Abstract
Objective: Build probabilistic topic model representations of hospital admissions processes and compare the ability of such models to predict clinical order patterns as compared to preconstructed order sets.
Materials and Methods: The authors evaluated the first 24 hours of structured electronic health record data for > 10 K inpatients. Drawing an analogy between structured items (e.g., clinical orders) to words in a text document, the authors performed latent Dirichlet allocation probabilistic topic modeling. These topic models use initial clinical information to predict clinical orders for a separate validation set of > 4 K patients. The authors evaluated these topic model-based predictions vs existing human-authored order sets by area under the receiver operating characteristic curve, precision, and recall for subsequent clinical orders.
Results: Existing order sets predict clinical orders used within 24 hours with area under the receiver operating characteristic curve 0.81, precision 16%, and recall 35%. This can be improved to 0.90, 24%, and 47% (P < 10−20) by using probabilistic topic models to summarize clinical data into up to 32 topics. Many of these latent topics yield natural clinical interpretations (e.g., “critical care,” “pneumonia,” “neurologic evaluation”).
Discussion: Existing order sets tend to provide nonspecific, process-oriented aid, with usability limitations impairing more precise, patient-focused support. Algorithmic summarization has the potential to breach this usability barrier by automatically inferring patient context, but with potential tradeoffs in interpretability.
Conclusion: Probabilistic topic modeling provides an automated approach to detect thematic trends in patient care and generate decision support content. A potential use case finds related clinical orders for decision support.
Keywords: clinical decision support systems, electronic health records, data mining, probabilistic topic modeling, clinical summarization, order sets
Background and Significance
High-quality and efficient medical care requires clinicians to distill and interpret patient information for precise medical decisions. This can be especially challenging when the majority of clinical decisions (e.g., a third of surgeries to place pacemakers or ear tubes) lack adequate evidence to support or refute their practice.1,2 Even after current reforms,3 evidence-based medicine from randomized control trials cannot keep pace with the perpetually expanding breadth of clinical questions, with only ∼11% of guideline recommendations backed by high-quality evidence.4 Clinicians are left to synthesize vast streams of information for each individual patient in the context of a medical knowledge base that is both incomplete and yet progressively expanding beyond the cognitive capacity of any individual.5,6 Medical practice is thus routinely driven by individual expert opinion and anecdotal experience.
The meaningful use era of electronic health records (EHRs)7 presents a potential learning health system solution.8–12 EHRs generate massive repositories of real-world clinical data that represent the collective experience and wisdom of the broad community of practitioners. Automated clinical summarization mechanisms are essential to organize such a large body of data that would otherwise be impractical to manually categorize and interpret.13,14 Applied to clinical orders (e.g., labs, medications, imaging), such methods could answer “grand challenges” in clinical decision support15 to automatically learn decision support content from clinical data sources.
The current standard for executable clinical decision support includes human-authored order sets that collect related orders around common processes (e.g., admission and transfusion) or scenarios (e.g., stroke and sepsis). Computerized provider order entry16 typically occurs on an “à la carte” basis where clinicians search for and enter individual computer orders to trigger subsequent clinical actions (e.g., pharmacy dispensation and nurse administration of a medication or phlebotomy collection and laboratory analysis of blood tests). Clinician memory and intuition can be error prone when making these ordering decisions; thus, health system committees produce order set templates as a common mechanism to distribute standard practices and knowledge (in paper and electronic forms). Clinicians can then search by keyword for common scenarios (e.g., “pneumonia”) and hope they find a preconstructed order set that includes relevant orders (e.g., blood cultures, antibiotics, chest X-rays).17–19 While these can already reinforce consistency with best practices,20–25 automated methods are necessary to achieve scalability beyond what can be conventionally produced through manual definition of clinical content 1 intervention at a time.26
Probabilistic topic modeling
Here we seek to algorithmically learn the thematic structure of clinical data with an application toward anticipating clinical decisions. Unlike a top-down rule-based approach to isolate preconceived clinical concepts from EHRs,27 this is more consistent with bottom-up identification of patterns from the raw clinical data.28 Specifically, we develop a latent Dirichlet allocation (LDA) probabilistic topic model29–33 to infer the underlying “topics” for hospital admissions, which can then inform patient-specific clinical orders. Most prior work in topic modeling focuses on the organization of text documents ranging from newspaper and scientific articles34 to clinical discharge summaries.35 More recent work has modeled laboratory results36 and claims data37 or used similar low-dimensional representations of heterogeneous clinical data sources for the unsupervised determination of clinical concepts.38–40 Here we focus on learning patterns of clinical orders, as these interventions are the concrete representation of a clinician’s decision making, regardless of what may (or may not) be documented in narrative clinical notes and diagnosis codes.
In the analogous text analysis context, probabilistic topic modeling conceptualizes documents as collections of words derived from underlying thematic topics that define a probability distribution over topic-relevant words. For example, we may expect our referenced article on the “Scientific Evidence Underlying the American College of Cardiology (ACC)/American Heart Association (AHA)”2 to be about the abstract topics of “cardiology” and “clinical practice guidelines,” weighted by respective conditional probabilities P(TopicCardiology|DocumentEvidenceACC/AHA) and P(TopicGuidelines|DocumentEvidenceACC/AHA). Words we may expect to be prominently associated with the “cardiology” topic would include heart, valve, angina, pacemaker, and aspirin, while the “clinical practice guideline” topic may be associated with words like evidence, recommendation, trials, and meta-analysis. The relative prevalence of each word in each topic is defined by conditional probabilities P(Wordi|Topicj) in a categorical probability distribution. With the article composed as a weighted mixture of multiple topics, the document contents are expected to be generated from a proportional mixture of the words associated with each topic as determined by the conditional probability:
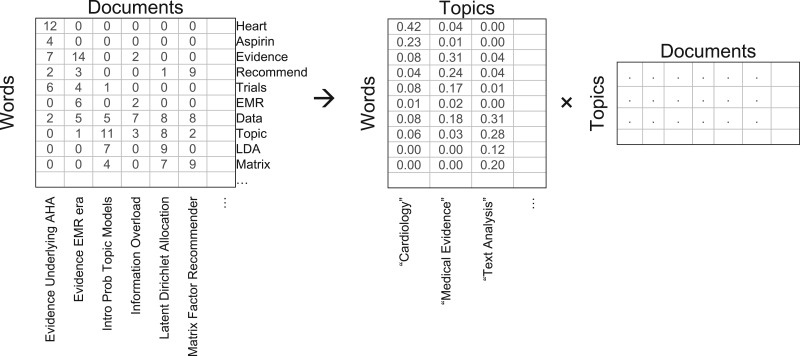
In practice, we are not actually interested in generating new documents from predefined word and topic distributions. Instead, we wish to infer the underlying topic and word distributions that generated a collection of existing documents. Such a body of documents can be represented as a word-document matrix where each document is a vector containing the frequencies of every possible word (Figure 1). Topic modeling methods factor this matrix based on the underlying latent topic structure that links associated words to associated documents. A precise solution to this inverted inference is not generally tractable, requiring iterative optimization solutions such as variational Bayes approximations29 or Gibbs sampling.31 This is closely related to other dimensionality-reduction techniques to provide low-rank data approximations,41–43 with the probabilistic LDA framework interpreting the interrelated structure as conditional probabilities P(Wordi|Topicj) and P(Topicj|Documentk). Once this latent topic structure is learned, it provides a convenient, efficient, and largely interpretable means of information retrieval, classification, and exploration of document data.
Figure 1.
Topic modeling as factorization of a word-document matrix. Simulated data in the top-left reflects that the word “Heart” appears 12 times in the article “Evidence Underlying AHA.” Factoring this full matrix into simpler matrices can discover a smaller number of latent dimensions that summarize the content. Topic modeling represents these latent dimensions as topics defining a categorical probability distribution of word occurrences in the topic-word matrix. This reveals the underlying statistical structure of the data, but an algorithmic process cannot itself provide meaning. By observing the most prevalent words in each topic axis, however, an underlying meaning is often interpretable (e.g., prevalence of the words “heart” and “aspirin” in the first topic axis implies a general topic of “Cardiology”).
Clinical data analogy
For our clinical context, we draw analogies between words in a document to clinical items occurring for a patient. The key clinical items of interest here are clinical orders, but other structured elements include patient demographics, laboratory results, diagnosis codes, and treatment team assignments. Modeling patient data as such allows us to learn topic models that relate patients to their clinical data. A patient receiving care for multiple complex conditions could then have his or her data separated out into multiple component dimensions (i.e., topics), as an “informative abstractive” approach to clinical summarization.14 For example, we might use this to describe a patient hospital admission as being “50% about a heart failure exacerbation, 30% about pneumonia, and 20% about mechanical ventilation protocols.” Prior work has accomplished similar goals of unsupervised abstraction of latent factors out of clinical records using varying methods.38–40 Based on the distribution of clinical orders associated within such low-dimensional representations, we aim to impute additional clinical orders for decision support.
Objective
Our objective is to evaluate the current real-world standard of care in terms of preauthored hospital order set usage during the first 24 hours of inpatient hospitalizations, build probabilistic topic model representations of clinical data to summarize the principal axes of clinical care underlying those same first 24 hours, and compare the ability of these models to anticipate relevant clinical orders as compared to existing order sets.
Methods
We extracted deidentified patient data from the (Epic) EHR for all inpatient hospitalizations at Stanford University Hospital in 2013 via the Stanford Translational Research Integrated Database Environment (STRIDE) clinical data warehouse.44 The structured data covers patient encounters from their initial (emergency room) presentation until hospital discharge. The dataset includes more than 20 000 patients with > 6.7 million instances of more than 23 000 distinct clinical items. Patients, items, and instances are respectively analogous to documents, words, and word occurrences in an individual document. The space of clinical items includes more than 6000 medication, more than 1500 laboratory, more than 1000 imaging and more than 1000 nursing orders. Nonorder items include more than 400 abnormal lab results, more than 7000 problem list entries, more than 5000 admission diagnosis ICD9 codes, more than 300 treatment team assignments, and patient demographics. Medication data was normalized with RxNorm mappings45 down to active ingredients and routes of administration. Numerical lab results were binned into categories based on “abnormal” flags established by the clinical laboratory or by deviation of more than 2 standard deviations from the observed mean if “high” and “low” flags were not prespecified. We aggregated ICD9 codes up to the 3-digit hierarchy such that an item for code 786.05 would be counted as 3 separate items (786.05, 786.0, 786). This helps compress the sparsity of diagnosis categories while retaining the original detailed codes if they are sufficiently prevalent to be useful. The above preprocessing models each patient as a timeline of clinical item instances, with each instance mapping a clinical item to a patient time point.
With the clinical item instances following the “80/20 rule” of a power law distribution,46 most items may be ignored with minimal information loss. Ignoring rare clinical items with fewer than 256 instances reduces the item vocabulary size from more than 23 000 to ∼3400 (15%), while still capturing 6 million (90%) of the 6.7 million item instances. After excluding common process orders (e.g., check vital signs, notify MD, regular diet, transport patient, as well as most nursing orders and PRN medications), 1512 clinical orders of interest remain.
LDA topic modeling algorithms infer topic structures from “bag of words” abstractions that represent each document as an unordered collection of word counts (i.e., 1 column of the word-document matrix in Figure 1). To construct an analogous model for our structured clinical data, we use each patient’s first 24 hours of data to populate an unordered “bag of clinical items,” reflecting the key initial information and decision making during a hospital admission. We randomly selected 10 655 (∼50%) patients to form a training set. We chose to use the GenSim package47 to infer topic model structure, given its convenient implementation in Python, streaming input of large data corpora, and parallelization to efficiently use multicore computing. Model inference requires an external parameter for the expected number of topics, for which we systematically generated models with topic counts ranging from 2 to 2048. Running the model training process on a single Intel 2.4 GHz core for 10 655 patients and 256 topics requires ∼1 GB of main memory and ∼2 minutes of training time. Maximum memory usage and training time increases proportionally to the number of topics modeled, while the streaming learning algorithm requires more execution time but no additional main memory when processing additional training documents.
Evaluation
To evaluate the utility of the generated clinical topic models and determine an optimal topic count range, we assessed their ability to predict subsequent clinical orders. For a separate random selection of 4820 (∼25%) validation patients, we isolated each use of a pre-existing human-authored order set within the first 24 hours of each hospitalization. We simulated production of an individually personalized, topic model-based “order set” at each such moment in time. To dynamically generate this content, the system evaluates the patient’s available clinical data to infer the relative weight of relevance for each clinical topic, P(Topicj|Patientk). With this patient topic distribution defined, the system can then score-rank a list of suggested orders by the probability of each order occurring for the patient:
We compared these clinical order suggestions against the “correct” set of orders that actually occurred for the patient within a followup verification time of t. Sensitivity analyses with respect to this followup verification time varied t from 1 minute (essentially counting only orders drawn from the immediate real order set usage) up to 24 hours afterwards. Prediction of these subsequent orders is evaluated by the area under the receiver operating characteristic curve (c-statistic) when considering the full score-ranked list of all possible clinical orders. Existing order sets will have N suggested orders to choose from, so we evaluated those N items vs the top N score-ranked suggestions from the topic models toward predicting subsequent orders by precision (positive predictive value) at N and recall (sensitivity) at N. We executed paired, 2-tailed t-tests to compare results with SciPy.48
Results
Table 1 reports the names of the most commonly used human-authored inpatient order sets, while Table 2 reports summary usage statistics during the first 24 hours of hospitalization. Table 3 illustrates example clinical topics inferred from the structured clinical data. Figure 2 visualizes additional example topics and how patient-topic weights can be used to predict additional clinical orders. Figures 3 and 4 summarize clinical order prediction rates using clinical topic models vs human-authored order sets.
Table 4.
Summary of relative tradeoffs between manually authored order sets vs algorithmically generated order suggestions
| Aspect | Order sets | Topic models |
|---|---|---|
| Production | Manual development | Automated generation |
| Construction | Preconceived concepts | Underlying data structure |
| Usability | Interruptive workflow | Passive dynamic adaption |
| Applicability | Isolated scenarios | Composite patient context |
| Interpretability | Annotated rationale | Numerical associations |
| Reliability | Clinical judgment | Statistical significance |
Table 1.
Most commonly used human-authored inpatient order sets
| Use rate (%) | Size | Description |
|---|---|---|
| 35.1 | 51 | Anesthesia—Post-Anesthesia (Inpatient) |
| 27.2 | 161 | Medicine—General Admit |
| 23.5 | 51 | General—Pre-Admission/Pre-Operative |
| 18.4 | 17 | Insulin–Subcutaneous |
| 15.7 | 28 | General—Transfusion |
| 9.3 | 13 | General—Discharge |
| 7.6 | 150 | Surgery—General Admit |
| 6.9 | 9 | Emergency—Admit |
| 6.1 | 224 | Intensive Care—General Admit |
| 5.9 | 147 | Orthopedics—Total Joint Replacement |
| 4.9 | 46 | Pain—Regional Anesthesia Admit |
| 4.4 | 80 | Emergency—General Complaint |
| 4.1 | 40 | Anesthesia—Post-Anesthesia (Outpatient) |
| 3.9 | 135 | Orthopedics Trauma |
| 3.9 | 9 | Pain—Patient Controlled Analgesia |
| 3.4 | 168 | Psychiatry—Admit |
| 3.3 | 132 | Neurosurgery–Intensive Care |
| 3.3 | 16 | General—Heparin Protocols |
| 3.0 | 39 | Pain—Epidural Analgesia Post-Op |
| 2.9 | 11 | Insulin—Subcutaneous Adjustment |
| 2.7 | 9 | Lab—Blood Culture and Infection |
| 2.6 | 155 | Neurology—General Admit |
| 2.5 | 169 | Intensive Care—Surgery/Trauma Admit |
| 2.4 | 9 | Pharmacy—Warfarin Protocol |
| 2.4 | 14 | Insulin—Intravenous Infusion |
| … | … | … |
Use rate reflects the percentage of validation patients for whom the order set was used within the first 24 hours of hospitalization. Size reflects the number of order suggestions available in each order set. Notably, these essentially all reflect nonspecific care processes, while scenario specific order sets (e.g., management of asthma, heart attacks, pneumonia, sepsis, or gastrointestinal bleeds) are rarely used.
Table 2.
Summary statistics for human-authored order set use within the first 24 hours of hospitalization for 4820 validation patients
| Metric (per first 24 hours of each hospitalization) | Mean (std dev) | Median interquartile range |
|---|---|---|
| A: Order sets used | 3.0 | 3 |
| (1.4) | (2, 4) | |
| B: Orders entered (including non-order set) | 32.7 | 30 |
| (15.7) | (22, 41) | |
| C: Orders entered from order sets | 13.3 | 12 |
| (7.9) | (8, 18) | |
| D: Orders available from used order sets | 129.0 | 130 |
| (47.5) | (102, 153) | |
| E: Order set precision = (C/D) | 11% | 9.5% |
| (7.2%) | (6.3%, 13.8%) | |
| F: Order set recall = (C/B) | 43% | 42% |
| (20%) | (28%, 58%) |
Metrics count only orders used in the final set of 1512 preprocessed clinical orders after normalization of medication orders and exclusion of rare orders and common process orders.
Table 3.
Example clinical topics generated when modeling 32 topics from training patient data
| Weight (%) | Clinical item | Weight | Clinical item | Weight | Clinical item |
|---|---|---|---|---|---|
| 2.37 | PoC Arterial Blood Gas | 1.56 | Insulin Lispro (Subcutaneous) | 2.98 | Culture + Gram Stain, Fluid |
| 1.39 | Team—Respiratory Tech | 1.26 | Metabolic Panel, Basic | 2.42 | Cell Count and Diff, Fluid |
| 1.38 | Lactate, Whole Blood | 1.09 | Dx—Diabetes mellitus (250) | 1.59 | Protein Total, Fluid |
| 1.32 | XRay Chest 1 View | 1.08 | Dx—DM w/o complication (250.0) | 1.51 | Albumin, Fluid |
| 1.14 | Blood Gases, Venous | 1.03 | Dx—DM not uncontrolled (250.00) | 1.24 | LDH Total, Fluid |
| 1.00 | Ventilator Settings Change | 1.01 | Hemoglobin A1c | 1.10 | Albumin (IV) |
| 0.97 | Blood Gases, Arterial | 0.99 | Diet—Low Carbohydrate | 1.09 | Glucose, Fluid |
| 0.96 | Vancomycin (IV) | 0.91 | CBC w/ Diff | 0.84 | Pathology Review |
| 0.92 | PoC Arterial Blood Gas B | 0.91 | Sodium Chloride (IV) | 0.68 | Team—Registered Nurse |
| 0.88 | Epinephrine (IV) | 0.89 | Diagnosis—Essential hypertension | 0.67% | CBC w/Diff |
| 0.88 | Norepinephrine (IV) | 0.82% | MRSA Screen | 0.61 | MRSA Screen |
| 0.87 | Central Line | 0.75 | Team—Registered Nurse | 0.60 | XRay Chest 1 View |
| 0.86 | Team—Medical ICU | 0.73 | Regular Insulin (Subcutaneous) | 0.53 | Albumin, Serum |
| 0.84 | Sodium Bicarbonate (IV) | 0.73 | XRay Chest 1 View | 0.52 | Metabolic Panel, Basic |
| 0.81 | MRSA Screen | 0.67 | Fungal Culture | 0.51 | Cytology |
| 0.79 | Hepatic Function Panel | 0.66 | Anaerobic Culture | 0.50 | Amylase, Fluid |
| 0.76 | Midazolam (IV) | 0.66 | Consult—Diabetes Team | 0.47 | Prothrombin Time (PT/INR) |
| 0.73 | Result—Lactate (High) | 0.65 | Team—Respiratory Tech | 0.47 | Midodrine (Oral) |
| 0.73 | Result—TCO2 (Low) | 0.63 | Admit—Thoracolumbar… (722.1) | 0.47 | Male Gender |
| 0.72 | NIPPVentilation | 0.63 | Admit—Lumbar Disp. (722.10) | 0.42 | Result—RBC (Low) |
| 0.69 | Result—pH (Low) | 0.62 | EKG 12-Lead | 0.41 | Sodium Chloride (IV) |
| … | … | … | |||
| 21 | “Intensive Care” | 21% | “Diabetes mellitus” | 16% | “Ascites/Effusion Workup” |
| Weight | Clinical item | Weight | Clinical item | Weight | Clinical item |
|---|---|---|---|---|---|
| 2.37 | Team—Respiratory Tech | 3.48 | Cell Count and Diff, CSF | 1.63 | Lupus Anticoagulant |
| 2.19 | Nebulizer Treatment | 3.28 | Glucose, CSF | 1.35 | Dx—Pulmonary emb… (415.1) |
| 1.64 | Respiratory Culture | 3.25 | Protein Total, CSF | 1.34 | Dx—Pulmonary heart Dz (415) |
| 1.29 | Blood Culture (An)Aerobic | 2.98 | Culture and Gram Stain, CSF | 1.34 | Factor V Leiden |
| 1.27 | Team—Registered Nurse | 0.95 | Enterovirus PCR, CSF | 1.33 | Dx—Other PEmbolism (415.19) |
| 1.26 | Blood Culture (Aerobic x2) | 0.74 | West Nile Virus AB, CSF | 1.16 | Prothrombin 20210A |
| 1.25 | Droplet Isolation | 0.63 | Coccidioides AB, CSF | 1.16 | Homocysteine |
| 1.20 | Respiratory DFA Panel | 0.61 | Cytology | 1.02 | Protein C Activity |
| 1.17 | CBC w/ Diff | 0.39 | Zonisamide (Oral) | 1.01 | Protein S Activity |
| 1.17 | Vancomycin (IV) | 0.34 | Team—Neurology | 0.72 | Admit—PEmbolism (415.1) |
| 1.08 | Gram Stain | 0.33 | Cytology Exam | 0.71 | Admit—Pulm heart Dz (415) |
| 1.01 | Albuterol-Ipratropium (Inh) | 0.24 | Result—WBC, CSF (High) | 0.69 | Admit—Other PE (415.19) |
| 0.98 | Metabolic Panel, Basic | 0.24 | HSV PCR, CSF | 0.66 | Anti-Phospholipid AB Panel |
| 0.90 | XRay Chest 2 View | 0.23 | Cryptococcal AG, CSF | 0.54 | Methylprednisolone (Oral) |
| 0.79 | Prednisone (Oral) | 0.20 | Fungal Culture | 0.52 | Rapid HIV-1/2 AB |
| 0.79 | Sodium Chloride (IV) | 0.13 | Valproic Acid, Serum | 0.33 | Dx—Osteomyelitis (730.2) |
| 0.77 | Levofloxacin (IV) | 0.10 | IgA, Serum | 0.29 | Dx—Bone Infection (730) |
| 0.77 | Prothrombin Time (PT/INR) | 0.08 | Team—Neurology Consult | 0.29 | Warfarin (Oral) |
| 0.72 | Azithromycin (Oral) | 0.08 | Clonazepam (Oral) | 0.29 | Team—Registered Nurse |
| 0.72 | Pantoprazole (Oral) | 0.07 | ANA (Anti-Nuclear AB) | 0.29 | Partial Thromboplastin Time |
| 0.71 | Magnesium, Serum | 0.07 | Levetiracetam (Oral) | 0.28 | Factor VIII Assay |
| … | … | … | |||
| 16% | “Pneumonia” | 13% | “Neuro CSF Workup” | 10% | “PE / Hypercoaguability Workup” |
The most prominent clinical items (e.g., medications, imaging, laboratory orders, and results) are listed for each example topic, with corresponding P(Itemi|Topicj) weights. The bottom rows reflect the percentage of validation patients with estimated P(Topicj|Patientk) > 1% along with our manually ascribed labels that summarize the largely interpretable topic contents.
Abbreviations: AB: Antibody, AG: Antigen, CBC: Complete blood count, CSF: Cerebrospinal fluid, Diff: Differential, Disp: Displacement, DFA: Direct fluorescent antibody, DM: Diabetes mellitus, Dx: Diagnosis, Dz: Disease, HSV: Herpes simplex virus, ICU: Intensive care unit, Inh: Inhaled, INR: International normalized ratio, IV: Intravenous, LDH: Lactate dehydrogenase, MRSA: Methicillin resistant Staphylococcus aureus, NIPPV: Noninvasive positive pressure ventilation, PE: Pulmonary embolism, PoC: Point-of-care, PCR: Polymerase chain reaction, RBC: Red blood cells, TCO2: Total carbon dioxide, WBC: White blood cells.
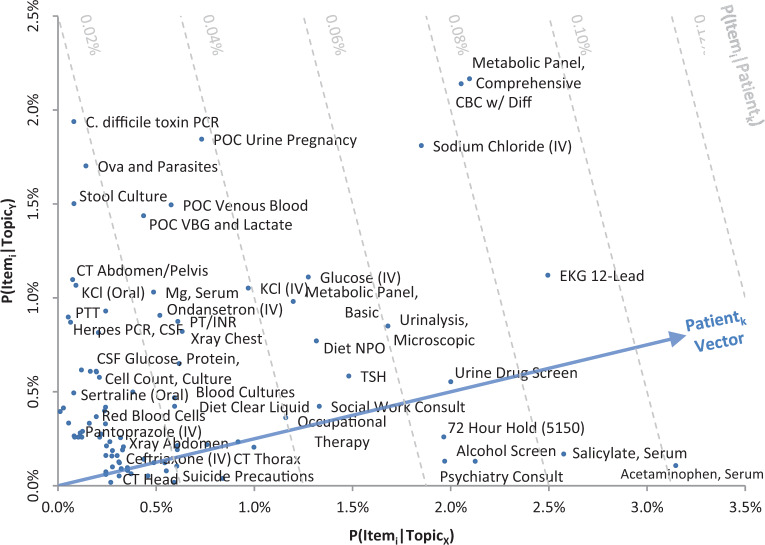
Figure 2.
Example of 2 generated clinical topics plotted in a 2-dimensional space. Only clinical orders are plotted, based on their prominence in each of the topics. The top left reflects clinical orders most associated with TopicY, with little association with TopicX, suggestive of a workup for diarrhea and abdominal pain. The bottom right reflects clinical orders associated with TopicX, suggestive of a workup for an intentional (medication) overdose and involuntary psychiatric hospitalization. The top right reflects common clinical orders that are associated with both topics. For legibility, items whose score is < 0.2% for both topics are omitted and only a subsample of the bottom-left items are labeled. The diagonal arrow represents a hypothetical patient inferred to have P(TopicX|Patientk) = 80% and P(TopicY|Patientk) = 20%. The dashed lines reflect orthogonal P(Itemi|Patientk) isolines to visually illustrate how clinical order suggestions can be made from such a topic inference. In this case, orders farthest along the projected patient vector (e.g., serum acetaminophen) are predicted to be most relevant for the patient.
Figure 3.
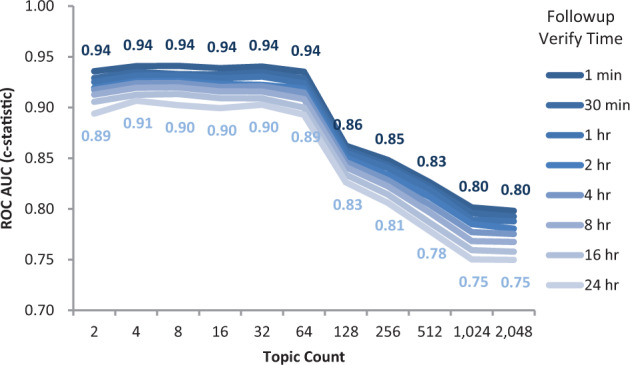
Topic count selection. Average discrimination accuracy (ROC AUC) when predicting additional clinical orders occurring within t followup verification time of the invocation of a pre-authored order set during the first 24 hours of hospitalization for 4820 validation patients. Predictions based on Latent Dirichlet allocation (LDA) topic models trained on 10 655 separate training patients. The standard LDA algorithm requires external specification of a topic count parameter to indicate the number of latent dimensions by which to organize the source data, which varies along this X-axis from 2 to 2048. Peak performance occurs around a choice of 32 topics, degrading once attempting to model > 64 topics.
Figure 4.
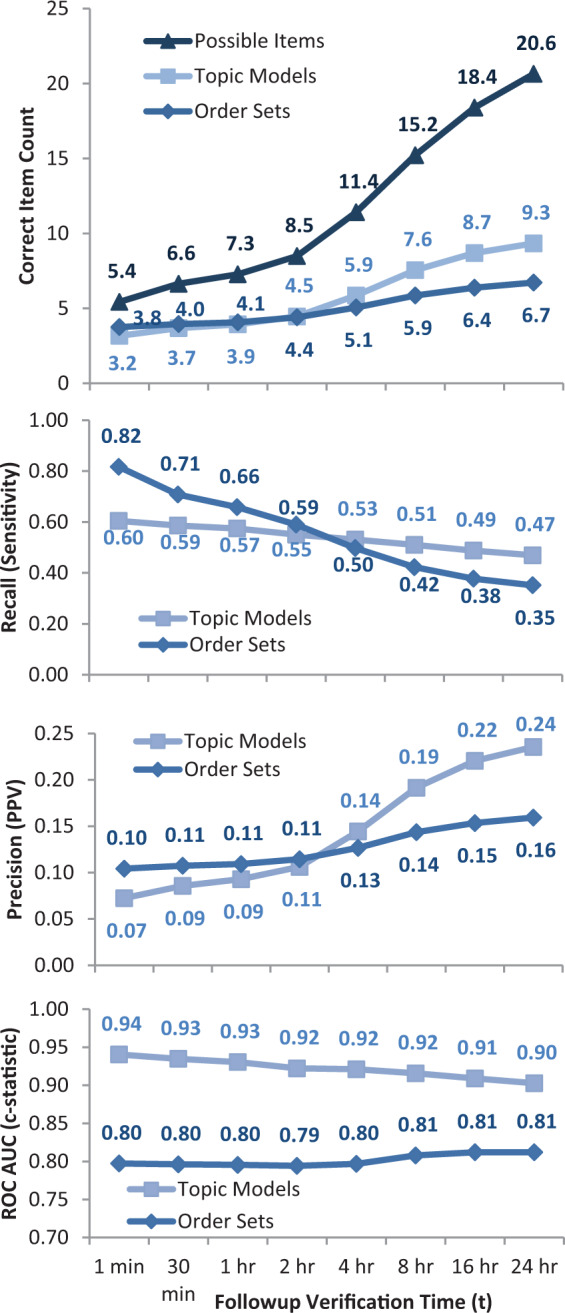
(A) Topic models vs order sets for different followup verification times. For each real use of a preauthored order set, either that order set or a topic model (with 32 trained topics) was used to suggest clinical orders. For longer followup times, the number of subsequent possible items considered correct increases from an average of 5.4–20.6. The average correct predictions in the immediate timeframe is similar for topic models (3.2) and order sets (3.8), but increases more for topic models (9.3) vs order sets (6.7) when forecasting up to 24 hours. At the time of order set usage, physicians choose an average of 3.8 orders out of 54.8 order set suggestions, as well as 1.6 = (5.4 – 3.8) a la carte orders. (B) Topic models vs order sets by recall at N. For longer followup verification times, more possible subsequent items are considered correct (see 4A). This results in an expected decline in recall (sensitivity). Order sets, of course, predict their own immediate use better, but lag behind topic model-based approaches when anticipating orders beyond 2 hours (P < 10−20 for all times). (C) Topic models vs order sets by precision at N. For longer followup verification times, more subsequent items are considered correct, resulting in an expected increase in precision (positive predictive value). Again, topic model-based approaches are better at anticipating clinical orders beyond the initial 2 hours after order set usage (P < 10−6 for all times). (D) Topic models vs order sets by ROC AUC (c-statistic), evaluating the full ranking of possible orders scored by topic models or included/excluded by order sets (P < 10−100 for all times).
Discussion
Complex clinical data like clinical orders, lab results, and diagnoses extracted from EHRs can be automatically organized into thematic structures through probabilistic topic modeling. These thematic topics can be used to automatically generate natural “order sets” of commonly co-occurring clinical data items, as illustrated in the examples in Table 3. Figure 2 visually illustrates how these latent topics can separate clinical items that are specific or general across varying scenarios, and how they can be used to generate personalized clinical order suggestions for individual patients. Suggestions have some interpretable rationale by indicating that a patient case in question appears to be “about” a given set of clinical topics (e.g., abdominal pain and involuntary psychiatric hold) and the suggested orders (e.g., serum acetaminophen level, Electrocardiogram (EKG) 12-Lead) are those that commonly occur for other patient cases involving those topics.
In the absence of a gold standard to define high-quality medical decision making, we must establish a benchmark to evaluate the quality of algorithmically generated decision support content. Human-authored order sets and alerts represent the current standard of care in clinical decision support. Figure 4 indicates that existing order sets are slightly better than topic model-generated order suggestions at anticipating physician orders within the immediate time period (< 2 h). This is, of course, biased in favor of the existing order sets since the evaluation time points were specifically chosen where an existing order set was used. This ignores other time points where the clinicians did not (or could not) find a relevant order set, but where an automated system could have generated personalized suggestions. Topic model-based methods consistently predict more future orders than the existing order sets when forecasting longer followup time periods beyond 2 hours.
On an absolute scale, it is interesting that manually produced content like order sets continues to demonstrate improvements in care21,23,24,49 despite what we have found to be a low “accuracy” of recommendations. Table 2 indicates that initial inpatient care on average involves a few order sets (3.0), with a preference for general order sets with a large number of suggested orders (> 100), resulting in higher recall (43%) but low precision (11%). This illustrates that such tools are decision aids that benefit clinicians who can interpret the relevance of any suggestions to their individual patient’s context.
Framed as an information retrieval problem in clinical decision support, retrieval accuracy may not even be as important as other aspects for real-world implementation (e.g., speed, simplicity, usability, maintainability).26 Even if algorithmically generated suggestions were only as good as the existing order sets, the more compelling implication is how this can alter the production and usability of clinical decision support. Automated approaches can generate content spanning any previously encountered clinical scenario. While this incurs the risk of finding “mundane” structure (e.g., the repeated sub-diagnosis codes for diabetes and pulmonary embolism in Table 3), it is a potentially powerful unsupervised approach to discovering latent structure that is not dependent on the preconceptions of content authors. The existing workflow for pre-authored order sets requires clinicians to previously be aware of, or spend their time searching for, order sets relevant to their patient’s care. Table 1 illustrates that clinicians favor a few general order sets focused on provider processes (e.g., admission, insulin, transfusion), while they rarely use order sets for patient-focused scenarios (e.g., stroke, sepsis). With the methods presented here, automated inference of patient context could overcome this usability barrier by inferring relevant clinical “topics” (if not specific clinical orders) based on information already collected in the EHR (e.g., initial orders, problem list, lab results). Such a system could present related order sets (human-authored or machine-learned) to the clinician without the clinician ever having to explicitly request or search for a named order set. The tradeoff for these potential benefits is that current physicians are more likely comfortable with the interpretability and human origin of manually produced content.
Most of the initial applications of topic modeling have been for text document organization.33–35 More recent work has applied topic modeling and similar low-dimensional representations to clinical data for the unsupervised determination of clinical phenotypes38 and concept embeddings,40 or as features toward classification tasks such as high-cost prediction.39 Other efforts to algorithmically predict clinical orders have mostly focused on problem spaces with dozens of possible candidate items.50–53 In comparison, the problem space in this manuscript includes over 1000 clinical items. This results in substantially different expected retrieval rates,54 even as the latent topics help address data interpretability, sparsity, and semantic similarity. While there is likely further room for improvement, perhaps with other graphical models specifically intended for recommender applications,55 our determination of order set retrieval rates contributes to the literature by defining the state-of-the-art real-world reference benchmark for this and any future evaluations.
Limitations of the LDA topic modeling approach include external designation of the topic count parameter. Similarly, while we used default model hyperparameters that assume a symmetric prior, this may affect the coherence of the model.56 Hierarchical Dirichlet process57 topic modeling is an alternative nonparametric approach that determines the topic count by optimizing observed data perplexity;58 however, this may not align with the application of interest. Validating against a held-out set of patients allowed us to optimize the topic count against an outcome measure like order prediction. Precision and recall is optimized in this case with approximately 32 topics of inpatient admission data. Another key limitation is that the standard LDA model interprets data as an unordered “bag of words,” which discards temporal data on the sequence of clinical data. Our prior work noted the value of temporal data toward improving predictions.59 This could potentially be addressed with alternative topic model algorithms that account for such sequential data.60
Another limitation of any unsupervised learning process is that it can yield content with variable interpretability. For example, while we manually ascribe labels to the topics in Table 3, the contents are ultimately defined by the underlying structure of the data and need not map to preconceived medical categorizations. This is reflected in the presence of items such as admission diagnoses of thoraco-lumbar disc displacement, osteomyelitis, and tests for rapid Human Immunodeficiency Virus (HIV) antibodies that do not seem to fit our artificial labels. From an exploratory data analysis perspective, however, this may actually be useful in identifying latent concepts in the clinical data that could not be anticipated prospectively. When we discarded rare clinical items (< 256 instances), we may also have lost precision on the most important data elements. As noted in our prior work, this design decision trades the potential of identifying rare but “interesting” elements in favor of predictions more likely to be generally relevant and that avoid statistically spurious cases with insufficient power to make sensible predictions.61
Organization of clinical data through probabilistic topic modeling provides an automated approach to detecting thematic trends in patient care. A potential use case illustrated here finds related clinical orders for decision support based on inferred underlying topics. This has the general potential for clinical information summarization13,62 that dynamically adapts to changing clinical practices,63 which would otherwise be limited to preconceived concepts manually abstracted out of potentially lengthy and complex patient chart reviews. Such algorithmic approaches are critical to unlocking the potential of large-scale health care data sources to impact clinical practice.
Contributors
JHC conceived the study and design, and implemented the algorithms, performed the analysis, and drafted the initial manuscript. MKG, SMA, LM, and RBA contributed to analysis design and manuscript revisions, and supervised the study.
Funding
This research was supported by the National Institute of Environmental Health Sciences of the National Institutes of Health under Award Number K01ES026837, the Stanford Translational Research and Applied Medicine program in the Department of Medicine, and the Stanford Learning Healthcare Systems Innovation Fund and the Stanford Clinical and Translational Science Award (CTSA) to Spectrum (UL1 TR001085). The CTSA program is led by the National Center for Advancing Translational Sciences at the National Institutes of Health. J.H.C. was supported in part by VA Office of Academic Affiliations and Health Services Research and Development Service Research funds. R.B.A. is supported by National Institutes of Health/National Institute of General Medical Sciences PharmGKB resource R24GM61374, as well as LM05652 and GM102365. Additional support is from the Stanford NIH/National Center for Research Resources CTSA award number UL1 RR025744. Patient data were extracted and de-identified by Tanya Podchiyska, Lee Ann Yasukawa, Gomathi Krishnan, and Susan Weber of the Stanford Translational Research Integrated Database Environment (STRIDE) project, a research and development project at Stanford University to create a standards-based informatics platform supporting clinical and translational research. The STRIDE project described was supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through grant UL1 RR025744. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH, VA, or Stanford Healthcare.
Competing Interests
The authors have no competing interests to declare.
REFERENCES
- 1. Richardson WC, Berwick DM, Bisgard JC. et al. . Crossing the Quality Chasm: A New Health System for the 21st Century. Washington DC: Natl Acad Press, Institute of Medicine, Committee on Quality of Health Care in America Committee on Quality of Health Care in America; 2001. [Google Scholar]
- 2. Tricoci P, Allen JM, Kramer JM, Califf RM, Smith CS. Scientific evidence underlying the ACC/AHA. J Am Med Inform Assoc. 2009;301:831–41. [DOI] [PubMed] [Google Scholar]
- 3. Lauer MS, Bonds D. Eliminating the ‘expensive’ adjective for clinical trials. Am Heart J. 2014;167:419–20. [DOI] [PubMed] [Google Scholar]
- 4. Tricoci P, Allen JM, Kramer JM. et al. . Scientific evidence underlying the ACC/AHA clinical practice guidelines. JAMA. 2009;301:831–41. [DOI] [PubMed] [Google Scholar]
- 5. Durack DT. The weight of medical knowledge. N Engl J Med. 1978;298:773–5. [DOI] [PubMed] [Google Scholar]
- 6. Alper J, Grossmann C. Health System Leaders Working Toward High-Value Care. Washington DC: The National Academies Press; 2014. [Google Scholar]
- 7. ONC. Health information technology: standards, implementation specifications, and certification criteria for electronic health record technology, 2014 edition; revisions to the permanent certification program for health information technology. Final rule. Fed Regist. 2012;77:54163–292. [PubMed] [Google Scholar]
- 8. Longhurst CA, Harrington RA, Shah NH. A ‘Green Button’ for using aggregate patient data at the point of care. Health Aff. 2014;33:1229–35. [DOI] [PubMed] [Google Scholar]
- 9. Frankovich J, Longhurst CA, Sutherland SM. Evidence-based medicine in the EMR era. N Engl J Med. 2011;365:1758–9. [DOI] [PubMed] [Google Scholar]
- 10. Smith M, Saunders R, Stuckhardt L, McGinnis JM. Best Care at Lower Cost: the Path to Continuously Learning Health Care in America. Washington DC: Institute of Medicine, Committee on the Learning Health Care System in America; 2012. [PubMed] [Google Scholar]
- 11. Krumholz HM. Big data and new knowledge in medicine: The thinking, training, and tools needed for a learning health system. Health Aff. 2014;33:1163–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Lissovoy G. Big data meets the electronic medical record: a commentary on ‘identifying patients at increased risk for unplanned readmission’. Med Care. 2013;51:759–60. [DOI] [PubMed] [Google Scholar]
- 13. Feblowitz JC, Wright A, Singh H, Samal L, Sittig DF. Summarization of clinical information: a conceptual model. J Biomed Inform. 2011;44:688–99. [DOI] [PubMed] [Google Scholar]
- 14. Pivovarov R, Elhadad N. Automated methods for the summarization of electronic health records: Table 1. J Am Med Inform Assoc. 2015;22:938–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sittig DF, Wright A, Osheroff JA. et al. . Grand challenges in clinical decision support. J Biomed Inform. 2008;41:387–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaushal R, Shojania KG, Bates DW. Effects of computerized physician order entry and clinical decision support systems on medication safety: a systematic review. Arch Intern Med. 2003;163:1409–16. [DOI] [PubMed] [Google Scholar]
- 17. Cowden D, Barbacioru C, Kahwash E, Saltz J. Order sets utilization in a clinical order entry system. AMIA Annu Symp Proc. 2003;819. [PMC free article] [PubMed] [Google Scholar]
- 18. Payne TH, Hoey PJ, Nichol P, Lovis C. Preparation and use of preconstructed orders, order sets, and order menus in a computerized provider order entry system. J Am Med Inform Assoc. 2003;10:322–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bobb AM, Payne TH, Gross PA. Viewpoint: controversies surrounding use of order sets for clinical decision support in computerized provider order entry. J Am Med Inform Assoc. 2007;14:41–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kaushal R, Shojania KG, Bates DW. Effects of computerized physician order entry and clinical decision support systems on medication safety: a systematic review. Arch Intern Med. 2003;163:1409–16. [DOI] [PubMed] [Google Scholar]
- 21. Overhage J, Tierney W. A randomized trial of ‘corollary orders’ to prevent errors of omission. J Am Med Inform Assoc. 1997;4:364–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ballard DW, Kim AS, Huang J. et al. . Implementation of computerized physician order entry is associated with increased thrombolytic administration for emergency department patients with acute ischemic stroke. Ann Emerg Med. 2015;1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ballesca MA, LaGuardia JC, Lee PC. et al. . An electronic order set for acute myocardial infarction is associated with improved patient outcomes through better adherence to clinical practice guidelines. J Hosp Med. 2014;9:155–61. [DOI] [PubMed] [Google Scholar]
- 24. Micek ST, Roubinian N, Heuring T. et al. . Before-after study of a standardized hospital order set for the management of septic shock. Crit Care Med. 2006;34:2707–13. [DOI] [PubMed] [Google Scholar]
- 25. Jacobs BR, Hart KW, Rucker DW. Reduction in clinical variance using targeted design changes in computerized provider order entry (CPOE) order sets: impact on hospitalized children with acute asthma exacerbation. Appl Clin Inform. 2012;3:52–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bates DW, Kuperman GJ, Wang Samuel. et al. . Ten commandments for effective clinical decision support: making the practice of evidence-based medicine a reality. J Am Med Inform Assoc. 2003;10:523–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Newton KM, Peissig PL, Kho AN. et al. . Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc. 2013;20:e147–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. J Am Med Inform Assoc. 2013;20:117–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Blei DM, Ng AY, Jordan MI. Latent Dirichlet allocation. J Mach Learn Res. 2003;3: 993–1022. [Google Scholar]
- 30. Blei DM. Introduction to probabilistic topic modeling. Commun ACM. 2012;55:77–84. [Google Scholar]
- 31. Steyvers M, Griffiths T. Probabilistic Topic Models. In Handbook of Latent Semantic Analysis. New York, NY, USA: Routledge; 2011. [Google Scholar]
- 32. Blei D, Carin L, Dunson D. Probabilistic topic models. IEEE Signal Process Mag. 2010;27:55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Blei DM, Lafferty JD. Topic models. In: Text Mining: Classification, Clustering, and Applications. Boca Raton, FL: Chapman & Hall/CRC; 2009. [Google Scholar]
- 34. Wang C, Blei DM. Collaborative topic modeling for recommending scientific articles. Proc 17th ACM SIGKDD Int Conf Knowl Discov data Min - KDD ’11. 2011;448.. Available at: http://dl.acm.org/citation.cfm?id=2020480. [Google Scholar]
- 35. Barajas KLC, Akella R. Incorporating statistical topic models in the retrieval of healthcare documents. CLEF eHealth. 2013 Proc 2013; 1–5. [PMC free article] [PubMed] [Google Scholar]
- 36. Huang Z, Dong W, Duan H. A probabilistic topic model for clinical risk stratification from electronic health records. J Biomed Inform. 2015;58:28–36. [DOI] [PubMed] [Google Scholar]
- 37. Lu H-M, Wei C-P, Hsiao F-Y. Modeling healthcare data using multiple-channel latent Dirichlet allocation. J Biomed Inform. 2016;60:210–23. [DOI] [PubMed] [Google Scholar]
- 38. Pivovarov R, Perotte AJ, Grave E, Angiolillo J, Chris H. Learning probabilistic phenotypes from heterogeneous EHR Data. J Biomed Inform. 2015;58:156–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ho J, Ghosh J, Sun J. Marble: high-throughput phenotyping from electronic health records via sparse nonnegative tensor factorization. Proc 20th ACM SIGKDD Int Conf Knowl Discov data Min - KDD ’14 2014;2014:115–24. [Google Scholar]
- 40. Choi Y, Chiu CY-I, Sontag D. Learning low-dimensional representations of medical concepts. AMIA Jt Summits Transl Sci Proc. 2016:41–50. [PMC free article] [PubMed] [Google Scholar]
- 41. Fodor IK. A survey of dimension reduction techniques. Library (Lond) 2002;18:1–18. [Google Scholar]
- 42. Koren Y, Bell R, Volinsky C. Matrix factorization techniques for recommender systems. Computer. 2009;42:30–7. [Google Scholar]
- 43. Stevens K, Kegelmeyer P, Andrzejewski D, Buttler D. Exploring topic coherence over many models and many topics. Proc 2012 Jt Conf Empir Methods Nat Lang Process Comput Nat Lang Learn. 2012;952–61. [Google Scholar]
- 44. Lowe HJ, Ferris TA, Hernandez PM, Weber SC. STRIDE–An integrated standards-based translational research informatics platform. AMIA Annu Symp Proc. 2009;2009:391–5. [PMC free article] [PubMed] [Google Scholar]
- 45. Hernandez P, Podchiyska T, Weber S, Ferris T, Lowe H. Automated mapping of pharmacy orders from two electronic health record systems to RxNorm within the STRIDE clinical data warehouse. AMIA Annu Symp Proc. 2009;2009:244–8. [PMC free article] [PubMed] [Google Scholar]
- 46. Wright A, Bates DW. Distribution of problems, medications and lab results in electronic health records: the pareto principle at work. Appl Clin Inform. 2010;1:32–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rehurek R, Sojka P. Software framework for topic modelling with large corpora. Proc Lr 2010 Work New Challenges NLP Fram. 2010;45–50. [Google Scholar]
- 48. Jones E, Oliphant T, Peterson P, Al E. SciPy: Open Source Scientific Tools for Python. Available at: http://www.scipy.org. Accessed September 2016. [Google Scholar]
- 49. Kawamoto K, Houlihan CA, Balas EA, Lobach DF. Improving clinical practice using clinical decision support systems: a systematic review of trials to identify features critical to success. BMJ. 2005;330:765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Klann JG, Szolovits P, Downs SM, Schadow G. Decision support from local data: creating adaptive order menus from past clinician behavior. J Biomed Inform. 2014;48:84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hasan S, Duncan GT, Neill DB, Padman R. Automatic detection of omissions in medication lists. J Am Med Inform Assoc. 2011;18:449–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wright AP, Wright AT, McCoy AB, Sittig DF. The use of sequential pattern mining to predict next prescribed medications. J Biomed Inform. 2014;53:73–80. [DOI] [PubMed] [Google Scholar]
- 53. Zhang Y, Padman R, Levin JE. Paving the COWpath: data-driven design of pediatric order sets. J Am Med Inform Assoc. 2014;21:e304–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schröder G, Thiele M, Lehner W. Setting goals and choosing metrics for recommender system evaluations. in CEUR Workshop Proc. 2011. [Google Scholar]
- 55. Gopalan P, Hofman JM, Blei DM. Scalable recommendation with poisson factorization. arXiv Prepr. 2013;1–10. http://arxiv.org/pdf/1311.1704. Accessed September 2016. [Google Scholar]
- 56. Wallach HM, Mimno DM, Mccallum A. In Bengio Y, Schuurmans D, Lafferty JD, Williams CKI, Culotta A. Adv Neural Inf Process Syst. Vol. 22. Curran Associates, Inc.; 2009;1973–81. [Google Scholar]
- 57. Wang C, Paisley J, Blei DM. Online variational inference for the hierarchical dirichlet process. Proc Fourteenth Int Conf Artif Intell Stat. 2011;15:752–60. [Google Scholar]
- 58. Wallach HM, Murray I, Salakhutdinov R, Mimno D. Evaluation methods for topic models. Int Conf Mach Learn. 2009;1–8. [Google Scholar]
- 59. Chen JH, Altman RB. Automated physician order recommendations and outcome predictions by data-mining electronic medical records. AMIA Jt Summits Transl Sci Proc AMIA Summit Transl Sci. 2014;2014:206–10. [PMC free article] [PubMed] [Google Scholar]
- 60. Barbieri N, Manco G, Ritacco E, Carnuccio M, Bevacqua A. Probabilistic topic models for sequence data. Mach Learn. 2013;93:5–29. [Google Scholar]
- 61. Chen JH, Altman RB. Mining for clinical expertise in (undocumented) order sets to power an order suggestion system. AMIA Jt Summits Transl Sci Proc AMIA Summit Transl Sci. 2013;2013:34–8. [PMC free article] [PubMed] [Google Scholar]
- 62. Pivovarov R, Elhadad N. Automated methods for the summarization of electronic health records. J Am Med Inform Assoc. 2015;22:938–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Blei DM, Lafferty JD. Dynamic topic models. Proc 23rd Int Conf Mach Learn (ICML 2006). 2006;113–20. [Google Scholar]