Publisher's Note: There is an Inside Blood Commentary on this article in this issue.
Key Points
Acquired expression of CblQ367P induces sustained proliferation of myelomonocytes, multilineage dysplasia, and splenomegaly resembling CMML.
Combined inhibition of PI3K and JAK2 efficiently suppressed the growth of CblQ367P-induced CMML cells.
Abstract
Chronic myelomonocytic leukemia (CMML) is a hematological malignancy characterized by uncontrolled proliferation of dysplastic myelomonocytes and frequent progression to acute myeloid leukemia (AML). We identified mutations in the Cbl gene, which encodes a negative regulator of cytokine signaling, in a subset of CMML patients. To investigate the contribution of mutant Cbl in CMML pathogenesis, we generated conditional knockin mice for Cbl that express wild-type Cbl in a steady state and inducibly express CblQ367P, a CMML-associated Cbl mutant. CblQ367P mice exhibited sustained proliferation of myelomonocytes, multilineage dysplasia, and splenomegaly, which are the hallmarks of CMML. The phosphatidylinositol 3-kinase (PI3K)-AKT and JAK-STAT pathways were constitutively activated in CblQ367P hematopoietic stem cells, which promoted cell cycle progression and enhanced chemokine-chemokine receptor activity. Gem, a gene encoding a GTPase that is upregulated by CblQ367P, enhanced hematopoietic stem cell activity and induced myeloid cell proliferation. In addition, Evi1, a gene encoding a transcription factor, was found to cooperate with CblQ367P and progress CMML to AML. Furthermore, targeted inhibition for the PI3K-AKT and JAK-STAT pathways efficiently suppressed the proliferative activity of CblQ367P-bearing CMML cells. Our findings provide insights into the molecular mechanisms underlying mutant Cbl-induced CMML and propose a possible molecular targeting therapy for mutant Cbl-carrying CMML patients.
Introduction
Myelodysplastic syndrome (MDS) is a hematopoietic disorder originating from hematopoietic stem/progenitor cells (HSPCs ) and is characterized by ineffective hematopoiesis, multilineage dysplasia, and a susceptibility to leukemia.1,2 Because subsets of MDS possess clinical and pathological features that overlap with those of myeloproliferative disorders, they were categorized as myelodysplasia/myeloproliferative neoplasms (MDS/MPN) by the 2008 World Health Organization classification.3
Chronic myelomonocytic leukemia (CMML) is a disease category belonging to MDS/MPN that exhibits an absolute increase of dysplastic myelomonocytes.4-6 Genetic analyses have identified a number of molecular aberrations in CMML patients, including mutations in signaling molecules (JAK2, KRAS, NRAS, CBL, FLT3, and RUNX1), epigenetic regulators (EZH2, ASXL1, TET2, DNMT3A, IDH1, and IDH2), and spliceosome components (SF3B1, SRSF2, U2AF1/U2AF35, ZRSR2, SF3A1, PRPF40B, U2AF2/U2AF65, and SF1), which suggest genetic diversity in the pathogenesis of the disease.4-6
We previously performed allele-specific copy number analysis in patients with MDS/MPN and identified mutations in the c-Cbl (Casitas B cell lymphoma, hereafter referred to as Cbl) gene in CMML samples with acquired uniparental disomy (aUPD) at chromosome 11q.7 Subsequent studies have identified homozygous mutations of the Cbl gene in MDS/MPN and related neoplasms, indicating that dysfunction of CBL contributes to the development of myeloid diseases.8-10
c-CBL (hereafter, referred to as CBL), a cellular homolog of v-CBL, functions as an E3 ubiquitin ligase that negatively regulates receptor tyrosine kinase–mediated intracellular signaling.8-10 Mutations in the Cbl gene are clustered within the linker and RING finger domains that are essential for E3 ubiquitin ligase activity,8-10 strongly suggesting that mutated forms of CBL contribute to the development of myeloid malignancies through the sustained intracellular signaling and deregulated proliferation of HSPCs. This idea is supported by the creation and analysis of Cbl mutant mice that exhibit lymphoid hyperplasia, enhanced hematopoietic signaling, and extramedullary hematopoiesis.11,12 In addition, mice carrying a mutation in the RING finger domain of the Cbl gene (C379A) with a Cbl-null background (CblC379A/–) develop myeloproliferation and exhibit spontaneous progression to acute myeloid leukemia (AML).13
These findings provide insights into a functional link between Cbl mutations and myeloid neoplasms. However, the precise molecular mechanism(s) by which mutated forms of CBL contribute to the development of CMML is not yet fully understood. To address this issue and to create an animal model for CMML, we generated and analyzed conditional knockin (cKI) mice that express wild-type Cbl at steady state and in turn express the Q367P mutation of Cbl (CblQ367P), which we identified in CMML patients.7
Materials and methods
Construction of the targeting vector and generation of cKI mice for a mutant Cbl
The detailed procedures for construction of the targeting vector and generation of the cKI mice are described in the supplemental Methods, available on the Blood Web site. CblWT/Q367P mice were crossed with MxCre+ mice14 to generate CblWT/Q367P, MxCre+ mice, which were intercrossed to generate CblQ367P/Q367P, MxCre−, and CblQ367P/Q367P, MxCre+ mice. Cre activation was achieved by intraperitoneal administration of 500 µg of polyinosinic-polycytidylic acid (pIpC; Sigma-Aldrich, St. Louis, MO) 3 times at 2-day intervals. Mice that had been backcrossed to the C57BL/6N-Ly5.2 background at least 7 times were used in this study. All of the experiments were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Hiroshima University Animal Research Committee (permission no. 27-52).
Statistics
Mouse survival curves were constructed by using the Kaplan-Meier methodology and compared by the log-rank test by using the GraphPad Prism software. Other statistical analyses were performed using the Student t test, unless otherwise stated.
A complete and detailed description of methods is available in the supplemental Methods.
Results
CblQ367P mice exhibited sustained proliferation of white blood cells with dysplasia, splenomegaly, and enhanced colony formation mimicking human CMML
To analyze the biological effect of CBLQ367P and to create a mouse model for human CMML, we attempted to generate cKI mice that express CblWT at steady state and inducibly express CblQ367P (supplemental Figure 1A). Correctly targeted embryonic stem cells (supplemental Figure 1B) were used to create chimeric mice, which transmitted the mutated allele through the germline. Mice carrying the CblWT/WT allele were crossed with MxCre+ mice that inducibly express Cre by pIpC administration. The pIpC-induced deletion of the CblWT complementary DNA was confirmed in the hematopoietic tissues of CblWT/WT, MxCre+ mice by Southern blot and direct sequencing, which led to the expression of the CblQ367P complementary DNA (supplemental Figure 1A and 1C). Enhanced tyrosine phosphorylation of CBL was detected in the bone marrow (BM) and spleen of pIpC-treated CblWT/WT, MxCre+ (ie, CblQ367P/Q367P) mice compared with those of pIpC-treated CblWT/WT, MxCre− mice (supplemental Figure 1D), indicating successful expressional conversion from CBLWT to CBLQ367P.
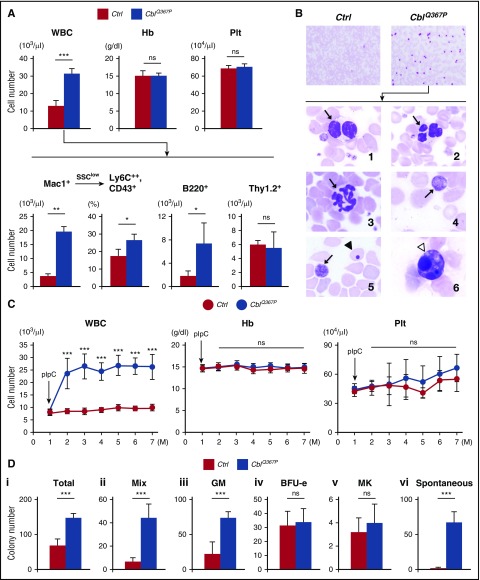
After pIpC treatment, CblWT/WT, MxCre+ (CblQ367P/Q367P) mice exhibited a marked elevation of white blood cells (WBCs) compared with that exhibited by CblWT/WT, MxCre− mice (Figure 1A-B, upper panels), which was composed mainly of Mac1+ myelomonocytic cells and partly of B220+ B-lymphocytes (Figure 1A, lower panels) (hereafter, pIpC-treated CblWT/WT, MxCre−, and CblWT/WT, MxCre+ [CblQ367P/Q367P] mice are referred to as control and CblQ367P mice, respectively). In addition, in the SSClow Mac1+ fraction, the ratio of Ly6C (equivalent to human CD14)++, CD43+ cells was significantly higher in CblQ367P mice than in control mice (Figure 1A, second lower panel), indicating that the increased myelomonocytic cells were classical monocytes as reported in human CMML.15 In CblQ367P mice, the spleen was markedly enlarged (indicated by an arrow in supplemental Figure 2A), and proliferation and infiltration of myeloid cells were observed in the BM, white pulp of the spleen, and perisinusoidal space of the liver (indicated by arrowheads in supplemental Figure 2B). In addition, peripheral blood (PB) smears of CblQ367P mice exhibited various types of morphological abnormalities (Figure 1B, lower panels). Time course analysis of the PB cells revealed sustained elevation of WBC numbers in CblQ367P mice for 6 months after pIpC induction (Figure 1C), indicating that the CblQ367P-induced hematopoietic disorder is chronic. During the 1-year observation period, although a small portion (<5%) of CblQ367P mice developed acute leukemia and died, other mice did not show any fatal phenotype (data not shown). To analyze the alteration(s) of cytokine-induced proliferative ability, colony formation assays were performed. CblQ367P cells exhibited markedly increased numbers in total, mix, and granulocyte/monocyte colonies compared with control cells (Figure 1Di-iii). Of note, a number of spontaneous colonies (ie, colonies without cytokines) were generated from CblQ367P cells, which was hardly detectable in control cells (Figure 1Dvi). Because the phenotypes in CblQ367P mice, such as sustained proliferation of WBCs mainly consisting of myelomonocytes, multilineage dysplasia, and hypersensitivity to cytokines, are characteristics of CMML,4-6,16,17 this allows us to consider CblQ367P mice as a model of human CMML.
Figure 1.
Analysis of PB parameters and colony formation assay. (A) PB parameters of control and CblQ367P mice. A higher total WBC count was observed in CblQ367P mice, which was mainly due to the proliferation of Mac1+ myelomonocytes, including Ly6C++, CD43+ monocytic cells, and B220+ B lymphoid cells. (B) Giemsa-stained PB smears from control and CblQ367P mice. The higher number of WBCs in the PB of CblQ367P mice and higher magnification of WBCs with abnormal morphologies are shown in the upper and lower panels, respectively. Panels 1 and 2: WBCs with pseudo–Pelger-Huet anomaly and abnormal nuclei (indicated by arrows); panel 3: a hypersegmented neutrophil (indicated by an arrow); and panels 4–6: giant platelets, an erythrocyte with a Howell-Jolly body, and an apoptotic cell (indicated by arrows, an arrowhead, and a white arrowhead, respectively). (C) Changes of PB parameters during the observation period. CblQ367P mice exhibited a sustained elevation of WBC numbers after pIpC stimulation. (D) Hematopoietic colony numbers of control and CblQ367P mice. CblQ367P hematopoietic cells exhibited a significant increase in total, mix, and GM colonies and generated a remarkable number of spontaneous colonies. ***P < .001; **P < .01; *P < .05. ns, not significant.
Increase of HSPCs and activation of AKT, STAT3, and STAT5 proteins in long-term hematopoietic stem cells in CblQ367P mice
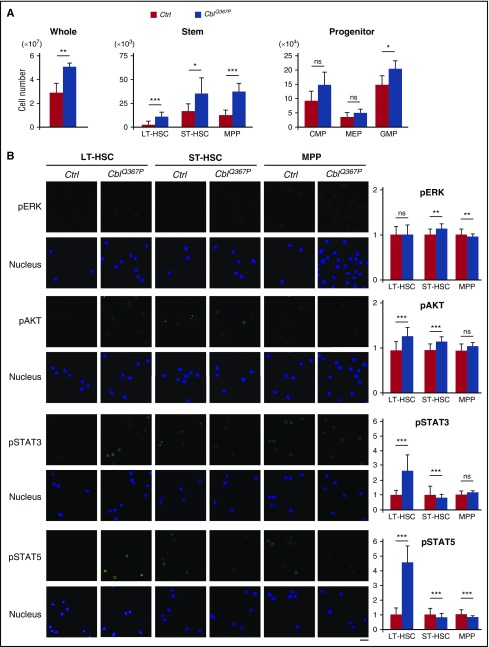
We next analyzed the numbers in HSPC fractions in the BM. The number of whole cells was significantly higher in CblQ367P mice than in the controls (Figure 2A, left panel). In addition, the cell numbers of total LSK and subfractions of LSK, including long-term hematopoietic stem cells (LT-HSCs), short-term hematopoietic stem cells (ST-HSCs), and multipotent progenitors (MPPs), were also markedly higher in CblQ367P mice (Figure 2A, middle panel). In the progenitor fractions, cell numbers in the common myeloid progenitors, megakaryocyte-erythrocyte progenitor, and granulocyte/macrophage progenitor fractions were elevated in CblQ367P mice, with a significant increase in the granulocyte/macrophage progenitor fraction (Figure 2A, right panel).
Figure 2.
Analysis of HSPC numbers and detection of activated signaling pathways. (A) Comparison of cell numbers of whole, stem, and progenitor fractions in the control and CblQ367P BM. (B) Activation of ERK, AKT, STAT3, and STAT5 proteins in the HSC subpopulations. Representative photos taken with a confocal microscope and relative fluorescent intensities to those of control LT-HSCs are shown in the left and right panels, respectively. Scale bar: 10 μm. ***P < .001; **P < .01, *P < .05. ns, not significant.
Previous studies have reported that mutated CBL proteins activate the RAS-MAPK, phosphatidylinositol 3-kinase (PI3K)-AKT, and/or JAK-STAT pathways.4,6,17-19 To clarify CBLQ367P-mediated proliferative signals, LT-HSCs, ST-HSCs, and MPPs were immunofluorescently stained with antibodies against phosphorylated (p) extracellular signal-regulated kinase (ERK), pAKT, pSTAT3, and pSTAT5 proteins. As shown in Figure 2B, whereas activation of ERK was observed in the ST-HSC and MPP fractions, those of AKT and STATs were detected in the more primitive LT-HSC fraction. These results strongly suggest that CBLQ367P induces CMML by activating these pathways in HSPCs.
Transfer of CblQ367P LT-HSCs recapitulates the CMML phenotype in recipient mice
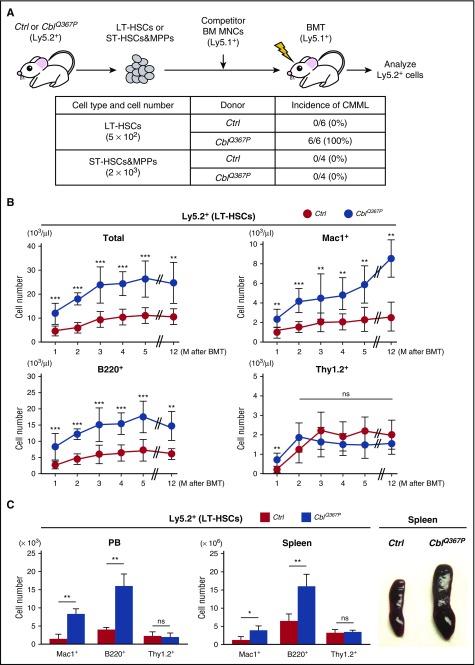
A previous report demonstrated that CMML arises from transformed hematopoietic cells at stem-multipotent progenitor stages.20 To test this idea in our model, we performed competitive repopulation assays using LT-HSCs as well as ST-HSCs and MPPs from control and CblQ367P mice (Figure 3A, upper panel). As shown in the lower panel of Figure 3A, in contrast with the finding that all the recipients transplanted with CblQ367P LT-HSCs developed CMML (6/6), none of the recipients transplanted with CblQ367P ST-HSCs and MPPs exhibited myeloid proliferation mimicking CMML (0/4). The CblQ367P LT-HSCs exhibited significantly higher PB chimerism than control cells, and the proliferated cells were positive for a myeloid (Mac1+) or a B-lymphoid (B220+) antigen (Figure 3B), which is reminiscent of the phenotypes in parental CblQ367P mice (Figure 1A, lower panels). At the end of the observation period (12 months after transplantation), significant proliferation of Mac1+ myeloid cells and B220+ B-lymphocytes was detected in the PB and in the enlarged spleen of recipients transplanted with CblQ367P LT-HSCs (Figure 3C). These results indicate that mutant CBL–induced CMML originates from transformed LT-HSCs, possibly through the constitutive activation of the PI3K-AKT and JAK-STAT pathways (Figure 2B).
Figure 3.
Competitive repopulation assay of LT-HSCs. (A) Experimental procedure and incidence of CMML. LT-HSCs or ST-HSCs and MPPs from control or CblQ367P mice (Ly5.2+) were transplanted into irradiated syngeneic mice (Ly5.1+) with competitor BM mononuclear cells (Ly5.1+), and Ly5.2+ cells in the recipient mice were analyzed. The incidence of CMML in the recipient mice is shown. (B) Changes of total and lineage marker–positive Ly5.2+ cells in the PB during the observation period. (C) Numbers of lineage marker–positive Ly5.2+ cells in the PB and the spleen and macroscopic appearances of the spleens in recipient mice at the endpoint of transplantation. ***P < .001; **P < .01, *P < .05. ns, not significant.
Transcriptome analysis of CblQ367P-expressing HSPCs
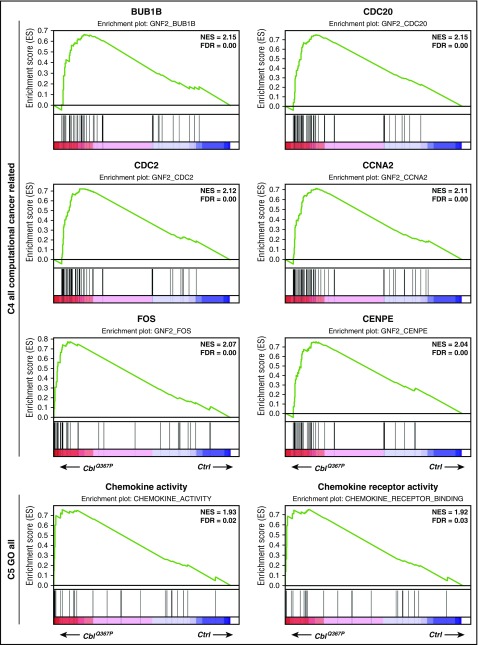
To elucidate the molecular mechanism(s) underlying CblQ367P-induced CMML, gene expression profiles in HSPCs were analyzed. LSK cells from control and CblQ367P mice were subjected to RNA sequencing, and the results were subjected to Gene Set Enrichment Analysis (http://software.broadinstitute.org/gsea/index.jsp). The Molecular Signatures Database identified pathways with significant enrichment in CblQ367P HSPCs (normalized enrichment scores = ∼2.0) in C4 computational cancer-related gene sets and C5 Gene Ontology gene sets. As shown in Figure 4, CblQ367P LSK cells exhibited significant enrichment in gene sets involved in cell cycle regulation and cell cycle checkpoints, such as BUB1B, CDC20, CDC2, CCNA2, FOS, and CENPE, and also in those governing chemokine-chemokine receptor activity. These findings indicate that induced expression of CBLQ367P activated cell cycle–regulating genes and chemokine pathways, which eventually expanded myeloid cells and, consequently, developed CMML.
Figure 4.
Results of the pathway analysis of the LSK cells. Pathways positively enriched in CblQ367P HSPCs compared to control cells are shown. The top 6 plots of C4 all computational cancer-related gene sets and the top 2 plots of C5 GO all gene sets are shown. FDR: false discovery rate; NES, normalized enrichment score.
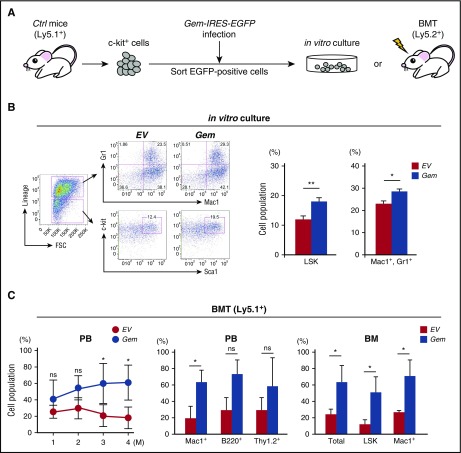
Enhanced expression of Gem increases hematopoietic stem cell activity and induces myeloid cell proliferation
Among the upregulated genes in CblQ367P LSK cells (DRX046249 and DRX046250, deposited in the DNA Data Bank of Japan), Gem (GTP-binding protein overexpressed in skeletal muscle) drew our attention, because it has been reported to be the most upregulated gene in hematopoietic stem cells (HSCs) in a mutant Nras (NrasG12D)-induced CMML model21 (supplemental Figure 3A). The expression analysis of Gem in HSPCs revealed that Gem is highly expressed in LT-HSC and MPP fractions (supplemental Figure 3B), leading us to the idea that enhanced expression of Gem may play a role in the development of CMML, mediated by CblQ367P and NrasG12D. To address this possibility, HSPCs were infected with Gem-expressing retrovirus and subjected to in vitro and in vivo assays (Figure 5A). In vitro culture of Gem-transduced c-kit+ cells showed significant proliferation of LSK and Mac1+, Gr1+ cells (Figure 5B). In addition, the bone marrow transplantation (BMT) experiment demonstrated that Gem-overexpressing HSPCs exhibited higher chimerism than control cells, with a significant increase at 3 and 4 months after transplantation (Figure 5C, left panel). Proliferation of Mac1+ cells in the PB and significant increase of total, LSK, and Mac1+ cells in the BM were observed at the end point of the transplantation (Figure 5C, middle and right panels). These results indicate that overexpression of Gem in HSPCs induces hematopoietic stemness and primed myeloid proliferation, suggesting that upregulation of GEM contributes, at least partly, to the pathogenesis of CMML.
Figure 5.
Forced expression of Gem in HSPCs. (A) Experimental procedure. The c-kit+ cells of control or CblQ367P mice (Ly5.1+) were infected with Gem-IRES-EGFP–expressing retrovirus, and sorted EGFP-positive cells were subjected to in vitro culture or BMT into recipient mice (Ly5.2+). (B) Results of the in vitro culture. Representative plots of flow cytometry and percentages of the LSK and Mac1+, Gr1+ cells are shown in the left and right panels, respectively. (C) Results of BMT. The changes of percentages of the Ly5.1+ cells in the PB, percentages of lineage marker–positive Ly5.1+ cells in the PB, and percentages of total, LSK, and Mac1+ Ly5.1+ cells in the BM at the end point of BMT are shown in the left, middle, and right panels, respectively. *P < .05. ns, not significant.
Secondary genetic events, including overexpression of Evi1, progress CMML to acute leukemia
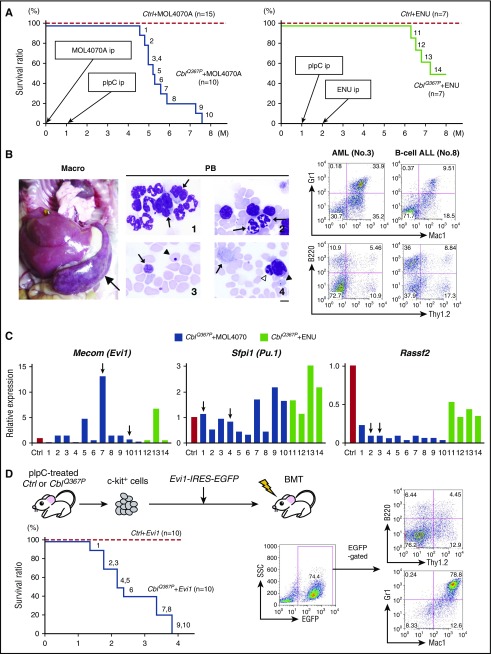
A significant portion (20%-30%) of CMML patients progress to AML.4,6 To elucidate the molecular mechanism(s) underlying the disease evolution, we applied retrovirus (MOL4070A)-mediated22 and chemical reagent (N-ethyl-N-nitrosourea [ENU])-induced mutagenesis23 to CblQ367P mice. During the 8-month observation period, all the MOL4070A-infected CblQ367P mice (CblQ367P + MOL4070A) and about half of the ENU-injected CblQ367P mice (CblQ367P + ENU) developed acute leukemia (blast count of >20%), whereas none of the MOL4070A-infected or ENU-injected control mice exhibited hematopoietic abnormalities (Figure 6A). Macroscopically, all the diseased mice exhibited massive splenomegaly and marked elevation of WBC counts (Figure 6B, left panel; supplemental Table 1). Dysplastic hematopoietic cells were evident in CblQ367P mice with the acute leukemias (Figure 6B, middle panels), indicating that the leukemias were acute transformation of CMML. Flow cytometric and gene rearrangement analyses of the leukemic cells revealed that most of the samples were diagnosed as AML, except 1 case that was classified as B-lineage acute lymphoblastic leukemia (ALL) (Figure 6B, right panels; supplemental Figure 4; supplemental Table 1). These results indicate that additional genetic aberrations cooperate with CblQ367P to develop acute leukemia, mainly of myeloid lineage.
Figure 6.
Analysis of progression from CMML to acute leukemia. (A) Survival curves of MOL4070A-infected (left panel) and ENU-injected (right panel) control and CblQ367P mice. The time points of intraperitoneal administration (ip) of pIpC, MOL4070A, and ENU are indicated by arrows, and the diseased mice are numbered. (B) Pathological findings of the CblQ367P mice with acute transformation. Macroscopic appearance, abnormal PB cells, and representative results of flow cytometric analyses of AML (no. 3) and B-cell ALL (no. 8) samples are shown in the left, middle, and right panels, respectively. In the left panel, the enlarged spleen is indicated by an arrow. In the middle panel, Giemsa-stained PB smears are shown: panel 1, WBCs with abnormal nuclei, including a pseudo–Pelger-Huet anomaly (indicated by arrows); panel 2, hypersegmented neutrophils (indicated by arrows); and panels 3 and 4, giant platelets, erythrocytes with a Howell-Jolly body, and an apoptotic cell (indicated by arrows, arrowhead, and a white arrowhead, respectively). Scale bar: 10 μm. (C) Expression levels of CIS genes, Mecom (Evi1), Sfpi1 (PU.1), and Rassf2 in the tumor tissues. The results are shown relative to those of a control spleen (Ctrl). Tumors with virus integration in the corresponding CIS gene are indicated by vertical arrows. (D) Retrovirus-mediated transfer of Evi1. Experimental procedure, survival curves of recipient mice, and a representative result of the flow cytometric analysis of a leukemic mouse are shown in the upper left, lower left, and right panels, respectively. In the lower left panel, diseased mice in the CblQ367P + Evi1 group are numbered.
We then analyzed retroviral integration sites of MOL4070A in the CblQ367P + MOL4070A leukemic tissues by inverse polymerase chain reaction.24-27 Among the integration sites detected, we identified 3 common integration sites (CISs): Mecom (Mds1 and Evi1 complex locus, also known as Evi1, ecotropic virus integration site 1),28 Sfpi-1 (spleen focus forming virus proviral integration oncogene, also known as PU.1),29 and Rassf2 (Ras association [RalGDS/AF-6] domain family member 2)30 (supplemental Table 2). Examination of the expressional alteration of these genes in CblQ367P + MOL4070A and CblQ367P + ENU tumor tissues revealed that, although the expression levels of Rassf2 and Sfpi-1 were not apparently increased, that of Mecom/Evi1 was upregulated more than fivefold in 3 tumors (no. 5, 7, and 13), including the virus-integrated case (no. 7) (Figure 6C). To confirm the contribution of overexpressed Evi1 to the disease progression of CblQ367P mice, control and CblQ367P c-kit+ cells were infected with Evi1-IRES-EGFP–expressing retrovirus and transplanted into irradiated syngeneic mice (Figure 6D, upper left panel). Within the 4-month observation period, all the recipients transplanted with Evi1-IRES-EGFP–expressing CblQ367P cells (CblQ367P + Evi1) developed acute leukemia, whereas no diseases were observed in those transplanted with Evi1-IRES-EGFP–expressing control cells (Ctrl + Evi1) (Figure 6D, lower left panel; supplemental Table 3). The leukemic cells were positive for EGFP and myeloid markers, indicating that the diseases originated from the transplanted CblQ367P + Evi1 cells, and were diagnosed as AML (Figure 6D, right panels; supplemental Table 3). These results indicate that EVI1 synergizes with CBLQ367P in vivo, which accelerates CMML and develops AML.
Combination therapy with JAK2 and PI3K inhibitors attenuated proliferation of CMML cells
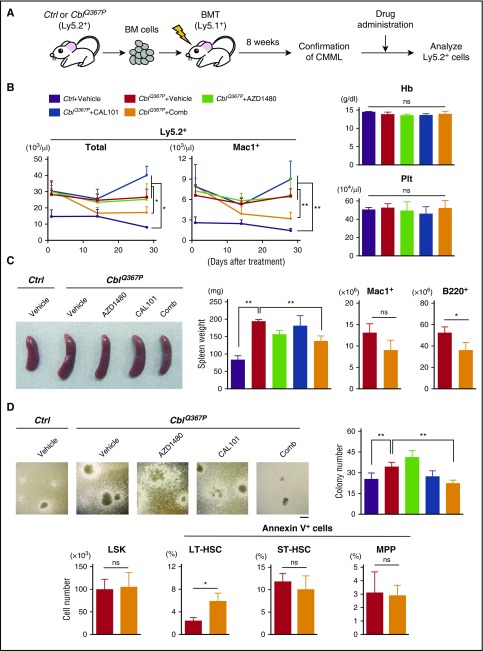
Finally, we investigated the possibility of molecular targeted therapy for CMML carrying a Cbl mutation. Given that the PI3K-AKT and JAK-STAT pathways are activated in disease-initiating LT-HSCs of CblQ367P mice (Figure 2), we tested the effect(s) of anti-Jak2 and anti-PI3K inhibitors on the growth of CblQ367P cells. Recipient mice transplanted with CblQ367P cells (CblQ367P recipients) were orally administered with a vehicle, a JAK2 inhibitor (AZD1480), a PI3K inhibitor (CAL101), or a combination of AZD1480 and CAL101 (Comb ) (Figure 7A). Mice transplanted with control cells and treated with a vehicle were used as a negative control. Although no decrease in WBC number was detected in CblQ367P recipients treated with vehicle (CblQ367P + Vehicle), AZD1480 alone (CblQ367P + AZD1480), and CAL101 alone (CblQ367P + CAL101), those treated with combination therapy of both AZD1480 and CAL101 (CblQ367P + Comb) exhibited a significant decrease in WBC counts (Figure 7B, first left panel). Notably, the number of Mac1+ cells was significantly lower in CblQ367P + Comb than in CblQ367P + Vehicle, CblQ367P + AZD1480, and CblQ367P + CAL101 recipients (Figure 7B, second left panel). No changes were observed in hemoglobin concentrations and platelet counts (Figure 7B, right panels), indicating that the combination therapy exerted its inhibitory effect mainly on proliferating malignant cells. In accordance with these observations, the spleen weights of CblQ367P + Comb recipient mice was significantly decreased with a reduction of Mac1+ and B220+ cells (Figure 7C). In the BM, colony formation activity was significantly decreased in CblQ367P + Comb (Figure 7D, upper panels), and although no significant change was observed in LSK cell numbers between CblQ367P + Vehicle and CblQ367P + Comb recipients (Figure 7D, lower left panel), the percentage of Annexin V+ cells in the LT-HSC fraction was significantly increased in CblQ367P + Comb compared with CblQ367P + Vehicle recipients (Figure 7D, second lower left panel). The immunofluorescent staining showed that activation of STAT5 was significantly inhibited in LT-HSCs of CblQ367P + Comb compared with those of CblQ367P + Vehicle recipients (supplemental Figure 5). Because the LT-HSC fraction contained disease-initiating cells (Figure 3A), these results indicate that the combination therapy of AZD1480 and CAL101 suppressed proliferation of CMML stem cells by inducing apoptosis through mainly inhibiting the JAK2-STAT5 pathway.
Figure 7.
Drug application to CblQ367PCMML cells. (A) Experimental procedure of drug application. BM cells of control or CblQ367P mice (Ly5.2+) were transplanted into irradiated syngeneic mice (Ly5.1+), and, after confirming the development of CMML, recipient mice were subjected to drug administration, and the Ly5.2+ cells were analyzed. (B) Left two panels: Changes in the numbers of total and Mac1+ Ly5.2+ cells in the PB of drug-treated recipients during the observation period. Right panels: hemoglobin (Hb) concentrations and platelet (Plt) count at 4 weeks after treatment. (C) Analyses of the spleen. Left two panels: Representative macroscopic appearances and weights of the spleens of drug-treated recipients. Right two panels: Mac1+ and B220+ cell numbers of CblQ367P+Vehicle and CblQ367P+Comb recipients. (D) Analyses of the BM. Upper panels: Representative photos taken with a phase-contrast microscope and total colony numbers (diameter, >500 µm) of drug-treated recipients. Scale bar: 500 μm. Lower panels: LSK cell numbers and percentages of Annexin V+ cells in the LT-HSC, ST-HSC and MPP fractions of CblQ367P+Vehicle and CblQ367P+Comb recipients. **P < .01, *P < .05. ns, not significant.
Discussion
CMML is the most commonly observed hematopoietic neoplasm in patients who fulfill the criteria of MDS/MPN.3 We previously identified somatic and uniparental mutations in the Cbl gene in a significant portion of CMML patients.7 To clarify the contribution of acquired mutation of Cbl to CMML pathogenesis, we generated CblQ367P mice that express normal CBL in a steady state and in turn express CBLQ367P upon induction. In the CblQ367P mice, both Cbl alleles were converted from CblWT to CblQ367P (supplemental Figure 1), which recapitulates the aUPD of 11q in CMML patients.7 CblQ367P mice exhibited hallmarks of CMML, including prolonged and sustained elevation of myelomonocytes, multilineage dysplasia, splenomegaly, and the formation of spontaneous colonies (Figure 1), demonstrating that acquired biallelic expression of mutant Cbl initiates and maintains the phenotypes of CMML. To analyze the effect of CblQ367P heterozygosity on hematopoiesis, we generated Cbl+/WT, Cbl+/Q367P, and CblQ367P/Q367P mice and analyzed the PB data. As shown in supplemental Figure 6, the WBC count of Cbl+/Q367P mice was slightly elevated compared with Cbl+/WT mice, but did not show a significant increase as observed in CblQ367P/Q367P mice. These data suggest that the heterozygous state of mutant Cbl is not sufficient to develop CMML and provide experimental evidence that CMML with Cbl mutations is almost exclusively associated with 11q aUPD.7
By using competitive repopulation assays, we demonstrated that CblQ367P-induced CMML originates from mutant LT-HSCs (Figure 3), as reported in CblC379A/–-induced myeloid diseases.13 In addition, we detected increased phosphorylation of AKT and STAT proteins in CblQ367P LT-HSCs (Figure 2). Gene expression analysis demonstrated significant enrichments of cell cycle–regulating and chemokine-chemokine receptor genes (Figure 4). These results indicate that CBLQ367P activates the PI3K-AKT and JAK-STAT pathways in LT-HSCs, which eventually promotes cell proliferation by upregulating cell cycle- and chemokine-related genes and consequently leads to the development of CMML. Studies have demonstrated that the chemokine pathway plays essential roles in HSC homing,31 raising the possibility that the enhanced repopulation activity of CblQ367P LT-HSCs (Figure 3) is attributed to the increased homing ability. To address this issue, we performed a BMT analysis, in which recipients were transplanted with CblWT/WT, MxCre− or CblWT/WT, MxCre+ LT-HSCs first, and then administered pIpC after confirmation of repopulation. As shown in supplemental Figure 7, CblWT/WT, MxCre+ LT-HSCs exhibited significantly higher chimerism than CblWT/WT, MxCre− cells after pIpC treatment, strongly suggesting that the activation of the chemokine pathway contributes to proliferation and survival rather than to the homing of the CblQ367P leukemic cells, as reported in a previous study.32
It is noteworthy that Gem was commonly upregulated in the HSPCs of 2 different CMML models induced by CblQ367P (DRX046249 and DRX046250) and NrasG12D21 (supplemental Figure 3A). We demonstrated that enhanced expression of Gem increased the percentages of HSPCs and myelomonocytes in both in vitro cell culture and in vivo BMT studies (Figure 5). GEM belongs to the Ras superfamily of small GTPases that are involved in various intracellular pathways.33-35 Studies have demonstrated that GEM contributes to cell invasion,36 mitotic progression,37 and actin remodeling.38 In addition, GEM has been reported to be an adverse prognostic factor in patients with bladder carcinoma39 and upregulated in mutant ALK-expressing neuroblastoma cells.40 In human hematopoietic malignancies, studies showed that the expression levels of Gem are higher in AML samples than B-cell and T-cell ALL samples41 and increase as chronic myeloid leukemia, another chronic hematologic disorder of the myeloid lineage, progresses from the chronic phase to blast crisis through the acute phase42 (supplemental Figure 8). Therefore, it is suggested that the expression of Gem is predominantly associated with myeloid malignancies, and its expression levels are correlated with the malignant progression of the diseases.
CMML frequently progresses to AML.4,6 Investigation of the mechanisms underlying the disease progression has identified genetic alterations, such as mutations in the Asxl1 (additional sex combs-like 1), Setbp1 (SET-binding protein 1), and NPM1 (nucleophosmin) genes43-46; however, little is known about genes whose expressional changes are responsible for disease evolution. By employing retrovirus-mediated mutagenesis, we identified Evi1, a gene encoding a transcription factor involved in normal hematopoiesis and leukemogenesis,28 as a cooperative gene with CblQ367P and demonstrated that overexpressed EVI1 synergized with CBLQ367P to develop AML. These findings support the idea that class I mutations (activation of signal transduction, mutant CBL) and class II mutations (deregulation of transcription factors, overexpression of EVI1) play a cooperative role in the development of AML, as proposed in previous studies.47,48
CMML is a difficult disease to cure, and BMT is so far the only curative therapy. However, for patients who are ineligible for BMT, other therapeutic approaches should be considered. Because mutant CBL induced hematopoietic diseases through increased tyrosine phosphorylation of downstream molecules, tyrosine kinase inhibitors (TKIs) are expected to be effective. However, although application of Dasatinib, a TKI, to CblC379A/− knockin mice suppressed the number of WBCs, the decreased cells were mainly B lymphocytes, and no apparent effect was observed in the myeloid and HSC populations.49 In our study, administration of Dasatinib to CblQ367P mice did not decrease, but occasionally increased, the WBC number, including myeloid cells (data not shown). These findings strongly suggest that dasatinib treatment is not effective for eradicating CMML cells expressing mutant CBL. On the other hand, several different therapeutic approaches for juvenile myelomonocytic leukemia and CMML have been reported, using anti-MEK-ERK and/or anti–JAK-STAT inhibitors.50-52 Given the results that the PI3K-AKT and JAK-STAT pathways are constitutively activated in CblQ367P LT-HSCs (Figure 2), we applied anti-PI3K and/or anti-JAK inhibitors to CBLQ367P cell–transplanted recipient mice. We found that combination therapy significantly reduced the WBC number, including myelomonocytes, regressed splenomegaly with reduction of myeloid and B lymphocytes, suppressed hematopoietic colony formation, and induced apoptosis of disease-initiating LT-HSCs (Figure 7). The analysis of pAKT and pSTAT proteins in LT-HSCs showed that, although phosphorylation of STAT5 was significantly inhibited in CblQ367P + Comb compared with CblQ367P + Vehicle recipients, those of STAT3 and AKT were unchanged (supplemental Figure 5). These results indicate that the JAK2-STAT5 pathway mainly promotes the proliferation of CMML stem cells, as proposed in a previous study,53 and suggest that the anti-PI3K inhibitor may exert its antiproliferative potential by other mechanism(s) than the phosphorylation of AKT. In human myeloid cell lines with or without Cbl mutations, treatment with anti-PI3K and/or anti-JAK inhibitors also showed that cell lines with Cbl mutations tended to be more sensitive to drug treatment than those without, especially when cultured with a cytokine (supplemental Figure 9). Our findings, together with the results of previous studies,50-52 suggest that combined targeting of the PI3K-AKT and JAK-STAT pathways might be promising for patients with CMML carrying a Cbl mutation who are not eligible for intensive chemotherapies and/or BMT.
In summary, by generating and analyzing CblQ367P mice, we demonstrated that (1) acquired expression of mutant CBL is sufficient for developing CMML, (2) the disease originates from mutant LT-HSCs, (3) the PI3K-AKT and JAK-STAT pathways are constitutively activated in CMML stem cells, (4) deregulated expression of GEM, a GTPase protein, at least partly participates in the phenotypes of CMML, (5) overexpressed EVI1 cooperates with mutant CBL and transforms CMML to AML, and (6) combined inhibition of the PI3K-AKT and JAK-STAT pathways may be effective for the suppression of mutant CBL–expressing cells (supplemental Fig. 10). Our CblQ367P model will be useful not only for further clarifying the molecular mechanism(s) underlying mutant Cbl–induced hematopoietic malignancy, but also for developing rational therapies for mutant CBL–carrying CMML patients.
Acknowledgments
The authors thank Yuki Sakai, Sawako Ogata, and Rika Tai for animal care, genotyping, and molecular experiments. The authors also thank Junji Takeda (Osaka University) and the RIKEN BioResource Center for providing us with KY1.1 ES cells and B6-Tg(CAG-FLPe)36 mice (RBRC01834), respectively.
This work was supported in part by a grant-in-aid from the Ministry of Education, Science and Culture of Japan and by the Tsuchiya Foundation.
Footnotes
The data reported in this article have been deposited in the DNA Data Bank of Japan (accession numbers DRX046249 and DRX046250).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: Y.N., A.N., M.S., S.O., Z.-i.H., T.S., T.I., and H.H. designed the research, generated the cKI mice, and wrote the manuscript; T.U., N.Y., K.-i.I., Y.S., and K. Takubo performed HSPC analyses; K. Tsuji and Y.E. participated in hematopoietic colony assays; A.K. performed RNA sequence analysis; H.O. analyzed pathological specimens; L.W. generated the MOL4070A retrovirus; and all of the authors checked the final version of the manuscript.
Conflict of interest disclosure: The authors declare no competing financial interests.
Correspondence: Hiroaki Honda, Department of Disease Model, Research Institute for Radiation Biology and Medicine, Hiroshima University, 1-2-3 Kasumi, Minami-ku, Hiroshima 734-8553, Japan; e-mail: hhonda@hiroshima-u.ac.jp.
References
- 1.Raza A, Galili N. The genetic basis of phenotypic heterogeneity in myelodysplastic syndromes. Nat Rev Cancer. 2012;12(12):849-859. [DOI] [PubMed] [Google Scholar]
- 2.Bejar R, Steensma DP. Recent developments in myelodysplastic syndromes. Blood. 2014;124(18):2793-2803. [DOI] [PubMed] [Google Scholar]
- 3.Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th ed, vol. 2 Geneva, Switzerland: WHO Press; 2008. [Google Scholar]
- 4.Itzykson R, Solary E. An evolutionary perspective on chronic myelomonocytic leukemia. Leukemia. 2013;27(7):1441-1450. [DOI] [PubMed] [Google Scholar]
- 5.Patnaik MM, Parikh SA, Hanson CA, Tefferi A. Chronic myelomonocytic leukaemia: a concise clinical and pathophysiological review. Br J Haematol. 2014;165(3):273-286. [DOI] [PubMed] [Google Scholar]
- 6.Patnaik MM, Tefferi A. Cytogenetic and molecular abnormalities in chronic myelomonocytic leukemia. Blood Cancer J. 2016;6:e393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanada M, Suzuki T, Shih LY, et al. Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature. 2009;460(7257):904-908. [DOI] [PubMed] [Google Scholar]
- 8.Nadeau S, An W, Palermo N, et al. Oncogenic signaling by leukemia-associated mutant Cbl proteins. Biochem Anal Biochem. 2012;S6:001. doi:10.4172/2161-1009.S6-001. [DOI] [PMC free article] [PubMed]
- 9.Katzav S, Schmitz ML. Mutations of c-Cbl in myeloid malignancies. Oncotarget. 2015;6(13):10689-10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohapatra B, Ahmad G, Nadeau S, et al. Protein tyrosine kinase regulation by ubiquitination: critical roles of Cbl-family ubiquitin ligases. Biochim Biophys Acta. 2013;1833(1):122-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy MA, Schnall RG, Venter DJ, et al. Tissue hyperplasia and enhanced T-cell signalling via ZAP-70 in c-Cbl-deficient mice. Mol Cell Biol. 1998;18(8):4872-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naramura M, Kole HK, Hu RJ, Gu H. Altered thymic positive selection and intracellular signals in Cbl-deficient mice. Proc Natl Acad Sci USA. 1998;95(26):15547-15552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rathinam C, Thien CB, Flavell RA, Langdon WY. Myeloid leukemia development in c-Cbl RING finger mutant mice is dependent on FLT3 signaling. Cancer Cell. 2010;18(4):341-352. [DOI] [PubMed] [Google Scholar]
- 14.Kühn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269(5229):1427-1429. [DOI] [PubMed] [Google Scholar]
- 15.Ziegler-Heitbrock L, Ancuta P, Crowe S, et al. Nomenclature of monocytes and dendritic cells in blood. Blood. 2010;116(16):e74-e80. [DOI] [PubMed] [Google Scholar]
- 16.Bowen DT. Chronic myelomonocytic leukemia: lost in classification? Hematol Oncol. 2005;23(1):26-33. [DOI] [PubMed] [Google Scholar]
- 17.Tefferi A. Novel mutations and their functional and clinical relevance in myeloproliferative neoplasms: JAK2, MPL, TET2, ASXL1, CBL, IDH and IKZF1. Leukemia. 2010;24(6):1128-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kales SC, Ryan PE, Nau MM, Lipkowitz S. Cbl and human myeloid neoplasms: the Cbl oncogene comes of age. Cancer Res. 2010;70(12):4789-4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naramura M, Nadeau S, Mohapatra B, et al. Mutant Cbl proteins as oncogenic drivers in myeloproliferative disorders. Oncotarget. 2011;2(3):245-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Itzykson R, Kosmider O, Renneville A, et al. Clonal architecture of chronic myelomonocytic leukemias. Blood. 2013;121(12):2186-2198. [DOI] [PubMed] [Google Scholar]
- 21.Li Q, Bohin N, Wen T, et al. Oncogenic Nras has bimodal effects on stem cells that sustainably increase competitiveness. Nature. 2013;504(7478):143-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolff L, Koller R, Hu X, Anver MR. A Moloney murine leukemia virus-based retrovirus with 4070A long terminal repeat sequences induces a high incidence of myeloid as well as lymphoid neoplasms. J Virol. 2003;77(8):4965-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Justice MJ, Noveroske JK, Weber JS, Zheng B, Bradley A. Mouse ENU mutagenesis. Hum Mol Genet. 1999;8(10):1955-1963. [DOI] [PubMed] [Google Scholar]
- 24.Yamasaki N, Miyazaki K, Nagamachi A, et al. Identification of Zfp521/ZNF521 as a cooperative gene for E2A-HLF to develop acute B-lineage leukemia. Oncogene. 2010;29(13):1963-1975. [DOI] [PubMed] [Google Scholar]
- 25.Nagamachi A, Matsui H, Asou H, et al. Haploinsufficiency of SAMD9L, an endosome fusion facilitator, causes myeloid malignancies in mice mimicking human diseases with monosomy 7. Cancer Cell. 2013;24(3):305-317. [DOI] [PubMed] [Google Scholar]
- 26.Ueda T, Nagamachi A, Takubo K, et al. Fbxl10 overexpression in murine hematopoietic stem cells induces leukemia involving metabolic activation and upregulation of Nsg2. Blood. 2015;125(22):3437-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ueda T, Nakata Y, Nagamachi A, et al. Propagation of trimethylated H3K27 regulated by polycomb protein EED is required for embryogenesis, hematopoietic maintenance, and tumor suppression. Proc Natl Acad Sci USA. 2016;113(37):10370-10375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nucifora G, Laricchia-Robbio L, Senyuk V. EVI1 and hematopoietic disorders: history and perspectives. Gene. 2006;368:1-11. [DOI] [PubMed] [Google Scholar]
- 29.Kastner P, Chan S. PU.1: a crucial and versatile player in hematopoiesis and leukemia. Int J Biochem Cell Biol. 2008;40(1):22-27. [DOI] [PubMed] [Google Scholar]
- 30.Richter AM, Pfeifer GP, Dammann RH. The RASSF proteins in cancer; from epigenetic silencing to functional characterization. Biochim Biophys Acta. 2009;1796(2):114-128. [DOI] [PubMed] [Google Scholar]
- 31.Suárez-Álvarez B, López-Vázquez A, López-Larrea C. Mobilization and homing of hematopoietic stem cells. Adv Exp Med Biol. 2012;741:152-170. [DOI] [PubMed] [Google Scholar]
- 32.Sison EA, Brown P. The bone marrow microenvironment and leukemia: biology and therapeutic targeting. Expert Rev Hematol. 2011;4(3):271-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olson MF. Gem GTPase: between a ROCK and a hard place. Curr Biol. 2002;12(14):R496-R498. [DOI] [PubMed] [Google Scholar]
- 34.Correll RN, Pang C, Niedowicz DM, Finlin BS, Andres DA. The RGK family of GTP-binding proteins: regulators of voltage-dependent calcium channels and cytoskeleton remodeling. Cell Signal. 2008;20(2):292-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sasson Y, Navon-Perry L, Huppert D, Hirsch JA. RGK family G-domain:GTP analog complex structures and nucleotide-binding properties. J Mol Biol. 2011;413(2):372-389. [DOI] [PubMed] [Google Scholar]
- 36.Suyama E, Kawasaki H, Nakajima M, Taira K. Identification of genes involved in cell invasion by using a library of randomized hybrid ribozymes. Proc Natl Acad Sci USA. 2003;100(10):5616-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andrieu G, Quaranta M, Leprince C, Hatzoglou A. The GTPase Gem and its partner Kif9 are required for chromosome alignment, spindle length control, and mitotic progression. FASEB J. 2012;26(12):5025-5034. [DOI] [PubMed] [Google Scholar]
- 38.Andrieu G, Quaranta M, Leprince C, Cuvillier O, Hatzoglou A. Gem GTPase acts upstream Gmip/RhoA to regulate cortical actin remodeling and spindle positioning during early mitosis. Carcinogenesis. 2014;35(11):2503-2511. [DOI] [PubMed] [Google Scholar]
- 39.Laurberg JR, Jensen JB, Schepeler T, Borre M, Ørntoft TF, Dyrskjøt L. High expression of GEM and EDNRA is associated with metastasis and poor outcome in patients with advanced bladder cancer. BMC Cancer. 2014;14:638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cazes A, Lopez-Delisle L, Tsarovina K, et al. Activated Alk triggers prolonged neurogenesis and Ret upregulation providing a therapeutic target in ALK-mutated neuroblastoma. Oncotarget. 2014;5(9):2688-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramaswamy S, Tamayo P, Rifkin R, et al. Multiclass cancer diagnosis using tumor gene expression signatures. Proc Natl Acad Sci USA. 2001;98(26):15149-15154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radich JP, Dai H, Mao M, et al. Gene expression changes associated with progression and response in chronic myeloid leukemia. Proc Natl Acad Sci USA. 2006;103(8):2794-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gelsi-Boyer V, Trouplin V, Roquain J, et al. ASXL1 mutation is associated with poor prognosis and acute transformation in chronic myelomonocytic leukaemia. Br J Haematol. 2010;151(4):365-375. [DOI] [PubMed] [Google Scholar]
- 44.Becker H, Yoshida K, Blagitko-Dorfs N, et al. Tracing the development of acute myeloid leukemia in CBL syndrome. Blood. 2014;123(12):1883-1886. [DOI] [PubMed] [Google Scholar]
- 45.Patnaik MM, Itzykson R, Lasho TL, et al. ASXL1 and SETBP1 mutations and their prognostic contribution in chronic myelomonocytic leukemia: a two-center study of 466 patients. Leukemia. 2014;28(11):2206-2212. [DOI] [PubMed] [Google Scholar]
- 46.Peng J, Zuo Z, Fu B, et al. Chronic myelomonocytic leukemia with nucleophosmin (NPM1) mutation. Eur J Haematol. 2016;96(1):65-71. [DOI] [PubMed] [Google Scholar]
- 47.Kelly LM, Gilliland DG. Genetics of myeloid leukemias. Annu Rev Genomics Hum Genet. 2002;3:179-198. [DOI] [PubMed] [Google Scholar]
- 48.Takahashi S. Current findings for recurring mutations in acute myeloid leukemia. J Hematol Oncol. 2011;4:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duyvestyn JM, Taylor SJ, Dagger SA, et al. Dasatinib targets B-lineage cells but does not provide an effective therapy for myeloproliferative disease in c-Cbl RING finger mutant mice. PLoS One. 2014;9(4):e94717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lyubynska N, Gorman MF, Lauchle JO, et al. A MEK inhibitor abrogates myeloproliferative disease in Kras mutant mice. Sci Transl Med. 2011;3(76):76ra27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang T, Krisman K, Theobald EH, et al. Sustained MEK inhibition abrogates myeloproliferative disease in Nf1 mutant mice. J Clin Invest. 2013;123(1):335-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kong G, Wunderlich M, Yang D, et al. Combined MEK and JAK inhibition abrogates murine myeloproliferative neoplasm. J Clin Invest. 2014;124(6):2762-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kato Y, Iwama A, Tadokoro Y, et al. Selective activation of STAT5 unveils its role in stem cell self-renewal in normal and leukemic hematopoiesis. J Exp Med. 2005;202(1):169-179. [DOI] [PMC free article] [PubMed] [Google Scholar]