Abstract
Hepatitis B virus (HBV) transcribes two subsets of 3.5-kb RNAs: precore RNA for hepatitis B e antigen (HBeAg) expression, and pregenomic RNA for core and P protein translation as well as genome replication. HBeAg expression could be prevented by mutations in the precore region, while an upstream open reading frame (uORF) has been proposed as a negative regulator of core protein translation. We employed replication competent HBV DNA constructs and transient transfection experiments in Huh7 cells to verify the uORF effect and to explore the alternative function of precore RNA. Optimized Kozak sequence for the uORF or extra ATG codons as present in some HBV genotypes reduced core protein expression. G1896A nonsense mutation promoted more efficient core protein expression than mutated precore ATG, while a +1 frameshift mutation was ineffective. In conclusion, various HBeAg-negative precore mutations and mutations affecting uORF differentially regulate core protein expression and genome replication.
Keywords: hepatitis B virus, hepatitis B e antigen, core protein, pregenomic RNA, precore RNA, precore mutant, translational control, genome replication
1. Introduction
Hepatitis B virus (HBV) has a small circular DNA genome of 3.2 kb. It produces two subsets of the 3.5-kb terminally redundant RNAs. The precore RNA (pcRNA) is the mRNA for precore protein, which following proteolytic cleavage is secreted as hepatitis B e antigen (HBeAg) (Milich and Liang, 2003; Tong et al., 2005). The slightly shorter pregenomic RNA (pgRNA) does not have the entire precore region at its 5′ end, and is hence used for the translation of core protein. The 3′ end of the core gene overlaps with the 5′ end of the polymerase (P) gene, and P protein is translated from pgRNA through ribosomal leaky scanning of the initiating (C) ATG codon as well as two internal (C1 and C2) ATGs of the core gene (Fouillot et al., 1993; Hwang and Su, 1998; Lin and Lo, 1992) (Fig. 1A). For wild-type virus only pgRNA is involved in HBV genome replication, because core protein translated from pgRNA is the building block for core particle, the site of HBV genome replication. pgRNA is encapsidated into core particles for reverse transcription into minus-stranded DNA. This step as well as subsequent pgRNA degradation and plus strand DNA synthesis are catalyzed by P protein, the other translation product of pgRNA. Selective encapsidation of the pgRNA is governed by a stem-loop structure located at its 5′ end (Junker-Niepmann et al., 1990). The pregenome encapsidation (s) signal is composed of the 3′ 2/3rds of the precore region and the first seven nucleotides (nts) of the core gene (Fig. 2). The slightly longer pcRNA is not packaged despite presence of such a sequence at its 5′ end due to melting of RNA secondary structure by the translating ribosomes (Nassal et al., 1990).
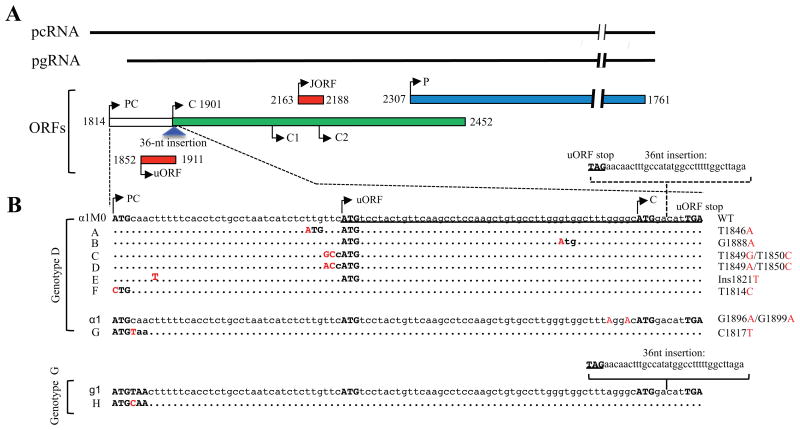
Figure 1.
The uORF and HBV DNA constructs used for the present study. (A) Schematic representation of the pcRNA and pgRNA, as well as P gene, precore/core gene, and uORF occupying three different open reading frames (ORFs). Arrowheads indicate translation initiation sites. C, C1, and C2 are initiating and two internal ATG codons of the core gene. The JORF helps ribosomal skipping of the C2 ATG, while location of the uORF makes it a negative regulator of core gene translation. The 36-nt insertion found in genotype G (blue triangle) is located at the 5′ end of core gene overlapping with the 3′ end of the uORF. (B) DNA sequences of the precore region and uORF of the HBV DNA constructs used for the present study. Genotype G (clone g1) has a 36-nt insertion at the 5′ end of core gene. Transferring that sequence into αlM0 of genotype D generated αlM0+36. Mutations A-F were introduced to αlM0 or αlM0+36 (in that case they are called A+36 -F+36). Mutants A and B have an extra ATG codon created for the uORF, while mutants C and D have the original ATG codon of the uORF optimized for translation initiation. Mutant E has a single nucleotide insertion to fuse the precore ORF with uORF. In mutant F, the precore ATG was mutated to make pcRNA virtually indistinguishable from pgRNA. αl differs from αlM0 in possessing the G1896A nonsense mutation at codon 28 as well as G1899A mutation, while mutant G differs from αl in having an extra nonsense mutation (C1817T) at codon 2. Mutation H was introduced to clone g1 of genotype G by ablating the C1817T mutation.
Figure 2.
RNA secondary structure of the ε signal. It covers the 3′ 2/3rds of the precore region and the first 7 nucleotides of the core gene, and contains two base-paired regions (stems), a bulge, and a loop. Translation initiation sites for the uORF and core gene are indicated, as is the site of the 36-nt insertion found in genotype G. Most HBV genotypes have T1858, and the G1896A nonsense mutation would improve a base pair from U:G to U:A. Genotypes A, C1, F2, and F3 have C1858 to preclude the rise of the G1896A mutation.
As a core particle is assembled from 180 or 240 copies of core protein but packages just one molecule of the P protein, balanced translation of core vs. P protein from pgRNA is essential for efficient genome replication. Previous work revealed a small open reading frame (ORF) inside the core gene but in a different frame as a facilitator of P gene expression. Translation of that 7-codon JORF enables ribosomes to bypass the C2 ATG (Fig. 1A), which has a good context for translation initiation, followed by termination and reinitiation at the P gene ATG (Fouillot et al, 1993; Hwang and Su, 1998). A subsequent study identified another out-of-frame ORF in the precore region (hereby referred to as uORF because it is upstream of the core gene) as a negative regulator of core protein translation (Chen et al, 2005). Translation initiation site of the uORF (nt 1852-1854) is the 5′ most ATG on pgRNA, and can generate a short peptide of 19 residues. Since its translational stop codon is downstream of the C ATG codon (ATGGACATTGA), the uORF most likely provides a mechanism to bypass translation initiation site of the core gene. However, functional characterization of the uORF was based primarily on reporter constructs rather than authentic pgRNA (Chen et al, 2005).
HBeAg functions as an immune toleragen to facilitate the establishment of chronic infection (Milich and Liang, 2003). Its presence often correlates with high viral load. Seroconversion from HBeAg to anti-HBe, a step towards the clearance of HBV replication, can be accompanied by the outgrowth of HBV mutants that no longer express HBeAg (Tong et al, 2005). The mutations occur in the precore region rather than the core gene so as to preserve core protein expression, which is essential for genome replication. The hot-spot HBeAg-abrogating mutation for most HBV genotypes is G1896A converting precore codon 28 from TGG to TAG (Carman et al, 1989; Okamoto et al., 1990; Tong et al, 1990), at least partly because it would improve a base pair at the lower stem of the ε signal from U:G to U:A (Fig. 2). The G1896A nonsense mutation rarely develops in genotypes A, H, C1, F2, or F3, for which a preexisting C:G pair with C1858 would be disrupted by the mutation (Li et al, 1993; Lok et al, 1994; Norder et al, 2003). In the absence of G1896A mutation, alternative mutations to abolish HBeAg expression include C1817T converting codon 2 from CAA to TAA, various frameshift mutations (Li et al, 1990; Tong et al, 1990), and mutated precore ATG codon (Gunther et al, 1992). Intriguingly, genotype G harbors both C1817T and G1896A nonsense mutations, and possesses a 36-nt insertion at the 5′ end of the core gene (Stuyver et al, 2000). The insertion increases the size of core protein from 183 aa to 195 aa, and shortens the uORF from 19 to 18 codons (Fig. 1B).
In the present study, we investigated the alternative function of pcRNA when HBeAg expression was prevented by naturally occurring mutations. We also verified the uORF as a negative regulator of core protein translation using the authentic HBV DNA replication constructs. Genotypes B and C have A1846 rather than T1846 found in other HBV genotypes (Chen et al, 2005; Yin et al., 2011), thus creating a new upstream ATG codon for the uORF. In subgenotype A1, a downstream ATG codon is created for the uORF due to A1888 (Kimbi et al, 2012). The impacts of A1846 and A1888 on core protein expression and genome replication were also examined by site-directed mutagenesis.
2. Materials and Methods
2.1. HBV DNA constructs and site-directed mutants
HBVαl (shortened as αl hereafter) is a naturally occurring HBeAg-negative precore mutant of genotype D capable of genome replication (Tong et al, 1990; 1991). αlM0 was derived from αl by A1896G and A1899G back mutations to restore HBeAg expression (Tong et al, 1992). The extra 36 nt found in genotype G was inserted between positions 1905 and 1906 of αl and αlM0, respectively to generate αl+36 and αlM0+36 (Gutelius et al, 2011). These HBV DNA constructs were cloned to pUC18 vector and converted to tandem dimer via the unique SphI site (SphI dimer). The SphI dimer can transcribe pgRNA under endogenous core promoter and enhancer elements, and is thus replication competent. Point mutations and insertions in the precore region were introduced by overlap extension PCR followed by exchange of the 0.6-kb NcoI - BglII restriction fragment. Since restriction fragment exchange reverted the HBV constructs into SphI monomer, they were reconverted to SphI dimer using a rapid method (Zong et al., 2016). Clone g1 of genotype G was derived from patient serum sample and also available as SphI dimer (Li et al., 2007). Mutations to clone g1 were introduced by overlap extension PCR followed by replacement of the 1.5-kb NcoI -XhoI restriction fragment.
2.2. Transient transfection and Western blot analysis of core protein expression
Huh7 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplement with 10% fetal bovine serum. Transient transfection was carried out on cells seeded in 6-well plates at a density of 60-80%, using LT1 reagent (Mirus). Each well of cells was transfected with 2 ug of HBV DNA. Cells were lyzed 3 days post-transfection in 100 ul of lysis buffer [10mM HEPES (pH 7.4), 100mM NaCl, 1mM EDTA, and 1% NP-40], and protein concentration was determined by BCA assay. Proteins (60 ug for genotype D-transfected cells and 100 ug for genotype G-transfected cells) were separated in 0.1% SDS-12% polyacrylamide gel and transferred to polyvinylidene difluoride membranes. The blots were blocked at room temperature for 1 h with 5% skimmed milk dissolved in Tris-buffered saline-0.05% Tween 20 (TBST) buffer. Blots of genotype D and genotype G were incubated at 4°C overnight with custom made rabbit polyclonal anti-core antibody (1: 1000 dilution) (Guo et al., 2010) and a commercial rabbit polyclonal antibody (Dako; 1:3000 dilution), respectively. The Dako polyclonal antibody failed to efficiently detect core protein from non-G genotypes by direct Western blot analysis, possibly due to much lower expression level (Gutelius et al., 2011; Li et al., 2007). The blots were washed with several changes of 1 × TBST buffer for a duration of 40 minutes, and subsequently incubated at room temperature for 1 hour with 1:40,000 dilution of anti-rabbit antibody conjugated to horseradish peroxidase (HRP). After further wash, signals were detected by Western Lighting chemiluminescence reagent (Perkin-Elmer). For loading control, the blots were stripped and incubated with 1:10000 dilution of a mouse antibody against β-actin, followed by HRP-conjugated anti-mouse secondary antibody.
2.3. Southern blot analysis of replicative HBV DNA
Replicative HBV DNA was extracted from cell lysate as previously described (Parekh et al., 2003). Briefly, one-third of the cell lysate was treated in a volume of 100 ul with 5U of mung bean nuclease and 1U of DNase I at 37°C for 15min. Core particles were precipitated with polyethylene glycol solution, resuspended, and further treated with mung bean nuclease and DNase I. HBV DNA was released from core particles by proteinase K digestion, extracted with phenol, and precipitated with ethanol. DNA samples for the two HBV genotypes were run in separate agarose gels and transferred to nylon membranes, and the blots were hybridized with 32P –labeled full-length HBV DNA probe of their respective genotype. The blots were washed at 65°C for 1 hour each with 2×SSC/0.1% SDS solution and 0.5×SSC/0.1% SDS solution. Retained 32P signals were revealed by autoradiography.
3. Results
3.1. Rationale
As is shown in Fig. 1B, various single or double nucleotide substitutions or a single nucleotide insertion were introduced to αlM0, a genotype D clone with wild-type precore sequence, to generate constructs A-F. The same set of mutations was introduced to αlM0+36, which had a 36-nucleotide insertion at the 5′ end of core gene. This insertion increased core protein size from 183 aa to 195 aa and markedly augmented its expression (Fig. 3A, compare lane 2 with lane 1) but suppressed genome replication (Fig. 3B) due to a too high core/P protein ratio (Gutelius et al, 2011). Use of αlM0+36 as an additional recipient of the mutations (referred to as A+36 - F+36) enabled us to estimate their impact on core/P protein ratio through Southern blot analysis. It was anticipated that in contrast to αlM0, a mild reduction in core protein expression for αlM0+36 would rather improve core/P protein ratio to increase genome replication.
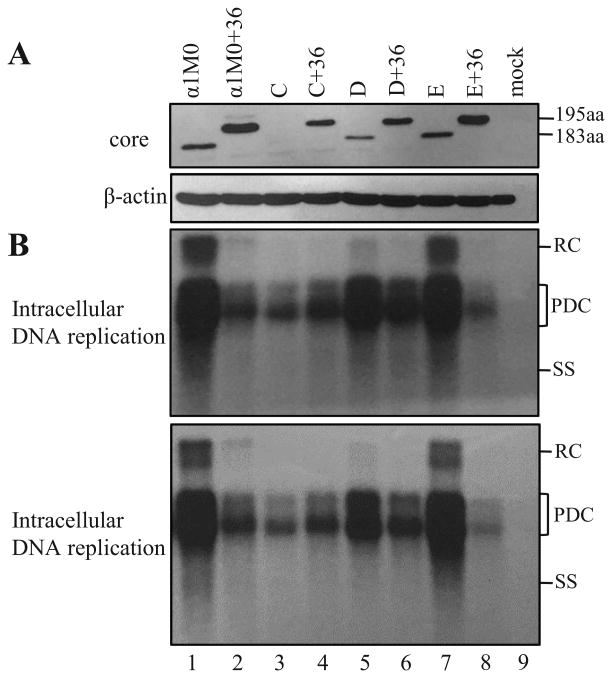
Figure 3.
Impact of adding an extra ATG codon to the uORF on core protein expression and genome replication. The T1846A mutation was introduced to α1M0 and α1M0+36 to generate mutant A and A+36, respectively. Similarly, G1888A was introduced to generate B and B+36. Huh7 cells were transiently transfected with the parental HBV DNA constructs and their site-directed mutants, and harvested three days later. (A) Western blot analysis of core protein expression using β-actin as a loading control. A total of 60 μg of cellular proteins was loaded. α1M0 and α1M0+36 express a core protein of 183aa and 195aa, respectively. (B) Southern blot analysis of intracellular replicative DNA. Results from two independent transfection experiments are presented. RC, relaxed circular; PDS, partially double stranded; SS, single stranded.
3.2. Extra AUG codons in the uORF reduced core protein expression with differential effects onreplication of αlM0 vs αlM0+36
The impact of extra ATG codons for the uORF, as found in genotypes B/C and subgenotype A1, was examined by site-directed mutagenesis. Core protein expression from αlM0 was slightly diminished by both T1846A and G1888A mutations (Fig. 3A, compare lanes 1, 3, and 5). This was accompanied by reduced replication capacity (Fig. 3B). When introduced to αlM0+36, both T1846A and G1888A also slightly reduced core protein expression (Fig. 3A, lanes 2, 4, and 6), but increased genome replication (Fig. 3B). This, as stated earlier, is consistent with a too high core/P protein ratio in αlM0+36.
3.3. Optimizing the initiation context for the uORF markedly suppressed core protein expression with differential effects on replication of αlM0 vs αlM0+36
Results so far supported a negative effect of the uORF on core protein translation, as has been proposed (Chen et al., 2005). An alternative approach is to optimize the context for translation initiation from the conserved ATG codon of the uORF, which is TTCATGT for αlM0. Considering that efficient translation initiation requires a purine at the -3 position, we converted the three nt immediately upstream of the ATG from TTC to GCC (mutant C or C+36) or ACC (mutant D or D+36). Core protein expression was reduced in mutant D and severely reduced in mutant C (Fig. 4A, lanes 1, 3, 5). Similarly, expression of the 195-aa core protein from αlM0+36 was reduced to greater extent in C+36 than in D+36 (Fig. 4A, lanes 2, 4, 6). Consistent with the profile of core protein, mutant C showed much greater reduction in genome replication than mutant D (Fig. 4B, lane 3 vs. lane 5). As for the αlM0+36 series, both C+36 and D+36 manifested enhanced genome replication, with D+36 achieving higher level of replication than C+36 (Fig. 4B, lanes 2, 4, 6).
Figure 4.
Impact of optimization of the Kozak sequence of the uORF on core protein expression and genome replication. In C and C+36, the T1849G/T1850C mutations were introduced to the -3/-2 positions of the uORF, while in D and D+36, T1849A/T1850C mutations were introduced. For E and E+36, a thymidine was inserted at position 1821 to fuse the precore ORF with the uORF. The mutant constructs as SphI dimers were transiently transfected to Huh7 cells, together with α1M0 and α1M0+36. Cells were harvested three days later. (A) Western blot analysis of intracellular core protein (upper panel) and β-actin (lower panel) from 60 μg of total cellular proteins. (B) Southern blot analysis of intracellular replicative DNA from two independent transfection experiments. RC, relaxed circular; PDS, partially double stranded; SS, single stranded.
3.4. Loss of the precore AUG codon rather than a +1 frameshift mutation upstream of the uORF increased core protein expression
HBeAg expression could be abolished by mutation of precore ATG, or a nonsense or frameshift mutation. The 5′ precore region has a run of five Ts (1821-1825), which can be expanded to six Ts (Tong et al, 1990), apparently due to polymerase stuttering during replication. We introduced this frameshift mutation as well as an A1814C mutation converting precore ATG to CTG, to αlM0 and αlM0+36. While loss of precore ATG clearly increased core protein expression (Fig. 5A, compare lane 1 with 3, and lane 2 with 4), the +1 frameshift mutation had little effect (Fig. 4A, lane 1 vs. 7, lane 2 vs. 8). Loss of the precore ATG would make the pcRNA similar to pgRNA and an extra source for core protein. In contrast, the +1 frameshift would fuse the precore ORF with the uORF. The very efficient translation initiation from the precore ATG combined with the function of the uORF as a negative regulator of core protein expression could explain why a +1 frameshift mutation failed to increase core protein expression. Consistent with Western blot data, replication of αlM0 was unaffected by the frameshift (Fig. 4B, lane 1 vs. 7) and increased by the loss of precore ATG (Fig. 5B, lane 1 vs. 3). Replication of αlM0+36 was unaffected or slightly increased by the loss of precore ATG codon (Fig. 5B, lane 2 vs. 4) and intriguingly, diminished by the +1 frameshift (Fig. 4B, lane 2 vs. 8).
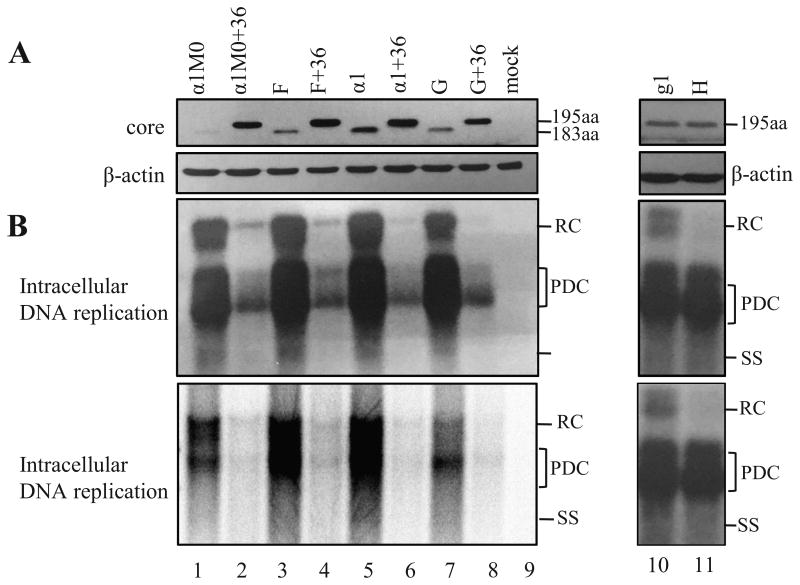
Figure 5.
Impact of the G1896A mutation alone or together with the C1817T mutation on core protein expression and genome replication. Mutants F and F+36 were derived from α1M0 and α1M0+36, respectively, by mutating precore ATG to CTG. α1 and α1+36 differ from α1M0 and α1M0+36, respectively, in having the G1896A/G1899A mutations. Constructs G and G+36 were derived from α1 and α1+36 by addition of the C1817T nonsense mutation. Conversely, construct H was originated from clone g1 of genotype G by abolishing the C1817T nonsense mutation. (A) Western blot analysis of core protein expression, with β-actin serving as loading control. Please note that for lanes 10 and 11, 100 μg of total cellular proteins were loaded to the gel and a commercial rabbit polyclonal antibody (Dako) was used. (B) Southern blot analysis of intracellular replicative DNA using a probe of genotype D (lanes 1-9) or genotype G (lanes 10, 11).
3.5. G1896A markedly increased core protein expression, which was largely blocked by concurrent C1817T mutation
αlM0 was derived from αl, a naturally occurring HBeAg-negative mutant, by introducing the A1896G and A1899G back mutations (Tong et al, 1992). Comparison of αlM0, F (αlM0 with mutated precore ATG), and αl in the same blot indicates that G1896A/G1899A were more effective than mutated precore ATG in augmenting core protein expression, although ol+36 did not show higher core protein level than F+36 (Fig. 5A). In this regard, G1896A converts precore codon 28 into a stop codon (Fig. 2). Most likely, the short (one codon) distance between this stop codon and initiating ATG of core gene makes efficient translational termination-reinitiation. Considering that genotype G harbors two nonsense mutations in the precore region (C1817T and G1896A), we introduced C1817T mutation to αl and αl+36, respectively to generate construct G and G+36. Construct G expressed less core protein than αl but still more core protein than αlM0 (Fig. 5A, lanes 7, 5, 1). Core protein expressed by G+36 was lower than al+36 and comparable to αlM0+36 (lanes 8, 6, 2). Therefore, by shifting the site of translational termination from codon 28 to codon 2, the extra C1817T mutation abolished or diminished the enhancing effect of G1896A mutation on core protein expression. In the reverse experiment, we introduced the T1817C back mutation to clone g1 of genotype G. However, that did not increase core protein expression (Fig. 5 A, lane 11 vs. 10).
In agreement with the core protein data, αl showed higher level of genome replication than αlM0 (Fig. 5B, lanes 5 vs. 1). Introduction of the additional C1817T mutation reverted genome replication to a low level (Fig. 5B, lane 7). As for the αlM0 +36 series, the replication impact of the G1896A mutation was mild. The T1817C back mutation did not alter DNA replication for clone g1 of genotype G, but specifically reduced relaxed circular (RC) DNA form (Fig. 5B, lane 11 vs. 10). That finding was highly reproducible (data not shown). Although the C1817T nonsense mutation should permit the packaging of pcRNA (Nassal et al., 1990), encapsidated pcRNA should generate double stranded linear DNA rather than RC DNA due to the inability of the RNA primer generated by RNase H digestion to translocate to DR2. Thus, reduced RC DNA production by the T1817C mutant was most likely attributable to the single nucleotide change in pgRNA.
4. Discussion
Chen and colleagues previously identified a conserved 19-codon ORF spanning nt.1852-1911 in the precore/core gene, but in a different reading frame than the core gene (Chen et al, 2005). That uORF has a poor context for translation initiation (T/CT/A/T/CATGT), and its translation efficiency from pgRNA was estimated at about 20% of the core gene according to luciferase reporter constructs. Ablating its ATG codon increased translation initiation from downstream ATG codons of the core gene and P gene by 40% and 70%, respectively, whereas optimizing the Kozak sequence for the uORF reduced translation from the core gene by 25 fold, yet increased translation from the P gene by 50% (Chen et al., 2005). These results are consistent with the uORF being a negative regulator of core protein translation, although its role in P gene expression was less than clear. Considering the long distance between the uORF and the P gene, its regulation of P protein translation is apparently indirect.
In the present study, we found that introduction of extra ATG codons to the uORF moderately diminished core protein expression for both αlM0 and αlM0+36 (Fig. 3A). Changing the -3/-2 positions of the uORF from TT to AC and especially GC, drastically reduced core protein expression (Fig. 4A). Therefore, our data provide direct evidence for the uORF to serve as a negative regulator of core protein expression from authentic pgRNA. Since we did not measure P protein level (which is technically challenging), we cannot establish whether the 19-codon uORF promotes P protein translation, and what the impact of the extra ATG codons found in genotypes B, C, and A1 would be on P protein expression. Among HBeAg-negative precore mutations tested, G1896A in conjunction with G1899A was more effective than mutated precore ATG at increasing core protein expression (Fig. 5A, compare αlM0, F, and αl). Although both pcRNA and pgRNA were transcribed from the SphI dimer used for the transfection experiments, the increased core protein level most likely originated from translational termination - reinitiation from the pcRNA. This interpretation was supported by the finding that adding an extra C1817T mutation, which shifted translational termination from codon 28 to codon 2, reverted core protein expression to a low level (Fig. 5A). A single nucleotide insertion at position 1821 did not increase core protein expression (Fig. 4A, mutant E and E+36). That insertion is predicted to fuse the precore ORF, which has a very strong efficiency of translation initiation, with the uORF. This result reinforces the negative effect of the uORF on core protein translation.
In general, replication impact of mutations introduced to α1M0 (not α1M0+36) correlated with their effect on core protein expression. Thus, converting the Kozak sequence of the uORF to GCC impaired genome replication to greater extent than conversion to ACC, or addition of extra ATG codons to the uORF. Similarly, α1 (which harbors G1896A mutation) had higher replication capacity than α1M0, which was nullified by addition of the C1817T mutation (mutant G) (Fig. 5B). One may argue that DNA replication was diminished rather by reduced stability of the ε signal, which is essential for pgRNA packaging. In this regard, nucleotide 1846 lies outside the ε signal, while the G1888A mutation converts a wobble U:G pair in stem II into a more stable U:A pair (see Fig. 2). Both the G1896A and G1899A mutations convert a U:G pair in stem I into a UA pair (Fig. 2). Optimization of the Kozak sequence of the uORF through the -3/-2 positions has complex impact: while the T1849A and T1849G mutations would both disrupt a U:A pair, the T1850C mutation converts a U:G pair into a much stronger C:G pair. The fact that C+36 and especially D+36 displayed much increased replication capacity than a1M0+36 (Fig. 4B) indicates that the T1849A/T1850C and T1849G/ T1850C double mutations did not impair the ε signal function. For the α1M0+36 series, reduced core protein expression often led to higher replication capacity due to the more physiological ratio between core and P proteins (Fig. 3B and 4B, compare α1M0+36 with A+36, B+36, and D+36).
Another consideration when analyzing the replication phenotype of the HBeAg-negative mutants is the contribution of pcRNA. The wild-type pcRNA is not a precursor to genome replication because the translating ribosomes would melt the secondary structure of the ε signal (Nassal et al, 1990). However, mutated precore ATG codon or the C1817T nonsense mutation at codon 2 will inactivate this mechanism to permit the packaging of pcRNA. We do not know whether such pcRNA was indeed encapsidated to contribute to progeny replicative DNA. Although the T1817C back mutation in clone g1 of genotype G was accompanied by selective loss of the RC DNA (Fig. 5), that is unlikely caused by the loss of pcRNA encapsidation.
HBV genotypes B and C are, due to their perinatal mode of transmission, most effective at establishing chronic infection. Similarly, subgenotype A1 is transmitted during early childhood. These HBV genotypes can persist in their host for up to several decades and are major causes of liver cirrhosis and hepatocellular carcinoma. From virological point of view, virus survival at the immune clearance phase relies on increased replication capacity to offset virus destruction. Thus, wild-type genotype C isolates may have lower replication capacity than genotype B isolates due to lower transcription of the pgRNA (Qin et al., 2011). They show higher prevalence of core promoter mutations later on than genotype B (Lindh et al., 1999; Wang et al., 2007; Yuen et al., 2003), which augment HBV genome replication through increased transcription of pgRNA (Baumert et al., 1998; Buckwold et al., 1996; Parekh et al., 2003; Tsai et al., 2009). In fact, genotype C isolates seroconvert from HBeAg to anti-HBe about 10 years later than genotype B isolates (Chu et al., 2002; Orito et al., 2001; Yuen et al., 2003). In addition, genotype B and subgenotype C2 can develop G1896A precore mutation, which was shown to increase core protein expression and genome replication. Finally, wild-type genotype B and genotype C isolates have A1846 (Chen et al., 2005; Yin et al., 2011) to minimize core protein expression at the immune tolerance phase, while wild-type subgenotype A1 has A1888 (Kimbi et al., 2012) to reduce core protein expression and possibly genome replication. At the immune clearance phase genotypes B and C could develop the A1846T mutation, which according to this study will increase core protein expression and genome replication (Table 1). A1846T has been associated with acute-on-chronic liver failure (Yan et al., 2011) and HCC development (Ahn et al., 2012; Chen et al., 2008), and can coexist with G1896A mutation (Cho et al., 1999; our unpublished observation). Since these two types of mutations augment core protein expression through different mechanisms, their impact on core protein expression is likely additive (Table 1). Thus, subgenotype C2 isolates can use several strategies to tune up their core protein expression and genome replication while reducing HBeAg expression during the transition from immune tolerance phase to immune clearance phase, thus prolonging their replicative phase.
Table 1. Biological consequences of core promoter and precore mutations.
| A1762T/G1764A | A1846T | G1896A | |
|---|---|---|---|
| level of pcRNA | reduced 1 | no effect | no effect |
| level of pgRNA | increased 1 | no effect | no effect |
| HBeAg expression | reduced 1 | no effect | abolished |
| core protein expression | increased 1 | increased 2 | increased 3 |
| genome replication | increased 1 | increased 4 | increased 4 |
Effect can be augmented by additional mutations such as T1753C and C1766T.
Through reduced suppression of core protein translation by the uORF.
Through translational termination – reinitiation from pcRNA.
If a proper core/P protein ratio is maintained.
Highlights.
An upstream open reading frame is indeed a negative regulator of core protein translation, and extra AUG codons found in some HBV genotypes reduce core protein expression.
Among precore mutations to abolish HBeAg expression, G1896A is more effective than mutated precore ATG at promoting core protein expression. A +1 frameshift mutation near the 5′ end of precore region has no effect.
An extra C1817T nonsense mutation can largely abrogate the G1896A effect on core protein expression, suggesting G1896A works by translational termination-reinitiation.
Acknowledgments
This study was supported by NIH grants AI103648, AI107618, AI113394, and AI116639, and also by a grant from National Science Foundation of China (81371822).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn SH, Yuen L, Han K, Littlejohn M, Chang H, Damerow H, Ayres A, Heo J, Locarnini S, Revill P. Molecular and Clinical Characteristics of Hepatitis B Virus in Korea. J Med Virol. 2012;82:1126–1134. doi: 10.1002/jmv.21844. [DOI] [PubMed] [Google Scholar]
- Baumert TF, Marrone A, Vergalla J, Liang TJ. Naturally occurring mutations define a novel function of the hepatitis B virus core promoter in core protein expression. J Virol. 1998;72:6785–6795. doi: 10.1128/jvi.72.8.6785-6795.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckwold VE, Xu Z, Chen M, Yen TS, Ou JH. Effects of a naturally occurring mutation in the hepatitis B virus basal core promoter on precore gene expression and viral replication. J Virol. 1996;70:5845–5851. doi: 10.1128/jvi.70.9.5845-5851.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman WF, Jacyna MR, Hadziyannis S, Karayiannis P, McGarvey MJ, Markris A, Thomas HC. Mutation preventing formation of hepatitis B e antigen in patients with chronic hepatitis B infection. Lancet. 1989;2:588–591. doi: 10.1016/s0140-6736(89)90713-7. [DOI] [PubMed] [Google Scholar]
- Chen A, Kao Y, Brown C. Translation of the first upstream ORF in the hepatitis B virus pregenomic RNA modulates translation at the core and polymerase initiation codons. Nucleic Acids Res. 2005;33:1169–1181. doi: 10.1093/nar/gki251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Changchien CS, Lee CM, Hung CH, Hu TH, Wang JH, Wang JC, Lu SN. Combined mutations in pre-s/surface and core promoter/precore regions of hepatitis B virus increase the risk of hepatocellular carcinoma: A case-control study. J Infect Dis. 2008;198:1634–1642. doi: 10.1086/592990. [DOI] [PubMed] [Google Scholar]
- Cho SW, Shin YJ, Hahm KB, Jin JH, Kim YS, Kim JH, Kim HJ. Analysis of the precore and core promoter DNA sequence in liver tissues from patients with hepatocellular carcinoma. J Korean Med Sci. 1999;14:424–430. doi: 10.3346/jkms.1999.14.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CJ, Hussain M, Lok AS. Hepatitis B virus genotype B is associated with earlier HBeAg seroconversion compared with hepatitis B virus genotype C. Gastroenterology. 2002;122:1756–1762. doi: 10.1053/gast.2002.33588. [DOI] [PubMed] [Google Scholar]
- Fouillot N, Tlouzeau S, Rossignol JM, Jean-Jean O. Translation of the hepatitis B virus P gene by ribosomal scanning as an alternative to internal initiation. J Virol. 1993;67:4886–4895. doi: 10.1128/jvi.67.8.4886-4895.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Mao R, Block TM, Guo JT. Production and function of the cytoplasmic deproteinized relaxed circular DNA of hepadnaviruses. J Virol. 2010;84:387–396. doi: 10.1128/JVI.01921-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther S, Meisel H, Reip A, Miska S, Kruger DH, Will H. Frequent and rapid emergence of mutated pre-C sequences in HBV from e-antigen positive carriers who seroconvert to anti-HBe during interferon treatment. Virology. 1992;187:271–279. doi: 10.1016/0042-6822(92)90315-g. [DOI] [PubMed] [Google Scholar]
- Gutelius D, Li J, Wands J, Tong S. Characterization of the pleiotropic effects of the genotype G-specific 36-nucleotide insertion in the context of other hepatitis B virus genotypes. J Virol. 2011;85:13278–13289. doi: 10.1128/JVI.05583-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WL, Su TS. Translational regulation of hepatitis B virus polymerase gene by termination-reinitiation of an upstream minicistron in a length-dependent manner. J Gen Virol. 1998;79:2181–2189. doi: 10.1099/0022-1317-79-9-2181. [DOI] [PubMed] [Google Scholar]
- Junker-Niepmann M, Bartenschlager R, Schaller H. A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J. 1990;9:3389–3396. doi: 10.1002/j.1460-2075.1990.tb07540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimbi G, Kew MC, Kramvis A. The effect of the G1888A mutation of subgenotype A1 of hepatitis B virus on the translation of the core protein. Virus Res. 2012;163:334–340. doi: 10.1016/j.virusres.2011.10.024. [DOI] [PubMed] [Google Scholar]
- Li JS, Tong S, Vitvitski L, Zoulim F, Trepo C. Rapid detection and further characterization of infection with hepatitis B virus variants containing a stop codon in the distal pre-C region. J Gen Virol. 1990;71:1993–1998. doi: 10.1099/0022-1317-71-9-1993. [DOI] [PubMed] [Google Scholar]
- Li JS, Tong S, Wen YM, Vitvitski L, Zhang Q, Trepo C. Hepatitis B virus genotype A rarely circulates as HBe-minus mutant: possible contribution of a single nucleotide in the precore region. J Virol. 1993;67:5402–5410. doi: 10.1128/jvi.67.9.5402-5410.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Zoulim F, Pichoud C, Kwei K, Villet S, Wands J, Li J, Tong S. Critical role of the 36-nucleotide insertion in hepatitis B virus genotype G on core protein expression, genome replication, and virion secretion. J Virol. 2007;81:9202–9215. doi: 10.1128/JVI.00390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CG, Lo SJ. Evidence for involvement of a ribosomal leaky scanning mechanism in the translation of the hepatitis B virus pol gene from the viral pregenomie RNA. Virology. 1992;188:342–352. doi: 10.1016/0042-6822(92)90763-f. [DOI] [PubMed] [Google Scholar]
- Lindh M, Hannoun C, Dhillon AP, Norkrans G, Horal P. Core promoter mutations and genotypes in relation to viral replication and liver damage in East Asian hepatitis B virus carriers. J Infect Dis. 1999;179:775–782. doi: 10.1086/314688. [DOI] [PubMed] [Google Scholar]
- Lok AS, Akarca U, Greene S. Mutations in the pre-core region of hepatitis B virus serve to enhance the stability of the secondary structure of the pre-genome encapsidation signal. Proc Natl Acad Sci USA. 1994;91:4077–4081. doi: 10.1073/pnas.91.9.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milich D, Liang TJ. Exploring the biological basis of hepatitis B e antigen in hepatitis B virus infection. Hepatology. 2003;38:1075–1086. doi: 10.1053/jhep.2003.50453. [DOI] [PubMed] [Google Scholar]
- Nassal M, Junker-Niepmann M, Schaller H. Translational inactivation of RNA function: discrimination against a subset of genomic transcripts during HBV nucleocapsid assembly. Cell. 1990;63:1357–1363. doi: 10.1016/0092-8674(90)90431-d. [DOI] [PubMed] [Google Scholar]
- Norder H, Arauz-Ruiz P, Blitz L, Pujol FH, Echevarria JM, Magnius LO. The T (1858) variant predisposing to the precore stop mutation correlates with one of two major genotype F hepatitis B virus clades. J Gen Virol. 2003;84:2083–2087. doi: 10.1099/vir.0.19034-0. [DOI] [PubMed] [Google Scholar]
- Okamoto H, Yotsumoto S, Akahane Y, Yamanaka T, Miyazaki Y, Sugai Y, Tsuda F, Tanaka T, Miyakawa Y, Mayumi M. Hepatitis B viruses with precore region defects prevail in persistently infected hosts along with seroconversion to the antibody against e antigen. J Virol. 1990;64:1298–1303. doi: 10.1128/jvi.64.3.1298-1303.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orito E, Ichida T, Sakugawa H, Sata M, Horiike N, Hino K, Okita K, Okanoue T, Iino S, Tanaka E, Suzuki K, Watanabe H, Hige S, Mizokami M. Geographic distribution of hepatitis B virus (HBV) genotype in patients with chronicHBV infection in Japan. Hepatology. 2001;34:590–594. doi: 10.1053/jhep.2001.27221. [DOI] [PubMed] [Google Scholar]
- Parekh S, Zoulim F, Ahn SH, Tsai A, Li J, Kawai S, Khan N, Trepo C, Wands J, Tong S. Genome replication, virion secretion, and e antigen expression of naturally occurring hepatitis B virus core promoter mutants. J Virol. 2003;77:6601–6612. doi: 10.1128/JVI.77.12.6601-6612.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Tang X, Garcia T, Hussain M, Zhang J, Lok A, Wands J, Li J, Tong S. Hepatitis B virus genotype C isolates with wild-type core promoter sequence replicate less efficiently than genotype B isolates but possess higher virion secretion capacity. J Virol. 2011;85:10167–10177. doi: 10.1128/JVI.00819-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuyver L, De Gendt S, Van Geyt C, Zoulim F, Fried M, Schinazi RF, Rossau R. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J Gen Virol. 2000;81:67–74. doi: 10.1099/0022-1317-81-1-67. [DOI] [PubMed] [Google Scholar]
- Tong S, Diot C, Gripon P, Li JS, Vitvitski L, Trepo C, Guguen-Guillouzo C. In vitro replication competence of a cloned hepatitis B virus variant with a nonsense mutation in the distal pre-C region. Virology. 1991;181:733–737. doi: 10.1016/0042-6822(91)90908-t. [DOI] [PubMed] [Google Scholar]
- Tong S, Kim KH, Chante C, Wands J, Li J. Hepatitis B virus e antigen variants. Int J Med Sci. 2005;2:2–7. doi: 10.7150/ijms.2.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Li J, Vitvitski L, Trepo C. Active hepatitis B virus replication in the presence of anti-HBe is associated with viral variants containing an inactive pre-C region. Virology. 1990;176:596–603. doi: 10.1016/0042-6822(90)90030-u. [DOI] [PubMed] [Google Scholar]
- Tong S, Li JS, Vitvitski L, Trepo C. Replication capacities of natural and artificial precore stop codon mutants of hepatitis B virus: relevance of pregenome encapsidation signal. Virology. 1992;191:237–245. doi: 10.1016/0042-6822(92)90185-r. [DOI] [PubMed] [Google Scholar]
- Tsai A, Kawai S, Kwei K, Gewaily D, Hutter A, Tong DR, Li J, Wands JR, Tong S. Chimeric constructs between two hepatitis B virus genomes confirm transcriptional impact of core promoter mutations and reveal multiple effects of core gene mutations. Virology. 2009;387:364–372. doi: 10.1016/j.virol.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Tanaka Y, Huang Y, Kurbanov F, Chen J, Zeng G, Zhou B, Mizokami M, Hou J. Clinical and virological characteristics of hepatitis virus subgenotypes Ba, C1, and C2 in China. J Clin Microbiol. 2007;45:1491–1496. doi: 10.1128/JCM.02157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan T, Li K, Li F, Su H, Mu J, Tong S, Patel M, Xia J, Wands J, Wang H. T1846 and A/G1913 are associated with acute on chronic liver failure in patients infected with hepatitis B virus genotypes B and C. J Med Virol. 2011;83:996–1004. doi: 10.1002/jmv.22067. [DOI] [PubMed] [Google Scholar]
- Yin J, Xie J, Liu S, Zhang H, Han L, Lu W, Shen Q, Xu G, Dong H, Shen J, Zhang J, Han J, Wang L, Liu Y, Wang F, Zhao J, Zhang Q, Ni W, Wang H, Cao G. Association between the various mutations in viral core promoter region to diferent stages of hepatitis B, ranging of asymptomtic carrier state to hepatocellular carcinoma. Am J Gastroenterol. 2011;106:81–92. doi: 10.1038/ajg.2010.399. [DOI] [PubMed] [Google Scholar]
- Yuen MF, Sablon E, Yuan HJ, Wong DK, Hui CK, Wong BC, Chan AO, Lai CL. Significance of hepatitis B genotype in acute exacerbation, HBeAg seroconversion, cirrhosis-related complications, and hepatocellular carcinoma. Hepatology. 2003;37:562–567. doi: 10.1053/jhep.2003.50098. [DOI] [PubMed] [Google Scholar]
- Zong L, Qin Y, Jia H, Zhou L, Chen C, Zhang J, Wang Y, Li J, Tong S. Two-way molecular ligation for efficient conversion of monomeric hepatitis B virus DNA constructs into tandem dimers. J Virol Methods. 2016;233:46–50. doi: 10.1016/j.jviromet.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]