Abstract
Flagellar length regulation provides a simple model system for addressing the general problem of organelle size control. Based on a systems-level analysis of flagellar dynamics, we have proposed a mechanism for flagellar length control in which length is set by the balance of continuous flagellar assembly and disassembly. The model proposes that the assembly rate is length dependent due to the inherent length dependence of intraflagellar transport, whereas disassembly is length independent, such that the two rates can only reach a balance point at a single length. In this report, we test this theoretical model by using three different measurements: 1) the quantity of intraflagellar transport machinery as a function of length, 2) the variation of flagellar length as a function of flagellar number, and 3) the rate of flagellar growth as a function of length. We find that the quantity of intraflagellar transport machinery is independent of length, that flagellar length is a decreasing function of flagellar number, and that flagellar growth rate in regenerating flagella depends on length and not on the time since regeneration began. These results are consistent with the balance-point model for length control. The three strategies used here are not limited to flagella and can in principle be adapted to probe size control systems for any organelle.
INTRODUCTION
A major unanswered question in cell biology is, How do cells regulate the size of organelles? Flagella provide an excellent model system for studying organelle size control, because flagellar length is so easy to measure. Flagellar length is important for proper motile function, and a number of human diseases seem to involve abnormal length flagella (Chemes et al., 1990; Toyama et al., 1996; Masyuk et al., 2003). Dynamic changes in flagellar length are also potentially important; for example, most cells remove their cilia or flagellar before cell division, by a combination of resorbtion and severing (Bloodgood, 1974; Parker and Quarmby, 2003; Quarmby, 2004). Most importantly, due to extensive prior knowledge on the mechanisms of flagellar assembly, this organelle provides the ideal test bed for developing new strategies for studying size control systems.
Genetic analysis of flagellar length control in Chlamydomonas reinhardtii, a unicellular green alga with genetics similar to budding yeast, has revealed a number of genes involved in flagellar length determination. These include the SHF genes (Jarvik et al., 1984; Kuchka and Jarvik, 1987), whose mutants cause abnormally short flagella, and the LF genes (McVittie, 1972; Barsel et al., 1988; Asleson et al., 1998), whose mutants result in aflagellate and flagellated cells with abnormally long flagella. The first flagellar length gene to be cloned was LF4, which encodes a mitogen-activated protein kinase (Berman et al., 2003). The substrates of LF4, and its functional role in length regulation, remain unknown. It seems likely that this genetic approach will yield important insights into the mechanism of flagellar length control.
As an alternative to the classical genetics approach, we have begun to take a systems-level approach to understanding flagellar length control, by asking what control elements would need to be added to the known biosynthetic machinery of flagella to enable a cell to construct a flagellum of defined length. This analysis was initially prompted by the observation, based on radioactive pulse-labeling experiments (Stephens, 1997; Song and Dentler, 2001) that the flagellum is dynamic and undergoes continuous turnover even after the flagellum has fully formed. Measurements of the incorporation of epitope-tagged tubulin into nongrowing flagella (Marshall and Rosenbaum, 2001) localized the site of the turnover to the distal end of the flagellar outer doublet microtubules. This showed that the length control system, whatever it is, must continuously act to balance the rates of assembly and disassembly to maintain a defined length. But how are the assembly and disassembly rates controlled to achieve this balance at the correct length?
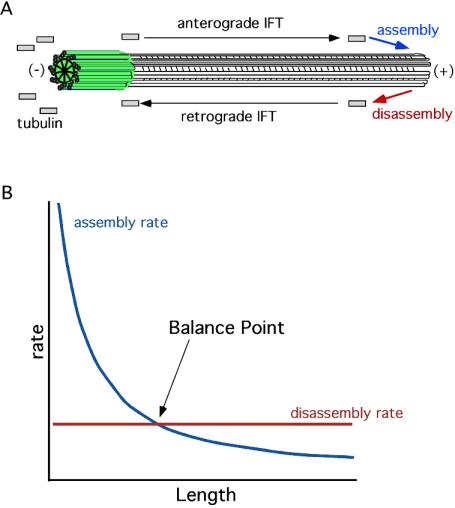
Steady-state assembly of tubulin onto the distal end of the flagellum requires intraflagellar transport (IFT), a kinesin-based motility that occurs within the flagellum and is required for flagellar assembly (Kozminski et al., 1993; Rosenbaum and Witman, 2002; Cole, 2003). The role of IFT in length maintenance (Figure 1A) is presumably to transport proteins to the flagellar tip (Qin et al., 2004), which is the site of assembly both during flagellar growth (Johnson and Rosenbaum, 1992) and at steady state (Marshall and Rosenbaum, 2001). IFT is mediated by complexes called IFT particles, and immunofluorescence analysis indicated that the number of these transport complexes is independent of length (Marshall and Rosenbaum, 2001). If the number of transport complexes is fixed, and the particles move at a fixed velocity, then the efficiency with which these IFT particles can transport flagellar precursors to the tip must decrease as length increases, hence the assembly rate will be length dependent. Thus, the rate of transport and assembly should decrease as the flagellar length increases (Marshall, 2002). In contrast to the inherent length dependence of assembly, measurements of flagellar resorbtion in the fla10-1 mutant (in which assembly is arrested at the nonpermissive temperature; Marshall and Rosenbaum, 2001) show that the shortening rate is constant during the course of resorbtion (Kozminski et al., 1995; Parker and Quarmby, 2003) and hence disassembly is length-independent. Because assembly decreases monotonically as a function of length, and disassembly is length independent, the assembly and disassembly rates will only balance at a single value of length, thus yielding a simple mechanism for flagellar length control (Figure 1B). The model does not require any assumptions about the site of flagellar disassembly, and whether it occurs at the tip as suggested by our pulse-labeling experiments (Marshall and Rosenbaum, 2001) or via a combination of disassembly at the tip and in the transition region (Parker and Quarmby, 2003), the length control model continues to work as long as the overall rate of disassembly is not length dependent. We have previously shown that this model can account for several phenomenological aspects of length control. For example, the ability of flagella to equalize their lengths when one flagellum is severed (Coyne and Rosenbaum, 1970) is completely explained by the balance-point model, as demonstrated by computer simulations (Marshall and Rosenbaum, 2001).
Figure 1.
Balance-point model for flagellar length control. (A) Intraflagellar transport and turnover. IFT particles move cargo such as tubulin from the cell body, where all protein is synthesized, out to the flagellar tip, where all assembly occurs. Simultaneously, axonemal proteins are continuously disassembled at the tip and returned to the cell body by retrograde IFT. Length maintenance requires that the rates of assembly and disassembly at the tip must be equal. + and - indicate microtubule plus and minus ends. Green indicates flagellar basal body located at cell surface. (B) Rates of assembly (blue curve) and disassembly (red curve) as a function of length. Disassembly is length independent, but transport-limited assembly is length dependent. The two curves only intersect at a single steady-state length.
However, alternatives to the balance-point model also have been proposed, including the idea that flagella contain a signal transduction pathway that acts as a feedback loop by changing its activation state in response to deviations in length (Asleson and Lefebvre, 1998; Berman et al., 2003) or the idea that calcium channels in the flagellum could regulate length via a length-dependent calcium current (Rosenbaum, 2003). Ultimately, to distinguish between these potential mechanisms, it is necessary to test each model by using experiments that have the capacity to potentially falsify the model. So far, such tests have not been reported for any of the flagellar length control models that have been proposed.
In this report, we test the balance-point model by using three different experimental strategies. These strategies were selected specifically for their ability to potentially invalidate the balance-point model that we have proposed. The results we observe are fully consistent with the balance-point model, suggesting that more elaborate models based on feedback control loops or ion-current length sensors may not be necessary to explain length regulation. Importantly, this report describes three strategies that can, in principle, be used to probe size control mechanisms in any organelle.
MATERIALS AND METHODS
Strains and Media
Chlamydomonas strains were obtained from the Chlamydomonas Genetics Center (Duke University, Durham, NC). The uni3-1 mutant strain was generously provided by Susan Dutcher (Washington University, St. Louis, MO). Construction of vfl2 ts10009 was discussed previously (Marshall et al., 2001). Gamete cultures were grown in M-N media (Harris, 1989) under continuous illumination, whereas vegetative cultures were grown in M1 (Sager and Granick Medium I) medium in a 14:10-h light/dark cycle and measured during the light part of the cycle to ensure a majority were in G1.
Analysis of IFT Protein Levels in Regenerating Flagella
Normally, flagellar assembly is induced by pH shock, which causes severing of the flagella and growth of new flagella. However, removal of the severed flagella would require time-consuming centrifugation steps, and we were concerned that residual severed flagella would continue to contaminate the cells, confusing subsequent biochemical analysis. As a simple way to induce flagellar assembly without having to worry about time-consuming separation steps or the presence of severed flagella in the media, we took advantage of the fact that Chlamydomonas cells lack flagella when grown on solid substrates. Cells grown on agar plates typically have either no flagella at all or else very short stumps, but they rapidly grow full-length flagella upon transfer to liquid medium (Figure 2A). Wild-type cells were grown on TAP plates at 21°C. Under these conditions, cells mostly lack flagella. Once each plate contained a dense lawn of cells, cells were scraped off of plates by using a rubber spatula into 10 mM HEPES at 4°C (cells were maintained at this temperature to prevent regeneration). Cells were stirred for 20 min at 4°C and then centrifuged at 4°C and resuspended in room temperature 10 mM HEPES. The time of the shift to room temperature medium was considered t = 0. At time points after the restoration to room temperature, an aliquot of cells from the culture was removed and deflagellated by pH shock. Flagella were harvested, and the final pellet of isolated flagella was resuspended in an equal volume of 1× HMDEK. Flagellar proteins were analyzed by running the isolated flagella on an 8% SDS-PAGE gel. Either equal numbers of flagella (based on the number of cells subjected to deflagellation) were loaded in each lane (Figure 2B), or equal quantities (5 μg) of total flagellar protein, as judged by Bradford assay, were loaded in each lane (Figure 2C).
Figure 2.
IFT protein content within the flagellum is independent of flagellar length. (A) Flagellar length plotted versus time in cells growing flagella after a shift from solid to liquid medium. (B) Western blot analysis of IFT proteins in flagella isolated from cells plotted in A in which an equal number of flagella were loaded per well. Radial spoke protein RspI content increases proportional to flagellar length during regeneration, whereas IFT protein content remains constant. (C) IFT proteins in flagella during regeneration in which an equal quantity of protein was loaded per well. (D) Number of IFT aggregates (rafts) plotted versus flagellar length. Each black circle represents data from a single cell. Variation of number versus length was insignificant over the length range examined. Inset, example immunofluorescence staining of IFT52. (E) Flagellar elongation rate calculated from flagellar regeneration data. Curve indicates a best fit to Eq. 3, indicating that elongation rate is proportional to 1/L as suggested by the balance-point model. (F) Reduction of IFT protein levels in fla10 temperature-sensitive mutant grown at 28°C, conditions which result in half-length flagella at steady-state, confirming a partial reduction in IFT protein content.
Flagellar Length Measurement
Cells were fixed by mixing an aliquot of culture with either an equal volume of 2% glutaraldehyde or an equal volume of 2× Lugol's iodine solution and imaged by phase contrast by using a 40× numerical aperture 0.75 lens with a 2× optivar and 10× ocular on a Zeiss Standard 16 microscope. For measurements on live cells, cells were embedded in M-N medium containing 0.7% agarose and imaged using the same optics as for the fixed cells. Length measurements were made visually using an ocular micrometer with 100 rulings corresponding to a spacing of 1.29 μm between rulings in the object plane. Spacing of the micrometer rulings was calibrated using an S12 stage micrometer with 2-μm rulings (Electron Microscopy Sciences). Calibration was confirmed using a different stage micrometer with 10 μm rulings (Bausch and Lomb). The calibration was further confirmed using distance measurements between gridlines on a photo-etched coverslip (Electron Microscopy Sciences, Hatfield, PA) and as well as a hemacytometer (Clay Adams, Parsippany, NJ). Error-bars for all length measurements report SE of the mean, a statistic chosen to allow the significance of differences in mean length to be assessed.
Length measurements in Figure 3 were made using three-dimensional (3D) differential interference contrast (DIC) imaging on a Deltavision Spectris four-dimensional (4D) microscopy system (Applied Precision, Issaquah, WA). Measurements of flagellar shortening in fla mutants were made by growing cells in TAP media under continuous illumination at 21°C and then fixing cells in 1% glutaraldehyde at time points after shifting to continuous illumination at 32°C in an illuminated incubator. Measurements of flagellar shortening before mitosis were done by synchronizing cells in M1 media in a 14:10-h light/dark cycle for 3 d and then shifting cells into TAP medium, mounting them in 0.1% agarose in TAP within a ring of petroleum jelly under a coverlsip, which was then sealed with VALAP (vaseline, lanolin, paraffin). Cultures were mounted and observation begun during the time the cultures would shift from light to dark. Observations were carried out using time-lapse 3D DIC imaging (i.e., 4D DIC), in which a 3D data set consisting of 24 sections spaced 0.5 μm apart were collected once every 10 min.
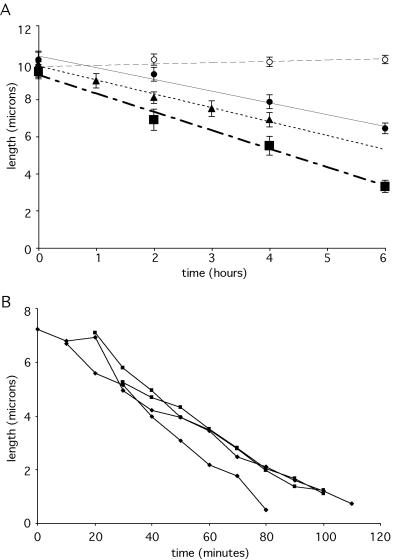
Figure 3.
Flagellar disassembly rate is length independent. (A) Flagellar length versus time after shift from 21°C (permissive temperature) to 32°C (nonpermissive temperature) in wild-type and IFT motor mutants. Wild type (○), fla1 mutant in motor subunit of heterotrimeric kinesin (•), fla10-1 mutant in motor subunit of heterotrimeric kinesin-II (▪), and fla3 mutant in nonmotor KAP subunit of heterotrimeric kinesin-II (▴). Data points show average length measured for 30 cells per time point for fla3, 12 per time point for fla1, and 15 cells per time point for fla10-1 and wild type. Best-fit lines are indicated as follows wild type (-----), fla1 mutant (―), fla3 mutant (······), and fla10-1 mutant (—·—·—). Error bars denote SEM. Only cells that had flagella were counted to avoid effects due to spontaneous deflagellation (Parker and Quarmby, 2003). In all three mutants, flagella shorten at a constant rate indicating disassembly in the absence of IFT is length independent. (B) Flagellar shortening before mitosis. Synchronized wild-type (cc-124) cells embedded in agarose were observed using 4D microscopy. Three-dimensional length measurement software was used to measure flagellar lengths at 10-min time points. Four individual flagella from different cells are shown in figure. In all cases, flagella shorten at a constant rate, indicating that predivision disassembly is length independent.
Regeneration of Partial Length Flagella
For the experiment of Figure 5C, wild-type cells were deflagellated in cycloheximide and allowed to regrow flagella as described previously (Marshall and Rosenbaum, 2001).
Figure 5.
Flagellar growth rate is length dependent. (A) Strategy for measuring flagellar elongation rate of partial-length flagella. Gametes with partial-length flagella are produced either by taking shf1 mutants that constitutively form short flagella, or by inducing flagellar regeneration in wild-type gametes in the absence of protein synthesis, under which conditions cells grow half-length flagella. Once cells with partial-length flagella are obtained, these are mated to wild-type cells of the opposite mating type having full-length flagella. Immediately after mating, the lengths of all four flagella are measured at 5- to 10-min time points. (B) Flagellar elongation in shf1 cells mated to wild type. Red circles, average length of short flagella after mating to wild-type cells (n = 56 flagella measured per time point). Green circles, average length of wild-type flagella regenerating in a control experiment (n = 30 flagella measured per time point). Error bars indicate SE of the mean length. (C) Flagellar elongation after mating of wild-type cells with partial-length flagella regenerated in cycloheximide (red circles; n = 10 flagella measured per time point) to wild-type cells with full-length flagella, compared with regeneration of flagella in completely deflagellated cells (green circles; n = 20 flagella measured per time point). Control regeneration kinetics and extent are different than in B because the mating experiment as well as the wild-type control were both done in the presence of cycloheximide.
Mathematical Analysis
According to the simplest implementation of this model (Marshall and Rosenbaum 2001) the rate of change of flagellar length is given by
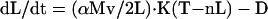 |
1 |
where D is the length-independent disassembly rate, M is the number of IFT particles moving within a given flagellum, v is the harmonic mean of the anterograde and retrograde velocities of IFT particle movement, K is a constant reflecting the equilibrium binding affinity of IFT particles for cargo proteins from the cytoplasmic pool, n is the number of flagella, and α is a proportionality constant that reflects the efficiency of incorporation of transported cargo proteins onto the flagellar tip. In this equation, we represent the free cytoplasmic pool by T-nL where n is the number of flagella and T is the total cellular pool of flagellar protein, expressed in terms of the length of axoneme that could be assembled if the entire pool was used. It is assumed in this model that the total pool of flagellar precursor protein is a constant. Variation of T from cell to cell could thus result in variations in steady-state length, but in the simplified analysis presented here we ignore such variation, which would only affect the outcome by increasing the variance in steady-state lengths in the population. The harmonic mean velocity v is the reciprocal of the mean of the reciprocals of the anterograde and retrograde velocities, and arises because the elongation rate is proportional to the reciprocal of the round-trip time for a particle, which is the sum of the times required to move out to the tip and the time required to move back, each of which is in turn proportional to the reciprocal of the respective velocities. We can simplify this equation by combining constants into a single constant A = αMvK/2. We then can find the steady-state length of a flagellum by setting dL/dt = 0 and solving for L, yielding Eq. 4. Curve fitting shown in Figure 4 was performed by recognizing that Eq. 4 implies n is proportional to 1/L, allowing a straight line to be fitted to n versus 1/L.
Figure 4.
Variation of flagellar length with flagellar number. Lengths were measured in vfl1 gametes having different numbers of flagella (see data from Table 2). Average length indicated by circles: cells fixed in glutaraldehyde (○); cells fixed with Lugol's iodine (•). Error bars indicate SEM. Lines represent best-fit curves from Eq. 4. Fitting parameters for glutaraldehyde fixed cells: T = 76.7, D/A = 5.7, rms fitting error = 0.36 μm. Fitting parameters for iodine fixed cells: T = 93.9, D/A = 8.5, rms fitting error 0.22 μm.
To derive Eq. 3, we start with Eq. 1 and carry through the multiplications in the first term. Rearranging, we obtain
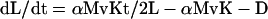 |
2 |
if we then let P = αMvKT/2 and Q = αMvK + D, we obtain Eq. 3.
RESULTS
IFT Protein Quantity versus Flagellar Length
One method for probing size control systems is to explore the relationship between organelle size and the quantity of biosynthetic machinery acting on the organelle. The balance-point model for flagellar length control proposes that as the flagellum elongates, an initially fixed quantity of IFT complexes are forced to travel longer and longer distances to deliver axonemal precursors to the tip, thus making transport and steady-state assembly inherently length dependent.
This model was initially prompted by immunofluorescence intensity measurements showing that the total integrated intensity of immunostaining with anti-IFT protein antibodies in the entire flagellum is independent of length (Marshall and Rosenbaum, 2001). However, such immunofluorescence-based measurements of IFT protein suffer from potential difficulties of quantifying protein levels based on immunofluorescence. We therefore measured the IFT protein content biochemically by isolating flagella during a regeneration time course (Figure 2A) and detecting IFT protein levels by immunoblotting. When equal numbers of isolated flagella were loaded per time point (Figure 2B), it was observed that the quantity of IFT proteins stayed constant during flagellar growth, as judged by Western blotting with antibodies specific for IFT139 and IFT81. In comparison with the IFT proteins, whose amount per flagellum does not change with flagellar length, the quantity of an axonemal protein, RspI, increases proportionally as the length increases, as expected for an axonemal component.
When equal quantity of total protein was loaded per time point in a separate experiment (Figure 2C), it was observed that the quantity of IFT protein decreases as the flagella elongate, in contrast to axonemal components RspI and dynein IC IC69, which stay at a constant level. This is the result that is expected if the quantity of IFT protein remains fixed as flagella elongate, because the quantity of axonemal proteins will continuously increase, whereas the quantity of IFT protein stays constant, so that IFT proteins constitute a continuously decreasing fraction of the total protein. These results confirm our previous immunofluorescence results that the total quantity of IFT protein per flagellum is independent of flagellar length. We do not presently have any information about how the quantity of IFT protein is kept constant.
IFT proteins within the flagellum self-associate into complexes of particles arranged in a row like cars in a freight train. Because this higher level organization could in principle affect transport capacity of the IFT system, we examined the distribution of IFT proteins into multiparticle complexes as a function of length. Cells undergoing flagellar regeneration were fixed and stained to detect the IFT protein IFT52 (Figure 2D, inset), and the number of visible IFT particle aggregates was counted in each image and plotted versus flagellar length (Figure 2D). For flagella that were 6 μm or less, it was impossible to count the number of complexes because they began to overlap and the resulting bright fluorescence made resolution of individual particles unreliable. Under normal steady-state conditions, flagella are always longer than 6 μm, so our analysis covers the length range that is relevant for length control. As seen in Figure 2D, over this entire range the average number of IFT particle complexes remained roughly constant. Numerical line-fitting indicated that the slope of the number versus length curve was just 0.063 aggregates/μm, with an intercept of 7.9 aggregates, indicating that from 6 to 12 μm of length, the number of IFT particle aggregates changes only by ∼0.3. We therefore conclude that not only IFT protein levels but also IFT protein organization into multiparticle complexes is effectively constant as a function of length.
Given that the total quantity of IFT protein per flagellum is in fact independent of length, the balance-point model can be used to predict the kinetics of flagellar regeneration (Marshall and Rosenbaum, 2001). This prediction is based on the idea that when flagella are severed, they begin to regenerate because the flagellar stubs are shorter than the balance-point length; hence, assembly predominates over disassembly, leading to outgrowth. Such considerations allow the balance-point model to predict the rate of flagellar growth as a function of length. As derived in Materials and Methods, it can be shown that the flagellar growth rate r given by the model is described by an equation of the form
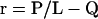 |
3 |
where L is the length and P and Q are constants. This indicates that the growth rate is a hyperbolic function of the length. As plotted in Figure 2E, actual regeneration data can be fit by just such a curve, suggesting that the balance-point model can account for the variation of growth rate as a function of length.
The derivation of the balance-point model assumes that transport of precursor proteins to the tip of the flagellum is rate-limiting when flagella approach full length. We have previously reported that a temperature-sensitive mutant of the IFT kinesin FLA10, when grown at temperatures between fully permissive (22°C) and fully nonpermissive (32°C), results in a decreased steady-state flagellar length (Marshall and Rosenbaum, 2001). For example, fla10-1 cells grown at 28°C had half-length flagella. We interpreted this result as reflecting a decreased transport capacity due to a partial reduction in the quantity of IFT particles in the flagellum, but this had not been directly verified. To confirm a reduction in IFT in fla10-1 mutants grown at 28°C, we examined IFT protein levels in isolated flagella under these conditions. It has been reported previously (Cole et al., 1998) that IFT proteins are rapidly lost from fla10-1 flagella grown at the nonpermissive temperature of 32°C. As seen in Figure 2F, IFT protein levels are reduced, but not eliminated, in fla10-1 cells grown at 28°C compared with the same cells grown at the permissive temperature of 22°C. The fact that a temperature-sensitive (ts) mutation grown under conditions that partially reduce IFT protein levels leads to a decrease in steady-state length is consistent with IFT being the rate-limiting step in steady-state assembly.
Flagellar Disassembly Is Length Independent
An important element of the balance-point length control mechanism is that the disassembly component of turnover should be length independent. In fact, the disassembly rate need not be strictly length independent, provided its length dependence is such that the disassembly rate versus length curve only intersects the assembly rate versus length curve at a single value of length. For instance, if the disassembly rate is an increasing function of length, it is obvious by inspection of Figure 1 that the system will still result in a stable, uniquely defined length. However, if the disassembly rate showed the same length dependence as the assembly rate, that is, if the disassembly rate were to vary as 1/L, this would not be guaranteed to result in a stable length control system. We therefore examined the dependence of disassembly rate on length. Previous measurements by using a particular allele of the FLA10 kinesin gene (fla10-1) indicated that when IFT was shut off by shifting this mutation to the nonpermissive temperature, flagella resorbed at a constant rate, indicating disassembly in the absence of IFT was length independent. As plotted in Figure 3A, we repeated this measurement for fla10-1 and obtained the same result: flagella shortened at a constant rate. To test whether this effect was unique to the particular FLA10 mutant used, we also examined shortening in conditional mutants of two other components of the heterotrimeric kinesin-II. fla3 mutants are defective in the nonmotor, KAP subunit of kinesin-II (Mueller et al., 2002), and we found that just as in fla10-1, a fla3 mutant showed length-independent flagellar shortening. The fla1 mutant is defective in the other motor subunit of kinesin-II (Miller et al., 2004) and this mutant also showed flagellar shortening at a constant rate [note that Miller et al. (2004) have shown that the fla1 and fla8 genes are allelic and have proposed that fla1 be designated fla8-2]. In all three mutants, the residual fitting error was within the SE of the mean for each data point, indicating that to within the error of measurement the shortening data matched a straight line. We therefore conclude that, when IFT is inactivated using mutations in three different components of the anterograde IFT motor kinesin-II, in all cases flagella shorten at a constant rate, confirming that disassembly in the absence of IFT is length independent and that this length independence is not a peculiar feature of the fla10-1 allele. We also note that another fla mutant, fla2, which is known to affect the velocity of IFT particle movement (Iomini et al., 2001), has recently been shown to lead to flagellar shortening at a constant rate after a shift to the nonpermissive temperature (Parker and Quarmby, 2003), suggesting this is a general feature of flagellar shortening processes that involve inhibition of IFT.
As an alternative approach to measuring flagellar disassembly kinetics, we examined flagellar resorbtion before mitosis. Although the mechanism of predivisional resorbtion is not yet understood, it at least provides a natural situation in which to observe flagellar shortening without using mutants. By observing synchronized cell populations embedded in agarose pads under a coverslip, we measured flagellar length versus time in cells immediately preceding division (Figure 3B). We found that flagella shortened at a constant rate, again suggesting that predivisional flagellar disassembly is length independent. This same constant-rate shortening of flagella before mitosis has been clearly demonstrated in the flagellate Paranema (Tamm, 1967), implying it may be a general feature of predivision flagellar resorbtion. We note that the rate of shortening before division (Figure 3B) is much higher than the rate of shortening following inhibition of IFT (Figure 3A), suggesting that the disassembly either occurs by a distinct mechanism (perhaps involving disassembly at the transition zone; Parker and Quarmby 2003) or else involves an up-regulation of the normal disassembly machinery.
Flagellar Length versus Flagellar Number
A general method to probe size control mechanisms is to ask how the size of an organelle changes as a function of copy number. For flagellar length control, the balance-point model implies that the tip of the flagellum is effectively in equilibrium with a pool of unassembled flagellar precursor protein in the cytoplasm, such that an increase or decrease in cytoplasmic protein concentration will result in increase or decrease in association of protein to the flagellar tip, producing a change in length. It has long been known that flagella regenerated in cells whose precursor pool is depleted grow to a shorter than normal length (Coyne and Rosenbaum, 1970), suggesting that length does indeed depend on the quantity of cytoplasmic precursor protein. If the cell produces a fixed quantity of precursor protein, and each flagellum consumes a certain quantity of precursor protein during its assembly, then the more flagella there are, the more precursor protein they will collectively consume per unit length, and the smaller the remaining free precursor pool will be in the cytoplasm. If the flagellar tips are in equilibrium with the cytoplasmic precursor pool as predicted by the balance-point model, then flagellar length should decrease with increasing flagellar number.
To measure variation of length with number, we exploited the vfl mutations of Chlamydomonas, in which a basal body segregation defect results in cells with variable numbers of flagella (Kuchka and Jarvik, 1982; Wright et al., 1983; Adams et al., 1985). We use three different vfl mutants, vfl1, vfl2, and vfl3, which are defective for different gene products, to be sure that results are dependent simply on the number of flagella and not on the particular vfl mutant used. The VFL1 gene encodes a novel protein that localizes to the interior of the basal body cylinder (Silflow et al., 2001). The VFL2 gene encodes the EF-hand protein centrin that localizes to several fibrous structures associated with basal bodies (Taillon et al., 1992). The VFL3 gene sequence has not yet been published. In mutants in all three genes, most cells have zero, one, two, or three flagella, and a small fraction of cells have more flagella, in some cases as many as six or more. In contrast, wild-type cells always have two flagella per cell during G1.
As recorded in Table 1 when flagellar lengths were measured as a function of flagellar number in vfl1, vfl2, and vfl3 mutant cells, in all three cases it was found that flagellar length decreases monotonically with increasing flagellar number, as predicted by the balance-point length control model. Because flagellar length changes during progression through G1, we restricted measurements to cell cycle-arrested vfl cells, either by examining gametes, cells grown in the dark in minimal medium, or for vfl2, in a double mutant with the cell cycle arrest mutation ts10009, which arrests in G1 at the restrictive temperature (Howell and Naliboff, 1973). As shown in Table 1, comparable results were obtained in all three mutants and under all fixation conditions (we compared Lugol's iodine and glutaraldehyde fixation because these two standard fixation methods sometimes result in different average flagellar lengths, possibly as an artifact of fixation rates, but in our hands choice of fixative does not affect the ultimate conclusion that length varies with flagellar number).
Table 1.
Flagellar length versus flagellar number in vfl mutants
| Gametes | n = 1 | n = 2 | n = 3 | n = 4 or more | |
|---|---|---|---|---|---|
| vfl1 | Iod | 10.0 ± 0.2 (44) | 8.6 ± 0.2 (47) | 8.3 ± 0.2 (47) | 7.3 ± 0.1 (104) |
| Glut | 11.1 ± 0.3 (40) | 9.9 ± 0.4 (40) | 8.7 ± 0.4 (40) | 8.1 ± 0.3 (50) | |
| Live | 12.2 ± 0.3 (30) | 11.3 ± 0.3 (30) | 9.9 ± 0.4 (30) | 8.0 ± 0.6 (9) | |
| vfl2 | Iod | 9.4 ± 0.3 (32) | 7.9 ± 0.2 (53) | 6.2 ± 0.6 (9) | 6.4 ± 0.0 (1) |
| vfl3 | Iod | 11.6 ± 0.2 (74) | 10.2 ± 0.2 (66) | 9.0 ± 0.2 (65) | 7.7 ± 0.3 (37) |
| uni3 | Iod | 10.0 ± 0.3 (66) | 9.1 ± 0.3 (71) | n.a. | n.a. |
| G1 arrest by growth in dark | |||||
| vfl1 | Glut | 11.1 ± 0.3 (40) | 8.9 ± 0.4 (40) | 8.3 ± 0.4 (40) | 6.9 ± 0.4 (29) |
| vfl2 | Glut | 9.2 ± 0.3 (60) | 8.8 ± 0.3 (60) | 7.5 ± 0.3 (49) | 6.4 ± 0.4 (20) |
| vfl3 | Glut | 9.9 ± 0.3 (40) | 8.7 ± 0.3 (40) | 7.6 ± 0.3 (40) | 6.0 ± 0.4 (11) |
| G1 arrested by ts10009 mutation | |||||
| vfl2 | Glut | 9.1 ± 0.3 (60) | 8.1 ± 0.3 (60) | 6.1 ± 0.4 (31) | 4.9 ± 0.5 (11) |
n.a., not applicable.
Measurements are grouped according to the number of flagella on the cell. For each type of cell, the average length of the flagella (one measured per cell) is given along with the SE of the mean and the total number of cell measured in parentheses. Iod, Glut, and live indicate fixation by Lugol's iodine, by 1% glutaraldehyde, and observation of unfixed live cells, respectively.
The results show that vfl mutant cells with more flagella have shorter flagella. However, vfl cells with increased numbers of flagella also have increased numbers of basal bodies, and basal bodies are known to recruit proteins involved in flagellar assembly (Deane et al., 2001). One could imagine that when additional basal bodies are present, they compete for a limited quantity of protein and become less effective at nucleating flagellar assembly. We therefore asked, Is the decrease in flagellar length due to increased numbers of flagella, as predicted by the balance-point model, or is it somehow due to increased numbers of basal bodies? To distinguish these two possibilities, we take advantage of the uni3 mutation, in which all cells have two basal bodies, but in which the basal bodies show variable function such that some basal bodies cannot template the assembly of a flagellum (Dutcher and Trabuco, 1998). As a result, some cells have only a single flagellum even though they have two basal bodies, whereas other cells are biflagellate. If flagellar length depended on number of basal bodies and not number of flagella, then in uni3 mutants flagellar length should not vary with flagellar number. However, as seen in Table 1, flagella are significantly shorter in uni3 cells having two flagella than in uni3 cells having one. This suggests that the variation in flagellar length is not due to variation in number of basal bodies (which are the same in all uni3 cells) but rather due to the variation in number of flagella.
The balance-point model leads to a quantitative prediction of the variation of length with number, which allows the model to be tested numerically. As derived in Materials and Methods, the steady-state flagellar length is expected to vary with flagellar number according to the relation
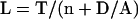 |
4 |
in which L is the steady-state flagellar length; D is the rate of the disassembly component of flagellar turnover; T is proportional to the total flagellar precursor pool in the cell, including that already assembled in flagella; and A is a constant representing the carrying capacity of IFT and the binding efficiency of axonemal subunits at the tip. The shape of the L versus N curve is fixed completely by just two parameters, the constant T and the ratio D/A. To test the ability of this model to fit the variation of length with number, lengths were measured in vfl1 cells with numbers of flagella ranging from 1 to 6 (Table 2). As shown in Figure 4, even over this large range in flagellar numbers, a curve derived from the balance-point model fits the data with a root mean square fitting error comparable with the SE of the data. This suggests that the balance-point model provides an adequate explanation sufficient to account for the variation of length with number in vfl mutants.
Table 2.
Length versus number from one to six flagella per cell in vfl1 gametes
| Glutaraldehyde | Lugol's iodine | |
|---|---|---|
| 1 | 11.1 ± 0.3 (40) | 10.0 ± 0.2 (44) |
| 2 | 9.9 ± 0.4 (40) | 8.6 ± 0.2 (47) |
| 3 | 8.7 ± 0.4 (40) | 8.3 ± 0.2 (47) |
| 4 | 8.6 ± 0.4 (40) | 7.4 ± 0.2 (46) |
| 5 | 7.0 ± 0.6 (7) | 7.3 ± 0.2 (46) |
| 6 | 4.3 ± 0.4 (3) | 6.4 ± 0.4 (12) |
Measurements list average length and SE, with number of flagella measured given in parentheses.
Growth Rate versus Length
A third general approach to probing size control mechanisms is to analyze the kinetics of organelle assembly, to determine the relationship between growth rate and size. When flagella regenerate, they do so with decreasing kinetics, such that the initial growth rate is very rapid and then gradually slows down, eventually reaching zero as the length approaches its final value. The balance-point model for length control predicts these kinetics (Marshall and Rosenbaum, 2001) because the rate of transport, and hence the rate of assembly, is predicted to be inherently length dependent. On the other hand, it is known that flagellar regeneration involves the execution of a complex program of gene expression, and it could be argued that the decreasing kinetics of regeneration simply reflect temporal changes in gene expression levels or translational rates after the initiation of the regeneration program. If in fact the flagellar growth rate variation seen during regeneration could be shown to depend only on time and not on length, then this would argue strongly against the balance-point model. A microsurgical study in Paranema (Tamm, 1967) showed a strong correlation between length and the rate at which a cut flagellum regenerated, with shorter flagella showing a much faster initial rate of growth than longer flagella. In contrast, a genetic experiment using rescue of short-flagella mutants in Chlamydomonas (Jarvik et al., 1984) indicated that partial length flagella grow at an initial rate equal to the initial growth rate of normal flagellar regeneration, implying that the growth rate is the same regardless of length and that the variation in growth rate during a regeneration time course reflects a dependence on time after regeneration initiation rather than a dependence on length. Do these differences reflect an inherent difference in length control mechanisms between Paranema and Chlamydomonas?
To resolve this controversy, we replicated the strategy developed by Jarvik et al. (1984). This strategy relies on short-flagella mutants, in which cells have abnormally short flagella, roughly half the length of flagella in wild-type cells. To probe growth rate versus length, shf1 mutants were mated with wild-type cells of the opposite mating type (Figure 5A). The resulting quadriflagellate dikaryon has two short flagella and two flagella of normal length. Because the shf1 mutation is recessive and can be rescued in the dikaryon by the wild-type gene product, these short flagella immediately begin to grow. If growth rate is a simple function of time after the initiation of regeneration, then the initial growth rate of these shf mutant flagella should be comparable with the initial growth rate seen when flagella are completely severed in standard regeneration experiments (Lefebvre et al., 1978). If, on the other hand, growth rate depends only on flagellar length, then the initial growth rate of the short flagella would not match the initial growth rate of regenerating flagella, but would instead match the growth rate that regenerating flagella attain when they reach the same length as the shf flagella. Jarvik et al. (1984) reported the former outcome, implying that growth rate is a function of time and not of length. However, when we repeated the experiment, we obtained the opposite result. As plotted in Figure 5B, the initial growth rate for the shf flagella was found to be 0.18 μm/min (measured over the first 10 min of growth), roughly half the initial growth rate of 0.35 μm/min seen in zero-length wild-type flagella, but almost identical to the growth rate that wild-type flagella reach when they have grown to a length similar to the initial length in shf1 (0.16 μm/min). This result suggests that flagellar growth rate depends strictly on flagellar length, such that a flagellum reaching a given length grows at a fixed rate regardless of whether it reached that length after regenerating from zero length or because of a constitutive short-flagella mutation.
We developed an alternative method for measuring growth rate in short flagella that does not rely on the use of shf mutations. As depicted in Figure 5A, wild-type cells were deflagellated and allowed to regenerate flagella in the presence of cycloheximide to block protein synthesis. In this case, flagellar assembly relies on the preexisting cytoplasmic precursor pool, which is sufficient only to regenerate flagella of roughly half length (Rosenbaum et al., 1969). These cells were then mated with wild-type cells having intact flagella, again producing a dikaryon with two full-length and two half-length flagella. As with the shf1 mating experiment, the additional precursor pool donated by the untreated cell allows the initially short flagella to grow. The kinetics of this outgrowth is plotted in Figure 5C, along with the kinetics of normal flagella regenerating from zero length after deflagellation in cycloheximide (to ensure comparable conditions). Analysis of growth kinetics in this experiment indicated that the initially half length flagella regenerated at an initial rate of 0.08 μm/min, whereas the zero-length control flagella regenerated at an initial rate of 0.64 μm/min. The initially zero-length flagella grew at a rate of 0.06 μm/min when they had reached a length comparable with the half-length flagella. Kinetics of regeneration from zero length in cycloheximide matched those reported previously (Lefebvre et al., 1978). Thus, we obtain the same result as with the shf1 mating experiment: flagella starting out at half length grow at a rate significantly slower than flagella starting out at zero length, but equivalent to the rate at which initially zero-length flagella elongate once they reach half length. We conclude that flagellar growth rate at any point during regeneration seems to depend on how long the flagellum is, rather than on how much time has elapsed since regeneration was triggered. This confirms the prediction of the balance-point model.
DISCUSSION
Comparison with Previous Studies on Flagellar Length Control
Two of the results reported in this article contradict previously published experiments. The variation of flagellar length with number (Figure 4) was previously tested by Kuchka and Jarvik (1982), who concluded no significant dependence of length on number in vfl2 cells. Our results presented here seem to conflict with those reported by Kuchka and Jarvik (1982). However, although Kuchka and Jarvik (1982) found that cells with one or two flagella had similar lengths, their own published data actually show that cells with three flagella have significantly shorter flagella than those with one or two. Moreover, Kuchka and Jarvik (1982) used vegetatively dividing cells that we have found to demonstrate a less dramatic dependence on length compared with gametes or G1-arrested cells, possibly due to increased length variability resulting from loss of cell cycle synchrony (our unpublished data). In any case, we see an unmistakable decrease of length with increasing number, in three different vfl mutants grown under different conditions, which is consistent with the balance-point model.
A second point of disagreement with previous work concerns the prediction that flagellar growth rate should be a strict function of length. Such a dependence was observed in Paranema (Tamm, 1967), whereas a lack of such dependence was reported in Chlamydomonas (Jarvik et al., 1984). When we repeated the experiment described by Jarvik by using the identical procedure (Figure 5), we obtained the opposite result. However, the kinetics that we observed in the shf1 mating experiment was virtually the same as that reported by Jarvik, who reported an initial growth rate of 0.15 μm/min for the shf1 flagella after mating, compared with our initial observed growth rate of 0.18 μm/min. In fact, the discrepancy between our results and those of Jarvik is due to a difference in the control experiments, in which flagella were detached by pH shock and regenerated from zero length. The rate of control regeneration measured by Jarvik and coworkers, and used for their comparison with shf1 flagellar outgrowth, was significantly slower than the regeneration rate typically observed (Lefebvre et al., 1978). We do not know why the wild-type flagella regenerated so slowly in the experiments of Jarvik et al., but in any case this seems to account for the apparent disagreement in our results.
Our results that IFT protein content is length independent, that flagellar length decreases with increased flagellar copy number, and that growth rate during regeneration depends on instantaneous flagellar length are all consistent with the balance-point model for length control. Whether these results are also consistent with other models that have been proposed is less clear, partly because these models have not yet been presented in a sufficiently precise form to allow definite predictions. We certainly would not propose to rule out any other length control mechanisms at this point. Indeed, there is no reason to assume that a single length control system is operating in a given cell type. For example, one could easily superimpose a feedback control loop onto the simple balance-point control system we propose. This would follow the well-established precedent in engineering control systems, where control systems of different types are frequently combined to yield improved performance over a wider range of regimes, for instance, in the well-known proportional-integral derivative control system architecture, which combines three types of control systems. In the balance-point model that we present, no assumptions are made about the mechanism by which the IFT protein content per flagellum is maintained at a constant, length-independent level. It is formally possible that a length sensor, such as one mediated by calcium channels or the LF4 MAP kinase cascade, might in fact be needed to regulate IFT protein content and thus act as a subsystem within the balance-point length control system. Thus, the balance-point system provided by the length dependence of IFT and the length independence of turnover might just be one loop in a much more complex control network. One approach to testing whether a given length control gene, for example, LF4, acts through the balance-point system or some other, parallel length-regulating feedback loop, will be to determine whether a given LF or SHF mutant alters any of the individual parameters of the balance-point model (for example, turnover rate) in a way consistent with the resulting length phenotype.
General Methods to Probe Size Control in Biological Systems
In the results stated above, we tested the flagellar length control system of Chlamydomonas by using three measurements: 1) the variation in quantity of organelle biosynthetic machinery as a function of size, 2) the dependence of size on copy number, and 3) the dependence of growth rate on size. These strategies are completely general in that, at least conceptually, they can be applied to any organelle in any organism. To implement these methods in practice may, however, require more detailed information and genetic tools. Analysis of biosynthetic capacity versus size requires prior knowledge of proteins required for organelle assembly and disassembly, so that their quantities can be measured. The second method, measuring size versus number, requires a way to vary the organelle number. Probably the easiest way to do this is using mutants that perturb organelle inheritance, as was done in this report, but it also could be accomplished by micromanipulation. Similarly, the third method, measuring growth rate versus size, requires a way to set an organelle to a defined size independently of its normal assembly program. Again, the practical application of this method requires appropriate mutants or microsurgical methods to produce a smaller than normal structure.
Fortunately, many organelles are becoming well enough understood that these simple methods could readily be applied. Thus, as we gradually refine our understanding of how microtubule turnover and intraflagellar transport cooperate in achieving a defined flagellar length, the path is clear to begin a similar analysis of size control systems in other organelles.
Acknowledgments
We thank Susan Dutcher and Doug Cole for strains, antibodies, and helpful discussions. We thank Bill Snell, Dennis Diener, Lotte Pedersen, Kim Wemmer, and Jessica Feldman for critical reading of the manuscript. This work was supported by a Leukemia and Lymphoma Society Special Fellowship (to W.F.M.), a Polycystic Kidney Disease Foundation fellowship (to H.Q.), National Science Foundation grant 0416210 (to W.F.M.), and National Institutes of Health grant GM-14642 (to J.L.R.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-07-0586. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-07-0586.
References
- Adams, G. M., Wright, R. L., and Jarvik, J. W. (1985). Defective temporal and spatial control of flagellar assembly in a mutant of Chlamydomonas reinhardtii with variable flagellar number. J. Cell Biol. 100, 955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asleson, C. M., and Lefebvre, P. A. (1998). Genetic analysis of flagellar length control in Chlamydomonas reinhardtii: a new long-flagella locus and extragenic suppressor mutations. Genetics 148, 693-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsel, S. E., Wexler, D. E., and Lefebvre, P. A. (1988). Genetic analysis of long-flagella mutants of Chlamydomonas reinhardtii. Genetics 118, 637-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman, S. A., Wilson, N. F., Haas, N. A., and Lefebvre, P. A. (2003). A novel MAP kinase regulates flagellar length in Chlamydomonas. Curr. Biol. 13, 1145-1149. [DOI] [PubMed] [Google Scholar]
- Bloodgood, R. A. (1974). Resorption of organelles containing microtubules. Cytobios 9, 143-161. [PubMed] [Google Scholar]
- Chemes, H. E., Morero, J. L., and Lavieri, J. C. (1990). Extreme asthenozoospermia and chronic respiratory disease: a new variant of the immotile cilia syndrome. Int. J. Androl. 13, 216-222. [DOI] [PubMed] [Google Scholar]
- Cole, D. G., Diener, D. R., Himelblau, A. L., Beech, P. L., Fuster, J. C., and Rosenbaum, J. L. (1998). Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory cilia. J. Cell Biol. 141, 993-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, D. G. (2003). The intraflagellar transport machinery of Chlamydomonas reinhardtii. Traffic 4, 435-442. [DOI] [PubMed] [Google Scholar]
- Coyne, B., and Rosenbaum, J. L. (1970). Flagellar elongation and shortening in Chlamydomonas. II. Re-utilization of flagellar proteins. J. Cell Biol. 47, 777-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane, J. A., Cole, D. G., Seeley, E. S., Diener, D. R., and Rosenbaum, J. L. (2001). Localization of intraflagellar transport protein IFT52 identifies basal body transitional fibers as the docking site for IFT particles. Curr. Biol. 11, 1586-1590. [DOI] [PubMed] [Google Scholar]
- Dutcher, S. K., and Trabuco, E. C. (1998). The UNI3 gene is required for assembly of basal bodies of Chlamydomonas and encodes delta-tubulin, a new member of the tubulin superfamily. Mol. Biol. Cell 9, 1293-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, E. H. (1989). The Chlamydomonas Sourcebook. San Diego: Academic Press.
- Howell, S. H., and Naliboff, J. A. (1973). Conditional mutants in Chlamydomonas reinhardtii blocked in the vegetative cell cycle. J. Cell Biol. 57, 760-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iomini, C., Babaev-Khaimov, V., Sassaroli, M., and Piperno, G. (2001). Protein particles in Chlamydomonas flagella undergo a transport cycle consisting of four phases. J. Cell Biol. 153, 13-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvik, J. W., Reinhardt, F. D., Kuchka, M. R., and Adler, S. A. (1984). Altered flagellar size control in shf-1 short flagellar mutants of Chlamydomonas reinhardtii. J. Protozool. 31, 100-104. [Google Scholar]
- Johnson, K. A., and Rosenbaum, J. L. (1992). Polarity of flagellar assembly in Chlamydomonas. J. Cell Biol. 119, 1605-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski, K. G., Beech, P. L., and Rosenbaum, J. L. (1995). The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J. Cell Biol. 131, 1517-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski, K. G., Johnson, K. A., Forscher, P., and Rosenbaum, J. L. (1993). A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. USA 90, 5519-5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchka, M. R., and Jarvik, J. W. (1987). Short-flagella mutants of Chlamydomonas. Genetics 115, 685-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchka, M. R., and Jarvik, J. W. (1982). Analysis of flagellar size control using a mutant of Chlamydomonas reinhardtii with a variable number of flagella. J. Cell Biol. 92, 170-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre, P. A., Nordstrom, S. A., Moulder, J. E., and Rosenbaum, J. L. (1978). Flagellar elongation and shortening in Chlamydomonas. IV. Effects of flagellar detachment, regeneration, and resorption on the induction of flagellar protein synthesis. J. Cell Biol. 78, 8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, W. F., and Rosenbaum, J. L. (2001). Intraflagellar transport balances continuous turnover of outer doublet microtubules: implications for flagellar length control. J. Cell Biol. 155, 405-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, W. F., Vucica, Y., and Rosenbaum, J. L. (2001). Kinetics and regulation of de novo centriole assembly: implications for the mechanism of centriole duplication. Curr. Biol. 111, 308-317. [DOI] [PubMed] [Google Scholar]
- Marshall, W. F. (2002). Size control in dynamic organelles. Trends Cell Biol. 12, 414-419. [DOI] [PubMed] [Google Scholar]
- Masyuk, T. V., et al. (2003). Defects in cholangiocyte fibrocystin expression and ciliary structure in the PCK rat. Gastroenterology 125, 1303-1310. [DOI] [PubMed] [Google Scholar]
- McVittie, A. C. (1972). Flagellum mutants of Chlamydomonas reinhardtii. J. Gen. Microbiol. 71, 525-540. [DOI] [PubMed] [Google Scholar]
- Miller, M. S., Lippa, A., Dutcher, S. K., and Cole, D. G. (2004). The Chlamydomonas FLA8 gene encodes a kinesin-II motor subunit that is associated with FLA10. Mol. Biol. Cell 15, meeting abstract B153.
- Mueller, J. J., Perrone, C. A., and Porter, M. E. (2002). Identification of a mutation in the KAP subunit of kinesin II that disrupts flagellar assembly in Chlamydomonas. Mol. Biol. Cell 13, meeting abstract B256.
- Parker, J.D.K., and Quarmby, L. M. (2003). Chlamydomonas fla mutants reveal a link between deflagellation and intraflagellar transport. BMC Cell Biol. 4, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarmby, L. M. (2004). Cellular deflagellation. Int. Rev. Cytol. 233, 47-91. [DOI] [PubMed] [Google Scholar]
- Qin, H., Diener, D. R., Geimer, S., Cole, D. G., and Rosenbaum, J. L. (2004). Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell Biol. 164, 255-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum, J. L. (2003). Organelle size regulation: length matters. Curr. Biol. 13, R506-R507. [DOI] [PubMed] [Google Scholar]
- Rosenbaum, J. L., Moulder, J. E., and Ringo, D. L. (1969). Flagellar elongation and shortening in Chlamydomonas. The use of cycloheximide and colchicine to study the synthesis and assembly of flagellar proteins. J. Cell Biol. 41, 600-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum, J. L., and Witman, G. B. (2002). Intraflagellar transport. Nat. Rev. Mol. Cell. Biol. 3, 813-825. [DOI] [PubMed] [Google Scholar]
- Silflow, C. D., LaVoie, M., Tam, L. W., Tousey, S., Sanders, M., Wu, W., Borodovsky, M., and Lefebvre, P. A. (2001). The Vfl1 protein in Chlamydomonas localizes in a rotationally asymmetric pattern at the distal end of the basal bodies. J. Cell Biol. 153, 63-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, L., and Dentler, W. L. (2001). Flagellar protein dynamics in Chlamydomonas. J. Biol. Chem. 276, 29754-29763. [DOI] [PubMed] [Google Scholar]
- Stephens, R. E. (1997). Synthesis and turnover of embryonic sea urchin ciliary proteins during selective inhibition of tubulin synthesis and assembly. Mol. Biol. Cell 8, 2187-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taillon, B. E., Adler, S. A., Suhan, J. P., and Jarvik, J. W. (1992). Mutational analysis of centrin: an EF-hand protein associated with three distinct contractile fibers in the basal body apparatus of Chlamydomonas. J. Cell Biol. 119, 1613-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm, S. L. (1967). Flagellar development in the protozoan Paranema trichophorum. J. Exp. Zool. 164, 163-186. [Google Scholar]
- Toyama, Y., Sumiya, H., Fuse, H., and Shimazaki, J. (1996). A case of an infertile man with short-tailed spermatozoa. Andrologia 28, 81-87. [DOI] [PubMed] [Google Scholar]
- Wright, R. L., Chojnacki, B., and Jarvik, J. W. (1983). Abnormal basal-body number, location, and orientation in a striated fiber-defective mutant of Chlamydomonas reinhardtii. J. Cell Biol. 96, 1697-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]