Abstract
Dictyostelium PakB, previously termed myosin I heavy chain kinase, is a member of the p21-activated kinase (PAK) family. Two-hybrid assays showed that PakB interacts with Dictyostelium Rac1a/b/c, RacA (a RhoBTB protein), RacB, RacC, and RacF1. Wild-type PakB displayed a cytosolic distribution with a modest enrichment at the leading edge of migrating cells and at macropinocytic and phagocytic cups, sites consistent with a role in activating myosin I. PakB fused at the N terminus to green fluorescent protein was proteolyzed in cells, resulting in removal of the catalytic domain. C-terminal truncated PakB and activated PakB lacking the p21-binding domain strongly localized to the cell cortex, to macropinocytic cups, to the posterior of migrating cells, and to the cleavage furrow of dividing cells. These data indicate that in its open, active state, the N terminus of PakB forms a tight association with cortical actin filaments. PakB-null cells displayed no significant behavioral defects, but cells expressing activated PakB were unable to complete cytokinesis when grown in suspension and exhibited increased rates of phagocytosis and pinocytosis.
INTRODUCTION
Members of the p21-activated kinase (PAK) family are key regulators of the actin cytoskeleton and cell motility in organisms ranging from yeast to mammals (Bokoch, 2003). PAKs are characterized by the presence of two conserved domains: a p21-binding domain (PBD) and a C-terminal Ser/Thr protein kinase catalytic domain. The PBD mediates interactions with active Cdc42 and Rac GTPases and encompasses an autoinhibitory sequence that potently suppresses the activity of the catalytic domain. The binding of GTPCdc42/Rac to the PBD disrupts the autoinhibitory interaction, permitting a series of autophosphorylation events that maximize kinase activity.
Studies on Dictyostelium discoideum have provided valuable insights into the signaling pathways that regulate cell polarization and chemotaxis (Merlot and Firtel, 2003). To date, three Dictyostelium PAKs have been identified: PakA (Chung and Firtel, 1999), PakB (Lee et al., 1996), and PakC (GenBank accession no. AF277804). The three PAK isoforms share between 50 and 70% sequence identity within the PBD and catalytic domains but exhibit no homology outside of these regions. Loss of PakA produces cytokinesis defects in cells grown in suspension culture and prevents cells from suppressing lateral pseudopod extension or from properly retracting the cell posterior during chemotaxis (Chung and Firtel, 1999; Chung et al., 2001). These authors conclude that PakA, which localizes to the posterior of migrating cells, functions to promote the assembly of myosin II into bipolar filaments. In a second study, PakA-null cells were found to display no detectable defects with respect to locomotion or cytokinesis (Muller-Taubenberger et al., 2002).
PakB was initially identified through its ability to phosphorylate and activate MyoD, a single-headed type I myosin (Lee and Côté, 1995; Lee et al., 1996). PakB was originally called myosin I heavy chain kinase, but it has been renamed to conform to the gene name (pakB) (Fey et al., 2004). PakB phosphorylates a site located in the MyoD motor domain designated the TEDS rule site due to the presence of a Thr, Glu, Asp, or Ser residue at this position in almost all myosins (Bement and Mooseker, 1995). Members of the PAK family in a variety of organisms display a conserved ability to activate myosins with Ser/Thr at the TEDS rule site (Lee et al., 1996; Wu et al., 1996; Brzeska et al., 1999; Yoshimura et al., 2001). Because all seven Dictyostelium myosin I isozymes (MyoA-F and MyoK) and MyoM (a myosin that exhibits guanine nucleotide exchange factor activity for Rac) have a Ser/Thr at the TEDS rule site, PAKs are likely to play a key role in regulating motile processes driven by these myosins (de la Roche and Côté, 2001). In vitro studies show that PakB is activated by human Cdc42 and Rac1, providing a direct mechanism to link Rho-related GTPases to the regulation of myosin-driven motility (Lee et al., 1996).
In this study, we have identified the Dictyostelium Rho-related proteins that bind to PakB and show that PakB is enriched at sites, including the leading edge of migrating cells, consistent with a role in the regulation of myosin I. Cells in which the pakB gene has been disrupted do not, however, show impaired myosin I-dependent functions. In contrast, a constitutively active mutant of PakB increases the rates of pinocytosis and phagocytosis, two myosin I-dependent processes, and disrupts cytokinesis. Constitutively active PakB is concentrated at the posterior of migrating cells, suggesting a model in which the N-terminal domain of activated PakB attaches tightly to cortical actin filaments that flow to the rear of the cell.
MATERIALS AND METHODS
Yeast Two-Hybrid Analysis
A DNA fragment encoding residues 313-411 of PakB was cloned into the yeast two-hybrid vector pACT2 (BD Biosciences Clontech, Palo Alto, CA). DNA fragments carrying the G12V or equivalent (constitutively active) mutation or the T17N or equivalent (dominant negative) mutation of human and Dictyostelium Rho GTPases were generated from wild-type cDNA by polymerase chain reaction-based site-directed mutagenesis. In all cases, the CAAX motif was either modified by mutagenesis (Cys to Ser) or removed by restriction enzyme digestion. Rho GTPases were cloned into the yeast two-hybrid vectors pGADT7, pAS2-1, or pGBKT7 (BD Biosciences Clontech). For RacA, only the GTPase domain was cloned, and for RacH a truncated protein (residues 1-163) was used because full-length constructs activated the β-galactosidase reporter. All products were verified by sequencing. The protocols of the Matchmaker Two-hybrid system from BD Biosciences Clontech were followed for all experiments dealing with two-hybrid assays. Constructs in pGADT7 were introduced into yeast strain Y187. Constructs in pAS2-1 or pGBKT7 were introduced into yeast strain Y190. After mating, interactions were estimated by colony-lift β-galactosidase filter assay.
Construction of Vectors
The full-length PakB coding sequence was cloned into the pNEB vector (New England Biolabs, Beverly, MA) to generate pNEB-PakB (Lee et al., 1996). A vector expressing green fluorescent protein (GFP) fused to the N terminus of PakB was constructed by inserting the PakB open reading frame (ORF) into the plasmid pDXA-GFP2 to generate pDXA-GFP-PakB (Levi et al., 2000). To generate a vector expressing GFP fused to the C terminus of PakB, the multiple cloning site downstream of the GFP coding sequence in pDXA-GFP2 was removed by digestion with BamHI and XbaI, treated with T4 DNA polymerase, and religated. A linker containing unique SacI, BamHI, and XhoI restriction sites was then inserted between the HindIII and KpnI sites of the vector to yield pDXA-CT-GFP. The pakB ORF was amplified using primers containing a 5′ SacI site and a 3′ XhoI site, digested at these sites, and inserted into pDXA-CT-GFP digested at the same sites. PakB-ΔN was constructed by digesting pNEB-PakB with BsaBI and XbaI and inserting the resulting fragment into pDXA-GFP2 cut with XhoI, treated with T4 DNA polymerase, and then digested with XbaI. GFP-PakB-ΔPL was constructed by digesting pDXA-GFP-PakB with BamHI followed by treatment with T4 DNA polymerase, digestion with BsaBI, and religation. N-terminal FLAG-tagged versions of PakB, PakB-ΔN, and PakB-ΔPL were produced by cloning inserts from the respective pDXA-GFP vectors into the vector pDXA-FLAG (Levi et al., 2000). An N-terminal 7 × His-tagged version of PakB was produced by cloning the pakB ORF into the expression vector pDXA-HC (Manstein et al., 1995). A pakB targeting vector was constructed in the plasmid pGem7Z (Promega, Madison, WI) by ligating the 3′ end of a DNA fragment comprising base pairs 360-1351 of the pakB ORF to the 5′ end of the 1.7-kb thy gene (Dynes and Firtel, 1989), excised using NsiI from the plasmid pThy-short (a gift of K. Ng and G. Nuckolls, Stanford University, Stanford, CA). A DNA fragment comprising base pairs 2060-2553 of the pakB ORF plus 118 base pairs of 3′ noncoding sequence was then ligated to the 3′ end of the thy gene. Before use, the plasmid was digested with XbaI and blunt-ended. All plasmids were verified by sequencing.
Culturing and Transformation of Dictyostelium
Cells of the Dictyostelium thymidine auxotroph strain JH10 were cultured at room temperature in HL5 medium (Sussman, 1987) supplemented with 200 μg/ml thymidine. JH10 cells were transformed by electroporation by using a Gene Pulser (Bio-Rad, Mississauga, Ontario, Canada). Transformants were selected in HL5 lacking thymidine and were subsequently cloned by plating at limiting dilution in 96-well dishes. Constructs expressing GFP- or FLAG-tagged versions of PakB were introduced into cells by electroporation, and transformants were selected for growth in HL5 medium containing 5 μg/ml G-418 (Geneticin). Transformants were cloned by plating at limiting dilution and were maintained in HL5 containing 20 μg/ml G-418. To initiate development cells were harvested during midlog phase growth, washed in starvation buffer (20 mM K/Na phosphate buffer, 1 mM MgCl2, pH 6.8), and plated at a density of 2 × 108 cells/ml on Whatman 50 filter papers.
Southern and Western Blotting, Immunoprecipitation, and Kinase Assays
Southern blot analysis was performed using genomic DNA isolated as described previously (Nellen et al., 1987). DNA was hybridized at 68°C in buffer containing 6× SSC, 5× Denhardts, 0.2% SDS, and 0.1 mg/ml salmon sperm DNA by using 32P-labeled probes generated using the random primer method (Sambrook et al., 1989). Extracts for immunoblotting were prepared by boiling cells in SDS sample buffer (6% sucrose, 1% SDS, and 40 mM β-mercaptoethanol). After SDS-PAGE proteins were transferred to Immobilon-P membranes (Millipore, Billerica, MA) and incubated with affinity-purified rabbit polyclonal antibody against PakB (Lee et al., 1996), a monoclonal antibody (mAb) to GFP (Santa Cruz Biotechnology, Santa Cruz, CA) or anti-FLAG M5 mAb (Sigma-Aldrich, Oakville, Ontario, Canada). Blots were visualized using horseradish peroxidase-coupled second antibodies and enhanced chemiluminescence (Amersham Biosciences, Piscataway, NJ). For immunoprecipitation experiments, cells were lysed in 1 ml of ice-cold buffer containing 25 mM HEPES, pH 6.8, 10% glycerol, 100 mM NaCl, 1 mM EDTA, 2 mM dithiothreitol (DTT), 0.2% Triton X-100, and a protease inhibitor cocktail (Sigma-Aldrich), centrifuged at 15,000 × g, and incubated with 20 μl of anti-FLAG M52 agarose (Sigma-Aldrich). The agarose beads were washed twice with lysis buffer, twice with 25 mM HEPES, pH 6.8, containing 100 mM NaCl, and resuspended in 50 μl of this buffer. Kinase assays were performed by adding 10 μl of bead suspension to a 40-μl reaction mix containing 20 mM TES (pH 7.5), 1 mg/ml bovine serum albumin, 1 mM DTT, 0.25 mM cold ATP, 0.2 μl of [γ-32P]ATP, 2 mM MgCl2, 100 μg/ml myelin basic protein, and 1 μg of GTP-bound human Rac1V12G. Reactions were stopped by the addition of SDS sample buffer, and radioactive proteins were visualized and quantified after SDS-PAGE by using a Bio-Rad Personal Molecular Imager FX.
Pinocytosis and Phagocytosis Assays
Quantitative fluid phase and bead uptake assays were conducted as described previously (Novak et al., 1995; Jung et al., 1996). For pinocytosis assays, HL5 medium containing 1.0 mg/ml Texas Red-conjugated dextran (70,000 kDa) (Molecular Probes, Eugene OR) was added to 5 × 106 cells attached to a plastic dish. For phagoctyosis assays, 1 mg/ml of 1 μm-diameter yellow-green fluorescence FluoSpheres (Molecular Probes) was added to cells in suspension culture. Cells were washed three times with ice-cold starvation buffer and lysed in 1 ml of starvation buffer containing 0.2% Triton X-100. Fluorescence was measured using a PerkinElmer LS50B spectrofluorometer. The results were normalized to total protein content measured using the Quick Start Bradford protein assay (Bio-Rad).
Microscopy
Cells expressing GFP fusion proteins were harvested from petri dishes, washed once with starvation buffer, and allowed to reattach to glass coverslips for 2 h in a moist chamber at room temperature before observation. Heat-killed Saccharomyces cerevisiae were added to cells at a concentration of 5 × 106 yeast per milliliter in starvation buffer. Cells were fixed in 2% paraformaldehyde, treated with 0.2% Triton X-100, washed with phosphate-buffered saline (PBS), blocked in 30% normal goat serum, and incubated with 15 μg/ml M5 anti-FLAG mAb, affinity-purified rabbit anti-PakB polyclonal antibody (1:1000 dilution), rabbit anti-Dictyostelium myosin II polyclonal antibody (1:20 dilution) (a gift of T. T. Egelhoff, Case Western Reserve University, Cleveland, OH), or tetramethylrhodamine B isothiocyanate (TRITC)-phalloidin (Sigma-Aldrich) (1:200 dilution). After three washes with PBS, cells were incubated with goat anti-rabbit or anti-mouse total IgG conjugated to Alexa Fluor 488 or 568 (Molecular Probes). Conventional transmission and fluorescence microscopy was performed using a Zeiss Axiovert 100 inverted microscope equipped with a Plan-Neofluar 40×/0.75 objective and a Plan-Apochromat 63×/1.4 oil immersion objective. Images were captured using a SensiCam cooled charge-coupled device videocamera (Cooke, Auburn Hills, MI) and processed using SlideBook software (Intelligent Imaging Innovations, Denver, CO). For the chemotaxis assay, cells were directionally stimulated using a glass capillary microtip (Eppendorf Femtotip) filled with 0.1 M cAMP.
RESULTS
Identification of Dictyostelium Rho Family GTPases That Bind PakB
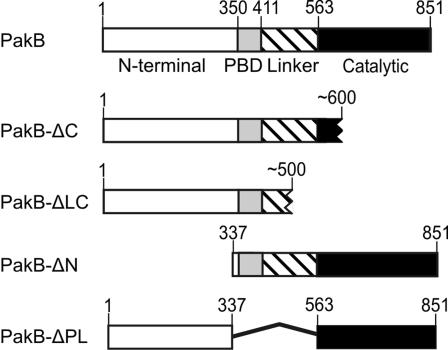
Dictyostelium PakB consists of a proline-rich N-terminal domain, a conserved PBD, a linker region rich in glutamine and asparagine residues, and a C-terminal Ser/Thr kinase domain (Figure 1). In vitro studies have shown that the catalytic activity of PakB can be stimulated by GTP-bound human Cdc42 and Rac1 (Lee et al., 1996). Yeast two-hybrid analyses were carried out using the isolated PBD of PakB to identify which of the 15 different Dictyostelium Rho-related GTPases bind to PakB (Rivero et al., 2001; Rivero and Somesh, 2002). In this assay, the PakB PBD interacted strongly with activated human Rac1 and Cdc42, used as positive controls, and with Dictyostelium Rac1a, b, and c; the GTPase domain of RacA (a member of the RhoBTB protein family); and RacB, RacC, and RacF1 (Table 1). The dominant negative forms of these GTPases scored negative. RacF2 was not tested but is expected to behave similarly to RacF1, with which it shares 94% sequence identity. Not unexpectedly, the Dictyostelium Rho-like GTPases that bind to PakB all have a GTPase domain that belongs to the Rac subclass of Rho GTPases (Rivero and Somesh, 2002).
Figure 1.
Schematic diagrams for PakB and mutants used in this study. PakB (GenBank accession no. AAC71063) is comprised of a proline-rich N-terminal domain, a PBD, a linker region in which almost one-half the residues are glutamines or asparagines, and a Ser/Thr kinase catalytic domain. The structures of the endogenously generated C-terminal truncation products (PakB-ΔC and PakB-ΔCL) are predicted based on the size of the proteins as determined by SDS-PAGE.
Table 1.
Yeast two-hybrid analysis of the interaction of the PBD domain of PakB with human (Hs) and Dictyostelium Rho family GTPases
| Rho GTPase | Strength of interaction |
|---|---|
| Rac1a Q61L | +++ |
| Rac1b G12V | +++ |
| Rac1c G12V | +++ |
| RacA G12V | +++ |
| RacB G12V | +++ |
| RacC G15V | +++ |
| RacD G17V | - |
| RacE G20V | + |
| RacF1 G12V | +++ |
| RacG G12V | + |
| RacH M13V | - |
| RacI S14V | - |
| RacJ D18V | - |
| RacL G12V | - |
| HsRac1 G12V | +++ |
| HsCdc42 G12V | +++ |
| HsRhoA G14V | - |
The strength of the interaction was estimated by colony-lift β-galactosidase filter assays. Color development was monitored 4 days after plating and judged to be negative (-), weak (+), medium (++), or strong (+++).
Generation of PakB-Null Cells
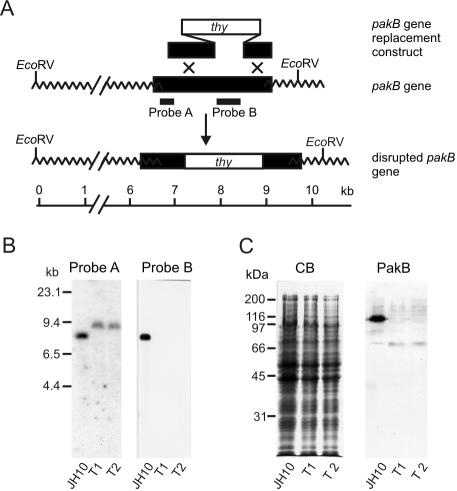
To evaluate the cellular role of PakB, the pakB gene was disrupted in the thymidine auxotroph JH10 cell line by using the gene targeting construct depicted in Figure 2A. Clonal cell lines were selected for growth in the absence of thymidine and disruption of the pakB gene was confirmed by Southern blot analysis in two independently isolated transformed clones (T1 and T2) (Figure 2B). Immunoblot analysis by using an anti-PakB polyclonal antibody was used to confirm the loss of PakB in the T1 and T2 cell lines. PakB migrates on SDS gels with an apparent size of 110 kDa, somewhat larger than its calculated mass of 93 kDa (Lee et al., 1996). A protein of 110 kDa was detected by the anti-PakB antibody in JH10 cells but not in T1 and T2 cells (Figure 2C). Although an N-terminal fragment of 50 kDa could potentially be expressed from the disrupted pakB gene, a protein of this size was not detected in T1 or T2 cells by using the anti-PakB antibody. In all subsequent studies, the T1 and T2 cell lines behaved identically and will henceforth be referred to as PakB-null cells.
Figure 2.
Gene replacement construct and targeted disruption of the pakB gene. (A) Schematic diagram shows the pakB gene replacement construct, the wild-type pakB locus and the altered structure of the pakB gene expected if a double crossover, gene replacement event occurs. Black segments correspond to pakB sequences, the white segment to the thy gene, and wavy lines to extragenic sequences. The locations of probes A and B used in the genomic Southern analysis are indicated. (B) Southern blot analysis of JH10 cells and two clones (T1 and T2) that grew in the absence of thymidine. Genomic DNA was digested with EcoRV and hybridized with either probe A or probe B. The increased size of the EcoRV fragment detected with Probe A and the absence of bands hybridizing to Probe B are consistent with the scheme illustrated in A. Molecular size standards are indicated in kilobases. An equal amount of DNA was loaded on each lane of the gel. (C) Extracts from 1 × 106 JH10, T1, or T2 cells were subjected to SDS-PAGE and stained with Coomassie Blue (CB) or transferred to Immobilon-P and probed with an anti-PakB antibody (PakB). Molecular size standards are indicated in kilodaltons.
PakB-Null Cells Display No Severe Phenotypic Defects
PakB-null cells grew at a slightly slower rate than JH10 cells in suspension culture (doubling time of 13.5 ± 0.6 h compared with 11.8 ± 0.5 h). After 3 d in suspension culture, PakB-null cells contained on average 2.1 ± 0.2 nuclei per cell, a number typical for Dictyostelium cells grown in suspension (Nagasaki et al., 2002). When attached to plastic, PakB-null and JH10 cells grew at identical rates (doubling time of 10.5 ± 0.5 h). On starvation, PakB-null cells aggregated, formed motile slugs, and differentiated into morphologically normal fruiting bodies with a time course comparable with JH10 cells, showing that PakB is not essential for development. PakB-null cells were able to chemotax toward a micropipette secreting cAMP and take up fluorescein isothiocyanate (FITC)-labeled dextran and FITC-labeled beads at rates equivalent to JH10 cells. Dictyostelium cells that lack two or three myosin I isoforms display severe defects in chemotaxis, pinocytosis, and phagocytosis (Novak et al., 1995; Jung et al., 1996), indicating that myosin I function is not significantly impaired in the absence of PakB. There were also no obvious defects in myosin II filament assembly in PakB-null cells. GFP-myosin II localized normally to the posterior of migrating PakB-null cells. Moreover, in response to cAMP stimulation myosin II transiently distributed into and out of the Triton X-100-insoluble fraction of developed PakB-null cells with a time course comparable with that observed for JH10 cells.
Full-Length PakB Localizes to the Leading Edge of Migrating Cells
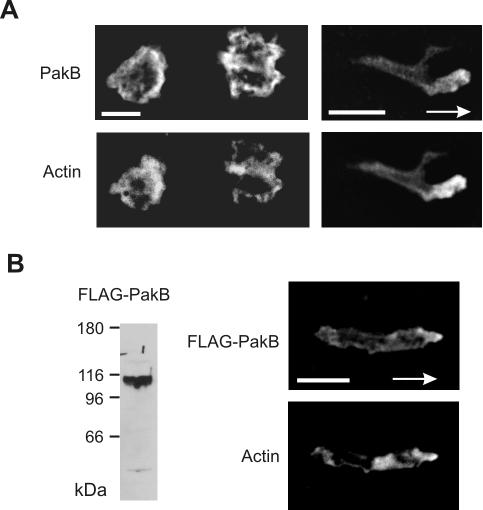
Immunofluorescence analysis showed that in stationary cells endogenous PakB was localized diffusely in the cytoplasm, with some enrichment at the cell cortex (Figure 3A). In elongated migrating cells, PakB was modestly enriched at the leading edge (Figure 3A). PakB with an N-terminal FLAG-tag displayed a similar distribution to endogenous PakB when expressed in PakB-null cells (Figure 3B). The relatively weak cortical distribution of PakB is consistent with protein purification and quantitative immunoblotting studies that show that PakB is predominately a cytosolic protein (Lee and Côté, 1995; Senda et al., 2001).
Figure 3.
PakB localizes to the leading edge of migrating cells. (A) Paired fluorescent images of resting growth-phase JH10 cells (left) and polarized aggregation phase JH10 cells (right) labeled with anti-PakB antibody (top) and TRITC-phalloidin (bottom). The arrow shows the direction of migration toward the aggregation center. (B) Immunoblot analysis (left) of whole cell lysates from PakB-null cell transformants expressing FLAG-PakB by using anti-FLAG M5 antibody. FLAG-PakB was expressed as a protein of ∼110 kDa. Paired fluorescent images of an aggregation stage amoeba stained with M5 anti-FLAG antibody (top) and TRITC-phalloidin (bottom right). Arrows show the direction of cell migration toward the aggregation center. Bars, 5 μm.
GFP-PakB Is Proteolytically Processed in Cells
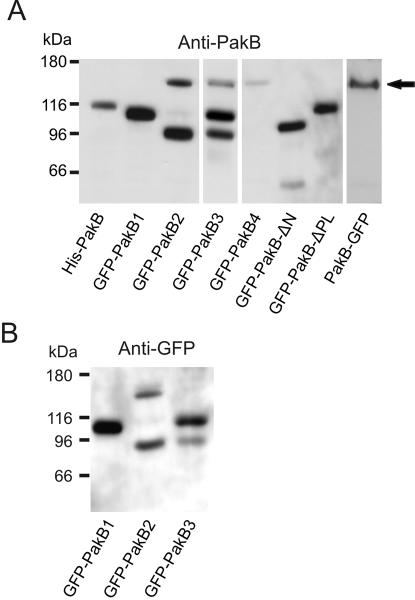
When PakB with GFP fused to the N terminus (GFP-PakB) was expressed in PakB-null cells, none of the brightly fluorescent clonal cell lines that were isolated expressed primarily the full-length GFP-PakB (predicted size of 140 kDa). Instead, immunoblot analysis with an anti-PakB antibody showed that the clonal cell lines contained a GFP-PakB fragment of either 110 or 95 kDa (Figure 4A). Interestingly, clonal cell lines were isolated that produced only the 110- or 95-kDa fragment and that maintained their distinctive pattern of GFP-PakB degradation through multiple passages (Figure 4A). The 110- and 95-kDa fragments had an intact N terminus as judged by their ability to react with an anti-GFP antibody (Figure 4B). Thus, it can be inferred from their sizes that the 110-kDa fragment is produced by cleavage at a site within the catalytic domain (PakB-ΔC), and the 95-kDa fragment is produced by cleavage within the linker region (PakB-ΔLC) (Figure 1). The fate of the released catalytic domain is not clear, but because a fragment in the 40- to 45-kDa size range was not detected by the polyclonal anti-PakB antibody, it seems probable that the catalytic domain is not stable and is further degraded.
Figure 4.
Fusion of GFP to the N terminus of PakB results in intracellular proteolysis. (A) Immunoblot analysis using an anti-PakB antibody of PakB-null cells expressing PakB constructs. Extracts were obtained from cells expressing PakB with an N-terminal 7 × His tag (His-PakB), PakB with GFP fused to the N terminus (4 clones labeled GFP-PakB1-4), PakB-ΔN with GFP fused to the N terminus (GFP-PakB-ΔN), PakB-ΔPL with GFP fused to the N terminus (GFP-PakB-ΔPL), and PakB with GFP fused to the C terminus (PakB-GFP). GFP-PakB, expected to have a size of 140 kDa (arrow), is degraded to products of 110 and 95 kDa in cell lines GFPPakB1-3. The other PakB fusion proteins exhibit electrophoretic mobilities consistent with their predicted molecular masses. (B) Immunoblot analysis using an anti-GFP antibody shows that the 110- and 95-kDa GFP-PakB degradation products retain an intact N terminus. For each lane in A and B, 20 μg of protein was loaded. Molecular mass standards are indicated in kilodaltons.
PakB with a 7 × His-tag or a FLAG-tag fused to the N terminus or with GFP fused to the C terminus (PakB-GFP) was not degraded in cells (Figures 3B and 4A). Possibly, the addition of GFP to the N terminus of PakB interferes with ability of PakB to adopt an autoinhibited conformation, resulting in an overactive form of the enzyme that is susceptible to proteolysis. It is interesting to note that in apoptotic mammalian cells, a similar cleavage of PAK2 into regulatory and catalytic domains is catalyzed by caspases (Lee et al., 1997; Rudel and Bokoch, 1997). The nature of the Dictyostelium protease that cleaves PakB, and the physiological relevance of the cleavage process, remain to be determined.
PakB-GFP Localizes to the Anterior of Migrating Cells
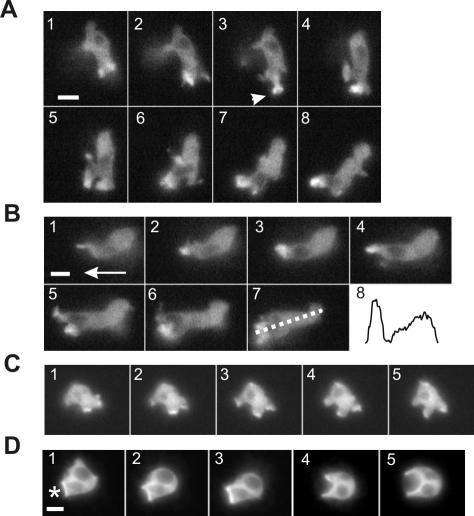
Time-lapse images of live cells showed that full-length PakB-GFP, like endogenous PakB and FLAG-PakB, was enriched within the leading edge of migrating amoebae. As amoebae sharply changed direction, PakB-GFP was rapidly lost from the existing leading edge and reappeared within the newly protruding leading edge (Figure 5, A and B, and Supplemental Movie S1 available at http://www.molbiolcell.org). PakB-GFP was highly concentrated at the tip of extended pseudopods, and in some instances could be observed to transiently adopt a circular or cup-shaped distribution, suggesting an involvement in some type of endocytic process (Figure 5A, 3, arrow). In stationary vegetative cells, PakB-GFP was enriched at the leading edge of large, dynamic cup-shaped structures that seemed to be involved in the process of fluid uptake by macropinocytosis (Figure 5C). PakB-GFP also accumulated within the actin-rich cup-shaped structure that cells protruded to surround and engulf heat-killed yeast particles (Figure 5D). In end-on views of the phagocytic cup, PakB-GFP formed a continuous ring that encircled the yeast particle.
Figure 5.
PakB-GFP localizes to the leading edge of migrating cells. Time-lapse fluorescent images of live PakB-null cells expressing PakB-GFP. Also see Supplemental Movie S1 available at http://www.molbiolcell.org. (A) Images taken at 10-s intervals of a 6-h starved amoeba that traveled from top to bottom (1-4) and then changed direction to migrate from right to left (5-8). PakB-GFP redistributes from the original leading edge to the new leading edge and in 3, momentarily shows up in a cup-shaped structure (arrow) at the tip of the extended pseudopod. (B) Images taken at 10-s intervals of the same cell at a later time migrating in the direction shown by the arrow in 1. Panel 8 shows the fluorescence intensity from the leading edge to the posterior of the cell measured as indicated by the dotted line in 7. (C) Images taken at 20-s intervals of a 2-h starved amoeba taking up fluids by macropinocytosis. (D) Images taken at 20-s intervals of a 2-h starved amoeba ingesting yeast particles. The asterisk in 1 shows the location of the yeast particle. Bars, 5 μm.
PakB-ΔC Localizes to the Posterior of Migrating Cells
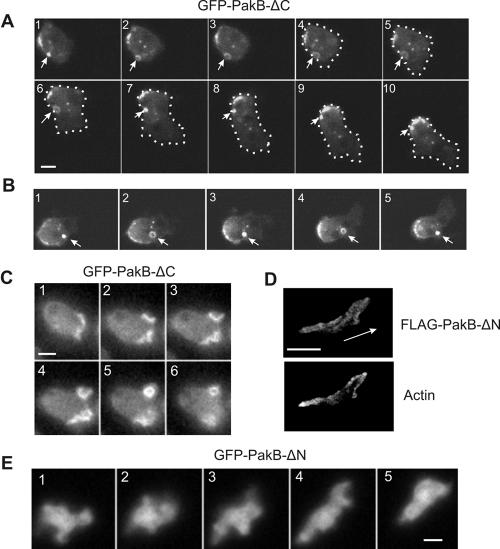
GFP-PakB-ΔC, the 110-kDa C-terminal truncation product produced by cleavage of GFP-PakB, was present on small, long-lived, dot-like particles or vesicles that localized predominately at the cell cortex (Figure 6, A and B). In some instances, the particles were observed to translocate through the cytoplasm. These dot-like particles may play a role in pinocytosis, because they could transiently open up into ring-shaped structures at the cell cortex (Figure 6, A and B). GFP-PakB-ΔC localized much more intensely to extending and retracting macropinocytic cups than did full-length GFP-PakB (compare Figure 6C with 5C). As stationary cells started to move, GFP-PakB-ΔC redistributed to the rear of the cell within less than a minute (Figure 6A, 6-10). When a migrating cell ceased to move, the GFP-PakB-ΔC again resolved into multiple patches and spots that dispersed around the cell cortex.
Figure 6.
PakB-ΔC accumulates at the posterior of migrating cells, whereas PakB-ΔN is diffusely distributed in the cell body. (A) Time lapse fluorescent images of a live 4-h starved amoeba from the GFP-PakB1 clonal cell line, containing GFP-PakB-ΔC. Images taken at intervals of 10 s show an amoeba that was initially stationary (1-3) and then migrated to the lower right (4-10). GFP-PakB-ΔC rapidly accumulated at the rear of the moving cell. The dot-like structures were frequently observed to expand into ring-shaped structures (arrows). The outline of the cell is indicated by the dotted white line. (B) Time-lapse fluorescent images taken at 10-s intervals show the opening and closing of a GFP-PakB-ΔC-containing structure (arrows). (C) Time-lapse fluorescent images taken at 10-s intervals of a live 2-h starved amoeba show that GFP-PakB-ΔC is associated with pinocytotic cups. (D) Paired fluorescent images of a PakB-null cell expressing FLAG-PakB-ΔN labeled with anti-FLAG antibody (top) and TRITC-phalloidin (bottom). The aggregation-phase cell was migrating in the direction indicated by the arrow. (E) Time-lapse fluorescent images taken at 10-s intervals of a live 6-h starved PakB-null cell expressing GFP-PakB-ΔN. Bar, 5 μm.
The strong cortical localization of PakB-ΔC might be explained by its inability to adopt the inactive, closed conformation characteristic of inactive PakB. As a result, the N-terminal domain would be constitutively available to participate in protein-protein interactions. To investigate the potential targeting role of the N-terminal domain, PakB lacking residues 1-336 (PakB-ΔN) was expressed in cells (Figures 1 and 4A). GFP- and FLAG-tagged PakB-ΔN were diffusely distributed throughout the cell body and exhibited no enrichment at the cell cortex or to the front or rear of migrating cells (Figure 6, D and E). The results indicate that PakB is targeted to the cell cortex by interactions mediated by the N-terminal domain.
Constitutively Active PakB
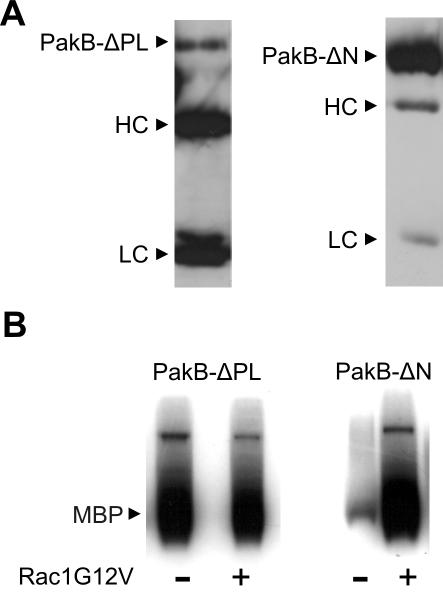
PAKs that express strong constitutive kinase activity have been created by mutating the autoinhibitory sequence or by deleting the entire PBD (Frost et al., 1998; Zhao et al., 1998; Tu and Wigler, 1999). To generate a constitutively active PakB, the PBD and linker regions were excised to give PakB-ΔPL (Figure 1). The linker region was removed along with the PBD in an attempt to minimize the possibility that PakB-ΔPL would be subject to proteolysis in cells. Indeed, GFP-PakB-ΔPL expressed in PakB-null cells exhibited an apparent molecular mass of 110 kDa, close to its predicted size of 98 kDa (Figure 4A). The kinase activity of FLAG-PakB-ΔPL and FLAG-PakB-ΔN was assessed after immunoprecipitation with an anti-FLAG antibody. Interestingly, at least 20 times as much FLAG-PakB-ΔN was recovered in the immunoprecipitates than FLAG-PakB-ΔPL (Figure 7A), even though both proteins were expressed in cells at nearly equivalent levels (Figure 4A). This large difference in the recovery of the protein likely reflects the fact that PakB-ΔN is located in the cytosol whereas, as shown below, PakB-ΔPL is tightly associated with the cortical cytoskeleton. FLAG-PakB-ΔPL displayed a high level of kinase activity in the presence and absence of activated human Rac1, whereas FLAG-PakB-ΔN was active only when GTP-Rac1 was included in the assay (Figure 7B). These results demonstrate that FLAG-PakB-ΔPL is constitutively active and that FLAG-PakB-ΔN, which retains the PBD domain, is still subject to regulation by Rho-family GTPases.
Figure 7.
PakB-ΔPL exhibits constitutive kinase activity. (A) Immunoblot analysis of FLAG-PakB-ΔPL (left) and FLAG-PakB-ΔN (right) immunoprecipitated from 2.5 × 107 cells by using anti-FLAG M2 agarose beads and probed with anti-FLAG M5 antibody. Immunoprecipitates were resuspended in 50 μl of 1× SDS sample buffer, and aliquots of 25 and 5 μl were loaded on the SDS gel for the PakB-ΔPL and PakB-ΔN samples, respectively. HC and LC refer to the antibody heavy chain and light chain, respectively. (B) In vitro kinase assays were performed using a 10-μl aliquot of the FLAG-PakB-ΔPL or FLAG-PakB-ΔN immunoprecipitate in the presence or absence of human Rac1G12V with myelin basic protein (MBP) as substrate. The autoradiograph shows the incorporation of 32P into MBP after a 60-min incubation.
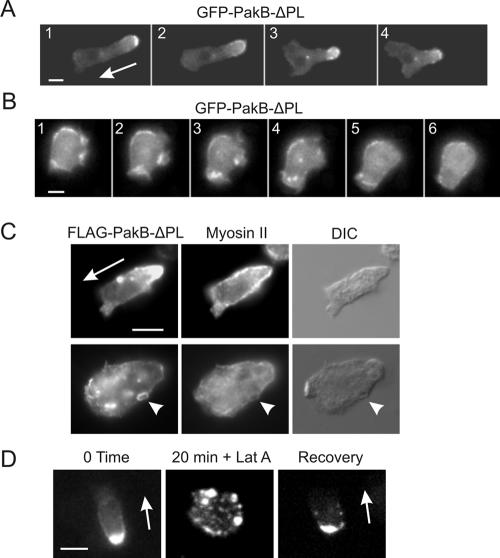
PakB-ΔPL Localizes to the Rear of Migrating Cells
FLAG- and GFP-tagged versions of PakB-ΔPL displayed a subcellular distribution similar to that of PakB-ΔC. In cells migrating toward a micropipette releasing the chemoattractant cAMP, GFP-PakB-ΔPL localized to small dot-like structures that clustered at the cell posterior (Figure 8A and Supplemental Movie S2 available at http://www.molbiolcell.org). GFPPakB-ΔPL also localized to the base of macropinocytic cups, although in a patchy and relatively disordered manner (Figure 8B). The accumulation of active PakB-ΔPL at the rear of the cell did not cause a noticeable disruption or alteration in the distribution of myosin II. As in wild-type cells, myosin II filaments were located at the cortex along the posterior portion of the cell (Figure 8C, top). In stationary cells, myosin II did not colocalize to the dot-like particles or ring-shaped pinocytic structures containing PakB-ΔPL (Figure 8C, bottom). These results support the view that PakB is not involved in the regulation of myosin II filament assembly.
Figure 8.
PakB-ΔPL localizes to the posterior of migrating cells and to sites of pinocytosis. (A) Time-lapse images taken at 20-s intervals of a live 6-h starved PakB-null cell expressing GFP-PakB-ΔPL. Also see Supplemental Movie S2 available at http://www.molbiolcell.org. The cell was migrating in the direction shown by the arrow in 1 toward a micropipette secreting cAMP. GFP-PakB-ΔPL localized in a punctate distribution to the posterior of the migrating cell. (B). Images of a live 2-h starved amoeba taken at 20-s intervals show that GFP-PakB-ΔPL accumulates in a fragmented manner within pinocytic cups. (C) PakB-null cells expressing FLAG-PakB-ΔPL were starved for 4 h, double-labeled using anti-FLAG (left) and anti-myosin II (middle) antibodies, and visualized by fluorescence and differential interference contrast microscopy (right). Shown are migrating (top) and a stationary (bottom) cells. The arrowheads (bottom) point out a ring shaped structure containing FLAG-PakB-ΔPL that corresponds to a depression in the cell surface. (D) Effects of latrunculin A on the distribution of GFP-PakB-ΔPL in a 6-h starved cell. GFP-PakB-ΔPL present at the rear of the cell (left) was dispersed throughout the cytoplasm 20 min after addition of latrunculin A (middle). Within 20 min of the removal of latrunculin A cells regained a motile phenotype with GFP-ΔPLPakB localized at the rear (right). Bar, 5 μm.
Treatment with the actin filament-depolymerizing drug latrunculin A caused migrating cells to round up and cease moving (Figure 8D). In cells treated with latrunculin A, the punctate GFP-PakB-ΔPL-containing structures remained intact but dispersed throughout the cell cytosol. These processes were completely reversible after removal of latrunculin A.
PakB-ΔPL and PakB-ΔC Impair Cytokinesis
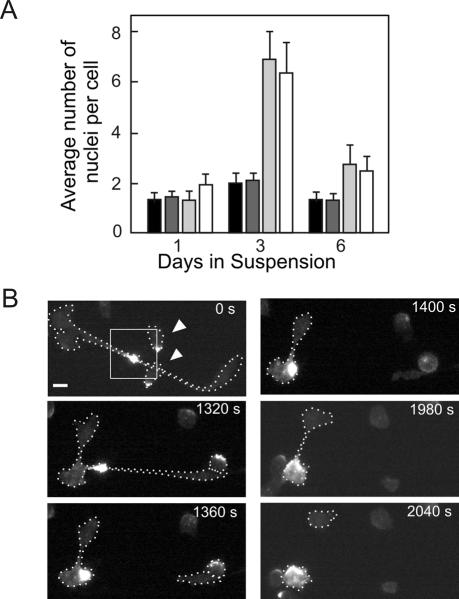
PakB-null cells expressing PakB-ΔPL or PakB-ΔC exhibited a severe defect in cytokinesis when grown in suspension culture. After 3 d in suspension, these cells contained an average of nearly seven nuclei per cell, compared with an average of 2.1 nuclei per cell for JH10 or PakB-null cells (Figure 9A). After 6 d of growth in suspension culture, most of the large multinucleated cells expressing GFP-PakB-ΔPL and GFP-PakB-ΔC had burst, resulting in a reduction in the average number of nuclei per cell.
Figure 9.
Cells expressing GFP-PakB-ΔPL and GFP-PakB-ΔC exhibit a defect in cytokinesis. (A) Average number of nuclei per cell was determined for JH10 cells (black bars), PakB-null cells (dark gray bars), and PakB-null cells expressing GFP-PakB-ΔC (light gray bars) or GFP-PakB-ΔPL (white bars) after 1, 3, and 6 d in suspension culture. (B) Time-lapse images of a dividing, adherent 2-h starved PakB-null cell expressing GFP-PakB-ΔPL. The cell was first imaged (0 s) after it had formed a long cytoplasmic bridge containing GFP-PakB-ΔPL. The white dotted lines indicate the outline of the dividing cell and two smaller cells (arrowheads) with GFP-PakB-ΔPL at the posterior that crawled over the cytoplasmic bridge. Cleavage of the first cytoplasmic bridge took more than 20 min to complete, at which time the cell had already started a second division that took more than 10 min to complete. After two divisions, one of the daughter cells retains the bulk of the GFP-PakB-ΔPL. Bar, 5 μm.
When cultured on plastic ∼5% of cells expressing PakB-ΔPL or PakB-ΔC were very large (diameter in excess of 30 μm) and highly multinucleated, suggesting a partial defect in cytokinesis. Examination of adherent cells undergoing cytokinesis showed that the particles containing GFP-PakB-ΔPL accumulated within the cytoplasmic bridge connecting two daughter cells (Figure 9B). Cytoplasmic bridges that contained large amounts of GFP-PakB-ΔPL were resistant to severing, often stretching up to 50 μm in length and persisting for >40 min. For comparison, cytokinesis in normal Dictyostelium cells is usually completed within 2-3 min (Konzok et al., 1999).
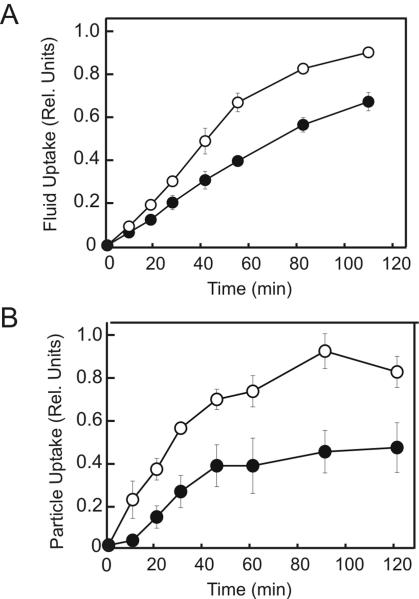
PakB-ΔPL Stimulates Phagocytosis and Pinocytosis
PakB-null cells exhibited rates of phagocytosis and pinocytosis identical to those of JH10 cells and these rates did not change when PakB-null cells were complemented with PakB, PakB-ΔC, or PakB-ΔN. Significant increase in the rates of phagocytosis and pinocytosis were observed when PakB-null cells were complemented with PakB-ΔPL (Figure 10, A and B). From the early portion of the uptake curves, expression of PakB-ΔPL was calculated to increase the rates of pinocytosis and phagocytosis 1.7- and 2.5-fold, respectively. Cells expressing PakB-ΔPL also ingested a larger total number of particles than did PakB-null cells. The finding that PakB-ΔPL but not PakB-ΔC effects pinocytosis and phagocytosis indicates that the stimulatory effects of PakB on these processes are a consequence of its catalytic activity.
Figure 10.
PakB-ΔPL increases the rates of pinocytosis and phagocytosis. The uptake of fluorescent dextran (A) and beads (B) was measured for PakB-null cells (black circles) and PakB-null cells expressing GFP-PakB-ΔPL (white circles). Data presented are the average of three independent determinations for each cell line. Uptake rates were normalized to cell number, but similar results were obtained when uptake was normalized to total protein content.
DISCUSSION
Dictyostelium PakA, PakB, and PakC share closely related PBD and catalytic domains but have highly divergent N-terminal domains. PakA localizes to the rear of migrating cells and has been implicated in the regulation of myosin II filament assembly (Chung and Firtel, 1999; Chung et al., 2001). In this report, we show that PakB is enriched at the leading edge of polarized migrating cells but that truncated forms of PakB that retain the N-terminal domain accumulate at the posterior of migrating cells. Cells lacking a functional PakB show no apparent defects in a variety of motile processes, whereas constitutively active PakB causes severe cytokinesis defects by entrapping actin filaments, while stimulating phagocytosis and pinocytosis. Overall, the studies reported here provide new insights into the role of PakB in cells and highlight significant differences between the properties of PakA and PakB.
Interaction with Dictyostelium Racs
PAKs are activated in response to signaling pathways that generate the active GTP-bound forms of Cdc42- or Rac-related GTPases (Bokoch, 2003). In this study, yeast two-hybrid experiments using all of the Dictyostelium Rho-related GTPases except RacF2 showed that Rac1a, b, and c; and RacA, RacB, RacC, and RacF1 have the potential to function as upstream activators of PakB (Table 1). These GTPases all exhibit a considerable degree of sequence identity to human Rac1, which is consistent with results showing that human Rac1 can activate PakB in vitro (Lee et al., 1996). The three closely related Dictyostelium Rac1 isoforms also bind to the PBD of PakA (Chung and Firtel, 1999; Muller-Taubenberger et al., 2002). The Rac1 isoforms, which are expressed at high levels at all stages of development (Bush et al., 1993), may therefore represent an important common pathway to activate both PakA and PakB.
The strong interaction detected between RacA and PakB is intriguing because RacA belongs to the novel family of RhoBTB proteins (Rivero et al., 2001). RhoBTB proteins are comprised of an N-terminal GTPase domain joined to two BTB domains. Some BTB domains function as adaptors in cullin-3-dependent ubiquitin ligase complexes, suggesting that RacA could function to target PakB to such complexes (Pintard et al., 2004). Members of the RhoBTB family are present in Drosophila and humans, but these proteins have GTPase domains that are divergent from Rac (Rivero et al., 2001; Ramos et al., 2002). Recent studies show that human RhoBTB1/2 does not interact with the PBD of Pak1 (Aspenstrom et al., 2004). Further analysis of PakA and PakC with the complete set of Dictyostelium Rho-related GTPases should provide interesting insights into the common and unique pathways responsible for regulation of each of the three Dictyostelium PAKs.
Implications of the Lack of a PakB-Null Cell Phenotype
Myosins that have a Ser/Thr residue at the TEDS site in the motor domain must be phosphorylated at this site to display motor activity (Bement and Mooseker, 1995). This mechanism of regulation is likely to be of great importance in Dictyostelium, because all seven myosin I isozymes (MyoA-F, K) and MyoM have a Ser/Thr at the TEDS site. PakB was identified on the basis of its ability to phosphorylate and activate MyoD, but in this study PakB-null cells displayed no significant defects in a variety of motile processes known to be dependent on myosin I, including cell migration, phagocytosis, and pinocytosis (Novak et al., 1995; Jung et al., 1996, 2001; Schwarz et al., 2000). One explanation for these results might be that the loss of PakB is compensated for by other Dictyostelium protein kinases. Yeast PAKs (Ste20p and Cla4p) and mammalian PAK3 are able to phosphorylate and activate Dictyostelium MyoD (Wu et al., 1996), so it is not unreasonable to expect that Dictyostelium PakA and/or PakC might also function as myosin I-activating kinases. On the other hand, we cannot rule out the possibility that one or more myosin I isozymes are indeed inactivated in PakB-null cells. For example, the inactivation of MyoD in PakB-null cells would not be detected, because MyoD-null cells show no noticeable defects in cell behavior (Jung et al., 1993). Moreover, the possibility must be considered that the loss of motor activity does not produce a phenotype as severe as that produced by the complete loss of myosin I. Although studies on yeast and Dictyostelium show that phosphorylation of the TEDS site is required for the cellular functions of myosin I (Novak and Titus, 1997, 1998; Wu et al., 1997), the essential function of the single class I myosin (MYOA) in Aspergillus nidulans is structural, requiring little, if any, actin-activated MgATPase or motor activity (Liu et al., 2001). Studies to quantify the phosphorylation state of individual myosin I isozymes in PakB-null cells and JH10 cells will be required to resolve this question.
PakA strongly localizes to the rear of polarized migrating cells (Chung and Firtel, 1999; Muller-Taubenberger et al., 2002), whereas in this study full-length versions of PakB (PakB, FLAG-PakB, and PakB with GFP fused to the carboxy terminus) were enriched at the leading edge. The opposing distributions of PakA and PakB strongly suggest that these two proteins do not have overlapping functions, at least in migrating cells. PakB is properly localized to regulate the activity of myosin I isozymes, which are concentrated within the leading edge of migrating Dictyostelium (Fukui et al., 1989; Novak et al., 1995; Jung et al., 1996; Novak and Titus, 1998). Although it is tempting to speculate that the role of PakB within the leading edge is to activate myosin I, PAKs also are found at the leading edge of polarized, migrating neutrophils (Dharmawardhane et al., 1999; Li et al., 2003) and Entamoeba histolytica (Labruyere et al., 2003), two organisms in which myosins I do not have a Ser/Thr at the TEDS site. Thus, it is possible that PAKs have a more generalized role in promoting or stabilizing actin filament assembly at the leading edge of cells.
The findings that PakA-null cells are multinucleated when grown in suspension culture, do not polarize properly in response to chemotactic gradients, and contain reduced levels of filamentous actin and myosin further support the view that PakA has unique cellular functions that cannot be compensated for by PakB (Chung and Firtel, 1999; Chung et al., 2001). It should be noted, though, that there are strains of Dictyostelium in which the loss of PakA produces no detectable defects (Muller-Taubenberger et al., 2002). Additional studies in which combinations of pakA, pakB, and pakC are subjected to targeted disruption are needed to conclusively identify the overlapping and unique functions of the three PAK isoforms.
A Model for the Localization of Truncated PakB at the Rear of Cells
Full-length versions of PakB (endogenous PakB, FLAG-PakB, and PakB-GFP) were distributed in a diffuse manner throughout the cell body but displayed a modest level of enrichment at the leading edge of migrating cells and within macropinocytic and phagocytic cups (Figures 3 and 5). This subcellular localization pattern is therefore typical of PakB that is properly regulated and able to switch reversibly between inactive and active conformations. We propose that the large soluble pool of PakB, whose existence is evident from both protein purification and subcellular fractionation experiments (Lee and Côté, 1995; Senda et al., 2001), represents the autoinhibited form of PakB, whereas the smaller fraction of PakB that dynamically localizes to the cortical region of the cell represents PakB that is transiently activated, for example, through interactions with active Rac GTPases. In this model, translocation to the cell cortex requires the activation of PakB, which would result in the unmasking of cortical targeting sequences within the N-terminal domain.
In contrast to full-length PakB, PakB-ΔPL and PakB-ΔC strongly localized to small dot-like structures that accumulated at the posterior of migrating cells, at the base of phagocytic and pinocytic cups, and at the cleavage furrow of dividing cells (Figures 6 and 8). The posterior localization of these proteins cannot be attributed to the presence of GFP, because FLAG-PakB-ΔPL also clustered at the rear of migrating cells (Figure 8C). Because PakB-ΔPL and PakB-ΔC each lack one of the complementary domains required to form the autoinhibited conformation, they are likely to both adopt an open conformation in which the cortical targeting sites within the N-terminal domain are always exposed. The cortical targeting sites must be located in the region encompassed by residues 1-337, because this is the only region shared in common by PakB-ΔPL and PakB-ΔC.
PakB-ΔC and PakB-ΔPL adopt a distribution in Dictyostelium amoeba remarkably similar to that observed for injected, fluorescent phalloidin (Fukui et al., 1999) and for a GFP fusion protein containing the C-terminal actin-binding fragment of talin A (TalC63) (Weber et al., 2002). Phalloidin and TalC63 bind tightly to cortical actin filaments and slow or prevent actin filament disassembly. As markers for the stabilized cortical actin filaments, phalloidin and TalC63 flow into the tail of migrating cells, to the base of phagocytic and macropinocytic cups, and into the cleavage furrow of dividing cells. It seems possible that PakB-ΔC and PakB-ΔPL, because they are permanently in an activated conformation might be locked onto cortical actin filaments and, like phalloidin and TalC63, act as markers for the transport of the cortical actin cytoskeleton. This would be in contrast to full-length PakB, which would be able to revert to an inactive state, dissociate from the cortical actin network, and recycle back to the leading edge of migrating cells. In a similar manner, full-length, regulated talin A is present at the leading edges of motile cells, whereas the constitutively active TalC63 actin-binding fragment accumulates as a cap or compact cluster at the tail of migrating cells (Kreitmeier et al., 1995; Weber et al., 2002). The mechanism by which the N-terminal domain of PakB binds actin filaments is currently under investigation.
Phenotypic Consequences of Activated PakB
Cells overexpressing both PakB-ΔC and PakB-ΔPL exhibited a severe cytokinesis defect when grown in suspension and a milder defect when grown on a solid surface. The impaired cell cleavage is therefore due to interactions mediated by the PakB N-terminal domain and not to PakB catalytic activity. The cytokinesis defects produced by PakB-ΔC or PakB-ΔPL on surface-grown cells closely resemble those that occur upon expression of TalC63 (Weber et al., 2002) and so might be a consequence of entrapping and sequestering actin filaments. Interestingly, the expression of active Rac1b-Q61L results in the formation of long, persistent cytoplasmic bridges during cell division on a substrate (Palmieri et al., 2000). Given the strong interaction between activated Rac1b and the PBD of PakB, it is possible that this effect of Rac1b is due to the overactivation of PakB.
PakB-ΔPL and PakB-ΔC were both enriched at cortical regions underlying phagocytic cups and macropinosomes (Figures 6C and 8, B and C), but only the catalytically active PakB-ΔPL stimulated the rates of these processes (Figure 10). Given the requirement for catalytic activity, it can be speculated that activation of myosin I by PakB may be the underlying mechanism by which phagocytosis and pinocytosis rates are enhanced. Analysis of Dictyostelium cells shows that myosin I plays a role in both phagocytosis and pinocytosis, most likely by driving the retraction of actin-rich projections (Novak et al., 1995; Jung et al., 1996; Novak and Titus, 1997). Dictyostelium expressing GFP-PakA exhibit a significantly enhanced rate of phagocytosis but not pinocytosis (Muller-Taubenberger et al., 2002), indicating that PakA and PakB cooperate to regulate this process but not pinocytosis. PAKs in mammalian cells and Entamoeba histolytica also promote phagocytosis (Diakonova et al., 2002; Labruyere et al., 2003) and macropinocytosis (Dharmawardhane et al., 2000). In conclusion, the results of this study are consistent with a role for PakB in the regulation of myosin I-mediated processes. This is very different from the role ascribed to PakA, which regulates myosin II filament assembly.
Supplementary Material
Acknowledgments
We are grateful to the Protein Function Discovery Centre at Queen's University for use of the microscope facilities and to Dr. T. Egelhoff for providing plasmids and advice. We thank Ann-Kathrin Meyer for invaluable technical assistance. This work was supported by grants from the Canadian Institutes of Health Research (#8603) to G.P.C. and from the Deutsche Forschungsgemeinschaft (RI 1034/2) and the Köln Fortune program to F.R.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-06-0534. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-06-0534.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Aspenstrom, P., Fransson, A., and Saras, J. (2004). Rho GTPases have diverse effects on the organization of the actin filament system. Biochem. J. 377, 327-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bement, W. M., and Mooseker, M. S. (1995). TEDS rule: a molecular rationale for differential regulation of myosins by phosphorylation of the heavy chain head. Cell Motil. Cytoskeleton 31, 87-92. [DOI] [PubMed] [Google Scholar]
- Bokoch, G. M. (2003). Biology of the p21-activated kinases. Annu. Rev. Biochem. 72, 743-781. [DOI] [PubMed] [Google Scholar]
- Brzeska, H., Young, R., Knaus, U., and Korn, E. D. (1999). Myosin I heavy chain kinase: cloning of the full-length gene and acidic lipid-dependent activation by rac and cdc42. Proc. Natl. Acad. Sci. USA 96, 394-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush, J., Franek, K., and Cardelli, J. (1993). Cloning and characterization of seven novel Dictyostelium discoideum rac-related genes belonging to the rho family of GTPases. Gene 136, 61-68. [DOI] [PubMed] [Google Scholar]
- Chung, C. Y., and Firtel, R. A. (1999). PAKa, a putative PAK family member, is required for cytokinesis and the regulation of the cytoskeleton in Dictyostelium discoideum cells during chemotaxis. J. Cell Biol. 147, 559-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, C. Y., Potikyan, G., and Firtel, R. A. (2001). Control of cell polarity and chemotaxis by Akt/PKB and PI3 kinase through the regulation of PAKa. Mol. Cell 7, 937-947. [DOI] [PubMed] [Google Scholar]
- de la Roche, M. A., and Côté, G. P. (2001). Regulation of Dictyostelium myosin I and II. Biochim. Biophys. Acta 1525, 245-261. [DOI] [PubMed] [Google Scholar]
- Dharmawardhane, S., Brownson, D., Lennartz, M., and Bokoch, G. M. (1999). Localization of p21-activated kinase 1 (PAK1) to pseudopodia, membrane ruffles, and phagocytic cups in activated human neutrophils. J. Leukoc. Biol. 66, 521-527. [DOI] [PubMed] [Google Scholar]
- Dharmawardhane, S., Schurmann, A., Sells, M. A., Chernoff, J., Schmid, S. L., and Bokoch, G. M. (2000). Regulation of macropinocytosis by p21-activated kinase-1. Mol. Biol. Cell 11, 3341-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diakonova, M., Bokoch, G., and Swanson, J. A. (2002). Dynamics of cytoskeletal proteins during Fcgamma receptor-mediated phagocytosis in macrophages. Mol. Biol. Cell 13, 402-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynes, J. L., and Firtel, R. A. (1989). Molecular complementation of a genetic marker in Dictyostelium using a genomic DNA library. Proc. Natl. Acad. Sci. USA 86, 7966-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey, P., Gaudet, P., Just, E. M., Pilcher, K. E., Dyck, P. A., Kibbe, W. A., and Chisholm, R. L. (2004). “dictyBase.” http://www.dictybase.org/.
- Frost, J. A., Khokhlatchev, A., Stippec, S., White, M. A., and Cobb, M. H. (1998). Differential effects of PAK1-activating mutations reveal activity-dependent and -independent effects on cytoskeletal regulation. J. Biol. Chem. 273, 28191-28198. [DOI] [PubMed] [Google Scholar]
- Fukui, Y., Kitanishi-Yumura, T., and Yumura, S. (1999). Myosin II-independent F-actin flow contributes to cell locomotion in Dictyostelium. J. Cell Sci. 112, 877-886. [DOI] [PubMed] [Google Scholar]
- Fukui, Y., Lynch, T. J., Brzeska, H., and Korn, E. D. (1989). Myosin I is located at the leading edges of locomoting Dictyostelium amoebae. Nature 341, 328-331. [DOI] [PubMed] [Google Scholar]
- Jung, G., Fukui, Y., Martin, B., Hammer, J. A., 3rd. (1993). Sequence, expression pattern, intracellular localization, and targeted disruption of the Dictyostelium myosin ID heavy chain isoform. J. Biol. Chem. 268, 14981-14990. [PubMed] [Google Scholar]
- Jung, G., Remmert, K., Wu, X., Volosky, J. M., Hammer, J. A., III (2001). The Dictyostelium CARMIL protein links capping protein and the Arp2/3 complex to type I myosins through their SH3 domains. J. Cell Biol. 153, 1479-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, G., Wu, X. F., Hammer, J .A., III (1996). Dictyostelium mutants lacking multiple classic myosin I isoforms reveal combinations of shared and distinct functions. J. Cell Biol. 133, 305-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konzok, A., Weber, I., Simmeth, E., Hacker, U., Maniak, M., and Muller-Taubenberger, A. (1999). DAip1, a Dictyostelium homologue of the yeast actin-interacting protein 1, is involved in endocytosis, cytokinesis, and motility. J. Cell Biol. 146, 453-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitmeier, M., Gerisch, G., Heizer, C., and Müller-Taubenberger, A. (1995). A talin homologue of Dictyostelium rapidly assembles at the leading edge of cells in response to chemoattractant. J. Cell Biol. 129, 179-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labruyere, E., Zimmer, C., Galy, V., Olivo-Marin, J. C., and Guillen, N. (2003). EhPAK, a member of the p21-activated kinase family, is involved in the control of Entamoeba histolytica migration and phagocytosis. J. Cell Sci. 116, 61-71. [DOI] [PubMed] [Google Scholar]
- Lee, N., MacDonald, H., Reinhard, C., Halenbeck, R., Roulston, A., Shi, T., and Williams, L. T. (1997). Activation of hPAK65 by caspase cleavage induces some of the morphological and biochemical changes of apoptosis. Proc. Natl. Acad. Sci. USA 94, 13642-13647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.-F., and Côté, G. P. (1995). Purification and characterization of a Dictyostelium protein kinase required for the actin-activated MgATPase activity of Dictyostelium myosin ID. J. Biol. Chem. 270, 11776-11782. [DOI] [PubMed] [Google Scholar]
- Lee, S.-F., Egelhoff, T. T., Mahasneh, A., and Côté, G. P. (1996). Cloning and characterization of a Dictyostelium myosin I heavy chain kinase activated by Cdc42 and Rac. J. Biol. Chem. 271, 27044-27048. [DOI] [PubMed] [Google Scholar]
- Levi, S., Polyakov, M., and Egelhoff, T. T. (2000). Green fluorescent protein and epitope tag fusion vectors for Dictyostelium discoideum. Plasmid 44, 231-238. [DOI] [PubMed] [Google Scholar]
- Li, Z., et al. (2003). Directional sensing requires G beta gamma-mediated PAK1 and PIX alpha-dependent activation of Cdc42. Cell 114, 215-227. [DOI] [PubMed] [Google Scholar]
- Liu, X., Osherov, N., Yamashita, R., Brzeska, H., Korn, E. D., and May, G. S. (2001). Myosin I mutants with only 1% of wild-type actin-activated MgATPase activity retain essential in vivo function(s). Proc. Natl. Acad. Sci. USA 98, 9122-9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manstein, D. J., Schuster, H. P., Morandini, P., and Hunt, D. M. (1995). Cloning vectors for the production of proteins in Dictyostelium discoideum. Gene 162, 129-134. [DOI] [PubMed] [Google Scholar]
- Merlot, S., and Firtel, R. A. (2003). Leading the way: directional sensing through phosphatidylinositol 3-kinase and other signaling pathways. J. Cell Sci. 116, 3471-3478. [DOI] [PubMed] [Google Scholar]
- Muller-Taubenberger, A., Bretschneider, T., Faix, J., Konzok, A., Simmeth, E., and Weber, I. (2002). Differential localization of the Dictyostelium kinase DPAKa during cytokinesis and cell migration. J. Muscle Res. Cell Motil. 23, 751-763. [DOI] [PubMed] [Google Scholar]
- Nagasaki, A., De Hostos, E. L., and Uyeda, T. Q. (2002). Genetic and morphological evidence for two parallel pathways of cell-cycle-coupled cytokinesis in Dictyostelium. J. Cell Sci. 115, 2241-2251. [DOI] [PubMed] [Google Scholar]
- Nellen, W., Datta, S., Reymond, C., Sivertsen, A., Mann, S., Crowley, T., and Firtel, R. A. (1987). Molecular biology in Dictyostelium: tools and applications. Methods Cell Biol. 28, 67-100. [DOI] [PubMed] [Google Scholar]
- Novak, K. D., Peterson, M. D., Reedy, M. C., and Titus, M. A. (1995). Dictyostelium myosin I double mutants exhibit conditional defects in pinocytosis. J. Cell Biol. 131, 1205-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak, K. D., and Titus, M. A. (1998). The myosin I SH3 domain and TEDS rule phosphorylation site are required for in vivo function. Mol. Biol. Cell 9, 75-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak, K. D., and Titus, M. A. (1997). Myosin I overexpression impairs cell migration. J. Cell Biol. 136, 633-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri, S. J., Nebl, T., Pope, R. K., Seastone, D. J., Lee, E., Hinchcliffe, E. H., Sluder, G., Knecht, D., Cardelli, J., and Luna, E. J. (2000). Mutant Rac1B. expression in Dictyostelium: effects on morphology, growth, endocytosis, development, and the actin cytoskeleton. Cell Motil. Cytoskeleton 46, 285-304. [DOI] [PubMed] [Google Scholar]
- Pintard, L., Willems, A., and Peter, M. (2004). Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. EMBO J. 23, 1681-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos, S., Khademi, F., Somesh, B. P., and Rivero, F. (2002). Genomic organization and expression profile of the small GTPases of the RhoBTB family in human and mouse. Gene 298, 147-157. [DOI] [PubMed] [Google Scholar]
- Rivero, F., Dislich, H., Glockner, G., and Noegel, A. A. (2001). The Dictyostelium discoideum family of Rho-related proteins. Nucleic Acids Res. 29, 1068-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero, F., and Somesh, B. P. (2002). Signal transduction pathways regulated by Rho GTPases in Dictyostelium. J. Muscle Res. Cell Motil. 23, 737-749. [DOI] [PubMed] [Google Scholar]
- Rudel, T., and Bokoch, G. M. (1997). Membrane and morphological changes in apoptotic cells regulated by caspase-mediated activation of PAK2. Science 276, 1571-1574. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, Plainview, NY: Cold Spring Harbor Laboratory.
- Schwarz, E. C., Neuhaus, E. M., Kistler, C., Henkel, A. W., and Soldati, T. (2000). Dictyostelium myosin IK is involved in the maintenance of cortical tension and affects motility and phagocytosis. J. Cell Sci. 113, 621-633. [DOI] [PubMed] [Google Scholar]
- Senda, S., Lee, S. F., Côté, G. P., and Titus, M. A. (2001). Recruitment of a specific amoeboid myosin I isoform to the plasma membrane in chemotactic Dictyostelium cells. J. Biol. Chem. 276, 2898-2904. [DOI] [PubMed] [Google Scholar]
- Sussman, M. (1987). Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. Methods Cell Biol. 28, 9-29. [DOI] [PubMed] [Google Scholar]
- Tu, H., and Wigler, M. (1999). Genetic evidence for Pak1 autoinhibition and its release by Cdc42. Mol. Cell. Biol. 19, 602-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, I., Niewohner, J., Du, A., Rohrig, U., and Gerisch, G. (2002). A talin fragment as an actin trap visualizing actin flow in chemotaxis, endocytosis, and cytokinesis. Cell Motil. Cytoskeleton 53, 136-149. [DOI] [PubMed] [Google Scholar]
- Wu, C., Lee, S.-F., Furmaniak-Kazmierczak, E., Côté, G. P., Thomas, D. Y., and Leberer, E. (1996). Activation of myosin-I by members of the Ste20p protein kinase family. J. Biol. Chem. 271, 31787-31790. [DOI] [PubMed] [Google Scholar]
- Wu, C., Lytvyn, V., Thomas, D. Y., and Leberer, E. (1997). The phosphorylation site for Ste20p-like protein kinases is essential for the function of myosin-I in yeast. J. Biol. Chem. 30623-30626. [DOI] [PubMed]
- Yoshimura, M., Homma, K., Saito, J., Inoue, A., Ikebe, R., and Ikebe, M. (2001). Dual regulation of mammalian myosin VI motor function. J. Biol. Chem. 276, 39600-39607. [DOI] [PubMed] [Google Scholar]
- Zhao, Z. S., Manser, E., Chen, X. Q., Chong, C., Leung, T., and Lim, L. (1998). A conserved negative regulatory region in alphaPAK: inhibition of PAK kinases reveals their morphological roles downstream of Cdc42 and Rac1. Mol. Cell. Biol. 18, 2153-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.