Abstract
Multiple sclerosis (MS) is an autoimmune disease with both genetic and environmental risk factors. Recent studies indicate that childhood and adolescent obesity double the risk of MS, but this association may reflect unmeasured confounders rather than causal effects of obesity. We used separate-sample Mendelian randomization to estimate the causal effect of body mass index (BMI) on susceptibility to MS. Using data from non-Hispanic white members of the Kaiser Permanente Medical Care Plan of Northern California (KPNC) (2006–2014; 1,104 cases of MS and 10,536 controls) and a replication data set from Sweden (the Epidemiological Investigation of MS (EIMS) and the Genes and Environment in MS (GEMS) studies, 2005–2013; 5,133 MS cases and 4,718 controls), we constructed a weighted genetic risk score using 97 variants previously established to predict BMI. Results were adjusted for birth year, sex, education, smoking status, ancestry, and genetic predictors of MS. Estimates in KPNC and Swedish data sets suggested that higher genetically induced BMI predicted greater susceptibility to MS (odds ratio = 1.13, 95% confidence interval: 1.04, 1.22 for the KPNC sample; odds ratio = 1.09, 95% confidence interval: 1.03, 1.15 for the Swedish sample). Although the mechanism remains unclear, to our knowledge, these findings support a causal effect of increased BMI on susceptibility to MS for the first time, and they suggest a role for inflammatory pathways that characterize both obesity and the MS disease process.
Keywords: body mass index, genetic instrumental variables, Mendelian randomization, multiple sclerosis, obesity
Multiple sclerosis (MS) is characterized as an immune system–mediated demyelinating disorder with widespread axonal degeneration occurring throughout the disease process (1), resulting in significant disability and decreased quality of life. Strong evidence supports the contribution of both genetic and environmental factors to MS susceptibility (2). Investigators have identified several genetic variants, including the *15:01 allele of the human leukocyte antigen (HLA) antigen D–related β1 subunit (DRB1) gene (HLA-DRB1*15:01) within the major histocompatibility complex (3, 4) and 110 non-HLA variants (4). Environmental risk factors associated with MS include exposure to tobacco smoke and low levels of vitamin D (5).
Recently, obesity has emerged as a significant risk factor for MS. Associations between MS and body mass index (BMI) at age 18 years (6), at age 20 years (7), and during the ages 20–29 years (8) have been observed; individuals with a BMI of at least 30 demonstrated a more than 2-fold increased risk of MS compared with persons with a BMI of at least 18.5 and less than 21 (within the normal weight range according to BMI). The relationship was also confirmed using retrospective assessments of body size, with the strongest association being observed at age 25 years (9). Additionally, childhood obesity and risks of both pediatric (10) and later-onset (11) MS have been reported. Research has also indicated that HLA genes interact with BMI during adolescence to increase the risk of MS (12). Although investigators have hypothesized about various mechanisms that might mediate the association between obesity and MS, a causal relationship remains to be confirmed. Observational findings cannot exclude the possibility that weight change preceding a diagnosis may be associated with a prodromal period of disease—that is, a decrease in physical activity due to MS symptoms before diagnosis may lead to increased weight and thus bias findings.
The genetic basis of obesity has been demonstrated by studies revealing multiple variants throughout the genome associated with obesity-related traits. A recent genome-wide association study (GWAS) identified 97 significant loci that explained 2.7% of the variance associated with BMI (13). To our knowledge, no study has examined the relationship between genes associated with BMI and MS. Using genetic predictors of an exposure of interest as independent variables in an observational study corresponds with a “Mendelian randomization” (MR) study design, which avoids bias from reverse causation and potential confounders, therefore strengthening causal inferences with complex exposures such as BMI.
We applied instrumental variable analysis in an MR framework to estimate the causal relationship between BMI and susceptibility to MS using a BMI genetic risk score (GRS) comprising 97 variants (13) in 2 populations. We further examined whether any variants appeared to directly influence MS via mechanisms unrelated to BMI.
METHODS
KPNC participants
MS cases and controls were recruited from members of Kaiser Permanente Medical Care Plan of Northern California (KPNC). KPNC is an integrated health care–services delivery system with 3.2 million members that covers about 25%–30% of the population of a 22-county service area and is the largest health-care provider in Northern California. Membership is objectively representative of the general population; however, persons in impoverished neighborhoods are underrepresented (14).
This study was restricted to individuals with self-identified white (non-Hispanic) race/ethnicity, the population with the highest prevalence of MS. Eligible KPNC cases were defined as individuals who had received a diagnosis of MS from a neurologist (International Classification of Diseases, Ninth Revision, code 340.x; 94.7% of cases had at least 2 MS diagnoses by a neurologist), were aged 18–69 years, and were members of KPNC at initial contact. Recruitment began in mid-2006; a total of 3,293 potential MS cases were reviewed by KPNC neurologists, who approved contact with 2,823 (86%) at the time of the data freeze (August 2014). Diagnoses were validated using published diagnostic criteria and review of electronic health records (15, 16).
Controls were current KPNC members without a diagnosis of MS or a related condition (optic neuritis, transverse myelitis, or demyelination disease), confirmed through electronic records, and who reported white (non-Hispanic) race/ethnicity. Controls were matched to cases on age, sex, and zip code. Potential study participants were contacted by mail, with a follow-up phone call to explain the study and procedures. The participation rate was approximately 80% for cases and 66% for controls. Genetic data were available for approximately 80% of study participants.
Additional controls included individuals in the Genetic Epidemiology Research on Aging (GERA) cohort who participated in the KPNC Research Program on Genes, Environment, and Health. The KPNC program was established to research genetic and environmental influences on health and disease and is described in more detail elsewhere (17). Recruitment began in 2007. GERA cohort members gave broad, written consent and provided a saliva sample for DNA extraction (17). Approximately 77% of the cohort returned completed consent forms for placement in the Database of Genotypes and Phenotypes, which resulted in a final sample size of 78,486 participants. From these participants, we selected a subset of 12,605 self-reported non-Hispanic white individuals without evidence of MS in their electronic health records and matched them to cases on sex and age (within 2 years) at a 10:1 ratio. Study protocols were approved by the institutional review boards of KPNC and the University of California, Berkeley.
Swedish participants
Data were collected from 2 population-based case-control studies, one a study of MS incidence (Epidemiological Investigation of MS (EIMS)) and the other a study of MS prevalence (Genes and Environment in MS (GEMS)). The EIMS Study (2005–2013) inclusion criteria were being aged 16–70 years, having been diagnosed with MS according to the McDonald criteria (15, 16) within the past 2 years, and having the ability to understand the Swedish language. GEMS Study participants were identified from the Swedish National MS registry, fulfilled the McDonald criteria (15, 16), and were recruited during 2009–2011. For both studies, controls were randomly chosen from the population register and matched to cases by sex, age at inclusion in the study, and region of residence. Two controls were matched to each case in the EIMS Study, and 1 control was matched to each case in the GEMS Study. All participants in the EIMS Study were distinct from those in the GEMS Study. Ethical approval for both studies was obtained from the Regional Ethical Review Board in Stockholm at the Karolinska Institutet, and participants provided written informed consent. Details on the study's design have been published elsewhere (7, 12). The participation rate in the EIMS Study was 92% for cases and 67% for controls, and participation in the GEMS Study was 82% for cases and 66% for controls. Genotyping data were available for 75% of EIMS participants and 91% of GEMS participants. Data for 6,335 cases and 5,762 controls were available from the Swedish studies.
Exposure assessment
KPNC participants completed a computer-assisted telephone interview administered by trained staff interviewers and composed of questions related to various events and exposures, as described elsewhere (8). GERA controls completed a survey related to health behaviors, sociodemographic information, and diagnoses. KPNC study participants reported their highest and lowest (nonpregnancy) weights at ages 20–29 years. The mean weights of the KPNC participants during their twenties were calculated by averaging the highest and lowest weights reported. GERA controls reported their weight at age 18 years.
Exposure assessment in the Swedish studies was done through an extensive questionnaire that participants completed at home. Incomplete questionnaires were completed by mail or telephone. The questionnaire covered demographic and environmental/lifestyle factors, including current height and weight at age 20 years. The details have been provided elsewhere (7, 12).
Each participant's BMI was calculated by dividing weight in kilograms (or mean weight for KPNC participants) by squared height in meters (reported at time of interview). KPNC and Swedish participants provided blood or saliva samples for genotyping. For details on platforms and quality control, see the Appendix.
Statistical analyses
A weighted genetic risk score (wGRS) for MS risk variants was calculated for each individual. The GRS weights risk alleles by the logarithm of the odds ratio for each of the 110 non-HLA loci for MS susceptibility identified through the most recent MS GWAS (4). The wGRS was calculated by multiplying the number of risk alleles for each locus by the weight for that variant, and then taking the sum across the 110 loci. One variant for KPNC (rs201202118) and 2 variants for the Swedish studies (rs2028597, rs6874308) were missing.
The BMI GRS was calculated by multiplying the number of risk alleles for each BMI-related locus (13) by the weight (defined as the beta coefficient from the BMI GWAS) for that variant and then taking the sum across the 97 loci. One variant was missing for the Swedish data set (rs2245368). To correct for the fact that the GWAS by Locke et al. (13) was based on a residualized BMI transformed into standard deviation units—and to express this weight in terms of BMI units—we multiplied by a constant of 4.95. This value was estimated using the regression analysis among persons of European descent in the Health and Retirement Study, as reported in the GWAS (13). With this transformation, each unit change in the weighted BMI GRS corresponds with an anticipated 1-unit change in BMI, allowing for direct interpretation of the estimates as the estimated effect of a unit increase in the genetically predicted BMI on the odds of MS (BMI GRS ranges: 8.64–14.52 in KPNC and 8.41–14.33 in Sweden).
After quality control and removal of population outliers, a total of 2,163 individuals (1,104 cases and 804 controls) with genetic data were available from the KPNC sample, as well as an additional 9,732 controls from the GERA Study, for a total of 1,104 cases and 10,536 controls. The final data set for Swedish participants included 5,133 cases and 4,718 controls. Demographic differences between cases and controls were compared using χ2 tests and independent sample t tests where appropriate. Linear regression analysis was used to demonstrate the association of each GRS with BMI during young adulthood (during ages 20–29 years for KPNC, at age 18 years for GERA, and at age 20 years for the Swedish studies) and to test the assumption that the BMI GRS is not associated with confounding factors. MR analysis was performed by regressing MS case status on BMI GRS.
In addition, we evaluated evidence that any of the BMI variants had a direct effect on MS, implying a violation of the MR assumption that there is no direct effect of the instrument on the outcome (18). Direct effects were analyzed using regression-based mediation to estimate the controlled direct effect for changes in exposure level (19). Analyses examined 97 BMI variants, measuring the effect on MS status (case/control) of having no alleles that increase BMI risk (a = 0) versus having 2 risk alleles (a = 1) at each locus. The mediator was specified as BMI during young adulthood and set to 22.0, or the mean of the “normal” healthy BMI range as defined by the World Health Organization. Analyses were bootstrapped with 100 replications, and estimates from both data sets (KPNC and Swedish studies) were used to conduct a random-effects meta-analysis.
Odds ratios with 95% confidence intervals were estimated. All analyses controlled for established risk factors associated with MS susceptibility that were measured in both the KPNC and Swedish populations, including sex, year of birth, smoking status, college education, HLA-DRB1*15:01 status, wGRS of non-HLA MS risk variants, and ancestry as derived from principal components analysis. Swedish analyses additionally controlled for study type (EIMS vs. GEMS). Analyses were conducted in PLINK (http://pngu.mgh.harvard.edu/~purcell/plink), STATA (StataCorp LP, College Station, Texas), and R (R Foundation for Statistical Computing, Vienna, Austria). This study was focused on a single hypothesis established a priori: BMI is causally associated with MS as represented by a BMI GRS. Therefore, we report 95% confidence intervals and use an α = 0.05 threshold for 2-sided tests of statistical significance.
RESULTS
There were significant differences between cases and controls with respect to smoking status, college graduation, HLA-DRB1*15:01 status, and wGRS in the KPNC and Swedish studies (Table 1). BMI during young adulthood was significantly higher in cases than in controls, as was the BMI GRS, in both data sets. The BMI GRS was associated with college education and wGRS in the KPNC members and with smoking in both populations. The association between the BMI GRS and BMI in young adulthood was similar in men and women (Table 2). For every unit increase in the GRS, BMI in young adulthood increased on average 0.64 units in the KPNC sample and 0.54 units in the Swedish data set, where a value of 1.0 would indicate that the effect of the variants on BMI in our populations was the same as the effect in the GWAS from which the weights were drawn.
Table 1.
Demographic and Disease Characteristics of Non-Hispanic White Persons Diagnosed With Multiple Sclerosis and Controls in Northern California (2006–2014) and Sweden (2005–2013)a
| Characteristic | KPNC | Sweden | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MS Cases (n = 1,104) | Controls (n = 10,536) | P Valueb | P Valuec | MS Cases (n = 5,133) | Controls (n = 4,718) | P Valueb | P Valuec | |||||||||
| No. | % | Mean (SD) | No. | % | Mean (SD) | No. | % | Mean (SD) | No. | % | Mean (SD) | |||||
| Birth year | 1958 (8.93) | 1958 (8.95) | 0.67 | 0.72 | 1960 (13.38) | 1961 (13.53) | <0.001 | 0.38 | ||||||||
| Sex | 0.29 | 0.83 | <0.001 | 0.27 | ||||||||||||
| Female | 882 | 79.89 | 8,555 | 81.20 | 3,741 | 73.17 | 3,592 | 76.13 | ||||||||
| Male | 222 | 20.11 | 1,981 | 18.80 | 1,392 | 27.22 | 1,126 | 23.87 | ||||||||
| Smoker | <0.001 | <0.001 | <0.001 | 0.003 | ||||||||||||
| Never smoker | 555 | 50.36 | 7,004 | 67.93 | 2,163 | 42.30 | 2,371 | 50.25 | ||||||||
| Ever smoker | 547 | 49.64 | 3,307 | 32.07 | 2,853 | 55.80 | 2,146 | 45.49 | ||||||||
| College graduate | <0.001 | 0.02 | 0.001 | 0.10 | ||||||||||||
| Yes | 488 | 44.20 | 3,677 | 35.84 | 3,725 | 72.85 | 3,279 | 69.50 | ||||||||
| No | 616 | 55.80 | 6,583 | 64.16 | 1,397 | 27.32 | 1,433 | 30.37 | ||||||||
| HLA-DRB1*15:01 status | <0.001 | 0.98 | <0.001 | 0.73 | ||||||||||||
| 0 alleles | 518 | 46.92 | 7,752 | 73.60 | 2,144 | 41.93 | 3,321 | 70.39 | ||||||||
| 1–2 alleles | 586 | 53.08 | 2,781 | 26.40 | 2,989 | 58.46 | 1,397 | 29.61 | ||||||||
| wGRS of non-HLA risk variants | 12.86 (0.68) | 12.47 (0.68) | <0.001 | 0.03 | 12.48 (0.67) | 12.08 (0.69) | <0.001 | 0.05 | ||||||||
| BMI in young adulthoodd | 22.97 (4.37) | 21.47 (3.30) | <0.001 | <0.001 | 21.97 (3.56) | 21.68 (3.20) | <0.001 | <0.001 | ||||||||
| BMI GRS | 11.55 (0.82) | 11.47 (0.81) | 0.002 | 11.56 (0.82) | 11.50 (0.81) | 0.0002 | ||||||||||
Abbreviations: BMI, body mass index; GRS, genetic risk score; HLA, human leukocyte antigen; HLA-DRB1*15:01, *15:01 allele of the human leukocyte antigen (HLA) antigen D–related β1 subunit (DRB1) gene; KPNC, Kaiser Permanente Medical Care Plan of Northern California; MS, multiple sclerosis; SD, standard deviation; wGRS, weighted genetic risk score.
a Race/ethnicity was defined by genetic ancestry. Percentages may not total 100 due to missing values.
bP value from a t test (continuous variables) or χ2 test (categorical variables) of the difference between MS cases and controls for each cohort (KPNC and Swedish studies).
cP value for linear regression analysis of BMI GRS and each variable.
d BMI in young adulthood was defined as BMI at age 18 years (Genetic Epidemiology Research on Aging), during ages 20–29 years (KPNC), or at age 20 years (Sweden) and was calculated as weight (kg)/height (m)2.
Table 2.
Association Between Body Mass Index–Related Genes and Self-Reported Body Mass Index in Young Adulthooda, Northern California (2006–2014) and Sweden (2005–2013)
| FTO and Genetic Risk Score | KPNC | Sweden | ||||
|---|---|---|---|---|---|---|
| Coefficient | 95% CI | R2, % | Coefficient | 95% CI | R2, % | |
| FTO only | 0.29 | 0.20, 0.39 | 0.4 | 0.27 | 0.18, 0.36 | 0.3 |
| Women | 0.23 | 0.13, 0.34 | 0.2 | 0.29 | 0.18, 0.40 | 0.4 |
| Men | 0.51 | 0.31, 0.72 | 1.2 | 0.22 | 0.06, 0.38 | 0.2 |
| BMI GRS | 0.64 | 0.56, 0.72 | 2.3 | 0.54 | 0.45, 0.63 | 1.5 |
| Women | 0.61 | 0.53, 0.70 | 2.1 | 0.51 | 0.40, 0.61 | 1.3 |
| Men | 0.74 | 0.57, 0.91 | 3.6 | 0.61 | 0.45, 0.76 | 2.3 |
Abbreviations: BMI, body mass index; CI, confidence interval; FTO, fat mass and obesity–associated gene; GRS, genetic risk score; KPNC, Kaiser Permanente Medical Care Plan, Northern California Region.
a The results of linear regression analyses of BMI-related genes and self-reported BMI in young adulthood were all significant (P < 0.001). BMI in young adulthood was defined as BMI at age 18 years (Genetic Epidemiology Research on Aging), during ages 20–29 years (KPNC), or at age 20 years (Sweden) and was calculated as weight (kg)/height (m)2.
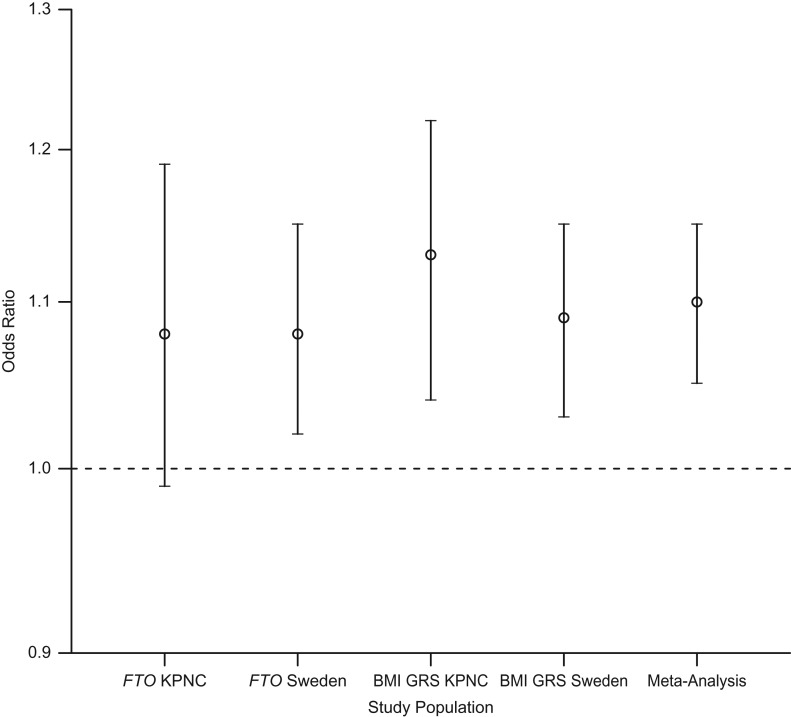
The BMI GRS significantly predicted odds of MS after controlling for sex, year of birth, education, ancestry, smoking status, wGRS, and number of HLA-DRB1*15:01 alleles in the KPNC sample participants (odds ratio (OR) = 1.13, 95% confidence interval (CI): 1.04, 1.22; Table 3 and Figure 1). Similar results were found for the Swedish studies after controlling for the same covariates plus study type (OR = 1.09, 95% CI: 1.03, 1.15) and in the meta-analysis of the findings from both studies (OR = 1.10, 95% CI: 1.05, 1.15). No evidence of heterogeneity between populations was observed (I2 = 0.0%; P = 0.47).
Table 3.
Results From a Multivariate Regression Analysis of the Effect of Body Mass Index on Susceptibility to Multiple Sclerosis Carried Out Using Body Mass Index Genetic Risk Scores, Northern California (2006–2014) and Sweden (2005–2013)
| Variable | KPNC | Sweden | ||||
|---|---|---|---|---|---|---|
| OR | 95% CI | P Valuea | OR | 95% CI | P Valuea | |
| BMI GRS | 1.13 | 1.04, 1.22 | 0.004 | 1.09 | 1.03, 1.15 | 0.002 |
| Female sex | 0.94 | 0.80, 1.12 | 0.50 | 0.80 | 0.72, 0.89 | <0.001 |
| Birth year | 1.00 | 1.00, 1.01 | 0.07 | 1.01 | 1.00, 1.01 | 0.006 |
| No college education | 0.77 | 0.67, 0.88 | <0.001 | 0.91 | 0.83, 1.01 | 0.07 |
| Ever smoker | 2.02 | 1.76, 2.31 | <0.001 | 1.49 | 1.36, 1.62 | <0.001 |
| HLA-DRB1*15:01 status | 3.55 | 3.10, 4.06 | <0.001 | 3.43 | 3.14, 3.76 | <0.001 |
| wGRS for non-HLA MS risk variants | 2.32 | 2.10, 2.57 | <0.001 | 2.36 | 2.20, 2.52 | <0.001 |
Abbreviations: BMI, body mass index; CI, confidence interval; GRS, genetic risk score; HLA, human leukocyte antigen; HLA-DRB1*15:01, *15:01 allele of the human leukocyte antigen (HLA) antigen D–related β1 subunit (DRB1) gene; KPNC, Kaiser Permanente Medical Care Plan of Northern California; MS, multiple sclerosis; OR, odds ratio; wGRS, weighted genetic risk score.
aP value for a logistic regression model that adjusted for ancestry using principal components. For the Swedish studies, the model additionally controlled for study type.
Figure 1.
Odds ratios for the effect of body mass index (BMI) on susceptibility to multiple sclerosis (MS), obtained using genetic variants as instrumental variables in populations in Northern California (2006–2014) and Sweden (2005–2013). Results were adjusted for smoking, education, birth year, HLA-DRB1*15:01 status, weighted genetic risk score (GRS) for non-HLA MS-risk variants, genetic ancestry, and sex. Results from Sweden were additionally adjusted for study type. Odds ratios were significant (P < 0.01) for fat mass and obesity–associated gene (FTO) Sweden, BMI GRS Kaiser Permanente Medical Care Plan of Northern California (KPNC), BMI GRS Sweden, and meta-analysis. Bars, 95% confidence interval. HLA-DRB1*15:01, *15:01 allele of the human leukocyte antigen (HLA) antigen D–related β1 subunit (DRB1) gene.
Results stratified by sex showed a significant association in women but not men among the KPNC participants (for women, OR = 1.14, 95% CI: 1.04, 1.25; for men, OR = 1.11, 95% CI: 0.92, 1.33) and in the Swedish studies (for women, OR = 1.09, 95% CI: 1.03, 1.16; for men, OR = 1.08, 95% CI: 0.97, 1.20). However, the wide confidence intervals suggest that this is consistent with chance given the smaller sample of male participants.
The meta-analysis results for the direct effect estimates showed that 5 variants associated with increased BMI in the literature exhibited a significant controlled direct effect on MS susceptibility after adjustment for covariates (Table 4). Four variants were positively associated with MS, and 1 variant was inversely associated with MS.
Table 4.
Significant Controlled Direct Effects of Gene Variants Related to Body Mass Index on Susceptibility to Multiple Sclerosis, Northern California (2006–2014) and Sweden (2005–2013)
| Single-Nucleotide Polymorphism | Chromosome | Gene | KPNC | Sweden | Meta-Analysis | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | P Valuea | OR | 95% CI | P Valuea | OR | 95% CI | |||
| Significant Controlled Direct Effect Associated With an Increased Risk of MS | ||||||||||
| rs11126666 | 2 | KCNK3 | 1.20 | 0.96, 1.48 | 0.002 | 1.13 | 0.93, 1.29 | 0.001 | 1.15 | 1.01, 1.30 |
| rs2112347 | 5 | POC5 | 1.05 | 0.83, 1.28 | 0.32 | 1.16 | 1.04, 1.30 | <0.001 | 1.13 | 1.02, 1.25 |
| rs1558902 | 16 | FTO | 1.13 | 0.94, 1.36 | 0.01 | 1.17 | 1.02, 1.35 | <0.001 | 1.16 | 1.03, 1.28 |
| rs7243357 | 18 | GRP | 1.14 | 0.80, 1.49 | 0.06 | 1.26 | 1.06, 1.46 | <0.001 | 1.23 | 1.06, 1.40 |
| Significant Controlled Direct Effect Associated With a Decreased Risk of MS | ||||||||||
| rs7599312 | 2 | ERBB4 | 0.77 | 0.62, 0.97 | <0.001 | 0.91 | 0.78, 1.09 | 0.01 | 0.85 | 0.71, 0.98 |
Abbreviations: BMI, body mass index; CI, confidence interval; HLA, human leukocyte antigen; HLA-DRB1*15:01, *15:01 allele of the human leukocyte antigen (HLA) antigen D–related β1 subunit (DRB1) gene; KPNC, Kaiser Permanente Medical Care Plan of Northern California; MS, multiple sclerosis; OR, odds ratio.
aP value for logistic regression analyses that adjusted for sex, smoking status, education, birth year, HLA-DRB1*15:01 status, weighted genetic risk score for 110 non-HLA variants related to MS risk, and genetic ancestry; for the Swedish studies, results were additionally adjusted for study type. BMI was calculated as weight (kg)/height (m)2, and the mediator was defined as BMI during young adulthood (set at 22.0). All analyses were bootstrapped with 100 replications. Meta-analysis was conducted with bootstrapped estimates, with adjustment for random effects.
Given the potential violation of the assumption that there is no direct effect of the instrument on the outcome, we repeated the MR analysis using a BMI GRS that excluded the 5 variants for which we found evidence of a possible direct effect on MS. The findings demonstrated a consistent, significant association between the revised 92-variant BMI GRS and MS after controlling for covariates in both KPNC and Swedish participants (for KPNC, OR = 1.13, 95% CI: 1.04, 1.23; for the Swedish studies, OR = 1.09, 95% CI: 1.03, 1.15).
We also adjusted the MR analysis of 92 variants for BMI during young adulthood to examine evidence that the GRS had effects on MS that were not mediated by BMI. This adjustment attenuated our findings and reduced the significance of the association in the KPNC participants (OR = 1.05, 95% CI: 0.97, 1.15) and the Swedish data set (OR = 1.06, 95% CI: 1.00, 1.12).
In order to test the assumptions of the MR model for the BMI GRS, we conducted overidentification tests to evaluate the null hypothesis that effect estimates from multiple instrumental variables are identical (18). The 92 variants without a direct effect on the outcome were randomly split into 5 separate instruments. Estimates suggested the same direction of causal effect, with odds ratios ranging from 1.04 to 1.22 in KPNC participants and from 1.01 to 1.25 in the Swedish studies.
DISCUSSION
To our knowledge, this study was the first to examine the relationship between BMI and MS using MR. We also present novel results suggesting that 5 variants previously established to predict BMI may have direct effects on MS. None of the direct effect variants or genes—and none of the variants or genes from the 97 BMI-related variants analyzed by Locke et al.—overlap with the established 110 non-HLA MS risk loci. Our results suggest a causal association between higher BMI and MS susceptibility, and they identify new genetic variants that may directly influence MS onset.
The most widely studied obesity-related genetic variant is the fat mass and obesity–associated gene (FTO) (20, 21). Variants in FTO have been found to be associated with various cancers (22–24), Alzheimer's disease and dementia (25), reduced brain volume in healthy elderly individuals (26), and cognitive decline in healthy adults (27). Although one study found that an FTO risk allele was correlated with significantly increased homocysteine levels in MS cases compared with controls (28), to our knowledge, no study has previously examined the relationship between BMI genes and MS. Our study found that FTO alone slightly increased the risk of MS and that there was some evidence of a direct effect of FTO on MS susceptibility.
Since the discovery of FTO, additional genes have been found to be associated with obesity-related traits. In a recent study, Locke et al. (13) reported 97 variants, of which 56 were novel, in the largest GWAS meta-analysis of BMI to date. Previous studies using a subset of these variants improved prediction of BMI and obesity beyond demographic, geographic, and socioeconomic status information (29). This subset of variants has also been found to have a significant influence on BMI during childhood, adolescence, and adulthood in a longitudinal cohort (30). Thus, the variants seem to confer a life-course risk of obesity rather than risk at one time point. We found that a score including the 97 BMI variants was significantly associated with MS in 2 populations. Effect estimates were consistent with estimates based on FTO only, as would be expected if all these variants influenced MS via a common pathway—for example, BMI. This perspective is also supported by our overidentification tests, which found no significant difference in effects across 5 arbitrary groupings. We also found very similar estimates when we used all 97 variants or excluded 5 with some evidence of a direct effect. These 3 lines of evidence suggest that although some of the genetic variants may have small direct effects on MS, there is probably a common pathway to MS mediated by BMI.
There are several hypotheses linking obesity and autoimmune diseases, including MS. T-helper 17 cells, which secrete interleukin-17, have recently been implicated in the pathogenesis of autoimmune disease, and obesity may predispose induction of T-helper 17 cells via an interleukin-6–dependent process leading to exacerbation of inflammatory diseases such as MS (31). The intestinal immune response has also been hypothesized to explain the association between obesity and MS; an imbalance of T-helper 17 and T-regulatory cells may lead to alteration of the intestinal microbiome (32, 33). It has also been shown that vitamin D deficiency is prevalent among obese individuals (34). Given that vitamin D regulates immune response (35), this may have implications for MS susceptibility. Additionally, white adipose tissue is an essential endocrine organ that secretes adipokines (e.g., interleukin-6, tumor necrosis factor alpha, leptin, adiponectin), which are involved in immune and inflammatory processes and contribute to a low-grade inflammatory state in obese individuals (36). Certain adiposity genes, some of which overlap with genes in the BMI GRS used in this study, have been shown to be associated with age of menarche (37), and adipokines such as leptin are up-regulated by ovarian sex steroids. Lastly, a recent study demonstrated genetic evidence for connections between motor deficits, obesity, and neurological disorders (38). More research is needed to identify the biological mechanisms mediating the association between BMI and MS, which has the potential to contribute to our understanding of other autoimmune and neurological diseases.
A major strength of this study was the large sample size and the statistical power to demonstrate an association using a relatively weak instrument (R2 for the BMI GRS was 1.5% in the Swedish studies and 2.3% in KPNC participants), which is in accordance with the GWAS identifying the 97 variants (reported R2 = 2.7%) (13). We were also able to demonstrate findings in 2 data sets and conduct rigorous quality control to account for population stratification. Additionally, we used the most recent data to build a BMI GRS of 97 variants.
As with any efforts to make a causal inference from observational data, MR analysis involves many assumptions, and we have addressed them to the best of our ability. We were able to meet most model assumptions by using a BMI GRS established to be associated with BMI in an independent population through a large GWAS, testing whether the BMI GRS was independent of measured confounders, and conducting a direct effect analysis to ensure that the BMI GRS did not contain any variants independently associated with MS. We additionally validated our BMI GRS by conducting overidentification tests and by adjusting for BMI in our model. However, associations of genetic variants with unmeasured or unknown confounders cannot be ruled out, leaving one assumption not fully testable (39).
Our study included non-Hispanic white persons, which limits the generalizability of our findings. Additional limitations of our study include a relatively small sample of men, an assumption of linearity, and possible pleiotropic effects of BMI GRS on MS (i.e., genes may influence phenotypes other than BMI that are associated with an increased risk of MS). We did not have serum vitamin D concentrations and thus were not able to include this variable in our statistical models. Further, the life-course specificity of the BMI GRS remains to be fully understood. Limitations of effects analysis include reliance on self-reported weight and height to calculate BMI, which may have biased the mediation results. We also used 2 slightly different assessments to calculate BMI during young adulthood for the KPNC (weight during ages 20–29 years) and GERA (weight at age 18 years) cohorts; however, repeated sampling procedures in the GERA data set indicated no significant deviations in BMI between the 2 control samples. Additional studies should aim to replicate these findings, specifically in populations of other race/ethnicities.
In conclusion, we found that BMI is causally associated with MS by using a BMI GRS of 97 genetic variants with an analytical approach that avoids bias from reverse causation and potential confounders. While BMI-related variants demonstrated indirect effects on MS through their association with increased BMI, we also found that certain variants may directly influence MS via independent mechanisms. These findings provide targets for future therapeutic interventions and aid in the understanding of the complex relationship between genetics, BMI, and the MS disease process.
ACKNOWLEDGMENTS
Author affiliations: Division of Epidemiology, School of Public Health, University of California, Berkeley, Berkeley, California (Milena A. Gianfrancesco, Xiaorong Shao, Hong Quach, Lisa F. Barcellos); Department of Epidemiology and Biostatistics, University of California, San Francisco, San Francisco, California (M. Maria Glymour, Stefan Walter); Center for Computational Biology, University of California, Berkeley, Berkeley, California (Brooke Rhead); Kaiser Permanente Division of Research, Oakland, California (Ling Shen, Catherine Schaefer, Lisa F. Barcellos); Division of Biostatistics, School of Public Health, University of California, Berkeley, Berkeley, California (Alan Hubbard); deCODE Genetics, Reykjavik, Iceland (Ingileif Jónsdóttir, Kári Stefánsson); Department of Clinical Neuroscience and Center for Molecular Medicine, Karolinska Institutet at Karolinska University Hospital, Stockholm, Sweden (Pernilla Strid, Lars Alfredsson); Institute of Environmental Medicine, Karolinska Institutet, Stockholm, Sweden (Jan Hillert, Anna Hedström, Tomas Olsson, Ingrid Kockum); Research Program on Genes, Environment, and Health, Kaiser Permanente, Oakland, California (Catherine Schaefer); and Centre for Occupational and Environmental Medicine, Stockholm County Council, Stockholm, Sweden (Lars Alfredsson).
This work was supported by the National Institute of Neurological Disorders and Stroke (grants R01 NS049510, R01 NS0495103, and F31 NS093832); the National Institute of Allergy and Infectious Diseases (grants R01 AI 076544 and RC2 AG036607); the Robert Wood Johnson Foundation; the Wayne and Gladys Valley Foundation; the Ellison Medical Foundation; the AFA Foundation; the Knut and Alice Wallenberg Foundation; the Swedish Brain Foundation; the Margareta af Ugglas Foundation; European Union Seventh Framework Programme NEURINOX (grant 2012-278611); the Swedish Medical Research Council (grant 521-2012-2917 to L.A.); and the Swedish Research Council for Health, Working Life, and Welfare (including grants 2012-0325 and 2015-00195 to L.A.).
We thank Dr. Allan Bernstein for his contributions to the Kaiser Permanente Multiple Sclerosis Research Program. We also thank all members and staff of the Kaiser Permanente Division of Research and the University of California, Berkeley, Genetic Epidemiology and Genomics Laboratory. We also thank the International Multiple Sclerosis Genetics Consortium for providing the genotype data for the body mass index–related genetic variants and variants used for human leukocyte antigen imputation for the Swedish population custom array.
Conflict of interest: T.O. has received lecture and/or advisory board honoraria and unrestricted grant support for multiple sclerosis research from Biogen, Novartis, Genzyme, and Merck. He has received academic grant support from the Swedish Research Council, the AFA Foundation, the Knut and Alice Wallenberg Foundation, and the Swedish Brain Foundation J.H. has received honoraria for serving on advisory boards for Biogen Inc. and Novartis and speaker's fees from Biogen Inc., Merck Serono, Bayer Schering Pharma, Teva Neuroscience, and Sanofi-Aventis and has served as principle investigator for projects by, or received unrestricted research support from, Biogen Inc., Merck Serono, Teva Neuroscience, Sanofi, Novartis, and Bayer Schering Pharma. His multiple sclerosis research is funded by the Swedish Research Council. I.K. has received speaker honoraria from Merck and has received research support the Neurologiskt Handikappades Riksforbund Foundation and the Swedish Childhood Diabetes Foundation. L.A. has received speaker honoraria from Teva. The other authors report no conflicts of interest.
APPENDIX
Kaiser Permanente genotyping and quality control
Whole blood was collected and processed and DNA was extracted using the Gentra Puregene (Qiagen, Hilden, Germany) protocol. Saliva was collected using Oragene kits (DNA Genotek Inc., Ottowa, Ontario, Canada). Medium-resolution human leukocyte antigen (HLA) antigen D–related β1 subunit (DRB1) gene (HLA-DRB1) and genome-wide single-nucleotide polymorphism (SNP) genotyping was performed as previously described (3, 40), using an Axiom (Affymetrix, Santa Clara, California) custom chip for Genetic Epidemiology Research on Aging controls and the Illumina Infinium 660K BeadChip Array (Illumina Inc., San Diego, California) and Human Omni Express (Illumina Inc.) for cases and controls from the Kaiser Permanente Medical Care Plan of Northern California. Low-quality SNPs were removed prior to imputation (<1% minor allele frequency, genotyped in <90% of individuals) as were samples with more than 10% failed genotype calls, duplicates, or related individuals. Imputation against reference haplotypes from the 1000 Genomes Project (http://www.internationalgenome.org/) was conducted using SHAPEIT (http://www.shapeit.fr/) and IMPUTE2 (https://mathgen.stats.ox.ac.uk/impute/impute_v2.html) (information score >0.8 on all 3 platforms; minor allele frequency in controls with standard deviation <3%). Cross-platform association tests were also conducted to remove SNPs associated with the genotype array (false-discovery-rate Q < 0.05). Population outliers were identified using multidimensional scaling and reference samples from the Human Genome Diversity Project (http://www.hagsc.org/hgdp) and were removed from the analyses.
Swedish genotyping and quality control
All participants were asked to give blood samples, which were genotyped on an Illumina custom array and on OmniExpress-24 (Illumina Inc.). HLA-DRB1 information was imputed with HLA*IMP02 (Affymetrix), using genotypes in the major histocompatibility complex region from the custom array. SNPs with less than a 2% minor allele frequency, genotyped in fewer than 98% of individuals, or not in Hardy-Weinberg equilibrium among controls (P < 0.0001) were removed from the analysis. Individuals with more than 2% failed genotype calls, related individuals, or population outliers identified using the SmartPCA program were also removed (41). Twelve SNPs related to body mass index were taken from the custom array, and the remaining genotypes were taken from the OmniExpress chip. Forty-three SNPs related to body mass index were not present on the array and were imputed using MaCH 1.0 (42) with standard settings and the Northern European 1000 Genomes Project reference panel. Seventeen markers used the August 2009 reference panel, 25 markers used the August 2010 panel, and 1 marker used the July 2011 panel.
REFERENCES
- 1. Su KG, Banker G, Bourdette D, et al. . Axonal degeneration in multiple sclerosis: the mitochondrial hypothesis. Curr Neurol Neurosci Rep. 2009;9(5):411–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Favorova OO, Kulakova OG, Boiko AN. Multiple sclerosis as a polygenic disease: an update [in Russian]. Genetika. 2010;46(3):302–313. [PubMed] [Google Scholar]
- 3. Barcellos LF, Sawcer S, Ramsay PP, et al. . Heterogeneity at the HLA-DRB1 locus and risk for multiple sclerosis. Hum Mol Genet. 2006;15(18):2813–2824. [DOI] [PubMed] [Google Scholar]
- 4. Beecham AH, Patsopoulos NA, Xifara DK, et al. . Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45(11):1353–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ascherio A, Munger K. Epidemiology of multiple sclerosis: from risk factors to prevention. Semin Neurol. 2008;28(1):17–28. [DOI] [PubMed] [Google Scholar]
- 6. Munger KL, Chitnis T, Ascherio A. Body size and risk of MS in two cohorts of US women. Neurology. 2009;73(19):1543–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hedström AK, Olsson T, Alfredsson L. High body mass index before age 20 is associated with increased risk for multiple sclerosis in both men and women. Mult Scler. 2012;18(9):1334–1336. [DOI] [PubMed] [Google Scholar]
- 8. Gianfrancesco MA, Acuna B, Shen L, et al. . Obesity during childhood and adolescence increases susceptibility to multiple sclerosis after accounting for established genetic and environmental risk factors. Obes Res Clin Pract. 2014;8(5):e435–e447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wesnes K, Riise T, Casetta I, et al. . Body size and the risk of multiple sclerosis in Norway and Italy: the EnvIMS study. Mult Scler. 2015;21(4):388–395. [DOI] [PubMed] [Google Scholar]
- 10. Langer-Gould A, Brara SM, Beaber BE, et al. . Childhood obesity and risk of pediatric multiple sclerosis and clinically isolated syndrome. Neurology. 2013;80(6):548–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Munger KL, Bentzen J, Laursen B, et al. . Childhood body mass index and multiple sclerosis risk: a long-term cohort study. Mult Scler. 2013;19(10):1323–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hedstrom AK, Lima Bomfim I, Barcellos L, et al. . Interaction between adolescent obesity and HLA risk genes in the etiology of multiple sclerosis. Neurology. 2014;82(10):865–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Locke AE, Kahali B, Berndt SI, et al. . Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Krieger N. Overcoming the absence of socioeconomic data in medical records: validation and application of a census-based methodology. Am J Public Health. 1992;82(5):703–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McDonald WI, Compston A, Edan G, et al. . Recommended diagnostic criteria for multiple sclerosis: guidelines from the International Panel on the Diagnosis of Multiple Sclerosis. Ann Neurol. 2001;50(1):121–127. [DOI] [PubMed] [Google Scholar]
- 16. Polman CH, Reingold SC, Banwell B, et al. . Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald Criteria. Ann Neurol. 2011;69(2):292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kvale MN, Hesselson S, Hoffmann TJ, et al. . Genotyping informatics and quality control for 100,000 subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort. Genetics. 2015;200(4):1051–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Glymour MM, Tchetgen Tchetgen EJ, Robins JM. Credible Mendelian randomization studies: approaches for evaluating the instrumental variable assumptions. Am J Epidemiol. 2012;175(4):332–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Valeri L, Vanderweele TJ. Mediation analysis allowing for exposure-mediator interactions and causal interpretation: theoretical assumptions and implementation with SAS and SPSS macros. Psychol Methods. 2013;18(2):137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Scuteri A, Sanna S, Chen WM, et al. . Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3(7):e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Loos RJ, Bouchard C. FTO: the first gene contributing to common forms of human obesity. Obes Rev. 2008;9(3):246–250. [DOI] [PubMed] [Google Scholar]
- 22. Kaklamani V, Yi N, Sadim M, et al. . The role of the fat mass and obesity associated gene (FTO) in breast cancer risk. BMC Med Genet. 2011;12:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lewis SJ, Murad A, Chen L, et al. . Associations between an obesity related genetic variant (FTO rs9939609) and prostate cancer risk. PLoS One. 2010;5(10):e13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lurie G, Gaudet MM, Spurdle AB, et al. . The obesity-associated polymorphisms FTO rs9939609 and MC4R rs17782313 and endometrial cancer risk in non-Hispanic white women. PLoS One. 2011;6(2):e16756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Keller L, Xu W, Wang HX, et al. . The obesity related gene, FTO, interacts with APOE, and is associated with Alzheimer's disease risk: a prospective cohort study. J Alzheimers Dis. 2011;23(3):461–469. [DOI] [PubMed] [Google Scholar]
- 26. Ho AJ, Stein JL, Hua X, et al. . A commonly carried allele of the obesity-related FTO gene is associated with reduced brain volume in the healthy elderly. Proc Natl Acad Sci USA. 2010;107(18):8404–8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bressler J, Fornage M, Demerath EW, et al. . Fat mass and obesity gene and cognitive decline: the Atherosclerosis Risk in Communities Study. Neurology. 2013;80(1):92–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Davis W, van Rensburg SJ, Cronje FJ, et al. . The fat mass and obesity-associated FTO rs9939609 polymorphism is associated with elevated homocysteine levels in patients with multiple sclerosis screened for vascular risk factors. Metab Brain Dis. 2014;29(2):409–419. [DOI] [PubMed] [Google Scholar]
- 29. Belsky DW, Moffitt TE, Sugden K, et al. . Development and evaluation of a genetic risk score for obesity. Biodemography Soc Biol. 2013;59(1):85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Choh AC, Lee M, Kent JW, et al. . Gene-by-age effects on BMI from birth to adulthood: the Fels Longitudinal Study. Obesity (Silver Spring). 2014;22(3):875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Winer S, Paltser G, Chan Y, et al. . Obesity predisposes to Th17 bias. Eur J Immunol. 2009;39(9):2629–2635. [DOI] [PubMed] [Google Scholar]
- 32. Brown K, DeCoffe D, Molcan E, et al. . Diet-induced dysbiosis of the intestinal microbiota and the effects on immunity and disease. Nutrients. 2012;4(8):1095–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Manzel A, Muller DN, Hafler DA, et al. . Role of “Western diet” in inflammatory autoimmune diseases. Curr Allergy Asthma Rep. 2014;14(1):404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Soskic S, Stokic E, Isenovic ER. The relationship between vitamin D and obesity. Curr Med Res Opin. 2014;30(6):1197–1199. [DOI] [PubMed] [Google Scholar]
- 35. Schoindre Y, Terrier B, Kahn JE, et al. . Vitamin D and autoimmunity. First part: fundamental aspects [in French]. Rev Med Interne. 2012;33(2):80–86. [DOI] [PubMed] [Google Scholar]
- 36. Cao H. Adipocytokines in obesity and metabolic disease. J Endocrinol. 2014;220(2):T47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fernandez-Rhodes L, Demerath EW, Cousminer DL, et al. . Association of adiposity genetic variants with menarche timing in 92,105 women of European descent. Am J Epidemiol. 2013;178(3):451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mao JH, Langley SA, Huang Y, et al. . Identification of genetic factors that modify motor performance and body weight using Collaborative Cross mice. Sci Rep. 2015;5:16247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Burgess S, Timpson NJ, Ebrahim S, et al. . Mendelian randomization: where are we now and where are we going. Int J Epidemiol. 2015;44(2):379–388. [DOI] [PubMed] [Google Scholar]
- 40. Barcellos LF, May SL, Ramsay PP, et al. . High-density SNP screening of the major histocompatibility complex in systemic lupus erythematosus demonstrates strong evidence for independent susceptibility regions. PLoS Genet. 2009;5(10):e1000696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li Y, Abecasis GR. Mach 1.0: rapid haplotype reconstruction and missing genotype inference [abstract]. Am J Hum Genet. 2006;S79:2290. [Google Scholar]
- 42. Price AL, Patterson NJ, Plenge RM, et al. . Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. [DOI] [PubMed] [Google Scholar]