new near-infrared window,” Nat. Photonics 8, 723–730 (2014). 10.1038/nphoton.2014.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lein E. S., et al. , “Genome-wide atlas of gene expression in the adult mouse brain,” Nature 445, 168–176 (2006). 10.1038/nature05453 [DOI] [PubMed] [Google Scholar]
- 25.Bergonzi K. M., et al. , “Mapping functional connectivity using cerebral blood flow in the mouse brain,” J. Cereb. Blood Flow Metab. 35, 367–370 (2015). 10.1038/jcbfm.2014.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunn A. K., et al. , “Spatial extent of oxygen metabolism and hemodynamic changes during functional activation of the rat somatosensory cortex,” NeuroImage 27, 279–290 (2005). 10.1016/j.neuroimage.2005.04.024 [DOI] [PubMed] [Google Scholar]
- 27.Konecky S. D., et al. , “Hyperspectral optical tomography of intrinsic signals in the rat cortex,” Neurophotonics 2, 045003 (2015). 10.1117/1.NPh.2.4.045003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hillman E. M. C., et al. , “Depth-resolved optical imaging and microscopy of vascular compartment dynamics during somatosensory stimulation,” NeuroImage 35, 89–104 (2007). 10.1016/j.neuroimage.2006.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adamczak J. M., et al. , “High field bold response to forepaw stimulation in the mouse,” NeuroImage 51, 704–712 (2010). 10.1016/j.neuroimage.2010.02.083 [DOI] [PubMed] [Google Scholar]
- 30.Schlegel F., Schroeter A., Rudin M., “The hemodynamic response to somatosensory stimulation in mice depends on the anesthetic used: implications on analysis of mouse fMRI data,” NeuroImage 116, 40–49 (2015). 10.1016/j.neuroimage.2015.05.013 [DOI] [PubMed] [Google Scholar]
- 31.Dehghani H., et al. , “Depth sensitivity and image reconstruction analysis of dense imaging arrays for mapping brain function with diffuse optical tomography,” Appl. Opt. 48, D137–D143 (2009). 10.1364/AO.48.00D137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keller P. J., et al. , “Fast, high-contrast imaging of animal development with scanned light sheet-based structured-illumination microscopy,” Nat. Methods 7, 637–642 (2010). 10.1038/nmeth.1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nikolenko V., et al. , “SLM microscopy: scanless two-photon imaging and photostimulation with spatial light modulators,” Front. Neural Circuits 2(5), 1–14 (2008). 10.3389/neuro.04.005.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cuccia D. J., et al. , “Quantitation and mapping of tissue optical properties using modulated imaging,” J. Biomed. Opt. 14, 024012 (2009). 10.1117/1.3088140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Konecky S. D., et al. , “Quantitative optical tomography of sub-surface heterogeneities using spatially modulated structured light,” Opt. Express 17, 14780–14790 (2009). 10.1364/OE.17.014780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bélanger S., et al. , “Real-time diffuse optical tomography based on structured illumination,” J. Biomed. Opt. 15, 016006 (2010). 10.1117/1.3290818 [DOI] [PubMed] [Google Scholar]
- 37.O’Sullivan T. D., et al. , “Diffuse optical imaging using spatially and temporally modulated light,” J. Biomed. Opt. 17, 071311 (2012). 10.1117/1.JBO.17.7.071311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abookasis D., et al. , “Imaging cortical absorption, scattering, and hemodynamic response during ischemic stroke using spatially modulated near-infrared illumination,” J. Biomed. Opt. 14, 024033 (2009). 10.1117/1.3116709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cuccia D. J., et al. , “Quantitative in vivo imaging of tissue absorption, scattering, and hemoglobin concentration in rat cortex using spatially modulated structured light,” Chapter 12 in In Vivo Optical Imaging of Brain Function, Frostig R. D., Ed., CRC Press/Taylor & Francis, Boca Raton, Florida: (2009). [PubMed] [Google Scholar]
- 40.Lin A. J., et al. , “In vivo optical signatures of neuronal death in a mouse model of Alzheimer’s disease,” Lasers Surg. Med. 46, 27–33 (2014). 10.1002/lsm.v46.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin A. J., et al. , “Differential pathlength factor informs evoked stimulus response in a mouse model of Alzheimer’s disease,” Neurophotonics 2, 045001 (2015). 10.1117/1.NPh.2.4.045001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mazhar A., et al. , “Wavelength optimization for rapid chromophore mapping using spatial frequency domain imaging,” J. Biomed. Opt. 15, 061716 (2010). 10.1117/1.3523373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weber J. R., et al. , “Multispectral imaging of tissue absorption and scattering using spatial frequency domain imaging and a computed-tomography imaging spectrometer,” J. Biomed. Opt. 16, 011015 (2011). 10.1117/1.3528628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nadeau K. P., Durkin A. J., Tromberg B. J., “Advanced demodulation technique for the extraction of tissue optical properties and structural orientation contrast in the spatial frequency domain,” J. Biomed. Opt. 19, 056013 (2014). 10.1117/1.JBO.19.5.056013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jacques S. L., “Optical properties of biological tissues: a review,” Phys. Med. Biol. 58, R37 (2013). 10.1088/0031-9155/58/11/R37 [DOI] [PubMed] [Google Scholar]
- 46.Taroni P., et al. , “In vivo absorption and scattering spectroscopy of biological tissues,” Photochem. Photobiol. Sci. 2, 124 (2003). 10.1039/b209651j [DOI] [PubMed] [Google Scholar]
- 47.van Leeuwen-van Zaane F., et al. , “In vivo quantification of the scattering properties of tissue using multi-diameter single fiber reflectance spectroscopy,” Biomed. Opt. Express 4, 696–708 (2013). 10.1364/BOE.4.000696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferradal S. L., et al. , “Atlas-based head modeling and spatial normalization for high-density diffuse optical tomography: in vivo validation against fMRI,” NeuroImage 85(1), 117–126 (2014). 10.1016/j.neuroimage.2013.03.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferradal S. L., et al. , “Functional imaging of the developing brain at the bedside using diffuse optical tomography,” Cereb. Cortex 26(4), 1–11 (2015). 10.1093/cercor/bhu320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gibson A. P., Hebden J. C., Arridge S. R., “Recent advances in diffuse optical imaging,” Phys. Med. Biol. 50, R1 (2005). 10.1088/0031-9155/50/4/R01 [DOI] [PubMed] [Google Scholar]
- 51.Gibson A. P., et al. , “Three-dimensional whole-head optical tomography of passive motor evoked responses in the neonate,” NeuroImage 30, 521–528 (2006). 10.1016/j.neuroimage.2005.08.059 [DOI] [PubMed] [Google Scholar]
- 52.Patwardhan S. V., Culver J. P., “Quantitative diffuse optical tomography for small animals using an ultrafast gated image intensifier,” J. Biomed. Opt. 13, 011009 (2008). 10.1117/1.2830656 [DOI] [PubMed] [Google Scholar]
- 53.Boas D. A., et al. , “Three dimensional Monte Carlo code for photon migration through complex heterogeneous media including the adult human head,” Opt. Express 10, 159–170 (2002). 10.1364/OE.10.000159 [DOI] [PubMed] [Google Scholar]
- 54.Fang Q., Boas D. A., “Monte Carlo simulation of photon migration in 3D turbid media accelerated by graphics processing units,” Opt. Express 17, 20178–20190 (2009). 10.1364/OE.17.020178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boas D. A., Dale A. M., Franceschini M. A., “Diffuse optical imaging of brain activation: approaches to optimizing image sensitivity, resolution, and accuracy,” NeuroImage 23(1), S275–S288 (2004). 10.1016/j.neuroimage.2004.07.011 [DOI] [PubMed] [Google Scholar]
- 56.Culver J. P., et al. , “Optimization of optode arrangements for diffuse optical tomography: a singular-value analysis,” Opt. Lett. 26, 701–703 (2001). 10.1364/OL.26.000701 [DOI] [PubMed] [Google Scholar]
- 57.Dehghani H., et al. , “Numerical modelling and image reconstruction in diffuse optical tomography,” Philos. Trans. R. Soc. London, Ser. A 367, 3073–3093 (2009). 10.1098/rsta.2009.0090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yodh A., Chance B., “Spectroscopy and imaging with diffusing light,” Phys. Today (2008). 10.1063/1.881445 [DOI] [Google Scholar]
- 59.Boas D. A., et al. , “Imaging the body with diffuse optical tomography,” IEEE Signal Process. Mag. 18, 57–75 (2001). 10.1109/79.962278 [DOI] [Google Scholar]
- 60.O’Leary M. A., et al. , “Experimental images of heterogeneous turbid media by frequency-domain diffusing-photon tomography,” Opt. Lett. 20, 426–428 (1995). 10.1364/OL.20.000426 [DOI] [PubMed] [Google Scholar]
- 61.Ntziachristos V., Weissleder R., “Experimental three-dimensional fluorescence reconstruction of diffuse media by use of a normalized born approximation,” Opt. Lett. 26, 893–895 (2001). 10.1364/OL.26.000893 [DOI] [PubMed] [Google Scholar]