Abstract
We have developed an interface to serve patient data from Informatics for Integrating Biology and the Bedside (i2b2) repositories in the Fast Healthcare Interoperability Resources (FHIR) format, referred to as a SMART-on-FHIR cell. The cell serves FHIR resources on a per-patient basis, and supports the “substitutable” modular third-party applications (SMART) OAuth2 specification for authorization of client applications. It is implemented as an i2b2 server plug-in, consisting of 6 modules: authentication, REST, i2b2-to-FHIR converter, resource enrichment, query engine, and cache. The source code is freely available as open source. We tested the cell by accessing resources from a test i2b2 installation, demonstrating that a SMART app can be launched from the cell that accesses patient data stored in i2b2. We successfully retrieved demographics, medications, labs, and diagnoses for test patients. The SMART-on-FHIR cell will enable i2b2 sites to provide simplified but secure data access in FHIR format, and will spur innovation and interoperability. Further, it transforms i2b2 into an apps platform.
Keywords: Database management systems, information storage and retrieval, medical informatics, systems integration, clinical decision support, user-computer interface
INTRODUCTION
We have developed an open-source interface that serves patient data from Informatics for Integrating Biology and the Bedside (i2b2) repositories in a Fast Healthcare Interoperability Resources (FHIR) format. The objective of this work is to enable simplified and secure access to patient data, in order to spur innovation of portable and scalable clinical apps by leveraging emerging health IT standards.
Informatics for Integrating Biology and the Bedside
I2b2 is an open source clinical data analytics platform originally developed under funding from the National Institutes of Health.1,2 I2b2 is used at over 140 sites for querying patient data to address research questions. I2b2 has been adapted to build multi-institutional networks to allow federated querying, and is a central component in the infrastructure for many institutions that have Clinical and Translational Science Awards and Patient-Centered Outcomes Research Institute Awards.3,4
I2b2 supports a sidecar approach wherein the software aggregates a copy of the patient data from the electronic health record (EHR) and provides query services in parallel to the EHR for research. While the EHR manages patient data for clinical workflow, the i2b2 sidecar hosts a data copy for secondary use. Each i2b2 installation (called a hive) comprises several i2b2 cells that provide different services, and the cells communicate with each other using XML web services.
I2b2 software has been extended to handle a variety of data models and functionalities. There are i2b2 extensions for importing C-CDAs,5 translating HQMF,6 managing images,7 doing federated querying, conducting data analysis,8 conducting studies in specific disease domains,9,10 and complying with meaningful use. I2b2 has an active community of developers and has become a popular component of health care research.
Fast Healthcare Interoperability Resources
FHIR is a new standard for exchanging health care information electronically.11 It is based on emerging industry approaches and builds on the lessons from previous standards including Health Level Seven’s v2 and v3, the Reference Information Model, and Clinical Document Architecture. FHIR uses a predominantly compositional approach, in contrast to the previous v3 approach of modeling by constraint, in that FHIR data is organized utilizing a combination of building blocks called resources. Furthermore, FHIR specifies a RESTful application programming interface (API) to access resources. Several projects are under way through standards organizations to facilitate adoption of FHIR, including the Argonaut Project,12 the Data Access Framework, and the Clinical Information Modeling Initiative.13
SMART Health IT
SMART is a project aimed at developing a health information technology platform for supporting “substitutable” modular third-party applications (SMART apps) constructed around a set of core services.14–16 These apps can run directly within EHRs; the 5 top EHR vendors have implemented SMART standards through a project called Argonaut.12 The same apps that run in the EHR context could also run on a “sidecar” such as i2b2 supporting the SMART-on-FHIR API. SMART Health IT transforms an EHR or its sidecar into an iPhone-like platform.17 Smartphone platforms support an ecosystem of applications that extend the capability of the phone beyond the core software. Similarly, SMART Health IT supports an “app store for health” of substitutable apps constructed around core services and is intended to drive down health care technology costs and spur innovation, by fostering market competition for the evolution of applications that better support care provider needs.
The SMART specification provides a protocol for apps to authenticate with and run against existing health information technology systems, and the SMART library uses a JSON format for payload representation. We have implemented FHIR and OAuth2 specifications, and have completely rewritten the i2b2 plug-in for SMART apps developed previously.18 Our new interface is referred to as the SMART-on-FHIR cell. Given the wide adoption and investment in the FHIR standard, the i2b2 SMART-on-FHIR cell will support a vast array of applications.
METHODS
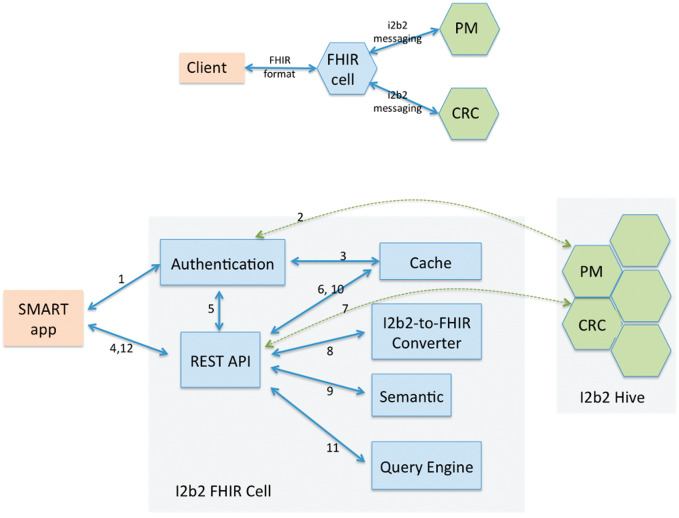
We designed and implemented the SMART-on-FHIR cell as an i2b2 server component consisting of 6 modules: authentication module, REST API, i2b2-to-FHIR converter, resource enrichment module, FHIR query engine, and a cache (Figure 1). The source code is freely available in Github (https://github.com/i2b2plugins/i2b2-fhir-cell). A technical reference is attached as a supplementary file.
Figure 1.
Modular architecture. The numbers denote the typical flow of control. PM is the core project management cell, and CRC is the core data cell.
The authentication module implements OAuth2 SMART specification to control access to the REST API. Specifically, it allows a client to request read-only access to data for a particular i2b2 user.
The REST-API allows a client to query and read FHIR resources, and provides metadata about the FHIR cell. On receiving a request from the web client, it communicates with the i2b2 instance to retrieve “i2b2 observation facts” for a particular patient and invokes the i2b2-to-FHIR conversion module to construct FHIR resources from the i2b2 facts. The conversion module constructs the following FHIR resources: patient-containing demographic information, observation for laboratory data, condition for diagnosis, and medication prescription information (Table 1).
Table 1.
Resources supported by the SMART-on-FHIR i2b2 cell
| FHIR Resource | Description |
|---|---|
| Patient | Demographic information for a patient |
| Medication | Drug information, using RxNorm |
| Medication Prescription | Record of prescription for a particular patient, including dosage, frequency, and route, along with a reference to the medication resource |
| Observation | Laboratory information using LOINC coding, including units |
| Condition | Diagnoses in ICD9 codes |
Additional semantic information is then added to these resources by the enrichment module, and then persisted on the disk cache. The enrichment/semantic module translates codes in the FHIR resources into preferred coding systems, and expands on codes in Systematized Nomenclature of Medicine-Clinical Terms (SNOMED-CT), Logical Observation Identifiers Names and Codes (LOINC), and International Statistical Classification of Diseases and Related Health Problems (ICD9), to add semantic information to the resources.
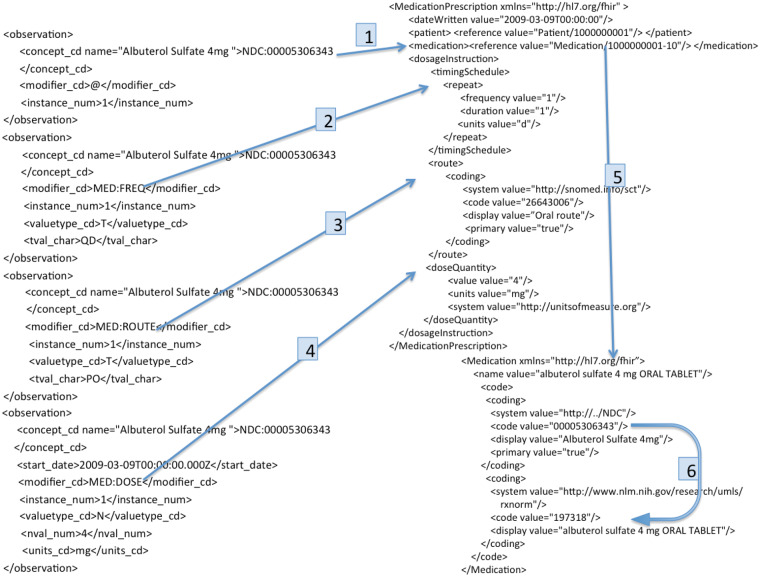
Finally, if the client request is a FHIR query, the Representational state transfer-Application Programming interface (REST-API) module invokes the query engine to execute the query for selection of a subset of FHIR resources, and sends them back to the client. The i2b2-to-FHIR converter translates the i2b2 observation facts to FHIR resources in EXtensible Markup Language (XML) format, using XQuery (Figure 2). The query engine implements the FHIR search specification. Its output is a set of FHIR resources in the form of a FHIR bundle. The cache persists FHIR resources between RESTful calls, thereby reducing redundant data retrieval requests to the i2b2 instance.
Figure 2.
XQuery-based mapping of i2b2 XML to FHIR resource, elucidating an example translation of i2b2 medication facts into medication prescription and medication resources in FHIR. Steps 1–4 show mapping of the i2b2 model to the FHIR model, and 5 denotes linking of medication to prescription; 6 is enrichment by adding RXNORM translation of the NDC code.
Figure 1 details the SMART-on-FHIR cell architecture, where the numbers denote the typical flow of control among the modules:
The client initiates authentication with the cell.
Using OAuth2 protocol, the authentication module seeks authentication credentials and verifies them with the project management cell of the I2b2 hive.
On successful authentication, the authentication module saves OAuth2 authentication and access tokens to disk.
Client makes HTTP requests including an access token in the header, which are handled by the REST API.
The REST API validates the access tokens with the authentication module.
Next it checks if the required FHIR resources (obtained in previous client requests) are present locally, on the disk.
If no resources are found on the disk, it connects to the i2b2 instance to retrieve i2b2 facts for the patient.
The i2b2-to-FHIR converter is invoked to translate retrieved i2b2 resources into FHIR resources.
Next the semantic module parses the FHIR resources to translate codes therein to preferred codes, and appends the code translations and human-readable code names to the resources.
The resources are then persisted to disk.
Finally, the query submitted by the client in step 4 is parsed and executed on the persisted FHIR resources.
The result is returned to the client.
The cell protects the password of the i2b2 user (from the client app) by directly communicating with the user using the OAuth2 SMART specification. It supports 2 types of FHIR query URLs: (1) retrieval of 1 particular FHIR resource by providing the resource ID, and (2) retrieval of a set of FHIR resources of a particular type, by specifying the resource type and query name value parameters (see Table 2). The cell does not support paging, versioning (history), or tagging.
Table 2.
Example of FHIR queries that can be executed on the cell
| Get all Patients: http://[FHIR-base]/Patient Get particular Patient: http://[FHIR-base]/1000000005 Get Prescriptions for particular Patient: http://[FHIR-base]/MedicationPrescription?patient=1000000005 |
We tested the cell by accessing FHIR resources from a demonstration installation of i2b2, and tested whether a SMART app could be executed on the cell to display FHIR resources for simulated patients.
RESULTS AND DISCUSSION
We successfully retrieved the demographics, medications, labs, and diagnoses for fake test patients. Table 2 shows example queries. The developed SMART app ran successfully, demonstrating a dashboard to summarize data. These results are demonstrated live at http://fhir.i2b2.org and http://fhir.i2b2.org/smartapp-demo.html.
Extension of the sidecar approach
I2b2 software has generally been deployed in a sidecar approach at most institutions, wherein a copy of the data from the EHR is stored in the i2b2 repository (Figure 3). The i2b2 client is used to interactively query the repository to identify patient cohorts for hypothesis testing. I2b2 software has evolved to provide a secure and scalable environment for such research operations. Another advantage of this approach is that the EHR system is not overloaded by research operations, allowing the EHR to focus on clinical operations.
Figure 3.
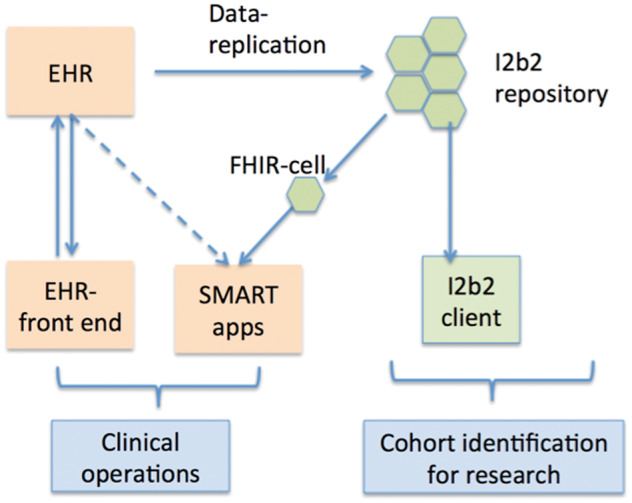
The SMART-on-FHIR i2b2 cell allows SMART apps to run on an i2b2 data warehouse, thereby supplementing data and computing resources to an EHR system for executing SMART apps.
The FHIR cell builds on this approach, providing an alternate channel to access the i2b2 data, which can be used to supplement clinical operations through use of apps.
OAuth2 authentication
The cell supports the SMART specification, which is based on the OAuth2 protocol.19,20 OAuth2 specifies a process for resource owners to authorize third-party access to their server resources without sharing their credentials. Hence the FHIR cell can issue access tokens to clients/apps, with the approval of users with i2b2 accounts. The tokens can then be used by the apps to access FHIR resources from the cell. Hence the cell provides an additional layer of security over existing i2b2 security.
SMART app development
The FHIR specification provides a simplified abstraction of a medical record with normalized data elements that app developers can understand. FHIR specifications are freely available and easy to understand and develop on.11
Currently the cell provides read-only access on a per-patient basis. Provision of per-patient data access itself enables implementation of a vast array of clinical decision support use cases. The cell is being used to develop SMART apps for decision support at Partners Healthcare and the Reachnet Consortium,21 and is expected to accelerate the development of SMART apps at PCORnet sites as an integral part of the architecture for Scalable Collaborative Infrastructure for a Learning Healthcare System (SCILHS).22 The SCILHS infrastructure supports apps to improve PCOR research methods and enables functions such as consent, enrollment, randomization, and outreach for patient-reported outcomes.
Rich query interface
The query module is implemented using XQuery, and is generated automatically from FHIR specification. Inclusion of patient ID is required in each query (see Table 2).
Institutional migration to FHIR
In addition to conventional methods for importing EHR data into i2b2, an i2b2 extension was recently released that provides a method for PCORnet’s patient-powered research to import their patients’ EHR data in CCD format into i2b2 instances.5,23 The FHIR cell can be used to expose this data in FHIR format. This pathway lowers the barrier for smaller practices to invest in making their data available in FHIR format. In addition to supporting SMART apps, the FHIR API can also be useful in importing data from a legacy EHR system into a new EHR system that supports the FHIR standard, potentially reducing the effort required for migrating data from the legacy system.
In summary, for institutions running i2b2 instances, the FHIR cell will facilitate (1) deployment of SMART apps, (2) an additional mechanism to search and access the i2b2 repository, and (3) migration of data from legacy EHR systems replicated with the sidecar approach into i2b2 instances into new FHIR-enabled EHRs.
Limitations and upcoming efforts
Currently the cell only supports read-only access on a per-patient basis. We are implementing additional FHIR resources and plan to extend the cell to allow population-level querying. Another planned feature is write capability to the cell, which will allow importing of data in FHIR format into i2b2. Including population queries and allowing apps to write data back to the i2b2 instance considerably increase the complexity of implementation, which was the rationale for currently limiting the scope to per-patient access.
CONCLUSIONS
We have developed an interface that provides patient data from i2b2 repositories in FHIR format on a per-patient basis. The source code is freely available online as open source. We were able to successfully retrieve the demographics, medications, labs, and diagnoses for random patients in a demonstration i2b2 instance, and were able to successfully run SMART apps to show a dashboard summary for these patients. The SMART-on-FHIR cell will facilitate i2b2 sites in providing simplified but secure data access in FHIR format, and will spur innovation and interoperability.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank member organizations of the SMART Advisory Committee, including the Hospital Corporation of America, Lilly, Surescripts, the Advisory Board Company, Centros, Polyglot System Inc., Premier, Blue Cross Blue Shield Association, and the BMJ Group, which provide philanthropic support to Boston Children’s Hospital, which funds SMART development.
DATA SHARING
Demo: http://fhir.i2b2.org, http://fhir.i2b2.org/smartapp-demo.html Source code: https://github.com/i2b2plugins/i2b2-fhir-cell
STUDY APPROVAL
This study was approved by the institutional review board at Massachusetts General Hospital, Boston.
CONTRIBUTORS
K.B.W. led the design and implementation of the FHIR cell and wrote the majority of the manuscript. J.C.M., J.G.K., N.W., M.M., and S.N.M. made key contributions to the architecture of the SMART-on-FHIR cell. S.N.M., K.D.M., and C.G.C. conceived of and initiated the project. All authors reviewed and approved the final version of the manuscript.
FUNDING
This work was supported by National Library of Medicine grant numbers K99LM011575 and R00LM011575, and National Institute of General Medical Sciences grant number R01GM104303.
COMPETING INTERESTS
The authors have no competing interests to declare.
REFERENCES
- 1. Murphy SN, Weber G, Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J Am Med Inform Assoc 2010;17(2):124–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Murphy S, Wilcox A. Mission and sustainability of informatics for integrating biology and the bedside (i2b2). EGEMS (Wash DC) 2014;2(2):1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McMurry AJ, Murphy SN, MacFadden D, et al. SHRINE: enabling nationally scalable multi-site disease studies. PLoS One 2013;8(3):e55811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. CTSActs Network. https://www.act-network.org/. Accessed December 15, 2015.
- 5. Klann JG, Mendis M, Phillips LC, et al. Taking advantage of continuity of care documents to populate a research repository. J Am Med Inform Assoc 2015;22(2):370–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Klann JG, Murphy SN. Computing health quality measures using Informatics for Integrating Biology and the Bedside. J Med Internet Res 2013;15(4):e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Murphy SN, Mendis ME, Grethe JS, Gollub RL, Kennedy D, Rosen BR. A web portal that enables collaborative use of advanced medical image processing and informatics tools through the Biomedical Informatics Research Network (BIRN). AMIA Annual Symposium Proceedings/AMIA Symposium 2006:579–583. [PMC free article] [PubMed] [Google Scholar]
- 8. Segagni D, Ferrazzi F, Larizza C, et al. R engine cell: integrating R into the i2b2 software infrastructure. J Am Med Inform Assoc 2011;18(3):314–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. London JW, Balestrucci L, Chatterjee D, Zhan T. Design-phase prediction of potential cancer clinical trial accrual success using a research data mart. J Am Med Inform Assoc 2013;20(e2):e260–e266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Segagni D, Tibollo V, Dagliati A, Napolitano C, Priori G, Bellazzi R. CARDIO-i2b2: integrating arrhythmogenic disease data in i2b2. Stud Health Technol Inform 2012;180:1126–1128. [PubMed] [Google Scholar]
- 11. Fast Healthcare Interoperability Resources: Draft Standards for Trial Use 2. 2015. https://www.hl7.org/fhir/2015May/index.html. Accessed December 15, 2015.
- 12. Argonaut Project. http://argonautwiki.hl7.org/. Accessed December 15, 2015.
- 13. Clinical Information Modeling Initiative. 2015. http://www.opencimi.org/. Accessed December 15, 2015.
- 14. Mandl KD, Mandel JC, Kohane IS. Driving innovation in health systems through an apps-based information economy. Cell Syst 2015;1(1):8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mandl KD, Mandel JC, Murphy SN, et al. The SMART Platform: early experience enabling substitutable applications for electronic health records. J Am Med Inform Assoc 2012;19(4):597–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mandel JC, Kreda DA, Mandl KD, Kohane IS, Ramoni RB. SMART on FHIR: a standards-based, interoperable apps platform for electronic health records. J Am Med Inform Assoc 2016. doi: 10.1093/jamia/ocv189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mandl KD, Kohane IS. No small change for the health information economy. N Engl J Med 2009;360(13):1278–1281. [DOI] [PubMed] [Google Scholar]
- 18. Wattanasin N, Porter A, Ubaha S, et al. Apps to display patient data, making SMART available in the i2b2 platform. AMIA Annual Symposium Proceedings/AMIA Symposium 2012;2012:960–969. [PMC free article] [PubMed] [Google Scholar]
- 19. Mandel J. SMART: Tech Stack for Health Apps http://docs.smarthealthit.org/. Accessed December 15, 2015. [Google Scholar]
- 20. Force IET. The OAuth 2.0 Authorization Framework, 2010. Report No. rfc6749.
- 21. Reachnet Codeathon on developing SMART apps on i2b2. 2015. http://connectathon.reachnet.org/technology. Accessed December 15, 2015.
- 22. Mandl KD, Kohane IS, McFadden D, et al. Scalable Collaborative Infrastructure for a Learning Healthcare System (SCILHS): architecture. J Am Med Inform Assoc 2014;21(4):615–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Abend A, Housman D, Johnson B. Integrating Clinical Data into the i2b2 Repository. Summit on Translat Bioinforma 2009;2009:1–5. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.