Abstract
Collapsin response mediator proteins (CRMPs) have been implicated in signaling of axonal guidance, including semaphorins. We have previously identified a unique member of this gene family, CRMP-associated molecule CRAM (CRMP-5), which is phylogenetically divergent from the other four CRMPs. In this study, we have examined the distribution and function of CRAM in developing neurons. Immunohistochemical analysis showed accumulation of CRAM in the filopodia of growth cones. Experiments using cytochalasin D indicated that filopodial localization of CRAM was independent of filamentous actin. Overexpression of CRAM in neuronal cells significantly promoted filopodial growth and led to the formation of supernumerary growth cones, which acquired resistance to semaphorin-3A stimulation. Finally, knockdown of CRAM by using RNA interference blocked filopodial formation and revealed an aberrant morphology of growth cones. We propose that CRAM regulates filopodial dynamics and growth cone development, thereby restricting the response of growth cone to repulsive guidance cues.
INTRODUCTION
Neuronal outgrowth and axonal guidance are important processes that are highly regulated through extracellular signaling (Tessier Lavigne and Goodman, 1996). Neuronal outgrowth is directed by attractive and repulsive factors such as netrins and members of the semaphorin protein family (Luo et al., 1993; Kolodkin, 1998). Neuropilins and Plexins have been identified as semaphorin receptors (Comeau et al., 1998; Winberg et al., 1998; Takahashi et al., 1999; Tamagnone et al., 1999), and several candidate downstream molecules of semaphorin receptor have been reported (Pasterkamp and Kolodkin, 2003). However much less is known about the intracellular molecules involved in the relay of those multiple signals toward the resulting cellular response.
Collapsin response mediator proteins (CRMPs) are a family of cytosolic phosphoproteins that are expressed exclusively in the nervous system (Minturn et al., 1995; Hamajima et al., 1996; Wang and Strittmatter, 1996; Byk et al., 1998). Chick CRMP-62 (CRMP-2) was first identified through its possible involvement in the collapsin-1/semaphorin-3A (Sema3A)-induced mediation of growth cone collapse in chick dorsal root ganglion (DRG) neurons (Goshima et al., 1995). Based on sequence similarity, four CRMP isoforms have been identified in rats. Certain of these CRMP homologues also have been identified by independent methods as TOAD-64 (Minturn et al., 1995), Ulip (Byk et al., 1996), and dihydropyrimidinase (DHPase)-related proteins. Previous studies showed that the expression of CRMPs is developmentally regulated and isoform specific within the nervous system. Importantly, CRMPs share homology with a nematode protein, unc-33, whose absence produces aberrant elongation of axons and uncoordinated worm movement (Hedgecock et al., 1985). These suggest that CRMPs are essential signaling molecules in axonal guidance, including Sema3A-induced growth cone collapse signaling. Furthermore, it has been reported that the overexpression of CRMP-2 in cultured hippocampal neurons led to the formation of supernumerary axons and that CRMP-2 binds to tubulin heterodimers to promote microtubule assembly (Inagaki et al., 2001; Fukata et al., 2002). Therefore, CRMPs seem to play a key role in the signaling not only of semaphorin-induced growth cone collapse but also various other intracellular responses during neural development.
We have previously identified a novel CRMP-associated protein, designated CRAM for CRMP-associated molecule that belongs to the unc-33 gene family (Inatome et al., 2000). The deduced amino acid sequence reveals that the CRAM gene encodes a protein of 563 amino acids and shows 57% identity with DHPase and 50–51% identity with CRMPs. A phylogenetic tree analysis indicates that CRAM sequence shows the greatest similarity with DHPase and divergence from the four CRMPs, indicating that CRAM is more closely related to DHPase than to the four CRMPs. The expression of CRAM is remarkable for brain, and especially high in fetal and neonatal, but decreases to very low levels in adult brain. Thus, CRAM is a unique new member of this gene family and may play a role in axonal guidance signaling distinct from other four CRMPs. Furthermore, we have recently reported that CRAM was associated with several cellular proteins, including tyrosine kinase Fes/Fps and mitochondrial septin (Mitsui et al., 2002; Takahashi et al., 2003a,b).
In this study, we examined the distribution and function of CRAM in primary cultured developing neurons. Here, we report that CRAM localizes intensely at filopodia of growth cone and plays an important role in filopodia and growth cone formation during neural development. The involvement of CRAM in axonal guidance signaling mediated by Sema3A is discussed.
MATERIALS AND METHODS
Antibodies and Reagents
Anti-FLAG M2 monoclonal antibody, anti-α-tubulin antibody, phalloidintetramethylrhodamine B isothiocyanate, cytochalasin D, and poly-d-lysine were purchased from Sigma-Aldrich (St. Louis, MO). Anti-tau-1 antibody and nerve growth factor were purchased from Chemicon International (Temecula, CA), and Alexa-conjugated secondary antibodies were from Molecular Probes (Eugene, OR). Anti-CRAM antibody was produced by immunizing a rabbit with synthetic peptide (residues 468–485). Laminin and Matrigel basement membrane matrix were purchased from BD Biosciences (San Jose, CA). Pregnant rats and mice were purchased from Japan SLC (Shizuoka, Japan). Semaphorin-3A/Fc chimera was purchased from Techne (Cambridge, United Kingdom).
Histology
Mice brains were postfixed in 10% buffered formalin for 7 d, embedded in paraffin, and serially sectioned in the frontal plane at 4 mm in thickness. The sections were stained with hematoxylin and eosin and anti-CRAM antibody.
Cell Culture
Murine E12 DRG were dissected and cultured in DME/10% fetal bovine serum containing 100 ng/ml nerve growth factor (NGF) on Matrigel-coated coverslips. Before stimulation, cells were cultured in serum-free DMEM containing 100 ng/ml NGF for 3 h.
Hippocampal neurons from embryonic day 18 (E18) rat embryos prepared by the use of papain as described previously (Berninger et al., 1993) were seeded on coverslips coated with poly-d-lysine and laminin in neurobasal medium (Invitrogen, Carlsbad, CA) with B-27 supplement (Invitrogen), 1 mM glutamine, and 2.5 μM cytosine β-d-arabinofuranoside (Sigma-Aldrich). Hippocampal neurons were transfected using the Amaxa Biosystems Nucleofector device (program O-03) following the manufacturer's instructions for primary rat neurons.
Small Interfering RNA (siRNA)
Sense and antisense oligonucleotides corresponding to the following cDNA sequences were purchased from Dharmacon (Boulder, CO): 5′-TCACCATTGCAAACAGGAC-3′ (no. 1, nucleotides 694–712), 5′-GACTTCACTAAGATCCCAC-3′ (no. 2, nucleotides 1014–1032), 5′-GAAGCCACAAAGACCATCT-3′ (no. 3, nucleotides 1218–1236), and 5′-GTTTCAGCCTCTCTGGTTC-3′ (no. 4, nucleotides 1591–1609). To reconstitute the siRNA-mediated phenotype by reexpression of CRAM, we constructed FLAG-CRAMm expression vector containing silent mismatches in the knockdown oligonucleotide sequence of no. 4 (5′-GcTTttcgtTgagcGGaag-3′ corresponding to nucleotides 1591–1609 (mutation points were indicated by small letters). Hippocampal neurons were transfected with Amaxa Nucleofector (Amaxa, Cologne, Germany) according to the manufacturer's protocol with siRNA. In these experiments cells were fixed and stained 72 h posttransfection with siRNA.
Immunofluorescence Microscopy
Neuronal cells were fixed with 4% paraformaldehyde/10% sucrose in phosphate-buffered saline (PBS) for 1 h at 37°C, washed twice with 0.2% Tween 20 in PBS, permeabilized with 0.2% Triton X-100 in PBS for 10 min, washed four times with PBS, and blocked with 3% bovine serum albumin in PBS, all at room temperature. For double staining, the cells were incubated with appropriate antibody for 1 h at room temperature, washed three times with 0.5% Triton X-100 in PBS, and then with appropriate secondary antibody (Alexa Fluor goat anti-rabbit IgG, Alexa Fluor goat anti-mouse IgG) for 30 min. The samples were washed as before, mounted using PermaFluor (Immunon, Pittsburgh, PA), and analyzed using Zeiss LSM510 confocal laser scanning microscope.
Immunoblotting Procedures
COS-7 cells were lysed in the lysis buffer (10 mM Tris-HCl, pH 7.8, 1% Nonidet P-40, 150 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, and 10 μg/ml leupeptin) at 4°C. The lysates from cultured cells were boiled with SDS-PAGE sample buffer for 3 min, separated by SDS-PAGE gels, and transferred to polyvinylidene difluoride membranes (Immobilon P; Millipore, Milford, MA) followed by detection with the appropriate antibody as described previously (Inatome et al., 2000).
RESULTS
Distributional Analysis of CRAM in Developing Neurons
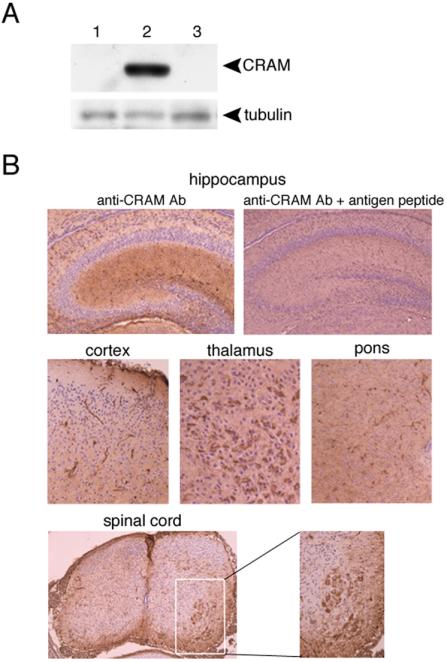
In our previous work, we reported by Northern and Western blot analyses that CRAM was specifically expressed in developing brain. Subsequently, in situ hybridization analysis demonstrated the specific expression of CRAM (CRMP-5) mRNA in the developing central and peripheral nervous system (Fukada et al., 2000). Because histological distribution of CRAM in developing brain has not been analyzed, we examined it in detail. The specificity of anti-CRAM antibody was demonstrated by the blocking effect of antigen peptides on antibody recognition with immunoblot analysis (Figure 1A). Immunohistochemical analysis by using anti-CRAM antibody (antibody) showed that CRAM was widely distributed in various rat brain regions, including hippocampus, cerebral cortex, thalamus, pons, and spinal cord during the neonatal development period (Figure 1B). On the other hand, any CRAM signal was undetectable in the adult brain (our unpublished data), consistent with our previous observation (Inatome et al., 2000). Thus, expression of CRAM protein appeared closely correlated with CRAM mRNA levels. On the basis of these data, we next investigated the intracellular distribution of CRAM in primary cultured rat hippocampal neurons at different stages, because polarity and morphological characterization of them are well established.
Figure 1.
Distribution of CRAM in the central and peripheral nervous system. (A) Characterization of anti-CRAM antibody. Lysates (1 mg) of rat brain (postnatal 1 d) were immunoblotted with preimmune or immune serum with CRAM antigen peptide in the presence or absence of antigen peptide (1 mM) provided from CRAM protein (amino acid 468–485). Deprobed membrane was reimmunoblotted with anti-tubulin antibody (bottom). (B) Distribution of CRAM in hippocampus and various brain regions. Each rat brain slice (postnatal 1 d) was immunostained with anti-CRAM antibody and subsequently stained with hematoxylin and eosin. The blocking effect of antigen peptide (1 mM) on CRAM staining indicates an antigen-specific recognition of anti-CRAM antibody. Note that CRAM is ubiquitously expressed in the brain and intense signals are detected in subsets of neurons such as anterior horn cells of the spinal cord.
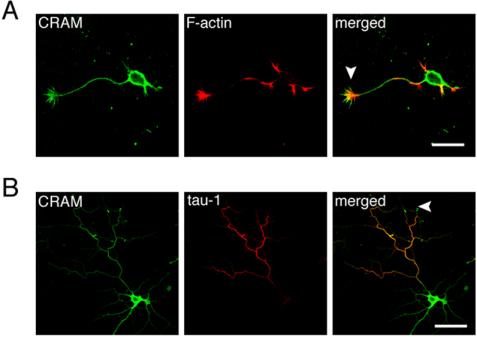
Rat hippocampal neurons (E18) were fixed after 24 h in culture (stage 3) and stained with anti-CRAM and phalloidin to label F-actin. At stage 3, CRAM was widely distributed in hippocampal neuron, including soma, neuritis, and growth cones (Figure 2A). Merged image indicated that CRAM was accumulated at the growth cone. Figure 2B shows hippocampal neuron at stage 5 when hippocampal neurons generally possess a polarized morphology, developing several dendrites and one axon. Even at this stage, high concentration of CRAM was detected at the growth cone. Therefore, CRAM was ubiquitously distributed in neurons during neural development, yet CRAM accumulation at the growth cone seemed to be a notable feature. Because CRMP family is believed to be involved in semaphorin signaling that leads to growth cone collapse, a role of CRAM in the regulation of axonal guidance at growth cone is extremely intriguing.
Figure 2.
Subcellular distribution of CRAM at different stages in hippocampal neurons. (A) CRAM distribution at stage 3. Cultured rat hippocampal neurons were fixed after 24 h in culture and stained with anti-CRAM (green) and phalloidin (red) to label F-actin. (B) CRAM distribution at stage 5. Hippocampal neurons were fixed after 7 d in culture and immunostained with anti-CRAM (green) and the axonal marker anti-tau-1 (red). CRAM was widely distributed in cell body and neurites. Merged images indicate the concentration of CRAM at the growth cones (arrow heads). Bars, 20 μm (A) and 50 μm (B).
CRAM Localization at Filopodia of Growth Cone in an Actin Assembly-independent Manner
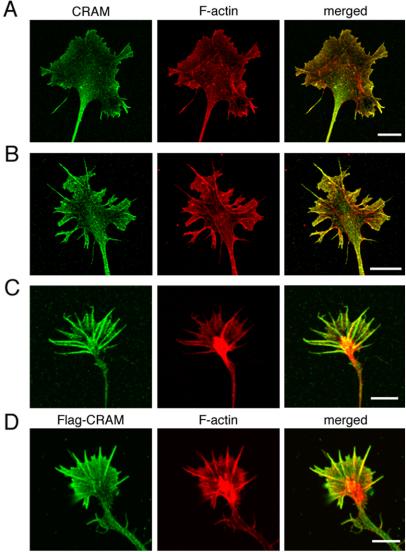
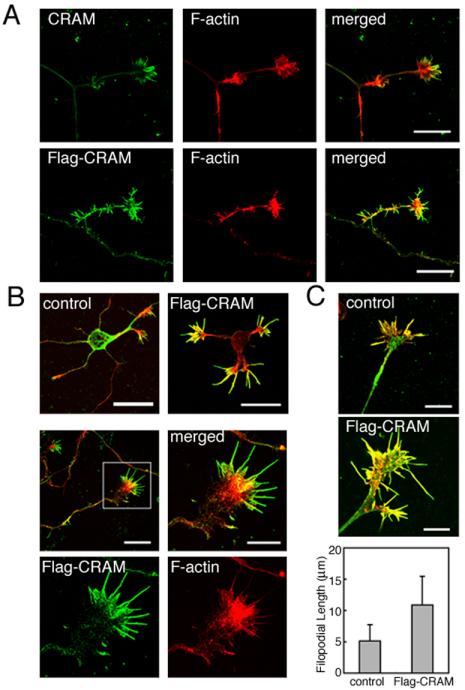
We examined CRAM localization at the growth cone in detail. Primary cultured DRG neurons derived from mouse embryos at E12 were stained with anti-CRAM antibody and phalloidin (Figure 3, A and B), and CRAM distribution at growth cone was examined by confocal microscopic analysis. Figure 3A shows a typical growth cone that did not possess apparent filopodia. In this growth cone, CRAM was lightly distributed at the peripheral region of growth cone but not in the actin meshwork of lamellipodia. On the other hand, Figure 3B shows a type of growth cone with filopodia. In this growth cone CRAM was accumulated in filopodia not in the actin meshwork of lamellipodia as shown in Figure 3A. To confirm filopodial localization of CRAM, we examined growth cone of hippocampal neurons that generally contain well developed filopodia (Figure 3C). As expected, a strong accumulation of CRAM at filopodia was clearly detected. Interestingly, CRAM also was distributed at the distal edges of filopodia lacking any F-actin. To further confirm CRAM localization in filopodia, FLAG-tagged CRAM (Flag-CRAM) was introduced into hippocampal neurons, and its distribution was analyzed by immunostaining with anti-FLAG antibody. As shown in Figure 3D, Flag-CRAM was found to be preferentially accumulated in filopodia. These results suggested the possible involvement of CRAM in filopodial development.
Figure 3.
Accumulation of CRAM at filopodia of growth cone. Endogenous CRAM (A–C) and transfected Flag-CRAM (D) were stained green, and F-actin was labeled with phalloidin (red). A and B and C and D are DRG and hippocampal neurons, respectively. (A) No localization of CRAM in the actin meshwork within lamellipodia. (B) CRAM accumulation at filopodia of DRG growth cone. (C) CRAM accumulation at filopodia of hippocampal growth cone. (D) Filopodial localization of Flag-CRAM is similar to endogenous CRAM. Flag-CRAM was immunostained with anti-FLAG antibody. Bars, 10 μm (A and B) and 5 μm (C and D).
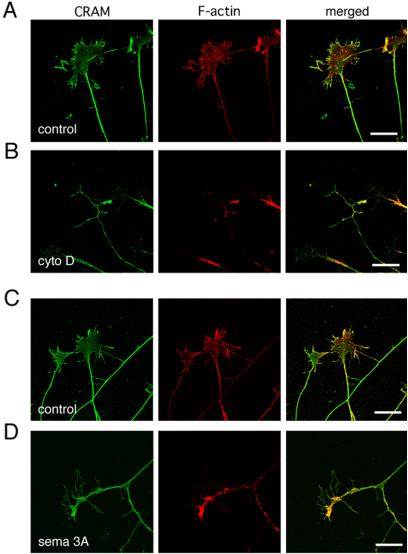
Because CRAM was colocalized with F-actin at filopodia, we investigated whether CRAM localization was dependent on actin structure. DRG neurons were treated with cytochalasin D, which induces actin disassembly, and changes in the distribution of CRAM and actin were compared. Figure 4B shows the growth cone structure after cytochalasin D treatment. Although a partial colocalization of CRAM with actin was observed, CRAM remained in filopodial terminal areas where actin structure had completely disappeared. These results suggested that CRAM localized at filopodia in an actin-independent manner and supported the observation of CRAM distribution at filopodial tips lacking in F-actin.
Figure 4.
Dissociation of CRAM from disassembled actin during growth cone collapse. Primary mouse DRG neurons were treated with solvents as control (A and C), cytochalasin D (cyto D) at 100 ng/ml (B), or Sema3A at 500 ng/ml (D) for 15 min and stained with anti-CRAM antibody (green) and phalloidin (red). Merged images indicate the dissociation of CRAM from actin structure. Bars, 20 μm.
CRMPs have been implicated in Sema3A signaling, and Sema3A mediates growth cone collapse with actin disassembly. Therefore, we next investigated the distributional changes in CRAM and actin in Sema3A-stimulated DRG neurons. As shown in Figure 4D, CRAM remained in filopodia in spite of actin disassembly with Sema3A stimulation. This result further shows that CRAM localized at filopodia in an actin structure-independent manner.
Overexpression of CRAM Promoted Filopodial Growth and Increased the Number of Growth Cones
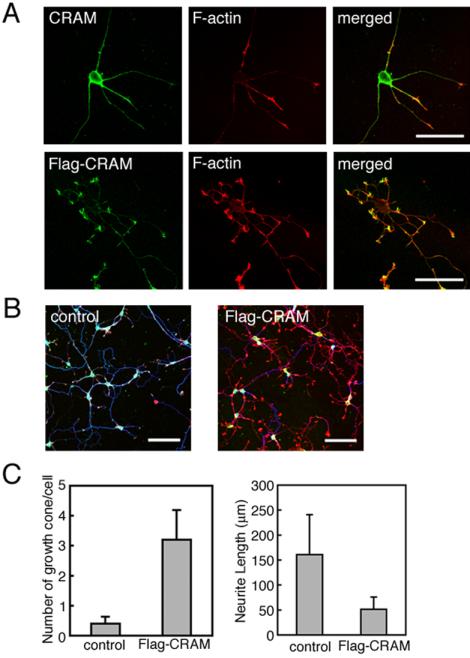
That CRAM localizes at filopodia prompted us to examine whether CRAM overexpression affects filopodial formation. As shown in Figure 5A, Flag-CRAM overexpression induced drastic morphological changes in hippocampal neurons such as excess filopodial induction on growth cone and extension of collateral axon branches of neurons. Almost all filopodia of cells expressing Flag-CRAM displayed a similar response.
Figure 5.
Effect of CRAM overexpression on filopodial formation. (A) Overexpression of CRAM promoted filopodial growth. Hippocampal neurons transfected with control vector were stained with anti-CRAM antibody and phalloidin. Hippocampal neurons expressing Flag-CRAM were stained with anti-FLAG antibody and phalloidin. (B) Enlarged filopodia after Flag-CRAM expression. Typical patterns are shown in high magnification images. (C) Filopodial induction and extension by Flag-CRAM in DRG neurons. DRG neurons transfected with control vector were stained with anti-CRAM antibody and phalloidin. DRG neurons expressing Flag-CRAM were stained with anti-FLAG antibody and phalloidin. Typical patterns of filopodia and growth cone observed in control and Flag-CRAM–expressing DRG neurons are shown in the merged images, and statistical analysis of filopodial length is indicated (bottom). Averaged filopodial length per growth cone was measured (control: 5.15 μm, SD ±2.59; Flag-CRAM: 10.9 μm, SD ±4.58; n = 100, p < 0.001). Bars, 20 μm (A); upper, 20 μm; middle left, 20 μm; right, 10 μm (B); and 10 μm (C).
To understand the effect of Flag-CRAM expression on filopodial morphology, we examined filopodia under high magnification. As shown in Figure 5B, huge and abnormally developed filopodia were produced by Flag-CRAM expression. Interestingly, Flag-CRAM was localized even at the distal ends of filopodia where F-actin signal was almost absent. This result also supported the possibility that CRAM distribution at filopodia does not depend on F-actin structure.
Figure 5C shows a typical image of DRG growth cone with extended filopodia by Flag-CRAM expression in comparison with control. Average filopodial length of Flag-CRAM–expressing cells was 10.9 μm and that of control was 5.15 μm, indicating that filopodia expressing Flag-CRAM extended ∼2.1-fold more than control filopodia (Figure 5C, bottom). These results demonstrated that Flag-CRAM expression promoted induction and growth of filopodia.
Furthermore, we noticed that not only filopodial overgrowth but also growth cone formation was induced by Flag-CRAM. Figure 6 shows hippocampal neurons cultured 3 d in vitro. Normal hippocampal neurons transfected with control vector usually extend one axon with growth cone (Figure 6A, top), whereas hippocampal neurons expressing Flag-CRAM produced supernumerary large growth cones (Figure 6A, bottom). Typical images of growth cone from control and Flag-CRAM–expressing hippocampal neurons are compared in Figure 6B. These growth cones were very large and associated with well-developed filopodia. Furthermore, these growth cones were immunostained with antibody against growth cone marker GAP43 (our unpublished data). This phenomenon was reproducibly observed in more than five independent experiments. Average number of growth cones per cell was measured from 150 cells. The statistical analysis demonstrated that Flag-CRAM–expressing hippocampal neurons produced 3.2 growth cones per cell compared with 0.4 growth cones per cell in control. In other words, an eightfold higher growth cone formation was induced in hippocampal neurons expressing Flag-CRAM compared with control cells (Figure 6C, left). On the other hand, we found that neurites were shortened by Flag-CRAM expression. The statistical analysis demonstrated that average neurite length expressing Flag-CRAM (51.3 μm) was reduced by ∼30% of control neurite (161 μm) (Figure 6C, right). Together, these observations mean that CRAM may be a growth cone inducer and also an inhibitor of neurite extension during neuronal development.
Figure 6.
Effect of CRAM overexpression on growth cone formation. (A) Supernumerary growth cone induction by Flag-CRAM overexpression in hippocampal neurons. Hippocampal neurons transfected with control vector were stained with anti-CRAM antibody and phalloidin (top). Hippocampal neurons expressing Flag-CRAM were stained with anti-FLAG antibody and phalloidin (bottom). (B) Comparison between control and Flag-CRAM expression cells. Hippocampal neurons were stained with anti-FLAG (green), anti-tubulin (blue), and phalloidin (red). (C) Statistical analysis of growth cone number and neurite length between control cells and cells expressing Flag-CRAM. Average number of growth cones per cell and neurite length were measured. Growth cone (control: 0.4, SD ±0.23; Flag-CRAM: 3.2, SD ±0.99; n = 150, p < 0.001). Neurite length (control: 161 μm, SD ±79.6; Flag-CRAM: 51.3 μm, SD ±24.8; n = 155, p < 0.001). Bars, 50 μm (A) and 100 μm (B).
Growth Cone Induced by Flag-CRAM Expression Exhibited Resistance to Sema3A
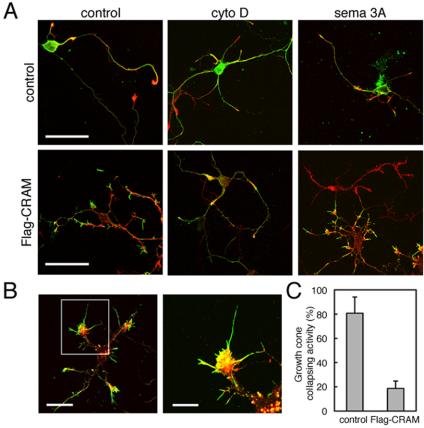
Because CRMPs are believed to be involved in Sema3A signaling, we investigated the role of CRAM in Sema3A-mediated growth cone collapse. Normal growth cones and Flag-CRAM–expressing growth cones of hippocampal neurons were treated with cytochalasin D or Sema3A, and the morphological changes in them were observed. As shown in Figure 7A, cytochalasin D induced the collapse both of control and Flag-CRAM–expressing growth cones, whereas Sema3A failed to collapse the growth cones expressing Flag-CRAM. In the high-magnification images, we found the well developed filopodia expressing Flag-CRAM without any obvious morphological changes, even after Sema3A stimulation (Figure 7B). The statistical analysis of growth conecollapsing activity indicated that only ∼18.7% of growth cones expressing Flag-CRAM were collapsed, whereas >80.8% of control growth cones were collapsed in response to Sema3A (Figure 7C). This result suggested a negative role for CRAM in Sema3A signaling. Because Sema3A was able to induce collapse of growth cones expressing FLAG-tagged CRMPs 1–4 (our unpublished data), this negative function of CRAM in Sema3A signaling may be unique among CRMP family proteins.
Figure 7.
Flag-CRAM expression blocked Sema3A-mediated growth cone collapse. (A) Collapse of Flag-CRAM–induced growth cone by cytochalasin D (cyto D), but not Sema3A. Control or Flag-CRAM–expressing hippocampal neurons were treated with or without cytochalasin D (100 ng/ml) for 15 min or Sema3A (500 ng/ml) for 30 min, and stained with anti-CRAM antibody (green) or anti-FLAG antibody (green) and phalloidin (red). Merged images are shown. (B) Sema3A failed to induce growth cone collapse in Flag-CRAM–expressing hippocampal neurons. These cells were treated with Sema3A as described above and magnified images of growth cone labeled with anti-FLAG (green) and phalloidin (red) are shown. (C) Statistical analysis of growth cone sensitivity to Sema3A. Percentage of growth cone collapse by Sema3A was compared between control vector-transfected cells and Flag-CRAM–expressing cells (control: 80.8%, SD ±13.3; Flag-CRAM: 18.7%, SD ±6.09; n = 3, p < 0.001). Bars, 50 μm (A); left, 20 μm; right, 10 μm (B).
Filopodial Defect and Aberrant Morphology of CRAM-deficient Growth Cone
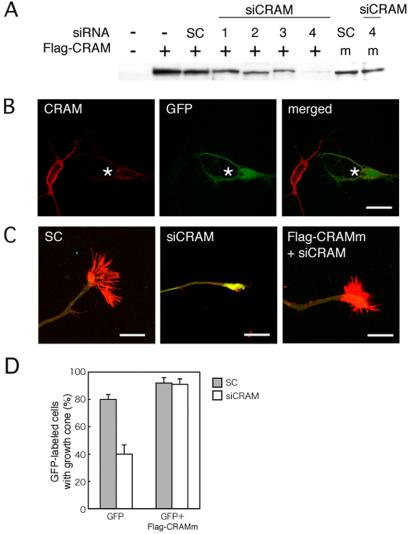
To understand the physiological relevance of endogenous CRAM, the effects of CRAM knockdown by small interfering RNA (siRNA) on growth cone formation of hippocampal neurons were investigated. Several candidate siRNAs were designed against CRAM sequence, and their inhibitory effect on Flag-CRAM expression in COS-7 cell was estimated by immunoblot analysis probed with anti-FLAG antibody. As shown in Figure 8A, all siRNA constructs except for scramble oligonucleotides exhibited the inhibitory effect on CRAM expression. The siRNA construct (no. 4), showing the strongest inhibitory effect was used in the following experiments. To reconstitute the siRNA-mediated phenotype by reexpression of CRAM, we constructed Flag-CRAMm expression vector containing silent mismatches in the knockdown oligonucleotide sequence of no. 4. No inhibitory effect of siRNA (no. 4) on Flag-CRAMm expression also was confirmed (Figure 8A). To test whether this siRNA can suppress the expression level of endogenous CRAM, hippocampal neurons cotransfected with siRNA (no.4) plus phosphorylated enhanced green fluorescent protein (pEGFP) vector as a marker were fixed at 72 h after transfection and immunostained with anti-CRAM antibody. As shown in Figure 8B, the merged image indicated the decrease in endogenous CRAM protein by siRNA (see cell labeled with an asterisk). Cotransfection with scramble oligonucleotides plus pEGFP vector into hippocampal neurons did not affect the expression level of CRAM at all (our unpublished data). Therefore, CRAM siRNA construct was found to be a powerful tool for analyzing CRAM function in developing neurons. Using this siRNA construct, morphological changes of CRAM-deficient hippocampal neurons were investigated. As a control, a typical growth cone of hippocampal neurons transfected with scramble oligonucleotides plus pEGFP vector is shown in Figure 8C, left. Compared with a normal growth cone, CRAM-depleted cell revealed a significant defect in filopodia and growth cone formation. Aggregated F-actin was accumulated at the tip of neurites of CRAM-deficient cell (Figure 8C, middle). Sema3A treatment did not affect on the growth cone morphology after siRNA-mediated CRAM knockdown, because CRAM depletion had already caused filopodia disappearance and growth cone collapse. In addition, no significant change in neurite length was observed in CRAM-deficient cells (our unpublished data). Reconstitution of the phenotype by reexpression of CRAM was achieved by use of Flag-CRAMm expression construct (Figure 8C, right). Furthermore, quantification of the effects of CRAM knockdown on growth cone formation indicated a significant defect in growth cone formation (Figure 8D). The reason for 40% growth cone formation in siRNA- and pEGFP-cotransfected cells may be due to incomplete CRAM deficiency because green fluorescent protein (GFP) signal was used as a transfection marker. On the other hand, the phenotype of siRNA-mediated CRAM knockdown was completely restored by expression of Flag-CRAMm. Finally, we indicated a gallery of images showing the effect of siRNA-mediated CRAM knockdown on growth cone morphology. Together, these results demonstrate that CRAM is required for filopodia and growth cone development.
Figure 8.
Knockdown of CRAM by siRNA revealed filopodial defect. (A) Effect of siRNA oligonucleotides on CRAM expression in COS-7 cells. Four siRNAs (1–4) against CRAM sequence were transfected with Flag-CRAM into COS-7 cells. Flag-CRAMm (m) expression construct contains silent mismatches in the knockdown oligonucleotide sequence of no. 4 as described in Materials and Methods. Scramble oligonucleotide (SC) was used as a negative control. Inhibitory effect on CRAM expression was estimated by immunoblot analysis probed with anti-FLAG antibody. (B) Knockdown of endogenous CRAM by siRNA. Hippocampal neurons were fixed 72 h after transfection of siRNA (no. 4) plus pEGFP vector and immunstained with anti-CRAM antibody (red). Cell labeled with an asterisk shows the decrease in endogenous CRAM by siRNA. (C) Defect in filopodial formation after siRNA-mediated CRAM depletion. Hippocampal neurons were fixed 72 h after transfection of scramble or siRNA (no.4) with pEGFP or pEGFP/Flag-CRAMm and stained with phalloidin (red). Reconstitution of the phenotype by reexpression of CRAM was achieved by use of Flag-CRAMm expression construct. Merged images of GFP (green) and phalloidin (red) are indicated. Bars, 50 μm (B) and 10 μm (C). (D) Quantification of the effects of CRAM knockdown on growth cone formation. Percentage of GFP-labeled cells with growth cone formation was measured between SC and siRNA (no. 4) with pEGFP or pEGFP/Flag-CRAMm–cotransfected cells (pEGFP/SC: 80.2%, SD ±3.5, pEGFP/siRNA: 39.8%, SD ±6.7, pEGFP/SC/Flag-CRAMm: 91.5%, SD ±4.0, pEGFP/siRNA/Flag-CRAMm: 91.2%, SD ±4.0; n = 3, p < 0.001).
DISCUSSION
Role of CRAM in Filopodia and Growth Cone Development
CRMP family has been implicated in the intracellular signaling of axonal guidance. Among CRMP family proteins, CRAM reveals phylogenetic divergence (Inatome et al., 2000), suggesting that CRAM plays a role distinct from the other four CRMPs. In this study, we found that CRAM was concentrated at filopodia and that siRNA-mediated CRAM knockdown blocked normal filopodial formation. Moreover, neurons overexpressing Flag-CRAM showed aberrant filopodial extension and formation of multiple growth cone. Because we did not observe similar phenomena in cells overexpressing the other four CRMPs, CRAM may have a very specific role in filopodial dynamics.
Filopodia play a critical role in the regulation of growth cone motility through a direct recognition of extracellular conditions such as repellent or attractant factors. Small GTP-binding proteins such as CDC42 have been considered to be involved in filopodial formation via actin reorganization (Meyer and Feldman, 2002); however, the molecular mechanisms underlying filopodial organization during neural development are largely unknown. It is known that at the filopodial tips there is a small space between filopodial membrane and the end of actin filaments. Therefore, it is possible that some proteins localized in this space may lead filopodial growth by controlling actin dynamics. Here, we found that CRAM localized in this space and that Flag-CRAM overexpression promoted actin assembly at filopodia. Moreover, experiments using cytochalasin D suggested that filopodial localization of CRAM did not depend on actin structure. Thus, we would like to propose that CRAM is one of potent candidates that leads to actin assembly and filopodial extension. We are currently studying the molecular mechanism by which CRAM induces filopodial formation, focusing on the relationship between CDC42 and CRAM.
Endogenous CRAM was found to be distributed not only at growth cones but also in cell body, axon, and dendrites. Compared with endogenous CRAM, Flag-CRAM preferentially accumulated at filopodia of growth cone. This suggested that Flag-CRAM may have a modification allowing to target filopodia. Previous work demonstrated that CRMPs and CRAM were phosphorylated during neural development. In addition, we reported that tyrosine kinase Fes/Fps was associated with CRAM and could phosphorylate CRAM in vitro (Mitsui et al., 2002). Therefore, we speculated that phosphorylation events may regulate CRAM localization at filopodia. Further study would be needed to identify phosphorylation sites of CRAM that may affect filopodial localization of CRAM.
In DRG neurons Flag-CRAM overexpression promoted the extension of collateral axon branches, but it did not exhibit multiple growth cone formation observed in hippocampal neurons. This means that CRAM may not determine the fate of growth cone induction. Preferably, CRAM may be involved in the maturation of growth cone buds. Actually, hippocampal neurons contain multiple moving growth cone-like structures referred to as “waves,” that emerge at the base of neurites and travel distantly to the tip (Ruthel and Banker, 1999). We speculate that these growth cone-like structures might be maturated by Flag-CRAM activity. Thus, we propose that CRAM is a potent maturation factor of growth cone.
On the other hand, Flag-CRAM overexpression in hippocampal neurons inhibited neurite extension. This fact seems to be a result of stabilizing filopodial actin structures. It has been believed that actin meshwork in the central region of growth cone limits the microtubule invasion to the peripheral region (Tanaka and Sabry, 1995). Indeed, actin disassembly by cytochalasin B treatment allowed microtubule invasion to the central region of growth cone of Aplysia neurons (Forscher and Smith, 1988). These observations suggested that actin dynamics such as turnover of actin filaments in growth cone are important for the microtubule translocation or assembly which leads to neurite extension. Certainly, actin structures in filopodia and growth cones were significantly enlarged by Flag-CRAM expression. In addition, these structures showed the resistance to Sema3A stimulation, although they were sensitive to cytochalasin D. It is therefore critical to examine the effect of CRAM on actin dynamics. Alternatively, the inhibition of neurite growth by Flag-CRAM may be due to the modulation of moving growth cone-like wave structures as described above. As a wave nears the tip, the neurite undergoes retraction, and when it reaches the tip, the neurite undergoes a burst of growth (Ruthel and Banker, 1999). Maturation of growth cone-like structures by Flag-CRAM may decrease moving speed of a wave that modulates regularly occurring retraction of growth cone and thus decrease average neurite outgrowth rates.
Previous work has suggested that increased turnover of actin filaments in growth cone is required for axonal formation (Bradke and Dotti, 1999). Because we observed the CRAM accumulation at the tip of dendrites, CRAM may suppress the conversion of dendrites to axon. It was reported that overexpression of CRMP-2 in hippocampal neurons led to multiple axonal formation and extension (Inagaki et al., 2001). Thus, CRAM could play an opposite role to CRMP-2 in the neural development. At present, however, we could not detect any inhibitory effect of Flag-CRAM on axonal formation.
Negative Role of CRAM in Sema3A Signaling
CRMP-2 was initially identified by its possible involvement in the Sema3A-induced mediation of growth cone collapse in chick DRG neurons (Goshima et al., 1995). The authors demonstrated that introduction of anti-CRMP antibody into chick DRG neurons blocked Sema3A-mediated growth cone collapse. However, this anti-CRMP antibody did not cross-react with CRAM protein. This means that there is no evidence that CRAM is a semaphorin response mediator protein. Here, we found that Sema3A failed to collapse growth cones overexpressing Flag-CRAM. Because this phenomenon could not be detected in neurons overexpressing the other four Flag-CRMPs, CRAM seemed to play a specific role in the negative regulation of Sema3A-mediated signaling among CRMP family proteins.
Immunohistochemical analysis indicated that neuropilin1 and plexinA1, a Sema3A receptor complex, were normally expressed in growth cones induced by Flag-CRAM. Thus, it is unlikely that this negative regulation by Flag-CRAM is due to the down-regulation of Sema3A receptor. In addition, collapse of Flag-CRAM–expressing growth cones by cytochalasin D suggested that this Flag-CRAM–mediated resistance to Sema3A may not be due to the F-actin stabilization such as cross-linking of actin filaments.
What is the molecular mechanism underlying the inhibition of Sema3A-mediated growth cone collapse by CRAM expression? CRAM must inhibit at an unknown step downstream event of Sema3A receptor activation. Recently, Terman et al. (2002) have demonstrated that MICAL, a putative monooxygenase, interacts with the neuronal plexinA and transmits the signal from the receptor plexin to the actin cytoskeleton through a redox mechanism. MICAL could act either indirectly, causing a local increase in the concentration of reactive oxygen species or directly, inducing redox changes in downstream molecules. Because previous work suggested that CRMP was associated with redox enzymes (Bulliard et al., 1997), it is possible that CRAM could block Sema3A-mediated growth cone collapse through a modification of redox changes induced by MICAL action. Alternatively, CRAM may block Sema3A-mediated growth cone collapse by inhibition of CRMP-2 function. Immununoprecipitation assay revealed the association of CRAM with CRMP-2 in DRG neurons (our unpublished data). Thus, distinct from four CRMPs, CRAM seems to play an opposite role in restricting the responsiveness to Sema3A. In conclusion, CRAM may control filopodial dynamics and growth cone development, thereby negatively regulating the sensitivity of growth cone to Sema3A.
Supplementary Material
Acknowledgments
We thank Drs. K. Itoh and S. Matsuyama for technical assistance in immunohistochemical analysis, and Dr. S. Jahangeer for critically reading the manuscript. This work was supported by a grant-in-aid for scientific research on priority areas (A) from the Ministry of Education, Science, Sports and Culture, Japan (to S.Y.). R.I. was supported by a Research Fellowship of the Japan Society for the Promotion of Science.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–08–0679. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-08-0679.
Abbreviations used: CRAM, CRMP-associated molecule; CRMP, collapsin response mediator protein; DHPase, dihydropyrimidinase; DRG, dorsal root ganglion; Flag-CRAM, FLAG-tagged CRAM; siRNA, small interfering RNA.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Berninger, B., Garcia, D. E., Inagaki, N., Hahnel, C., and Lindholm, D. (1993). BDNF and NT-3 induce intracellular Ca2+ elevation in hippocampal neurones. Neuroreport 4, 1303-1306. [DOI] [PubMed] [Google Scholar]
- Bradke, F., and Dotti, C. G. (1999). The role of local actin instability in axon formation. Science 283, 1931-1934. [DOI] [PubMed] [Google Scholar]
- Bulliard, C., Zurbriggen, R., Tornare, J., Faty, M., Dastoor, Z., and Dreyer, J. L. (1997). Purification of a dichlorophenol-indophenol oxidoreductase from rat and bovine synaptic membranes: tight complex association of a glyceraldehyde-3-phosphate dehydrogenase isoform, TOAD64, enolase-gamma and aldolase C. Biochem. J. 324, 555-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byk, T., Dobransky, T., Cifuentes-Diaz, C., and Sobel, A. (1996). Identification and molecular characterization of Unc-33-like phosphoprotein (Ulip), a putative mammalian homolog of the axonal guidance-associated unc-33 gene product. J. Neurosci. 16, 688-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byk, T., Ozon, S., and Sobel, A. (1998). The Ulip family phosphoproteins–common and specific properties. Eur. J. Biochem. 254, 14-24. [DOI] [PubMed] [Google Scholar]
- Comeau, M. R., et al. (1998). A poxvirus-encoded semaphorin induces cytokine production from monocytes and binds to a novel cellular semaphorin receptor, VESPR. Immunity 8, 473-482. [DOI] [PubMed] [Google Scholar]
- Forscher, P., and Smith, S. J. (1988). Actions of cytochalasins on the organization of actin filaments and microtubules in a neuronal growth cone. J. Cell Biol. 107, 1505-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukada, M., Watakabe, I., Yuasa-Kawada, J., Kawachi, H., Kuroiwa, A., Matsuda, Y., and Noda, M. (2000). Molecular characterization of CRMP5, a novel member of the collapsin response mediator protein family. J. Biol. Chem. 275, 37957-37965. [DOI] [PubMed] [Google Scholar]
- Fukata, Y., et al. (2002). CRMP-2 binds to tubulin heterodimers to promote microtubule assembly. Nat. Cell Biol. 4, 583-591. [DOI] [PubMed] [Google Scholar]
- Goshima, Y., Nakamura, F., Strittmatter, P., and Strittmatter, S. M. (1995). Collapsin-induced growth cone collapse mediated by an intracellular protein related to UNC-33. Nature 376, 509-514. [DOI] [PubMed] [Google Scholar]
- Hamajima, N., Matsuda, K., Sakata, S., Tamaki, N., Sasaki, M., and Nonaka, M. (1996). A novel gene family defined by human dihydropyrimidinase and three related proteins with differential tissue distribution. Gene 180, 157-163. [DOI] [PubMed] [Google Scholar]
- Hedgecock, E. M., Culotti, J. G., Thomson, J. N., and Perkins, L. A. (1985). Axonal guidance mutants of Caenorhabditis elegans identified by filling sensory neurons with fluorescein dyes. Dev. Biol. 111, 158-170. [DOI] [PubMed] [Google Scholar]
- Inagaki, N., Chihara, K., Arimura, N., Menager, C., Kawano, Y., Matsuo, N., Nishimura, T., Amano, M., and Kaibuchi, K. (2001). CRMP-2 induces axons in cultured hippocampal neurons. Nat. Neurosci. 4, 781-782. [DOI] [PubMed] [Google Scholar]
- Inatome, R., Tsujimura, T., Hitomi, T., Mitsui, N., Hermann, P., Kuroda, S., Yamamura, H., and Yanagi, S. (2000). Identification of CRAM, a novel unc-33 gene family protein that associates with CRMP3 and protein-tyrosine kinase(s) in the developing rat brain. J. Biol. Chem. 275, 27291-27302. [DOI] [PubMed] [Google Scholar]
- Kolodkin, A. L. (1998). Semaphorin-mediated neuronal growth cone guidance. Prog. Brain Res. 117, 115-132. [DOI] [PubMed] [Google Scholar]
- Luo, Y., Raible, D., and Raper, J. A. (1993). Collapsin: a protein in brain that induces the collapse and paralysis of neuronal growth cones. Cell 75, 217-227. [DOI] [PubMed] [Google Scholar]
- Meyer, G., and Feldman, E. L. (2002). Signaling mechanisms that regulate actin-based motility processes in the nervous system. J. Neurochem. 83, 490-503. [DOI] [PubMed] [Google Scholar]
- Minturn, J. E., Fryer, H. J., Geschwind, D. H., and Hockfield, S. (1995). TOAD-64, a gene expressed early in neuronal differentiation in the rat, is related to unc-33, a C. elegans gene involved in axon outgrowth. J. Neurosci. 15, 6757-6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui, N., Inatome, R., Takahashi, S., Goshima, Y., Yamamura, H., and Yanagi, S. (2002). Involvement of Fes/Fps tyrosine kinase in semaphorin3A signaling. EMBO J. 21, 3296-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasterkamp, R. J., and Kolodkin, A. L. (2003). Semaphorin junction: making tracks toward neural connectivity. Curr. Opin. Neurobiol. 13, 79-89. [DOI] [PubMed] [Google Scholar]
- Ruthel, G., and Banker, G. (1999). Role of moving growth cone-like “wave” structures in the outgrowth of cultured hippocampal axons and dendrites. J. Neurobiol. 39, 97-106. [DOI] [PubMed] [Google Scholar]
- Takahashi, S., Inatome, R., Hotta, A., Qin, Q., Hackenmiller, R., Simon, M. C., Yamamura, H., and Yanagi, S. (2003a). Role for Fes/Fps tyrosine kinase in microtubule nucleation through is Fes/CIP4 homology domain. J. Biol. Chem. 278, 49129-49133. [DOI] [PubMed] [Google Scholar]
- Takahashi, S., Inatome, R., Yamamura, H., and Yanagi, S. (2003b). Isolation and expression of a novel mitochondrial septin that interacts with CRMP/CRAM in the developing neurones. Genes Cells 8, 81-93. [DOI] [PubMed] [Google Scholar]
- Takahashi, T., Fournier, A., Nakamura, F., Wang, L. H., Murakami, Y., Kalb, R. G., Fujisawa, H., and Strittmatter, S. M. (1999). Plexin-neuropilin-1 complexes form functional semaphorin-3A receptors. Cell 99, 59-69. [DOI] [PubMed] [Google Scholar]
- Tamagnone, L., et al. (1999). Plexins are a large family of receptors for transmembrane, secreted, and GPI-anchored semaphorins in vertebrates. Cell 99, 71-80. [DOI] [PubMed] [Google Scholar]
- Tanaka, E., and Sabry, J. (1995). Making the connection: cytoskeletal rearrangements during growth cone guidance. Cell 83, 171-176. [DOI] [PubMed] [Google Scholar]
- Terman, J. R., Mao, T., Pasterkamp, R. J., Yu, H. H., and Kolodkin, A. L. (2002). MICALs, a family of conserved flavoprotein oxidoreductases, function in plexin-mediated axonal repulsion. Cell 109, 887-900. [DOI] [PubMed] [Google Scholar]
- Tessier Lavigne, M., and Goodman, C. S. (1996). The molecular biology of axon guidance. Science 274, 1123-1133. [DOI] [PubMed] [Google Scholar]
- Wang, L. H., and Strittmatter, S. M. (1996). A family of rat CRMP genes is differentially expressed in the nervous system. J. Neurosci. 16, 6197-6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winberg, M. L., Noordermeer, J. N., Tamagnone, L., Comoglio, P. M., Spriggs, M. K., Tessier-Lavigne, M., and Goodman, C. S. (1998). Plexin A is a neuronal semaphorin receptor that controls axon guidance. Cell 95, 903-916. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.