Abstract
Polymorphonuclear myeloid-derived suppressor cells (PMN-MDSC) are important regulators of immune responses in cancer and have been directly implicated in promotion of tumor progression. However, the heterogeneity of these cells and lack of distinct markers hampers the progress in understanding of the biology and clinical importance of these cells. Using partial enrichment of PMN-MDSC with gradient centrifugation we determined that low density PMN-MDSC and high density neutrophils from the same cancer patients had a distinct gene profile. Most prominent changes were observed in the expression of genes associated with endoplasmic reticulum (ER) stress. Surprisingly, low-density lipoprotein (LDL) was one of the most increased regulators and its receptor oxidized LDL receptor 1 OLR1 was one of the most overexpressed genes in PMN-MDSC. Lectin-type oxidized LDL receptor 1 (LOX-1) encoded by OLR1 was practically undetectable in neutrophils in peripheral blood of healthy donors, whereas 5–15% of total neutrophils in cancer patients and 15–50% of neutrophils in tumor tissues were LOX-1+. In contrast to their LOX-1− counterparts, LOX-1+ neutrophils had gene signature, potent immune suppressive activity, up-regulation of ER stress, and other biochemical characteristics of PMN-MDSC. Moreover, induction of ER stress in neutrophils from healthy donors up-regulated LOX-1 expression and converted these cells to suppressive PMN-MDSC. Thus, we identified a specific marker of human PMN-MDSC associated with ER stress and lipid metabolism, which provides new insight to the biology and potential therapeutic targeting of these cells.
Introduction
The accumulation of relatively immature and pathologically activated myeloid-derived suppressor cells (MDSC) with potent immunosuppressive activity is common in tumors. MDSC have the ability to support tumor progression by promoting tumor cell survival, angiogenesis, invasion of healthy tissue by tumor cells, and metastases (reviewed in (1)). There is now ample evidence of the association of accumulation of immune suppressive MDSC with negative clinical outcomes in various cancers (2). MDSC have been implicated in resistance to anticancer therapies with kinase inhibitor (3), chemotherapy (4–7), and immune therapy (8–12). Two large populations of MDSC are currently described: polymorphonuclear (PMN-MDSC) and monocytic cells (M-MDSC) (13). PMN-MDSC is the most abundant population of MDSC in most types of cancer, phenotypically and morphologically similar to neutrophils (PMN)(14). These cells share the CD11b+CD14−CD15+/CD66b+ phenotype. Currently, these cells can be separated only in peripheral blood (PB) and only by density gradient. Distinction between PMN-MDSC and PMN in tumor tissues is not possible. Since gradient centrifugation may enrich not only for true PMN-MDSC but also for activated PMN without suppressive activity the heterogeneity of PMN-MDSC population raised the questions of whether PMN-MDSC and PMN are truly cells with distinct features. It is not clear what defines the specific functional state of human PMN-MDSC vis-à-vis PMN in the same patient. More importantly, the mechanisms responsible for acquisition of pathological activity by human neutrophils in cancer remained unclear. In this study we attempted to address these questions by evaluating populations of PMN-MDSC and PMN from the same patients. We identified genomic signature of PMN-MDSC and surface marker specific for these cells. We found that induction of ER stress response was sufficient to convert neutrophils to PMN-MDSC.
Results
Human PMN-MDSC have a distinct gene expression profile from neutrophils
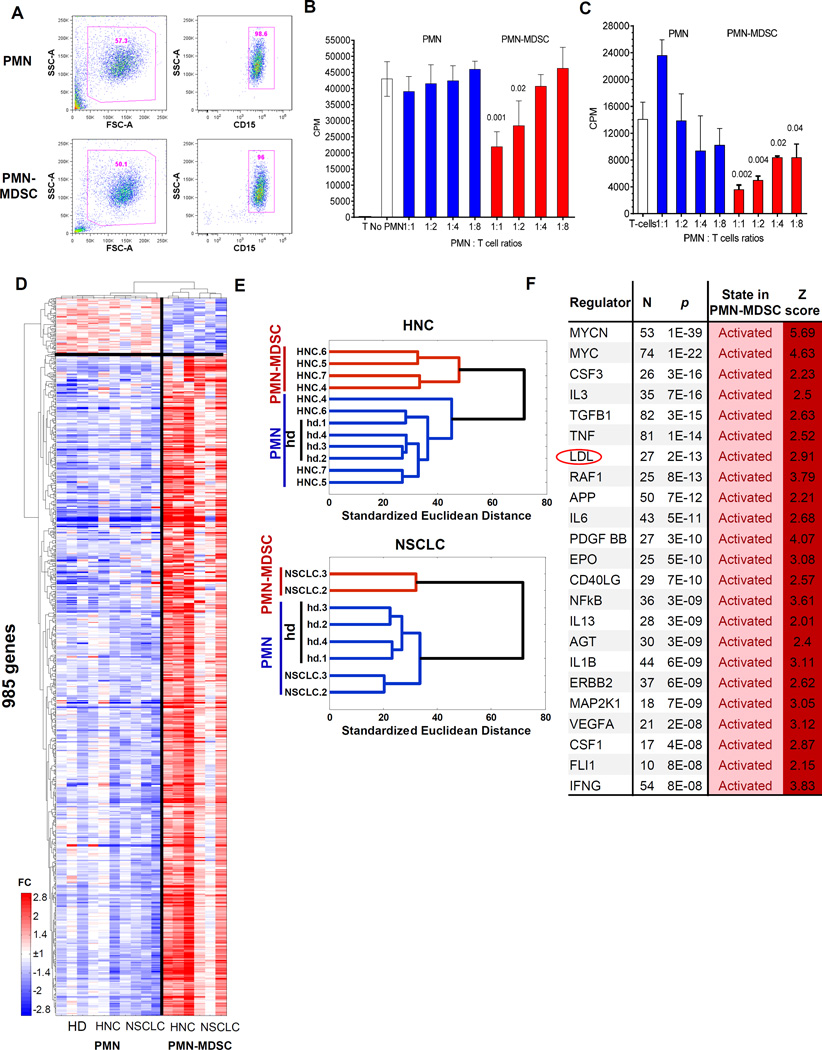
To compare PMN-MDSC and PMN from PB of the same patients with non-small cell lung cancer (NSCLC) and head and neck cancer (HNC) we used dual-density Histopaque gradient, the standard method of isolation of PMN-MDSC (15). Low density PMN-MDSC are co-purified with PBMC, whereas high density PMN are collected from lower gradient (16). As a control, PMN from healthy donors were used. Both, low-density PMN-MDSCs and high-density PMN were purified further with CD15 magnetic beads to achieve similar high purity of both cell populations (Fig. 1A). Immune suppressive activity of PMN-MDSC, the main characteristics of these cells, was confirmed in allogeneic mixed leukocyte reaction (MLR) (Fig. 1B) and in autologous system with T cells activated by CD3/CD28 antibodies (Fig. 1C). As expected, PMN were not suppressive (Fig. 1B,C)
Figure 1. PMN-MDSC have distinct genomic signature.
A. Typical phenotype of PMN-MDSC and PMN isolated from peripheral blood of cancer patients using gradient centrifugation and CD15 beads; B. Suppression assay of PMN and PMN-MDSC isolated from the same patient with HNC. Allogeneic mixed leukocyte reaction was performed as described in Methods. Cell proliferation was evaluated in triplicates using 3H-thymidine uptake. Mean and SD are shown. p values are calculated in t-test from control – T cell proliferation without the presence of PMN or PMN-MDSC and shown on the graph, n=3 different patients. C. Suppression assay of PMN and PMN-MDSC isolated from the same patient with NSCLC. T-cell proliferation in response to CD3/CD28 was performed as described in Methods. Cell proliferation was evaluated in triplicates using 3H-thymidine uptake. Mean and SD are shown. p values are calculated in t-test from control – T cell proliferation without the presence of PMN or PMN-MDSC and shown on the graph, n=3 different patients. D. Relative expression heatmap and gene/sample clustering based on expression of 985 genes significantly differentially expressed (p<0.05, fold>2) between cancer patients’ PMN, PMN-MDSCs and PMN of healthy donors. E. Hierarchical clustering of PMN-MDSCs from HNC and NSCLC cancer patients indicates gene expression signature specific to PMN-MDSCs and similarities of PMN from cancer patients and PMN from healthy donors. F. Upstream Regulators identified by Ingenuity Pathway Analysis (IPA) among genes significantly differentially expressed between PMN-MDSC and PMN cells. N=number of genes from the category, Z=z-score of predicted activation state calculated by IPA.
In order to study overall differences and similarities between patients’ PMN and PMN-MDSC as well as PMNs from healthy donors, we performed whole-genome analysis using Illumina HumanHT-12 v4 bead arrays. The direct pair-wise comparison identified 1870 array probes significantly differentially expressed (FDR<5%) between PMN-MDSC and PMN in the same patients (Fig. S1A) and 36 probes showed difference of at least 5 fold (Fig. S1B). Hierarchical clustering of the samples using expression of the 985 most differentially expressed genes (fold>2, p<0.05, Fig. 1D) revealed that PMN-MDSC samples have a unique expression profile and PMN from cancer patients are very similar to healthy donor PMN samples, as they grouped within the same cluster for HNC and NSCLC patients (Figure 1E). Specifically, of the 985 genes different between any pair of groups, the majority (74%) showed significant differences (false discovery rate, FDR<5%) between patients’ PMN-MDSC and PMN, while no genes were significantly different when corrected for multiple testing (best FDR=19%) between PMN from healthy donors and PMN from cancer patients, with only 12% of the genes significantly different at nominal p<0.05, which indicates a high similarity of PMN samples between cancer patients and healthy donors.
Using Ingenuity Pathway Analysis, we identified 14 pathways significantly enriched in PMN-MDSCs, including eukaryotic Translation Initiation Factors 2 and 4 (eIF2 and eIF4) pathways and mTOR signaling (Table. S1). The regulators of genes enriched in PMN-MDSC included regulators of ER stress response, MAPK pathway, CSF1, IL-6, IFN-γ, NF-κB. Interestingly, these molecules were previously directly implicated in MDSC biology, primarily PMN-MDSC (reviewed in (17)). Surprisingly, one of the most significant changes was associated with low-density lipoprotein (LDL) (Fig. 1F). Thus, PMN-MDSC had a distinct genomic profile from PMN isolated from the same cancer patients and PMN from healthy donors. Genes associated with ER stress and immune responses were among the most up-regulated in PMN-MDSC.
LOX-1 is differentially expressed on PMN-MDSC and PMN
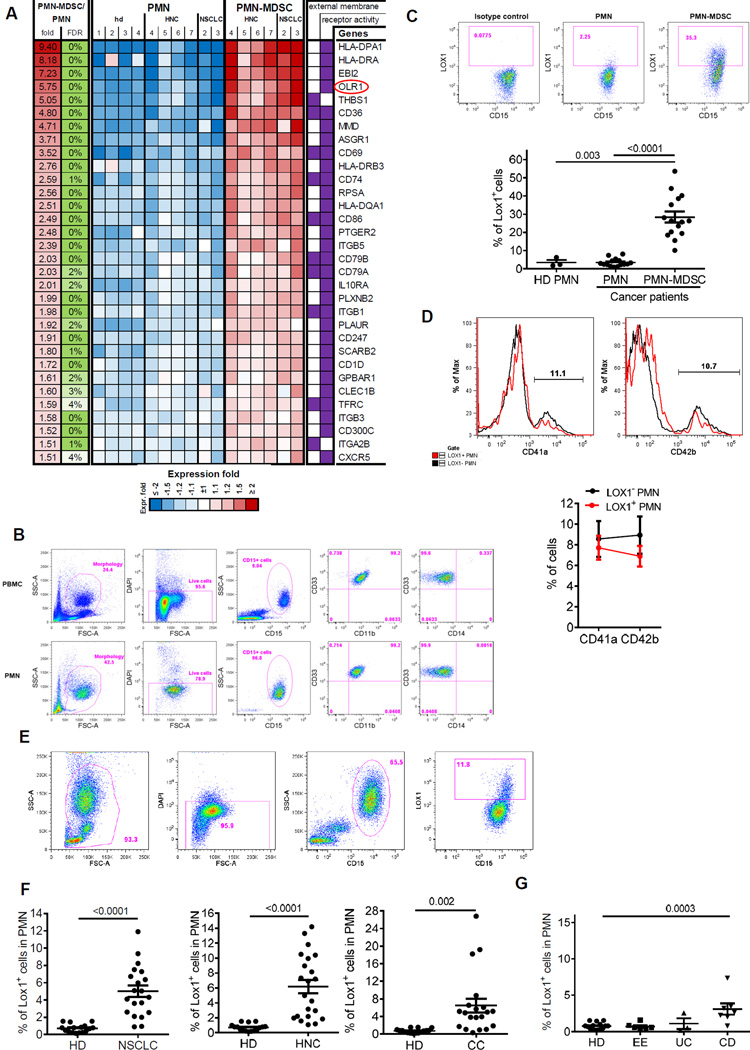
To search for potential markers of PMN-MDSC we evaluated differentially expressed genes, which encoded surface molecules and compared expression of various surface molecules between PMN-MDSC and PMN from the same patients and PMN from healthy donors. More than 20 genes encoded surface molecules were found to be differentially expressed in PMN-MDSC and PMN (Fig. 2A). In an attempt to validate these observations we tested surface expression of some of the proteins using available antibodies and flow cytometry. Unexpectedly, the differences were found in the expression of lectin-like oxidized low-density lipoprotein receptor-1 (LOX-1), a 50kDa transmembrane glycoprotein encoded by the OLR1 gene (oxidized LDL receptor 1) (18). In our analysis OLR1 was one of the mostly up-regulated genes in PMN-MDSC (Fig. S1B). LOX-1 is one of the main receptors for oxidized- LDL (oxLDL) (19). It also binds other ligands including other modified lipoproteins, advanced glycation end products, aged red blood cells, apoptotic cells and activated platelets (20). LOX-1 is expressed on endothelial cells, macrophages, smooth muscle cells, and some intestinal cell lines (18). However, it has not been associated with neutrophils.
Figure 2. LOX-1 as a marker of PMN-MDSC.
A. List and a heatmap of relative expression of candidate surface markers specific to the PMN-MDSCs. B. Typical phenotype of high density PMN and low density PMN-MDSC in cancer patient. C. Proportion of LOX-1 positive PMN and PMN-MDSC in peripheral blood of 15 cancer patients. Top panel - example of staining of PMN-MDSC and PMN with LOX-1 antibody. Cells were isolated using density gradient as described in Methods and the proportion of LOX-1+ cells was calculated among CD15+ cells. Bottom panel - individual results for each patient are shown as well as Mean and SE. p values (t-test) are shown. D. Example of staining with CD41a and CD42b antibody (top panel). Cumulative results of 7 patients with NSCLC (bottom panel). Mean and SD are shown. E. Typical example of the analysis of PMN in cancer patient. F. Proportion of LOX-1+ cells among PMN in unseparated PB from 16 healthy donors (HD), 20 patients with NSCLC, 21 patients with HNC, and 19 patients with CC. p values in t-test are shown. G. Proportion of LOX-1+ cells among PMN in unseparated PB from 16 healthy donors (HD), 6 patients with eosinophilic colitis (EE), 3 patients with ulcerative colitis (UC), and 7 patients with Crohn’s Disease (CD). p values in t-test are shown.
We evaluated LOX-1 expression in high density PMN and low-density PMN-MDSC in cancer patients (Fig. 2B). LOX-1 was practically undetectable in PMN but expressed in about 1/3 of PMN-MDSC fraction (Fig. 2C). Since LOX-1 can be expressed on platelets (21) and it is known that platelets can adhere to activated PMN we asked whether increased expression of LOX-1 in PMN-MDSC fraction was the result of increased adherence of platelets. However, LOX-1− and LOX-1+ cells in low density PMN-MDSC population had the same small proportion of cells that express platelets markers CD41a and CD42b (Fig. 2D). We asked whether LOX-1 expression on PMN correlated with CD16 expression associated with neutrophil activation (22, 23). Almost all PMN in cancer patients were CD16hi and expression of LOX-1 on PMN was not associated with CD16 expression (Fig. S2).
These results suggested that LOX-1 could be associated with PMN-MDSC. We asked whether LOX-1 can be a marker of PMN-MDSC. Cell density as a criterion for separation of PMN-MDSC from PMN has serious limitations. It may be changed during handling of cells. In addition, gradient centrifugation may enrich not only for PMN-MDSC but also for activated PMN without suppressive activity, which contribute to well established heterogeneity of PMN-MDSC population. Therefore, we avoided the use of gradient centrifugation and labeled cells in PB directly with granulocyte-specific CD15 antibody and evaluated expression of LOX-1 among all CD15+ cells (Fig. 2E). In preliminary experiments we found no differences in the results obtained with CD15 or CD66b antibodies. We referred to CD15+ cells as PMN since Siglec-8+ eosinophils represented very small proportion of CD15+ cells and no differences in the presence of eosinophils between CD15+LOX-1+ and CD15+LOX-1− cells was seen (Fig. S3). The proportion of LOX-1+ cells among all PMN in healthy donors was very low (range 0.1–1.5, mean 0.7%) (Fig. 2F and Fig. S4). In patients with NSCLC it increased to 4.9% (p<0.001), in patients with HNC to 6.4% (p<0.0001), and in patients with colon cancer (CC) to 6.5% (p=0.0035) (Fig. 2F). In all three types of cancer >75% of patients had proportion of LOX-1+ PMN higher than the range established for healthy donors. We also assessed the changes in LOX-1+PMN in tumor-free patients with inflammatory conditions: eosinophilic esophagitis, ulcerative colitis and Crohn’s disease. Only patients with Crohn’s disease had a small increase in the proportion of these cells (Fig. 2G). Thus, LOX-1 expression defined distinct population of neutrophils in cancer patients and was associated with accumulation of PMN-MDSC.
LOX-1 defines the population of PMN-MDSC among neutrophils
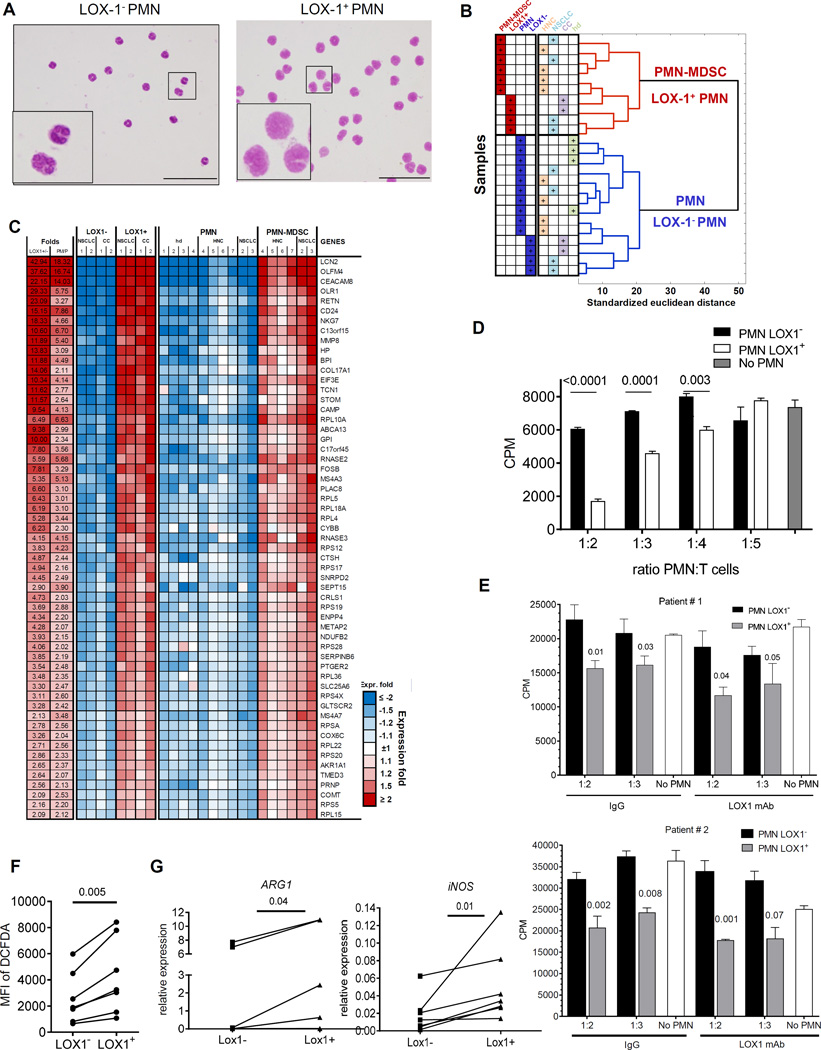
Next, we addressed the question whether LOX-1 can be considered as marker of human PMN-MDSC. LOX-1+ and LOX-1− PMN were sorted directly from PB of the same patients. LOX-1− PMN had the typical morphology of mature neutrophils, whereas LOX-1+ PMN displayed more immature morphology with band shape nuclei (Fig. 3A). Whole gene expression array was performed on LOX-1+ and LOX-1− PMN and compared with that of PMN and PMN-MDSC. Analysis of gene expression revealed 639 genes significantly different between LOX-1+ and LOX-1− (FDR<5%, fold>2) and based on expression of those genes LOX-1+ PMN clustered together with PMN-MDSC, whereas LOX-1− PMN were very similar to patients’ and healthy donor’s PMN (Fig. 3B). Overall, 92% of those genes had the same direction of change between LOX1+/LOX1− as between PMN-MDSC/PMN with 93 probes significantly upregulated (FDR<5%) at least 2 fold or more in both PMN-MDSC and LOX-1+ PMN (Fig. 3C and Fig. S5). Thus, LOX-1+ PMN from cancer patients had gene expression profile similar to that of PMN-MDSC.
Figure 3. LOX-1 expression defines bona-fide PMN-MDSC.
A. Typical morphology of sorted LOX-1+ and Lox-1− PMN from patient with HNC. Scale bar = 20 µm. B. Hierarchical clustering of samples based on expression levels of genes differentially expressed between LOX-1+ and LOX-1− PMN. C. List and relative expression values of the most changed known genes overlapped between LOX-1+ and PMN-MDSC cells. D. Suppressive activity of LOX-1+ and LOX-1− PMN isolated from peripheral blood of patient with HNC in allogeneic MLR. Cell proliferation was evaluated in triplicates using 3H-thymidine uptake. Mean and SE are shown. p values in t-test from T cell proliferation without the presence of PMN are shown. Experiments with similar results were performed with samples from 9 patients with HNC and NSCLC. E. Effect of neutralizing LOX-1 antibody on suppressive activity of LOX-1+ PMN. PMN were isolated from two patients with NSCLC. LOX-1+ and LOX-1− PMN were isolated as described in Material and Methods and then added to allogeneic MLR in the presence of 10 µg/ml neutralizing mouse anti-human LOX-1 antibody or mouse IgG. Cell proliferation was evaluated in triplicates using 3H-thymidine uptake. Mean and SE are shown. p values in t-test from T cell proliferation without the presence of PMN are shown. F. ROS production in LOX-1+ and LOX-1− PMN from 7 patients with HNC and NSCLC. ROS production was measured by staining with DCFDA. G. Expression of ARG1 and NOS2 in LOX-1+ and LOX-1− PMN from 6 patients with HNC and MM measured by qPCR. P values in t-test are shown.
The hallmark of PMN-MDSC is their ability to suppress T-cell function. We isolated LOX-1− and LOX-1+ PMN directly from PB of cancer patients and used them in T-cell suppression assay. LOX-1+ PMN suppressed T-cell proliferation, whereas LOX-1− PMN did not (Fig. 3D). We asked whether the LOX-1 antibody used for isolation of PMN-MDSC could directly affect the functional activity of PMN. PMN isolated from cancer patients were cultured with T cells in the presence of LOX-1 antibody or IgG isotype control. LOX-1 antibody did not make PMN to acquire immune suppressive function (Fig. S6). We asked whether LOX-1 neutralizing antibody (R&D Systems) could block suppressive activity of LOX-1+ PMN. LOX-1+ and LOX-1− PMN were isolated from two patients with NSCLC and added to MLR in the presence LOX-1 antibody at concentration (10 µg/ml) that block more than 90% of LOX-1 binding or the same amount of mouse IgG. In both experiments LOX-1 antibody did not abrogate suppressive activity of LOX-1+ PMN. LOX-1− PMN had no suppressive activity in any experiments. (Fig. 3E).
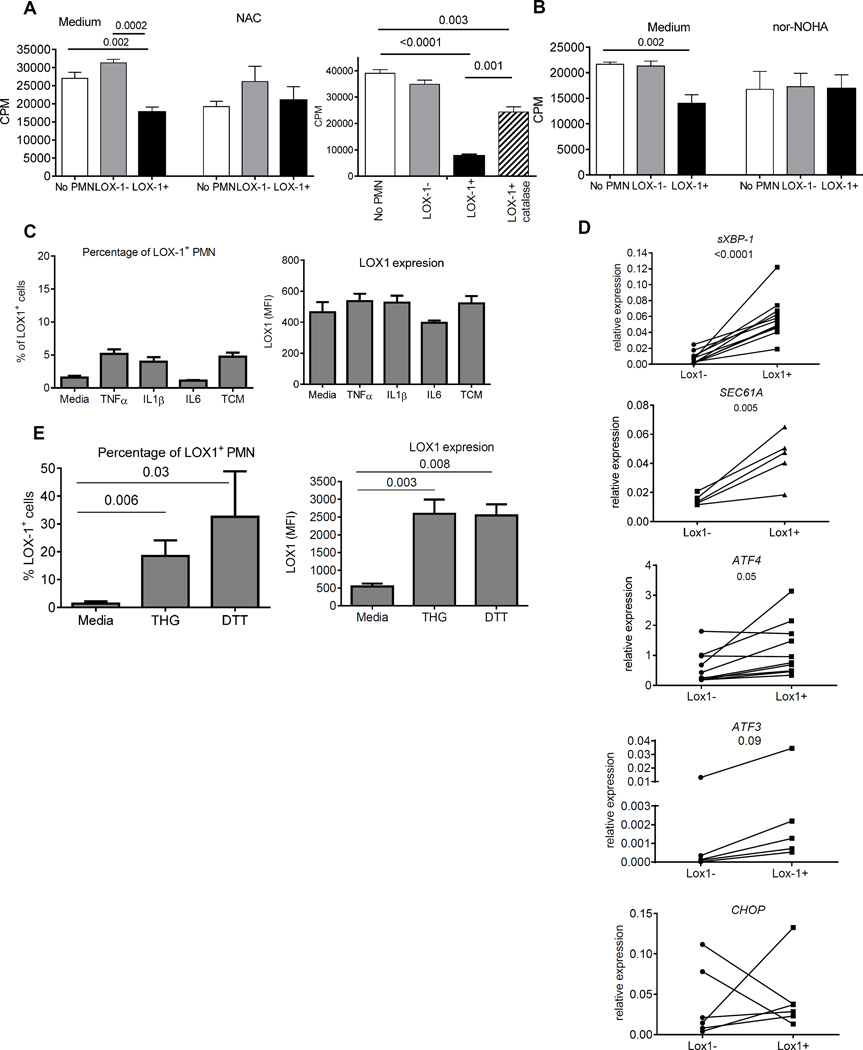
We then evaluated possible mechanisms responsible for LOX-1+ PMN-MDSC suppression. We tested several common mechanisms implicated in PMN-MDSC function. LOX-1+ PMN-MDSC had significantly higher production of reactive oxygen species (ROS) than LOX-1− PMN (Fig. 3F). Whole genome array showed that LOX-1+ PMN-MDSC had higher expression of ARG1, the gene directly associated with PMN-MDSC function, than LOX-1− PMN. These differences were not statistically significant (FDR=7%). However, direct evaluation of ARG1 expression by qPCR revealed significantly higher expression of this gene in LOX-1+ PMN-MDSC than in LOX-1− PMN (Fig. 3G). Expression of NOS2 in PMN was much lower than that of ARG1. However, it was still significantly higher in LOX-1+ PMN-MDSC than in LOX-1− PMN (Fig. 3G). We tested the contribution of ROS and ARG1 to immune suppression mediated by LOX-1+ PMN-MDSC. Both, ROS scavenger N-acetylcysteine (NAC) and catalase significantly decreased suppressive activity of LOX-1+ PMN (Fig. 4A). Inhibitor of Arg1 Nor-NOHA abrogated suppressive activity of these cells (Fig. 4B). Thus, taken together these data demonstrate that LOX-1+ PMN indeed represent a population of PMN- MDSC. Therefore, for clarity in this study we further refer to these cells as LOX-1+ PMN-MDSC.
Figure 4. Mechanism regulating LOX-1 expression on PMN-MDSC.
A.B. Effect of 1 µM of N-acetyl L-cysteine (NAC) (left panel), 1000 U/ml of catalase (right panel) (A) and 20 µM Nor-NOHA (B) on immune suppressive activity of LOX-1+ PMN-MDSC. Allogeneic MLR was used in all experiments. Cell proliferation was measured in triplicates by 3H-thymidine incorporation. 1:2 PMN : T cell ratio was used in all experiments. Three experiments with similar results were performed. P values in t-test are shown. C. Percentage of LOX-1+ PMN and expression of LOX-1 in PMN isolated from 4 healthy donors and treated with indicated cytokines. Range of concentrations based on reported data were tested and only one for each cytokine is shown. Conditioned medium from PCI30 tumor cells (TCM) was used at 20% v/v concentration. Mean and SD are shown. D. Expression of genes involved in ER stress response in PMN from 8 patients with HNC and NSCLC. P values in t-test (for ATF3 in Munn-Whitney test) are shown. E. Percentage of LOX-1+ PMN and expression of LOX-1 in PMN isolated from 4 healthy donors and treated with 1µM THG and 1 mM DTT. Mean and SD are shown. P values calculated in t-test.
We investigated the possible role of LOX-1 as marker of mouse PMN-MDSC. Similar to human PMN, CD11b+Ly6CloLy6G+ mouse PMN had very low expression of LOX-1. However, in contrast to human PMN-MDSC, spleen, BM, or tumor PMN-MDSC from mice bearing EL-4 lymphoma or Lewis Lung Carcinoma (LLC) did not up-regulate LOX-1 expression (Fig. S7). To evaluate the possible role of LOX-1 in PMN-MDSC function we used bone marrow (BM) cells from LOX-1 knockout olr1−/) mice (24). Lethally irradiated wild-type recipients were reconstituted with congenic bone marrow cells isolated from wild-type or olr1−/− mice. Ten weeks after reconstitution donor’s cells represented more than 95% of all myeloid cells. LLC tumor was implanted s.c. and mice evaluated 3 weeks later. No differences in the presence of PMN-MDSC in spleens or tumors were observed between mice reconstituted with WT and LOX-1 KO BM (Fig. S8A). WT and olr1−/− PMN-MDSC suppressed T-cell proliferation equally well (Fig. S8B). Gene expression profile demonstrated no differences between WT and olr1−/− PMN-MDSC. WT PMN-MDSC had the same undetectable level of olr1 expression as olr1−/− PMN-MDSC. Thus, in contrast to humans, mouse LOX-1 is not associated with PMN-MDSC.
ER stress regulates LOX-1 expression in PMN-MDSC
What could induce LOX-1 up-regulation in PMN-MDSC? Based on the fact that in endothelial cells LOX-1 can be induced by pro-inflammatory cytokines (25), we tested the effect of several cytokines as well as tumor-cell conditioned medium (TCM) on LOX-1 expression in PMN isolated from healthy donors. None of the tested pro-inflammatory cytokines (IL-1β, TNFα, IL-6) or TCM induced upregulation of LOX-1 in PMN after 24 hr culture (GM-CSF was added to protect PMN viability) (Fig. 4C).
Our previous observations (16) and data obtained in this study demonstrated that PMN-MDSC in cancer patients displayed signs of ER stress response. LOX-1+ and LOX-1− PMN were isolated from PB of cancer patients and expression of genes associated with ER-stress were evaluated. LOX-1+PMN-MDSC had significantly higher expression of sXBP1 and its target gene SEC61a than LOX-1− PMN. Expression of ATF4 and its target gene ATF3 was also higher in LOX-1+ PMN-MDSC. No changes in the expression of CHOP were observed (Fig. 4D). To test the effect of ER-stress on expression of LOX-1, PMN from healthy donors were treated with ER stress inducers: thapsigargin (THG) or dithiothreitol (DTT) overnight in the presence of GM-CSF. At selected doses (THG -1 µM, DTT – 1mM) cell viability remained above 95%. Both, THG and DTT caused dramatic up-regulation of LOX-1 expression in PMN (Fig. 4E).
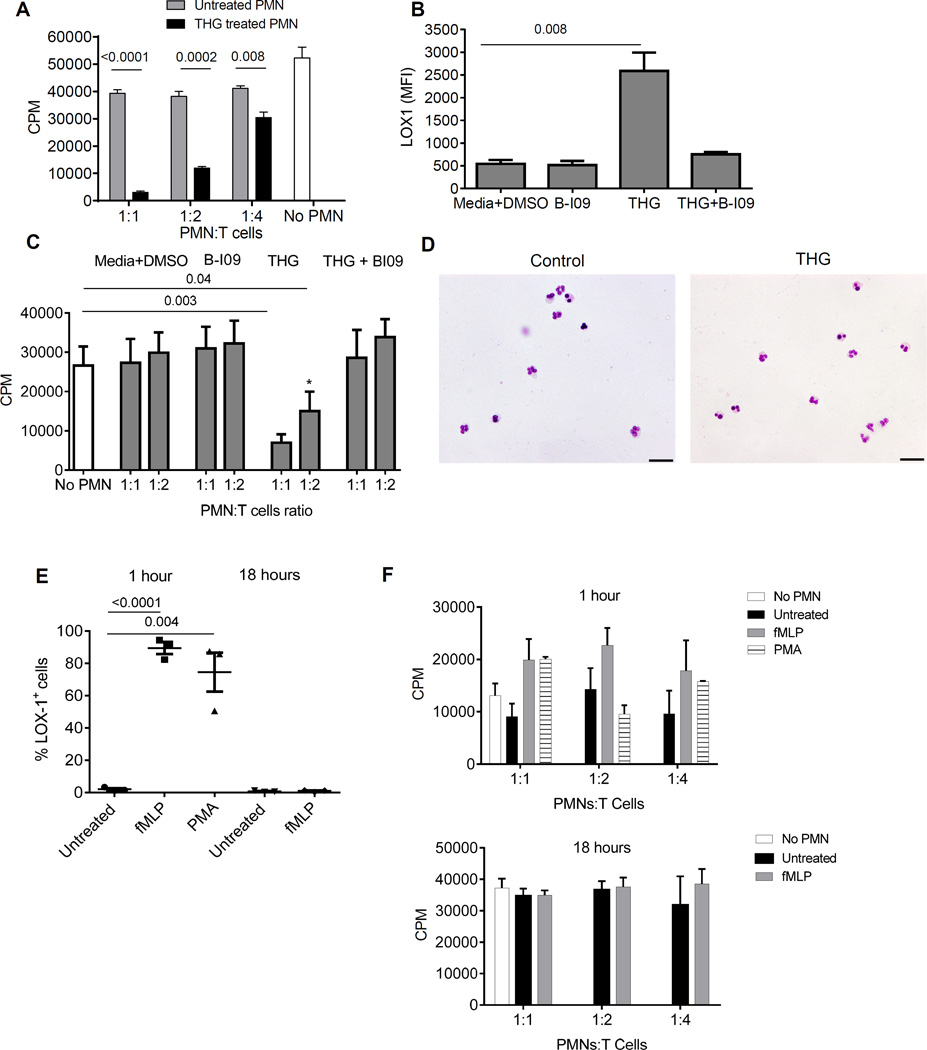
Overnight THG treatment of PMN caused acquisition of potent immune suppressive activity by the PMN (Fig. 5A). Since LOX-1+ PMN-MDSC have increased expression preferentially of one of ER stress sensors – sXBP1, we verified the role of ER stress using recently developed selective inhibitor of sXBP1 – B-I09 (26). In the presence of B-I09 THG failed to induce up-regulation of LOX-1 (Fig. 5B) and immune suppressive activity of PMN (Fig. 5C). THG treatment did not affect typical polymorphonuclear morphology of these cells (Fig. 5D). Thus, induction of ER stress in control neutrophils converted these cells to immune suppressive PMN-MDSC, which was associated with up-regulation of LOX-1 expression.
Figure 5. ER stress induce LOX-1 expression and suppressive activity in PMN.
A. ER stress inducer THG converted PMN to PMN-MDSC. PMN isolated from healthy donors were treated overnight with 1µM THG, extensively washed and then used in CD3/CD28 induced T-cell proliferaiton. T cell proliferation was measured in triplicates by 3H-thymidine uptake. Three experiments with similar results were performed. P values in t-test are shown, n=3. B,C. sXBP1 inhibitor B-IO9 abrogated THG inducible up-regulation of LOX-1 and T cells suppression in PMN from healthy donors. PMN were incubated together with 20 µM B-IO9 and THG overnight followed by evaluation of LOX-1 expression (B) or suppression activity (C). PMN from three healthy donors were used in these experiments. P values between treated and untreated PMN (t-test, n=3). D. Morphology of heathy donor’s PMN after 18 hr incubation with 10 ng/ml GM-CSF and THG. Giemsa stain. Scale bar = 20 µm. E. Healthy donor’s PMN were cultured for 1 hr or 18 hrs with 10 nM fMLP or 20 nM PMA in the presence of 10 ng/ml GM-CSF. The proportion of LOX-1+ cells was evaluated. P values were calculated in t-test (n=3). Analysis of PMA effect after 18 hrs was not performed because of undetectable number of viable cells. F. PMN treated with fMLP or PMA as described in Fig. 5E were extensively washed after 1 hr or 18 hr (fMLP only) incubation and tested in T-cell suppression assay. Each experiment was performed in triplicates. Three donors were tested.
Up-regulation of intracellular Ca2+ and reactive oxygen (ROS) are among of the consequences of THG treatment. We asked whether activation of neutrophils with agents that cause up-regulation of intracellular Ca2+ and ROS may also result to similar up-regulation of LOX-1 and suppressive activity as THG. We used two compounds: N-Formylmethionyl-leucyl-phenylalanine (fMLP) and phorbol 12-myristate 13-acetate (PMA). Both, fMLP and PMA caused massive up-regulation of LOX-1 expression on control PMN within 60 min after the start of the treatment (Fig. 5E). However, in contrast to THG, expression of LOX-1 in fMLP treated PMN returned to control (untreated cells) level after overnight culture despite the presence of fMLP in the culture (Fig. 5E). PMN did not survive overnight treatment with PMA. PMN treated for 60 min with fMLP or PMA were washed and used in T-cell suppression assay. No suppressive activity was observed (Fig. 5F). In another set of experiments PMN were cultured with fMLP for 18 hr before used in suppressive assay. However, this treatment also did not result in the development of suppressive activity by PMN (Fig. 5F). These data indicate that transient up-regulation of LOX-1 as a result of PMN activation was not associated with immune suppressive activity.
LOX-1 defines PMN-MDSC in tumor tissues
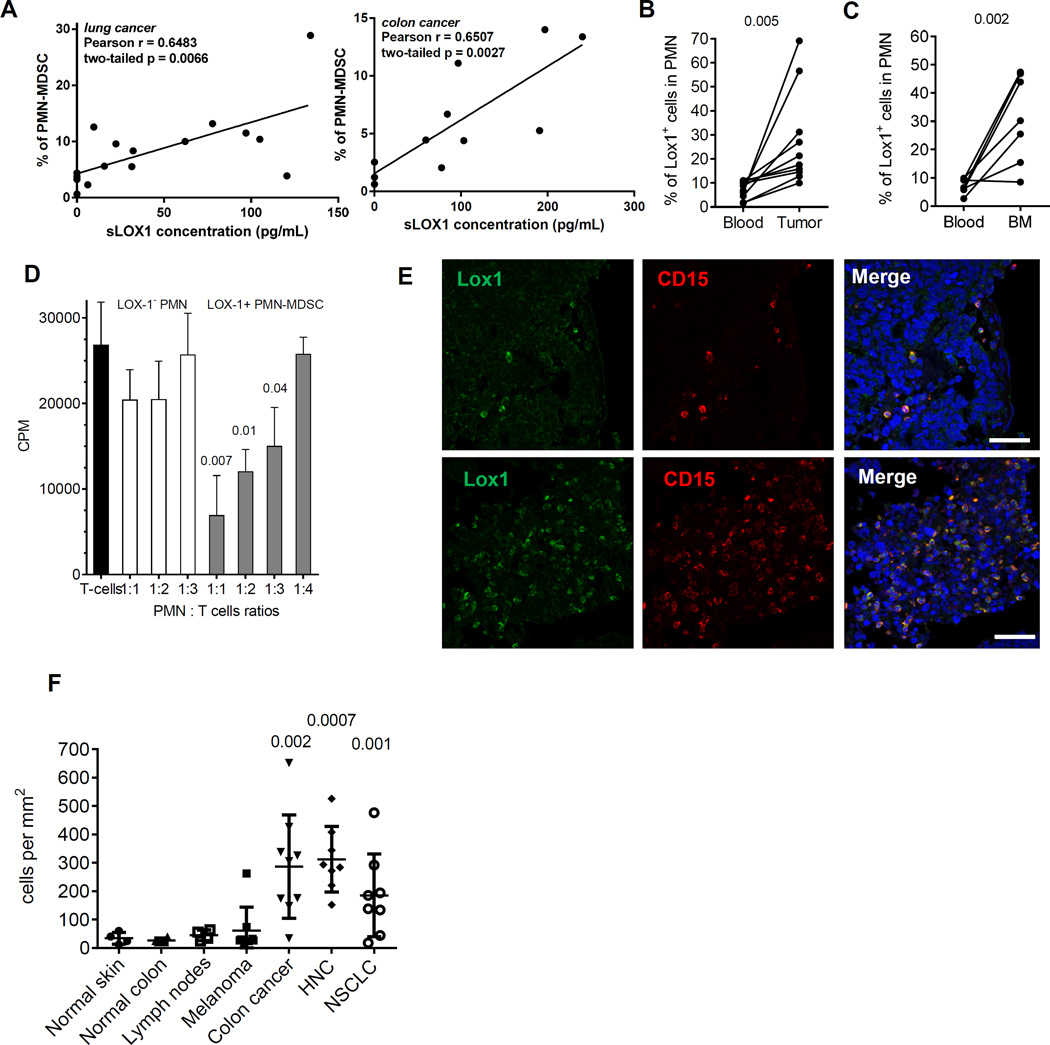
It is known that LOX-1 is shed from the surface of the cells and can be detected in plasma (27). We evaluated correlation between the presence of PMN-MDSC in cancer patients and soluble LOX-1 in plasma. In NSCLC and CC patients, the proportion of PMN-MDSC was associated with soluble LOX-1 (Fig. 6A) suggesting that these cells may be an important source of LOX-1 in plasma of cancer patients.
Figure 6. LOX-1+ PMN-MDSC in tumor tissues.
A. Correlation of soluble LOX1 in plasma of NSCLC and HNC with the presence of PMN-MDSC in PBMC fraction of PB. B. Presence of LOX-1+ PMN in PB and tumor tissues of 10 patients with HNC and NSCLC. P values are calculated in t-test. C. Presence of LOX-1+ PMN in PB and BM of 7 patients with MM. P values are calculated in t-test. D. Suppressive activity of LOX-1+ PMN in BM of patient with MM tested in allogeneic MLR. P (in t-test) from values without the presence of PMN are shown. Three patients were tested with the same results. E. Typical staining of tumor tissues. Scale bar = 100µm. F. LOX-1+ PMN in tissues from 4 normal skin samples, 4 samples of tumor-free lymph nodes, 4 samples of normal colon, as well as tumor tissues from 8 patients with melanoma, 8 patients with HNC patients, 8 patients with NSCLC, and 9 patients with CC. Mean and SD shown. P values calculated in t-test show difference between tumor samples and samples from control tissues.
There is now sufficient evidence demonstrating that tumor MDSC are more suppressive than cells in PB (reviewed in(28)). We asked whether the population of PMN-MDSC is more prevalent among all PMN in tumors than in PB. The proportion of LOX-1+ cells in CD15+ PMN isolated from tumors of patients with HNC and NSCLC was > 3-fold higher than in CD15+ PMN from PB of the same patients (p<0.001) (Fig. 6B). Cells in blood and tumor tissues were subjected to the same digestion protocol. However, to exclude possible effect of tissue digestions and isolation on LOX-1 expression, we also evaluated patients with multiple myeloma (MM) where the tumor is located in bone marrow (BM). We previously have shown substantial increase of PMN-MDSC in both BM and PB of MM patients (29). Similar to solid tumors, the proportion of LOX-1+ PMN-MDSC in BM was 3–4-fold higher than in PB of the same patients (p=0.004) (Fig. 6C). LOX-1+ PMN-MDSC isolated from BM of patients with MM had profound immune suppressive activity, whereas LOX-1− PMN did not suppress T cells (Fig. 6D) supporting the conclusion that LOX-1+ PMN represent PMN-MDSC at the tumor site.
To evaluate the presence of LOX-1+ PMN-MDSC in tumor tissues, we have developed a method of immune fluorescent staining of paraffin-embedded tissues with combination of LOX-1 and CD15 antibody (Fig. 6E). Control tissues from normal skin, colon and lymph nodes had similar low numbers of LOX-1+CD15+ PMN-MDSC (Fig. 6F). No statistical differences were found in the presence of these cells in melanoma samples, which is consistent with findings that M-MDSC but not PMN-MDSC are the predominant population of MDSC in these patients (2). The number of LOX-1+CD15+ PMN-MDSC in colon carcinoma increased more than 8-fold, in HNC more than 10-fold, and in NSCLC almost 8-fold (Fig. 6F). Thus, LOX-1 expression defines the population of PMN-MDSC in tumor tissues.
OLR-1 expression and the presence of LOX-1+PMN-MDSC are associated with clinical parameters
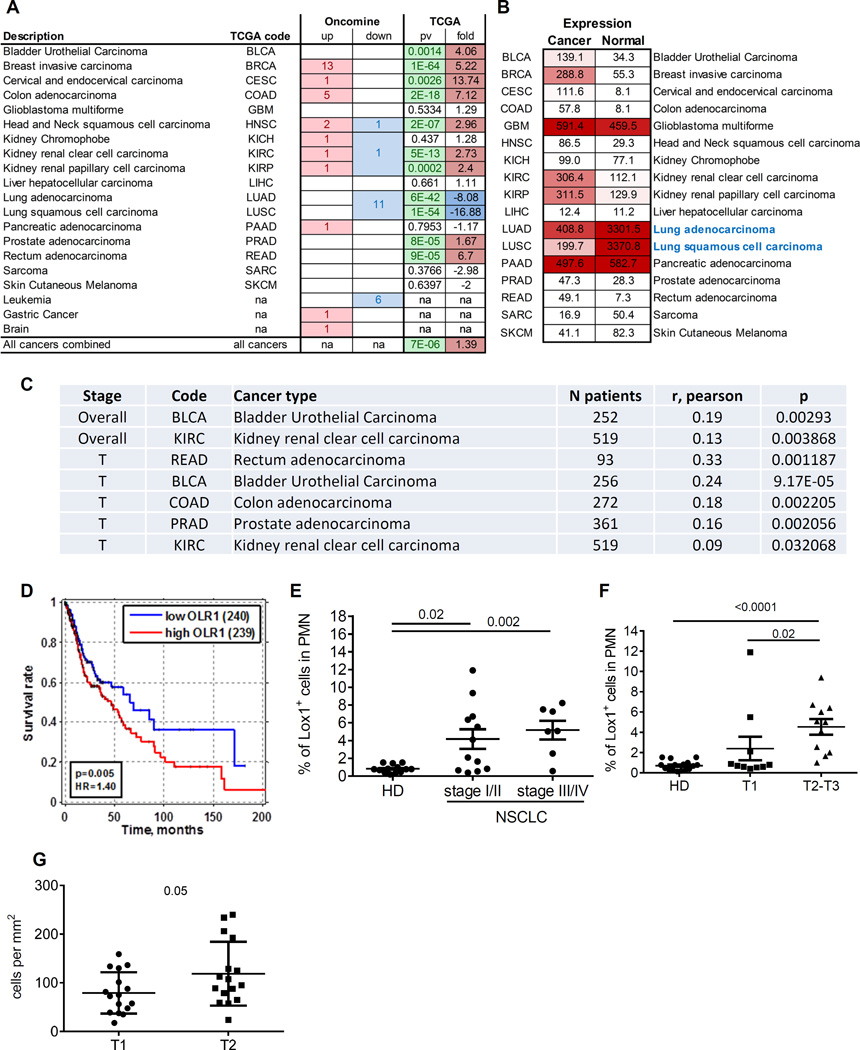
Using ONCOMINE and TCGA databases we evaluated the association of OLR1 expression in tumor tissues with clinical parameters in different types of cancer. Significant upregulation of OLR1 was observed in many types of cancer (Fig. 7A). The notable exception was lung cancer where normal lung tissues showed dramatically higher expression of OLR1 than other normal tissues (Fig. 7B), apparently due to cells with high expression of OLR1 (possibly lung epithelium). OLR1 expression positively correlated with clinical stage in patients with bladder cancer and clear cell kidney cancer. The positive correlation with tumor size was found in patients with prostate adenocarcinoma, colon adenocarcinoma, bladder cancer, and rectum adenocarcinoma (Fig. 7C). Higher expression of OLR1 was associated with worse survival in patients with HNC (Fig. 7D). Although these results are suggestive, their interpretation as reflecting PMN-MDSC presence has some limitation due to the fact that OLR1 can be expressed on different cells in the tumor microenvironment. We focused on evaluation of LOX-1+ PMN-MDSC in tumor tissues and PB.
Figure 7. Clinical association of OLR1 expression and LOX-1+ PMN-MDSC accumulation in cancer patients.
A. Number of independent data sets from Oncomine database that showed OLR1 upregulated (up) or downregulated (down) in Cancer vs Normal tissues. P-value and fold change for Cancer/Normal comparison from TCGA database. na=data not available; B. Average expression values (FPKM values) for cancer and normal tissues for different cancers indicates high baseline expression level of OLR1 in normal lung tissues. C. Association of OLR1 expression with clinical stage (overall) or tumor size (T) in patients with different types of cancer. D. Kaplan-Meier survival curves for HNC squamous cell carcinoma patients survival stratified by median OLR1 expression indicates decreased survival for subjects with high OLR1 expression. p=cox-regression p value, HR=hazard ratio, E. Proportion of LOX-1+ PMN-MDSC in PB of 12 patients with stage I-II; 7 patients with stage III-IV of NSCLC, and 16 healthy donors. P values calculated in Mann-Whitney test. F. Proportion of LOX-1+ PMN-MDSC in PB of NSCLC patients segregated based on tumor size. P values calculated in Mann-Whitney test. F. Amount of LOX-1+ CD15+ PMN-MDSC in tumor tissues from NSCLC patients segregated based on tumor size. P values calculated in t-test.
In patients with NSCLC we evaluated the possible link between stage of the disease and the proportion of LOX-1+ PMN-MDSC in PB. Patients with both early (I/II) and late (III/IV) stages of NSCLC had significantly higher proportion of LOX-1+ PMN-MDSC than in healthy donors. There was no statistical significant difference between these two groups of patients (Fig. 7E). However, whereas 85.7% of all patients with late stages of NSCLC had an increase in LOX-1+ PMN-MDSC population, only 50% of patients with early stages showed elevated level of these cells (Fig. 7E). Significant association of the presence of LOX-1+ PMN-MDSC in PB of cancer patients was found with size of the tumors. Only patients with large tumors (T2-T3) had significantly higher proportion of LOX-1+ PMN-MDSC than healthy donors, whereas patients with small tumors (T1) had similar very low level of LOX-1+ PMN-MDSC as healthy donors. Patients with large tumors had significantly more LOX-1+ PMN-MDSC than patients with small tumors (Fig. 7F). Using a NSCLC adenocarcinoma tissue array, we evaluated the association between the presence of LOX-1+ PMN-MDSC in tumor tissues and tumor size. Similar to the data obtained in PB no significant association was found with stage of the disease. However, the number of LOX-1+ PMN-MDSC was higher in larger (T2 vs. T1) tumors (Fig. 7G).
Discussion
The goal of this study was to address the issue of heterogeneity of human PMN-MDSC, which limits the progress of our understanding of the biology of these cells. Currently, only partially enriched population of PMN-MDSC isolated on Ficoll gradient can be evaluated. This population contains not only suppressive PMN-MDSC but also non-suppressive activated PMN. Separation of PMN-MDSC and PMN in tumor tissues is not possible, which further complicates the analysis of these cells. Here, we report that PMN-MDSC have a unique gene expression profile, which is substantially different from that of PMN from the same patients and from healthy donors. This directly supports the notion that PMN-MDSC represent a distinct functional state of pathological activation of neutrophils in cancer (30, 31) and is consistent with the analysis of gene expression performed in mice, which demonstrated differences in transcriptome between granulocytes isolated from naïve mice and PMN-MDSC from tumor-bearing mice (32). Up-regulation of genes associated with ER stress response was prominent feature of PMN-MDSC. The ER stress response is developed to protect cells from various stress conditions including hypoxia, nutrient deprivation, low pH and includes three major signaling cascades initiated by three protein sensors: PERK(protein kinase RNA (PKR)-like ER kinase), IRE-1 (inositol-requiring enzyme 1) and ATF6 (activating transcription factor 6) (33). PERK phosphorylates eukaryotic protein synthesis initiation factor 2 alpha (eIF2α), which controls the initiation of mRNA translation and inhibits the flux of synthesized proteins. eIF2α induces the expression of ATF4 and its downstream targets, including the pro-apoptotic transcription factor CHOP. IRE1 cleaves the mRNA encoding for the transcription factor X-box-binding protein-1 (XBP1) (34). Spliced XBP1 (sXBP1) mRNA is then ligated by a RNA ligase and translated to produce sXBP1 transcription factor that regulates many target genes including SEC61a (35).
ER stress response was previously shown to be transmitted to dendritic cells and macrophages from tumor cells and was associated with up-regulation of arginase-1 in macrophages (36–38). Constitutive activation of XBP1 in tumor-associated dendritic cells promoted ovarian cancer progression by blunting anti-tumor immunity (39). We have recently found activation of the ER stress response in MDSC by demonstrating that MDSC isolated from tumor-bearing mice or cancer patients overexpress sXBP1 and CHOP and displayed an enlarged endoplasmic reticulum, one of the hallmarks of the ER stress (16). Another study implicated CHOP in the suppressive activity of MDSC in tumor site (40). Consistent with these observations, administration of an ER stress inducer to tumor-bearing mice increased the accumulation of MDSC and their suppressive activity (41).
In this study, we have discovered that expression of LOX-1 receptor was associated with PMN-MDSC. LOX-1 is a class E scavenger receptor expressed on macrophages and chondrocytes, as well as endothelial and smooth muscle cells (42). Expression of this receptor on neutrophils has not been previously described. We have found that neutrophils from healthy donors and cancer patients have practically undetectable expression of LOX-1. Our data suggested that LOX-1 expression is not just associated but actually defines the population of PMN-MDSC in cancer patients. This conclusion is supported by several lines of evidence: 1) LOX-1+ PMN had gene expression profile similar to that of enriched PMN-MDSC isolated using gradient centrifugation, while LOX-1− PMN had profile similar to neutrophils; 2) LOX-1+ but not LOX-1− PMN potently suppressed T-cell responses; 3) LOX-1+ PMN had significantly higher expression of ARG1 and production of ROS, typical characteristics of PMN-MDSC. We also found that in tumor tissues, only LOX-1+ PMN were immune suppressive and could be considered as PMN-MDSC. This opens an opportunity for a direct identification of PMN-MDSC in PB and tumor tissues.
These observations, although unexpected, fit overall concept of the critical role of ER stress response in MDSC biology. It was recently demonstrated that in human endothelial cells oxLDL induced expression of LOX-1 through activation of ER stress sensors IRE1 and PERK (43). In contrast, ER stress induced by tunicamycin in hepatic L02 cells caused down-regulation of LOX-1. Knock down of IRE1 or XBP-1 restored LOX-1 expression in these cells (44). In our experiment we found that induction of ER stress in neutrophils caused dramatic up-regulation of LOX-1. More importantly, this was associated with acquisition of immune suppressive activity by these cells, indicating that induction of ER stress response could convert neutrophils to PMN-MDSC. MDSC accumulate as the result of convergences of two only partially overlapping groups of signals: the signals that promote myelopoiesis (primarily via growth factors and cytokines produced by tumors) and the signals that induce suppressive activity in these cells (believed to be associated with pro-inflammatory cytokines) (45). ER stress emerged as important factor in the second group. We did not find morphological evidence that PMN de-differentiate during the culture with THG. However, in our experiments we targeted mature neutrophils. It is possible that ER stress may have much broader effect on precursor or progenitor cells.
In our study we did not investigate whether signaling through LOX-1 is responsible for acquisition of immune suppressive activity by neutrophils. However, it is likely that it contributes to this process. Engagement of LOX-1 can lead to induction of oxidative stress, apoptosis, and activation of the NF-κB pathway (18). These pathways are known to be important for PMN-MDSC function. The ER stress response pathway has been shown to regulate inflammation by activating the NF-κB pathway (35, 46, 47). LOX-1 up-regulation has been observed during cellular transformation into cancer cell and can have a pro-oncogenic effect by activating the NF-κB pathway, by increasing DNA damage through increase ROS production and by promoting angiogenesis and cell dissemination (48, 49). It is possible that LOX-1 signaling may drive pathological activation of PMN towards PMN-MDSC. Cell surface LOX-1 expression can be elevated by multiple stimuli including reactive oxygen species (ROS), inflammatory cytokines (TNF-α, TGF-β) as well as oxLDL (50). These factors are produced in cancer and it is possible that they can affect differentiation of granulocytes from precursors leading to acquisition of LOX-1 expression. The question is why only 4–7% of neutrophils acquire LOX-1 expression and suppressive activity to become PMN-MDSC. This is unresolved issue at this moment mainly because the nature of factors that convert neutrophils to PMN-MDSC remains unclear. Based on mouse studies it is possible that PMN-MDSC may have different precursors than most of neutrophils and those precursors are more sensitive to ER stress inducible factors.
Our study has obvious limitations derived from the nature of human studies and the nature of neutrophils as short lived differentiated cells. This restrained our ability to provide deeper insight to mechanism of the observed phenomenon of LOX-1 up-regulation. More information can be generated from the experiments studying progenitors and precursors of neutrophils. For example, we used neutralizing LOX-1 antibody in an attempt to block signaling through this receptor. This approach may not be sufficient due to the fact that PMN-MDSC in peripheral blood may not be amendable for such regulation. However, LOX-1 can be involved in regulation of PMN precursors and antibody may have effect. PMN progenitors could be manipulated genetically, which would allow for more precise analysis of this pathway.
We could not find an association between LOX-1 expression and PMN-MDSC in two mouse models. These results are rather unexpected, since these models (EL and LLC) are associated with inflammation and expansion of PMN-MDSC. It is possible that such association can be found in other tumor model. However, it is more likely expression of LOX-1 in mice and humans is regulated differently. The mechanism remains unclear.
Combination of neutrophil markers with LOX-1 potentially allow for detection of PMN-MDSC in tissues. We observed dramatic increase in the number of PMN-MDSC in tumors of patients with HNC, colon cancer, and NSCLC. Interestingly, no such increase was observed in melanoma patients. It is possible that relatively low accumulation of PMN-MDSC in melanoma patients may contribute to this observation. Our data demonstrated that patients have variable amount of LOX-1+ PMN-MDSC, which at least in patients with NSCLC was associated with size of the tumors. Further prospective studies are needed to determine whether the presence of these cells in tumor tissues can predict clinical outcome. Expression of LOX-1 on PMN-MDSC opens an opportunity for selective targeting of these cells, since antibody targeting LOX-1 have been already tested in cardiovascular diseases in mice (51, 52).
Material and Methods
Study Design
The purpose of this study was to better characterize human PMN-MDSC and to identify specific marker that allow distinguishing these cells from neutrophils. We performed whole gene expression array using triplicate of samples. LOX-1 expression was measured by flow cytometry in variable number of replicates indicated in figure legends. Experiments were performed in a controlled and non-blinded manner. No randomization was performed because of the observational nature of the study. Sample size of patient cohorts was determined based on the results of initial experiments with LOX-1 expression and calculations of expected differences between mean and expected standard deviation.
Human Samples
Samples of peripheral blood and tumor tissues were collected from patients at Helen F. Graham Cancer Center and University of Pennsylvania. The study was approved by Institutional Review Boards of the Christiana Care Health System at the Helen F. Graham Cancer Center, University of Pennsylvania and The Wistar Institute. All patients signed approved consent forms. Peripheral blood was collected from:
-
1).
26 patients with different stages of non-small cell lung cancer (NSCLC) – 12 females, 18 males, age 59–79 (median 69). 13 patients had squamous cell carcinoma, 13 patients - adenocarcinoma;
-
2).
21 patients with head and neck cancer (HNC) – 8 females, 13 males, age 32–82 (median 65). 19 patients had squamous cell carcinoma and 2 patients - adenocarcinoma;
-
3).
38 patients with colon cancer (CC) (adenocarcinoma) – 20 females, 18 males, age 28–88 (median 58).
-
4).
6 patients with multiple myeloma (MM) – 1 female, 5 males, age 58–81 (median 75).
All patients were either previously untreated or received treatment (chemotherapy or radiation therapy) at least 6 months before collection of blood. In some patients tumor tissues were collected during the surgery. In addition 6 patients with eosinophilic colitis, 3 patients with ulcerative colitis, and 8 patients with Crohn’s disease were evaluated.
Peripheral samples of blood from 18 healthy volunteers 12 females, 6 males age 35–56 (median 42 years) were used as control.
All samples were evaluated within 3 hours after collection.
Lung cancer tumor microarrays were produced from formalin-fixed paraffin embedded tissue. Each block was examined by a pathologist and three cores were obtained from tumor-containing areas and three blocks from non-tumor involved lung regions. Samples were obtained from 32 patients with adenocarcinoma. Clinical data obtained included tumor histology, size, stage, and time to recurrence (all patients followed for 5 years).
De-identified samples from normal colonic biopsies colon were obtained from St. Mark’s Hospital, Harrow, UK. Samples were taken from patients after obtaining informed consent and with the approval of the Outer West London Research Ethics Committee (UK). Paraffin-embedded tissues blocks of samples of normal skin, lymph nodes, and melanoma were retrieved using an approved IRB protocol for de-identifed archived skin biopsies through the Department of Dermatology NIH SDRC Tissue Acquisition Core (P30-AR057217), Perelman School of Medicine, University of Pennsylvania, Philadelphia, USA.
Cell isolation and culture
PMN-MDSC and PMN were isolated by centrifugation over a double density gradient Histopaque (Sigma) (1.077 to collect PBMC and 1.119 to collect PMN) followed by labeling with CD15-PE mAb (BD Biosciences) and then separated using anti-PE beads and MACS column (Miltenyi).
Tissues were first digested with human tumor dissociation kit (Miltenyi) and then red blood cell lysed. Cells were then culture in RPMI (Biosource International) supplemented with 10% FBS, 5 mM glutamine, 25 mM HEPES, 50µM β-mercaptoethanol and 1% antibiotics (Invitrogen). In some experiments, recombinant GM-CSF (Peprotech) was added to the culture media at a concentration of 10 ng/mL
Isolation of Lox1+ PMN from peripheral blood and suppression assay
Whole blood was enriched for PMNs using MACSxpress® Neutrophil Isolation Kit (Miltenyi) following the protocol provided by the manufacturer. Cells were then labeled with anti-Lox1-PE mAb (Biolegend) and then separated using anti-PE beads and MACS column (Miltenyi). For the three-way allogenic MLR suppression assay, T lymphocytes from one healthy donor were purified using the Human CD3+ T Cell Enrichment Column Kit (R&D Systems) and used as responder cells. Dendritic cells were generated from adherent monocytes from another healthy donor in the presence of 25 ng/mL GM-CSF and 25 ng/mL IL-4 (Peprotech) for 6 days and used as stimulator cells. Responder and stimulator cells were then mixed at a 10:1 ratio followed by the addition of Lox1+ or Lox1− PMNs. T lymphocyte proliferation was assessed after 5 days of culture by thymidine incorporation.
Concurrently, T-lymphocytes were isolated from the PBMC of the same patient as LOX-1+ PMN using human CD3+ T Cell Enrichment Column Kit. PMNs were plated at different ratios with 105 T lymphocytes in a 96-well plate coated with 10 µg/ml anti-CD3 (clone UCHT1; BD Biosciences) followed by the addition of 1 µg/ml of soluble anti-CD28 (clone CD28.2; BD Biosciences). T lymphocyte proliferation was assessed after 3 days of culture by thymidine incorporation. In some experiments, 1 µM N-acetyl cysteine (NAC; Sigma) or 20 µM of Nω- hydroxy-nor-arginine (nor-NOHA; Cayman Chemical) or 10 µg/ml of monoclonal human LOX-1 antibody (R&D Systems) was added to the culture media to block ROS or agrinase I activity, respectively. T lymphocyte proliferation was assessed after 5 days of culture by thymidine incorporation.
In vitro PMN Lox-1 induction
PMNs from healthy donors were isolated on a Histopaque gradient. 5×105 cells/ml were cultured for 12 hrs with 10 ng/ml of GM-CSF in the presence of dithiothreitol (DTT) (0.5, 1, 2 mM; Sigma), tunicamycin (0.5, 1, and 2 µg/ml; Sigma-Aldrich), or thapsigargin (0.5, 1, and 2 µM; Sigma). In some instances, 20 µM of the XBP-1 inhibitor B-IO9 was added 3 hours prior to culture. Cells were then stained for flow cytometry or used for functional assays as described above. For fMLP and PMA stimulation, PMN were isolated from healthy donors and cultured for 1 or 18 hours with either 10 nM fMLP (Sigma Alrich) or 20 nM PMA (Sigma Aldrich). Cells were then washed and LOX1 expression was measured by flow cytometry or used for functional assays as described above. For overnight cultures, 10 ng/ml of GM-CSF was added to the culture.
Flow cytometry
Flow cytometry data were acquired using a BD LSR II flow cytometer and analyzed using FlowJo software (Tree Star).
Immunofluorescent microscopy
Following deparaffinization and rehydration, heat induced antigen retrieval was performed using Tris-EDTA buffer pH 9. Followed by blocking with 5% BSA, tissues were stained with Lox1 antibody (Abcam; Cat no ab126538) and CD15 antibody (BD biosciences; Cat no 555400) at 1: 200 dilution in 5% BSA each for 1 hour at room temperature. The following secondary antibodies were used Alexa Fluor anti-rabbit A647 (1: 200 dilution in 5% BSA, Life technologies) for Lox1 and anti-mouse A514 (1: 400 dilution in 5% BSA, Life technologies) for CD15 staining. CD15 staining was pseudo colored red and Lox1 staining was pseudo colored green. Nucleus was stained using DAPI (1:5000 dilution in PBS, Life technologies). Images were obtained using Leica TCS SP5 Confocal microscope. Cell counts from 16 frames were used to calculate counts per sq. mm.
Microarray analysis
For sample preparation and hybridization, total RNA from purified cells was isolated with TRIzol reagent according to the manufacturer’s recommendations. RNA quality was assessed with a Bioanalyzer (Agilent). Only samples with RIN numbers>8 were used. Equal amount (400 ng) of total RNA was amplified as recommended by Illumina and was hybridized to the Illumina HumanHT-12 v4 human whole-genome bead arrays.
For data preprocessing, Illumina GenomeStudio software was used to export expression values and calculated detection p-values for each probe of each sample. Signal-intensity data were log2 transformed and quantile-normalized. Only probes with a significant detection p-value (p < 0.05) in at least one of sample were considered. The data was submitted to GEO and is accessible using accession number GSE79404.
Differential expression for probes was tested using SAM ('significance analysis of microarrays') method (53). Multiple groups were compared using “Multiclass” option and matched patient samples groups were compared using “Two sample paired” option. False discovery rate was estimated using Storey et. al procedure (54). Genes with a false-discovery rate of <5% were considered significant unless stated otherwise. Hierarchical cluster was performed using standardized Euclidean distance with average linkage. Genes that had GO annotation GO:0005886 (plasma membrane) and either GO:0004872 (receptor activity) or GO:0009897 (external side of plasma membrane) were considered as a candidate for a surface molecule marker. For expression heatmaps samples from the same patient were additionally normalized to the average between them, and samples from healthy donors were normalized to average between all patient samples. Enrichment analyses were done using QIAGEN’s Ingenuity Pathway Analysis software (IPA®, QIAGEN Redwood City, www.qiagen.com/ingenuity). Pathway results with FDR<5% and p<10−5 were considered significant. Only regulators that passed p<10−8 threshold with significantly predicted (Z>2) activation state in PMN-MDSCs were reported. For OLR1 gene expression association with cancer, Oncomine (https://www.oncomine.org) was used with “Cancer vs. Normal” gene report without any additional filters. Additionally, TCGA RNAseqV2 level 3 data (https://tcga-data.nci.nih.gov) was used and RPKM expression values were compared between cancer and normal tissues (where available) using t-test. Association with survival was done using univariate cox regression and Kaplan-Meier curves were plotted for patients split into two groups using median expression. Results with p<0.05 were considered significant.
qRTPCR
Total RNA was prepared with E.Z.N.A total RNA isolation Kit I (Omega Biotek) and cDNA was synthesized with High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative RT-PCR was performed with Power SYBR Green PCR Master Mix (Applied Biosystems). The relative amount of mRNA was estimated by the comparative threshold cycle method with gapdh as reference gene. For the analysis of gene expression the following primers were used:
sXBP-1 – 5′-CTGAGTCCGCAGCAGGTG-3′; 5′-AGTTGTCCAGAATGCCCAACA-3′
DDIT3 (CHOP) – 5’-GCACCTCCCAGAGCCCTCACTCTCC-3’; 5’- GTCTACTCCAAGCCTTCCCCCTGCG-3’,
ATF4 – 5’-TTCCTGAGCAGCGAGGTGTTG-3’; 5’- TCCAATCTGTCCCGGAGAAGG-3’,
ATF3 – 5'-TGCCTCGGAAGTGAGTGCTT-3'; 5'-GCAAAATCCTCAAACACCAGTG-3',
SEC61A – 5'-GGATGTATGGGGACCCTTCT-3'; 5'-CTCGGCCAGTGTTGACAGTA-3',
ARGI–5’-CTTGTTTCGGACTTGCTCGG-3’; 5’-CACTCTATGTATGGGGGCTTA-3’,
NOS2–5’- CAGCGGGATGACTTTCCAA-3’; 5’- AGGCAAGATTTGGACCTGCA-3’,
GADPH– 5’- GGAGTCAACGGATTTGGTCGTA-3’; 5’- GGCAACAATATCCACTTTACCAGAGT-3’.
Statistics
Statistical analysis was performed using a 2-tailed Student’s t-test or Mann-Whitney test after the analysis of distribution of variables. Significance determined at p < 0.05 with normal-based 95% CI mean ± 2SD. Analysis of gene expression was adjusted for multiple variables and false discovery rate (FDR) was estimated as described (54). All calculations were made using GraphPad Prism 5 software (GraphPad Software Inc.).
Supplementary Material
Acknowledgments
This work was supported in part by NIH grants CA084488, CA100062, and Janssen Pharmaceutical to D.I.G; NCI P01 CA 098101, American Cancer Society RP-10-033-01-CCE to AR. This work has been supported in part by the flow cytometry and bioinformatics core facilities at the Wistar Institute. The authors declare no competing interests. We thank Dr. Liu for help with statistical analysis. The data for this study have been deposited in the database GEO and are accessible using accession number GSE79404.
TC – designed and performed most of the experiments, and analyzed data; GAD – performed experiments, analyzed data, and wrote manuscript, JIY, SM, KAT, ET, AH, CL, SP –performed experiments, AVK – performed bioinformatics analysis, YN – analyzed data, wrote manuscript, AG, DTV, XX, SCK, GM, GHL, EE, SMA, MB, AR, NH, RW, GM, BN – provided clinical material and analyzed clinical correlates, XW, JLM – provided LOX-1−/− bone marrow for the experiment, DS, MAS – designed experiments and analyzed data, DIG – designed the concept of the study and experiments, analyzed data, and wrote paper.
Footnotes
Supplemental material
Eight supplemental figures and one table describing details results of microarray analysis and gating strategies by flow cytometry.
Figure S1. Gene expression differences between PMN-MDSCs and PMN cells from cancer patients and healthy donors
Figure S2. CD16 and LOX-1 staining in PMN
Figure S3. Siglec-8 staining of CD15+ PMN
Figure S4. Typical staining of healthy donors neutrophils
Figure S5. List of genes and their relative expression targeted by 93 microarray probes shown to be highly differentially expressed (FDR<5%, fold>2) for both LOX-1+/LOX-1- and PMN-MDSC/PMN comparisons
Figure S6. LOX-1 antibody does not induce immune suppressive activity of PMN
Figure S7. LOX-1 expression in mouse PMN-MDSC
Figure S8. LOX-1 expression is not associated with mouse PMN-MDSC
Table S1. Canonical pathways identified by Ingenuity Pathway Analysis (IPA) among genes significantly differentially expressed between PMN-MDSC and PMN cells.
References
- 1.Condamine T, Ramachandran I, Youn JI, Gabrilovich DI. Regulation of tumor metastasis by myeloid-derived suppressor cells. Annu Rev Med. 2015;66:97–110. doi: 10.1146/annurev-med-051013-052304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Messmer MN, Netherby CS, Banik D, Abrams SI. Tumor-induced myeloid dysfunction and its implications for cancer immunotherapy. Cancer Immunol Immunother. 2015;64:1–13. doi: 10.1007/s00262-014-1639-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finke J, Ko J, Rini B, Rayman P, Ireland J, Cohen P. MDSC as a mechanism of tumor escape from sunitinib mediated anti-angiogenic therapy. Int Immunopharmacol. 2011;11:856–861. doi: 10.1016/j.intimp.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng PH, Lee KY, Chang YL, Chan YF, Kuo LW, Lin TY, Chung FT, Kuo CS, Yu CT, Lin SM, Wang CH, Chou CL, Huang CD, Kuo HP. CD14(+)S100A9(+) monocytic myeloid-derived suppressor cells and their clinical relevance in non-small cell lung cancer. American journal of respiratory and critical care medicine. 2012;186:1025–1036. doi: 10.1164/rccm.201204-0636OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vetsika E-K, Koinis F, Gioulbasani M, Aggouraki D, Koutoulaki A, Skalidaki E, Mavroudis D, Georgoulias V, Kotsakis A. A Circulating Subpopulation of Monocytic Myeloid-Derived Suppressor Cells as an Independent Prognostic/Predictive Factor in Untreated Non-Small Lung Cancer Patients. Journal of Immunology Research. 2014;2014:659294. doi: 10.1155/2014/659294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu CY, Wang YM, Wang CL, Feng PH, Ko HW, Liu YH, Wu YC, Chu Y, Chung FT, Kuo CH, Lee KY, Lin SM, Lin HC, Wang CH, Yu CT, Kuo HP. Population alterations of L-arginase- and inducible nitric oxide synthase-expressed CD11b+/CD14(−)/CD15+/CD33+ myeloid-derived suppressor cells and CD8+ T lymphocytes in patients with advanced-stage non-small cell lung cancer. J Cancer Res Clin Oncol. 2010;136:35–45. doi: 10.1007/s00432-009-0634-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diaz-Montero CM, Salem ML, Nishimura MI, Garrett-Mayer E, Cole DJ, Montero AJ. Increased circulating myeloid-derived suppressor cells correlate with clinical cancer stage, metastatic tumor burden, and doxorubicin-cyclophosphamide chemotherapy. Cancer Immunol Immunother. 2009;58:49–59. doi: 10.1007/s00262-008-0523-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer C, Cagnon L, Costa-Nunes CM, Baumgaertner P, Montandon N, Leyvraz L, Michielin O, Romano E, Speiser DE. Frequencies of circulating MDSC correlate with clinical outcome of melanoma patients treated with ipilimumab. Cancer Immunol Immunother. 2014;63:247–257. doi: 10.1007/s00262-013-1508-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tarhini AA, Edington H, Butterfield LH, Lin Y, Shuai Y, Tawbi H, Sander C, Yin Y, Holtzman M, Johnson J, Rao UNM, Kirkwood JM. Immune Monitoring of the Circulation and the Tumor Microenvironment in Patients with Regionally Advanced Melanoma Receiving Neoadjuvant Ipilimumab. PloS one. 2014;9:e87705. doi: 10.1371/journal.pone.0087705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, Zhang Y, Liu Y, Wang L, Zhao L, Yang T, He C, Song Y, Gao Q. Association of myeloid-derived suppressor cells and efficacy of cytokine-induced killer cell immunotherapy in metastatic renal cell carcinoma patients. J Immunother. 2014;37:43–50. doi: 10.1097/CJI.0000000000000005. [DOI] [PubMed] [Google Scholar]
- 11.Finkelstein SE, Carey T, Fricke I, Yu D, Goetz D, Gratz M, Dunn M, Urbas P, Daud A, DeConti R, Antonia S, Gabrilovich D, Fishman M. Changes in dendritic cell phenotype after a new high-dose weekly schedule of interleukin-2 therapy for kidney cancer and melanoma. J Immunother. 2010;33:817–827. doi: 10.1097/CJI.0b013e3181ecccad. [DOI] [PubMed] [Google Scholar]
- 12.Kimura T, McKolanis JR, Dzubinski LA, Islam K, Potter DM, Salazar AM, Schoen RE, Finn OJ. MUC1 vaccine for individuals with advanced adenoma of the colon: a cancer immunoprevention feasibility study. Cancer Prev Res. 2013;6:18–26. doi: 10.1158/1940-6207.CAPR-12-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Youn JI, Nagaraj S, Collazo M, Gabrilovich DI. Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J Immunol. 2008;181:5791–5802. doi: 10.4049/jimmunol.181.8.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poschke I, Kiessling R. On the armament and appearances of human myeloid-derived suppressor cells. Clinical immunology. 2012;144:250–268. doi: 10.1016/j.clim.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 15.Mandruzzato S, Brandau S, Britten CM, Bronte V, Damuzzo V, Gouttefangeas C, Maurer D, Ottensmeier C, Burg SH, Welters MJP, Walter S. Toward harmonized phenotyping of human myeloid-derived suppressor cells by flow cytometry: results from an interim study. Cancer Immunology, Immunotherapy. 2016;65:161–169. doi: 10.1007/s00262-015-1782-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Condamine T, Kumar V, Ramachandran IR, Youn JI, Celis E, Finnberg N, El-Deiry WS, Winograd R, Vonderheide RH, English NR, Knight SC, Yagita H, McCaffrey JC, Antonia S, Hockstein N, Witt R, Masters G, Bauer T, Gabrilovich DI. ER stress regulates myeloid-derived suppressor cell fate through TRAIL-Rmediated apoptosis. J Clin Invest. 2014;124:2626–2639. doi: 10.1172/JCI74056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Condamine T, Mastio J, Gabrilovich DI. Transcriptional regulation of myeloid-derived suppressor cells. J Leukoc Biol. 2015;98:913–922. doi: 10.1189/jlb.4RI0515-204R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Al-Banna N, Lehmann C. Oxidized LDL and LOX-1 in experimental sepsis. Mediators of inflammation. 2013;2013:761789. doi: 10.1155/2013/761789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sawamura T, Kume N, Aoyama T, Moriwaki H, Hoshikawa H, Aiba Y, Tanaka T, Miwa S, Katsura Y, Kita T, Masaki T. An endothelial receptor for oxidized low-density lipoprotein. Nature. 1997;386:73–77. doi: 10.1038/386073a0. [DOI] [PubMed] [Google Scholar]
- 20.Taye A, El-Sheikh AA. Lectin-like oxidized low-density lipoprotein receptor 1 pathways. Eur J Clin Invest. 2013;43:740–745. doi: 10.1111/eci.12092. [DOI] [PubMed] [Google Scholar]
- 21.Yoshimoto R, Fujita Y, Kakino A, Iwamoto S, Takaya T, Sawamura T. The discovery of LOX-1, its ligands and clinical significance. Cardiovascular drugs and therapy / sponsored by the International Society of Cardiovascular Pharmacotherapy. 2011;25:379–391. doi: 10.1007/s10557-011-6324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moldovan I, Galon J, Maridonneau-Parini I, Roman Roman S, Mathiot C, Fridman WH, Sautes-Fridman C. Regulation of production of soluble Fc gamma receptors type III in normal and pathological conditions. Immunol Lett. 1999;68:125–134. doi: 10.1016/s0165-2478(99)00041-3. [DOI] [PubMed] [Google Scholar]
- 23.Banerjee A, Mondal NK, Das D, Ray MR. Neutrophilic inflammatory response and oxidative stress in premenopausal women chronically exposed to indoor air pollution from biomass burning. Inflammation. 2012;35:671–683. doi: 10.1007/s10753-011-9360-2. [DOI] [PubMed] [Google Scholar]
- 24.Mehta JL, Sanada N, Hu CP, Chen J, Dandapat A, Sugawara F, Satoh H, Inoue K, Kawase Y, Jishage K, Suzuki H, Takeya M, Schnackenberg L, Beger R, Hermonat PL, Thomas M, Sawamura T. Deletion of LOX-1 reduces atherogenesis in LDLR knockout mice fed high cholesterol diet. Circ Res. 2007;100:1634–1642. doi: 10.1161/CIRCRESAHA.107.149724. [DOI] [PubMed] [Google Scholar]
- 25.Pirillo A, Norata GD, Catapano AL. LOX-1, OxLDL, and atherosclerosis. Mediators Inflamm. 2013;2013:152786. doi: 10.1155/2013/152786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang CH, Ranatunga S, Kriss CL, Cubitt CL, Tao J, Pinilla-Ibarz JA, Del Valle JR, Hu CC. Inhibition of ER stress-associated IRE-1/XBP-1 pathway reduces leukemic cell survival. J Clin Invest. 2014;124:2585–2598. doi: 10.1172/JCI73448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sawamura T, Kakino A, Fujita Y. LOX-1: a multiligand receptor at the crossroads of response to danger signals. Current opinion in lipidology. 2012;23:439–445. doi: 10.1097/MOL.0b013e32835688e4. [DOI] [PubMed] [Google Scholar]
- 28.Kumar V, Patel S, Tcyganov E, Gabrilovich D. The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol. 2016 doi: 10.1016/j.it.2016.01.004. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramachandran IR, Martner A, Pisklakova A, Condamine T, Chase T, Vogl T, Roth J, Gabrilovich D, Nefedova Y. Myeloid-derived suppressor cells regulate growth of multiple myeloma by inhibiting T cells in bone marrow. J Immunol. 2013;190:3815–3823. doi: 10.4049/jimmunol.1203373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12:253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marvel D, Gabrilovich DI. Myeloid-derived suppressor cells in the tumor microenvironment: expect the unexpected. J Clin Invest. 2015;125:3356–3364. doi: 10.1172/JCI80005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fridlender ZG, Sun J, Mishalian I, Singhal S, Cheng G, Kapoor V, Horng W, Fridlender G, Bayuh R, Worthen GS, Albelda SM. Transcriptomic analysis comparing tumor-associated neutrohpils with granulocytic myeloid-derived suppressor cells and normal neutrophils. PloS one. 2012;7(2):e31524. doi: 10.1371/journal.pone.0031524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nature reviews. Molecular cell biology. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 34.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nature reviews. Molecular cell biology. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 35.Claudio N, Dalet A, Gatti E, Pierre P. Mapping the crossroads of immune activation and cellular stress response pathways. The EMBO Journal. 2013;32:1214–1224. doi: 10.1038/emboj.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahadevan NR, Anufreichik V, Rodvold JJ, Chiu KT, Sepulveda H, Zanetti M. Cell-extrinsic effects of tumor ER stress imprint myeloid dendritic cells and impair CD8(+) T cell priming. PloS one. 2012;7:e51845. doi: 10.1371/journal.pone.0051845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahadevan NR, Rodvold J, Sepulveda H, Rossi S, Drew AF, Zanetti M. Transmission of endoplasmic reticulum stress and pro-inflammation from tumor cells to myeloid cells. Proc Natl Acad Sci U S A. 2011;108:6561–6566. doi: 10.1073/pnas.1008942108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahadevan NR, Zanetti M. Tumor stress inside out: cell-extrinsic effects of the unfolded protein response in tumor cells modulate the immunological landscape of the tumor microenvironment. J Immunol. 2011;187:4403–4409. doi: 10.4049/jimmunol.1101531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 40.Thevenot PT, Sierra RA, Raber PL, Al-Khami AA, Trillo-Tinoco J, Zarreii P, Ochoa AC, Cui Y, Del Valle L, Rodriguez PC. The stress-response sensor chop regulates the function and accumulation of myeloid-derived suppressor cells in tumors. Immunity. 2014;41:389–401. doi: 10.1016/j.immuni.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee BR, Chang SY, Hong EH, Kwon BE, Kim HM, Kim YJ, Lee J, Cho HJ, Cheon JH, Ko HJ. Elevated endoplasmic reticulum stress reinforced immunosuppression in the tumor microenvironment via myeloid-derived suppressor cells. Oncotarget. 2014;5:12331–12345. doi: 10.18632/oncotarget.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taye A, El-Sheikh AA. Lectin-like oxidized low-density lipoprotein receptor 1 pathways. European journal of clinical investigation. 2013;43:740–745. doi: 10.1111/eci.12092. [DOI] [PubMed] [Google Scholar]
- 43.Hong D, Bai YP, Gao HC, Wang X, Li LF, Zhang GG, Hu CP. Ox- LDL induces endothelial cell apoptosis via the LOX-1-dependent endoplasmic reticulum stress pathway. Atherosclerosis. 2014;235:310–317. doi: 10.1016/j.atherosclerosis.2014.04.028. [DOI] [PubMed] [Google Scholar]
- 44.Hong D, Li LF, Gao HC, Wang X, Li CC, Luo Y, Bai YP, Zhang GG. High-Density Lipoprotein Prevents Endoplasmic Reticulum Stress-Induced Downregulation of Liver LOX-1 Expression. PloS one. 2015;10:e0124285. doi: 10.1371/journal.pone.0124285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Condamine T, Mastio J, Gabrilovich DI. Transcriptional regulation of myeloid-derived suppressor cells. J Leukoc Biol. 2015 doi: 10.1189/jlb.4RI0515-204R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bettigole SE, Glimcher LH. Endoplasmic Reticulum Stress in Immunity. Annu Rev Immunol. 2014 doi: 10.1146/annurev-immunol-032414-112116. [DOI] [PubMed] [Google Scholar]
- 47.Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008;454:455–462. doi: 10.1038/nature07203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirsch HA, Iliopoulos D, Joshi A, Zhang Y, Jaeger SA, Bulyk M, Tsichlis PN, Shirley Liu X, Struhl K. A transcriptional signature and common gene networks link cancer with lipid metabolism and diverse human diseases. Cancer Cell. 2010;17:348–361. doi: 10.1016/j.ccr.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu J, Mitra S, Wang X, Khaidakov M, Mehta JL. Oxidative stress and lectinlike ox-LDL-receptor LOX-1 in atherogenesis and tumorigenesis. Antioxid Redox Signal. 2011;15:2301–2333. doi: 10.1089/ars.2010.3792. [DOI] [PubMed] [Google Scholar]
- 50.Wang X, Phillips MI, Mehta JL. LOX-1 and angiotensin receptors, and their interplay. Cardiovascular drugs and therapy / sponsored by the International Society of Cardiovascular Pharmacotherapy. 2011;25:401–417. doi: 10.1007/s10557-011-6331-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Siqueira J, Abdul Zani I, Russell DA, Wheatcroft SB, Ponnambalam S, Homer-Vanniasinkam S. Clinical and Preclinical Use of LOX-1-Specific Antibodies in Diagnostics and Therapeutics. Journal of cardiovascular translational research. 2015;8:458–465. doi: 10.1007/s12265-015-9655-z. [DOI] [PubMed] [Google Scholar]
- 52.Mehta JL, Khaidakov M, Hermonat PL, Mitra S, Wang X, Novelli G, Sawamura T. LOX-1: a new target for therapy for cardiovascular diseases. Cardiovascular drugs and therapy / sponsored by the International Society of Cardiovascular Pharmacotherapy. 2011;25:495–500. doi: 10.1007/s10557-011-6325-5. [DOI] [PubMed] [Google Scholar]
- 53.Zhang S. A comprehensive evaluation of SAM, the SAM R-package and a simple modification to improve its performance. BMC bioinformatics. 2007;8:230. doi: 10.1186/1471-2105-8-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.