Abstract
We recently identified ERdj3 as a component of unassembled immunoglobulin (Ig) heavy chain:BiP complexes. ERdj3 also associates with a number of other protein substrates, including unfolded light chains, a nonsecreted Ig light chain mutant, and the VSV-G ts045 mutant at the nonpermissive temperature. We produced an ERdj3 mutant that was unable to stimulate BiP's ATPase activity in vitro or to bind BiP in vivo. This mutant retained the ability to interact with unfolded protein substrates, suggesting that ERdj3 binds directly to proteins instead of via interactions with BiP. BiP remained bound to unfolded light chains longer than ERdj3, which interacted with unfolded light chains initially, but quickly disassociated before protein folding was completed. This suggests that ERdj3 may bind first to substrates and serve to inhibit protein aggregation until BiP joins the complex, whereas BiP remains bound until folding is complete. Moreover, our findings support a model where interactions with BiP help trigger the release of ERdj3 from the substrate:BiP complex.
INTRODUCTION
The endoplasmic reticulum (ER) represents the entry site into the secretory pathway. Nascent secretory proteins are translocated into a crowded, calcium-rich, oxidizing environment where they must fold and assemble in the presence of many other unfolded proteins. ER molecular chaperones and folding enzymes associate with the newly synthesized proteins to prevent their aggregation and help them fold and assemble correctly. Through a process called ER quality control, misfolded proteins are retained in the ER and eventually targeted for ER-associated degradation (ERAD) via the action of the chaperones (Ellgaard et al., 1999). The mammalian ER possesses two major classes of chaperones, the lectin-like calnexin/calreticulin proteins and their supporting cofactors or enzymes and the Hsp70 orthologue BiP and its cofactors.
BiP has been best characterized for its role in protein folding and assembly (Simons et al., 1995; Hendershot et al., 1996), and its up-regulation during ER stress is a hallmark of the unfolded protein response (UPR) (Lee, 1992). In addition, BiP plays an essential role in maintaining the permeability barrier of the ER translocon during early stages of protein translocation (Hamman et al., 1998), targeting misfolded proteins for proteasomal degradation (Skowronek et al., 1998; Brodsky et al., 1999), serving as a sensor for ER stress (Bertolotti et al., 2000; Shen et al., 2002a), and contributing to the ER calcium stores (Lievremont et al., 1997). All except the last of these activities require BiP's ATPase activity, a feature that is conserved within Hsp70 family members. Like all Hsp70 proteins, BiP fluctuates between ATP- and ADP-bound states to bind and release substrates. In the ATP-bound state, BiP is in an “open” form, allowing it to bind and release unfolded substrates rapidly. Hydrolysis of ATP drives it to the ADP-bound or “closed” state, thus stabilizing its association with unfolded proteins, which serves to inhibit their aggregation. The release of ADP and rebinding of ATP reopens the substrate-binding domain leading to release and folding of the nascent protein. BiP's ATPase cycle is regulated by DnaJ-like proteins, which stimulate ATP hydrolysis, and proteins that regulate nucleotide exchange, like BAP in mammalian cells (Chung et al., 2002) and Sls1p/Sil1p (Kabani et al., 2000; Tyson and Stirling, 2000) and Lhs1p (Steel et al., 2004) in yeast.
DnaJ was first identified as a cofactor of DnaK, the bacterial Hsp70 homologue, which stimulates the ATPase activity of DnaK and helps replicate λ phage DNA in host cells (Yochem et al., 1978; Liberek et al., 1988). Since then, a large number of DnaJ homologues have been identified in most species and organelles. They possess a highly conserved ∼70 amino acid J domain, which contains the hallmark His-Pro-Asp (HPD) motif. Mutations of this sequence abolish the functional interaction between DnaJs and their Hsp70 partners (Cheetham and Caplan, 1998). Based on their domain structure, DnaJ homologues have been divided into three subgroups (Cheetham and Caplan, 1998): type I proteins possess all three domains found in the Escherichia coli DnaJ protein, including the J domain, a glycine/phenylalanine-rich domain, and a cysteine-rich Zn2+ binding domain; type II proteins contain the first two; and type III proteins have only a J domain. Some type I and type II DnaJ proteins bind directly to unfolded substrates through their zinc finger domain and/or a C-terminal region and may serve to target Hsp70s to these substrates. For example, several type I DnaJ proteins, including E. coli DnaJ (Langer et al., 1992; Banecki et al., 1996; Szabo et al., 1996), the cytosolic yeast DnaJ protein Ydj1p (Cyr, 1995; Lu and Cyr, 1998b); and the mammalian mitochondrial DnaJ protein Mdj1p (Prip-Buus et al., 1996) have been shown to bind to substrates through this region and can inhibit protein aggregation in vitro. Sis1p, a type II yeast DnaJ protein interacts with substrates via its C-terminal region, although it is not as efficient at catalyzing protein folding as Ydj1, which also possesses a zinc finger domain (Lu and Cyr, 1998a). It is presently unclear whether type III DnaJ proteins also have the ability to interact directly with polypeptides or only do so through interactions with their Hsp70 partner.
Three ER-localized DnaJ homologues have been identified in the yeast ER where they serve as cofactors for the various functions of yeast BiP, Kar2p. Sec63p is an essential transmembrane protein that assists Kar2p in translocating nascent proteins into the ER (Feldheim et al., 1992) and also may be a component of the retrograde translocon (Plemper et al., 1997). Scj1p cooperates with Kar2p to fold and assemble proteins in the ER lumen (Schlenstedt et al., 1995), and Jem1p interacts with Kar2p to mediate nuclear membrane fusion during mating (Nishikawa and Endo, 1997). Both Scj1p and Jem1p may facilitate the retrotranslocation of ERAD substrates to the cytosol by preventing their aggregation in the ER (Nishikawa et al., 2001). Currently, five mammalian ER DnaJ homologues have been identified. Using a hybrid of the two nomenclatures proposed previously (Bies et al., 1999; Ohtsuka and Hata, 2000), we have suggested that they be named ERdj1-n according to their order of discovery. Thus, they are ERdj1/Mtj1 (Brightman et al., 1995), ERdj2/hSec63 (Skowronek et al., 1999; Tyedmers et al., 2000), ERdj3/HEDJ/ERj3/ABBP-2 (Bies et al., 1999; Yu et al., 2000; Lau et al., 2001), ERdj4/Mdg1 (Prols et al., 2001; Shen et al., 2002b), and ERdj5/JPDI (Cunnea et al., 2003; Hosoda et al., 2003), all of which bind BiP in vitro and stimulate its ATPase activity. However, very little data are available to assess which BiP functions the various mammalian ERdjs assist.
We demonstrate here that ERdj3 is a soluble ER luminal glycoprotein and a component of unassembled Ig heavy chain:BiP complexes. We found that ERdj3 was ubiquitously expressed with the highest levels of expression occurring in secretory tissues and was transcriptionally up-regulated by ER stress conditions. In addition to its association with unassembled Ig heavy chains, ERdj3 bound directly to a number of nascent unfolded and mutant secretory pathway protein substrates, demonstrating that it serves as a cofactor for BiP's functions in protein folding and assembly. Finally, our data suggest that ERdj3 binds first to target proteins and recruits BiP onto the unfolded substrates and that the interaction between BiP and ERdj3 might play a role in releasing ERdj3 from the substrate:BiP complex.
MATERIALS AND METHODS
DNA Constructs
Expressed sequence tag (EST) clone #AA282838 containing the complete human ERdj3 cDNA was obtained from Incyte Genomics Systems (St. Louis, MO), and the sequence was confirmed by DNA sequencing. To create a hemagglutinin (HA)-epitope-tagged version of ERdj3, the stop codon was removed by polymerase chain reaction (PCR) amplification with Tag polymerase (Roche Diagnostics, Indianapolis, IN) by using 5′-CGGAATTCGTGTGGAACAGGACCCG-3′ as the forward primer and 5′-ATAAGAATGCGGCCGCCAATATCCTTGCAGTCCATTGT-3′ as the reverse primer, and the PCR product was subcloned into the 3HA-DSL vector. To produce recombinant His-tagged J domains (aa 23-94), ERdj3 was PCR-amplified using 5′-CGGGATCCGGACGAGATTTCTATAAG-3′ as the forward primer and 5′-CCCAAGCTTCATAATCCTTCTTCACCATAAG-3′ as the reverse primer, and the PCR product was subcloned into the QE-30 (Qiagen QIAexpress System; QIAGEN, Valencia, CA) vector. To produce an ERdj3 J domain mutant, H53 was changed to Q by site-directed mutagenesis by using QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The forward primer was 5′-CCTGCAGCTTCAGCCCGACCGGAACC-3′, and the reverse primer was 5′-GGTTCCGGTCGGGCTGAAGCTGCAGG-3′. The γ heavy chain (Gaut and Hendershot, 1993), NS-1 κ light chain (Skowronek et al., 1998), VSV-G ts045 (Gallione and Rose, 1985), and λ light chain and BiP (Hendershot et al., 1996) vectors have been described.
Protein Expression and Purification
The recombinant proteins were expressed in E. coli M15 cells under nondenaturing conditions according to the manufacturer's protocol (Qiagen QIAexpress System). The final J domain products were tested for residual ATPase activity and found to be negative, demonstrating that they were not contaminated with copurifying ATPases or kinases. All three proteins were stored at -20°C in 25 mM sodium phosphate, pH 7.0, containing 150 mM NaCl and 50% glycerol.
In Vitro Translation and Proteinase K treatment
ERdj3 mRNA was transcribed from the T7 promoter of pT7T3D-PAC-ERdj3 (EST #AA282838) by using the mCAP RNA Capping kit (Stratagene) and translated using [35S]methionine (Amersham Biosciences, Piscataway, NJ) and TNT-coupled reticulocyte lysate (Promega, Madison, WI) in the presence of rat liver microsomes produced as described previously (Walter and Blobel, 1983). The protein product was either left untreated or digested with 150 μg/ml proteinase K (Roche Diagnostics) for 1 h in the presence or absence of 1% NP-40, which disrupts microsomes. Yeast prepro-α-factor mRNA (catalog no. Y4070; Promega) was used as a positive control for protein translocation and glycosylation.
Endoglycosidase H (Endo H) Digestion and Tunicamycin Treatment
To remove N-linked glycans, the in vitro-translated protein or immunoprecipitated protein samples were denatured by adding 15 μl of freshly made denaturing buffer (0.5%SDS, 1% 2-mercapto-ethanol) and heated to 95°C for 15 min. The denatured samples were diluted with 10 μl of 0.5 M sodium citrate, pH5.5, 50 μl of H2O, 2 μl of 100 mM phenylmethylsulfonyl fluoride, and 3 mU of Endo H (Roche Diagnostics) and then incubated at 37°C for 2 h. De novo glycosylation was inhibited by labeling cells in the presence of 10 μg/ml tunicamycin for 6 h.
ATPase Assay
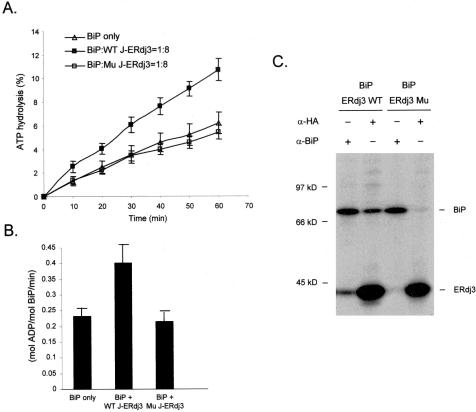
ATPase assays were performed as described previously (Shen et al., 2002b). The recombinant BiP (0.5 μM) was incubated alone or with either purified wild-type J-ERdj3 or mutant J-ERdj3 (4 μM) at 30°C. Samples were removed at the indicated time points, spotted onto thin layer chromatography plates (Sigma-Aldrich, St. Louis, MO), and chromatographed in 1 M formic acid, 0.5 M LiCl. The radioactive ATP and free phosphate signals were quantified by PhosphorImager analysis (Amersham Biosciences) and Image Quant software. The statistical data were deduced from three independent experiments, and the error bars in Figure 4, A and B, represent SD.
Figure 4.
ERdj3 interacts with BiP and HPD motif is indispensable for the interaction. (A) Recombinant BiP was incubated alone (▵) or with wild-type (▪, WT J-ERdj3) or mutant (□, Mu J-ERdj3) J domains in an ATPase assay mixture for the indicated times. ATP hydrolysis was determined by chromatographic separation and quantified by PhosphorImager. (B) The ATP turnover rate was deduced from data obtained in A. (C) COS-1 cells were cotransfected with the indicated cDNAs clones, metabolically labeled and incubated with DSP to crosslink proteins. Cell lysates were prepared and immunoprecipitated with the indicated antisera, and precipitated proteins were analyzed by SDS-PAGE.
Northern Analysis
HepG2 cells were incubated in the presence or absence of 10 μg/ml tunicamycin or 2 μM thapsigargin for 6 h. Total RNA was purified using the RNeasy mini prep kit (QIAGEN), and 20 μg of RNA was applied on a formaldehyde gel and transferred for Northern blotting as described previously (Lawson et al., 1998). The probes used for Northern blotting include a 1.5-kb fragment of human ERdj2 isolated with XhoI and EcoRI from EST clone #BE255328 (Incyte Genomics Systems) and an 840-base pair fragment (corresponding to aa 51-330) from human ERdj3 isolated with PstI and HincII from EST clone #AA282838. The BiP and GRP94 probes have been described previously (Lawson et al., 1998), and the human G3PDH and β-actin probes were purchased (catalog nos. 636830 and 636828; BD Biosciences Clontech, Palo Alto, CA). All the radioactive probes were labeled by using the Prime-it II kit (Stratagene) and purified by NucTrap columns (Stratagene). A human multitissue blot containing 1 μg of polyA+ RNA per lane from 12 different human tissues was purchased from BD Biosciences Clontech (catalog no. 7780-1).
Cell Lines and Eukaryotic Expression
Cell Lines. COS-1, a monkey kidney fibroblast line, and HeLa, a human epithelial line, were maintained in DMEM, and HepG2, a human hepatocarcinoma line; Ag8(8)/Ag8.653, murine plasmacytoma lines; and Ramos, a human Burkitt lymphoma, were maintained in RPMI 1640 medium. All media were supplemented with 10% fetal calf serum, 2 mM l-glutamine, and 1% Fungizone (Biowhittaker, Walkersville, MD).
Antibodies. Polyclonal anti-calnexin and anti-BiP antisera were described previously (Lawson et al., 1998). An anti-HA monoclonal antibody was kindly provided by Dr. Al Reynolds (Vanderbilt University, Nashville, TN). A polyclonal rabbit anti-ERdj3 antiserum was raised against full-length recombinant human ERdj3 protein, which lacked the signal sequence. Goat anti-human Ig heavy chain, goat anti-mouse κ, goat anti-mouse λ, goat anti-mouse Ig, and goat anti-rabbit Ig antibodies were purchased from Southern Biotechnology Associates (Birmingham, AL). The antiserum against VSV-G was a generous gift from Dr. Mike Witte (University of Tennessee, Health Science Center, Nashville, TN).
Microsomes Preparation and Solubilization of ER Proteins. Ag8.653 cells (8 × 107) were homogenized, and microsomes were prepared as described previously (Vanhove et al., 2001). The crude homogenates were centrifuged at 500 × g to pellet cell debris and nuclei. The supernatant consisting of ER microsomes and cytosol was divided into four aliquots and centrifuged at 10,000 × g to pellet the microsomes. The ER microsomes were then resuspended in 100 μl of phosphate-buffered saline (PBS) buffer alone or PBS containing either 0.1% digitonin, 0.2% digitonin, or 1% deoxycholic acid. After rocking at 4°C for 1 h, samples were centrifuged at 10,000 × g for 5 min to sediment residual membranes (Yu et al., 2000). The supernatants and pellets were separately loaded onto SDS-gels and analyzed by Western blotting.
Transient Expression and Immunoprecipitation. COS-1 cells were transfected with the indicated vectors by using the FuGENE 6 transfection reagent (Roche Diagnostics). For in vivo binding experiments, labeled cells were treated with 150 μg/ml 3,3′-dithio-bis (propionic acid N-hydroxysuccinimide ester) (DSP), a membrane permeable cross-linking reagent (Sigma-Aldrich), for 1 h at 4°C. After quenching with 100 mM glycine, cell lysates were prepared and immunoprecipitated with the indicated antibodies or antisera followed by protein A-Sepharose. Precipitated proteins were analyzed on SDS-gels under reducing conditions, and the signal was enhanced with Amplify (Amersham Biosciences) for radiographic visualization. For the light chain folding and secretion experiments, cells were labeled in the presence of dithiothreitol (DTT) and carried out as described previously (Lee et al., 1999). The samples were separated on SDS-gels under nonreducing conditions to visualize the folding intermediates of the light chain.
RESULTS
ERdj3 Is a Soluble ER Luminal Protein
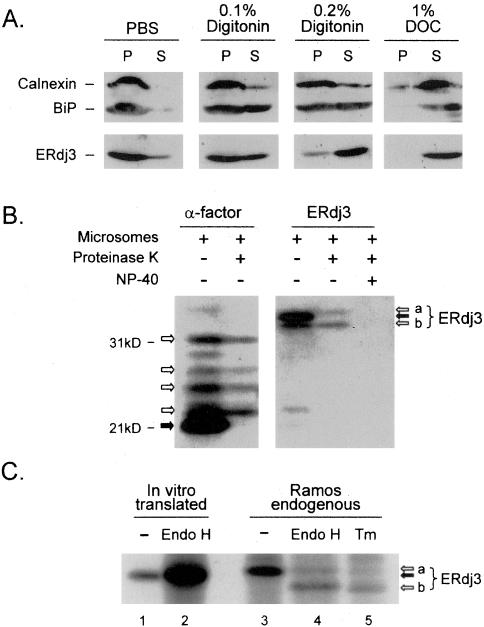
ERdj3 was independently identified by three groups who reported opposing data. The first group observed that native ERdj3 existed in the soluble fraction of dog pancreas microsomes (Bies et al., 1999), another group identified ERdj3/HEDJ as a shiga toxin binding protein and concluded that their exogenously produced protein was a membrane-anchored protein (Yu et al., 2000), and a third group identified it in a two-hybrid screen as a binding partner of the cytosolic protein apobec-1 and demonstrated cytosolic and nuclear localization patterns for a green fluorescent protein (GFP)-tagged version (Lau et al., 2001). To determine the cellular localization and topology of ERdj3 and to better predict its function in vivo, we first examined the endogenous protein in the mouse plasmacytoma cell line Ag8.653 by using a detergent release methodology, similar to that used for HEDJ. ER resident proteins calnexin and BiP were used as controls for the method. As a membrane integral protein, calnexin largely remained associated with ER vesicles in both 0.1 and 0.2% digitonin, whereas BiP, a soluble ER luminal protein, was partially released into supernatant by these same conditions (Figure 1A). Because BiP also interacts with the translocon (Hamman et al., 1998) and the UPR transmembrane signal transducers (Bertolotti et al., 2000; Shen et al., 2002a), it was not completely surprising that a portion of BiP remained associated with ER membranes even in 0.2% digitonin. Both calnexin and BiP were completely released into the supernatant when microsomes were dispersed with 1% deoxycholic acid. When ERdj3 was similarly analyzed, we found that it was partially released from microsomes with 0.1% digitonin, similar to the pattern observed with BiP, and almost completely released with 0.2% digitonin (Figure 1A), suggesting that it was even more freely soluble than BiP.
Figure 1.
ERdj3 is a soluble, resident ER glycoprotein. (A) Microsomes purified from Ag8.653 cells were incubated with the indicated detergents. Samples were centrifuged, and both pellets (P) and supernatants (S) were analyzed by Western blotting. (B) ERdj3 was in vitro translated in the presence of rat liver microsomes and then incubated with (+) or without (-) proteinase K or NP-40 lysing buffer as indicated. The precursor proteins that were not transported into the ER are indicated with solid arrows, whereas proteins that were translocated and protected are indicated with empty arrows. The two species of translocated ERdj3 represent signal-peptide-free ERdj3 with (a) or without (b) glycosylation. Yeast prepro α-factor was used as a positive control for translocation and glycosylation. (C) ERdj3 was in vitro translated in the absence of microsomes or immunoprecipitated from Ramos cells metabolically labeled in the presence or absence of tunicamycin (Tm). Samples in lanes 2 and 4 were further treated with Endo H before analysis by SDS-PAGE. The sample in lane 2 is overloaded.
Because the N-terminal signal peptide of ERdj3 represents the only hydrophobic stretch of amino acids in the protein long enough to serve as a membrane anchor, we translated ERdj3 mRNA in the presence of microsomes in vitro and examined the translocation of the nascent protein and the cleavage of the signal peptide. Our experiments revealed that only a small portion of ERdj3 was translocated into microsomes and protected from proteinase K digestion when the microsomes were intact (Figure 1B), indicating the rather poor efficiency of our in vitro translocation system, an observation that was confirmed when the yeast ER protein prepro-α-factor was similarly analyzed. Two forms of ERdj3 that were protected from protease digestion and therefore translocated into the ER, which migrated at ∼41 and 43 kDa. The two bands collapsed to a single ∼41 kDa band when samples were treated with Endo H (our unpublished data), which thus represented glycosylated and nonglycosylated forms of ERdj3. Similarly, our microsomes yielded several differentially glycosylated forms of α-factor. The nontranslocated ERdj3 represented the full-length protein with an intact signal peptide and migrated slower than the translocated but nonglycosylated protein, indicating that the signal sequence was removed once ERdj3 was translocated into the ER lumen. This result was confirmed by comparing the size of the endogenous ERdj3 protein without sugars (Figure 1C, lanes 4 and 5) and the ERdj3 protein translated in vitro in the absence of microsomes (Figure 1C, lanes 1 and 2). The in vitro translated, nonglycosylated form of ERdj3 migrates slower than the deglycosylated protein isolated from mammalian cells, which indicates that the endogenous ERdj3 (Figure 1C, lane 3) has had its signal peptide removed. Together, these data confirm that ERdj3 possesses a cleavable ER-targeting sequence and exists in the ER as a soluble luminal glycoprotein.
ERdj3 Shows the Same Expression Pattern as BiP in Tissues and Is Up-Regulated by ER Stress
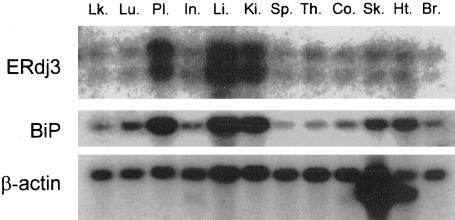
Previously, ERdj3/HEDJ was shown to be ubiquitously expressed with the highest expression levels occurring in tissues like heart, pancreas, and testis (Yu et al., 2000). To serve as a cofactor for BiP in protein folding, we reasoned that its pattern of expression should be similar to that of BiP. A blot containing polyA+ RNA isolated from various human tissues was probed for ERdj3 and BiP. As reported previously (Yu et al., 2000), two transcripts for ERdj3 (∼1.9 and ∼1.6 kb) were detected in all human tissues examined (Figure 2), with the highest levels of expression occurring in tissues that produce large quantities of secretory proteins, such as the liver, placenta, and kidney. When the membrane was hybridized with a BiP probe, we observed a very similar expression pattern (Figure 2), consistent with ERdj3 serving as a cofactor for BiP in some aspect of the maturation of secretory pathway proteins.
Figure 2.
Tissue distribution of ERdj3. A multitissue blot containing 12 human tissues was purchased from BD Biosciences Clontech (catalog no. 7780-1), hybridized with indicated probes, and transcripts were detected by autoradiography. Lk., peripheral blood leukocyte; Lu., lung; Pl., placenta; In., small intestine; Li., liver; Ki., kidney; Sp., spleen; Th., thymus; Co., colon (no mucosa); Sk., skeletal muscle; Ht., heart; Br., brain. Skeletal muscle and heart tissues have two isoforms of β-actin; normalization is based on the top band (BD Biosciences Clontech).
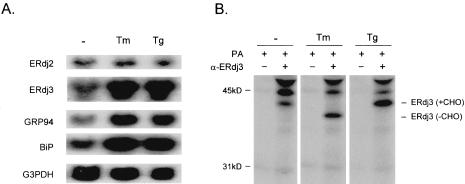
Most of the ER molecular chaperones are transcriptionally up-regulated during ER stress, through a signaling cascade known as the UPR pathway. This serves to alleviate the aggregation of misfolded proteins in the ER and to promote protein refolding when the stress subsides. The elevated expression of ERdj3 in secretory tissues inspired us to investigate whether ERdj3 was induced during ER stress along with BiP and other ER chaperones. HepG2 cells were treated with the ER stress-inducing agents thapsigargin and tunicamycin, and RNA was extracted for Northern blot analyses. Compared with unstressed cells, ERdj3 mRNA levels were dramatically induced by both tunicamycin and thapsigargin treatment (Figure 3A). ERdj2, which is a homologue of the yeast translocon subunit Sec63p, was not induced by ER stress. Thus, not all ER-localized J domain-containing proteins are up-regulated during UPR activation. The ER chaperones BiP and GRP94 were both transcriptionally elevated by ER stress as expected. To determine whether the ERdj3 protein level also was increased in response to ER stress, we performed metabolic labeling experiments on HeLa cells. The endogenous ERdj3 protein was up-regulated by both ER stress conditions (Figure 3B). The increase in mobility observed in the presence of tunicamycin confirmed that native ERdj3 possesses a single N-linked glycan, which further verified its luminal localization. The identity of the other two bands recognized by anti-ERdj3 antiserum has not been determined.
Figure 3.
ERdj3 is up-regulated during ER stress. (A) HepG2 cells were treated for 6 h in the absence (-) or presence of tunicamycin (Tm) or thapsigargin (Tg). RNA was extracted and subjected to Northern blotting with the indicated probes. (B) HeLa cells were treated as described above and then pulse-labeled for 30 min. Cell lysates were immunoprecipitated with anti-ERdj3 antiserum followed by protein A-Sepharose (PA) as indicated and samples were subjected to SDS-PAGE.
The HPD Motif of ERdj3/HEDJ J Domain Is Indispensable for Stimulating the ATPase Activity of BiP
The J domains of DnaJ proteins are highly conserved and serve to stimulate the ATPase activity of their partner Hsp70 proteins. Within the J domain is a conserved HPD sequence that coordinates the interaction of the J domain with the ATPase domain of the Hsp70. Mutations in this sequence of other DnaJ proteins have profound effects on their activity. In an attempt to produce an ERdj3 mutant, we changed H53→ Q in the HPD motif, produced recombinant protein, and investigated the effects of the mutation on its ability to stimulate BiP's ATPase activity. BiP was incubated alone or with either wild-type (WT J-ERdj3) or mutant (Mu J-ERdj3) J domain as indicated. The rate of ATP hydrolysis by BiP was in the linear range during the time course of the experiments (Figure 4A). In the absence of ERdj3, BiP's ATP hydrolysis rate was 0.23 mol/molBiP/min (Figure 4B), which was within the range previously reported for BiP (Wei and Hendershot, 1995). Addition of the wild-type ERdj3 J domain to the hydrolysis reaction elevated the rate of ATP hydrolysis by approximately twofold to 0.41 mol/mol BiP/min (Figure 4B). Mutation of the HPD motif in the J domain abolished its ability to activate the ATPase activity of BiP.
Mutation of the HPD Motif Abolishes the Ability of ERdj3 to Interact with BiP In Vivo
The data presented above indicated that ERdj3 functionally interacted with BiP in vitro and that the HPD motif was critical for the stimulation of the BiP's ATPase activity. The J domain of ERdj3 has been shown to physically associate with BiP in vitro by using a glutathione S-transferase pull-down assay (Yu et al., 2000). To determine whether ERdj3 binds to BiP in vivo, we cotransfected COS-1 cells with hamster BiP and tagged versions of wild-type and mutant ERdj3s. Cells were metabolically labeled and then incubated with DSP to stabilize naturally existing protein complexes. Coexpression of the tagged wild-type ERdj3 with hamster BiP resulted in a complex between the two proteins that could be recognized by either an antirodent BiP antiserum or anti-HA antibody (Figure 4C). Interactions between BiP and wild-type ERdj3 also could be detected without using the covalent cross-linker; however, this interaction was somewhat sensitive to the lysing conditions used (our unpublished data). In contrast, the mutant ERdj3 did not seem to form stable complexes with BiP, because neither the anti-BiP nor the anti-HA antibody was able to coprecipitate appreciable amounts of the other protein. Thus, in addition to functionally disabling ERdj3, the H→ Q mutation abrogates the physical interaction between BiP and ERdj3 in vivo.
Unassembled Ig Heavy Chains Are a Natural Substrate of ERdj3
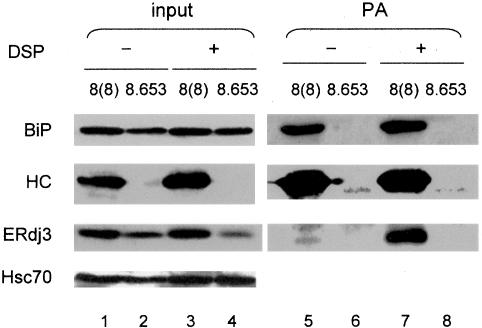
We next investigated the involvement of ERdj3 as a potential cochaperone in BiP:substrate complexes. We previously reported that ERdj3 is a component of a multichaperone complex that binds to unassembled Ig heavy chains in vivo (Meunier et al., 2002). Using our anti-ERdj3 antiserum, we were able to confirm this observation by Western blot. Ag8(8) and Ag8.653 cells were directly lysed or first treated with DSP to stabilize chaperone complexes. The Ag8(8) cells accumulate unassembled, nonsecreted γ heavy chains that can be isolated with protein A-Sepharose, and Ag8.653 cells, which are a derivative of the Ag8(8) cells that no longer synthesize heavy chains, served as a control. Isolation of heavy chains from Ag8(8) cells resulted in coprecipitation of BiP whether or not cross-linker was added, because the BiP:heavy chain complex is stable to normal lysis conditions (Figure 5). When the heavy chain complex was examined for ERdj3, we found that in the absence of cross-linker only small amounts of ERdj3 were detected. However, when cells were treated with DSP before lysing, ERdj3 was readily isolated with heavy chains (Figure 5, lanes 5 and 7). When the cell lysates from both cell lines were directly analyzed, we found that the protein levels of ERdj3 and BiP were higher in Ag8(8) than in Ag8.653 cells (Figure 5, lanes 1-4), which suggests that the accumulation of unfolded heavy chains in these cells triggers the UPR and ERdj3 up-regulation. Because the samples used for direct blotting represented one-ninth of the amount used for isolating heavy chains in the right panel, it was clear that a substantial proportion of the ERdj3 pool was associated with heavy chains in this cell line.
Figure 5.
ERdj3 associates with unassembled Ig heavy chains along with BiP in a plasmacytoma cell line. Ag8(8) and Ag8.653 cells were incubated with (+) or without (-) DSP. Cells were lysed and 1/10 of each lysate was removed for direct blotting, whereas the remaining 9/10 of the lysate was incubated with protein A-Sepharose (PA) to isolate γ heavy chains and coprecipitating proteins. The precipitated protein complexes were reduced, separated by SDS-PAGE, and transferred for Western blotting with the indicated antisera. The Hsc70 signal served as a loading control.
ERdj3 Associates Directly with a Number of Unfolded Proteins in the ER
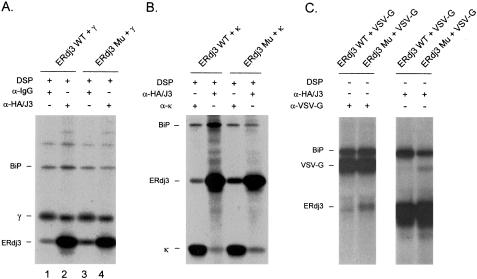
In addition to their ability to serve as cochaperones for their Hsp70 partners, some type I and type II DnaJ homologues are considered to be molecular chaperones themselves, because they bind directly to unfolded substrates through their cysteine-rich and/or C-terminal domains and inhibit protein aggregation in vitro. Because ERdj3 has both features, we speculated that it might bind to unfolded substrates directly. The ERdj3 mutant that had an impaired association with BiP provided us with a tool to investigate this point. Cotransfection experiments revealed that endogenous BiP and transfected wild-type ERdj3 formed a complex with the γ heavy chains (Figure 6A, lanes 1 and 2) and demonstrated that the transient expression of these proteins recapitulated the results obtained in plasmacytoma cells. When cells expressing the ERdj3 mutant were similarly examined, we found that not only did the three-protein complex still form but also that increased amounts of mutant ERdj3 were coprecipitated with the Ig heavy chains (lanes 1 and 3). The presence of heavy chains in the anti-HA immunoprecipitations (Figure 6A, lanes 2 and 4) is not informative, because they bind directly to protein A-Sepharose. Because the mutant ERdj3 does not interact with BiP (Figure 4C), we assume that its inclusion in the complex is due to it binding directly to the heavy chain. There was no apparent difference in the association of the multichaperone complex (Meunier et al., 2002) with the γ heavy chains when wild-type and mutant ERdj3-expressing cells were compared (Figure 6A; our unpublished data), suggesting that this aspect of chaperone biology was unaffected.
Figure 6.
ERdj3 interacts with three unfolded substrates. COS-1 cells were cotransfected with human Igγ heavy chain (A), NS-1 nonsecreted κ light chain (B), and VSV-G ts045 (C) along with the indicated cDNAs. Cells were metabolically labeled for 2 h (VSV-G transfectants were incubated at 39°C for 1 h before labeling at 39°C for 30 min) and then incubated with DSP. Cell lysates were prepared and immunoprecipitated with the indicated antisera and analyzed by reducing SDS-PAGE.
To determine whether ERdj3 also associated with other unfolded protein substrates, we performed similar experiments using a nonsecreted κ light chain (Figure 6B) and a temperature-sensitive mutant of VSV-G, ts045 (Figure 6C), as the substrates. In both cases, we found that although wild-type ERdj3 formed complexes with the unfolded protein, the ERdj3 mutant actually bound better as indicated by more of the mutant coprecipitating with the unfolded substrate. Again because the mutant ERdj3 does not interact stably with BiP, the data suggest that it must be binding directly to these substrate proteins. Because the H→ Q mutation is in the BiP interaction domain and not in the substrate-binding domain, there is no reason a priori to believe that it should directly alter the association of ERdj3 with substrates.
Mutant ERdj3 Expression Does Not Affect Ig Light Chain Folding
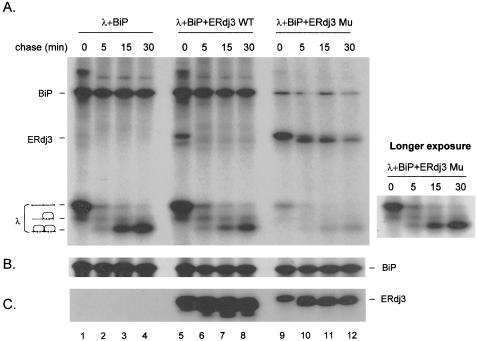
Because ERdj3 is a soluble luminal protein that associates with unfolded substrates and is induced by ER stress, we hypothesized that ERdj3 might play a role in protein folding. To test this possibility, COS-1 cells were transfected with cDNAs encoding λ light chain, BiP, and HA-tagged versions of wild-type and mutant ERdj3 and then labeled in the presence of DTT to inhibit cotranslational disulfide bond formation in the light chain. Cells were then washed to remove DTT and chased for the indicated times to allow light chain folding. We found that regardless of whether wild-type or mutant ERdj3 was expressed, there was no obvious change in the rate of light chain folding compared with cells in which no additional ERdj3 was expressed (Figure 7A). We consistently found that much less λ light chain was expressed in the presence of mutant ERdj3. However, this seemed to be specific to the λ light chain when it was expressed in the presence of DTT and did not seem to be due to rapid turnover or gross insolubility (our unpublished data). We found that less BiP was coprecipitated with light chains synthesized in the presence of mutant ERdj3, which was to be expected, because the level of light chain expression was so much lower. The most dramatic difference was in the coprecipitation of ERdj3 with the light chains. Wild-type ERdj3 was coprecipitated with the reduced light chains, but quickly disappeared in the chase, even before light chain folding was complete (Figure 7A). When immunoprecipitates from the ERdj3 mutant transfectant were examined, we observed first that a much stronger signal was observed for the ERdj3 mutant when it was coprecipitated with the λ light chain (Figure 7A). This was not because mutant ERdj3 labeled better or was produced at a much higher level. In fact, immunoprecipitation of ERdj3 mutant from these samples revealed that the level of expression was actually much lower than wild-type ERdj3 (Figure 7C). Given the fact that there are fewer light chains synthesized in these samples, the difference between this and the wild-type ERdj3 transfectant is even more dramatic. The second observation was that the kinetics of ERdj3 release were dramatically delayed in the ERdj3 mutant cultures, a finding consistent with the increased binding of ERdj3 mutant to γ heavy chains, NS-1 κ light chains, and VSV-G ts045.
Figure 7.
ERdj3 mutant interacts with BiP:light chain complex more stably. COS-1 cells were cotransfected with the indicated cDNA clones. Transfectants were labeled with [35S]methionine for 45 min in the presence of DTT, washed, and then chased in complete media devoid of DTT for the indicated times. Cell lysates were divided equally and immunoprecipitated with antisera specific for mouse λ (A), rodent BiP (B), or HA-tagged ERdj3 (HA/J3) (C) and subjected to nonreducing SDS-PAGE. The three light chain folding intermediates are indicated on the left (A). A longer exposure of light chain folding in the λ + BiP + mutant ERdj3 transfectant is shown to the right for comparison with the other transfectants (A).
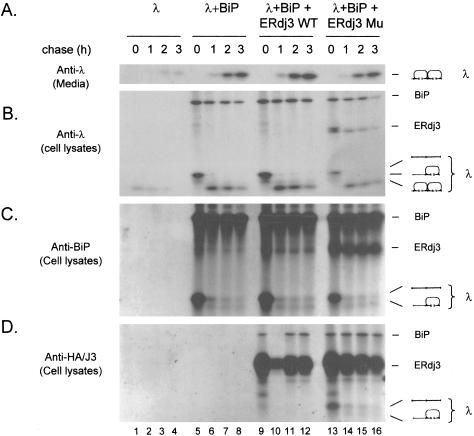
These results lead us to examine which folding intermediates of λ light chains ERdj3 associated with and to determine whether the prolonged binding of the mutant to light chains had any effect on their secretion. COS-1 cells were transfected and labeled as in Figure 7. The levels of λ synthesis in the absence of BiP coexpression was consistently very low (Figure 8, A and B), a finding that we have observed with a number of BiP substrates. Again, the folding rate of λ light chains seemed unaffected by either wild-type or mutant ERdj3 coexpression, although once again the expression level of λ was lower in the presence of mutant ERdj3 (Figure 8, A and B). Thus, the prolonged association of mutant ERdj3 with λ light chains did not delay or prevent their secretion (Figure 8A). When BiP was immunoprecipitated from these samples, we detected coprecipitation of the completely reduced light chain in the 0 time point sample for all transfectants (Figure 8C). As the light chains folded, BiP coprecipitated much less of the light chains, but in all cases, it continued to bind the completely reduced and partially oxidized intermediates that remained. When ERdj3 was isolated, we detected only very small amounts of the completely reduced light chain coprecipitating with wild-type ERdj3 in the t = 0 sample (Figure 8D). Thus, even though large quantities of the reduced light chain can be coprecipitated with BiP, very little of it is associated with ERdj3 under these conditions. When the cells coexpressing mutant ERdj3 were similarly examined, we found that much larger quantities of the reduced λ coprecipitated with ERdj3 in the 0 time point sample, which is all the more remarkable given the fact that the total amount of light chain synthesis is much lower in this sample (Figure 8B). Examination of the samples obtained after chasing without DTT revealed that mutant ERdj3 continued to associate with the remaining folding intermediates in a manner similar to that observed in the anti-BiP precipitated material. These data demonstrate that although both wild-type and mutant ERdj3 can bind to the λ light chains, wild-type ERdj3 binds much weaker or more transiently, and unlike BiP, leaves the complex long before folding is complete. On the other hand, the mutant ERdj3, which is completely capable of binding to the light chain, either binds more strongly or more stably to the unfolded substrate, and similar to BiP, continues to bind as long as unfolded substrate remains.
Figure 8.
ERdj3 interacts with light chain intermediates transiently, whereas BiP shows prolonged binding. COS-1 cells were transiently transfected with the indicated cDNAs and labeled as in Figure 7. Media from each of the various time points were immunoprecipitated with anti-λ antiserum (A). The cell lysates were divided and immunoprecipitated with antisera specific for mouse λ (B), rodent BiP (C), or HA-tagged ERdj3 (HA/J3) (D) and analyzed by nonreducing SDS-PAGE. Nearly half of the sample for lane 10 in D was lost.
DISCUSSION
To date, five ER localized DnaJ homologues have been reported in mammalian cells and three in yeast. Because the yeast ER DnaJ family members are each specific for regulating a different function of Kar2p, it is reasonable to hypothesize that the mammalian ER-associated DnaJ domain proteins also will be dedicated to different BiP functions. Presently, there are very little functional data available for any of the mammalian proteins. ERdj1/Mtj1 and ERdj2/hSec63 seem to be yeast Sec63p counterparts. ERdj1 possesses a ribosome binding site (Dudek et al., 2002), which should position it near translocating chains, and ERdj2 can be cross-linked to the Sec61 component of the translocon (Meyer et al., 2000), suggesting they may both play roles in translocation. Our finding that neither ERdj1 nor 2 is induced during UPR activation (Shen et al., 2002b; our unpublished data) is consistent with such a role, because protein translation is inhibited early in the stress response (Brostrom et al., 1996). For the remaining ERdjs, even less functional data are available.
ERdj3 was independently identified by three different groups who all assigned different subcellular locations or properties for the protein. One group examined the endogenous protein in dog pancreatic microsomes and concluded that ERdj3/ERj3 was a soluble ER luminal protein because it was released with 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate (Bies et al., 1999). The other two groups used transient expression of epitope-tagged versions of the protein for their analyses. When a GFP tag was placed at the N terminus, ERdj3/ABBP-2 was expressed in the cytosol and the nucleus (Lau et al., 2001), whereas when a V5-His tag was placed at the C terminus, ERdj3/HEDJ was targeted to the ER lumen but seemed to be membrane anchored (Yu et al., 2000). Our data showing that the signal sequence is cleaved from in vitro translated/translocated ERdj3 and that the endogenous protein is released from ER vesicles with very mild detergent treatment support the original description of ERdj3 as a soluble ER protein (Bies et al., 1999). It seems that epitope tagging interferes with ER targeting in the first case and signal sequence cleavage in the latter.
The unassembled Ig heavy chains represent a natural substrate for ERdj3 in plasmacytoma cell lines that no longer produce Ig light chains but maintain heavy chains in the ER bound to BiP (Haas and Wabl, 1983). We found that three other unfolded model proteins interacted with ERdj3, including the nonsecreted NS-1 κ light chain, a λ light chain synthesized in the presence of DTT to accumulate unfolded precursors, and the ts045 mutant of VSV-G that reversibly unfolds at the nonpermissive temperature. To this list, we might add shiga toxin, based on a previous report where a partial ERdj3 cDNA was identified in a genetic screen for proteins that provided resistance to shiga toxin (Yu et al., 2000). Shiga toxin binds to cell surface receptors, enters cells by endocytosis, and relocalizes to the ER where it is recognized as a substrate for retrotranslocation, allowing it to inactivate ribosomes and kill cells. Although we were able to detect binding of ERdj3 to a number of substrates, in no case was this binding as dramatic as it was with Ig heavy chains, suggesting that either more ERdj3 binds or that its binding to this particular substrate is more stable. It has been shown that BiP must release from ERAD substrates for them to be retrotranslocated (Knittler et al., 1995), and γ heavy chains have a rather long half-life for an unfolded protein (Vanhove et al., 2001). The prolonged association of ERdj3 with γ heavy chains might serve to keep BiP in the ADP-bound form and therefore stably bound to the heavy chains, which would delay their degradation. This enhanced binding of ERdj3 with γ heavy chains may further explain earlier experiments that revealed that BiP does not cycle on and off unfolded heavy chains in the absence of light chains (Vanhove et al., 2001).
Although endogenous BiP was observed in the complex of protein substrates and ERdj3, the interaction between ERdj3 and substrates does not seem to occur through BiP, because the mutant ERdj3 protein is impaired in its ability to associate with BiP but still binds to the substrates. This suggests that ERdj3 is binding directly to the unfolded proteins and at a site that is distinct from that of BiP, at least for the γ heavy chain. In the case of the λ light chains, our pulse-chase experiments revealed that wild-type ERdj3 was rapidly released after DTT was removed and well before folding was complete. This is in contrast to BiP, which remained bound as long as partially folded intermediates existed. We previously reported that BiP exists in a complex with a subset of ER molecular chaperones that does not include ERdj3 but that ERdj3 binds along with the chaperone complex to heavy chains (Meunier et al., 2002). Together, these data imply that ERdj3 binds directly to unfolded substrates and may serve to recruit BiP and the other proteins in the chaperone complexes, as has been suggested for other Hsp70 -DnaJ pairs (Hendrick et al., 1993; Rudiger et al., 2001). If this is indeed true, it is somewhat surprising that we saw no decrease in BiP association with our substrates when the ERdj3 mutant was coexpressed. It is possible that additional interactions occur between the C-terminal domains of BiP and ERdj3 (as has been shown for other Hsp70 -DnaJ pairs (Karzai and McMacken, 1996; Qian et al., 2002) that are sufficient to allow even the mutant ERdj3 to recruit BiP to the substrate but that this interaction is too transient or unstable to observe in pull-down assays or that the endogenous wild-type ERdj3 could serve to recruit BiP to the substrate. Unlike most genetic studies that have expressed DnaJ family members in the absence of wild-type protein, it is significant that our studies are all done in the presence of the endogenous wild-type protein. Finally, it is possible that our substrates have a sufficiently high-affinity for BiP, particularly under conditions of BiP overexpression, that ERdj3 is not needed.
It is surprising that we observed no delay in the folding or secretion of λ light chain when they were coexpressed with mutant ERdj3, even though the mutant remained bound to the substrate much longer than wild-type ERdj3. Our data show that wild-type ERdj3 is lost from BiP:substrate complexes well before folding is complete. This strongly suggests that, although ERdj3 may help recruit BiP to the substrate, BiP plays the major role in suppressing protein aggregation and allowing proper maturation. It is important to point out that the QPD mutant should be viewed as inactive in terms of its association with BiP but not as a dominant negative mutant. In this regard, our results are compatible with data obtained in yeast where deletion of SCJ1 only slightly reduced the transport rate of a nonglycosylated mutant carboxypeptidase Y and had no effect on the maturation of glycosylated carboxypeptidase Y, apparently because a second ER DnaJ protein, Jem1p, was able to compensate (Silberstein et al., 1998). Given that at least four other ERdjs are present in the mammalian ER, it is possible that one of these, or perhaps more likely the endogenous wildtype ERdj3, may be compensating for the loss of ERdj3 function. Alternatively, it is possible that the levels of BiP overexpression are sufficiently high to minimize any potential effects. We estimated from Western blot experiments coupled with immunofluorescence staining that BiP expression is approximately sixfold above normal levels and ERdj3 is increased by ∼12-fold (our unpublished data) in transfected cells. Although both of these possibilities would explain why folding of our substrate is not affected in the presence of mutant expression, they do not explain why the prolonged binding of the mutant does not have any obvious effect on light chain folding. It is possible that instead of mutant ERdj3 release being required to allow the light chain to fold; light chain folding could drive the release of mutant ERdj3 along with BiP. Although this interpretation of chaperone-mediated folding may be a bit antidogmatic, it is compatible with our failure to observe reiterative cycles of BiP binding to substrates in vivo (Vanhove et al., 2001) and much more in keeping with the fact that the amount of light chain associated with both mutant ERdj3 and BiP decreases dramatically after DTT removal when the majority of the light chains fold but that both chaperones remain associated with a smaller pool of intermediates that seem to remain blocked in folding.
If, as has been proposed for other Hsp70:DnaJ pairs, ERdj3 binds to substrates and transfers them to BiP, the question arises as to how ERdj3 is released. We consistently found that the mutant ERdj3 bound better or longer to unfolded substrates. Because mutations in the N-terminal J domain of ERdj3 would not be expected to influence the substrate-binding domain (and our preliminary data indicate that both wild-type and mutant ERdj3 have similar affinities for the unfolded protein in vitro), we hypothesize that the functional interaction between BiP and ERdj3 is essential for freeing ERdj3 from the substrate complex. The binding of BiP to ERdj3 or the ERdj3-enhanced hydrolysis of ATP by BiP may induce a reciprocal conformational change in ERdj3, which reduces the affinity of ERdj3 for the substrates. In this model, ERdj3 would bind first to substrates and recruit the ATP-bound form of BiP. Interaction of ERdj3 with BiP would enhance ATP hydrolysis thus inducing a conformational change in BiP locking it onto the unfolded protein. Concomitantly, BiP would induce a conformational change in ERdj3 to signal the successful recruitment and transfer of BiP to the substrate. Current studies are underway to test this hypothesis.
In summary, we have demonstrated that ERdj3, a soluble resident ER glycoprotein, served as a cochaperone of BiP both in vivo and in vitro. The HPD motif of ERdj3 is indispensable for its physical and functional interaction with BiP in both situations. ERdj3 binds directly to both unfolded proteins that are substrates for ERAD and nascent unfolded proteins, suggesting that either it plays a role in both pathways or the ERAD substrates bind to ERdj3 before they are targeted for degradation. Disruption of the association of BiP and ERdj3 does not impair the ability of ERdj3 to bind to substrates but does prolong this interaction, which may imply a mechanism for ERdj3 release from substrates.
Acknowledgments
This work was supported by National Institutes of Health grant GM-54068 (to L.M.H.), the Cancer Center CORE grant CA21765, and the American Lebanese Syrian Associated Charities of St. Jude Children's Research Hospital.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-05-0434. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-05-0434.
Abbreviations used: DSP, 3,3′-dithio-bis (propionic acid N-hydroxysuccinimide ester); Endo H, endoglycosidase H; ERAD, endoplasmic reticulum-associated degradation; HPD, His-Pro-Asp; UPR, unfolded protein response; Ig, immunoglobulin.
References
- Banecki, B., Liberek, K., Wall, D., Wawrzynow, A., Georgopoulos, C., Bertoli, E., Tanfani, F., and Zylicz, M. (1996). Structure-function analysis of the zinc finger region of the DnaJ molecular chaperone. J. Biol. Chem. 271, 14840-14848. [DOI] [PubMed] [Google Scholar]
- Bertolotti, A., Zhang, Y., Hendershot, L. M., Harding, H. P., and Ron, D. (2000). Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2, 326-332. [DOI] [PubMed] [Google Scholar]
- Bies, C., Guth, S., Janoschek, K., Nastainczyk, W., Volkmer, J., and Zimmermann, R. (1999). A Scj1p homolog and folding catalysts present in dog pancreas microsomes. Biol. Chem. 380, 1175-1182. [DOI] [PubMed] [Google Scholar]
- Brightman, S. E., Blatch, G. L., and Zetter, B. R. (1995). Isolation of a mouse cDNA encoding MTJ1, a new murine member of the DnaJ family of proteins. Gene 153, 249-254. [DOI] [PubMed] [Google Scholar]
- Brodsky, J. L., Werner, E. D., Dubas, M. E., Goeckeler, J. L., Kruse, K. B., and McCracken, A. A. (1999). The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation demonstrates that protein export and import are mechanistically distinct. J. Biol. Chem. 274, 3453-3460. [DOI] [PubMed] [Google Scholar]
- Brostrom, C. O., Prostko, C. R., Kaufman, R. J., and Brostrom, M. A. (1996). Inhibition of translational initiation by activators of the glucose-regulated stress protein and heat shock protein stress response systems. Role of the interferon-inducible double-stranded RNA-activated eukaryotic initiation factor 2alpha kinase. J. Biol. Chem. 271, 24995-25002. [DOI] [PubMed] [Google Scholar]
- Cheetham, M. E., and Caplan, A. J. (1998). Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones 3, 28-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, K. T., Shen, Y., and Hendershot, L. M. (2002). BAP, a mammalian BiP associated protein, is a nucleotide exchange factor that regulates the ATPase activity of BiP. J. Biol. Chem. 277, 47557-47563. [DOI] [PubMed] [Google Scholar]
- Cunnea, P. M., et al. (2003). ERdj5, an endoplasmic reticulum (ER)-resident protein containing DnaJ and thioredoxin domains, is expressed in secretory cells or following ER stress. J. Biol. Chem. 278, 1059-1066. [DOI] [PubMed] [Google Scholar]
- Cyr, D. M. (1995). Cooperation of the molecular chaperone Ydj1 with specific Hsp70 homologs to suppress protein aggregation. FEBS Lett. 359, 129-132. [DOI] [PubMed] [Google Scholar]
- Dudek, J., et al. (2002). A novel type of co-chaperone mediates transmembrane recruitment of DnaK-like chaperones to ribosomes. EMBO J. 21, 2958-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard, L., Molinari, M., and Helenius, A. (1999). Setting the standards: quality control in the secretory pathway. Science 286, 1882-1888. [DOI] [PubMed] [Google Scholar]
- Feldheim, D., Rothblatt, J., and Schekman, R. (1992). Topology and functional domains of Sec63p, an endoplasmic reticulum membrane protein required for secretory protein translocation. Mol. Cell. Biol. 12, 3288-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallione, C. J., and Rose, J. K. (1985). A single amino acid substitution in a hydrophobic domain causes temperature-sensitive cell-surface transport of a mutant viral glycoprotein. J. Virol. 54, 374-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, J. R., and Hendershot, L. M. (1993). Mutations within the nucleotide binding site of Ig-binding protein inhibit ATPase activity and interfere with release of Ig heavy chain. J. Biol. Chem. 268, 7248-7255. [PubMed] [Google Scholar]
- Haas, I. G., and Wabl, M. (1983). Ig heavy chain binding protein. Nature 306, 387-389. [DOI] [PubMed] [Google Scholar]
- Hamman, B. D., Hendershot, L. M., and Johnson, A. E. (1998). BiP maintains the permeability barrier of the ER membrane by sealing the lumenal end of the translocon pore before and early in translocation. Cell 92, 747-758. [DOI] [PubMed] [Google Scholar]
- Hendershot, L., Wei, J., Gaut, J., Melnick, J., Aviel, S., and Argon, Y. (1996). Inhibition of Ig folding and secretion by dominant negative BiP ATPase mutants. Proc. Natl. Acad. Sci. USA 93, 5269-5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrick, J. P., Langer, T., Davis, T. A., Hartl, F. U., and Wiedmann, M. (1993). Control of folding and membrane translocation by binding of the chaperone DnaJ to nascent polypeptides. Proc. Natl. Acad. Sci. USA 90, 10216-10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda, A., Kimata, Y., Tsuru, A., and Kohno, K. (2003). JPDI, a novel endoplasmic reticulum-resident protein containing both a BiP-interacting J-domain and thioredoxin-like motifs. J. Biol. Chem. 278, 2669-2676. [DOI] [PubMed] [Google Scholar]
- Kabani, M., Beckerich, J. M., and Gaillardin, C. (2000). Sls1p stimulates Sec63p-mediated activation of Kar2p in a conformation-dependent manner in the yeast endoplasmic reticulum. Mol. Cell Biol. 20, 6923-6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karzai, A. W., and McMacken, R. (1996). A bipartite signaling mechanism involved in DnaJ-mediated activation of the Escherichia coli DnaK protein. J. Biol. Chem. 271, 11236-11246. [DOI] [PubMed] [Google Scholar]
- Knittler, M. R., Dirks, S., and Haas, I. G. (1995). Molecular chaperones involved in protein degradation in the endoplasmic reticulum: quantitative interaction of the heat shock cognate protein BiP with partially folded Ig light chains that are degraded in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 92, 1764-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer, T., Lu, C., Echols, H., Flanagan, J., Hayer, M. K., and Hartl, F. U. (1992). Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature 356, 683-689. [DOI] [PubMed] [Google Scholar]
- Lau, P. P., Villanueva, H., Kobayashi, K., Nakamuta, M., Chang, B. H., and Chan, L. (2001). A DnaJ protein, apobec-1-binding protein-2, modulates apolipoprotein B mRNA editing. J. Biol. Chem. 276, 46445-46452. [DOI] [PubMed] [Google Scholar]
- Lawson, B., Brewer, J.W., and Hendershot, L. M. (1998). Geldanamycin, an HSP90/GRP94-binding drug, induces increased transcription of ER chaperones via the ER stress pathway. J. Cell Physiol. 174, 170-178. [DOI] [PubMed] [Google Scholar]
- Lee, A. S. (1992). Mammalian stress response: induction of the glucose-regulated protein family. Curr. Opin. Cell Biol. 4, 267-273. [DOI] [PubMed] [Google Scholar]
- Lee, Y.-K., Brewer, J. W., Hellman, R., and Hendershot, L. M. (1999). BiP and Ig light chain cooperate to control the folding of heavy chain and ensure the fidelity of Ig assembly. Mol. Biol. Cell 10, 2209-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberek, K., Georgopoulos, C., and Zylicz, M. (1988). Role of the Escherichia coli DnaK and DnaJ heat shock proteins in the initiation of bacteriophage lambda DNA replication. Proc. Natl. Acad. Sci. USA 85, 6632-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lievremont, J. P., Rizzuto, R., Hendershot, L., and Meldolesi, J. (1997). BiP, a major chaperone protein of the endoplasmic reticulum lumen, plays a direct and important role in the storage of the rapidly exchanging pool of Ca2+. J. Biol. Chem. 272, 30873-30879. [DOI] [PubMed] [Google Scholar]
- Lu, Z., and Cyr, D. M. (1998a). Protein folding activity of Hsp70 is modified differentially by the hsp40 co-chaperones Sis1 and Ydj1. J. Biol. Chem. 273, 27824-27830. [DOI] [PubMed] [Google Scholar]
- Lu, Z., and Cyr, D. M. (1998b). The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone Ydj1 assist Hsp70 in protein folding. J. Biol. Chem. 273, 5970-5978. [DOI] [PubMed] [Google Scholar]
- Meunier, L., Usherwood, Y. K., Chung, K. T., and Hendershot, L. M. (2002). A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol. Biol. Cell 13, 4456-4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, H. A., Grau, H., Kraft, R., Kostka, S., Prehn, S., Kalies, K. U., and Hartmann, E. (2000). Mammalian Sec61 is associated with Sec62 and Sec63. J. Biol. Chem. 275, 14550-14557. [DOI] [PubMed] [Google Scholar]
- Nishikawa, S., and Endo, T. (1997). The yeast JEM1p is a DnaJ-like protein of the endoplasmic reticulum membrane required for nuclear fusion. J. Biol. Chem. 272, 12889-12892. [DOI] [PubMed] [Google Scholar]
- Nishikawa, S. I., Fewell, S. W., Kato, Y., Brodsky, J. L., and Endo, T. (2001). Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J. Cell Biol. 153, 1061-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka, K., and Hata, M. (2000). Mammalian HSP40/DNAJ homologs: cloning of novel cDNAs and a proposal for their classification and nomenclature. Cell Stress Chaperones 5, 98-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemper, R. K., Bohmler, S., Bordallo, J., Sommer, T., and Wolf, D. H. (1997). Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature 388, 891-895. [DOI] [PubMed] [Google Scholar]
- Prip-Buus, C., Westerman, B., Schmitt, M., Langer, T., Neupert, W., and Schwarz, E. (1996). Role of the mitochondrial DnaJ homologue, Mdj1p, in the prevention of heat-induced protein aggregation. FEBS Lett. 380, 142-146. [DOI] [PubMed] [Google Scholar]
- Prols, F., Mayer, M. P., Renner, O., Czarnecki, P. G., Ast, M., Gassler, C., Wilting, J., Kurz, H., and Christ, B. (2001). Upregulation of the cochaperone Mdg1 in endothelial cells is induced by stress and during in vitro angiogenesis. Exp. Cell Res. 269, 42-53. [DOI] [PubMed] [Google Scholar]
- Qian, X., Hou, W., Zhengang, L., and Sha, B. (2002). Direct interactions between molecular chaperones heat-shock protein (Hsp) 70 and Hsp 40, yeast Hsp70 Ssa1 binds the extreme C-terminal region of yeast Hsp40 Sis1. Biochem. J. 361, 27-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudiger, S., Schneider-Mergener, J., and Bukau, B. (2001). Its substrate specificity characterizes the DnaJ co-chaperone as a scanning factor for the DnaK chaperone. EMBO J. 20, 1042-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenstedt, G., Harris, S., Risse, B., Lill, R., and Silver, P. A. (1995). A yeast DnaJ homologue, Scj1p, can function in the endoplasmic reticulum with BiP/Kar2p via a conserved domain that specifies interactions with Hsp70s. J. Cell Biol. 129, 979-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, J., Chen, X., Hendershot, L., and Prywes, R. (2002a). ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell 3, 99-111. [DOI] [PubMed] [Google Scholar]
- Shen, Y., Meunier, L., and Hendershot, L. M. (2002b). Identification and characterization of a novel endoplasmic reticulum (ER) DnaJ homologue, which stimulates ATPase activity of BiP in vitro and is induced by ER stress. J. Biol. Chem. 277, 15947-15956. [DOI] [PubMed] [Google Scholar]
- Silberstein, S., Schlenstedt, G., Silver, P. A., and Gilmore, R. (1998). A role for the DnaJ homologue Scj1p in protein folding in the yeast endoplasmic reticulum. J. Cell Biol. 143, 921-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, J. F., Ferro-Novick, S., Rose, M. D., and Helenius, A. (1995). BiP/Kar2p serves as a molecular chaperone during carboxypeptidase Y folding in yeast. J. Cell Biol. 130, 41-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronek, M. H., Hendershot, L. M., and Haas, I. G. (1998). The variable domain of non-assembled Ig light chains determines both their half-life and binding to BiP. Proc. Natl. Acad. Sci. USA 95, 1574-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronek, M. H., Rotter, M., and Haas, I. G. (1999). Molecular characterization of a novel mammalian DnaJ-like Sec63p homolog. Biol. Chem. 380, 1133-1138. [DOI] [PubMed] [Google Scholar]
- Steel, G. J., Fullerton, D. M., Tyson, J. R., and Stirling, C. J. (2004). Coordinated activation of Hsp70 chaperones. Science 303, 98-101. [DOI] [PubMed] [Google Scholar]
- Szabo, A., Korszun, R., Hartl, F. U., and Flanagan, J. (1996). A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured protein substrates. EMBO J. 15, 408-417. [PMC free article] [PubMed] [Google Scholar]
- Tyedmers, J., Lerner, M., Bies, C., Dudek, J., Skowronek, M. H., Haas, I. G., Heim, N., Nastainczyk, W., Volkmer, J., and Zimmermann, R. (2000). Homologs of the yeast Sec complex subunits Sec62p and Sec63p are abundant proteins in dog pancreas microsomes. Proc. Natl. Acad. Sci. USA 97, 7214-7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson, J. R., and Stirling, C. J. (2000). LHS1 and SIL1 provide a lumenal function that is essential for protein translocation into the endoplasmic reticulum. EMBO J. 19, 6440-6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhove, M., Usherwood, Y.-K., and Hendershot, L. M. (2001). Unassembled Ig heavy chains do not cycle from BiP in vivo, but require light chains to trigger their release. Immunity 15, 105-114. [DOI] [PubMed] [Google Scholar]
- Walter, P., and Blobel, G. (1983). Preparation of microsomal membranes for cotranslational protein translocation. Methods Enzymol. 96, 84-93, 84-93. [DOI] [PubMed] [Google Scholar]
- Wei, J.-Y., and Hendershot, L. M. (1995). Characterization of the nucleotide binding properties and ATPase activity of recombinant hamster BiP purified from bacteria. J. Biol. Chem. 270, 26670-26677. [DOI] [PubMed] [Google Scholar]
- Yochem, J., Uchida, H., Sunshine, M., Saito, H., Georgopoulos, C. P., and Feiss, M. (1978). Genetic analysis of two genes, dnaJ and dnaK, necessary for Escherichia coli and bacteriophage lambda DNA replication. Mol. Gen. Genet. 164, 9-14. [DOI] [PubMed] [Google Scholar]
- Yu, M., Haslam, R. H., and Haslam, D. B. (2000). HEDJ, an Hsp40 cochaperone localized to the endoplasmic reticulum of human cells. J. Biol. Chem. 275, 24984-24992. [DOI] [PubMed] [Google Scholar]