Abstract
Although it is known that the spatial coordination of Rac and Rho activity is essential for cell migration, the molecular mechanisms regulating these GTPases during migration are unknown. We found that the expression of constitutively activated R-Ras (38V) blocked membrane protrusion and random migration. In contrast, expression of dominant negative R-Ras (41A) enhanced migrational persistence and membrane protrusion. Endogenous R-Ras is necessary for cell migration, as cells that were transfected with siRNA for R-Ras did not migrate. Expression of R-Ras (38V) decreased Rac activity and increased Rho activity around the entire cell periphery, whereas expression of dominant negative R-Ras (41A) showed the converse, suggesting that R-Ras can spatially activate Rho and inactivate Rac. Consistent with this role, endogenous R-Ras localized and was preferentially activated at the leading edge of migratory cells in response to adhesion. The effects of R-Ras on cell migration are mediated by PI3-Kinase, as an effector mutant that uncouples PI3-Kinase binding from R-Ras (38V) rescued migration. From these data, we hypothesize that R-Ras plays a key role in cell migration by locally regulating the switch from Rac to Rho activity after membrane protrusion and adhesion.
INTRODUCTION
Cell migration is an essential event in several physiological processes. During cancer progression, it is the ability of a cell to metastasize to other areas of the body that causes 90% of cancer deaths (Sporn, 1996). Most work characterizing cell migration has been done in fibroblasts, and relatively less is known about epithelial cell migration. Although epithelial cells do migrate, they do so more slowly than fibroblasts (Walpita and Hay, 2002). Because human cancers are typically epithelial in origin, it is important to elucidate the mechanisms driving epithelial cell motility.
Members of the Rho family of GTPases are emerging as key regulators of cell migration. Specifically, Rac activity is increased at the leading edge of a migrating cell (Kraynov et al., 2000). This activity drives the actin polymerization that underlies lamellipodia formation and subsequent forward protrusions (Raftopoulou and Hall, 2004). Rac activity also directs the formation of focal complexes (Rottner et al., 1999) that provide the traction force needed to tether the cell to the extracellular matrix (ECM) during the contractile events of migration (Beningo et al., 2001; Munevar et al., 2001). In contrast, Rho activity has been correlated with decreased protrusion and migration (Arthur and Burridge, 2001; Wang et al., 2003; Worthylake and Burridge, 2003). In addition, in leukocytes and monocytes, Rho and its effector, ROCK, are necessary for rear detachment (Alblas et al., 2001; Worthylake et al., 2001). As the cell migrates forward, Rac-dependent focal complexes mature into Rho-dependent focal adhesions that provide the cell with anchorage behind the lamellipodium (Rottner et al., 1999). However, a fundamental unanswered question is, what are the exact molecular mechanisms coordinating Rac and Rho activities? Specifically, it is not known what lies upstream of these GTPases to control when each is spatially and temporally activated or inactivated during migration.
Because integrins can activate both Rho and Rac (Giancotti and Ruoslahti, 1999) and are needed to form new contacts to the substrate during migration, integrins are proposed to play an important regulatory role in cell migration (Huttenlocher et al., 1995). Because transformation of cells by the small GTPase R-Ras increases integrin affinity and avidity (Zhang et al., 1996; Sethi et al., 1999) and enhances focal adhesion formation (Kwong et al., 2003), we hypothesized that R-Ras may also be an important regulator of cell migration.
R-Ras is a member of the Ras superfamily of small GTPases and shares several effectors with H-, N-, and K-Ras, including PI3-Kinase, RalGDS, and Raf. Despite this similarity, R-Ras has biological functions distinct from classic Ras. Notably, H-Ras inhibits integrin function (Hughes et al., 1997; Kinbara et al., 2003), whereas R-Ras enhances integrin function (Zhang et al., 1996; Sethi et al., 1999; Kwong et al., 2003). We investigated the role of R-Ras in cell motility. Here we present evidence that R-Ras plays a key role in cell migration by modulating Rho and Rac activities in order to regulate membrane protrusion and migration.
MATERIALS AND METHODS
Tissue Culture
T47D cells were obtained from ATCC and maintained as previously described (Keely et al., 1999). T47D R-Ras (38V), (41A), and (38V/61S) cells were generated as pools of stable transfectants and have been previously described (Keely et al., 1999; Kwong et al., 2003).
R-Ras expression was knocked down by transient transfection of oligos directed against R-Ras (short interfering RNA [siRNA]). The oligos were made to target 5′-CCG GAA ATA CCA GGA ACA AGA-3′ (sense). A nonspecific control pool of 4 oligo duplexes was also provided by Dharmacon (Boulder, CO). Untagged oligos were made by Dharmacon and oligos tagged on the 3′ end with Alexa Fluor 488 were made by Qiagen (Valencia, CA).
Time-lapse Microscopy and Kymography
Subconfluent control T47D cells or cells expressing activated R-Ras (38V), dominant negative R-Ras (41A), or an effector mutant of activated R-Ras (38V/61S), were harvested in 0.5 mM EDTA in phosphate-buffered saline (PBS) and resuspended in 50 mM HEPES in T47D media (RPMI with l-glutamine, 0.2% insulin, 10% fetal bovine serum), pH 7.4. Cells were then plated on collagen-coated (0.1–300 μg/ml) or fibronectin-coated (3–30 μg/ml) Petri plates for 1 h. For inhibitor studies, inhibitors were added to the cells 10 min before time-lapse sequences were acquired and left on for the duration of the experiment. Inhibitors used were 10 μg/ml C3 exoenzyme (purified in bacteria) and 10 μM Y27632 (Calbiochem, San Diego, CA). For siRNA transfection, cells were transfected with 3 nM R-Ras siRNA using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. For other transfection studies, cells were transfected with constructs encoding GPF (eGFP C1 MCS vector, Clontech, Palo Alto, CA), GFP Rac (17N), or GFP Rac (61L) using Fugene (Roche, Indianapolis, IN). The transfection was performed according to the manufacturerer's instructions. All time-lapse images were taken 48 h after transfection.
Time-lapse images were acquired at 37° with a 20× objective on a TE300 Nikon inverted microscope (Garden City, NY) equipped with a Photometrics CoolSnap fx CCD camera (Tucson, AZ) and The Cube temperature controller (Life Imaging Services, Reinach, Switzerland). Images were captured with E-See Inovision Software (Inovision, Raleigh, NC), with one image taken per minute, for 90 min. For quantitating relative migration on varying collagen concentrations, the number of cells migrating in a field was counted and this number expressed as a percentage of migrating cells/total number of cells. For quantitating speed and persistence, 20 individual cells from 4 separate experiments were analyzed using I-See software (Inovision) and speed determined with Nanotrack (Inovision). Persistence was determined using the random walk equation (Maheshwari and Lauffenburger, 1998).
Kymography was performed using a 40× objective and images were collected with Metamorph software (Universal Imaging Corp, West Chester, PA), with one image taken every 3 s, for 10 min. Kymography was performed and protrusive velocity calculated as described (Bear et al., 2002). Statistical analysis was performed using Prism GraphPad software (San Diego, CA).
Immunofluorescence
Immunofluorescence was performed as previously described (Kwong et al., 2003). Antibodies used were anti-p16 (a generous gift from Dr. William Bement, 1:400 dilution), anti-FAK phosphorylated at Y397 (Biosource, Camarillo, CA; 1:100), anti-R-Ras (Santa Cruz Biotechnology, Santa Cruz, CA; 1:100), and anti-PI3-Kinase (Santa Cruz Biotechnology, 1:200). Staining for Rho and Rac activity using RBD:GST and PBD:GST is described in the text. Briefly, PBD:GST and RBD:GST were purified in bacteria and eluted off GS beads. Eluted protein, 100 μg, was incubated with cells overnight at 4°C. Anti-GST (Amersham Biosciences, Piscataway, NJ; 1:1000) and anti-goat FITC (Santa Cruz Biotechnology, 1:100) were then used to visualize active Rac and Rho. Immunostaining was analyzed by epifluorescence and images collected and deconvolved using E-See software (Inovision). For quantitation of cells based on p16 staining, 100 cells from three individual experiments were counted and assigned “polar” (p16 staining along one side of the cell), “nonpolar” (p16 staining along the entire periphery of the cell), or “round” (cells that were not spread). Statistical analysis was performed using Prism GraphPad software.
Rho, Rac, and R-Ras Activity Assays
Subconfluent T47D cells were harvested in 0.5 mM EDTA in PBS, resuspended in 5 mg/ml fatty acid–free bovine serum albumin (MP Biomedicals, Irvine, CA) in RPMI, and equal numbers of cells plated on collagen-coated Petri plates (3 μg/ml collagen) for 45 min. For inhibitor studies, cells were pretreated with 10 μM Y27632 for 15 min at 37°C before collagen stimulation. Cells were lysed and RBD-GST and PBD-GST pulldown assays were performed as previously described (Wozniak et al., 2003). Samples were run on a SDS-PAGE gel and transferred to a PVDF membrane, and the membrane was probed with anti-Rho (Santa Cruz, 1:250) or anti-Rac (1:500) followed by anti-mouse HRP (Jackson ImmunoResearch Laboratories, West Grove, PA; 1:5000) and ECL substrate (Amersham Biosciences).
For R-Ras activity assays, GST:Raf RBD was purified in bacteria and glutathione eluted from glutathione beads. Cells were lysed in an equal volume of 2× lysis buffer (50 mM HEPES, 150 mM NaCl, 2 mM MgCl, 2% NP-40, 0.5% deoxycholate, 2 mM EDTA, 2 mM NaF, 0.2% SDS, 1 mM sodium vanadate, and protease inhibitor cocktail; Sigma) containing 50 μg eluted GST:Raf RBD. Lysates were cleared by centrifugation, and glutathione beads were added to the lysates for 30 min, at 4°C, with rotating. The beads were then washed three times in lysis buffer without detergents, and Laemmli sample buffer added. Samples were run on a SDS-PAGE gel and transferred to a PVDF membrane, and the membrane was probed with anti-R-Ras (Santa Cruz, 1:1000 or PharMingen, San Diego, CA; 1:1000) followed by anti-rabbit HRP (Jackson ImmunoResearch Laboratories, 1:5000).
For activity assays using pseudopodial and cell body extracts, Cos7 cells were allowed to extend into 3.0-μm pores, and pseudopodia and cell bodies were isolated as previously described (Cho and Klemke, 2002). Fresh lysates were used, and the assay was performed as described above. After the experiment was performed, the membrane was stripped and reprobed for anti-ERK (Upstate Biotechnology, Lake Placid, NY; 1:1000) as a loading control. R-Ras and ERK were detected using ECL substrate (Amersham Biosciences).
For quantitation, the bands from three to four individual experiments (pairs of active and total Rho, Rac or R-Ras) were quantitated using ImageQuant software (Molecular Dynamics, Sunnyvale, CA). These data were statistically analyzed using Prism GraphPad software.
Online Supplementary Material
Seven videos demonstrating T47D cell migration on collagen. Supplementary Videos 1, 2, and 3 show the random migration of T47D cells stably expressing control plasmid, constitutively activated R-Ras (38V), or dominant negative R-Ras (41A), respectively. Supplementary Video 4 shows the random migration of T47D cells transiently transfected with a pool of four control siRNA oligos. Supplementary Video 5 shows the migration of T47D cells transfected with Alexa Fluor–labeled siRNA to knockdown R-Ras expression. Supplementary Videos 6 and 7 show the random migration of T47D cells stably expressing constitutively activated R-Ras (38V) upon Rho inhibition with C3 exoenzyme, or ROCK inhibition with Y27632, respectively.
RESULTS
R-Ras Regulates Random Migration across Collagen
We recently demonstrated that expression of constitutively activated R-Ras (38V) increased the migration of T47D breast epithelial cells in haptotactic transwell assays in which the underside of the filter was coated with collagen (Keely et al., 1999). This was also observed in cervical epithelial cells expressing activated R-Ras (87L; Rincon-Arano et al., 2003). However, in these assays, a directional signal is provided such that cells migrate toward collagen, making this migration different from random migration. Because we wanted to examine the effects of R-Ras on random migration, we determined the optimal collagen concentration for T47D cell migration. Control T47D cells were plated on Petri plates coated with varying concentrations of collagen (0.1–300 μg/ml), and random migration was determined by time-lapse video microscopy. Cell morphology was observed during migration and motility analyzed by counting the number of cells that migrated in the observation field. Because ∼100% of the cells in the field migrated when plated on 3 μg/ml (Figure 1, solid line), this concentration was used for the remaining time-lapse movies. Although most cells migrated on both low and high concentrations of collagen (Figure 1), cells plated on increased collagen concentrations showed an enhancement of long cell tails and slower migration (unpublished data).
Figure 1.
Determination of the optimum collagen concentration for T47D random cell migration. Control T47D cells and cells expressing activated R-Ras (38V) were plated on various concentrations of collagen-coated Petri plates for 1 h. Time-lapse microscopy sequences were acquired for 30 min, with one image collected per minute. The movies were then analyzed by counting the number of cells migrating in the field and relative migration expressed as the number of migrating cells/total number of cells. Altering collagen concentration had no effect on cell migration by R-Ras (38V)-expressing cells. Control cells, solid line; R-Ras (38V)-expressing cells, dashed line.
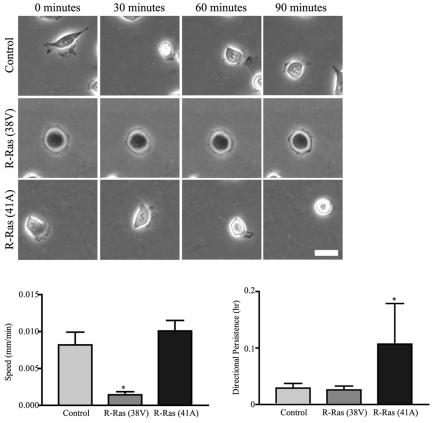
Next, control cells, cells stably expressing constitutively activated R-Ras (38V) or cells stably expressing dominant negative R-Ras (41A) were plated on collagen (3 μg/ml), and random migration was observed. Control cells were motile, forming lamellipodia at the leading edge and contracting their cell bodies as they migrated across the collagen substratum (Figure 2, Supplementary Video 1). In contrast, the expression of activated R-Ras (38V) blocked random migration. Although R-Ras (38V)-expressing cells ruffled along their periphery, they were unable to form stable forward protrusions or translocate their cell bodies (Figure 2, Supplementary Video 2). Cells expressing dominant negative R-Ras (41A) also showed extensive ruffling, yet these cells were able to migrate across collagen (Figure 2, Supplementary Video 3). This migration was not the same as control cells, however, because dominant negative R-Ras (41A)-expressing cells migrated with a slightly enhanced speed and a significantly increased persistence compared with control cells (Figure 2). These results demonstrate that misregulation of R-Ras alters cell migration.
Figure 2.
R-Ras regulates random cell migration across collagen. Control T47D cells, cells stably expressing activated R-Ras (38V), or cells stably expressing dominant negative R-Ras (41A) were plated on collagen-coated Petri plates (3 μg/ml) for 1 h. Time-lapse microscopy sequences were acquired for 90 min, with one image collected per minute (see Supplementary Videos 1, 2, and 3). Shown here is a representative cell at 0, 30, 60, and 90 min during the observation period. Scale bar, 25 μm. Expression of activated R-Ras significantly (*p < 0.05 vs. control) decreases cell speed, whereas expression of dominant negative R-Ras (41A) increases (p = 0.2) speed. Twenty individual cells from three experiments were tracked using a cell tracking program (Nanotrack, Inovision Software) and cell speed (mm/min) was calculated using the random walk equation (Maheshwari and Lauffenburger, 1998). Expression of dominant negative R-Ras (41A) increases directional persistence (*p < 0.05 vs. control).
Knockdown of R-Ras Expression Blocks Random Cell Migration
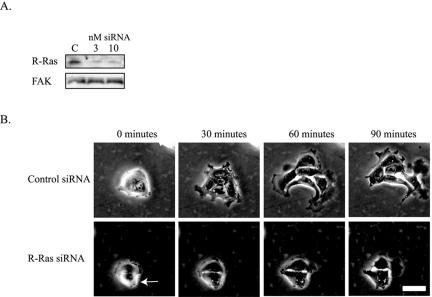
To determine if endogenous R-Ras plays a role in cell migration, T47D cells were transiently transfected with short interfering RNA oligos (siRNA) directed against R-Ras. To assess the knockdown efficiency of this siRNA oligo, T47D cells were transfected, grown for 2 d, lysed, and R-Ras expression was determined. Cells transfected with 3 or 10 nM siRNA showed decreased endogenous R-Ras protein compared with cells transfected with a pool of nonspecific siRNA (Figure 3A). T47D cell migration on collagen (3 μg/ml) was then analyzed using time-lapse microscopy. Cells transfected with a pool of control, nonspecific oligos were able to migrate (Figure 3B, Supplementary Video 4), although they did so in groups of cells, because transfection causes these cells to group together. However, cell migration was blocked when 3 nM R-Ras was transfected into the cells (Figure 3B, Supplementary Video 5). These data suggest that endogenous R-Ras plays a key role in regulating cell migration.
Figure 3.
Transfection of siRNA toward R-Ras blocks cell migration. (A) T47D cells were transfected with a pool of four nonspecific control siRNAs (labeled C) or with 3 or 10 nM siRNA directed against R-Ras. After 2 days, the cells were lysed and immunoblotted for R-Ras or for FAK as a control. The R-Ras siRNA knocks down the majority of endogenous R-Ras. (B) Two days after transfection of 3 nM control or R-Ras siRNA, T47D cells were plated on collagen (3 μg/ml) for 1 h, and then time-lapse microscopy sequences were acquired for 90 min, with one image taken per minute (see Supplementary Videos 4 and 5). Shown is a representative cell. The arrow indicates which cell was transfected with R-Ras siRNA, which was determined by using fluorescently labeled Alexa-Fluor oligos. Scale bar, 25 μm.
R-Ras Regulates Protrusive Activity during Migration
Because cells expressing activated R-Ras appeared unable to protrude and migrate (Figure 2) and R-Ras was necessary for migration (Figure 3), we analyzed cell protrusion using kymography (Hinz et al., 1999; Bear et al., 2002). Kymography is a technique used to analyze the activity of a single point along the cell membrane. Because only one section of the membrane is analyzed over the time interval, this method allows the visualization of lamellipodial protrusion and retraction dynamics. Cells were plated on collagen-coated plates (3 μg/ml) and time-lapse sequences were acquired, collecting images every 3 s for 10 min. To generate the kymograph, a 1-pixel-wide line was drawn along a protrusion event. Individual lines were then stacked in series, creating a kymograph with the x-axis representing time and the y-axis representing distance. Analysis of control cells shows they protruded and retracted continuously during migration and advanced within 10 min (Figure 4). In contrast, cells expressing activated R-Ras (38V) showed greatly diminished protrusion and retraction, forming only small ruffles and migrating little over the 10-min observation period (Figure 4). Cells expressing dominant negative R-Ras (41A) formed mainly stable protrusions with few retractions, migrating further within 10 min than control cells (Figure 4). Protrusion velocity was not greater than control cells (Figure 4), but rather dominant negative R-Ras (41A)-expressing cells protruded more and retracted less, consistent with their increased persistence (Figure 2). These results suggest that R-Ras regulates protrusion during migration.
Figure 4.
R-Ras modulates protrusive activity during cell migration. Control cells protrude and retract, whereas cells expressing activated R-Ras (38V) show little protrusive activity. Cells expressing dominant negative R-Ras (41A) protrude continuously, consistent with the observed increased persistence. T47D cells were plated on collagen (3 μg/ml) for 1 h and then time-lapse microscopy sequences were acquired for 10 min, with one image collected every 3 s. A one pixel-wide line was drawn along a protrusion in order to generate a kymograph using Metamorph software (Bear et al., 2002). Fifty protrusions from two separate experiments were analyzed, per cell type, and protrusive velocity was then calculated as previously described (Bear et al., 2002), *p < 0.05 versus control. Scale bars: x, 2.5 min; y, 15 μm.
R-Ras enhances integrin avidity and cell adhesion (Zhang et al., 1996; Sethi et al., 1999; Kwong et al., 2003), both of which affect random cell migration (Palecek et al., 1997, Cox et al., 2001). To address the possibility that R-Ras inhibits protrusion and migration through effects on cell adhesion, we plated cells expressing activated R-Ras (38V) on various collagen concentrations. Lower ECM concentrations have been shown to enhance cell migration by reducing integrin-mediated adhesion, whereas higher concentrations decrease motility (Huttenlocher et al., 1996; Palecek et al., 1997; Cox et al., 2001). However, altering collagen concentration from 0.1 to 300 μg/ml could not rescue the migratory defect in R-Ras (38V)-expressing cells (Figure 1, dashed line). In addition, we recently found that R-Ras enhances adhesion and haptotactic migration on collagen, but not on fibronectin (Keely et al., 1999). We reasoned that because R-Ras does not enhance adhesion and migration on fibronectin, R-Ras (38V)-expressing cells plated on fibronectin should be able to migrate if R-Ras is affecting migration solely through enhancing α2β1 integrin-mediated adhesion. However, R-Ras (38V)-expressing cells were also unable to migrate on fibronectin-coated plates (3, 10, or 20 μg/ml; unpublished data). These results suggest that R-Ras may regulate membrane protrusion and random migration in a manner that is separate from its effects on strengthening integrin-mediated adhesion.
R-Ras Regulates the Establishment of Cellular Polarity
Cellular protrusions arise from an ability of the cell to locally polymerize actin in a polarized manner. To determine if R-Ras regulates local actin dynamics, we compared the polarization of control cells and cells expressing constitutively activated (38V) and dominant negative (41A) R-Ras. When cells migrate, they polarize by forming lamellipodial protrusions driven by the force of actin polymerization (Welch et al., 1997; Svitkina and Borisy, 1999). The cytoskeleton in lamellipodia is organized as a dense, branching actin network. This organization of actin is mediated, in part, by the Arp2/3 complex, which regulates actin nucleation (Mullins and Pollard, 1999; Svitkina and Borisy, 1999; Welch, 1999).
Control cells and cells expressing activated or dominant negative R-Ras were costained for p16, a subunit of the Arp2/3 complex (Welch, 1999), and actin in order to determine if R-Ras regulates cytoskeletal polarity. In control cells, p16 localized to areas of dense actin networks at the leading edge of the lamellipodia (Figure 5). Cells expressing dominant negative R-Ras (41A) also localized p16 to lamellipodia, yet these cells often had an increase in p16 compared with control cells and had an exaggerated actin network at the leading edge (Figure 5). In contrast, p16 was very faint at the leading edge of R-Ras (38V)-expressing cells, which predominantly form stress fibers (Figure 5). We quantified the localization of p16 by classifying cells as localizing p16 in a polar manner (along one side of the cell), nonpolar (all along the periphery), or diffuse. Expression of R-Ras (38V) significantly decreased the number of polar cells, while expression of dominant negative R-Ras (41A) significantly increased the number of polar cells (Figure 5). These results demonstrate that misregulation of R-Ras alters the polarization of the actin cytoskeleton during cell migration.
Figure 5.
R-Ras regulates cell polarity. T47D cells were plated on collagen-coated coverslips (3 μg/ml) for 45 min and then coimmunostained for p16, a component of the arp2/3 complex, and actin. The merge shows actin in green and p16 in red. Control cells localize p16 and a rich actin network to the leading edge. This is enhanced in cells expressing dominant negative R-Ras (41A) and diminished in cells expressing activated R-Ras (38V). Scale bar, 25 μm. One hundred cells from three individual experiments were counted, and p16 staining was scored as polar, nonpolar, or diffuse. The percent of polar cells ± SEM are graphed. Expression of activated R-Ras (38V) significantly (*p < 0.05) decreases the number of polar cells, whereas expression of dominant negative R-Ras (41A) significantly (**p < 0.05) increases the number of polar cells.
R-Ras Regulates Rac and Rho Activity
Because the expression of activated R-Ras (38V) disrupted membrane protrusion and migration, we sought to determine the signals downstream of R-Ras that regulate these processes. The Rho family of GTPases are important regulators of focal adhesion assembly, cytoskeletal rearrangement, and motility. Because expression of activated or dominant negative R-Ras affected migration, we analyzed whether R-Ras can modulate Rho and Rac activity.
GTP-bound Rac and Rho were assessed in pulldown assays using GST fusion proteins of the GTPase-binding domains of Pak (PBD:GST) and Rhotekin (RBD:GST), which will bind to activated Rac and Rho, respectively. Rac and Rho activity levels were compared in cells adherent to collagen (3 μg/ml) or a nonadherent control for 45 min. This time point was chosen because it represents a time at which Rac activity is increased in control cells adherent to collagen (Figure 6A), consistent with published results (del Pozo et al., 2000). Expression of R-Ras (38V) prevented this increase on collagen, whereas expression of dominant negative R-Ras (41A) slightly enhanced Rac activity compared with control cells (Figure 6A). Moreover, cells expressing dominant negative R-Ras (41A) had increased Rac activity even when nonadherent, suggesting that R-Ras may regulate Rac activity independent of integrin ligation. Together, these results suggest that R-Ras inhibits Rac activity upon adhesion to collagen.
Figure 6.
R-Ras enhances Rho activity and decreases Rac activity. T47D cells were plated on a collagen-coated plate (3 μg/ml) for 45 min. (A) Cells were lysed and 30 μg PBD:GST was incubated with the lysates to pull down active Rac. Active and total protein was detected by Western blotting. Expression of activated R-Ras (38V) decreases (p = 0.15) Rac activity, whereas expression of dominant negative Rac slightly enhances (p = 0.72) Rac activity. (B) RBD:GST was used to determine Rho activity. Expression of activated R-Ras (38V) significantly enhances Rho activity, whereas expression of dominant negative R-Ras (41A) decreases (p = 0.08) Rho activity. Quantification was performed on three individual experiments and is shown in the bar graphs on the right (±SEM; *p < 0.05 vs. Control, + collagen).
In contrast to Rac signaling, when control cells were plated on collagen, at the 45-min time point, Rho activity was decreased (Figure 6B), again consistent with published results for Rho regulation (Ren et al., 1999; Arthur et al., 2000). Cells expressing activated R-Ras (38V) plated on collagen significantly increased Rho activity compared with control cells, whereas dominant negative R-Ras–expressing cells had a slight decrease in Rho activity (Figure 6B). This suggests that R-Ras activation may increase Rho activity in a regulated manner. It is interesting that cells expressing dominant negative R-Ras (41A) had both high Rac and Rho activity in nonadherent conditions (Figures 6, A and B), because these GTPases often act antagonistically to each other. However, this pulldown assay only measures total, and not local, GTPase activity. In nonadherent dominant negative R-Ras (41A)-expressing cells, active Rho and active Rac may be localized in different areas of the cell so their activities will still be antagonistic. In addition, it is possible that dominant negative R-Ras may alter a cell's response to nonadherent, suspension conditions. Taken together, the above data indicate that R-Ras can down-regulate Rac activity and up-regulate Rho activity, which may contribute to the inhibitory effect of activated R-Ras on cell migration.
R-Ras Alters the Localization of Active Rac and Rho
The local activation of Rac and the inactivation of Rho at the leading edge are essential for membrane protrusion that coordinates cell motility (Kraynov et al., 2000; Arthur and Burridge, 2001; Wang et al., 2003; Worthylake and Burridge, 2003). Because R-Ras activation enhances Rho activity and reduces Rac activity, we determined whether R-Ras does this in a spatial manner in migrating cells. In cells expressing activated R-Ras (38V), R-Ras is likely active in an unrestricted manner at the entire plasma membrane, thus increasing Rho activity and decreasing Rac activity in a nonpolarized manner.
To determine if expression of activated or dominant negative R-Ras affects the localization of active Rac and Rho, Rac and Rho activity were detected in situ using GST fusion proteins previously used in the pulldown assays, in a manner similar to previous work done in Xenopus brain (Li et al., 2002), rat spinal cord (Dubreuil et al., 2003) retinal growth cones (Gehler et al., 2004), and fibroblast podosomes (Berdeaux et al., 2004). Cells were attached to collagen-coated coverslips (3 μg/ml), fixed, and then incubated with 100 μg PBD:GST or RBD:GST, followed by an anti-GST primary antibody and a fluorescent secondary antibody. To ensure that this method was capable of detecting Rac GTPase activity in cells, we assayed positive control cells transiently transfected with constitutively activated and dominant negative Rac (61L and 17N). The same was used to assay Rho activity in cells either transfected with activated Rho (63L) or treated with C3 exoenzyme, which inhibits Rho activity. Cells which were transfected with constitutively activated Rac or Rho showed greatly enhanced PBD:GST or RBD:GST staining, respectively, at the membrane (unpublished data). Cells transfected with dominant negative Rac or treated with C3 exoenzyme showed little membrane staining (unpublished data). These controls indicate that this method is capable of detecting the localization of active GTPase in cells. In situ analysis of Rac and Rho activity was next performed in control cells, cells expressing activated R-Ras (38V), and cells expressing dominant negative R-Ras (41A). In control cells, active Rac was localized to the lamellipodium of migrating cells (Figure 7), consistent with previous findings (Kraynov et al., 2000). In migrating control cells, active Rho is diffuse through the cell (Figure 7). Expression of constitutively activated R-Ras (38V) altered the coordinate spatial distribution of active Rac and Rho. R-Ras (38V)-expressing cells, which form stress fibers preferentially over lamellipodia (Figure 5), lost active Rac at the membrane and, instead, had active Rho concentrated at the membrane in an unpolarized manner (Figure 7). In contrast, cells expressing dominant negative R-Ras (41A) showed a dramatic increase in active Rac at the leading edge (Figure 7), consistent with the increased lamellipodia formation observed in Figures 4 and 5. An important consideration is that PBD:GST can bind to both Rac and Cdc42 and it has been proposed that, in some systems, PBD:GST preferentially binds Rac, and not Cdc42 (Li et al., 2002). However, it has not been determined whether this occurs in T47D cells, so it is possible that staining with PBD:GST is detecting both Rac and Cdc42 activity (Figure 7). Taken together, these data suggest that R-Ras down-regulates Rac and up-regulates Rho in a spatially restricted manner and that this pattern of activity may regulate cell migration.
Figure 7.
R-Ras alters the spatial localization of active Rac and Rho. After cell fixation and permeabilization, 100 μg/ml GBD: GST was incubated with the cells, followed by an anti-GST antibody, and then secondary antibody. Expression of activated R-Ras (38V) caused an increase in active Rho along the periphery of cells and a loss of active Rac (see arrows). Expression of dominant negative R-Ras (41A) caused an increase in the amount of activated Rac localized to lamellipodia (see arrows). Scale bar, 25 μm.
R-Ras Is Localized to and Activated at the Leading Edge during Migration
Our results suggest a model in which activated R-Ras locally increases Rho activity and decreases Rac activity. If true, our model would predict that, like Rac and Rho, R-Ras is active in a spatially and temporally restricted manner in motile cells. To examine if R-Ras localizes in a polar manner during cell migration, we compared the amount of endogenous R-Ras in Cos7 lysates that were allowed to protrude into a transwell filter (“pseudopod”) with the remaining cell body. This method allows the quantification and comparison of the cellular components that are localized in different regions of polarized cells during protrusion and migration (Cho and Klemke, 2002). The expression and/or phosphorylation of molecules associated with cell migration, including the p130Cas/Crk complex and Rac-GTP, are enhanced in the pseudopod (Cho and Klemke, 2002). In the protruding pseudopod, total R-Ras was increased relative to the amount of R-Ras in the cell body (Figure 8B). This data indicates that endogenous R-Ras is located in a polar manner at the leading edge. This polar localization can also be seen using immunofluorescence, as staining for endogenous R-Ras in control T47D migratory cells is increased at the leading edge (Figure 8A). In cells that are not migratory, R-Ras is diffuse through the cell (unpublished data), which is consistent with our previous findings (Kwong et al., 2003).
Figure 8.
R-Ras is activated at the leading edge and upon adhesion to collagen. (A) T47D control cells and cells expressing activated R-Ras (38V) were plated on collagen-coated coverslips and immunostained for R-Ras, which localizes to membranes (right panel). Control motile cells also localize endogenous R-Ras to the leading edge (see arrows in left panel). Scale bar, 25 μm. (B) Cell lysates from pseudopodia and cell bodies were prepared as previously described (Cho and Klemke, 2002). Raf RBD:GST, 50 μg, was used to pulldown GTP-bound R-Ras from lysates as detailed in the Materials and Methods. ERK was also analyzed as a loading control (Brahmbhatt and Klemke, 2003). R-Ras activity is significantly increased (*p < 0.05 vs. cell body) in the protruding pseudopod. (C) T47D cells were plated on collagen or BSA-coated plates and the R-Ras activity assay performed. Cells plated on collagen showed increased (p = 0.12 vs. no collagen) R-Ras activity. All quantification was performed on three individual experiments.
We next wanted to determine if R-Ras is activated at the leading edge. To address this, pseudopod and cell body lysates were made and pulldown assays performed using a GST protein fused to the Ras-binding domain of Raf, which will bind to active, GTP-bound R-Ras. Cos-7 cells were allowed to extend pseudopodia toward an LPA gradient for 1 h and then cells lysed, and the assay was performed. R-Ras activity was significantly enhanced in the protruding pseudopodia compared with the cell body (Figure 8B). Together, these data demonstrate that R-Ras preferentially localizes to, and is activated at, the leading edge during membrane protrusion and migration.
Because R-Ras is activated preferentially at the leading edge and because R-Ras enhances Rho activity and decreases Rac activity (Figures 6 and 7), we hypothesized that R-Ras was endogenously activated upon adhesion to collagen, where it could then activate Rho to stabilize cell protrusions. To test this hypothesis, T47D cells were plated on collagen or BSA-coated plates and the R-Ras activity assay was performed. Cells plated on collagen showed higher R-Ras activity compared with nonadherent cells (Figure 8C), indicating that adhesion activates R-Ras. We envision a model in which active Rac directs cell protrusion. The cell protrudes forward and adheres to the ECM, which then activates R-Ras to activate Rho and stabilize cell protrusion to promote efficient cell migration.
Inhibition of ROCK in Cells Expressing Activated R-Ras (38V) Rescues Rac Activity and Cell Migration
Recent work has identified a mechanism by which active Rho activates its effector, ROCK, which then inhibits Rac (Katsumi et al., 2002; Tsuji et al., 2002). To determine if a similar mechanism controls Rac activity downstream of R-Ras, cells were pretreated with the ROCK inhibitor Y27632 and assayed for Rac activation. Normally, expression of activated R-Ras decreases Rac activation (Figure 6), but when ROCK was inhibited, Rac activation increased to similar baseline levels observed in control cells (Figure 9A). These results suggest that R-Ras regulation of Rac occurs through its effects on Rho-ROCK signaling. When Rho was inhibited by C3 exoenzyme, cells expressing activated R-Ras (38V) did not migrate randomly (Figure 9B, Supplementary Video 6). This result was not unexpected because Rho has several effectors, such as mDia, that also contribute to normal cell behavior and migration (Bishop and Hall, 2000; Ridley, 2001a). However, when ROCK was inhibited by Y27632, cells expressing activated R-Ras began to form forward protrusions and migrated across collagen (Figure 9B, Supplementary Video 7). The ability of R-Ras(38V)-expressing cells to form protrusions upon ROCK inhibition is consistent with a recently reported role for Rho and ROCK in limiting membrane protrusions during migration (Worthylake and Burridge, 2003).
Figure 9.
Inhibition of ROCK in cells expressing activated R-Ras rescues Rac activity, focal complex formation and cell migration. (A) Control cells and cells expressing activated R-Ras were pretreated with Y27632 for 15 min, plated on collagen (3 μg/ml) for 45 min, and then the Rac activity assay was performed. Inhibition of ROCK in cells expressing activated R-Ras (38V) restores Rac activity. Quantification was performed on three individual experiments (*p < 0.05 vs. R-Ras (38V)). (B) Cells expressing activated R-Ras (38V) were treated with 10 μg/ml C3 exoenzyme, to inhibit Rho, or 10 μM Y27632, to inhibit ROCK, 10 min before time-lapse sequences acquired and left on for the duration of the time-lapse series. Time-lapse sequences were acquired for 90 min, with one image collected per minute. Inhibition of ROCK with Y27632 in cells expressing activated R-Ras (38V) enables the cells to migrate with significantly (*p < 0.05 vs. control) enhanced speed and persistence, whereas inhibition of Rho significantly increases cell persistence (**p < 0.05 vs. control). Scale bar, 25 μm. (C) T47D cells were treated with 10 μg/ml C3 exoenzyme, to inhibit Rho, or 10 μM Y27632, to inhibit ROCK, for 15 min and then plated on collagen coated coverslips (3 μg/ml) for 45 min. Cells were then immunostained for FAK phosphorylated at Y397, a marker of focal adhesions. Scale bar, 25 μm.
In addition to regulating cell migration, Rac and Rho are also important regulators of focal adhesion formation and dynamics. Specifically, Rac mediates the formation of focal complexes (Rottner et al., 1999), some of which mature into focal adhesions dependent on Rho activity (Rottner et al., 1999). Cell migration is a cyclical process that is dependent on the ability of a cell to continually assemble and disassemble its focal adhesions (Sheetz et al., 1998, 1999; Horwitz and Parsons, 1999; Munevar et al., 2001; Webb et al., 2002). Therefore, we analyzed focal adhesions in control T47D cells and cells expressing activated R-Ras (38V) using immunofluorescence. As previously described, expression of R-Ras (38V) enhances focal adhesion formation (Kwong et al., 2003; Figure 9C), consistent with increased Rho activity (Figures 6 and 7). C3 exoenzyme, which inhibits Rho, and Y27632, an inhibitor of ROCK, both decreased focal adhesion formation in control cells and cells expressing activated R-Ras (38V) (Figure 9C). However, these cells were still able to form small focal complexes, which is consistent with active Rac (Rottner et al., 1999). Taken together, these results suggest that activated R-Ras (38V) excessively activates Rho at the leading edge, which decreases Rac activity and inhibits migration. Blocking the Rho effector, ROCK, relieves Rac inhibition, allowing cells to form smaller focal complexes that favor cell migration.
In an effort to further verify our hypothesis that active R-Ras inhibits Rac activity through Rho and ROCK, control cells and cells expressing activated R-Ras (38V) were transiently transfected with dominant negative (17N) or constitutively active (61L) Rac:GFP and the effects on random cell migration were examined using time-lapse video microscopy. If the expression of activated R-Ras inhibits Rac activity, then expression of dominant negative Rac should not further affect cell migration. Transient transfection of Rac (17N):GFP in cells stably expressing R-Ras (38V) caused a decrease in ruffle activity, but did not render the cells able to migrate (unpublished data), consistent with our model. Although transient transfection of Rac (61L): GFP caused a dramatic increase in ruffle activity, cells still could not migrate (unpublished data). However, we do not believe this result is inconsistent with our model because expression of constitutively activated Rac can both increase and decrease migration, depending on the cell type and assay used (Leng et al., 1999; Banyard et al., 2000; Ridley, 2001b). An explanation for our result may be that expression of activated Rac localizes Rac in a nonpolar manner, so the cell cannot move because it is trying to protrude in every direction (Allen et al., 1998).
R-Ras Regulates Cell Migration through its Effector, PI3-Kinase
The mechanism by which R-Ras can modulate Rho and Rac activity is unknown. Because PI3-Kinase is the major effector of R-Ras (Marte et al., 1996), we wanted to determine if R-Ras regulates membrane protrusion and migration through this effector. To analyze this possibility, time-lapse video microscopy was performed on cells that stably expressed a mutant of R-Ras (38V/61S), that uncouples activated R-Ras from binding to PI3K, but not Raf or Ral-GDS. This mutant is the same as the activated R-Ras (38V) mutant, except it cannot bind to PI3-Kinase (Self et al., 2001). These cells were plated on collagen (3 μg/ml) and random migration determined. Cells that expressed activated R-Ras that could not bind to PI3-Kinase were able to protrude and migrate across collagen (Figure 10A, Supplementary Video 8). This data suggests that R-Ras can exert its regulatory effects on membrane protrusion and cell migration through its effector, PI3-Kinase.
Figure 10.
R-Ras regulates cell migration through the spatial localization of PI3-Kinase. (A) PI3-Kinase is needed for R-Ras to exert its effects on cell migration. Expression of an effector mutant of R-Ras that uncouples binding to PI3-Kinase rescues migration. Cells stably expressing the effector mutant of activated R-Ras (38V/61S) were plated on collagen-coated Petri plates (3 μg/ml) for 1 h. Time-lapse microscopy sequences were acquired for 45 min, with one image collected per minute (see Supplementary Video 8). Shown here is a representative cell at 0, 15, 30, and 45 min during the observation period. Scale bar, 25 μm. (B) Localization of PI3-Kinase is dependent on R-Ras activity. T47D cells were plated on collagen-coated coverslips (3 μg/ml) for 45 min and then immunostained for PI3-Kinase. PI3-Kinase is localized to the leading edge of control cells (see arrows) and expression of activated R-Ras (38V) disrupts this polar localization. Scale bar, 25 μm.
Lipid products of PI3-Kinase are localized to the leading edge during chemotactic migration (Parent et al., 1998; Wang et al., 2002). Because PI3-Kinase is an effector of R-Ras, we analyzed the localization of PI3-Kinase in control cells and cells expressing activated R-Ras (38V) in order to determine if R-Ras can regulate the localization of PI3-Kinase. Control cells localized PI3-Kinase to the leading edge of migrating cells (Figure 10B). In contrast, cells expressing activated R-Ras localized PI3-Kinase to the entire cell periphery (Figure 10B). These data indicate that activated R-Ras can direct the localization, and likely, the signaling downstream of PI3-Kinase in migrating cells.
DISCUSSION
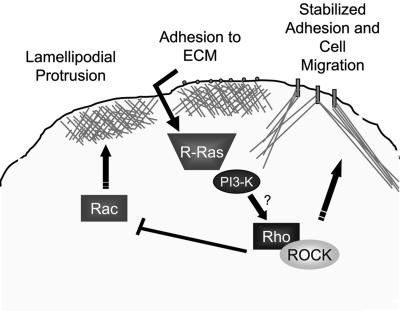
Here we present the novel observation that the small GTPase R-Ras is needed for epithelial cell migration. R-Ras is localized to and activated at the leading edge, where it regulates membrane protrusion and modulates Rho and Rac activities. Although many studies have elucidated the mechanisms by which cells use Rho family GTPases to establish migratory polarity, little is known about the regulatory cues governing these GTPases. We propose that R-Ras plays a key role in cell motility by activating Rho and ROCK, which leads to the inactivation of Rac, thus directing the correct spatial signals needed for membrane protrusion and migration to occur (Figure 11).
Figure 11.
Model for the regulation of cell migration by R-Ras. Rac is activated at the leading edge to promote lamellipodial formation and membrane protrusion. On protrusion and subsequent integrin-mediated adhesion to the ECM, R-Ras becomes activated. GTP-bound R-Ras can then spatially activate Rho by an unknown mechanism that involves PI3-Kinase, which will inactivate Rac, leading to stabilization of the protrusion. Cycles of such events ultimately result in productive cell migration.
From the work of others, a general model of GTPase regulation during random cell migration has emerged, in which Rac is activated and Rho is inactivated at the leading edge. Rac activity leads to lamellipodia formation and forward protrusion. Rho activity is hypothesized to be required later to stabilize the cell during the contractile events of migration. However, the regulation of Rho activity during migration has not been studied in detail (Raftopoulou and Hall, 2004), leaving open the possibility that this model may be too simplistic. In addition, Rottner et al. (1999) has proposed that Rac activity leads to the formation of small focal complexes. However, some of these complexes develop into larger, Rho-dependent focal adhesions that provide the cell with anchorage directly behind the lamellipodia. In this model, it is likely that Rac and Rho activities are cycling at the front of the cell in order to support sustained motility. The exact molecular mechanism regulating this switch from Rac to Rho is not fully understood.
Our data suggest that R-Ras is a key regulator of this switch from Rac to Rho. We found that constitutively activated R-Ras enhances Rho activity and decreases Rac activity. Conversely, expression of dominant negative R-Ras enhances Rac activity and decreases Rho activity. More importantly, however, is the observation that R-Ras can do this in a spatially relevant manner (Figure 7). GTPase activities are carefully regulated during motility, and if this regulation is altered, motility is compromised. Because R-Ras can regulate the localization of Rac and Rho and is itself activated at the leading edge, R-Ras is poised to play an important role in migration (Figure 11). We envision that R-Ras activation is tightly regulated spatially and temporally in the cell during the establishment of cell polarity leading to migration. As the cell initially protrudes, R-Ras activity will be low or preferentially inactivated, whereas Rac will be activated so that the cell has decreased contractility in order to spread. Once a protrusion adheres to the substratum via integrin engagement, R-Ras becomes activated at the leading edge, which then increases Rho activity and inhibits Rac activity to locally enhance focal adhesion formation. This provides the cell with stronger adhesion to the substrate and allows migration to occur. Under normal conditions, R-Ras activity is likely regulated at the leading edge both spatially and temporally in order for the cell to establish polarity to migrate and to help regulate the balance between Rac and Rho.
It is clear that GTPase cross-talk in migration is complex. For example, activated Rac has been reported to decrease or increase Rho activity in different cell types (Sander et al., 1999; Zondag et al., 2000; Cox et al., 2001; Li et al., 2002). In addition, although activated Rac can lead to increased Rho activity in some cells, this newly active Rho then inhibits Rac activation, suggesting a negative feedback loop by which Rho and Rac activity are precisely regulated (Li et al., 2002). In addition to this work, Tsuji et al. (2002) have revealed that downstream of activated Rho, its effectors ROCK and mDia antagonize each other so that ROCK inhibits, and mDia promotes, Rac activation. They also propose that Rho-dependent Rac activation is controlled spatially so that mDia activation of Rac may occur preferentially at the leading edge, whereas ROCK's inhibition of Rac may occur at the trailing edge (Tsuji et al., 2002). Because the cell needs to be stabilized after membrane protrusion, it is also possible that Rho preferentially activates ROCK at the leading edge after protrusion. We find that inhibition of ROCK in cells expressing activated R-Ras (38V) increases Rac activity to baseline levels (Figure 9). This demonstrates a mechanism by which R-Ras can regulate both Rho and Rac activity and also implicates R-Ras as an upstream regulator of this pathway in motility.
Our results also demonstrate an important role for R-Ras as a regulator of membrane protrusion. Constitutively active R-Ras altered protrusion such that the membrane ruffles, but has no persistence. In contrast, dominant negative R-Ras enhanced the persistence of membrane protrusion. A role for R-Ras in regulating membrane protrusion was also apparent from the siRNA results, as protrusive events were also inhibited. The observation that inhibition of ROCK both restores membrane protrusion and increases directional persistence also suggests that R-Ras regulation of protrusion is in part downstream of its activation of Rho.
Although R-Ras is an important regulator of integrin avidity and affinity, our data suggest that R-Ras regulates migration in a manner that is not completely dependent on its effects on α2β1 integrin-mediated adhesion. Altering adhesion (by varying collagen concentration) did not rescue the migratory defect induced by R-Ras. Furthermore, R-Ras enhances adhesion and haptotactic migration on collagen, but not fibronectin (Keely et al., 1999). Because cells expressing activated R-Ras (38V) were still unable to migrate on fibronectin, this suggests that R-Ras does not regulate cell migration solely through regulation of integrin-mediated cell adhesion. However, because activation of Rac and Rho are regulated by integrin-mediated adhesion, it is possible that local effects of R-Ras on integrin avidity may also contribute to Rac and Rho regulation and migration.
PI3-Kinase is emerging as a key regulator of cell polarity and chemotactic migration (reviewed by Iijima et al., 2002). PI3-Kinase is the major effector of R-Ras (Marte et al., 1996) and uncoupling of this effector from R-Ras rescued membrane protrusion and migration across collagen (Figure 10). This suggests that it is through PI3-Kinase that R-Ras can regulate Rho activity to modulate cell migration. So far, PI3-Kinase has been shown to predominantly activate Rac or Cdc42 (Wang et al., 2002; Weiner et al., 2002; Srinivasan et al., 2003). PI3-Kinase can activate several molecules through specialized signaling motifs, including Dbl-homology (DH) domains (reviewed in Zheng, 2001). Because guanine nucleotide exchange factors for Rho family GTPases include DH domains, activation of PI3-Kinase downstream of R-Ras could preferentially activate distinct exchange factors to activate Rho instead of Rac. Because R-Ras can alter the localization of PI3-Kinase (Figure 10B), it is likely that R-Ras can do this in a spatially relevant manner.
Expression of activated R-Ras (38V) enhances haptotactic migration (Keely et al., 1999) and blocks random migration (Figure 2). Others have found similar differences in haptotactic and random migration (Huttenlocher et al., 1996). To account for these differences, it is important to consider that random and haptotactic migration measure different properties. Random migration measures the ability of a cell to move on a two-dimensional surface in the absence of an ECM gradient. Haptotactic migration, however, is a directional assay that measures the ability of a cell to sense and respond to an ECM gradient. It is possible that expression of activated R-Ras allows cells to better sense a concentration gradient. In addition, it has been proposed that haptotactic migration is more dependent on the ability of the cell to make strong adhesions at its leading edge. The ability of a cell to migrate randomly, therefore, may be regulated by the ability of the cell to release strong adhesions at the rear (Huttenlocher et al., 1996). R-Ras enhances focal adhesion formation (Kwong et al., 2003) so it is likely that R-Ras is advantageous in a haptotactic assay because it provides the cell with strong adhesion. R-Ras enhances focal adhesions all over the cell periphery, however, so in a random migration assay, these large adhesions are inhibitory to migration.
The observation that R-Ras regulates cell polarity is relevant not only to the field of cell migration but also to the understanding of the Ras superfamily of small GTPases. Although H-, N-, and K-Ras are well studied and characterized, this is not the case for R-Ras. Although R-Ras was first characterized 16 years ago (Lowe et al., 1987), few biological functions other than increasing integrin affinity and avidity have been described. Here we show that R-Ras plays a key regulatory role in the random migration of breast epithelial cells.
H-Ras enhances motility (Walsh and Bar-Sagi, 2001) and has been proposed to regulate focal adhesion turnover in migration (Schlaepfer and Hunter, 1998; Nobes and Hall, 1999). However, mechanistic roles for Ras in migration have not been described. Although H-Ras can regulate Rac and Rho activity, it is unknown how this plays a role in migration. H-Ras activation can lead to enhancement of Rac activity (Walsh and Bar-Sagi, 2001). The regulation of Rho by Ras is not well understood. Oncogenic Ras has been reported to enhance Rho activity, both through the MAP Kinase pathway (Chen et al., 2003) and through the down-regulation of Rac (which feeds back to down-regulate Rho; Zondag et al., 2000). In contrast to this, Izawa et al. (1998) has also proposed the possible inactivation of Rho and its effector ROCK in Ras-induced transformation. Here we show another biological difference between R-Ras and Ras in that R-Ras increases Rho and ROCK activity, and this feeds back to down-regulate Rac and inhibit membrane protrusion and migration.
In conclusion, here we present evidence that R-Ras plays a key role in the regulation of the Rho GTPases during membrane protrusion and breast epithelial cell migration. Additional studies will be necessary to determine the precise mechanism by which R-Ras regulates Rac and Rho through PI3-Kinase. Moreover, it will be important to determine more precisely how and when R-Ras is activated during migration.
Supplementary Material
Acknowledgments
The authors are grateful to Santos Franco, Benjamin Perrin, and Drs. Mary Lokuta and Anna Huttenlocher (University of Wisconsin) for technical assistance on cell persistence quantitation, kymography, and siRNA; Dr. Alan Hall (University College London) for the R-Ras (38V/61S) effector loop construct; Dr. William Bement (University of Wisconsin) for providing the p16 antibody; and Estuardo Robles and Dr. Timothy Gomez (University of Wisconsin) for critically reading the manuscript. This work was supported by the National Institutes of Health (CA076537 to P.J.K., GM068487 to R.L.K., and CA75924 to D.C.), the American Cancer Society (RPG-00–339 to P.J.K.), and a Shaw Scientist Award from the Greater Milwaukee Foundation (to P.J.K.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–04–0277. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-04-0277.
Abbreviations used: ECM, extracellular matrix; PI3-Kinase, phosphatidylinositol 3′-Kinase; ROCK, Rho Kinase.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Alblas, J., Ulfman, L., Hordijk, P., and Koenderman, L. (2001). Activation of Rhoa and ROCK are essential for detachment of migrating leukocytes. Mol. Biol. Cell 12, 2137-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, W. E., Zicha, D., Ridley, A. J., and Jones, G. E. (1998). A role for Cdc42 in macrophage chemotaxis. J. Cell Biol. 141, 1147-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur, W. T., Petch, L. A., and Burridge, K. (2000). Integrin engagement suppressed RhoA activity via a c-Src-dependent mechanism. Curr. Biol. 10, 719-722. [DOI] [PubMed] [Google Scholar]
- Arthur, W. T., and Burridge, K. (2001). RhoA inactivation by p190RhoGAP regulates cell spreading and migration by promoting membrane protrusion and polarity. Mol. Biol. Cell 12, 2711-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banyard, J., Anand-Apte, B., Symons, M., and Zetter, B. R. (2000). Motility and invasion are differentially modulated by Rho family GTPases. Oncogene 19, 580-591. [DOI] [PubMed] [Google Scholar]
- Bear, J. E. et al. (2002). Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 109, 509-521. [DOI] [PubMed] [Google Scholar]
- Beningo, K. A., Dembo, M., Kaverina, I., Small, J. V., and Wang, Y. L. (2001). Nascent focal adhesions are responsible for the generation of strong propulsive forces in migrating fibroblasts. J. Cell Biol. 153, 881-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdeaux, R. L., Diaz, B., Kim, L., and Martin, G. S. (2004). Active Rho is localized to podosomes induced by oncogenic Src and is required for their assembly and function. J. Cell Biol. 166, 317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, A. L., and Hall, A. (2000). Rho GTPases and their effector proteins. Biochem. J. 348(Pt 2), 241-255. [PMC free article] [PubMed] [Google Scholar]
- Brahmbhatt, A. A., and Klemke, R. L. (2003). ERK and RhoA differentially regulate pseudopodia growth and retraction during chemotaxis. J. Biol. Chem. 278, 13016-13025. [DOI] [PubMed] [Google Scholar]
- Chen, J. C., Zhuang, S., Nguyen, T. H., Boss, G. R., and Pilz, R. B. (2003). Oncogenic Ras leads to Rho activation by activating the mitogen-activated protein kinase pathway and decreasing Rho-GTPase-activating protein activity. J. Biol. Chem. 278, 2807-2818. [DOI] [PubMed] [Google Scholar]
- Cho, S. Y., and Klemke, R. L. (2002). Purification of pseudopodia from polarized cells reveals redistribution and activation of Rac through assembly of a CAS/Crk scaffold. J. Cell Biol. 156, 725-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, E. A., Sastry, S. K., and Huttenlocher, A. (2001). Integrin-mediated adhesion regulates cell polarity and membrane protrusion through the Rho family of GTPases. Mol. Biol. Cell 12, 265-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo, M. A., Price, L. S., Alderson, N. B., Ren, X. D., and Schwartz, M. A. (2000). Adhesion to the extracellular matrix regulates the coupling of the small GTPase Rac to its effector PAK. EMBO J. 19, 2008-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil, C. I., Winton, M. J., and McKerracher, L. (2003). Rho activation patterns after spinal cord injury and the role of activated Rho in apoptosis in the central nervous system. J. Cell Biol. 162, 233-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehler, S., Gallo, G., Veien, E., and Letourneau, P. C. (2004). p75 Neurotrophin receptor signaling regulates growth cone filopodial dynamics through modulating RhoA activity. J. Neurosci. 24(18): 4363-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giancotti, F. G., and Ruoslahti, E. (1999). Integrin signaling. Science 285, 1028-1032. [DOI] [PubMed] [Google Scholar]
- Hinz, B., Alt, W., Johnen, C., Herzog, V., and Kaiser, H. W. (1999). Quantifying lamella dynamics of cultured cells by SACED, a new computer-assisted motion analysis. Exp. Cell Res. 251, 234-243. [DOI] [PubMed] [Google Scholar]
- Horwitz, A. R., and Parsons, J. T. (1999). Cell migration—movin'on. Science 286, 1102-1103. [DOI] [PubMed] [Google Scholar]
- Hughes, P. E., Renshaw, M. W., Pfaff, M., Forsyth, J., Keivens, V. M., Schwartz, M. A., and Ginsberg, M. H. (1997). Suppression of integrin activation: a novel function of a Ras/Raf-initiated MAP kinase pathway. Cell 88, 521-530. [DOI] [PubMed] [Google Scholar]
- Huttenlocher, A., Ginsberg, M. H., and Horwitz, A. F. (1996). Modulation of cell migration by integrin-mediated cytoskeletal linkages and ligand-binding affinity. J. Cell Biol. 134, 1551-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenlocher, A., Sandborg, R. R., and Horwitz, A. F. (1995). Adhesion in cell migration. Curr. Opin. Cell Biol. 7, 697-706. [DOI] [PubMed] [Google Scholar]
- Iijima, M., Huang, Y. I., and Devreotes, P. (2002). Temporal and spatial regulation of chemotaxis. Dev. Cell 3, 469-478. [DOI] [PubMed] [Google Scholar]
- Izawa, I., Amano, M., Chihara, K., Yamamoto, T., and Kaibuchi, K. (1998). Possible involvement of the inactivation of the Rho-Rho-kinase pathway in oncogenic Ras-induced transformation. Oncogene 17, 2863-2871. [DOI] [PubMed] [Google Scholar]
- Katsumi, A., Milanini, J., Kiosses, W. B., del Pozo, M. A., Kaunas, R., Chien, S., Hahn, K. M., and Schwartz, M. A. (2002). Effects of cell tension on the small GTPase Rac. J. Cell Biol. 158, 153-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keely, P. J., Rusyn, E. V., Cox, A. D., and Parise, L. V. (1999). R-Ras signals through specific integrin alpha cytoplasmic domains to promote migration and invasion of breast epithelial cells. J. Cell Biol. 145, 1077-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinbara, K., Goldfinger, L. E., Hansen, M., Chou, F. L., and Ginsberg, M. H. (2003). Ras GTPases: integrins' friends or foes? Nat. Rev. Mol. Cell. Biol. 4, 767-776. [DOI] [PubMed] [Google Scholar]
- Kraynov, V. S., Chamberlain, C., Bokoch, G. M., Schwartz, M. A., Slabaugh, S., and Hahn, K. M. (2000). Localized Rac activation dynamics visualized in living cells. Science 290, 333-337. [DOI] [PubMed] [Google Scholar]
- Kwong, L., Wozniak, M. A., Collins, A. S., Wilson, S. D., and Keely, P. J. (2003). R-Ras promotes focal adhesion formation through focal adhesion kinase and p130(Cas) by a novel mechanism that differs from integrins. Mol. Cell. Biol. 23, 933-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng, J., Klemke, R. L., Reddy, A. C., and Cheresh, D. A. (1999). Potentiation of cell migration by adhesion-dependent cooperative signals from the GTPase Rac and Raf kinase. J. Biol. Chem. 274, 37855-37861. [DOI] [PubMed] [Google Scholar]
- Li, Z., Aizenman, C. D., and Cline, H. T. (2002). Regulation of rho GTPases by crosstalk and neuronal activity in vivo. Neuron 33, 741-750. [DOI] [PubMed] [Google Scholar]
- Lowe, D. G., Capon, D. J., Delwart, E., Sakaguchi, A. Y., Naylor, S. L., and Goeddel, D. V. (1987). Structure of the human and murine R-ras genes, novel genes closely related to ras proto-oncogenes. Cell 48, 137-146. [DOI] [PubMed] [Google Scholar]
- Maheshwari, G., and Lauffenburger, D. A. (1998). Deconstructing (and reconstructing) cell migration. Microsc. Res. Tech. 43, 358-368. [DOI] [PubMed] [Google Scholar]
- Marte, B. M., Rodriguez-Viciana, P., Wennstrom, S., Warne, P. H., and Downward, J. (1996). R-Ras can activate the phosphoinositide 3-kinase but not the MAP kinase arm of the Ras effector pathways. Curr. Biol. 7, 63-70. [DOI] [PubMed] [Google Scholar]
- Mullins, R. D., and Pollard, T. D. (1999). Structure and function of the Arp2/3 complex. Curr. Opin. Struct. Biol. 9, 244-249. [DOI] [PubMed] [Google Scholar]
- Munevar, S., Wang, Y. L., and Dembo, M. (2001). Distinct roles of frontal and rear cell-substrate adhesions in fibroblast migration. Mol. Biol. Cell 12, 3947-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobes, C. D., and Hall, A. (1999). Rho GTPases control polarity, protrusion, and adhesion during cell movement. J. Cell Biol. 144, 1235-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palecek, S. P., Loftus, J. C., Ginsberg, M. H., Lauffenburger, D. A., and Horwitz, A. F. (1997). Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness. Nature 385, 537-540. [DOI] [PubMed] [Google Scholar]
- Parent, C. A., Blacklock, B. J., Froehlich, W. M., Murphy, D. B., and Devreotes, P. N. (1998). G protein signaling events are activated at the leading edge of chemotactic cells. Cell 95, 81-91. [DOI] [PubMed] [Google Scholar]
- Raftopoulou, M., and Hall, A. (2004). Cell migration: Rho GTPases lead the way. Dev. Biol. 265, 23-32. [DOI] [PubMed] [Google Scholar]
- Ren, X. D., Kiosses, W. B., and Schwartz, M. A. (1999). Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 18, 578-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley, A. J. (2001a). Rho family proteins: coordinating cell responses. Trends Cell Biol. 11, 471-477. [DOI] [PubMed] [Google Scholar]
- Ridley, A.J. (2001b). Rho GTPases and cell migration. J. Cell Sci. 114, 2713-2722. [DOI] [PubMed] [Google Scholar]
- Rincon-Arano, H., Rosales, R., Mora, N., Rodriguez-Castaneda, A., and Rosales, C. (2003). R-Ras promotes tumor growth of cervical epithelial cells. Cancer 97, 575-585. [DOI] [PubMed] [Google Scholar]
- Rottner, K., Hall, A., and Small, J. V. (1999). Interplay between Rac and Rho in the control of substrate contact dynamics. Curr. Biol. 9, 640-648. [DOI] [PubMed] [Google Scholar]
- Sander, E. E., ten Klooster, J.P., van Delft, S., van der Kammen, R. A., and Collard, J. G. (1999). Rac downregulates Rho activity: reciprocal balance between both GTPases determines cellular morphology and migratory behavior. J. Cell Biol. 147, 1009-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaepfer, D. D., and Hunter, T. (1998). Integrin signalling and tyrosine phosphorylation: just the FAKs? Trends Cell Biol. 8, 151-157. [DOI] [PubMed] [Google Scholar]
- Self, A. J., Caron, E., Paterson, H. F., and Hall, A. (2001). Analysis of R-Ras signalling pathways. J. Cell Sci. 114, 1357-1366. [DOI] [PubMed] [Google Scholar]
- Sethi, T., Ginsberg, M. H., Downward, J., and Hughes, P. E. (1999). The small GTP-binding protein R-Ras can influence integrin activation by antagonizing a Ras/Raf-initiated integrin suppression pathway. Mol. Biol. Cell 10, 1799-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheetz, M. P., Felsenfeld, D., Galbraith, C. G., and Choquet, D. (1999). Cell migration as a five-step cycle. Biochem. Soc. Symp. 65, 233-243. [PubMed] [Google Scholar]
- Sheetz, M. P., Felsenfeld, D. P., and Galbraith, C. G. (1998). Cell migration: regulation of force on extracellular-matrix-integrin complexes. Trends Cell Biol. 8, 51-54. [DOI] [PubMed] [Google Scholar]
- Sporn, M. B. (1996). The war on cancer. Lancet 347, 1377-1381. [DOI] [PubMed] [Google Scholar]
- Srinivasan, S., Wang, F., Glavas, S., Ott, A., Hofmann, F., Aktories, K., Kalman, D., and Bourne, H. R. (2003). Rac and Cdc42 play distinct roles in regulating PI(3,4,5)P3 and polarity during neutrophil chemotaxis. J. Cell Biol. 160, 375-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina, T. M., and Borisy, G. G. (1999). Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol. 145, 1009-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji, T. et al. (2002). ROCK and mDia1 antagonize in Rho-dependent Rac activation in Swiss 3T3 fibroblasts. J. Cell Biol. 157, 819-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walpita, D., and Hay, E. (2002). Studying actin-dependent processes in tissue culture. Nat. Rev. Mol. Cell. Biol. 3, 137-141. [DOI] [PubMed] [Google Scholar]
- Walsh, A. B., and Bar-Sagi, D. (2001). Differential activation of the Rac pathway by Ha-Ras and K-Ras. J. Biol. Chem. 276, 15609-15615. [DOI] [PubMed] [Google Scholar]
- Wang, F., Herzmark, P., Weiner, O. D., Srinivasan, S., Servant, G., and Bourne, H. R. (2002). Lipid products of PI(3)Ks maintain persistent cell polarity and directed motility in neutrophils. Nat. Cell Biol. 4, 513-518. [DOI] [PubMed] [Google Scholar]
- Wang, H. R., Zhang, Y., Ozdamar, B., Ogunjimi, A. A., Alexandrova, E., Thomsen, G. H., and Wrana, J. L. (2003). Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science 302, 1775-1779. [DOI] [PubMed] [Google Scholar]
- Webb, D. J., Parsons, J. T., and Horwitz, A. F. (2002). Adhesion assembly, disassembly and turnover in migrating cells— over and over and over again. Nat. Cell Biol. 4, E97-E100. [DOI] [PubMed] [Google Scholar]
- Weiner, O. D., Neilsen, P. O., Prestwich, G. D., Kirschner, M. W., Cantley, L. C., and Bourne, H. R. (2002). A PtdInsP3-and Rho GTPase-mediated positive feedback loop regulates neutrophil polarity. Nat. Cell Biol. 4, 509-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch, M.D. (1999). The world according to Arp: regulation of actin nucleation by the Arp2/3 complex. Trends Cell Biol. 9, 423-427. [DOI] [PubMed] [Google Scholar]
- Welch, M. D., Mallavarapu, A., Rosenblatt, J., and Mitchison, T. J. (1997). Actin dynamics in vivo. Curr. Opin. Cell Biol. 9, 54-61. [DOI] [PubMed] [Google Scholar]
- Worthylake, R. A., and Burridge, K. (2003). RhoA and ROCK promote migration by limiting membrane protrusions. J. Biol. Chem. 278, 13578-13584. [DOI] [PubMed] [Google Scholar]
- Worthylake, R. A., Lemoine, S., Watson, J. M., and Burridge, K. (2001). RhoA is required for monocyte tail retraction during transendothelial migration. J. Cell Biol. 154, 147-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak, M. A., Desai, R., Solski, P. A., Der, C. J., and Keely, P. J. (2003). ROCK-generated contractility regulates breast epithelial cell differentiation in response to the physical properties of a three-dimensional collagen matrix. J. Cell Biol. 163, 583-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Vuori, K., Wang, H., Reed, J. C., and Ruoslahti, E. (1996). Integrin activation by R-ras. Cell 85, 61-69. [DOI] [PubMed] [Google Scholar]
- Zheng, Y. (2001). Dbl family guanine nucleotide exchange factors. Trends Biochem. Sci. 26, 724-732. [DOI] [PubMed] [Google Scholar]
- Zondag, G. C., Evers, E. E., ten Klooster, J. P., Janssen, L., van der Kammen, R. A., and Collard, J. G. (2000). Oncogenic Ras down-regulates Rac activity, which leads to increased Rho activity and epithelial-mesenchymal transition. J. Cell Biol. 149, 775-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.