Abstract
Numerous proteins targeted for the secretory pathway are increasingly implicated in functional or pathological roles at alternative cellular destinations. The parameters that allow secretory or membrane proteins to reside in intracellular locales outside the secretory pathway remain largely unexplored. In this study, we have used an extremely sensitive and quantitative assay to measure the in vivo efficiency of signal sequence-mediated protein segregation into the secretory pathway. Our findings reveal that segregation efficiency varies tremendously among signals, ranging from >95 to <60%. The nonsegregated fraction is generated by a combination of mechanisms that includes inefficient signal-mediated translocation into the endoplasmic reticulum and leaky ribosomal scanning. The segregation efficiency of some, but not other signal sequences, could be influenced in cis by residues in the mature domain or in trans by yet unidentified cellular factors. These findings imply that protein compartmentalization can be modulated in a substrate-specific manner to generate biologically significant quantities of cytosolically available secretory and membrane proteins.
INTRODUCTION
In mammalian cells, an estimated one-fifth to one-third of all proteins reside in or transit through the secretory pathway. The decisive step in segregating these proteins from the cytosol is their cotranslational targeting to and translocation across the endoplasmic reticulum (ER) membrane (reviewed by Walter and Johnson, 1994; Rapoport et al., 1996; Johnson and van Waes, 1999). Both targeting and initiation of translocation are dependent on a signal sequence, often at the N terminus, encoded in secretory and membrane proteins. Although the primary sequence of signals varies broadly (von Heijne, 1985), they contain shared features (such as hydro-phobicity) that allow their common recognition by a ubiquitous machinery for ER translocation. Thus, proteins containing signal sequences are generally considered to have their intended functions at destinations accessed via the secretory pathway.
Increasingly, however, many signal-containing proteins have been implicated in functional, pathological, or yet unknown roles at sites outside the secretory pathway. For example, the ER lumenal chaperone calreticulin (CRT) was identified completely independently as a cytosolic integrin-binding protein (Leung-Hagesteijn et al., 1994; Coppolino et al., 1995, 1997), a nuclear export factor (Holaska et al., 2001), a regulator of steroid hormone receptor activity (Burns et al., 1994; Dedhar et al., 1994), and an mRNA binding protein that regulates p21 translation (Iakova et al., 2004). In each of these instances, both functional and physical interactions with the respective cytosolic components have been demonstrated. Similarly, a different ER resident protein, the β subunit of glucosidase II (Trombetta et al., 1996), was originally discovered (and given the name 80K-H) as a target of protein kinase C (Sakai et al., 1989; Hirai and Shimizu, 1990). More recently, it has been demonstrated to interact with the cytosolic adaptor protein Grb3 during signal transduction (Goh et al., 1996; Kanai et al., 1997), shuttle between the nucleus and cytosol (Forough et al., 2003), and modulate calcium homeostasis by binding to the cytosolic tail of the TRP5 channel (Gkika et al., 2004). Other secretory pathway proteins with alternative localizations include mitochondrial transforming growth factor-β (Heine et al., 1991), mitochondrial Slit3 (Little et al., 2001), mitochondrial cytochrome p450 (Addya et al., 1997; Anandatheerthavarada et al., 1999), cytosolic IL15 (Tagaya et al., 1997; Kurys et al., 2000), nucleocytoplasmic cathepsin L (Goulet et al., 2004), and nuclear HBV precore protein (Garcia et al., 1988; Ou et al., 1989).
Alternative localizations of secretory pathway proteins also are associated with various disease states. Although some observations of unusual localizations simply correlate with a disease state (Ramsamooj et al., 1995; Yoon et al., 2000; Brunagel et al., 2003), other examples have been mechanistically implicated in contributing to pathogenesis. For example, the secretory protein apolipoprotein E (apoE) has been suggested to interact with the neurofilament Tau in the cytosol to induce neurofibrillary tangles (Huang et al., 2001; Ljungberg et al., 2002; Harris et al., 2003; Brecht et al., 2004), a common feature of Alzheimer's disease. That only the disease-associated polymorphic form (apoE4) but not protective form (apoE3) had these effects further supports such a role in the cytosol. Similarly, another secretory pathway protein involved in Alzheimer's disease, APP, was shown to be capable of translocating partially into mitochondria and inducing apoptosis (Anandatheerthavarada et al., 2003). Furthermore, a pathogenic fragment of APP, a-β, seems to interact with and inhibit a key mitochondrial enzyme (Lustbader et al., 2004). Cytosolically localized prion protein (PrP) also has been suggested by recent studies to be harmful and associated with neurodegeneration (Ma et al., 2002). Yet other studies have observed PrP in the cytosol under non-pathogenic conditions (Mironov et al., 2003), where it may interact with and protect against Bax-mediated cell death (Bounhar et al., 2001; Roucou et al., 2003).
For most of these signal sequence-containing proteins, the mechanisms or extent to which they arrive at their alternative localizations remains largely a matter of speculation (for a notable exception, see Goulet et al., 2004). It is therefore important to know, with a high degree of sensitivity and precision, the efficiency by which signal sequence-containing substrates are segregated to the ER in vivo. Although small differences in efficiency (for example 95 vs. 99%) would have little or no consequences for the secretory pathway form of the substrate, it would have substantial implications for the alternative form (which in this example, would differ by a factor of 5). Such seemingly small proportions (e.g., even 1% or less) of abundant secretory pathway proteins (such as CRT, 80K-H, or PrP, among others) would still easily be within the range of physiologically relevant protein abundances. This is particularly true when the putative alternative form has proposed functions, such as transcriptional regulation (Burns et al., 1994; Dedhar et al., 1994) or signal transduction (Goh et al., 1996; Kanai et al., 1997) that are generally associated with low abundance proteins. Thus, an accurate measure of inefficiency, and the mechanisms that contribute to it, is an absolute prerequisite for rationally evaluating and modulating the proposed alternative localizations of various secretory pathway proteins.
Signal-mediated segregation into the mammalian ER involves several steps that have been well characterized for model substrates in vitro (reviewed in Walter and Johnson, 1994; Rapoport et al., 1996; Johnson and van Waes, 1999). On emergence from the ribosome, the signal sequence is recognized and bound by the signal recognition particle (SRP). For some substrates, this binding results in a transient slowdown of translation to facilitate targeting of the ribosomenascent chain-SRP complex to its receptor at the ER membrane (Wolin and Walter, 1989; Mason et al., 2000). Once at the ER, the ribosome-nascent chain complex is transferred, by a yet unknown mechanism, to a translocon whose central component is composed of the Sec61 complex (Song et al., 2000). The Sec61 complex, possibly with assistance from the TRAM and/or TRAP proteins (Voigt et al., 1996; Fons et al., 2003), mediates a second signal sequence recognition event (Jungnickel and Rapoport, 1995). If the signal is successfully recognized by the translocon, the nascent chain is inserted into the protein conducting channel (Jungnickel and Rapoport, 1995), which is then opened toward the ER lumen (Crowley et al., 1994) to allow the productive initiation of substrate translocation. Despite these mechanistic insights into the early steps of protein translocation, the level of fidelity for any individual step or for the entire process in vivo is not known. We have therefore devised a specific and sensitive assay to measure the in vivo inefficiencies in protein segregation that would result in alternative localizations of secretory pathway proteins. Our goal was to begin addressing several basic questions about the fidelity of compartmentalization in mammalian cells: How efficient is signal-mediated segregation into the secretory pathway? Can biologically significant amounts of such substrates exist in a nonsecretory pathway compartment? If so, what are the mechanisms and parameters that influence how much is localized to the alternative compartment? Is inefficiency of compartmentalization an inherent property of any given substrate or can it be selectively modulated? The answers to these questions may help provide a common framework for understanding the otherwise disparate observations of unexpected or aberrant locales of many physiologically important proteins.
MATERIALS AND METHODS
DNA Constructs
The coding region for the heterologous Gal4-nulcear factor-κB (NF-κB) transcription factor (TF) was amplified by polymerase chain reaction (PCR) from plasmid pBD-NF-κB (Stratagene, La Jolla, CA) and subcloned in frame into a calreticulin expression construct at the unique NotI site just preceding the coding region for the C-terminal KDEL ER retention sequence. The region coding for TF, together with the KDEL sequence, was PCR amplified and subcloned into a blunted PstI site and a downstream XbaI site of cassettes containing different signal sequences. Such signal sequence cassettes have been described previously (Kim et al., 2002). Changes to the region between the signal and mature domain (such as the inclusion of 10 amino acids from the respective mature domain, or various mutations as described in the text) were encoded within the oligonucleotides used for PCR amplification of TF. Some point mutants were generated by site-directed mutagenesis by using the QuikChange method (Stratagene). The vesicular stomatitis virus glycoprotein (VSVG)-green fluorescent protein (GFP) construct comprising the ts045 temperature-sensitive VSVG with a C-terminally appended enhanced green fluorescent protein (EGFP) has been described previously (Presley et al., 1997). The mature domain of this plasmid was PCR amplified and subcloned downstream of the Prl or PrP signal sequences by using the cassettes described above. A similar strategy was used to replace the PrP signal sequence with those from Osteopontin (Opn) or Angiotensinogen (Ang). All constructs were verified by automated sequencing. The luciferase reporter containing the Gal4 promoter (pLuc) was obtained from Stratagene. Enhanced yellow fluorescent protein (EYFP) and enhanced cyan fluorescent protein expression plasmids were from BD Biosciences Clontech (Palo Alto, CA).
Cell Culture and Luciferase
HeLa, Cos-7, N2a, and Madin-Darby canine kidney (MDCK) cells (clone T23.1; Lai et al., 2003) were cultured in DMEM containing 10% fetal bovine serum. Between 5000 and 10,000 cells were plated per well of a 96-well plate between 24 and 36 h before transfection, at which time they were ∼40-80% confluent. Unless specifically noted otherwise, transfections with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) were performed in 96-well plates with a total of 200 ng of plasmid DNA per well. Of this 200 ng, the luciferase reporter plasmid was always kept constant at 160 ng. The remaining 40 ng constituted a mixture of varying amounts of the TF-containing plasmid of interest and an EYFP expression plasmid. In this manner, TF expression could be changed systematically without altering other parameters of the experiment, whereas EYFP expression allowed routine monitoring of transfection efficiency (which remained constant at ∼70%). In independent experiments, we confirmed that the use of less plasmid for one of the components (while keeping the total amount of DNA constant) results in lower expression of only that component in individual cells (Supplementary Figure S1). Unless otherwise noted in the figure legend, the cells were analyzed 18-24 h after transfection. For the luciferase assays, the media were aspirated, the cells recovered in 100 μl per well of freshly prepared luciferase reporter assay substrate (Roche Diagnostics, Indianapolis, IN), and measured in a tube luminometer. For parallel Western blot analysis, the cells in replicate samples were harvested in 40 μl of 2× SDS-PAGE sample buffer, of which 10 μl was analyzed per lane on 12% Tris-tricine mini-gels. For experiments comparing multiple cell lines, the plating, growth, transfection, and analysis of the cells were done identically and in parallel. Importantly, each transfection condition used a single master mix of DNA-Lipofectamine complexes for all of the different cell lines to minimize variability in the relative amounts of transfected DNA. Comparable transfection efficiencies were confirmed with parallel analysis of fluorescent protein expression. In experiments where multiple cell lines were followed over multiple days, a single larger transfection was performed for each condition with 4 μg of total DNA per well of a six-well dish before replating 8 h later into multiple 96-well plates for subsequent growth and analyses.
Hippocampal Neuronal Culture
Cultures of rat hippocampal neurons, grown on top of a feeder layer of glial cells, were prepared as described previously (Mayer et al., 1989; Lu et al., 1998). Briefly, suspensions of a mixed population of brain cells prepared by papain dissociation of hippocampi from 21-d embryonic Sprague-Dawley rats were plated onto 2-wk-old glial feeder layers at a density of 250,000 per 35-mm dish. Cells were fed twice weekly with half changes of minimal essential medium supplemented with 5% (vol/vol) heat-inactivated horse serum, 1% (vol/vol) fetal bovine serum, 2 mM glutamax (all from Invitrogen), 136 μM uridine, 54 μM 2-deoxy-5-fluoro-uridine, and the growth factor mixture N3 (Mayer et al., 1989). Cells were maintained in a 37°C incubator with 10% CO2. All procedures were in accordance with the NIH Guide for the Care and Use of Laboratory Animals and were approved by the National Institutes of Health Animal Care and Use Committee. Neuronal cultures were transfected after ∼8-12 d in culture with Effectene transfection reagent (QIAGEN, Valencia, CA), following the protocol recommended by the manufacturer. Transfection efficiency was typically 1%. Imaging was performed on live cultures 16-21 d old. Images were captured with a 63× numerical aperture 1.4 objective on a LSM510 laser scanning confocal microscope (Carl Zeiss, Thornwood, NY) with 488-nm excitation. The temperature of cells before and at the time of imaging (which influences ER-Golgi trafficking of the temperature-sensitive vesicular stomatitis virus (VSV)-GFP mutant used in these studies) are described in the figure legends.
Biochemical Analyses
In vitro translation and analysis of protein translocation into rough microsomal membranes was performed as described previously (Fons et al., 2003). Immunoblotting used antibodies against the C terminus of NF-κB p65 (Santa Cruz Biotechnology, Santa Cruz, CA), protein disulfide isomerase (StressGen Biotechnologies, San Diego, CA), GFP (BD Biosciences Clontech), TRAP-α, and Sec61-β (Fons et al., 2003). Fractionation of cells by selective extraction was performed on cells growing in six-well dishes. The plate of cells was placed on a bed of ice, and after removal of the media, incubated for 5 min in 1 ml of ice-cold 50 mM HEPES, pH 7.4, 100 mM KAc, 2.5 mM MgAc2, 150 μg/ml digitonin. This digitonin extract was reserved in a separate tube on ice, and the cells extracted subsequently with 1 ml of ice-cold 50 mM HEPES, pH 7.4, 500 mM KAc, 5 mM MgAc2, 1% Triton X-100. After removal of the Triton extract to a separate tube, the remaining cellular material was solubilized in 100 μl of 100 mM Tris, pH 8.9, 1% SDS. The proteins in the digitonin and Triton extracts were collected by precipitation with trichloroacetic acid, washed in acetone, and dissolved in SDS-PAGE sample buffer before analysis. The SDS extract was analyzed directly. Equivalent proportions of each fraction were used for SDS-PAGE and immunoblotting. Treatment of cell lysates with peptide:N-glycosidase and endoglycosidase H (both from New England Biolabs, Beverly, MA) was performed as recommended by the manufacturer. Pulse labeling was performed in six-well dishes on cells at ∼70-80% confluence 24 h after transfection. Cells were pretreated for 30 min with methionine/cysteine-free media and labeled for 15 min with 150 μCi/ml 35S-Translabel (MP Biomedicals, Irvine, CA). The media were removed and the cells harvested in 1% SDS, 0.1 M Tris, pH 8. After heating to denature the proteins, PrP was immunoprecipitated with the 3F4 monoclonal antibody (Signet Laboratories, Dedham, MA) as described previously (Kim et al., 2002).
RESULTS
Experimental Strategy
To measure the efficiency of protein compartmentalization, an assay is required to detect very small populations of nonlocalized substrate with high specificity. Ideally, “noise” from correctly localized protein (representing the major population) would be invisible in the assay, whereas “signal” from any minor population of interest would be amplified. Neither conventional subcellular fractionation nor direct observation (e.g., of GFP-tagged substrate) afford such advantages. Hence, their reliability is limited to situations where the minor population represents at least ∼10-20% of the total amount. Furthermore, detection of the alternative fraction often requires overexpression of the substrate of interest, making extrapolation to physiological conditions difficult.
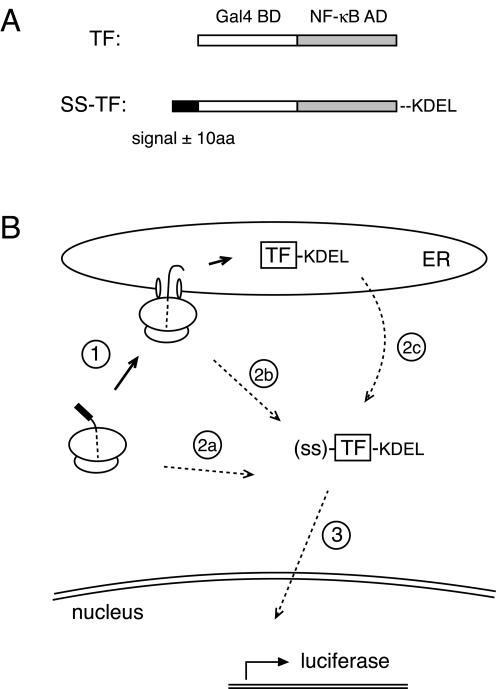
To circumvent these limitations, we devised an assay for compartmentalization (Figure 1) that is conceptually analogous to the selection scheme used to isolate yeast mutants defective in protein translocation (Deshaies and Schekman, 1987). In this assay, a protein that is only functional in the cytosol is fused to a signal sequence. Lack of compartmentalization into the ER allows the protein to function as a marker for its presence in the cytosol, whereas properly compartmentalized protein is effectively invisible. To maximally amplify the cytoplasmic signal, we used a heterologous TF as the marker protein. Even small amounts of TF in the cytosol can be detected because its function generates multiple copies of reporter mRNA, each of which makes multiple reporter proteins (in this case luciferase), each of which acts enzymatically to generate a detectable signal (light) that is both sensitive and quantifiable. The total amount of TF (regardless of cellular locale) can be measured in parallel by immunoblotting to determine a specific activity that is a measure of compartmentalization efficiency. Furthermore, if the TF-sequestration assay can detect very few molecules of alternatively-localized protein with high specificity, measurements can be made on substrates expressed at physiological levels to avoid potential artifacts associated with overexpression.
Figure 1.
A reporter assay for the measurement of protein compartmentalization. (A) Diagram of the heterologous TF composed of the DNA binding domain of Gal4 (white) fused to the transcriptional activation domain of NF-κB (gray). Constructs to measure TF segregation into the secretory pathway contain an N-terminal signal sequence with or without 10 additional amino acids of the respective mature domain (black) and the KDEL sequence for ER retention appended to the C terminus. (B) In the TF sequestration assay, nascent TF containing a signal sequence is cotranslationally targeted to ER translocons (step 1), through which TF is subsequently translocated into the lumen where it is retained by the KDEL sequence. Cytosolic TF can be generated by nascent chains that fail to target (step 2a), fail to initiate translocation (step 2b), or escape degradation after retrotranslocation from the ER lumen (step 2c). The presence of cytosolic TF, but not ER-localized TF, can be detected and quantified by its activation of a luciferase reporter preceded by a Gal4 binding element (step 3).
Sensitive Detection of noncompartmentalized TF
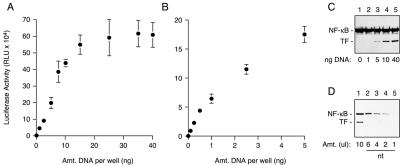
We first measured the sensitivity and linear range for detection of cytoplasmic TF. A heterologous TF composed of the Gal4 DNA-binding domain fused to the transcriptional activation domain of NF-κB (Figure 1A) was cotransfected into cultured cells with a luciferase reporter preceded by Gal4 binding elements (Figure 2, A and B). We found that transfection with increasing amounts of TF resulted in a roughly linear increase in luciferase activity up to ∼10-15 ng of plasmid per well of a 96-well plate, above which activity reached a plateau (Figure 2A). This plateau is likely due to saturation of transcription from the luciferase reporter plasmid because neither TF expression (Figure 2C) nor luciferase substrate was limiting. Luciferase activity ∼150-fold above background could be detected with as little as 0.1 ng of transfected plasmid (Figure 2B). By contrast, detection of TF by immunoblotting required transfection with at least 1-5 ng of plasmid (Figure 2C). Thus, using luciferase as a reporter, cytoplasmic TF can be detected and quantified with at least 10- to 100-fold better sensitivity than immunoblotting.
Figure 2.
Sensitivity and linearity of cytosolic TF detection. (A and B) Cells cotransfected with the luciferase reporter plasmid and varying amounts (up to 40 ng) of a TF expression plasmid were assayed for luciferase activity (mean ± SD; n = 3). The TF expressed from as little as 0.1 ng of transfected plasmid is readily detected as a luciferase signal ∼150- to 200-fold above background. (C) Anti-NF-κB immunoblot of selected cell lysates from A reveals endogenous NF-κB (which serves as a loading control) and the exogenously expressed TF. The minimum detectable amount of TF upon gross overexposure of the blot was the 1 ng of sample. (D) The amount of exogenous TF expression in cells transfected with 40 ng of plasmid (lane 1) was compared with endogenous NF-κB levels in nontransfected cells (nt; lanes 2-5). Note that the amount of endogenous NF-κB in ∼4-6 μl of cell lysate is comparable with the exogenous TF in 10 μl of transfected cells.
Importantly, the changes in expression level observed by the luciferase assays or immunoblots upon varying the amount of transfected plasmid occurred on a per cell basis and was not due to fewer transfected cells. This was confirmed in independent experiments using two different fluorescent proteins whose expression levels could be quantitatively monitored in individual live cells (Supplementary Figure S1). In this experiment, we demonstrate that for the cotransfection procedures used in this study, reducing the amount of one of the plasmids (while keeping total transfected DNA constant) results in the specific reduction of expression for the corresponding protein in individual cells. No effect is observed on either the expression of the other product or on the total number of cells that are cotransfected. This modulation of expression on a per cell basis occurred over a wide range (at least 20-fold; Supplemental Figure S1) and corresponded well with the analysis of expression on the whole cell population (e.g., by immmunoblotting). Thus, we are able to express our substrate over a broad range of physiological levels while still retaining the capability to detect small populations of cytosolically localized protein within a linear range that spans at least 2 orders of magnitude.
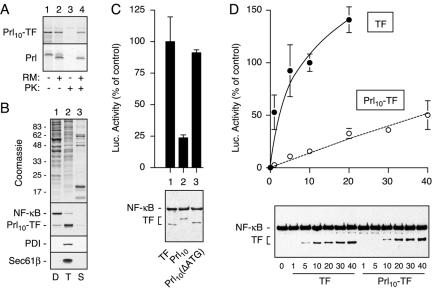
To assess compartmentalization efficiency, TF was inserted between the first 40 residues of prolactin (Prl) at the N terminus and a KDEL sequence for ER retention at the C terminus to generate Prl10-TF (Figure 1A). In vitro translation of Prl10-TF showed that this heterologous protein could be translocated into microsomal membranes as is observed for native Prl (Figure 3A). As with Prl (Lingappa et al., 1977), translocation of Prl10-TF only proceeded cotranslationally, with cytosolic Prl10-TF remaining nontranslocated even after prolonged posttranslational incubation with microsomal membranes (our unpublished data). Prl10-TF also was overexpressed in cultured cells and its localization assessed by subcellular fractionation using selective detergent extraction. A digitonin extract containing cytosolic proteins included the majority of endogenous NF-κB (Figure 3B) as well as several other cytosolic markers such as heat shock protein (HSP)70 and HSP90 (our unpublished data). By contrast, ER membrane and lumenal proteins such as components of the protein translocon (Sec61-β) and protein disulfide isomerase were not extracted unless Triton X-100 was used (Figure 3B). Consistent with an ER localization for Prl10-TF, the majority of it was found in the Triton X-100 fraction (Figure 3B). Similar analysis of TF lacking the Prl signal sequence showed it to be in the cytosolic fraction (our unpublished data).
Figure 3.
Detection and quantitation of signal-mediated sequestration in the ER. (A) Prl10-TF and Prl were translated in vitro with or without rough microsomal membranes (RM). Equal aliquots of translation products were either analyzed directly (lanes 1 and 2) or after digestion with proteinase K (PK) to assess translocation (lanes 3 and 4). (B) Cells expressing Prl10-TF were sequentially extracted with digitonin (D), Triton X-100 (T), and SDS (S). The respective fractions were analyzed for total protein with Coomassie staining (top) or specific components by immunoblotting (bottom). (C) Ten nanograms per well of plasmids encoding TF, Prl10-TF, or Prl10-TF(ΔATG) (lanes 1-3, respectively) were transfected into cells and analyzed in parallel for luciferase activity (top; mean ± SD; n = 3) and total expression (bottom; anti-NF-κB immunoblot). (D) Varying amounts (from 0 to 40 ng) of plasmids encoding TF (closed circles) or Prl10-TF (open circles) were analyzed for luciferase activity (top) and total expression (bottom) as in C. Luciferase measurements in C and D were normalized to the value for 10 ng of TF, which was set to 100%.
Analysis of Prl10-TF by the luciferase reporter assay showed ∼4- to 5-fold lower luciferase activity than TF, despite identical levels of expression (Figure 3C). Prl10-TF was often observed to migrate slightly slower by SDS-PAGE, as expected due to the presence of additional residues at the N and C termini. On mutation of the translation initiation codon of Prl10-TF from ATG to ATC [Prl10-TF(ΔATG); Figure 4A], the expressed product is now only slightly larger than TF and results in the restoration of full luciferase activity (Figure 3C). The consequence of this mutation is the initiation of translation at the next suitable start codon, which in this instance is the beginning of TF (Figure 4A). Thus, appending the first 40 residues from Prl to TF results in markedly lower luciferase activity without any effect on the expression level, consistent with the majority of Prl10-TF being sequestered into a noncytoplasmic compartment where it cannot function to activate transcription of the reporter.
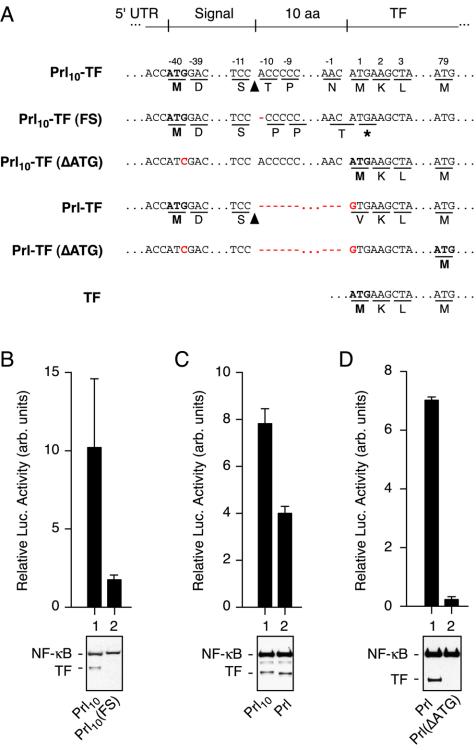
Figure 4.
Analysis of translational and posttranslational contributions to cytosolic TF. (A) Sequences of the indicated constructs are shown. The first potential start codon in each construct is indicated in bold. The site of signal sequence cleavage is indicated with an arrowhead. Stop codons are indicated by an asterisk. Mutations are indicated in red. (B-D) Various sets of constructs (indicated below each immunoblot) were compared for luciferase activity (top) and total expression (bottom). Constructs within each panel were analyzed in parallel, whereas the absolute luciferase values between panels are not directly comparable. Luciferase measurements were between three and five replicates. Amount of plasmid transfected per well was 40 ng in B and 10 ng in C and D.
Comparison of luciferase activity for TF vs. Prl10-TF over a broad range of expression levels (Figure 3D) consistently showed the same qualitative result observed in Figure 3C. The fold difference in relative luciferase activity was underestimated at expression levels above 10 ng/well of plasmid due to some saturation of the TF signal. Within the linear range of ∼1-10 ng/well plasmid, the relative luciferase signals from multiple independent experiments showed that Prl10-TF consistently displays ∼10% luciferase activity per unit protein than observed for TF. Thus, at low-to-moderate expression levels in vivo at steady state, ∼90% of an ER-targeted substrate is segregated from the cytosol, whereas the remainder is available for reporter activation.
Multiple Mechanisms Contribute to TF Noncompartmentalization
Cytosolic Prl10-TF can potentially be generated in multiple ways. One possibility is that initiation of translation from Prl10-TF mRNA at an internal AUG codon results in the synthesis of functional TF lacking the Prl signal sequence. This could occur if, despite an ideal Kozak's consensus sequence (Kozak, 1992), the first AUG codon was skipped due to “leaky” ribosomal scanning (Kozak, 2002). To determine the contribution of such a translational mechanism to the luciferase activity observed for Prl10-TF, we analyzed the effect of two mutants. In the first mutant, Prl10-TF(FS), a single nucleotide was deleted between the first and second ATG codons, causing a frameshift after synthesis of the Prl signal sequence (Figure 4A). Thus, initiation of translation at the first AUG would result in the synthesis of a shortened 40 amino acid product, which cannot activate the luciferase reporter. Functional TF containing both the Gal4 and NF-κB domains can only be generated from Prl10-TF(FS) if translation initiates at the second AUG (which is beyond the frameshift mutation) in which case full-length TF lacking a signal sequence would be synthesized. We found that compared with Prl10-TF, the Prl10-TF(FS) mutant generated ∼20% of the luciferase activity (and hence ∼2% of the activity for TF lacking a signal sequence). A concomitant decrease in the expression of TF-containing products by immunoblot also was observed as expected (Figure 4B). Similar results were obtained when two nucleotides were deleted to generate a different frameshift mutant of Prl10-TF. These results indicate that of the total cytoplasmic population of Prl10-TF, roughly 20% is generated by translational mechanisms that involve skipping of the first AUG codon. Conversely, the majority of cytosolic TF is therefore generated by mechanisms that involve a lack of compartmentalization despite proper synthesis of a protein containing a functional N-terminal signal sequence.
A similar conclusion was reached by a different approach in which only one in-frame AUG codon was made available for generating TF-containing translation product. In this construct (termed Prl-TF; Figure 4A), the signal of Prl (without the additional 10 amino acids of the mature domain) was fused directly to TF in which the first codon (ATG) was changed to GTG. Comparison with Prl10-TF showed that Prl-TF had ∼50% luciferase activity, suggesting that removal of the option for internal initiation results in a fewer proportion of molecules in the cytosol. As expected, changing the initiating ATG codon to ATC of Prl-TF [to generate Prl-TF(ΔATG)] resulted in an ∼30- to 50-fold drop in luciferase activity (Figure 4D) due to the lack of a suitable in-frame start codon capable of generating a functional TF. The results with Prl-TF indicate that when translation is limited to initiating at the beginning of the signal sequence, ∼50% less cytosolic TF is generated, again suggesting that a proportion of Prl10-TF is cytosolic due to translational mechanisms. The somewhat different proportions attributed to translational effects by the two approaches (∼20 vs. ∼50%) seems to be due in part to differences in the first few residues of the mature domain, which we found in subsequent experiments to influence signal sequence function (see below).
It is clear from Figure 4 that a majority of the cytosolic pool of Prl10-TF is generated despite proper translation of a product containing the N-terminal signal sequence of Prl. This could arise by either inefficiencies in targeting to the ER (Figure 1B, step 2a), inefficiencies in translocation across the ER membrane (Figure 1B, step 2b), or escape from degradation after retrotranslocation from the ER lumen (Figure 1B, step 2b). At present, it is unclear how one might directly and quantitatively distinguish the relative contributions each of these pathways makes to the total cytosolic Prl10-TF in vivo. However, we reasoned that comparisons of the compartmentalization efficiencies for a variety of signal sequences may provide some insight. If in vivo, targeting and translocation efficiencies of all signal sequences are generally high (i.e., >95%), the main contribution to cytosolic TF would be retrotranslocation from the ER lumen. In this case, the luciferase activities would vary little among signal sequences because the substrate for retrotranslocation is identical in each case, thereby generating similar amounts of total cytosolic TF. By contrast, significant differences in targeting and translocation efficiencies among signals would result in wider variation in cytosolic TF, with the most efficient signals generating luciferase activities that asymptotically approach the contribution generated by retrotranslocation alone.
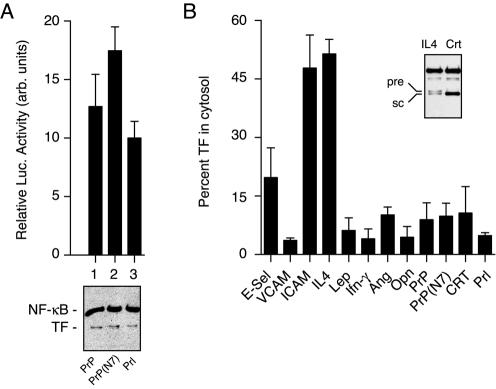
We first tested whether small differences in translocation efficiency can be discriminated by the TF-sequestration assay by comparing the luciferase activities generated by Prl-TF, PrP-TF, and PrP(N7)-TF. Based on in vitro assays (Kim et al., 2002), the PrP signal sequence is predicted to be slightly less efficient at a posttargeting step of initiating translocation than the Prl signal, whereas PrP(N7) is a point mutant of the PrP signal that further decreases its translocation efficiency slightly. When compared in vivo, we found that they showed the expected small but statistically significant differences in luciferase activity (Figure 5A). On analysis of a wider range of signal sequences by this assay, we observed remarkable variability in the luciferase activity generated per unit protein (Figure 5B). In these experiments, the constructs contained direct fusions between the signal and a version of TF in which its initiating ATG codon was changed to GTG (e.g., as for Prl-TF). Thus, the luciferase activity observed cannot be attributed to translational effects, indicating that essentially all of it derives from signal-TF that failed to be compartmentalized properly.
Figure 5.
Compartmentalization efficiencies vary broadly among signal sequences. (A) Ten nanograms per well of plasmids encoding PrP-TF, PrP(N7)-TF, and Prl-TF were analyzed for luciferase activity (top; n = 5) and total expression (bottom). Differences among each construct were statistically significant (p < 0.01 by Student's t test). (B) Ten nanograms per well of constructs encoding the indicated signal sequences fused to TF were analyzed for luciferase activity and total expression. The data from three independent experiments (with at least three replicates for the luciferase measurements in each experiment) were normalized and averaged to derive an estimate of the percentage of TF in the cytosol (see Materials and Methods). The inset shows an immunoblot of the least efficient signal sequence (IL4) in which approximately equal amounts of precursor (pre) and signal-cleaved (sc) TF are resolved and detected. A substantially more efficient signal sequence (CRT) is shown for comparison. Abbreviations used for the signals include E-selectin (E-sel), Leptin (Lep), Ang, and Opn.
The luciferase values obtained for the various signal-TF constructs (Figure 5B) were normalized relative to Prl-TF, for which ∼5% of total synthesized protein is cytosolic (i.e., half of the value observed for Prl10-TF; Figure 4C). Based on these values, the different constructs contained between 5 and 50% of total synthesized TF in the cytosol at steady state. For IL4-TF, which based on the luciferase measurements should contain ∼50% cytosolic protein, we could on some immunoblots resolve two bands of comparable intensity corresponding to precursor and signal-cleaved products (Figure 5B, inset). Parallel analysis of CRT (which should have ∼10% cytosolic based on the luciferase assay) showed predominantly the lower, signal-cleaved band. Not all proteins could be resolved reliably into precursor and processed bands to allow similar analysis (our unpublished data); nonetheless, the observation with IL4-TF provides further corroboration that the estimates of the cytosolic proportion based on the luciferase reporter measurements are accurate.
Substrate-specific Modulation of TF Compartmentalization in cis
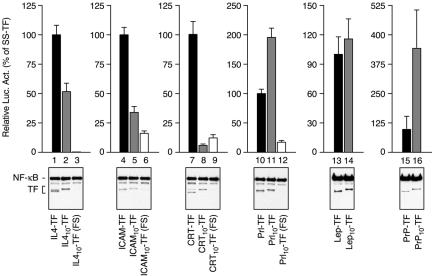
The observation in Figure 4 that Prl10-TF and Prl-TF generate somewhat different levels of cytosolic forms that cannot be fully explained by translational effects raised the possibility that residues immediately after the signal sequence influence the efficiency of protein compartmentalization. Although the information for targeting and translocation is generally considered to be encoded entirely within the signal sequence, evidence from in vitro studies has suggested that regions of the mature domain may influence signal function (Andrews et al., 1988; Kim et al., 2002). Whether cis-acting modulatory effects on signal function are similarly recapitulated in vivo in a relevant manner is not clear. To address this, we compared the level of cytosolic TF generated for signal-TF versus signal10-TF constructs (Figure 6). The extra 10 residues of the mature domain were found to significantly increase the cytosolic population in some instances (e.g., PrP and to a lesser extent Prl), decrease it in others (e.g., IL4, ICAM, and CRT) and have little effect on the leptin signal. Frameshift experiments similar to that in Figure 4B indicated that as for Prl, the contribution of translational effects on cytosolic populations of most constructs was detectable, but generally a relatively minor contributor (Figure 6). Thus, the region of the mature domain immediately after the signal sequence can substantially influence the amount of cytosolically available protein.
Figure 6.
Modulation of signal-mediated sequestration in cis. Sets of constructs containing the indicated signal sequences, with or without 10 amino acids from the respective mature domains (gray and black bars, respectively), were analyzed for luciferase activity and total expression. Except for the Lep and PrP signals, a frameshift mutation (Figure 4A) was analyzed in parallel (white bars) to assess the contribution of leaky ribosomal scanning to the luciferase activity observed for the respective Signal10-TF constructs. Ten nanograms of plasmid per well was analyzed in each instance. Due to the large differences in absolute luciferase activity among different signals (Figure 5B), each set of constructs was plotted separately and normalized to the respective SS-TF construct (set to 100%). Note that the 10 amino acids from the mature domain can increase, decrease, or have little effect on the efficiency of TF sequestration.
Substrate-specific Modulation of TF Compartmentalization in trans
The observation of various secretory pathway proteins in alternative locations has often been controversial and not universally observed. One possibility for such discrepancies might be that compartmentalization efficiency can vary depending on cellular conditions, cell type, or method of analysis. If this were true, the effect would presumably have to be substrate specific, because a global breakdown of proper targeting and translocation pathways is incompatible with cell viability. To determine whether such substrate-specific effects are plausible, we examined whether the luciferase assay could be used to detect relative changes in compartmentalization efficiencies among various substrates as a function of either different cell types or different growth and culture conditions.
In the first set of experiments, we sought to determine whether the choice of signal sequence on a substrate can influence compartmentalization efficiency in a cell type-specific manner. Five different signal10-TF constructs were transfected in parallel into four different cell lines and the respective luciferase activities determined. When normalized to one of the substrates (Prl10-TF), differences in the relative luciferase activities for the other substrates could be observed in a cell type-specific manner (Figure 7A). For example, the normalized luciferase activity for VCAM10-TF was the same or higher than Prl10-TF in N2a cells, but substantially lower than Prl10-TF in Cos-7, HeLa, or MDCK cells. Similar variations were seen for each of the constructs despite all of them having the same promoter, 5′ and 3′ untranslated regions, and polyA-addition signal. Thus, the relative compartmentalization efficiencies for five signal10-TF substrates differ among cell types.
Figure 7.
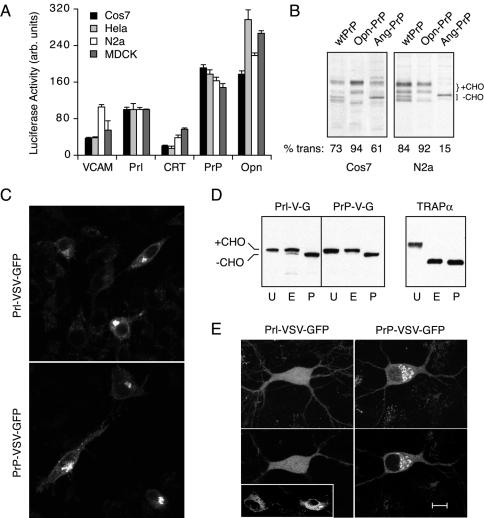
Influence of cell type on substrate-specific differences in compartmentalization. (A) Four different cell types (Cos-7, HeLa, N2a, and MDCK) grown under identical conditions were transfected in parallel with 10 ng of plasmid encoding each of five signal10-TF constructs and analyzed by the standard luciferase assay 24 h later. All of the luciferase measurements for each cell type were normalized to the value obtained for Prl10-TF in that cell type, which was arbitrarily set at 100. The average of five replicates ± SD is plotted. (B) Cos-7 (left) or N2a (right) cells were transfected with the indicated PrP constructs and analyzed 24 h later by pulse labeling and immunoprecipitation of PrP. The positions of the glycosylated (+CHO) and nonglycosylated (-CHO) species are indicated. The percentage of translocated PrP, defined as species that have undergone signal sequence cleavage and/or glycosylation, is quantified in each lane and indicated below the autoradiographs. (C) VSV-GFP containing the Prl or PrP signal sequences (Supplementary Figure S2) were transfected into N2a cells and the localization patterns visualized by confocal microscopy ∼20-30 min after shift from 40 to 25°C. The staining pattern in each case is characteristic for compartments of the secretory pathway and was confirmed by comparison to ER, Golgi, and cell surface markers expressed in this same cell type (our unpublished data). (D) Lysates from cells in C were analyzed by SDS-PAGE and immunoblotting with antibodies against GFP. Samples were either left untreated (U) or digested with endoglycosidase H (E) or PNGase F (P) before analysis. The positions of glycosylated and unglycosylated VSV-GFP are indicated. The blot also was probed with antibodies specific to TRAPα, an ER-resident glycoprotein that was deglycosylated efficiently with either enzyme (right). (E) Primary hippocampal neurons growing on a feeder layer of glial cells were transfected with Prl-VSV-GFP or PrP-VSV-GFP and visualized by confocal microscopy ∼20-30 min after shift from 40 to 25°C. Shown on the top panels are projection images of typical localization patterns seen in the neurons. The bottom panels show a single confocal slice taken at the level of the nucleus to illustrate the different patterns of localization. The inset shows transfected glial cells in the same culture dish, which showed secretory pathway localization (mostly ER) for both constructs. Bar, 10 μm in the neuron and N2a cell images, and 20 μm in the glial cell inset.
To provide additional support for this idea of signal-specific cell type differences in compartmentalization, we sought to find instances in which such differences are sufficiently dramatic to be detected by other methods. During the course of several other trafficking studies in our laboratory, two particularly striking examples were noted. In the first example, PrP containing different signal sequences was analyzed by pulse labeling after tranfection into different cell types. PrP contains two sites for N-linked glycosylation that together with signal sequence cleavage provide markers for translocation. We found in Cos-7 cells that, as with studies in vitro (Kim et al., 2002), the efficiencies of the Opn and Ang signals in directing PrP translocation are higher and lower, respectively, than the wild-type PrP signal (Figure 7B, left). In N2a cells (Figure 7B, right), translocation of Opn-PrP was essentially identical. By contrast, translocation of wtPrP was noticeably more efficient in N2a cells than in Cos-7 cells, whereas Ang-PrP was significantly less efficient. Thus, as observed for the TF reporter, the compartmentalization efficiency of PrP changes in different cell types in a manner influenced by the choice of signal sequence. Such cell type differences may help to partly explain the currently controversial and apparently contradictory observations on the generation and source of PrP in the cytosol (Yedidia et al., 2001; Ma and Lindquist, 2002; Drisaldi et al., 2003; Mironov et al., 2003; Roucou et al., 2003).
This substrate specificity was so dramatic in one example that it could be directly visualized. A GFP-tagged glycoprotein from the VSV containing either the Prl or PrP signal sequence (Prl-VSV-GFP and PrP-VSV-GFP, respectively; Supplementary Figure S2) was analyzed in multiple cell types. The VSV protein used in these experiments contains a temperature-sensitive mutation (ts045) that prevents its trafficking from the ER to Golgi at 40°C. Thus, cells grown at 40°C and subsequently released at lower temperatures show synchronous trafficking from the ER through the secretory pathway (Presley et al., 1997), a process that can be visualized in live cells. In N2a cells viewed ∼20-30 min after release from 40°C, both constructs showed identical localization patterns and were distributed throughout the secretory pathway, including the ER, Golgi, and cell surface (Figure 7C). Consistent with this localization, biochemical analysis (Figure 7D) demonstrated that in each case, all of the VSV-GFP is glycosylated, most of which is resistant to endoglycosidase H digestion (an indicator of trafficking to the Golgi, where trimming of N-linked glycans occurs).
Transfection of the identical DNA preparations, on the same day, into primary hippocampal neurons revealed a marked difference in localization: whereas PrP-VSV-GFP localized within the secretory pathway (ER and Golgi under the conditions in Figure 7E), Prl-VSV-GFP showed a diffuse pattern throughout the cell. Confocal slices at the level of the nucleus illustrated this difference particularly well (Figure 7E, bottom): PrP-VSV-GFP was completely excluded from the nucleus and concentrated in perinuclear structures consistent with the Golgi, whereas Prl-VSV-GFP displayed a diffuse nucleocytoplasmic localization. Remarkably, glial cells in the same culture expressing Prl-VSV-GFP showed clear secretory pathway localization (Figure 7E, inset), exactly as was observed for glial cells in the PrP-VSV-GFP culture (our unpublished data).
Because of the low transfection efficiency into primary neurons and because they are cocultured with glial cells, a parallel biochemical analysis was not feasible. However, we have analyzed the trafficking in live cells of Prl-VSV-GFP and PrP-VSV-GFP under various temperature growth conditions and at various times after release from 40°C. We consistently observe transport of PrP-VSV-GFP in live cells through the secretory pathway from its localization in the ER, to transport carriers (Supplementary Figure S3B), to Golgi (Figure 7E), and eventually to punctate structures within and on the surface of axons and dendrites (Supplementary Figure S3C). By contrast, Prl-VSV-GFP remains primarily diffuse and nucleocytoplasmic regardless of the temperature manipulations or time of visualization (Figure 7E, Supplementary Figure S3A, and our unpublished data). These data are consistent with comparably efficient compartmentalization to the secretory pathway for both Prl-VSV-GFP and PrP-VSV-GFP in N2a cells (Figure 7, C and D) and glial cells (Figure 7E, inset), but markedly different efficiencies in primary hippocampal neurons (Figure 7E and Supplementary Figure S3).
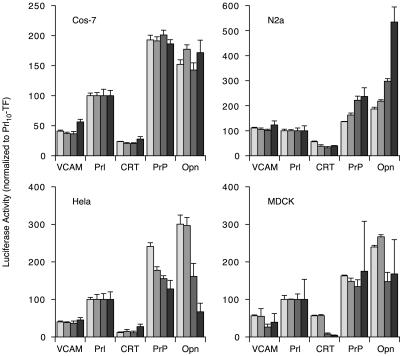
In a final experiment, we returned to the luciferase assay to determine whether, even within a single cell type, changes in compartmentalization could be observed in response to changes in culture conditions. Here, we performed an experiment similar to that in Figure 7A, but instead of analyzing the cells 24 h after transfection, they were analyzed at four separate times over several days. During this time, numerous conditions change such as the growth state of the cells, metabolic activity, and levels of nutrients and growth factors in the culture media. Analysis of luciferase activity during this time frame revealed that in some cell types for some substrates, relative compartmentalization efficiencies changed (Figure 8). At one end of the spectrum were Cos-7 cells, in which little or no changes were observed over time for any of the five substrates analyzed. By contrast, parallel analyses of the identical substrates (and in fact, transfected from the same transfection mixtures as used for Cos-7 cells) showed systematic changes over time in the other cell types. For example, luciferase activity from PrP10-TF increased steadily over time (relative to Prl10-TF, to which everything was normalized) in N2a cells, but decreased steadily in HeLa cells (Figure 8). Similar variations (albeit with different patterns of change) were seen for several constructs, indicating that yet unidentified factors, perhaps related to cellular growth and/or metabolic conditions, can influence compartmentalization in trans in a substrate-selective and cell type-specific manner. Defining the identity of these factors and the mechanisms by which they exert their effects remain challenges for future studies.
Figure 8.
Modulation of substrate compartmentalization by changes in cellular metabolism. Four different cell types transfected with the indicated constructs as in Figure 7A were plated into four separate sets of 96-well plates. The plates were analyzed for luciferase activity on four successive days, beginning on the day of replating (after being allowed 6 h to recover). The results for each cell type are on separate graphs. In each instance, the measurements from any one day were normalized to the activity observed for Prl10-TF in that cell type, which was arbitrarily set to 100. The normalized activities for each of the 4 d are shown in bars of increasing darkness of gray (darkest bar is day 4). Thus, by definition, Prl10-TF values do not vary on these graphs from day to day. Relative to this arbitrary standard, changes for the other constructs can be observed in some, but not all instances in a cell-type and substrate-specific manner. In this experiment, cells began at ∼50% density (i.e., actively growing and dividing) and became confluent between the second and third set of measurements.
DISCUSSION
In this study, we have measured the steady-state fidelity of compartmentalization for a substrate directed to the secretory pathway in mammalian cells. These experiments were motivated by the goal of developing a coherent framework for an otherwise disparate set of observations implicating roles for secretory pathway proteins at alternative cellular locations. As an important first step toward this aim, we developed a reporter-based assay to specifically and sensitively detect the nonsegregated population of a signal sequence-containing protein in vivo. Several conclusions can be drawn from the observations by using this approach. First, readily detectable and physiologically relevant amounts of a signal sequence containing protein can reside in a functional state in the cytosol at steady state. Second, the relative amount of this cytosolic population varies substantially, dependent in part on the particular signal sequence used. Third, both translational and posttranslational mechanisms can contribute to cytosolic localization of a signal-containing protein. And fourth, although a mechanistic basis remains to be determined, changes in cell type and metabolic conditions of growth can cause a substrate-specific change in compartmentalization efficiency that can be modulated by the choice of signal sequence. Based on these findings, we propose that “inefficiencies” in compartmentalization may be one mechanism to generate alternative forms of secretory pathway proteins that are used for cellular benefit.
The Efficiency of Segregation
Despite encoding an N-terminal signal sequence, the percentage of TF that remains cytosolic in vivo ranges surprisingly widely between different signals. Among the most efficient by this assay is the well characterized Prl signal sequence, which nonetheless permits ∼5% noncompartmentalized Prl-TF or Prl10-TF under the conditions tested. Most other signals result in between ∼5 and 20% cytosolic TF, with some generating a surprising ∼50% in the cytosol. Given both the high baseline (∼5%) and maximal (∼50%) amounts in the cytosol, how can the results of the TF sequestration assay be reconciled with currently available information from native proteins? After all, the ad hoc characterization of the subcellular localization for a tremendous variety of proteins has led to the generally accepted (albeit formally untested) notion that compartmentalization into the secretory pathway is highly efficient.
Nearly all studies of subcellular localization have relied on biochemical fractionation or direct visualization with either immunofluorescent methods or more recently, fluorescent protein tags. Between 5 and 15% of a protein in an alternative compartment would easily be within the experimental limitations of most such methods; hence, only the major population would have been considered. Even 50% of a protein in the cytosol could be easily missed by visual methods because the cytosol occupies severalfold more volume than all compartments of the secretory pathway combined. Thus, the effective “dilution” of proteins in the cytosol could lead to a severalfold lower signal below that arising from concentration in a smaller compartment. For example, predominantly cytosolic proteins such as components of SRP or the COP I coat can seem by immunofluorescence to be localized to the ER and Golgi, respectively, due to a population that is associated at steady state. Furthermore, compartments such as the ER and plasma membrane can be difficult to definitively resolve from the cytosol by imaging methods due to their spatial overlap. Only if the cytosolic population also were concentrated, perhaps by subsequent targeting to another compartment, would it be easily recognized. Indeed, in nearly all cases where alternative populations of a protein have been observed, the second locale is a noncytosolic compartment such as mitochondria or nucleus (see Introduction). Thus, the inability to infer the existence of significant amounts of alternatively localized populations for many secretory pathway proteins based on conventional methodology is not in itself surprising nor worrisome.
A related issue is whether the results from the TF reporter used in this study can be corroborated with some of the respective native substrates. Among the various substrates whose signal sequences were analyzed, we have studied both PrP and CRT in greater detail in vivo (to be described elsewhere; see Rane et al., 2004, for an example with PrP). Based on the results of the normalized TF sequestration assay (Figure 5B), both of these are slightly less efficiently compartmentalized than Prl and generate ∼10-15% cytosolic population in MDCK cells under the conditions analyzed. When native PrP was analyzed in vivo by using pulse-chase analysis, we found that ∼10-20% of it was unglycosylated and subsequently degraded by the proteasome (Rane et al., 2004), suggesting that it is inefficiently translocated into the ER. Importantly, this unglycosylated population could be reduced to at least half when the Prl or Opn signal sequences were used (Figure 7B), suggesting that the relative differences in compartmentalization efficiencies observed for TF are recapitulated similarly with native PrP. For CRT, ∼10-20% of the total has been suggested to reside in the cytosol based on fractionation studies (Holaska et al., 2001; our unpublished data). Again, replacement of the CRT signal with the Prl signal, but not the PrP signal, reduced this cytosolic population for native CRT under some conditions in at least some cell types (Hegde, unpublished data). These results with PrP and CRT lend support to the existence of a cytosolic population for native proteins that corroborates the TF-based assay by using their isolated signal sequences.
Although the percentage of noncompartmentalized TF has been quantified with reasonable accuracy for several signal sequences and verified by other studies to apply to at least some of the native substrates, it would be premature to assume that the TF assay for every signal can be directly extrapolated similarly. This is because the steady-state distribution of substrate is presumably influenced substantially by features of the substrate itself, most notably its half-life in each of its locales. For example, although both native PrP and PrP-TF seem to generate ∼10-15% of a cytosolic population at the time of synthesis, a cytosolic pool of native PrP is essentially undetectable at steady state because of its short half-life (<30 min; Drisaldi et al., 2003) relative to cell surface PrP (>3 h half-life). Thus, although the results of the TF assay have provided insight into the range of efficiencies for signal-mediated compartmentalization, specific efficiencies for individual native substrates will still require analysis on a case-by-case basis. Nonetheless, the surprisingly high diversity of efficiencies among substrates helps to explain the various in vivo observations of alternative localizations for a growing number of secretory pathway proteins.
Mechanisms for Alternative Localizations
A majority of the nonsegregated population for many substrates is likely to result from inefficiencies in its targeting and translocation into the ER. This conclusion is derived indirectly from two observations. First, the frameshift mutation analysis suggests that for most of the substrates analyzed, the efficiency of translational initiation at the first start codon is very high, with only ∼1-2% of the total substrate deriving from initiation at the second potential start codon. Second, because the same TF reporter is used for each signal, the substrate becomes essentially identical to each other after the signal sequences are removed during translocation. Mechanisms for generating cytosolic TF after this point (such as retrotranslocation from the ER lumen) would therefore be common to all constructs. This means that all such mechanisms can together contribute at most the amount observed for the most efficiently compartmentalized substrate. Hence, the high degree of variability among the different signal sequences suggests that for most of them, the majority of the noncompartmentalized population was determined before the cotranslational removal of the signal sequence by signal peptidase. We therefore suggest that events occurring between the initiation of translation at the proper start codon and the completion of translocation most influence signal-mediated compartmentalization efficiency.
These events include SRP-mediated targeting of ribosome-nascent chains to the ER, transfer to the translocon, signal recognition by the Sec61 complex, insertion of the substrate into the translocation channel, opening of the channel toward the ER lumen, and productive folding and/or interactions with lumenal chaperones that commits the nascent chain to forward transport (Rapoport et al., 1996; Johnson and van Waes, 1999). Comparative studies among signals at each of these steps are largely lacking, particularly in vivo. In vitro studies have recently suggested some variability in signal-translocon interactions (Rutkowski et al., 2001; Kim et al., 2002), an observation consistent with the finding that some signal sequences have different requirements for the translocon components TRAM and TRAP (Voigt et al., 1996; Fons et al., 2003). The available in vivo studies have suggested that signals can differ in their kinetics of targeting to the ER membrane (Johnsson and Varshavsky, 1994; Goder et al., 2000). This is particularly interesting because excessive polypeptide elongation before targeting or insertion into the translocon can preclude subsequent translocation. Thus, it is feasible that one mechanism for generating different compartmentalization efficiencies among substrates is by differential kinetics of signal interactions with either SRP and/or the Sec61 complex. Because the amount of polypeptide that can be synthesized while still allowing translocation differs among substrates, the relationship between the targeting kinetics and translocation efficiency also may differ from one protein to another. This may have to do, in part, with the folding properties of the mature domain that immediately follows the signal, a notion consistent with our finding that including even 10 residues of the matched mature domain after the signal can influence segregation efficiency. Future work will be required to elucidate the relationship between signal sequence function and the mature domain to which it is attached. It is nonetheless already clear that this relationship is potentially complex, suggesting that signals cannot be viewed as either readily interchangeable or as the only determinant influencing translocation efficiency.
At present, the exact contribution of retrotranslocated TF to the cytosolic population observed at steady state is not known. Our data does, however, put an upper limit on the amount of cytosolic TF generated by such a mechanism at ∼5% of the total TF; this is the amount of cytosolic TF observed for the most efficient signal sequences by this assay (e.g., Prl, Opn, and VCAM). Cytosolic TF generated by this mechanism would need to escape degradation, which presumably would involve removal of any ubiquitin modifications that are attached during retrotranslocation, and subsequent refolding into a functional state in the cytosol. Although some bacterial toxins can take such a route to reside in the cytosol after having been in the secretory pathway (Tsai et al., 2002), it is not yet known whether other proteins can use similar mechanisms. Because this mechanism is dependent on features of the mature protein, the TF-based assay does not necessarily provide insight into the use of such a putative pathway for generating a cytosolic population of native secretory pathway proteins.
Physiological and Pathological Consequences
It is likely that for numerous secretory pathway proteins, a biologically significant proportion (e.g., at least 1-2%) of the total synthesized material can have access to the cytosol. Even if substrates underwent highly efficient translocation into the ER, they could still access the cytosol via translational mechanisms. For example, the experiment in Figure 4 demonstrates that an ideally designed translational start site is still bypassed by ∼2% of scanning ribosomes. In most instances, initiation at the next suitable start codon would result in synthesis of a signal-lacking substrate residing in the cytosol. This mechanism, together with our observation that many signals are in fact not maximally efficient and the fact that many secretory pathway proteins are abundant suggests that the cytosolic proportions are well within normal protein abundances. This cytosolic access has several potential nonmutually exclusive implications, three of which are particularly noteworthy.
First, for many proteins, the nonsegregated population may serve as a source of peptides for major histocompatibility complex class I antigen presentation. Indeed, it is estimated by some recent studies that up to ∼30% of all proteins are degraded very shortly after their synthesis and may be the major source of peptide antigens (Reits et al., 2000; Schubert et al., 2000; Turner and Varshavsky, 2000). Although these studies did not distinguish between the types of proteins being degraded, it is reasonable to postulate that a similarly large proportion of secretory pathway proteins also are converted into antigens. If this were the case, moderately inefficient step(s) of compartmentalization into the ER, as is suggested by the present study, would generate a cytosolic pool of substrates that can be converted by proteasomes into an abundant source of antigenic peptides. Such a mechanism is more appealing and cost-effective than a pathway using two translocation steps (one into and another out of the ER) simply for the purpose of constitutive generation of self-antigens.
Second, certain proteins that are inefficiently segregated may have independent functions in the cytosol (or other compartments accessed via the cytosol, such as mitochondria or the nucleus; see Introduction). This would provide a means for generating increased functional diversity from a single mRNA. For such alternative functions, the nonsegregated fraction could be a very small proportion (even substantially <1%) of the total synthesized product while still being well within the realm of normal cellular protein levels. Importantly, small modulations in compartmentalization inefficiency (e.g., from 1 to 5%) could be used by the cell to significantly impact the cytosolic function essentially independently of the major population (which would be nearly unchanged). Evidence that the cell has mechanisms to modulate compartmentalization in a substrate-specific manner is provided by the experiments in Figures 7 and 8. Thus, it is attractive to postulate that for some proteins, slight inefficiencies in translocation are functionally regulated for cellular benefit. The evolutionary conservation of many signal sequences (Kim et al., 2002) and their differential requirements for components of the translocation machinery (Ng et al., 1996; Voigt et al., 1996; Dunnwald et al., 1999; Wittke et al., 2002; Fons et al., 2003) is consistent with such a view.
Third, the constant presence of secretory pathway proteins in the cytosol also may have pathological consequences in some instances. This would be expected if the secretory pathway protein, or a portion of it, had the potential for adverse effects such as aggregation, misfolding, or inappropriate interactions with other proteins. Such possibilities have been suggested for several secretory pathway proteins such as PrP, APP, and ApoE (as discussed in Introduction), but it has been unclear whether sufficient amount of these proteins could ever escape their cotranslational segregation into the ER to have these effects. The present study suggests that the opportunity for cytosolic access of these proteins exists. Although under normal conditions, this population of potentially harmful proteins is efficiently degraded, the adverse consequences may be manifest under circumstances where the degradative capacity is partially compromised. Indeed, for PrP, inhibition of the proteasome pathway leads to a rapid increase in an aggregation-prone, potentially toxic cytosolic population of PrP that can be avoided by the use of a more efficient signal sequence (Rane et al., 2004).
Future studies will be aimed at determining both the physiological and pathological consequences of the inefficiencies of protein compartmentalization highlighted in the present study. It will be particularly important to determine the mechanistic basis of substrate-specific modulation of compartmentalization in different cell types and in response to changes in cellular conditions (e.g., as observed in Figure 7 and 8). It is anticipated that the small cytosolic populations of certain secretory pathway proteins are not inconsequential, but rather play important physiological and potentially pathological roles under certain circumstances.
Supplementary Material
Acknowledgments
We thank Christine Winters (National Institute of Child Health and Human Development, National Institutes of Health) for preparing the primary neuronal cultures, Erik Snapp and Jennifer Lippincott-Schwartz (National Institute of Child Health and Human Development, National Institutes of Health) for initially bringing the unexpected behavior of Prl-VSV-GFP in neurons to our attention, and Soo Jung Kim for first noticing translocation differences for PrP constructs in different cell types.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-06-0508. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-06-0508.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Addya, S., Anandatheerthavarada, H. K., Biswas, G., Bhagwat, S. V., Mullick, J., and Avadhani, N. G. (1997). Targeting of NH2-terminal-processed microsomal protein to mitochondria: a novel pathway for the biogenesis of hepatic mitochondrial P450MT2. J. Cell Biol. 139, 589-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anandatheerthavarada, H. K., Biswas, G., Mullick, J., Sepuri, N.B.V., Otvos, L., Pain, D., and Advadhani, N. G. (1999). Dual targeting of cytochrome P4502B1 to endoplasmic reticulum and mitochondria involves a novel signal activation by cyclic AMP-dependent phosphorylation at Ser128. EMBO J. 18, 5494-5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anandatheerthavarada, H. K., Biswas, G., Robin, M. A., and Avadhani, N. G. (2003). Mitochondrial targeting and a novel transmembrane arrest of Alzheimer's amyloid precursor protein impairs mitochondrial function in neuronal cells. J. Cell Biol. 161, 41-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, D. W., Perara, E., Lesser, C., and Lingappa, V. R. (1988). Sequences beyond the cleavage site influence signal peptide function. J. Biol. Chem. 263, 15791-15798. [PubMed] [Google Scholar]
- Bounhar, Y., Zhang, Y., Goodyer, C. G., and LeBlanc, A. (2001). Prion protein protects human neurons against Bax-mediated apoptosis. J. Biol. Chem. 276, 39145-39149. [DOI] [PubMed] [Google Scholar]
- Brecht, W. J., et al. (2004). Neuron-specific apolipoprotein E4 proteolysis is associated with increased tau phosphorylation in brains of transgenic mice. J. Neurosci. 24, 2527-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunagel, G., Shah, U., Schoen, R. E., and Getzenburg, R. H. (2003). Identification of calreticulin as a nuclear matrix protein associated with human colon cancer. J. Cell. Biochem. 89, 238-243. [DOI] [PubMed] [Google Scholar]
- Burns, K., Duggan, B., Atkinson, E. A., Famulski, K. S., Nemer, M., Bleackley, R. C., and Michalak, M. (1994). Modulation of gene expression by calreticulin binding to the glucocorticoid receptor. Nature 367, 476-480. [DOI] [PubMed] [Google Scholar]
- Coppolino, M., Leung-Hagesteinjn, C., Dedhar, S., and Wilkins, J. (1995). Inducible interaction of integrin alpha2beta1 with calreticulin. J. Biol. Chem. 270, 23132-23138. [DOI] [PubMed] [Google Scholar]
- Coppolino, M. G., Woodside, M. J., Demaurex, N., Grinstein, S., St-Arnaud, R., and Dedhar, S. (1997). Calreticulin is essential for integrin-mediated calcium signalling and cell adhesion. Nature 386, 843-847. [DOI] [PubMed] [Google Scholar]
- Crowley, K. S., Liao, S., Worrell, V. E., Reinhart, G. D., and Johnson, A. E. (1994). Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore. Cell 78, 461-471. [DOI] [PubMed] [Google Scholar]
- Dedhar, S., et al. (1994). Inhibition of nuclear hormone receptor activity by calreticulin. Nature 367, 480-483. [DOI] [PubMed] [Google Scholar]
- Deshaies, R. J., and Schekman, R. (1987). A yeast mutant defective at an early stage in import of secretory protein precursors into the endoplasmic reticulum. J. Cell Biol. 105, 633-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drisaldi, B., Stewart, R. S., Adles, C., Stewart, L. R., Quaglio, E., Biasini, E., Fioriti, L., Chiesa, R., and Harris, D. A. (2003). Mutant PrP. is delayed in its exit from the endoplasmic reticulum, but neither wild-type nor mutant PrP. undergoes retrotranslocation prior to proteasomal degradation. J. Biol. Chem. 278, 21732-21743. [DOI] [PubMed] [Google Scholar]
- Dunnwald, M., Varshavsky, A., and Johnsson, N. (1999). Detection of transient in vivo interactions between substrate and transporter during protein translocation into the endoplasmic reticulum. Mol. Biol. Cell 10, 329-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fons, R. D., Bogert, B. A., and Hegde, R. S. (2003). Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J. Cell Biol. 160, 529-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forough, R., Lindner, L., Partridge, C., Jones, B., Guy, G., and Clark, G. (2003). Elevated 80K-H protein in breast cancer: a role for FGF-1 stimulation of 80K-H. Int. J. Biol. Markers 18, 89-98. [DOI] [PubMed] [Google Scholar]
- Garcia, P. D., Ou, J., Rutter, W. J., and Walter, P. (1988). Targeting of the hepatitis B virus precore protein to the endoplasmic reticulum membrane: after signal peptide cleavage translocation can be aborted and the product released into the cytoplasm. J. Cell Biol. 106, 1093-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkika, D., Mahieu, F., Nilius, B., Hoenderop, J. G., and Bindels, R. J. (2004). 80K-H as a new Ca2+ sensor regulating the activity of the epithelial Ca2+ channel transient receptor potential cation channel V5 (TRPV5). J. Biol. Chem. 279, 26351-26357. [DOI] [PubMed] [Google Scholar]
- Goder, V., Crottet, P., and Spiess, M. (2000). In vivo kinetics of protein targeting to the endoplasmic reticulum determined by site-specific phosphorylation. EMBO J. 19, 6704-6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh, K. C., Lim, Y. P., Ong, S. H., Siak, C. B., Cao, X., Tan, Y. H., and Guy, G. R. (1996). Identification of p90, a prominent tyrosine-phosphorylated protein in fibroblast growth factor-stimulated cells, as 80K-H. J. Biol. Chem. 271, 5832-5838. [DOI] [PubMed] [Google Scholar]
- Goulet, B., Baruch, A., Moon, N., Poirier, M., Sansregret, L. L., Erickson, A., Bogyo, M., and Nepveu, A. (2004). A cathepsin L isoform that is devoid of a signal peptide localizes to the nucleus in S phase and processes the CDP/Cux transcription factor. Mol. Cell 14, 207-219. [DOI] [PubMed] [Google Scholar]
- Harris, F.M., et al. (2003). Carboxyl-terminal-truncated apolipoprotein E4 causes Alzheimer's disease-like neurodegeneration and behavioral deficits in transgenic mice. Proc. Natl. Acad. Sci. USA 100, 10966-10971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine, U., Burmester, J. K., Flanders, K. C., Danielpour, D., Munoz, Roberts, A. B., and Sporn, M. B. (1991). Localization of transforming growth factor-β1 in mitochondria of murine heart and liver. Cell Regul. 2, 467-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai, M., and Shimizu, N. (1990). Purification of two distinct proteins of approximate Mr 80,000 from human epithelial cells and identification as proper substrates for protein kinase C. Biochem. J. 270, 583-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holaska, J. M., Black, B. E., Love, D. C., Hanover, J. A., Leszyk, J., and Paschal, B. M. (2001). Calreticulin is a receptor for nuclear export. J. Cell Biol. 2001 152, 127-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y., Lui, X. Q., Wyss-Coray, T., Brecht, W. J., Sanan, D. A., and Mahley, R. W. (2001). Apolipoprotein E fragments present in Alzheimer's disease brains induce neurofibrillary tangle-like intracellular inclusions in neurons. Proc. Natl. Acad. Sci. USA 98, 8838-8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakova, P., Wang, G., Timchenko, L., Michalak, M., Pereira-Smith, O. M., Smith, J. R., and Timchenko, N. A. (2004). Competition of CUGBP1 and calreticulin for the regulation of p21 translation determines cell fate. EMBO J. 23, 406-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, A. E., and van Waes, M. A. (1999). The translocon: a dynamic gateway at the ER membrane. Annu. Rev. Cell Dev. Biol. 15, 799-842. [DOI] [PubMed] [Google Scholar]
- Johnsson, N., and Varshavsky, A. (1994). Ubiquitin-assisted dissection of protein transport across membranes. EMBO J. 13, 2686-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungnickel, B., and Rapoport, T. A. (1995). A post-targeting signal sequence recognition event in the endoplasmic reticulum membrane. Cell 71, 489-503. [DOI] [PubMed] [Google Scholar]
- Kanai, M., Goke, M., Tsunekawa, S., and Podolsky, D. K. (1997). Signal transduction pathway of human fibroblast growth factor receptor 3. Identification of a novel 66-kDa phosphoprotein. J. Biol. Chem. 272, 6621-6628. [DOI] [PubMed] [Google Scholar]
- Kim, S. J., Mitra, D., Salerno, J. R., and Hegde, R. S. (2002). Signal sequences control gating of the protein translocation channel in a substrate-specific manner. Dev. Cell 2, 207-217. [DOI] [PubMed] [Google Scholar]
- Kozak, M. (2002). Pushing the limits of the scanning mechanism for initiation of translation. Gene 299, 1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak, M. (1992). Regulation of translation in eukaryotic systems. Annu. Rev. Cell Biol. 8, 197-225. [DOI] [PubMed] [Google Scholar]
- Kurys, G., Tagaya, Y., Bamford, R., Hanover, J. A., and Waldmann, T. (2000). The long signal peptide isoform and its alternative processing direct the intracellular trafficking of interleukin-15. J. Biol. Chem. 275, 30653-30659. [DOI] [PubMed] [Google Scholar]
- Lai. J. F., Juang, S. H., Hung, Y. M., Cheng, H. Y., Cheng, T. L., Mostov, K. E., and Jou, T. S. (2003). An ecdysone and tetracycline dual regulatory expression system for studies on Rac1 small GTPase-mediated signaling. Am. J. Physiol. 285, C711-C719. [DOI] [PubMed] [Google Scholar]
- Leung-Hagesteijn, C. Y., Milankov, K., Michalak, M., Wilkins, J., and Dedhar, S. (1994). Cell attachment to extracellular matrix substrates is inhibited upon downregulation of expression of calreticulin, an intracellular integrin alpha-subunit-binding protein. J. Cell Sci. 107, 589-600. [PubMed] [Google Scholar]
- Lingappa, V. R., Devillers-Thiery, A., and Blobel, G. (1977). Nascent prehormones are intermediates in the biosynthesis of authentic bovine pituitary growth hormone and prolactin. Proc. Natl. Acad. Sci. USA 74, 2432-2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little, M. H., Wilkinson, L., Brown, D. L., Piper, M., Yamada, T., and Stow, J. L. (2001). Dual trafficking of Slit3 to mitochondria and cell surface demonstrates novel localization for Slit protein. Am. J. Physiol. 281, C486-C495. [DOI] [PubMed] [Google Scholar]
- Ljungberg, M. C., Dayanadan, R., Asuni, A., Rupniak, T. H., Anderton, A., and Lovestone, S. (2002). Truncated apoE forms tangle-like structures in a neuronal cell line. Mol. Neurosci. 13, 867-870. [DOI] [PubMed] [Google Scholar]
- Lu, Z. M., McLaren, R. S., Winters, C. A., and Ralston, E. (1998). Ribosome association contributes to restricting mRNAs to the cell body of hippocampal neurons. Mol. Cell. Neurosci. 12, 363-375. [DOI] [PubMed] [Google Scholar]
- Lustbader, J. W., et al. (2004). ABAD Directly Link Aβ to mitochondrial toxicity in Alzheimer's disease. Science 304, 448-452. [DOI] [PubMed] [Google Scholar]
- Ma, J., Wollmann, R., and Lindquist, S. (2002). Neurotoxicity and neurodegeneration when PrP accumulates in the cytosol. Science 298, 1781-1785. [DOI] [PubMed] [Google Scholar]
- Mason, N., Ciufo, L. F., and Brown, J. D. (2000). Elongation arrest is a physiologically important function of signal recognition particle. EMBO J. 19, 4164-4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, M. L., Vyklicky, Jr., L., and Westbrook, G. L. (1989). Modulation of excitatory amino acid receptors by group IIIB metal cations in cultured mouse hippocampal neurones. J. Physiol. 415, 320-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov, A., Jr., Latawiec, D., Wille, H., Bouzamondo-Bernstein, E., Legname, G., Williamson, R. A., Burton, D., DeArmond, S. J., Prusiner, S. B., and Peters, P. J. (2003). Cytosolic prion protein in neurons. J. Neurosci. 23, 7183-7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, D. T., Brown, J. D., and Walter, P. (1996). Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J. Cell Biol. 134, 269-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou, J., Yeh, C., and Yen, B. (1989). Transport of hepatitis B virus precore protein into the nucleus after cleavage of its signal peptide. J. Virol. 63, 5238-5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presley, J. F., Cole, N. B., Schroer, T. A., Hirschberg, K., Zaal, K. J., and Lippincott-Schwartz, J. (1997). ER-to-Golgi transport visualized in living cells. Nature 389, 81-85. [DOI] [PubMed] [Google Scholar]
- Ramsamooj, P., Notario, V., and Dritschilo, A. (1995). Enhanced expression of calreticulin in the nucleus of radioresistant squamous carcinoma cells in response to ionizing radiation. Cancer Res. 55, 3016-3021. [PubMed] [Google Scholar]
- Rane, N. S., Yonkovich, J. L., and Hegde, R. S. (2004). Protection from cytosolic prion protein toxicity by modulation of protein translocation. EMBO J. (in press.) [DOI] [PMC free article] [PubMed]
- Rapoport, T. A., Jungnickel, B., and Kutay, U. (1996). Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu. Rev. Biochem. 65, 271-303. [DOI] [PubMed] [Google Scholar]
- Reits, E. A., Vos, J. C., Gromme, M., and Neefjes, J. (2000). The major substrates for TAP in vivo are derived from newly synthesized proteins. Nature 404, 774-778. [DOI] [PubMed] [Google Scholar]
- Roucou, X., Guo, Q., Zhang, Y., Goodyer, C. G., and LeBlanc, A. C. (2003). Cytosolic prion protein is not toxic and protects against Bax-mediated cell death in human primary neurons. J. Biol. Chem. 278, 40877-40881. [DOI] [PubMed] [Google Scholar]
- Rutkowski, D. T., Lingappa, V. R., and Hegde, R.S. (2001). Substrate-specific regulation of the ribosome-translocon junction by N-terminal signal sequences. Proc. Natl. Acad. Sci. USA 98, 7823-7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai, K., Hirai, M., Minoshima, S., Kudoh, J., Fukuyama, R., and Shimizu, N. (1989). Isolation of cDNAs encoding a substrate for protein kinase C: nucleotide sequence and chromosomal mapping of the gene for a human 80K protein. Genomics 5, 309-315. [DOI] [PubMed] [Google Scholar]
- Schubert, U., Anton, L. C., Gibbs, J., Norbury, C. C., Yewdell, J. W., and Bennink, J. R. (2000). Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404, 770-774. [DOI] [PubMed] [Google Scholar]
- Song, W., Raden, D., Mandon, E. C., and Gilmore, R. (2000). Role of Sec61-alpha in the regulated transfer of the ribosome-nascent chain complex from the signal recognition particle to the translocation channel. Cell 100, 333-343. [DOI] [PubMed] [Google Scholar]
- Tagaya, Y., Kurys, G., Thies, T. A., Losi, J. M., Azimi, N., Hanover, J. A., Bamford, R. N., and Waldmann, T. A. (1997). Generation of secretable and nonsecretable interleukin 15 isoforms through alternate usage of signal peptides. Proc. Natl. Acad. Sci. USA 94, 14444-14449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta, E. S., Simons, J. F., and Helenius, A. (1996). Endoplasmic reticulum glucosidase II is composed of a catalytic subunit, conserved from yeast to mammals and a tightly bound noncatalytic HDEL-containing subunit. J. Biol. Chem. 271, 27509-27516. [DOI] [PubMed] [Google Scholar]
- Tsai, B., Ye, Y., and Rapoport, T. A. (2002). Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat. Rev. Mol. Cell. Biol. 3, 246-255. [DOI] [PubMed] [Google Scholar]
- Turner, G. C., and Varshavsky, A. (2000). Detecting and measuring cotranslational protein degradation in vivo. Science 289, 2117-2120. [DOI] [PubMed] [Google Scholar]
- Voigt, S., Jungnickel, B., Hartmann, E., and Rapoport, T. A. (1996). Signal sequence-dependent function of the TRAM protein during early phases of protein transport across the endoplasmic reticulum membrane. J. Cell Biol. 134, 25-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne, G. (1985). Signal sequences. The limits of variation. J. Mol. Biol. 184, 99-105. [DOI] [PubMed] [Google Scholar]
- Walter, P., and Johnson, A. E. (1994). Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell Biol. 10, 87-119. [DOI] [PubMed] [Google Scholar]
- Wittke, S., Dunnwald, M., Albertsen, M., and Johnsson, N. (2002). Recognition of a subset of signal sequences by Ssh1p, a Sec61p-related protein in the membrane of endoplasmic reticulum of yeast Saccharomyces cervisiae. Mol. Biol. Cell 13, 2223-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin, S. L., and Walter, P. (1989). Signal recognition particle mediates a transient elongation arrest of preprolactin in reticulocyte lysate. J. Cell Biol. 109, 2617-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yedidia, Y., Horonchik, L., Tzaban, S., Yanai, A., and Taraboulos, A. (2001). Proteasomes and ubiquitin are involved in the turnover of the wild-type prion protein. EMBO J. 20, 5383-5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, G., Lee, H., Jung, Y., Yu, E., Moon, H., Song, K., and Lee, I. (2000). Nuclear matrix of calreticulin in hepatocellular carcinoma. Cancer Res. 60, 1117-1120. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.