Abstract
Cellular behavior can be considered to be the result of a very complex spatial and temporal integration of intracellular and extracellular signals. These signals arise from serum-soluble factors as well as from cell–substrate or cell–cell interactions. The current approach in mitogenesis studies is generally to analyze the effect of a single growth factor on serum-starved cells. In this context, a metabolic hormone such as insulin is found to be a mitogenic agent in many cellular types. In the present study, we have considered the effect of insulin stimulation in platelet-derived growth factor (PDGF)-activated NIH-3T3 and C2C12 cells. Our results show that insulin is able to inhibit strongly both NIH-3T3 and C2C12 cell growth induced by PDGF, one of the most powerful mitotic agents for these cell types. This inhibitory effect of insulin is due primarily to a premature down-regulation of the PDGF receptor. Thus, when NIH-3T3 or C2C12 cells are stimulated with both PDGF and insulin, we observe a decrease in PDGF receptor phosphorylation with respect to cells treated with PDGF alone. In particular, we find that costimulation with insulin leads to a reduced production of H2O2 with respect to cell stimulation with PDGF alone. The relative low concentration of H2O2 in PDGF/insulin-costimulated cell leads to a limited down-regulation of protein tyrosine phosphatases, and, consequently, to a reduced PDGF receptor phosphorylation efficiency. The latter is very likely to be responsible for the insulin-dependent inhibition of PDGF-receptor mitogenic signaling.
INTRODUCTION
Extracellular signaling molecules control many of the activities of vertebrate cells. The signals are often transduced across the cellular membrane by transmembrane receptors. Receptor tyrosine-kinases (RTKs) are a large family of cell surface receptors that possess intrinsic protein tyrosine kinase activity. RTKs play an important role in the control of many fundamental cellular processes, including the cell cycle, cell migration, cell metabolism and survival, as well as cell differentiation.
With the exception of the insulin receptor (IR) family, all known RTKs are monomers in the cell membrane. Ligand binding induces dimerization of these receptors, resulting in autophosphorylation of their cytoplasmic domains (Lemmon and Schlessinger, 1994; Jiang and Hunter, 1999). This dimerization activates receptor catalytic activity and generates phosphorylated tyrosine residues that mediate the specific binding of cytoplasmic signaling proteins containing Src homology-2 (SH2) or protein tyrosine-binding domains (Schlessinger, 2000).
Platelet-derived growth factor (PDGF) is a major mitogen for mesenchymal cells, smooth muscle cells, and glial cells (Heldin and Westermark, 1999). The PDGF α- and β-receptors contain five extracellular immunoglobulin-like domains and an intracellular tyrosine kinase domain with a characteristic inserted sequence (Claesson-Welsh et al., 1989). On stimulation PDGF receptor (PDGF-R) becomes highly phosphorylated on specific tyrosine residues. More than 10 different SH2 domain-containing molecules have been shown to bind to different autophosphorylation sites in both the PDGF α and β receptors, including proteins with enzymatic activity, such as phosphatidylinositol 3-phosphate kinase (PI-3K), phospholipase C-γ (PLC-γ), the Src family of tyrosine kinases, the SHP-2 tyrosine phosphatase, GTPase-activating protein for Ras, as well as adaptor molecules such as Grb2, Shc, Nck, Grb7, and Crk, and signal transducers and activators of transcription.
PDGF-R activation induces various cell responses, depending on cellular type and local environment. In general, PDGF elicits a mitogenic and antiapoptotic action, and in addition, promotes cell migration.
IR is a heterotetrameric receptor composed of two extracellular α subunits and two transmembrane β subunits. Insulin binding to the IR extracellular domain induces a modification in the quaternary structure that leads to increased autophosphorylation of the cytoplasmic domain. Autophosphorylation on tyrosine residues leads to an increase of IR tyrosine kinase activity and results in the phosphorylation of various cytosolic docking proteins, most notably Shc and the insulin receptor substrate (IRS) protein family (Claesson-Welsh et al., 1989; Pronk et al., 1993; Skolnik et al., 1993; White, 1998). In fact, unlike other growth factor RTKs, the IR uses the IRS docking proteins to initiate its signaling program. Tyrosine phosphorylated IRS proteins subsequently recruit several effector proteins containing specific SH2 domains, such as PI-3K, SHP-2 phosphatase, Grb2, and Csk (Sun et al., 1993; Tobe et al., 1996; Liu and Roth, 1998; Ogawa et al., 1998; White, 1998). Ultimately, these proteins transmit the mitogenic (e.g., DNA synthesis), metabolic (e.g., glucose transport), as well as cytoskeleton reorganization signals elicited by insulin. Another difference between IR and the other RTKs is that although RTKs regulate long-term events such as cell differentiation and mitogenesis, a major function of the IR is the acute regulation of glucose metabolism in muscle and fat cells. Despite these important differences RTKs and IR share essentially the same transduction machinery (e.g., PI-3K, Ras, and Src pathways) to elicit, in many cases, different cellular response.
In this work, we have studied the effect of simultaneous PDGF and insulin stimulation on NIH-3T3 cells overexpressing the insulin receptor (NIH-3T3-IR) and on C2C12 cells. Surprisingly, we have found that insulin, widely reported to promote cell growth in many cell types (Chiarugi et al., 1997; Wang et al., 1999), is able to inhibit cell proliferation induced by PDGF, one of the most powerful mitogenic agents for both these cell types. Our findings indicate that insulin inhibits PI-3K pathway/H2O2 production in PDGF-stimulated cells, leading to an up-regulation of intracellular protein tyrosine phosphatase (PTP) activity with respect to cells stimulated with PDGF alone. As a consequence, we found a reduced time course of PDGF-R phosphorylation in PDGF/insulin-costimulated cells with respect to cells treated with PDGF alone that is responsible of the insulin-induced cell growth inhibition of PDGF-stimulated cells.
MATERIALS AND METHODS
Materials
Unless specified, all reagents were obtained from Sigma-Aldrich (St. Louis, MO). NIH-3T3-IR fibroblast were kindly provided from Alan Saltiel (University of Michigan, Ann Arbor, MI), C2C12 mouse myoblasts were purchased at EACC (European Collection of Cell Culture), human recombinant PDGF was from PeproTech (Rocky Hill, NJ), and the enhanced chemiluminescence kit was from Amersham Biosciences (Piscataway, NJ). All antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA), except anti-phospho-extracellular signal-regulated kinase (ERKs) that were from New England Biolabs (Beverly, MA), anti-c-Src that were from Upstate Biotechnology (Lake Placid, NY). BCA protein assay reagent was from Pierce Chemical (Rockford, IL). [γ-32P]ATP was from Amersham Biosciences.
Cell Culture
NIH-3T3-IR and C2C12 cells were routinely cultured in DMEM with 10% fetal calf serum (FCS) in 5% CO2 humidified atmosphere.
Crystal Violet Staining
NIH-3T3-IR or C2C12 cells (2 × 104) were seeded in 24-multiwell plates and serum starved for 24 h before receiving 30 ng/ml PDGF and 10 nM insulin, either separately or in combination, for 24 and 48 h. Fresh growth factors were added daily. Cellular growth was stopped by removing the medium and by the addition of a 0.5% crystal violet solution in 20% methanol. After 5 min of staining, the fixed cells were washed with phosphate-buffered saline (PBS) and solubilized with 200 μl/well of 0.1 M sodium citrate, pH 4.2. The absorbance at 595 nm was evaluated using a microplate reader.
Thymidine Incorporation
NIH-3T3-IR or C2C12 cells (2 × 104) were seeded in 24-multiwell plates and serum starved for 24 h before receiving 30 ng/ml PDGF and 10 nM insulin, either separately or in combination, for the indicated times. Then, 1 μCi/ml3 [H]thymidine was added in each well, and the incubation was prolonged for 1 h. The wells were washed with PBS, and DNA was precipitated with ice-cold 5% trichloroacetic acid. The cells were then lysed in 1 N NaOH, and the [3H]thymidine incorporated was measured in a scintillation counter. All experiments were performed in triplicate.
Immunoprecipitations and Western Blot Analysis
Cells (1 × 106) were seeded in 10-cm plates in DMEM supplemented with 10% FCS. Cells were serum starved for 24 h before receiving 30 ng/ml PDGF and 10 nM insulin either separately or in combination. Cells were then lysed for 20 min on ice in 500 μl of RIPA lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Nonidet P-40, 2 mM EGTA, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 10 μg/ml leupeptin). Lysates were clarified by centrifugation and immunoprecipitated for 4 h at 4°C with 0.1 μg of the specific antibodies. Immune complexes were collected on protein A-Sepharose (Sigma Aldrich, St. Louis, MO), separated by SDS-PAGE, and transferred onto polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA). Immunoblots were incubated in 1% bovine serum albumin, 10 mM Tris-HCl, pH 7.5, 1 mM EDTA, and 0.05% Tween 20 for 1 h at room temperature, probed first with specific antibodies, and then with secondary antibodies conjugated with horseradish peroxidase, washed, and developed with the enhanced chemiluminescence kit. Chemiluminescence was detected by the Chemidoc apparatus (Bio-Rad, Hercules, CA). Quantity-One software (Bio-Rad) was used to perform the quantitative analysis of the spots.
Determination of Endocytotic PDGF-R
PDGF-R internalization was determined by measuring the percentage of receptors that were resistant to trypsinization according to Ceresa et al. (1998). Briefly, 1 × 106 cells were serum starved for 24 h and then stimulated with 30 ng/ml PDGF and 10 nM insulin either separately or in combination. Cells were washed with cold PBS and incubated on ice for 3 min with 20 mM sodium acetate, pH 3.7. Then, cells were washed with ice-cold PBS and incubated with trypsin (1 mg/ml in PBS) in ice for 30 min with occasional rocking. The reaction was stopped adding the same volume of soybean trypsin inhibitor (1 mg/ml) in PBS. Cells were then washed and solubilized in complete RIPA (cRIPA) buffer, which is RIPA buffer plus 0.5% Na-deoxycholate and 0.1% sodium-SDS, for 10 min at 4°C. Cell lysates were immunoprecipitated with anti-platelet–derived growth factor-R antibodies and subjected to SDS-PAGE. The trypsin-resistant 190-kDa PDGF-R represents the endosomic receptor.
PDGF-R Kinase Activity
Cells (7.5 × 105) were plated in 10-cm plates in complete medium. Cells were serum starved for 24 h before receiving 30 ng/ml PDGF and 10 nM insulin, either separately or in combination, for the indicated times. Cell lysates in cRIPA buffer were subjected to immunoprecipitation with anti PDGF-R antibodies (Santa Cruz Biotechnology). The immunoprecipitated proteins were washed twice in lysis buffer and twice in 50 mM Tris-HCl, pH 7.4, containing 1 mM sodium orthovanadate, and finally resuspended in 50 mM HEPES at pH 7.4, 10 mM MnCl2. The reaction was started with the addition of 10 μCi of [γ-32P]ATP (3000 Ci/mmol) to all samples, which were then incubated at 4°C for 10 min. The beads were then washed once with 50 mM Tris-HCl, pH 7.4, and finally resuspended in 20 μl of Laemmli's sample buffer, boiled for 5 min, and separated by a 7.5% SDS-PAGE. The autoradiogram was scannerized and then quantified using Quantity-One software (Bio-Rad). An aliquot of each sample was blotted on PVDF membrane and subjected to anti-platelet–derived growth factor-R immunoblot to obtain a normalized value for the amount of PDGF-R.
Src Activity Assay
Cells (7.5 × 105) were plated in 10-cm plates in complete medium. Cells were serum starved for 24 h before receiving 30 ng/ml PDGF and 10 nM insulin, either separately or in combination, for the indicated times and lysed in RIPA buffer, clarified by centrifugation, and assayed for protein content. Anti-Src antibodies (5 μg) were added to 500 μg of total proteins. The reaction mixtures were incubated overnight at 4°C. Immune complexes were collected using 20 μl of protein A-Sepharose resin that had been preincubated with 5 μg of rabbit anti-mouse IgG. The beads were incubated in 20 μl of reaction mixture (20 mM HEPES, pH 7.5, 5 μCi of [γ-32P]ATP [3000 Ci/mmol], 10 mM MnCl2, 20 μg/ml aprotinin) containing 20 μg of enolase for 10 min. at 30°C. The reaction was stopped by adding 10 μl of 4× SDS-PAGE sample buffer, and the samples were loaded onto 12% SDS-PAGE. The gel was then stained with Coomassie Blue, and the bands corresponding to enolase were cut and then radioactivity was quantitated in a scintillation counter. The amount of immunoprecipitated c-Src from each sample was evaluated by anti-c-Src immunoblot. Densitometric analysis of the immunoblot was used for normalization of the immunokinase assay.
Phosphatidylinositol 3-Kinase Activity
PI-3K assay was performed as described previously (Choudhury et al., 1991). Briefly, serum-starved cells were incubated with 30 ng/ml PDGF and 10 nM insulin, either separately or in combination, for the indicated times and then lysed in RIPA buffer. Equal amounts of protein were immunoprecipitated using agarose-conjugated anti-phosphotyrosine antibodies. After washing, the immunobeads were resuspended in 50 μl of 20 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 0.5 mM EGTA. Then, 10 μg of phosphatidylinositol was added, mixed, and incubated at 25°C for 10 min. Next, 1 μl of 1 M MgCl2 and 10 μCi of [γ-32P]ATP (3000 Ci/mmol) were added simultaneously and incubated at 25°C for additional 10 min. The reaction was stopped by the addition of 150 μl of chloroform/methanol/37% HCl (10:20:0.2). Samples were extracted with chloroform and dried. Radioactive lipids were separated by thin layer chromatography (TLC) by using chloroform/methanol/30% ammonium hydroxide/water (46:41:5:8). After drying, plates were autoradiographed. The identity of the 3-OH–phosphorylated lipids after separation by TLC has been previously confirmed using high-pressure liquid chromatography (Choudhury et al., 1991). The radioactive spots corresponding to phosphatidylinositol phosphate were scraped and counted in a liquid scintillation counter.
Determination of Intracellular H2O2
Intracellular production of H2O2 was assayed as described previously (Bae et al., 1997). Briefly, cells were starved for 24 h and then incubated for the indicated times with 30 ng/ml PDGF and 10 nM insulin either separately or in combination. 5 min before the end of incubation time 2′,7′-dichlorofluorescin diacetate was added to a final concentration of 5 μM. This compound is converted by intracellular esterases to 2′,7′-dichlorofluorescein, which is then oxidized by H2O2 to give a highly fluorescent compound. Cell were then detached from the substrate by trypsinization and analyzed immediately by flow cytometry by using a BD Biosciences FACScan flow cytometer equipped with an argon laser lamp (FL-1; emission, 488 nm; band pass filter, 530 nm).
PTP Activity Assay
Cells (7.5 × 105) were serum starved for 24 h before receiving 30 ng/ml PDGF or 30 ng/ml PDGF together with 10 nM insulin for the indicated times. Cells were collected in 300 μl of 0.05 M Tris-HCl, pH 7.4, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 10 μg/ml leupeptin buffer, and sonicated for 10 s. The lysates were clarified by centrifugation, and 10 μl of lysates was mixed with 40 μl of 0.05 M Tris-HCl, pH 7.4, buffer containing 1 mM [pTyr1146]-insulin receptor fragment 1142–1153 as substrate and kept at 37°C for 30 min. The production of Pi was measured colorimetrically at 620 nm by means of Malachite Green assay. Alternatively, 50 μl of lysate was mixed with 1 ml of 0.05 M Tris-HCl, pH 7.4, buffer containing 10 mM p-nitrophenylphosphate (pNPP) as substrate and kept at 37°C for 30 min. The production of p-nitrophenol was measured colorimetrically at 400 nm. The PTP specific activity was taken as the ratio between PTP activity and the total protein content of the lysate.
In Gel Tyrosine-Phosphatase Assay
For the detection of in gel PTP activity, we prepared a 10% SDS-polyacrylamide gel containing 105 cpm/ml 32P-labeled substrate as described previously Burridge and Nelson, 1995). Cells were lysed in 25 mM sodium acetate buffer, pH 5.5, 1% Nonidet P-40, 150 mM NaCl, 10% glycerol, 100 μg/ml catalase, and protease inhibitor cocktail as described above. In the modified version of the technique, which is used to detect oxidized PTPs, 10 mM iodoacetic acid was added to the samples, after degassing the buffer (Meng et al., 2002). Total proteins (15 μg) were then loaded onto the radiolabeled gel. After the electrophoresis run, the gels were sequentially washed for the indicated times at room temperature in ∼200 ml of the following buffers: buffer 1 (overnight): 50 mM Tris, pH 8.0, and 20% isopropanol; buffer 2 (twice for 30 min): 50 mM Tris, pH 8.0, and 0.3% β-mercaptoethanol; buffer 3 (90 min): 50 mM Tris, pH 8.0, 0.3% β-mercaptoethanol, 6 M guanidine hydrochloride, and 1 mM EDTA; buffer 4 (three washes for 1 h each): 50 mM Tris, pH 7.4, 0.3% β-mercaptoethanol, 1 mM EDTA, and 0.04% Tween 20; and buffer 5 (overnight): 50 mM Tris, pH 7.4, 0.3% β-mercaptoethanol, 1 mM EDTA, 0.04% Tween 20, and 4 mM dithiothreitol. The gels were then dried and analyzed using a PhosphorImager system.
RESULTS
Insulin Inhibits PDGF-induced Cell Growth in Both NIH-3T3-IR and C2C12 Cells
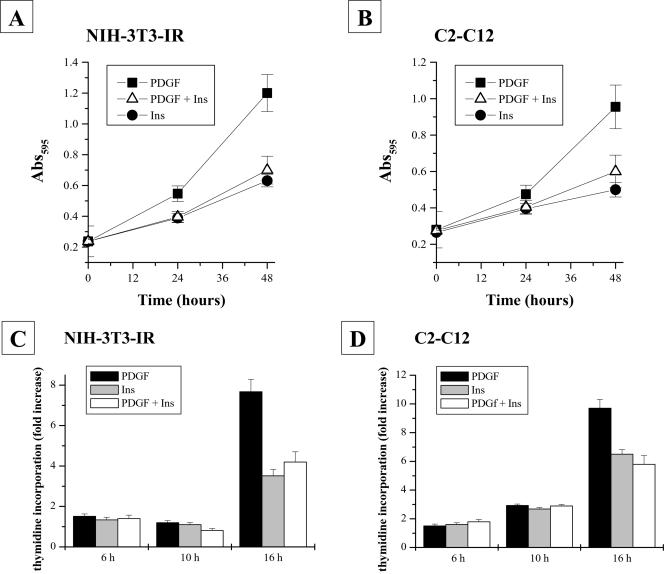
Insulin is able to transduce both metabolic and mitogenic signals in many cell types. We have chosen NIH-3T3 cells overexpressing the insulin receptor (NIH-3T3-IR) and C2C12 cells to study the cooperation of PDGF and insulin in mitogenesis induction. We used (if not otherwise stated) exponentially growing NIH-3T3-IR and C2C12 cells that were starved for 24 h and then stimulated with 30 ng/ml PDGF and 10 nM insulin (which represents the saturating dose of both hormones for this kind of cells), either separately or in combination, for 24 and 48 h. The cell growth rate was estimated both by means of both crystal violet staining and 3[H]thymidine incorporation (Figure 1B). As expected, PDGF is able to induce a powerful mitogenic induction in both NIH-3T3-IR (Figure 1, A–C) and C2C12 cells (Figure 1, B–D), whereas insulin is able to promote only limited cell growth. Surprisingly, cells simultaneously treated with insulin and PDGF exhibited growth rates that resemble that elicited by insulin alone. Hence, PDGF and insulin do not synergize, but, instead, insulin strongly inhibits PDGF-induced cell growth.
Figure 1.
NIH-3T3-IR and C2C12 cell growth rates after stimulation with platelet-derived growth factor, insulin, or platelet-derived growth factor together with insulin. Cells (2 × 104) were seeded in 24-multiwell plates and serum starved for 24 h. Cells were then treated with 30 ng/ml platelet-derived growth factor and 10 nM insulin either separately or in combination for the times indicated. A and B show crystal violet staining experiments for NIH-3T3-IR and C2C12 cells, respectively. C and D report [3H]thymidine incorporation experiments in NIH-3T3-IR and C2C12 cells, respectively. All experiments were performed in triplicate.
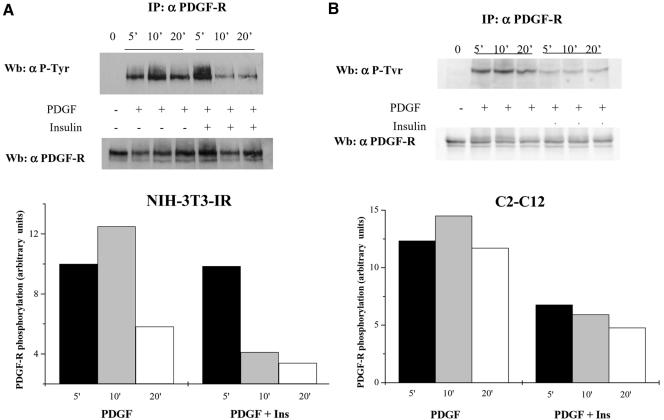
Insulin Cotreatment Inhibits PDGF-R Phosphorylation
IR and PDGF-R belong to the RTK family and consequently share most of the mitogenic signaling pathways. To study at a molecular level the inhibitory effect of insulin on PDGF-induced cell growth, we have evaluated the time course of PDGF-R phosphorylation in various conditions. NIH-3T3-IR and C2C12 cells were treated for the times indicated as described above, and the results of anti-phosphotyrosine and anti-platelet–derived growth factor-R immunoblots are shown in Figure 2. Insulin, when added simultaneously to PDGF, is able to reduce PDGF-R phosphorylation with respect to stimulation with PDGF alone both in NIH-3T3-IR (Figure 2A) and in C2C12 cells (Figure 2B). This evidence is in good agreement with recent findings (Chiarugi et al., 2002) that have established a close relationship between the time course of PDGF-R phosphorylation and the strength of the mitogenic stimuli elicited by this growth factor. In fact, Chiarugi et al. (2002) have shown that a prolonged phosphorylation of PDGF-R leads to a reduced G1/S phase and hence to an increased cell growth rate. In our case, the premature PDGF-R dephosphorylation in PDGF/insulin-costimulated cells leads to a weaker mitogenic stimulation with respect to cell treated with PDGF alone.
Figure 2.
Platelet-derived growth factor-R phosphorylation time course in NIH-3T3-IR (A) and C2C12 (B) cells stimulated with platelet-derived growth factor, insulin, or platelet-derived growth factor together with insulin. Cells (1 × 106) were serum starved for 24 h before being treated with 30 ng/ml platelet-derived growth factor and 10 nM insulin or 30 ng/ml platelet-derived growth factor together with 10 nM insulin. At the indicated times, cells were lysed in RIPA buffer and immunoprecipitated with anti-platelet–derived growth factor-R antibodies. Immunoblots were first probed with anti-phosphotyrosine antibodies and then the blot was stripped and reprobed with anti-platelet–derived growth factor-R antibodies. The diagrams report the normalized platelet-derived growth factor-R phosphorylation under the various experimental conditions. The result is representative of three independent experiments with similar results.
Analysis of PDGF-R Down-Regulation Pathways in PDGF/Insulin Costimulation
The level and extent of PDGF-R phosphorylation reflects the balance between PDGF-R kinase activity and PTP activity. In addition, the overall PDGF phosphorylation status also depends on the rate of PDGF-R internalization and degradation.
First, we tested whether insulin could inhibit PDGF-R kinase activity by using an in vitro kinase assay. NIH-3T3 cells were treated for the times indicated as described above and were then lysed and immunoprecipitated with anti PDGF-R antibodies. The kinase assay was initiated by the addition of [γ-32P]ATP and stopped with SDS sample buffer after 10 min of incubation at 4°C. Figure 3A shows the autoradiography of the gel after SDS-PAGE. The results clearly indicate that the time course of PDGF-R kinase activity after stimulation with either PDGF alone or with PDGF together with insulin is comparable.
Figure 3.
Analysis of platelet-derived growth factor-R down-regulation pathways. (A) Platelet-derived growth factor-R kinase assay: 7.5 × 105 NIH-3T3-IR cells were serum starved for 24 h before receiving 30 ng/ml platelet-derived growth factor and 10 nM insulin either separately or in combination for the indicated times. Samples were immunoprecipitated with platelet-derived growth factor-R antibodies and resuspended in 50 mM HEPES, pH 7.4, and 10 mM MnCl2. Kinase reaction was started with the addition of [γ-32P]ATP to all samples. The beads were resuspended in Laemmli's sample buffer, separated by SDS-PAGE, and autoradiographed. An aliquot of each sample was blotted onto PVDF membrane and subjected to anti-platelet–derived growth factor-R immunoblot to normalize the amount of platelet-derived growth factor-R. The result is representative of three independent experiments with similar results. (B) Platelet-derived growth factor-R internalization rate: NIH-3T3-IR cells were serum starved for 24 h and then stimulated with 30 ng/ml platelet-derived growth factor and 10 nM insulin either separately or in combination. Cells were treated according to Materials and Methods to detect the endosomic platelet-derived growth factor-R. The result is representative of three independent experiments. (C) Platelet-derived growth factor-R ubiquitination rate: NIH-3T3-IR cells were serum starved for 24 h and then stimulated with 30 ng/ml platelet-derived growth factor and 10 nM insulin either separately or in combination. At the indicated times, cells were lysed in cRIPA buffer, immunoprecipitated with anti-platelet–derived growth factor-R antibodies, and probed with anti-ubiquitin antibodies. The blot was then stripped and reprobed with anti-platelet–derived growth factor-R antibodies. All experiments were performed in triplicate.
Next, we studied two of the main PDGF-R down-regulation pathways, internalization and ubiquitin-mediated proteolysis, after stimulation of cells with PDGF/insulin or PDGF alone. NIH-3T3-IR cells were serum starved for 24 h and then stimulated with PDGF and insulin in combination. At the times indicated, cells were washed with and acidic solution to induce a rapid detachment of the hormone and thus the termination of signaling. Intact cells were then treated with trypsin for 30 min, to digest the exposed PDGF-R molecules, and then lysed and immunoprecipitated with anti-PDGF-R antibodies. The anti-platelet–derived growth factor-R immunoblot shown in Figure 3B reveals a band of 190 kDa that represents the intact receptor, i.e, the fraction of PDGF-R that has been internalized after growth factor stimulation. Figure 3B reveals that there is no marked difference in the PDGF-R internalization pattern between NIH-3T3-IR stimulated with PDGF and insulin with respect to cells stimulated with PDGF alone, indicating that insulin does not interfere with PDGF-R internalization rate.
Protein ubiquitination is a mechanism of controlled intracellular proteolysis (Hershko and Ciechanover, 1998). The ubiquitin system adds a polymer of ubiquitin to the target proteins. The ubiquitin-modified proteins are then rapidly destroyed by the 26S proteasome. We have measured the PDGF-R ubiquitination rate after stimulation of cells with PDGF, insulin, or PDGF and insulin together. Our results (Figure 3C) indicate that insulin does not interfere with the rate of PDGF-R ubiquitination, because the PDGF-R ubiquitination level of cells treated with PDGF/insulin it is not significantly different from that of cell stimulated with PDGF alone.
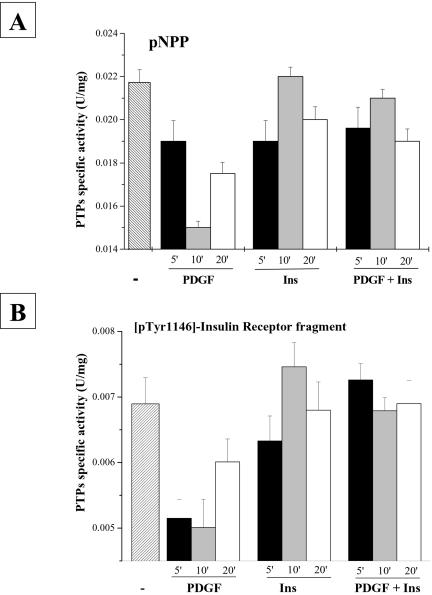
Finally, we treated NIH-3T3-IR cells as described above, and at the indicated times, cells were lysed and assayed for total PTP-specific activity by using both pNPP and [pTyr1146]-insulin receptor fragment 1142–1153 as substrates (Figure 4, A and B). The results show that in PDGF/insulin-costimulated cells the overall PTP activity is higher with respect to cells stimulated with PDGF alone.
Figure 4.
Intracellular PTP activity assay. NIH-3T3-IR cells (7.5 × 105) were serum starved for 24 h before being treated with 30 ng/ml platelet-derived growth factor, 10 nM insulin, or 30 ng/ml platelet-derived growth factor together with 10 nM insulin for the indicated times. Cells were lysed in 0.1 M sodium acetate buffer, pH 5.5, and sonicated for 10 s and then were assayed for PTP activity by using either pNPP (A) or [pTyr1146]-insulin receptor fragment 1142–1153 (B). All experiments were performed in triplicate.
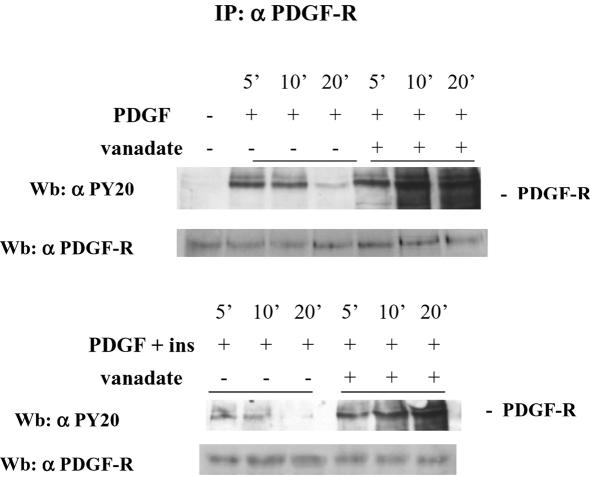
To confirm a role for PTPs in insulin inhibition of PDGF-R activation, we have treated NIH-3T3-IR cells as described above but preincubated them with vanadate, a specific inhibitor of PTP activity. The results show (Figure 5) that in cells preincubated with vanadate the difference in PDGF-R phosphorylation between PDGF or PDGF/insulin-treated cells is lost. Together, these results strongly suggest an involvement of PTPs in the inhibitory action of insulin in PDGF-R activation.
Figure 5.
Effect of vanadate pretreatment on platelet-derived growth factor-R phosphorylation state after platelet-derived growth factor/insulin stimulation. Cells (1 × 106) were serum starved for 24 h pretreated for 1 min with 0.1 mM vanadate and then stimulated with 30 ng/ml platelet-derived growth factor and 10 nM insulin either separately or in combination. At the indicated times, cells were lysed in RIPA buffer, immunoprecipitated with anti-platelet–derived growth factor-R antibodies. Immunoblots were first probed with anti-phosphotyrosine antibodies and then the blot was stripped and reprobed with anti-platelet–derived growth factor-R antibodies. The result is representative of three independent experiments with similar results.
Analysis of PDGF-R–activated Pathways after PDGF/Insulin Costimulation
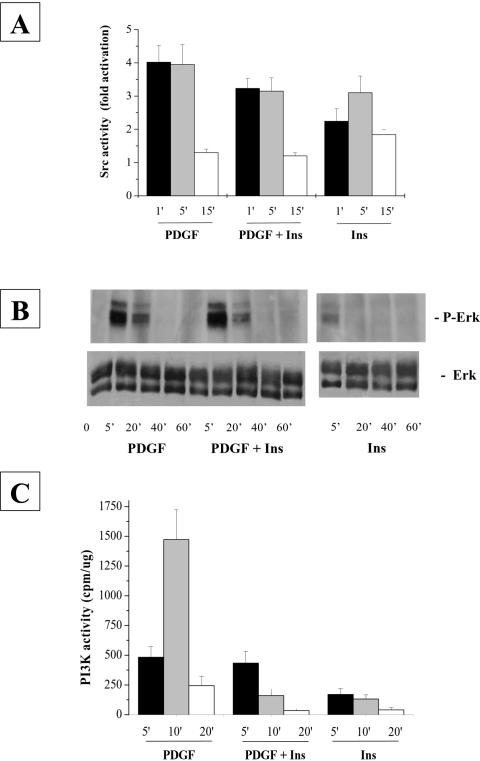
Insulin-dependent inhibition of platelet-derived growth factor-R phosphorylation is the cause of the reduced cell proliferation of NIH-3T3-IR cells simultaneously treated with the two hormones with respect to cells grown in the presence of platelet-derived growth factor alone. To investigate further the mechanism of this inhibition, we studied the downstream signaling pathways that arise from platelet-derived growth factor-R stimulation that are primarily involved in the induction of mitogenesis. First, we analyzed the activation of the intracellular tyrosine-kinase c-Src. Cells were treated with various growth factors in combination and then lysed at different times after stimulation. Immunoprecipitated Src from NIH-3T3-IR lysates was subjected to in vitro kinase assay by using enolase as substrate. The incorporated radioactivity of phosphorylated enolase was measured by scintillation counting. The results (Figure 6A) indicate that although platelet-derived growth factor induces a stronger activation of Src activity in NIH-3T3-IR with respect to insulin stimulation, costimulation with platelet-derived growth factor/insulin leads only to a slight decrease of Src activity with respect to the stimulation with platelet-derived growth factor alone. This effect, in our opinion, is not sufficient to justify the strong inhibition of insulin on platelet-derived growth factor-R–induced cell growth.
Figure 6.
Time course of Src kinase activity (A) and ERK phosphorylation (B) and PI-3K activity (C) after platelet-derived growth factor/insulin stimulation. (A) Src kinase activity assay: NIH-3T3-IR cells were serum starved for 24 h and then stimulated with 30 ng/ml platelet-derived growth factor and 10 nM insulin either separately or in combination. At the indicated times, cells were lysed in RIPA and immunoprecipitated with anti-Src antibodies and subjected to kinase assay as described in Materials and Methods. The result is representative of three independent experiments with similar results. (B) ERK phosphorylation: NIH-3T3-IR cells were treated as described above and lysed in RIPA buffer. ERK phosphorylation was determined resolving by 20 μg of lysates on a 12% SDS-PAGE. The proteins were transferred to PVDF membranes and probed with anti-phospho-ERK1/2 and anti-ERK1/2 antibodies The result is representative of three independent experiments with similar results. (C) PI-3K kinase activity assays: NIH-3T3-IR cells were treated as described above and lysed in RIPA buffer. PI-3K kinase activity was assayed as described in Material and Methods. The result is representative of three independent experiments with similar results.
Next, we studied the time course of ERK activation, which represents another important pathway for platelet-derived growth factor-R–mediated mitogenic signaling. NIH-3T3 cells were treated for the indicated times as described above, and an equal amount of each cell lysate was resolved on SDS-PAGE and probed with anti-phospho-ERK1–2 monoclonal antibodies. The results (Figure 6B) indicate that ERKs activation, which is proportional to their phosphorylation state, is higher in platelet-derived growth factor-stimulated cells than in cells treated with insulin, but the platelet-derived growth factor/insulin costimulation does not lead to a decrease of ERKs activation with respect to platelet-derived growth factor alone. Finally, we examined the time course of PI-3K activity. Cells were treated for the indicated times as described above and equal amounts of protein were immunoprecipitated using anti-phosphotyrosine antibodies. The activity of phosphorylated PI-3K was tested by an in vitro kinase assay by using phosphatidylinositol as substrate. Platelet-derived growth factor-R stimulation induces a much stronger PI-3K activation (Figure 6C) than insulin in NIH-3T3-IR cells. Surprisingly, simultaneous stimulation with platelet-derived growth factor and insulin results in a strong reduction of PI-3K activity (Figure 6C) compared with treatment with platelet-derived growth factor alone.
Together, our results indicate that the insulin-dependent inhibition of platelet-derived growth factor-induced mitogenesis is caused by the inhibition of platelet-derived growth factor-R phosphorylation. However, this fact does not result in a generic down-regulation of the platelet-derived growth factor-R signaling pathway but rather in a specific inhibition of the PI-3K pathway.
Analysis of Reactive Oxygen Species (ROS) Production after Platelet-derived Growth Factor/Insulin Stimulation
Ligation of a variety of cell surface receptors, including those for growth factors and cytokines, induces a transient production of H2O2 in mammalian cells (Sundaresan et al., 1995; Bae et al., 1997). This transient increase in intracellular H2O2 concentration leads to a reversible inactivation of PTPs (Lee et al., 1998; Chiarugi et al., 2001; Ostman and Bohmer, 2001) during the early phase of receptor signaling. This transient inhibition of PTP activity is necessary to reach the correct phosphorylation level of many proteins and hence to propagate the signaling started with growth factor engagement. Recently, Bae et al. (2000) have established that the activation of PI-3K is necessary for platelet-derived growth factor-induced hydrogen peroxide (H2O2) production. Our data indicate that NIH-3T3-IR cells costimulated with insulin and platelet-derived growth factor show a significant down-regulation of PI-3K activity with respect to cells stimulated with platelet-derived growth factor alone (Figure 5C). Analysis of H2O2 production after receptor activation (Figure 7) shows that the amount of H2O2 produced in cells simultaneously stimulated with both platelet-derived growth factor and insulin is lower than in cells treated with platelet-derived growth factor alone, as indicated by the reduced PI-3K activation in costimulated cells. The decreased H2O2 production in platelet-derived growth factor/insulin-costimulated cells compared with cells stimulated with platelet-derived growth factor alone could account for the reduced time course of platelet-derived growth factor-R phosphorylation in platelet-derived growth factor/insulin costimulation, because in this condition, PTP activity is less inhibited. This finding suggests that different levels of ROS production in platelet-derived growth factor/insulin versus platelet-derived growth factor-treated cells are responsible for the different PDFG-R phosphorylation level through a differential inhibitory action on PTPs (Figure 4).
Figure 7.
Time course of H2O2 production after platelet-derived growth factor/insulin stimulation. Cells were serum starved for 24 h and then incubated for the indicated times with 30 ng/ml platelet-derived growth factor and 10 nM insulin, either separately or in combination. Five minutes before the end of incubation time 2′,7′-dichlorofluorescin diacetate was added to a final concentration of 5 μM. Cells were then detached from the substrate by trypsinization and analyzed immediately by flow cytometry. The diagram reports the mean of the relative DFC fluorescence after 10 and 20 min from cell stimulation.
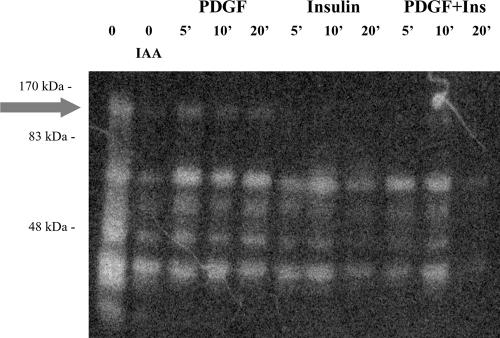
PTP Redox Pattern after Platelet-derived Growth Factor/Insulin Stimulation
To investigate further the relationship between platelet-derived growth factor/insulin stimulation and PTP inhibition, we performed an in gel PTP assay (Figure 8). NIH-3T3-IR cells were treated as described above and then lysed with 0.1 M acetate buffer, pH 5.5, containing 1% NP-40 and 5 mM iodoacetic acid. Equivalent amount of the samples were then subjected to SDS-PAGE onto a gel that was polymerized in the presence of a 32P-phosphorylated peptide. The gels were treated as described in Materials and Methods and then analyzed with a PhosphorImager apparatus. The white bands of Figure 8 represent PTPs in an oxidative/inactive state at the time of cell lysis. We observe that platelet-derived growth factor costimulation leads to an overall higher level of PTP inhibition compared with that of cells that were treated with platelet-derived growth factor and insulin together, confirming the results obtained with PTP assay (Figure 4). In addition, we have identified (Figure 8, arrow) a PTP of ∼120 kDa that is differentially redox-regulated in costimulation conditions. In fact, this PTP seems to be in an oxidative/inactive state during platelet-derived growth factor stimulation, whereas in cells cotreated with platelet-derived growth factor/insulin is in its reduced/active state. This PTP could therefore represent a major effector of insulin-mediated, platelet-derived growth factor-R phosphorylation inhibition.
Figure 8.
In gel tyrosine-phosphatases assay. Cells were serum starved for 24 h, incubated for the indicated times with 30 ng/ml platelet-derived growth factor and 10 nM insulin either separately or in combination, and then lysed in a buffer containing 10 mM iodoacetic acid as described in Materials and Methods. Total proteins (15 μg) for each sample were then loaded onto the radiolabeled gel. After the electrophoresis run the gels were sequentially washed for the times indicated at room temperature to allow protein renaturation. Gels were then dried and analyzed using a PhosphorImager system. The white bands of the gel represent the PTPs that have reacquired the catalytic activity. These PTPs are those that in vivo have been oxidized by ROS produced by RTK activation and hence that were not irreversibly modified by iodoacetic acid.
DISCUSSION
In physiological conditions, cells are continuously subjected to a variety of extracellular stimuli from soluble factors as well as from cell-cell and cell-substrate contact. The resulting cell response is the consequence of a complex integration of these different signals. In most cases, studies of cellular physiology have been done using very simplified models. Typically, responses of cells stimulated with a single growth factor are compared with those of serum-deprived cells. Under these conditions, a metabolic hormone such as insulin can induce a mitogenic response in many cell types.
In this work, we have chosen an experimental model in which two different growth factors, platelet-derived growth factor and insulin, are added simultaneously to NIH-3T3-IR cells and C2C12 cells. Both platelet-derived growth factor-R and IR belong to the RTKs family and share a common activation mechanism as well as most of the signaling transduction molecules, with the notable exception of the IRS proteins. In addition, both platelet-derived growth factor and insulin have been shown to be mitogenic for these cell lines (Chiarugi et al., 1995; Chiarugi et al., 1997; Wang et al., 1999). Our data confirm that both these hormones are able to induce mitogenesis in both NIH-3T3-IR and C2C12 cells, although insulin is a much less powerful mitogenic agent than platelet-derived growth factor (Figure 1). Surprisingly, when NIH-3T3-IR and C2C12 cells are stimulated simultaneously with platelet-derived growth factor and insulin, we observe a strong inhibition of cell growth rate with respect to that of cells treated with platelet-derived growth factor alone (Figure 1). In a recent work, Chiarugi et al. (2002) showed a correlation between the magnitude of platelet-derived growth factor-R phosphorylation, measured as the integral as a function of time of platelet-derived growth factor-R phosphorylation, and the strength of the mitogenic signaling elicited by this growth factor. This correlation is also found in the present case. In fact, our data show that the insulin-dependent inhibition of platelet-derived growth factor-induced cell growth is correlated with a premature down-regulation of platelet-derived growth factor-R phosphorylation (Figure 2).
The tyrosine phosphorylation level of platelet-derived growth factor-R depends on the relative activity of platelet-derived growth factor-R kinase and of PTPs. In addition, after stimulation platelet-derived growth factor-R undergoes internalization and ubiquitination, processes that both contribute to the decrease of platelet-derived growth factor-R concentration at the plasma membrane. In fact, ligand binding induces internalization of ligand–receptor complexes into endosomes (Sorkin et al., 1991). The platelet-derived growth factor-R/ligand complex then dissociates, and the receptor recycles to the cell membrane, or, alternatively, the ligand–receptor complex is degraded upon fusion of the endosomes with lysosomes. Platelet-derived growth factor-R also undergoes cytoplasmic degradation in proteasomes after ubiquitination (Mori et al., 1992, 1993, 1995).
To clarify the mechanism by which insulin induces a premature down-regulation of platelet-derived growth factor-R phosphorylation, we have first studied the relative platelet-derived growth factor-R tyrosine kinase activity in cells after stimulation with platelet-derived growth factor together with insulin or with platelet-derived growth factor alone (Figure 3A). Our results indicate that insulin does not interfere with the intrinsic platelet-derived growth factor-R kinase activity. In addition, neither the platelet-derived growth factor-R internalization rate (Figure 3B) nor the platelet-derived growth factor-R ubiquitination rate (Figure 3C) is significantly influenced by costimulation with insulin. Instead, Figure 4 shows that the total intracellular PTP activity is higher in costimulated cells compared with cells stimulated with platelet-derived growth factor alone.
The involvement of PTP regulation in insulin-mediated inhibition of platelet-derived growth factor-R activation also is confirmed by the evidence that in cells pretreated with vanadate, a specific inhibitor of PTPs, the difference in platelet-derived growth factor-R phosphorylation between platelet-derived growth factor or platelet-derived growth factor/insulin-stimulated cells is lost (Figure 5). In conclusion, the premature down-regulation of platelet-derived growth factor-R phosphorylation in platelet-derived growth factor/insulin-stimulated cells (Figure 2) could be explained by the higher PTP activity compared with that of cells treated with only with platelet-derived growth factor.
The reduced time-dependent platelet-derived growth factor-R phosphorylation in platelet-derived growth factor/insulin-costimulated cells is very likely the cause of the reduced growth rate of NIH-3T3-IR and C2C12 cells treated simultaneously with these two hormones respective to cells grown in the presence of platelet-derived growth factor alone. Chiarugi et al. (2002) have shown a correlation between the magnitude of platelet-derived growth factor-R phosphorylation and cell growth rate. In fact, under conditions in which platelet-derived growth factor-R phosphorylation is prolonged, they observed a shorter G1/S phase with respect to control cells (Chiarugi et al., 2002) and thus an increased cell growth rate. In our case, insulin induces an early down-regulation of platelet-derived growth factor-R phosphorylation and consequently inhibits cell growth.
Analysis of the major signaling pathways involved in platelet-derived growth factor-induced mitogenesis reveals that cells costimulated with insulin and platelet-derived growth factor show a remarkable reduction of PI-3K activity with respect to cells treated with platelet-derived growth factor alone (Figure 6C), whereas neither Src nor Ras pathways are influenced by the inhibitory action of insulin (Figure 6, A and B).
In recent work, Bae et al. (2000) have found that PI-3K activation is necessary for platelet-derived growth factor-induced H2O2 production. H2O2 is a small and diffusible molecule that acts as a versatile intracellular messenger because its intracellular concentration can be varied in response to external stimuli such as cytokines and peptide growth factors. Receptor-mediated generation of H2O2 has been linked to the activation of transcription factors (Schreck et al., 1991; Pahl and Baeuerle, 1994), mitogen-activated protein kinases (Sundaresan et al., 1995), phospholipase A2 (Zor et al., 1993), protein kinase C (Konishi et al., 1997), and phospholipase D (Min et al., 1998); and, in addition, it has been linked to the triggering of apoptosis (Jacobson, 1996) and to the inhibition of protein-tyrosine phosphatases (Lee et al., 1998; Chiarugi et al., 2001). In particular, it has been proposed by Lee et al. (1998) that H2O2 production during RTKs signaling could be essential in increasing the steady-state level of protein-tyrosine phosphorylation consequent to growth factor stimulation. The overall tyrosine-phosphorylation level of stimulated RTKs, as well as of other downstream signaling proteins, is not only due to the kinase activity of RTK itself but also to the concurrent inhibition of PTPs by H2O2 (Ostman and Bohmer, 2001). Hence, we measured the H2O2 level during stimulation with insulin, platelet-derived growth factor or platelet-derived growth factor/insulin costimulation. We find that insulin is able to reduce substantially the H2O2 production induced by platelet-derived growth factor stimulation (Figure 7), in agreement with the reduced PI-3K activity that we find in costimulated cells (Figure 6C). The mechanism by which, in platelet-derived growth factor/insulin costimulated cells, cross talk between platelet-derived growth factor-R and IR activation leads to a down-regulation of PI-3K activity is not yet clear. Levy-Toledano et al. (1995) report that, in NIH-3T3-IR cells, insulin stimulation competitively displaces the p85 subunit of PI-3K from its binding site on activated platelet-derived growth factor-R, possibly leading to a PI-3K inhibition with respect to cells stimulated with platelet-derived growth factor alone. More recently, Staubs et al., (1998) have shown that platelet-derived growth factor stimulation in 3T3-L1 fibroblast leads to inhibition of insulin-stimulated PI-3K activity by mean of a platelet-derived growth factor-induced serine-threonine phosphorylation of IRS1. Both these articles (Levy-Toledano et al., 1995; Staubs et al., 1998) demonstrate a cross talk between platelet-derived growth factor and insulin signaling pathways at the level of PI-3K activation, even if with substantial inconsistency that may be due both to the different cell types and/or to the different timing of the experiments. We now propose H2O2 production and PTP redox regulation as an additional mediator of platelet-derived growth factor-R/insulin-R cross talk. In fact, we have shown here that insulin inhibits PI-3K pathway/H2O2 production (Figures 6C and 7) in platelet-derived growth factor-stimulated cells. H2O2 produced during the early phases of RTK stimulation is responsible for the reversible down-regulation of PTP intracellular activity by inducing the oxidation of the reactive cysteine of the active site (Kamata and Hirata, 1999; Ostman and Bohmer, 2001; Meng et al., 2002). Our data are in agreement with this observation, because we have found that PTP activity is relatively higher in cells costimulated with platelet-derived growth factor/insulin than in cells stimulated with platelet-derived growth factor alone (Figure 4). We have found by means of in gel PTP assay that several PTPs are differentially oxidized during platelet-derived growth factor or platelet-derived growth factor/insulin stimulation (Figure 8). In particular, we have established that a PTP (Figure 8, arrow) of ∼120 kDa is present in its oxidized/inactive form during platelet-derived growth factor stimulation, and instead is reduced/active during platelet-derived growth factor/insulin costimulation. We can argue that this yet unidentified 120-kDa PTP is one of the main targets of differential regulation exerted by insulin costimulation in platelet-derived growth factor signaling.
In conclusion, we have studied a simplified model of combinatorial regulation of cell metabolism that has revealed an unexpected inhibitory role of insulin in platelet-derived growth factor-induced NIH-3T3 and C2C12 cell growth. We have found that this inhibitory effect of insulin is mediated by a down-regulation of platelet-derived growth factor-induced H2O2 production that leads to a relative up-regulation of PTP activity during platelet-derived growth factor/insulin costimulation that inhibits platelet-derived growth factor-R phosphorylation.
On the basis of our results, we can hypothesize that, at least for fibroblast and muscle cells, insulin is essentially a prosurvival/differentiating hormone, and hence insulin stimulation is able of counteract the proliferative signals elicited by serum-soluble growth factors such as platelet-derived growth factor. This work shows that the study of the effect of different growth factor can reveal new insights into the cooperative interactions involved in the regulation of cellular behavior.
Acknowledgments
This work was supported by Consiglio Nazionale delle Ricerche (target project on Biotechnology, Strategic project “Controlli posttrascrizionali dell'espressione genica”), in part by the Associazione Italiana Ricerca sul Cancro, and in part by the Ministero della Università e Ricerca Scientifica e Tecnologica (MIUR-PRIN 2000 and MURST-CNR Biotechnology program L.95/95), and Cassa di Risparmio di Firenze.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–01–0011. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-01-0011.
Abbreviations used: ERK, extracellular signal-regulated kinase; IR, insulin receptor; PDGF, platelet-derived growth factor, PDGF-R, platelet-derived growth factor-receptor; PI-3K, phosphatidylinositol 3-phosphate kinase; PTB, protein tyrosine-binding; PTK, protein tyrosine kinase; PTP, protein tyrosine phosphatase; ROS, reactive oxygen species; SH2, Src homology-2.
References
- Bae, Y. S., Kang, S. W., Seo, M. S., Baines, I. C., Tekle, E., Chock, P. B., and Rhee, S. G. (1997). Epidermal growth factor (EGF)-induced generation of hydrogen peroxide. Role in EGF receptor-mediated tyrosine phosphorylation. J. Biol. Chem. 272, 217-221. [PubMed] [Google Scholar]
- Bae, Y. S., Sung, J. Y., Kim, O. S., Kim, Y. J., Hur, K. C., Kazlauskas, A., and Rhee, S. G. (2000). Platelet-derived growth factor-induced H(2)O(2) production requires the activation of phosphatidylinositol 3-kinase. J. Biol. Chem. 275, 10527-10531. [DOI] [PubMed] [Google Scholar]
- Burridge, K. and Nelson, A. (1995). An in-gel assay for protein tyrosine phosphatase activity: detection of widespread distribution in cells and tissue. Anal. Biochem. 232, 56-64. [DOI] [PubMed] [Google Scholar]
- Ceresa, B. P., Kao, A. W., Santeler, S. R., and Pessin, J. E. (1998). Inhibition of clathrin-mediated endocytosis selectively attenuates specific insulin receptor signal transduction pathways. Mol. Cell Biol. 18, 3862-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarugi, P., Cirri, P., Marra, F., Raugei, G., Camici, G., Manao, G., and Ramponi, G. (1997). LMW-PTP is a negative regulator of insulin-mediated mitotic and metabolic signalling. Biochem. Biophys. Res. Commun. 238, 676-682. [DOI] [PubMed] [Google Scholar]
- Chiarugi, P., Cirri, P., Raugei, G., Camici, G., Dolfi, F., Berti, A., and Ramponi, G. (1995). platelet-derived growth factor receptor as a specific in vivo target for low M(r) phosphotyrosine protein phosphatase. FEBS Lett. 372, 49-53. [DOI] [PubMed] [Google Scholar]
- Giannoni, E., Camici, G., Raugei, G., and Ramponi, G. (2002). New perspectives in platelet-derived growth factor receptor downregulation: the main role of phosphotyrosine phosphatases. J. Cell Sci. 115, 2219-2232. [DOI] [PubMed] [Google Scholar]
- Chiarugi, P., Fiaschi, T., Taddei, M. L., Talini, D., Giannoni, E., Raugei, G., and Ramponi, G. (2001). Two vicinal cysteines confer a peculiar redox regulation to low molecular weight protein tyrosine phosphatase in response to platelet-derived growth factor receptor stimulation. J. Biol. Chem. 276, 33478-33487. [DOI] [PubMed] [Google Scholar]
- Choudhury, G. G., Wang, L. M., Pierce, J., Harvey, S. A., and Sakaguchi, A. Y. (1991). A mutational analysis of phosphatidylinositol-3-kinase activation by human colony-stimulating factor-1 receptor. J. Biol. Chem. 266, 8068-8072. [PubMed] [Google Scholar]
- Claesson-Welsh, L., Eriksson, A., Westermark, B., and Heldin, C. H. (1989). cDNA cloning and expression of the human A-type platelet-derived growth factor receptor establishes structural similarity to the B-type platelet-derived growth factor receptor. Proc. Natl. Acad. Sci. USA 86, 4917-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldin, C. H., and Westermark, B. (1999). Mechanism of action and in vivo role of platelet-derived growth factor. Physiol. Rev. 79, 1283-1316. [DOI] [PubMed] [Google Scholar]
- Hershko, A., and Ciechanover, A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67, 425-479. [DOI] [PubMed] [Google Scholar]
- Jacobson, M. D. (1996). Reactive oxygen species and programmed cell death. Trends Biochem. Sci. 21, 83-86. [PubMed] [Google Scholar]
- Jiang, G., and Hunter, T. (1999). Receptor signaling: when dimerization is not enough. Curr. Biol. 9, R568-R571. [DOI] [PubMed] [Google Scholar]
- Kamata, H., and Hirata, H. (1999). Redox regulation of cellular signalling. Cell Signal. 11, 1-14. [DOI] [PubMed] [Google Scholar]
- Konishi, H., Tanaka, M., Takemura, Y., Matsuzaki, H., Ono, Y., Kikkawa, U., and Nishizuka, Y. (1997). Activation of PKC by tyrosine phosphorylation in response to H2O2. Proc. Natl. Acad. Sci. USA 94, 11233-11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. R., Kwon, K. S., Kim, S. R., and Rhee, S. G. (1998). Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with EGF. J. Biol. Chem. 273, 15366-15372. [DOI] [PubMed] [Google Scholar]
- Lemmon, M. A., and Schlessinger, J. (1994). Regulation of signal transduction and signal diversity by receptor oligomerization. Trends Biochem. Sci. 19, 459-463. [DOI] [PubMed] [Google Scholar]
- Levy-Toledano, R., Blaettler, D. H., LaRochelle, W. J., and Taylor, S. I. (1995). Insulin-induced activation of phosphatidylinositol (PI) 3-kinase. Insulin-induced phosphorylation of insulin receptors and insulin receptor substrate-1 displaces phosphorylated platelet-derived growth factor receptors from binding sites on PI 3-kinase. J. Biol. Chem. 270, 30018-30022. [DOI] [PubMed] [Google Scholar]
- Liu, F., and Roth, R. A. (1998). Binding of SH2 containing proteins to the insulin receptor: a new way for modulating insulin signalling. Mol. Cell Biochem. 182, 73-78. [PubMed] [Google Scholar]
- Meng, T. C., Fukada, T., and Tonks, N. K. (2002). Reversible oxidation and inactivation of protein tyrosine phosphatases in vivo. Mol. Cell 9, 387-399. [DOI] [PubMed] [Google Scholar]
- Min, D. S., Kim, E. G., and Exton, J. H. (1998). Involvement of tyrosine phosphorylation and PKC in the activation of phospholipase D by H2O2 in Swiss 3T3 fibroblasts. J. Biol. Chem. 273, 29986-29994. [DOI] [PubMed] [Google Scholar]
- Mori, S., Heldin, C. H., and Claesson-Welsh, L. (1992). Ligand-induced polyubiquitination of the platelet-derived growth factor beta-receptor. J. Biol. Chem. 267, 6429-6434. [PubMed] [Google Scholar]
- Mori, S., Heldin, C. H., and Claesson-Welsh, L. (1993). Ligand-induced ubiquitination of the platelet-derived growth factor beta-receptor plays a negative regulatory role in its mitogenic signaling. J. Biol. Chem. 268, 577-583. [PubMed] [Google Scholar]
- Mori, S., Tanaka, K., Omura, S., and Saito, Y. (1995). Degradation process of ligand-stimulated platelet-derived growth factor beta-receptor involves ubiquitin-proteasome proteolytic pathway. J. Biol. Chem. 270, 29447-29452. [DOI] [PubMed] [Google Scholar]
- Ogawa, W., Matozaki, T., and Kasuga, M. (1998). Role of binding proteins to IRS-1 in insulin signalling. Mol. Cell Biochem. 182, 13-22. [PubMed] [Google Scholar]
- Ostman, A., and Bohmer, F. D. (2001). Regulation of receptor tyrosine kinase signaling by protein tyrosine phosphatases. Trends Cell Biol. 11, 258-266. [DOI] [PubMed] [Google Scholar]
- Pahl, H. L., and Baeuerle, P. A. (1994). Oxygen and the control of gene expression. Bioessays 16, 497-502. [DOI] [PubMed] [Google Scholar]
- Pronk, G. J., McGlade, J., Pelicci, G., Pawson, T., and Bos, J. L. (1993). Insulin-induced phosphorylation of the 46- and 52-kDa Shc proteins. J. Biol. Chem. 268, 5748-5753. [PubMed] [Google Scholar]
- Schlessinger, J. (2000). Cell signaling by receptor tyrosine kinases. Cell 103, 211-225. [DOI] [PubMed] [Google Scholar]
- Schreck, R., Rieber, P., and Baeuerle, P. A. (1991). Reactive oxygen intermediates as apparently widely used messengers in the activation of the NF-kappa B transcription factor and HIV-1. EMBO J 10, 2247-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skolnik, E. Y., Lee, C. H., Batzer, A., Vicentini, L. M., Zhou, M., Daly, R., Myers, M. J., Jr., Backer, J. M., Ullrich, A., and White, M. F. (1993). The SH2/SH3 domain-containing protein GRB2 interacts with tyrosine-phosphorylated IRS1 and Shc: implications for insulin control of ras signalling. EMBO J. 12, 1929-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin, A., Westermark, B., Heldin, C. H., and Claesson-Welsh, L. (1991). Effect of receptor kinase inactivation on the rate of internalization and degradation of platelet-derived growth factor and the platelet-derived growth factor beta-receptor. J. Cell Biol. 112, 469-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staubs, P. A., Nelson, J. G., Reichart, D. R., and Olefsky, J. M. (1998). Platelet-derived growth factor inhibits insulin stimulation of insulin receptor substrate-1-associated phosphatidylinositol 3-kinase in 3T3–L1 adipocytes without affecting glucose transport. J. Biol. Chem. 273, 25139-25147. [DOI] [PubMed] [Google Scholar]
- Sun, X. J., Crimmins, D. L., Myers, M. G., Jr., Miralpeix, M., and White, M. F. (1993). Pleiotropic insulin signals are engaged by multisite phosphorylation of IRS-1. Mol. Cell Biol. 13, 7418-7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan, M., Yu, Z. X., Ferrans, V. J., Irani, K., and Finkel, T. (1995). Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science 270, 296-299. [DOI] [PubMed] [Google Scholar]
- Tobe, K., et al. (1996). Csk enhances insulin-stimulated dephosphorylation of focal adhesion proteins. Mol. Cell Biol. 16, 4765-4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Dai, H., Yousaf, N., Moussaif, M., Deng, Y., Boufelliga, A., Swamy, O. R., Leone, M. E., and Riedel, H. (1999). Grb10, a positive, stimulatory signaling adapter in platelet-derived growth factor BB-, insulin-like growth factor I-, and insulin-mediated mitogenesis. Mol. Cell Biol. 19, 6217-6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, M. F. (1998). The IRS-signaling system: a network of docking proteins that mediate insulin and cytokine action. Recent Prog. Horm. Res. 53, 119-138. [PubMed] [Google Scholar]
- Zor, U., Ferber, E., Gergely, P., Szucs, K., Dombradi, V., and Goldman, R. (1993). Reactive oxygen species mediate phorbol ester-regulated tyrosine phosphorylation and phospholipase A2 activation: potentiation by vanadate. Biochem. J. 295, 879-888. [DOI] [PMC free article] [PubMed] [Google Scholar]