Abstract
Synaptonemal complexes (SCs) are evolutionary conserved, meiosis-specific structures that play a central role in synapsis of homologous chromosomes, chiasmata distribution, and chromosome segregation. However, it is still for the most part unclear how SCs do assemble during meiotic prophase. Major components of mammalian SCs are the meiosis-specific proteins SCP1, 2, and 3. To investigate the role of SCP1 in SC assembly, we expressed SCP1 in a heterologous system, i.e., in COS-7 cells that normally do not express SC proteins. Notably, under these experimental conditions SCP1 is able to form structures that closely resemble SCs (i.e., polycomplexes). Moreover, we show that mutations that modify the length of the central α-helical domain of SCP1 influence the width of polycomplexes. Finally, we demonstrate that deletions of the nonhelical N- or C-termini both affect polycomplex assembly, although in a different manner. We conclude that SCP1 is a primary determinant of SC assembly that plays a key role in synapsis of homologous chromosomes.
INTRODUCTION
During the extended prophase of the first meiotic division homologous chromosomes pair, synapse, and recombine. These processes are essential to ensure a correct chromosome segregation during the reductional first meiotic division. Synaptonemal complexes (SCs) are meiosis-specific structures that play a key role during these processes (von Wettstein et al., 1984; Zickler and Kleckner, 1998; Page and Hawley, 2004).
When completely assembled at pachytene stage of meiotic prophase I each SC extends all along a chromosome bivalent (Wettstein and Sotelo, 1971; von Wettstein et al., 1984; Zickler and Kleckner, 1998). SCs exhibit a typical tripartite ladder-like organization that is characterized by the presence of two lateral elements, to which the chromatin of the homologous chromosomes is attached, and the central region that is located in the gap between lateral elements. The central region is composed of a central element and numerous perpendicularly oriented transverse filaments that link the lateral elements with the central element (Wettstein and Sotelo, 1971; Schmekel and Daneholt, 1995).
Three major SC protein components have been described in mammals, the meiosis-specific proteins SCP1, 2, and 3 (Heyting, 1996). Although SCP2 and SCP3 are components of the lateral elements (Lammers et al., 1994; Dobson et al., 1994; Offenberg et al., 1998), previous findings indicate that SCP1 is the major component of transverse filaments, i.e., the structures playing an important role in interconnecting the homologous chromosomes (Meuwissen et al., 1992; Dobson et al., 1994; Liu et al., 1996; Schmekel et al., 1996).
SCP1 is a large fibrillar molecule (997 amino acids in the rat) composed of a central α-helical domain (699 amino acids long), flanked by globular N- and C-termini (Meuwissen et al., 1992). Available evidence indicates that two parallelly oriented SCP1 molecules interact with each other via the α-helical domain, forming a coiled-coil rod structure (Liu et al., 1996; Schmekel et al., 1996; see also Lupas et al., 1991). The C-terminus of SCP1 is embedded in one of the two lateral elements, where it may interact with chromatin through putative DNA-binding motifs (Meuwissen et al., 1992). The N-terminus is localized to the central element where it appears to interact head-to-head with the N-termini of SCP1 molecules anchored to the opposite lateral element (Liu et al., 1996; Schmekel et al., 1996). It is worth mentioning that putative transverse filament protein components of evolutionarily distant species lack sequence homologies, but show close similarities at the level of secondary structure and organization (Meuwissen et al., 1992; Sym et al., 1993; Dong and Roeder, 2000; Page and Hawley, 2001; MacQueen et al., 2002; Colaiácovo et al., 2003).
To investigate the contribution of SCP1 to the assembly of mammalian SCs, we expressed rat SCP1 as well as selected SCP1 mutants in a heterologous system, i.e., the cell line COS-7, which normally does not express SC proteins (Yuan et al., 1996, 1998). We found that under these experimental conditions SCP1 self-assembled to form structures that closely resemble SCs.
MATERIALS AND METHODS
Constructs
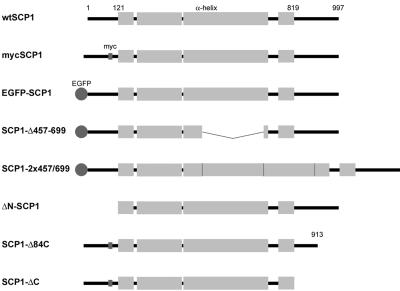
SCP1 cDNA of the rat (NM_012810; Meuwissen et al., 1992; Sage et al., 1995) was obtained by RT-PCR. As a template we used total RNA isolated from pachytene spermatocytes of Wistar rats that were highly enriched by centrifugal elutriation (Meistrich, 1977). For expression studies in COS-7 cells, the coding region was cloned into vector pEGFP-N3 (Clontech, Heidelberg, Germany) to express the wild-type protein and alternatively inserted into vector pEGFP-C3 to express the EGFP-SCP1 fusion protein. To create a myc-tagged SCP1 construct (mycSCP1), a double-stranded oligonucleotide encoding the myc-epitope leaded by 5′ CA (CA GAA CAA AAA CTC ATC TCA GAA GAG GAT CTG) with a single-stranded AATT-overhang at each side was inserted into the above described construct at the EcoRI restriction site at base pair position 402 of the cDNA (i.e., between amino acids 99 and 100). To delete the sequence coding for the first 120 amino acids of SCP1 (ΔN-SCP1), a PCR reaction was performed using a 5′-primer that starts at base pair 469 and is headed by a Kozak consensus sequence (Kozak, 1987) and the start codon ATG and a 3′-primer ending at position 3102. To delete the sequence corresponding to the last 84 or 178 amino acids of SCP1 (SCP1-Δ84C and SCP1-ΔC, respectively), we used the construct coding for myc-SCP1 as template and performed a PCR by using a 5′-primer starting at position 84 and a 3′-primer ending at position 2847 or 2565, each followed by a stop codon. For reduction of the length of the coiled-coil domain of SCP1 (amino acids 121–819; Lupas et al., 1991) the sequence between the two HindIII restriction sites (positions 1474 and 2203) was deleted (SCP1-Δ457-699). Instead, this sequence was duplicated in order to obtain an extension of the coiled-coil (SCP1–2x457-699).
Transfection and Microinjection
COS-7 cells were grown on coverslips according to standard procedures. The cells were transfected for 2–2.5 h with 2–4 μg of vector DNA and lipofectamin 2000 in 1 ml according to the instructions of the supplier (Invitrogen, Karlsruhe, Germany). Needle microinjection of cells (175 ng/μl vector-DNA) grown on CELLocate coverslips (Eppendorf, Hamburg, Germany) was performed according to standard procedures using Femtotips (Eppendorf) and a Microinjector 5242 (Eppendorf). The cells were then incubated for 24 h and processed for immunofluorescence or transmission electron microscopy.
Antibodies
For detection of SCP1 we generated a polyclonal serum by immunizing a rabbit with a peptide corresponding to the last 76 amino acids of SCP1. Before use the antibody was affinity purified. Myc-tagged SCP1 constructs were detected by monoclonal mouse-α-myc antibody purchased from Invitrogen. Secondary antibodies coupled to fluorochromes or 6-nm gold particles were purchased from Dianova (Hamburg, Germany).
Immunofluorescence Microscopy
Cells were fixed for 3–5 min in PBS containing 2% formaldehyde and permeabilized for 10 min in PBS-T (PBS plus 0.1% Triton X-100). After blocking for 10 min in PBS containing 0.1 M glycine, the cells were incubated with the primary antibody, washed in PBS, and then incubated with the secondary antibody coupled to Texas red (20 min). DNA in the preparations was visualized with the fluorochrome Hoechst 33258 (Hoechst, Frankfurt, Germany). After washing in PBS, the coverslips were mounted on glass slides with mowiol (Alsheimer et al., 2000).
Electron Microscopy
Cells were fixed for 30 min in 2.5% glutaraldehyde and postfixed in 2% osmium tetroxide. Afterward, the cells were washed in water and incubated overnight in 0.5% aqueous uranyl acetate. After dehydration in ethanol series, the cells were embedded in Epon. Ultrathin sections were stained with uranyl acetate and lead citrate according to standard procedures (Liebe et al., 2004). For the immunogold localization of the C-terminus of SCP1, the cells were processed as described above for the immunofluorescence, with the exception that gold-coupled secondary antibody (2 h) were used.
RESULTS AND DISCUSSION
SCP1 Forms Polycomplexes in Transfected COS-7 Cells
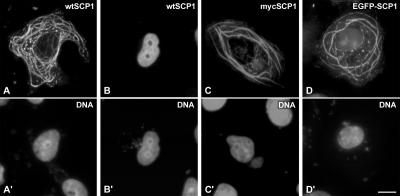
In a first set of experiments we expressed wild-type (wt) SCP1 in cultured COS-7 cells (Figures 1 and 2). When analyzed by immunofluorescence microscopy, 41% (n = 424) of the transfected cells showed an extensive network of cytoplasmic fibers and aggregates (Figure 2A) that are selectively recognized by SCP1 antibodies. In contrast, cells without such cytoplasmic structures (59%, n = 424) displayed no cytoplasmic fluorescence, but a homogeneous labeling of nuclei (Figure 2B). Figure 2 also shows that SCP1 tagged with a myc epitope (Figure 2C) or expressed as a EGFP fusion protein (Figure 2D) was able to form cytoplasmic structures that were virtually identical to those formed by wtSCP1.
Figure 1.
Schematic representation of rat SCP1 and SCP1 mutants expressed in COS-7 cells. Wild-type SCP1 of the rat is 997 amino acids long; 121 and 819 indicate the first and last amino acids of the α-helical domain.
Figure 2.
Expression of wtSCP1, myc-tagged SCP1 and EGFP-SCP1 fusion protein in cultured COS-7 cells as shown by fluorescence microscopy (A–D). The same cells were incubated with the DNA-specific fluorochrome Hoechst 33258 (A′–D′). The distribution of wtSCP1 (A and B) and myc-tagged SCP1 (C) was visualized with the aid of specific antibodies. Scale bar, 10 μm.
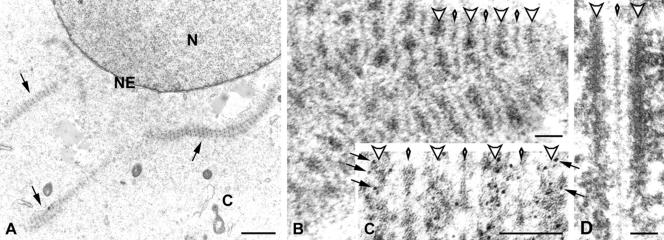
By the use of electron microscopy (EM) we obtained more detailed information about the cytoplasmic structures formed in SCP1-transfected cells. At this level of resolution fibers and aggregates showed a conspicuous organization. Remarkably, they resembled stacks of SCs arranged in parallel and were indistinguishable from the so-called polycomplexes (Figure 3, A and B; Sotelo and Trujillo-Cenóz, 1960; for review see Goldstein, 1987). Polycomplexes have been described previously in normal germ line cells of different species and are thought to be large assemblies of SC proteins that are not associated with chromosomes. The function of polycomplexes is not known, but they might represent storaged SC proteins (Goldstein, 1987). Like the germ line polycomplexes, those assembled by SCP1 in COS-7 cells showed a sequence of dense lines (resembling the lateral elements of SCs) alternating with lines of lesser density (the central element of SCs), which were connected to each other via transverse filaments (Figure 3, A and B).
Figure 3.
(A and B) Electron microscopy showing polycomplexes expressed in COS-7 cells. (A) Polycomplexes are denoted by arrows. N, cell nucleus; C, cytoplasm; NE, nuclear envelope. (B) At higher magnification polycomplexes are visualized as a sequence of dense lines (large arrowheads) that alternate with less dense lines (small arrowheads). Transverse filaments connect dense and less dense lines with each other. (C) Electron microscopic immunolocalization of SCP1 using a primary antibody specific for the C-terminal end of the molecules and secondary antibodies conjugated to 6-nm gold particles. Gold labeling (some of the particles are denoted by arrows) is distributed over the dense lines (large arrowheads), whereas the lines of lesser density (small arrowheads) and transverse filaments are free of gold particles. (D) For comparison, a rat SC is shown at the same final magnification as the polycomplexes in B. The SC lateral elements are denoted by large arrowheads and the central element by a small arrowhead. Scale bars, 1 μm (A), 0.1 μm (B–D).
In previous studies designed to determine the orientation of SCP1 molecules in SCs, the authors (Liu et al., 1996; Schmekel et al., 1996) used antibodies against SCP1 domains and immunogold EM. Using this experimental approach and an antibody specific for the C-terminus of SCP1, we found a selective labeling of the dense lines, whereas the lines of lesser density and the transverse filaments were virtually free of gold particles (Figure 3C). This result leads to the conclusion that SCP1 molecules are highly organized within polycomplexes and oriented in the same manner as in SCs.
Interestingly, polycomplexes were located in the cytoplasm of COS-7 cells, despite the fact that SCP1 is a nuclear protein. This feature could be related to the situation in which CHO cells were transfected with the major lateral element protein SCP3 (Yuan et al., 1998). Under these conditions, most of the fibers assembled by SCP3 were cytoplasmic. Most likely, self-assembly in the cytoplasm of culture cells overexpressing SCP1 or SCP3 is a consequence of the relative high concentrations reached at the sites of synthesis. Why SCP1 does not assemble polycomplexes in the nuclear compartment of COS-7 cells cannot be answered at present. However, this may be related to the fact that we are dealing with somatic, but not with meiotic nuclei. Furthermore, it is conceivable that overexpressed SCP1 interacts with chromatin via its C-terminal DNA binding motifs (Meuwissen et al., 1992), which, in turn, may interfere with self-assembly of this protein.
What are the mechanisms in meiotic cells that modulate SCP1 polymerization promoting the assembly of the central region of SCs instead of nonchromosomal polycomplexes? At present, there is no answer to this question, and to our knowledge, no SCP1-interacting protein partners have been described. Available evidence appears to exclude the other two major SC-proteins SCP2 and SCP3 as possible candidates, because biochemical and genetic approaches failed to demonstrate a direct interaction between SCP1 and SCP2 or SCP3 (Liu et al., 1996; Tarsounas et al., 1997; Pelttari et al., 2001). In support of these observations, we have recently shown that mice spermatocytes lacking SCP3 do assemble SC-like structures on meiotic chromosomes (Liebe et al., 2004). These structures that contain SCP1, but not SCP2 (Yuan et al., 2000; Pelttari et al., 2001), were found to mediate synapsis between homologous chromosomes (Liebe et al., 2004). At the ultrastructural level, they lack lateral elements and are composed of a central element and transverse filaments (Liebe et al., 2004). In other terms, the structures resulting from the assembly of SCP1 (central element and transverse filaments, see above) can be formed between homologous chromosomes in the absence of SCP2 and SCP3.
When overexpressed during yeast meiosis, yeast transverse filament-protein Zip1 became integrated into SCs and polycomplexes were also formed (Sym and Roeder, 1995). Our observation that SCP1 formed polycomplexes in a nonmeiotic milieu provides further support to the notion, that the polycomplexes formed during yeast meiosis resulted from the self-assembly of overexpressed Zip1 without the aid of other SC proteins (Sym and Roeder, 1995). Furthermore, the fact that both SCP1 and Zip1 form polycomplexes strongly suggests that these proteins are functional homologues, despite the fact that they display no primary amino acid sequence homologies.
The experiments described in Figures 2 and 3 do not allow the conclusion that SCP1 is the only central region protein of the mammalian SC. However, the fact that SCP1 can form central region structures on its own clearly shows that SCP1 is a major determinant of central region assembly. Therefore, we propose that SCP1 constitutes the backbone of the central region (transverse filaments and central element) of mammalian SCs and functions as a molecular framework to which other proteins attach.
Protein Domains of SCP1 and Polycomplex Assembly
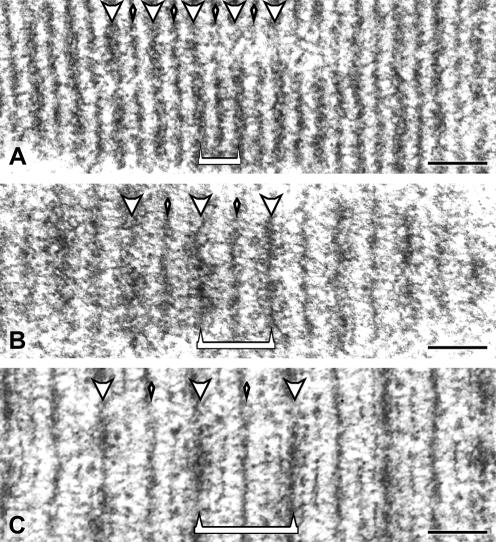
To investigate the contribution of the different protein domains of SCP1 in the process of polycomplex assembly, we transfected COS-7 cells with plasmids coding for mutated forms of SCP1. First, we investigated possible effects caused by changes in the length of the rod domain (see also Sym and Roeder, 1995). To this end, COS-7 cells were transfected with construct SCP1-Δ457-699, which lacks the segment coding for amino acids 457–699 (243 amino acids), or with construct SCP1–2x457-699 carrying a duplication of the same segment (Figure 1). In comparison to wtSCP1, the rod domain of these mutant proteins is expected to be 36 nm shorter or longer, respectively (i.e., 1.485 Å per amino acid; Fraser and MacRae, 1973). Interestingly, at the EM level polycomplexes revealed differences in size that are correlative to the length of the rod domain (Figure 4). In polycomplexes assembled by EGFP-SCP1 (Figure 4B), the distance from the middle of a dense line to the middle of the next one was 123 ± 17 nm (n = 30). Virtually the same distance was measured in the case of wtSCP1 (120 ± 9 nm, n = 19; Figure 3B). In the case of SCP1-Δ457-699 polycomplexes, the distance was only 69 ± 3 nm (n = 20; Figure 4A), whereas in polycomplexes assembled by SCP1–2x457-699 the width was increased (164 ± 9 nm, n = 18). That the difference between polycomplexes formed by SCP1 (Figure 4B) and the mutant forms (Figure 4, A and C) was smaller than the predicted 72 nm (SCP1 molecules transverse only half of the central region) is not surprising because it is known that transverse filaments are not straight but rippled structures (Liu et al., 1996; Schmekel et al., 1996). From these experiments it can be concluded that the length of the rod domain of SCP1 is a primary determinant of the length of transverse filaments and accordingly, of the width of the central region.
Figure 4.
Polycomplexes assembled by SCP1-Δ457-699 (A), EGFP-SCP1 (B), and SCP1–2x457-699 (C) are shown at the same final magnification. Dense lines (large arrowheads) and less dense lines (small arrowheads) are indicated. The distance from the middle of a dense line to the middle of the next one is indicated by a bracket (A–C). Scale bars, 0.1 μm.
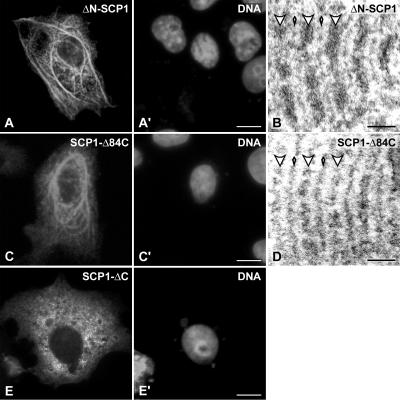
By using polyclonal antibodies against amino acid residues 53–128, the N-terminus of SCP1 was localized to the central element of the SC, where it is thought to interact head to head with the N-termini of SCP1 molecules that are anchored in the opposite lateral element (Liu et al., 1996). To examine the contribution of the nonhelical N-terminus of SCP1 in polycomplex assembly, we transfected COS-7 cells with construct ΔN-SCP1 (Figure 1). The ability to form cytoplasmic structures was strongly reduced in comparison to wtSCP1 (9%, n = 666 instead of 41%, n = 424; Figure 5A). Correspondingly, the vast majority of ΔN-SCP1 transfected cells (91%) showed a diffuse intranuclear fluorescence. This result points to an important role of the nonhelical N-terminus in the process of polycomplex assembly and, by extension, in that of the central region as previously suggested (Liu et al., 1996; Schmekel et al., 1996). Regardless, the EM analyses revealed that the cytoplasmic structures formed by ΔN-SCP1 (Figure 5B) were quite similar to those formed by wtSCP1. These results are remarkable and show that the coiled-coil domains from opposing SCP1 molecules can interact to assemble the central region. In other terms, the experiment of Figure 5, A and B, indicates that the lack of the N-terminus does not impair central region assembly, but reduces its efficiency. We speculate that during the process of SC assembly the nonhelical N-termini would be important in establishing the contact between opposing SCP1 molecules which would facilitate the interaction between the heads of opposing rod domains.
Figure 5.
Analysis of mutant SCP1 constructs lacking the N-terminus (A and B), the 84 C-terminal amino acids (C and D) or the whole C-terminus (E). The structures formed by ΔN-SCP1 or SCP1-Δ84C are shown by immunofluorescence (A and C) or electron microscopy (B and D). SCP1-ΔC is diffusely distributed in the cytoplasm as shown by immunofluorescence microscopy (E). The cells were incubated with the DNA-specific fluorochrome Hoechst 33258 (A′,C′, E′). Scale bars, 10 μm (A–A′, C–C′, E–E′), 0.1 μm (B and D).
Finally, we investigated the fate of SCP1 molecules lacking the nonhelical C-terminal domain. Cells expressing protein SCP1-ΔC revealed a homogeneous labeling of the cytoplasm. This observation clearly contrasts with the results described above, as the other SCP1 mutants were able to form polycomplexes, and indicates that the C-terminus of SCP1 is required for the assembly of higher order structures (Figure 5E). Removal of three of four putative nuclear localization signals of SCP1, which are according to PSORT II analysis located within the last 84 amino acids, has no apparent effect on polycomplex assembly (construct SCP1-Δ84; Figure 5, C and D). As expected, virtually no cells were found showing a homogeneous nuclear fluorescence in contrast to transfection with wtSCP1 (see Figure 2B). This experiment makes it very unlikely that SCP1 requires modifications by COS-7 nuclear factors in order to form polycomplexes. Summarizing, Figure 5E provides evidence for the first time that during the process of SC assembly the direct interaction between transverse filaments via the C-termini of SCP1 molecules is an essential step. We speculate that, in an early phase of central region assembly, the future transverse filaments (i.e., SCP1 dimers) have to associate at the level of the lateral element with other future transverse filaments (and other components of the lateral element) before the N-termini and rod domains of opposing SCP1 dimers can interact to form the central element.
CONCLUSIONS
At present, two bona fide structural components of mammalian SCs have been identified: the proteins SCP1 and SCP3. Both have the capability to self-assemble in the absence of additional SC proteins, forming thereby higher order structures that resemble subcomponents of the SC, i.e., the lateral elements in the case of SCP3 (Yuan et al., 1998) and the central region as shown here for SCP1. On the other hand, biochemical and genetical approaches failed to demonstrate a direct interaction between these two structural proteins (Liu et al., 1996; Tarsounas et al., 1997; Yuan et al., 1998; Pelttari et al., 2001). Notwithstanding, SCs can be isolated from meiotic cells as discrete entities that contain SCP1 and SCP3 among other few proteins (Heyting and Dietrich, 1991). Further investigations on interaction partners of SCP1 as well as SCP3 are now required to better understand the molecular architecture of the synaptonemal complex.
Acknowledgments
We thank Wolfgang Schütz for advice; Natalia Prikhodovski, Elisabeth Meyer-Natus, and Daniela Bunsen for excellent technical assistance; and Rosie Rudd and Robert Hock for correcting the manuscript. This work was supported by grants of the Graduiertenkolleg 639 and the Deutsche Forschungsgemeinschaft (Be 1168/6-1) to R.B.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–09–0771. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-09-0771.
This article is dedicated to Professor Omar Trujillo-Cenóz (IIBCE, Montevideo, Uruguay) on the occasion of his 70th birthday. Together with the late Prof. J. R. Sotelo, he published in 1960 the first description of polycomplexes.
Abbreviations used: SC, synaptonemal complex; EM, electron microscopy.
References
- Alsheimer, M., von Glasenapp, E., Schnölzer, M., Heid, H., and Benavente, R. (2000). Meiotic lamin C2, the unique amino-terminal hexapeptide GNAEGR is essential for nuclear envelope association. Proc. Natl. Acad. Sci. USA 97, 13120-13125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiácovo, M. P., MacQueen, A. J., Martínez-Pérez, E., McDonald, K., Adamo, A., La Volpe, A., and Villeneuve, A. M. (2003). Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell 5, 463-474. [DOI] [PubMed] [Google Scholar]
- Dobson, M. J., Pearlman, R. E., Karaiskakis, A., Spyropoulos, B., and Moens, P. B. (1994). Synaptonemal complex proteins: occurrence, epitope mapping and chromosome disjunction. J. Cell Sci. 107, 2749-2760. [DOI] [PubMed] [Google Scholar]
- Dong, H., and Roeder, G. S. (2000). Organization of the yeast Zip1 protein within the central region of the synaptonemal complex. J. Cell Biol. 148, 417-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, R.D.B., and MacRae, R. P. (1973). Conformation in Fibrous Proteins and Related Synthetic Polypeptides. New York: Academic Press.
- Goldstein, P. (1987). Multiple synaptonemal complexes (polycomplexes): origin, structure and function. Cell Biol. Int. Rep. 11, 759-796. [DOI] [PubMed] [Google Scholar]
- Heyting, C. (1996). Synaptonemal complexes: structure and function. Curr. Opin. Cell Biol. 8, 389-396. [DOI] [PubMed] [Google Scholar]
- Heyting, C., and Dietrich, A. J. (1991). Meiotic chromosome preparation and protein labeling. Methods Cell Biol. 35, 177-202. [DOI] [PubMed] [Google Scholar]
- Kozak, M. (1987). An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 15, 8125-8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammers, J. H., Offenberg, H. H., van Aalderen, M., Vink, A. C., Dietrich, A. J., and Heyting, C. (1994). The gene encoding a major component of the lateral elements of synaptonemal complexes of the rat is related to X-linked lymphocyte-regulated genes. Mol. Cell. Biol. 14, 1137-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebe, B., Alsheimer, M., Höög, C., Benavente, R., and Scherthan, H. (2004). Telomere attachment, meiotic chromosome condensation, pairing, and bouquet stage duration are modified in spermatocytes lacking axial elements. Mol. Biol. Cell 15, 827-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. G., Yuan, L., Brundell, E., Björkroth, B., Daneholt, B., and Höög, C. (1996). Localization of the N-terminus of SCP1 to the central element of the synaptonemal complex and evidence for direct interactions between the N-termini of SCP1 molecules organized head-to-head. Exp. Cell Res. 226, 11-19. [DOI] [PubMed] [Google Scholar]
- Lupas, A., Van Dyke, M., and Stock, J. (1991). Predicting coiled coils from protein sequences. Science 252, 1162-1164. [DOI] [PubMed] [Google Scholar]
- MacQueen, A. J., Colaiácovo, M. P., McDonald, K., and Villeneuve, A. M. (2002). Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 16, 2428-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meistrich, M. L. (1977). Separation of spermatogenic cells and nuclei from rodent testes. Methods Cell Biol. 15, 15-54. [DOI] [PubMed] [Google Scholar]
- Meuwissen, R. L., Offenberg, H. H., Dietrich, A. J., Riesewijk, A., van Iersel, M., and Heyting, C. (1992). A coiled-coil related protein specific for synapsed regions of meiotic prophase chromosomes. EMBO J. 11, 5091-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offenberg, H. H., Schalk, J. A., Meuwissen, R. L., van Aalderen, M., Kester, H. A., Dietrich, A. J., and Heyting, C. (1998). SCP2, a major protein component of the axial elements of synaptonemal complexes of the rat. Nucleic Acids Res. 26, 2572-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, S. L., and Hawley, R. S. (2001). c(3)G encodes a Drosophila synaptonemal complex protein. Genes Dev. 15, 3130-3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, S. L., and Hawley, R. S. (2004). The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 20, 525-558. [DOI] [PubMed] [Google Scholar]
- Pelttari, J. et al. (2001). A meiotic chromosomal core consisting of cohesin complex proteins recruits DNA recombination proteins and promotes synapsis in the absence of an axial element in mammalian meiotic cells. Mol. Cell. Biol. 21, 5667-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage, J., Martin, L., Cuzin, F., and Rassoulzadegan, M. (1995). cDNA sequence of the murine synaptonemal complex protein 1 (SCP1). Biochim. Biophys. Acta 1263, 258-260. [DOI] [PubMed] [Google Scholar]
- Schmekel, K., and Daneholt, B. (1995). The central region of the synaptonemal complex revealed in three dimensions. Trends Cell Biol. 5, 239-242. [DOI] [PubMed] [Google Scholar]
- Schmekel, K., Meuwissen, R. L., Dietrich, A. J., Vink, A. C., van Marle, J., van Veen, H., and Heyting, C. (1996). Organization of SCP1 protein molecules within synaptonemal complexes of the rat. Exp. Cell Res. 226, 20-30. [DOI] [PubMed] [Google Scholar]
- Sotelo, J. R., and Trujillo-Cenóz, O. (1960). Electron microscope study on spermatogenesis. Chromosome morphogenesis at the onset of meiosis (cyte l) and nuclear structure of early and late spermatids. Z. Zellforsch. 51, 243-277. [PubMed] [Google Scholar]
- Sym, M., Engebrecht, J. A., and Roeder, G. S. (1993). ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72, 365-378. [DOI] [PubMed] [Google Scholar]
- Sym, M., and Roeder, G. S. (1995). Zip1-induced changes in synaptonemal complex structure and polycomplex assembly. J. Cell Biol. 128, 455-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarsounas, M., Pearlman, R. E., Gasser, P. J., Park, M. S., and Moens, P. B. (1997). Protein-protein interactions in the synaptonemal complex. Mol. Biol. Cell 8, 1405-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wettstein, D., Rasmussen, S. W., and Holm, P. B. (1984). The synaptonemal complex in genetic segregation. Annu. Rev. Genet. 18, 331-413. [DOI] [PubMed] [Google Scholar]
- Wettstein, R., and Sotelo, J. R. (1971). The molecular architecture of synaptonemal complexes. Adv. Cell Mol. Biol. 1, 109-152. [Google Scholar]
- Yuan, L., Brundell, E., and Höög, C. (1996). Expression of the meiosis-specific synaptonemal complex protein 1 in a heterologous system results in the formation of large protein structures. Exp. Cell Res. 229, 272-275. [DOI] [PubMed] [Google Scholar]
- Yuan, L., Liu, J. G., Zhao, J., Brundell, E., Daneholt, B., and Höög, C. (2000). The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol. Cell 5, 73-83. [DOI] [PubMed] [Google Scholar]
- Yuan, L., Pelttari, J., Brundell, E., Björkroth, B., Zhao, J., Liu, J. G., Brismar, H., Daneholt, B., and Höög, C. (1998). The synaptonemal complex protein SCP3 can form multistranded, cross-striated fibers in vivo. J. Cell Biol. 142, 331-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler, D., and Kleckner, N. (1998). The leptotene-zygotene transition of meiosis. Annu. Rev. Genet. 32, 619-697. [DOI] [PubMed] [Google Scholar]