Abstract
Piwi-interacting RNAs (piRNAs) are 26–30-nucleotide germ line-specific small non-coding RNAs that have evolutionarily conserved function in mobile genetic element (transposons) silencing and maintenance of genome integrity. Drosophila Hsp70/90-organizing protein homolog (Hop), a co-chaperone, interacts with piRNA-binding protein Piwi and mediates silencing of phenotypic variations. However, it is not known whether Hop has a direct role in piRNA biogenesis and transposon silencing. Here, we show that knockdown of Hop in the germ line nurse cells (GLKD) of Drosophila ovaries leads to activation of transposons. Hop GLKD females can lay eggs at the same rate as wild-type counterparts, but the eggs do not hatch into larvae. Hop GLKD leads to the accumulation of γ-H2Av foci in the germ line, indicating increased DNA damage in the ovary. We also show that Hop GLKD-induced transposon up-regulation is due to inefficient piRNA biogenesis. Based on these results, we conclude that Hop is a critical component of the piRNA pathway and that it maintains genome integrity by silencing transposons.
Keywords: DNA damage response, heat shock protein (HSP), heat shock protein 90 (Hsp90), RNA, RNA interference (RNAi)
Introduction
The piRNA2 pathway differentiates itself from other RNA interference pathways through its action exclusively in the germ line. It works to suppress transposable elements (TE) through sequence-specific targeting using 23–29-nucleotide (nt) RNA molecules called piRNAs (1–5). In Drosophila, three PIWI clade proteins, Piwi, Aubergine (Aub), and Argonaute 3 (Ago3), bind piRNAs. All three proteins have a non-redundant function in silencing TE.
piRNAs originate from large genomic elements called piRNA clusters and active transposons (6). In the germ line nurse cells, piRNA biogenesis occurs in a perinuclear RNA-rich granule called the nuage. Unlike miRNAs and siRNAs, piRNA biogenesis does not involve a double-stranded precursor (7–9). The piRNA biogenesis process gives piRNA molecules their two distinct features: a first position U or a 10th position A, which signify origins in the primary processing pathway and secondary pathway, respectively (10–13). The primary pathway involves actions of Piwi and Aub, and the secondary pathway, also known as “Ping-Pong,” involves the co-function of Aub-antisense piRNAs and Ago3-sense piRNA complexes (6, 14, 15). In the primary pathway, the 5′ end of piRNAs is defined by Zucchini (a mitochondrion-associated endonuclease), and in the secondary pathway, it is defined by either Aub or Ago3 (6, 14, 16, 17). Two distinct mechanisms define the 3′ end of mature piRNAs: one dependent on Zucchini and the other dependent on the exonuclease Nibbler (18–24).
The relationship between heat shock proteins (Hsps) and the Piwi-piRNA pathway provides an exciting new piece to the puzzle of how the PIWI family proteins maintain genomic integrity. To deconstruct the relationship between the Piwi-piRNA pathway and heat shock proteins, the germ line is an essential point of concern. Heat shock, cellular stress, and aging impede ribonucleoprotein (RNP) assembly. There is evidence that the nuage is specifically affected and hindered during these cellular conditions (25). Further, these conditions also hinder Yb bodies, cytoplasmic RNPs found in Drosophila somatic follicle cells. The response to these stressors has been shown to induce transient RNP granules, as well as significantly yet reversibly modifying existing complexes (26, 27). The conserved effect stressors have on the germ line regulatory organelles warrants further study into whether regulatory changes in the piRNA pathway occur under these conditions and whether these incidences have lasting hereditary effects.
Chaperone proteins function to fold and unfold proteins and other macromolecular structures. Heat shock proteins, Hsp90 and Hsp70 in particular, are the most abundant class of chaperones and function under conditions of cellular stress and or elevated temperature. Thus, they play an important role in cancer, neurological disorders, oxidative stress, and other forms of non-optimal cellular conditions (28). Hsp90 is a well characterized chaperone protein; it is responsible for the stabilization and activation of ∼200 cellular proteins. The majority of these proteins are involved in cellular signaling processes. Hsp90 functions along with Hsp70, through a co-chaperone and physical linker, Hsp70-Hsp90-organizing protein (Hop), which together make up the Hsp90 chaperone machine (29). Recent work has explored epigenetic functions of Hsp90. Hsp90 acts as an evolutionary capacitor of phenotypic variation. In Drosophila, inhibition of Hsp83, the Hsp90 ortholog, along with environmental stress both similarly induce morphological changes. Hsp83's role in the maintenance of normal morphology was first deemed a result of Hsp83's stabilization of early development transcription factors and signaling clientele (30). The evolutionary conservation of the Hsp90-Hsp70 chaperone machinery reflects the complexity and specificity of this system's regulatory function. Hundreds of client proteins interact and depend on this system, and it has emerged as an important regulator of epigenetic processes.
Recent studies have shown that Hsp90 and its associated co-chaperones regulate small non-coding RNA pathways. Mutations of the Drosophila orthologs of Hsp90 and Hsp70 have been shown to reduce assembly of RNA-induced silencing complexes (RISCs) (31). Our previous work has shown that Hsp90 and Hop interact with Piwi, mediate its phosphorylation, and silence phenotypic variations (32). Hsp90 mediates accurate loading of piRNA precursors into piRNA-binding proteins, and the absence of Hsp90 leads to inefficient piRNA biogenesis with a concurrent increase in TE mobility (33, 34). Further, Shutdown (encoded by shu), a member of the FKBP family of immunophilins and an interacting partner of Hsp90, was shown to be required for both primary and secondary piRNA biogenesis (35, 36). The Shutdown ortholog in mice, FKBP6, is required for secondary piRNA biogenesis (37).
In this study, we show that germ line knockdown (GLKD) of Hop, a Hsp90 co-chaperone, leads to significant up-regulation of transposons, showing that Hop is essential for silencing TE. Like other piRNA pathway mutants (38), Hop GLKD leads to induction of phosphorylation of histone γ-H2Av, a marker for double-strand DNA breaks. Further, Hop GLKD leads to a sterile phenotype where females can lay eggs, but none of the eggs hatch into larvae. We also show that Hop is dispensable for either the stability or the cellular localization of key piRNA pathway proteins, Piwi, Aubergine, Ago3, and Vasa, indicating that Hop functions downstream of these proteins. We finally show that Hop is essential for efficient piRNA biogenesis. Based on these results, we conclude that Hop is critical for the maintenance of genome integrity and that it does so by silencing transposons and regulating piRNA biogenesis.
Results and discussion
Hop (STIP1 in humans) is the co-chaperone responsible for the transfer of client proteins between Hsp70 and Hsp90. Hop is evolutionarily conserved in eukaryotes, and it localizes to both the nucleus and the cytoplasm (39). Hop is a monomeric protein that consists of three tetratricopeptide repeat domain regions (TPR1, TPR2A, and TPR2B) and one aspartic acid-proline repeat dipeptide domain (Fig. 1A). The TPR domains interact with the C termini of Hsp90 and Hsp70, with TPR1 and TPR2B binding to Hsp70 and TPR2A binding preferentially to Hsp90. The intermediate structures of heat shock machinery are difficult to characterize completely because of the transient and fast-paced nature of chaperone function (40). The physical transfer process and the conformations involved in Hop's function have yet to be solved.
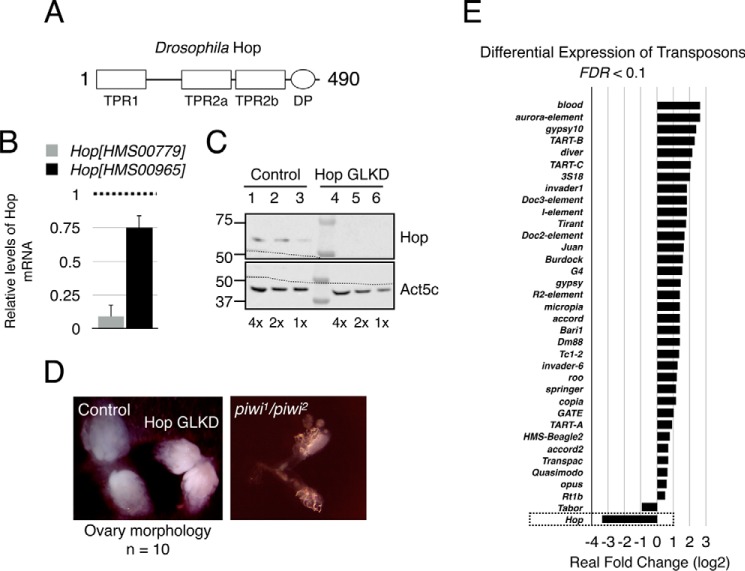
Figure 1.
Germ line knockdown of Hop leads to transposon up-regulation. A, domain organization of Hop. DP, dipeptide. B, quantitative PCR results showing the extent of Hop mRNA knockdown in two different RNAi lines. Act5c (β-actin) was used as internal control, and Oregon-R was used as wild-type control. The dotted line represents arbitrary levels of Hop mRNA in control ovaries. Error bars represent mean ± S.E. C, immunoblotting analysis demonstrating the efficiency of germ line knockdown of Hop. Molecular weight markers are shown on the left. Act5c (β-actin) was used as loading control. Two-fold serial dilutions of ovary lysates from control and Hop GLKD ovaries were used. The dotted lines across the immunoblots show the region where nitrocellulose membrane was cut prior to exposure to primary antibody. D, morphology of control and Hop GLKD ovaries. Ovaries containing nos.Gal4 driven Luciferase were used as control. For comparison of ovary morphology, ovary from piwi1/piwi2 flies is shown on the right. E, bar plot showing log2 value of real -fold change from EBSeq output. False discovery rate (FDR) cutoff was 0.1. Relative expression of Hop is also shown (box). Consistent with Fig. 1B, Hop mRNA levels were significantly reduced.
Previous work had demonstrated that co-chaperone Hop interacts with Piwi and functions in preventing phenotypic variations (32). Further, several heat shock proteins and FKBP6 were shown to interact with mammalian PIWI protein orthologs Miwi and Mili. These results gave us a clue that Hop could potentially play a role in the piRNA pathway. Hence, we decided to characterize the potential role of Hop in the piRNA pathway. To this end, we used RNA interference to knock down Hop specifically in the germ line cells of Drosophila ovary using nos.Gal4 (germ line knockdown or GLKD). We tested two different Hop RNAi lines, HopHMS00779 and HopHMS00965, that were generated by the Transgenic RNAi Project (TRiP, Harvard Medical School) (41, 42). Both RNAi lines target Hop mRNA using 21-nt shRNA. Analysis using the Updated Targets of RNAi Reagents (UP-TORR) Fly tool (43) showed that both shRNAs have zero off-target effects. Knockdown using HopHMS00779 and HopHMS00965 produced 91 and 25% reduction in Hop mRNA, respectively, when compared with Hop mRNA levels in Oregon-R (wild type) (Fig. 1B). Because HopHMS00779 was highly efficient in eliminating Hop mRNA, we used this line to characterize the role of Hop in the piRNA pathway. From here on, Hop GLKD refers to HopHMS00779.
Hop GLKD greatly reduced the levels of Hop protein in the germ line (Fig. 1C, compare lanes 1–3 with lanes 4–6). We next tested whether Hop GLKD impacted ovary development. Hop GLKD ovaries looked like wild-type counterparts, suggesting that germ line Hop is dispensable for general ovary development (Fig. 1D). However, mothers with Hop GLKD could lay eggs at the same rate as the control, but none of the eggs hatched into larvae (data not shown). Such a phenotype is reminiscent of piRNA pathway mutants where transposon up-regulation leads to severe DNA damage and activation of Chk2 DNA damage checkpoint (38, 44). So we tested whether Hop GLKD leads to activation of transposons. To this end, we performed mRNA-seq with RNA from control and Hop GLKD ovaries. Reads mapping to mRNAs from transposons and genes were quantified using RSEM (45). Differential expression of transposons was analyzed using EBSeq (46). Differentially expressed transposons and genes with false discovery rate < 0.1 were deemed statistically significant. Hop GLKD resulted in statistically significant differential expression of 35 transposons. Of these, 34 were up-regulated and 1 (Tabor) was down-regulated (Fig. 1E). This result shows that Hop is essential for transposon silencing. Of the 35 differentially expressed transposons, 24 (∼70%) were Group 1 transposons that require Ago3 for silencing (15). These results are not surprising considering that Hop mRNA was knocked down specifically in the germ line but not in the soma. Consistent with this, soma dominant transposons such as ZAM and Idefix did not make the list of differentially expressed transposons.
Transposon up-regulation in piRNA pathway mutants results in DNA double-strand breaks (DSBs), leading to induction of γ-H2Av (phosphorylated form of histone H2Av) foci (38, 44). DSBs induce phosphorylation of a conserved SQ motif in the C-terminal tail of H2Av (47, 48), and γ-H2Av accumulates near DSBs (49, 50). Because Hop GLKD leads to transposon up-regulation, we tested whether it also leads to induction and accumulation of γ-H2Av foci in the nuclei. Immunofluorescence microscopy showed that, upon Hop GLKD, γ-H2Av foci significantly accumulate in region 3/stage 1 of germarium, and these persist into stage 2 egg chambers and beyond (Fig. 2A, compare white arrows in Control and Hop GLKD panels). These results show that Hop is critical for maintenance of genome integrity and that its absence leads to activation of DNA damage response, presumably due to transposon activation.
Figure 2.
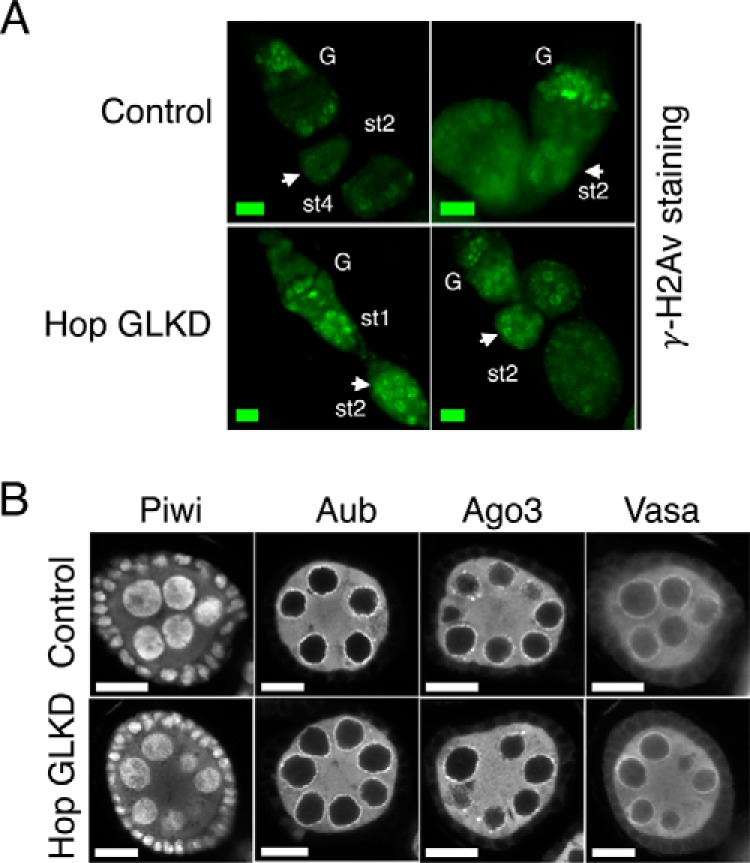
Effect of Hop GLKD on γ-H2Av accumulation and localization of piRNA pathway proteins. A, confocal images of two independent ovarioles from control and Hop GLKD flies stained for γ-H2Av. Scale bars are 10 μm. G, germarium; st, stage. White arrows point at stage two egg chambers. B, confocal images of stage two egg chambers from control and Hop GLKD ovaries staining for Piwi, Aub, Ago3, and Vasa. Scale bars are 10 μm.
We next tested whether Hop GLKD derails transposon silencing by affecting proper cellular localization of key piRNA pathway components, Piwi, Aub, Ago3, and Vasa. In control ovaries, Piwi localized to the nucleus; Aub, Ago3, and Vasa were cytoplasmic and localized to the perinuclear region called the nuage (Fig. 2B, upper panel). This is consistent with earlier findings (6, 15, 51–53). Proper localization of these proteins to their designated cellular spots is critical for both piRNA biogenesis and transposon silencing (11). However, localization of Piwi, Aub, Ago3, and Vasa did not change upon Hop GLKD (Fig. 2B, compare lower panel with upper panel). These results show that Hop is dispensable for proper cellular localization of key piRNA pathway components. Presumably, Hop functions downstream of Piwi, Aub, Ago3, and Vasa. These results are in contrast to the findings in earlier studies where either germ line clones of shu or knockdown of shu led to significant decrease in the levels and localization of Piwi, Aub, and Ago3 (35, 36). However, this is consistent with qin mutants where transposon silencing was derailed without affecting the levels and cellular localization of Piwi, Aub, Ago3, and Vasa proteins (44). Please note that Shutdown (encoded by shu) is an Hsp90 co-chaperone like Hop. Qin (encoded by qin) is a Tudor domain- and E3 ligase domain-containing protein.
We next tested whether transposon up-regulation in Hop GLKD is due to defective piRNA biogenesis. We deep-sequenced and analyzed small RNAs from Hop GLKD and control ovaries. We first analyzed the composition of genome mapping reads (Fig. 3A). Specifically, we quantified reads that mapped to coding and non-coding regions of the genome. In control flies, 14 and 86% of the reads mapped to coding and non-coding regions of the genome, respectively. In contrast, in Hop GLKD ovaries, 41 and 59% of the reads mapped to the coding and non-coding regions, respectively. Thus, there is an ∼3-fold increase in the number of small RNAs that map to the coding region of the genome in Hop GLKD when compared with the control. These results show that small RNA biogenesis from the non-coding portion of the genome decreases drastically in Hop GLKD.
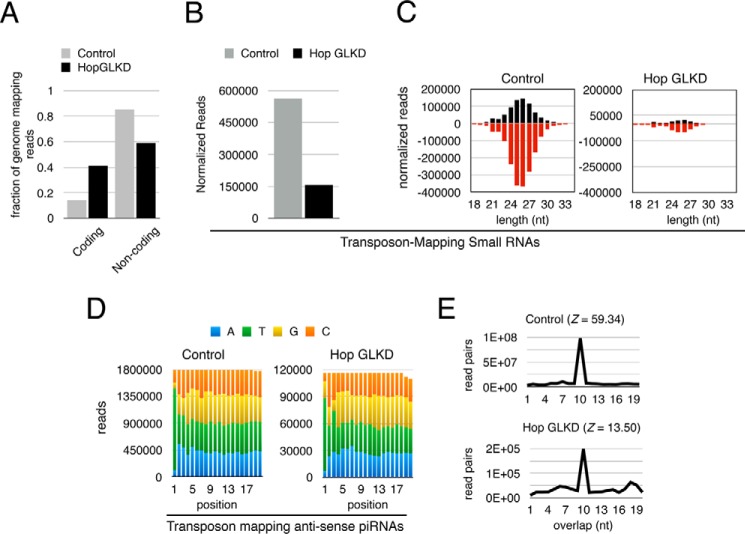
Figure 3.
Hop is required for efficient piRNA biogenesis. A, quantitation of small RNA reads mapping to coding and non-coding portion of the genome in control and Hop GLKD ovaries. B, normalized numbers of reads uniquely mapping to transposons are plotted. Reads were normalized to the number of reads mapping to the coding portion of the genome. C, length and strand orientation analysis of normalized reads mapping to transposons. Reads were normalized to the number of reads mapping to the coding portion of the genome. Positive and negative numbers indicate sense and antisense strands, respectively. D, nucleotide composition of first 20 nt of antisense small RNAs mapping to transposons in control and Hop GLKD ovaries. E, Ping-Pong analysis for small RNA reads mapping to transposons. Z-score is shown in parentheses. Notice the preference for 10-nt overlap between sense and antisense reads. Shown on the y axis is the number of read pairs that can be formed between sense and antisense reads mapping to transposons.
Next, we tested whether Hop GLKD affects biogenesis of transposon-mapping small RNAs. We normalized the small RNA libraries in two ways: (a) based on the number of reads that uniquely map to the genome and (b) based on the number of reads that map to coding regions (supplemental Fig. S1). Both normalization methods showed that Hop GLKD drastically reduced the number of transposon-mapping small RNAs. Specifically, when we normalized based on uniquely mapping genomic reads, we noticed a 50% drop in the number of transposon-mapping small RNAs, and a 72% drop when we normalized based on reads mapping to the coding portion of the genome (Fig. 3B). Both sense and antisense transposon-mapping reads showed drastic reduction when compared with the control (Fig. 3C). Based on these results, we conclude that Hop is critical for biogenesis of transposon-mapping small RNAs. Because the majority of transposon-mapping small RNAs are 23–29-nt piRNAs, we conclude that Hop is essential for piRNA biogenesis.
To get clues into how Hop regulates piRNA biogenesis, we analyzed the signature of residual piRNAs in Hop GLKD ovaries. We noticed that preference for U as the first nucleotide at the 5′ end does not change, showing that 5′ end processing of piRNAs remains active in Hop GLKD (Fig. 3D, compare green bars in left and right panels). Further, Ping-Pong cycle mediated by Aub and Ago3 remains active in Hop GLKD as evidenced by the preference for 10-nt overlap in sense and antisense piRNAs (Fig. 3E). However, Z-score, which is a relative measure of the preference for 10-nt overlap over other length overlaps, significantly decreases from 59.34 in control to 13.50 in Hop GLKD (Fig. 3E). The decrease in Z-score suggests that Hop GLKD affects the efficiency of Ping-Pong, which can, in turn, affect the accumulation of piRNAs. Inefficient piRNA accumulation, in turn, affects the piRNA pathway's ability to target and destroy transposons.
In summary, we have shown that Hop is essential for transposon silencing and efficient piRNA biogenesis. Hop GLKD leads to transposon activation and induction of DNA damage signaling due to the significant reduction in piRNA levels. Reduction in the number of piRNAs is due neither to the loss of Piwi, Aub, Ago3, and Vasa nor to the loss of Ping-Pong cycle. It is intriguing to note that Shutdown and Hop, both being TPR domain-containing Hsp90 co-chaperones, exhibit distinct effects upon GLKD. Shutdown GLKD not only affected Piwi proteins' levels and cellular localization, but it also affected fecundity (35, 36). Shutdown GLKD females lay far fewer eggs than control, showing that Shutdown is essential for general ovary development and that piRNA biogenesis is perhaps one of the many functions it performs during ovary development. On the other hand, upon Hop GLKD, ovary development is normal, females lay eggs at the same rate as control, and Piwi proteins' localization is not affected. However, transposon silencing and piRNA biogenesis are affected. In other words, Hop is dispensable for ovary development but is indispensable for transposon silencing and piRNA biogenesis. Perhaps Shutdown and Hop function at distinct stages of the piRNA pathway. We propose that Hop regulates the efficient accumulation of piRNAs. It can function at two distinct stages: (a) it could mediate efficient exchange of sense and antisense precursors between Aub and Ago3, which in turn increases the rate of Ping-Pong cycle, and/or (b) it could function in coordinating post-piRNA biogenesis targeting of transposon mRNAs. Derailment of either of these steps leads to defects in piRNA biogenesis and transposon silencing. Further biochemical and genetic dissection will precisely define the function of Hop in transposon silencing, efficient piRNA biogenesis, and maintenance of genome integrity.
Experimental procedures
Fly stocks and handling
All Drosophila strains were maintained at 25 °C. The nos-Gal4 driver fly strain (stock number 32563; y1 w*; P{GAL4-nos.NGT}A), Hop shRNA lines HopHMS00779 (stock number 32979) and HopHMS00965 (stock number 34002), and control line (stock number 35788, P{UAS-LUC.VALIUM10}attP2) came from the Bloomington Drosophila Stock Center, Bloomington, IN. We refer to the control line as UAS-Luc. The control line had firefly luciferase (Luc) coding sequence inserted at the attP2 site, the same site of insertion as in Hop shRNAs. Both Hop RNAi lines used in Fig. 1B are short hairpin RNAs and are not expected to have any off-target effects. To make sure that both shRNA lines do not have any off-target effects, we used the UP-TORR Fly tool (43). UP-TORR showed that both RNAi lines HMS00779 and HMS00965 do not have any off-target effects. Knockdowns were achieved by crossing 2–5 virgin females from the RNAi lines for Hop with 1–2 males from nos.Gal4. Control cross was set up with 1–2 males from nos.Gal4 and 2–5 virgin females of UAS-Luc. The progeny from the control cross are referred to as nos.Luc. The parental generation was then transferred to new fly food every 4 days. As the F1 generation was born, they were collected, and then put on yeast for 1–2 days for preparation for ovary dissection. Egg hatch rate calculation was performed as described earlier (36) except that apple juice-agar plates were used.
Ovary lysate preparation and Western blotting analysis
Ten pairs of ovaries from 2–3-day-old yeast-fed females were dissected and homogenized in an Eppendorf tube in 50 μl of lysis buffer (20 mm HEPES, pH 7.5, 100 mm KCl, 5 mm MgCl2, 0.1% SDS, 0.1% sodium deoxycholate, 1% Triton X-100, 1 mm DTT, and 5% glycerol). The homogenate was then mixed with 50 μl of 2× Laemmli Sample Buffer (Bio-Rad product number 1610737), boiled at 95 °C for 3 min, and then processed for Western blotting analysis using standard molecular biology techniques.
Immunostaining and microscopy
Drosophila ovaries were manually dissected in ice-cold PBS and fixed with 4% formaldehyde (Electron Microscopy Sciences, Hatfield, PA) as described before (54). Fixed ovary tissues were incubated at 4 °C overnight with primary antibodies at the following concentrations: anti-Piwi (1:500), anti-Ago3 (1:250), anti-Aub (1:1000), anti-Vasa (1:50), and anti-γ-H2Av (1:500). Piwi, Aub, and Ago3 antibodies were kind gifts from Dr. Mikiko Siomi (Keio University, Minato, Tokyo, Japan) and Haifan Lin (Yale University). Anti-vasa antibody was deposited to the Developmental Studies Hybridoma Bank (DSHB) by Spradling, A. C./Williams, D, (DSHB Hybridoma Product anti-vasa). Anti-γ-H2Av (UNC93-5.2.1) was deposited to the DSHB by Hawley, R. S. (DSHB Hybridoma Product UNC93-5.2.1). Alexa Fluor-conjugated secondary antibodies from Thermo Fisher Scientific were used at 1:500. Confocal images were captured using a Zeiss LSM 880 NLO microscope and Plan-Apochromat 63×/1.40 Oil differential interference contrast objective with identical settings for GLKD and control fly lines. Images were analyzed using Fiji (ImageJ) (55). Ovary morphology images shown in Fig. 1D were captured using a Moticam 10 camera mounted on a Zeiss Stemi 2000-C stereo microscope.
mRNA-seq
Total RNA was isolated using TRIzol Reagent (Ambion) from 10–15 pairs of manually dissected ovaries and stored at −80 °C until use. Two biological repeats of nos.Luc (control) and Hop GLKD were used. RNA integrity was verified on an Agilent 2200 TapeStation (Agilent Technologies, Palo Alto, CA). 100–200 ng of total RNA was used to prepare RNA-Seq libraries using the TruSeq RNA Sample Prep Kit following the protocol described by the manufacturer (Illumina, San Diego, CA). Please note that poly(A)-containing RNA was first purified prior to the library preparation step. Single end 50-bp sequencing was performed on an Illumina HiSeq2500. Sequencing was performed at the Hollings Cancer Center Genomics Core Lab at Medical University of South Carolina (MUSC).
Small RNA-seq
Thirty μg of total RNA from nos.Luc (control) and Hop GLKD ovaries was resolved on a 15% denaturing polyacrylamide gel containing 7 m urea. Small RNAs ranging from 20 to 40 nt were purified without oxidation step as described elsewhere (15). Small RNA sequencing libraries were prepared using the TruSeq Small RNA Library Preparation Kit (Illumina) with one minor modification. A terminator block oligonucleotide (5′-TAC AAC CCT CAA CCA TAT GTA GTC CAA GCA/3SpC3/-3′) was added prior to 5′ adapter ligation as described elsewhere (56). This step specifically blocked reverse transcription of 30-nt-long 2S rRNA and hence substantially reduced rRNA contamination in the final sequenced reads. Sequencing libraries were size-selected with a Pippin Prep (Sage Science, Beverly, MA). Single end 36- or 50-bp sequencing was performed on a HiSeq2500 (Illumina). Sequencing was performed at the Hollings Cancer Center Genomics Core Lab at MUSC.
Quantitative PCR
cDNA preparation was performed using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific) using the manufacturer's protocol. Quantitative PCR was performed using iQ SYBR Green Supermix (Bio-Rad) and a CFX384 Touch Real-Time PCR Detection System (Bio-Rad). Data analysis was performed using Apple Numbers. The following primers were used: (a) 5′-AAGTTGCTGCTCTGGTTGTCG-3′ and 5′-GCCACACGCAGCTCATTGTAG-3′ for Act5c and (b) 5′-CATTCGCCAAGGCTGGAAAG-3′ and 5′-GGGATCGTACTTGAGACCCTC-3′ for Hop.
Bioinformatics
mRNA-seq analysis
Our mRNA-seq analysis was inspired by the piPipes workflow (57). Raw mRNA-seq reads from Illumina HiSeq 2500 were aligned to Drosophila (version 5, dm3) transcriptome and reference transposon sequences using Bowtie 2 (58, 59) and then quantified by RSEM (45). dm3 transcriptome (mRNA sequences) was obtained from the UCSC table browser (60), and reference transposon sequences were obtained from FlyBase. Differential expression analysis of transcripts was then performed using EBSeq (46). Differentially expressed transcripts with a false discovery rate < 0.1 were deemed significant.
Small RNA-seq analysis
Raw small RNA sequencing reads were processed by cutadapt to remove the 3′-sequencing adapter (TGGAATTCTCGGGTGCCAAGG). Trimmed reads smaller than 18 nt and also the reads without sequencing adapter were discarded. Reads were then depleted of rRNA reads and then aligned to dm3 genome using Bowtie (59). rRNA mapping allowed two mismatches, and genomic alignment allowed no mismatches. Only reads that perfectly matched the genome were collected and further analyzed. Reads were then collapsed using Perl script TBr2_collapse.pl available as a part of NGS Toolbox (61). Reads were then mapped to mRNA sequences of RefSeq genes without any mismatches using Bowtie and then plotted as fraction of genome mapping reads in Fig. 3A. To compare reads between different samples, the normalization factor was calculated in two ways: (a) based on the number of uniquely mapping genomic reads and (b) based on the number of reads mapping to mRNA sequences of RefSeq genes (protein-coding portion of the genome). Reads were then mapped to transposon consensus sequences using Bowtie 2, and the numbers of reads that align uniquely were noted. The numbers of transposon-mapping reads in Hop GLKD were then multiplied by normalization factor and plotted as shown in Fig. 3B. To analyze size distribution and nucleotide composition of reads, we mapped reads to transposon consensus sequences using sRNAmapper (62) with default parameters. Under default parameters, sRNAmapper does not allow any mismatch in the first 18 nt from the 5′ end of piRNAs and at most allows one mismatch in the rest of the sequence. Size distribution, orientation, and nucleotide composition of such aligned reads were analyzed using Perl script checkmap.pl. Output was then exported to Apple Numbers and plotted as shown in Fig. 3, C and D. Ping-Pong signature was analyzed using Perl script TBr2_pingpong.pl. TBr2_pingpong.pl performs Ping-Pong signature analysis and Z-score calculation in the same way as described elsewhere (44). The output of TBr2_pingpong.pl was exported to Apple Numbers and plotted as shown in Fig. 3E. All Perl scripts are available upon request.
Author contributions
J. A. K. and V. K. G. conceived the project and wrote the manuscript. J. A. K. performed most of the experiments. R. Y. P. performed immunostaining of Piwi proteins and Vasa. D. N. performed immunostaining of γ-H2Av. D. R. wrote all Perl scripts used in this study. V. K. G. performed bioinformatic analysis with help from D. R.
Supplementary Material
Acknowledgments
We thank Mikiko Siomi and Haifan Lin for antibodies, members of the Gangaraju lab for critical reading of the manuscript, Robert Wilson and Jennifer Schulte for preparing mRNA and small RNA sequencing libraries, and Philip Zamore and Bo Han for answering our queries during installation and execution of piPipes. We acknowledge support of the Genomics/shRNA Shared Resource and National Institutes of Health Grant P30 CA138313 to Cell & Molecular Imaging Shared Resource, Hollings Cancer Center, Medical University of South Carolina.
This work was supported in part by National Institutes of Health Pathway to Independence Award R00ES021736 and Hollings Cancer Center Support Grant P30CA138313 (to V. K. G.). This work was also supported by the Natural and Medical Sciences Research Center of the University Medicine Mainz (NMFZ) (to D. R.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Fig. S1.
The Gene Expression Omnibus (GEO) accession number GSE93934 reported in work can be accessed through the NCBI GEO database.
- piRNA
- Piwi-interacting RNA
- Hsp
- heat shock protein
- Hop
- Hsp70-Hsp90-organizing protein
- FKBP
- FK506-binding protein
- GLKD
- germ line knockdown
- RNP
- ribonucleoprotein
- TPR
- tetratricopeptide repeat
- nt
- nucleotide(s)
- TE
- transposable element(s)
- DSB
- double-strand break
- Luc
- luciferase
- seq
- sequencing
- 3SpC3
- 3′ spacer C3 phosphoramidite.
References
- 1. Ghildiyal M., and Zamore P. D. (2009) Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 10, 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ishizu H., Siomi H., and Siomi M. C. (2012) Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes Dev. 26, 2361–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Juliano C., Wang J., and Lin H. (2011) Uniting germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms. Annu. Rev. Genet. 45, 447–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim V. N., Han J., and Siomi M. C. (2009) Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10, 126–139 [DOI] [PubMed] [Google Scholar]
- 5. Siomi M. C., Sato K., Pezic D., and Aravin A. A. (2011) PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 12, 246–258 [DOI] [PubMed] [Google Scholar]
- 6. Brennecke J., Aravin A. A., Stark A., Dus M., Kellis M., Sachidanandam R., and Hannon G. J. (2007) Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103 [DOI] [PubMed] [Google Scholar]
- 7. Das P. P., Bagijn M. P., Goldstein L. D., Woolford J. R., Lehrbach N. J., Sapetschnig A., Buhecha H. R., Gilchrist M. J., Howe K. L., Stark R., Matthews N., Berezikov E., Ketting R. F., Tavaré S., and Miska E. A. (2008) Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol. Cell 31, 79–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Houwing S., Kamminga L. M., Berezikov E., Cronembold D., Girard A., van den Elst H., Filippov D. V., Blaser H., Raz E., Moens C. B., Plasterk R. H., Hannon G. J., Draper B. W., and Ketting R. F. (2007) A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129, 69–82 [DOI] [PubMed] [Google Scholar]
- 9. Vagin V. V., Sigova A., Li C., Seitz H., Gvozdev V., and Zamore P. D. (2006) A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313, 320–324 [DOI] [PubMed] [Google Scholar]
- 10. Pane A., Wehr K., and Schüpbach T. (2007) zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev. Cell 12, 851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Malone C. D., Brennecke J., Dus M., Stark A., McCombie W. R., Sachidanandam R., and Hannon G. J. (2009) Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137, 522–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Czech B., Preall J. B., McGinn J., and Hannon G. J. (2013) A transcriptome-wide RNAi screen in the Drosophila ovary reveals factors of the germline piRNA pathway. Mol. Cell 50, 749–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Handler D., Meixner K., Pizka M., Lauss K., Schmied C., Gruber F. S., and Brennecke J. (2013) The genetic makeup of the Drosophila piRNA pathway. Molecular cell 50, 762–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gunawardane L. S., Saito K., Nishida K. M., Miyoshi K., Kawamura Y., Nagami T., Siomi H., and Siomi M. C. (2007) A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315, 1587–1590 [DOI] [PubMed] [Google Scholar]
- 15. Li C., Vagin V. V., Lee S., Xu J., Ma S., Xi H., Seitz H., Horwich M. D., Syrzycka M., Honda B. M., Kittler E. L., Zapp M. L., Klattenhoff C., Schulz N., Theurkauf W. E., et al. (2009) Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137, 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nishimasu H., Ishizu H., Saito K., Fukuhara S., Kamatani M. K., Bonnefond L., Matsumoto N., Nishizawa T., Nakanaga K., Aoki J., Ishitani R., Siomi H., Siomi M. C., and Nureki O. (2012) Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature 491, 284–287 [DOI] [PubMed] [Google Scholar]
- 17. Ipsaro J. J., Haase A. D., Knott S. R., Joshua-Tor L., and Hannon G. J. (2012) The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature 491, 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hayashi R., Schnabl J., Handler D., Mohn F., Ameres S. L., and Brennecke J. (2016) Genetic and mechanistic diversity of piRNA 3′-end formation. Nature 539, 588–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Han B. W., Wang W., Li C., Weng Z., and Zamore P. D. (2015) Noncoding RNA. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 348, 817–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mohn F., Handler D., and Brennecke J. (2015) Noncoding RNA. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science 348, 812–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Han B. W., Hung J. H., Weng Z., Zamore P. D., and Ameres S. L. (2011) The 3′-to-5′ exoribonuclease Nibbler shapes the 3′ ends of microRNAs bound to Drosophila Argonaute1. Curr. Biol. 21, 1878–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu N., Abe M., Sabin L. R., Hendriks G. J., Naqvi A. S., Yu Z., Cherry S., and Bonini N. M. (2011) The exoribonuclease Nibbler controls 3′ end processing of microRNAs in Drosophila. Curr. Biol. 21, 1888–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang H., Ma Z., Niu K., Xiao Y., Wu X., Pan C., Zhao Y., Wang K., Zhang Y., and Liu N. (2016) Antagonistic roles of Nibbler and Hen1 in modulating piRNA 3′ ends in Drosophila. Development 143, 530–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Feltzin V. L., Khaladkar M., Abe M., Parisi M., Hendriks G. J., Kim J., and Bonini N. M. (2015) The exonuclease Nibbler regulates age-associated traits and modulates piRNA length in Drosophila. Aging Cell 14, 443–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Eddy E. M. (1974) Fine structural observations on the form and distribution of nuage in germ cells of the rat. Anat. Rec. 178, 731–757 [DOI] [PubMed] [Google Scholar]
- 26. Qi H., Watanabe T., Ku H. Y., Liu N., Zhong M., and Lin H. (2011) The Yb body, a major site for Piwi-associated RNA biogenesis and a gateway for Piwi expression and transport to the nucleus in somatic cells. J. Biol. Chem. 286, 3789–3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schisa J. A. (2014) Effects of stress and aging on ribonucleoprotein assembly and function in the germ line. Wiley Interdiscip. Rev. RNA 5, 231–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Trepel J., Mollapour M., Giaccone G., and Neckers L. (2010) Targeting the dynamic HSP90 complex in cancer. Nat. Rev. Cancer 10, 537–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Alvira S., Cuéllar J., Röhl A., Yamamoto S., Itoh H., Alfonso C., Rivas G., Buchner J., and Valpuesta J. M. (2014) Structural characterization of the substrate transfer mechanism in Hsp70/Hsp90 folding machinery mediated by Hop. Nat. Commun. 5, 5484. [DOI] [PubMed] [Google Scholar]
- 30. Queitsch C., Sangster T. A., and Lindquist S. (2002) Hsp90 as a capacitor of phenotypic variation. Nature 417, 618–624 [DOI] [PubMed] [Google Scholar]
- 31. Iwasaki S., Sasaki H. M., Sakaguchi Y., Suzuki T., Tadakuma H., and Tomari Y. (2015) Defining fundamental steps in the assembly of the Drosophila RNAi enzyme complex. Nature 521, 533–536 [DOI] [PubMed] [Google Scholar]
- 32. Gangaraju V. K., Yin H., Weiner M. M., Wang J., Huang X. A., and Lin H. (2011) Drosophila Piwi functions in Hsp90-mediated suppression of phenotypic variation. Nat. Genet. 43, 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Izumi N., Kawaoka S., Yasuhara S., Suzuki Y., Sugano S., Katsuma S., and Tomari Y. (2013) Hsp90 facilitates accurate loading of precursor piRNAs into PIWI proteins. RNA 19, 896–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Specchia V., Piacentini L., Tritto P., Fanti L., D'Alessandro R., Palumbo G., Pimpinelli S., and Bozzetti M. P. (2010) Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature 463, 662–665 [DOI] [PubMed] [Google Scholar]
- 35. Olivieri D., Senti K. A., Subramanian S., Sachidanandam R., and Brennecke J. (2012) The cochaperone shutdown defines a group of biogenesis factors essential for all piRNA populations in Drosophila. Mol. Cell 47, 954–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Preall J. B., Czech B., Guzzardo P. M., Muerdter F., and Hannon G. J. (2012) shutdown is a component of the Drosophila piRNA biogenesis machinery. RNA 18, 1446–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xiol J., Cora E., Koglgruber R., Chuma S., Subramanian S., Hosokawa M., Reuter M., Yang Z., Berninger P., Palencia A., Benes V., Penninger J., Sachidanandam R., and Pillai R. S. (2012) A role for Fkbp6 and the chaperone machinery in piRNA amplification and transposon silencing. Mol. Cell 47, 970–979 [DOI] [PubMed] [Google Scholar]
- 38. Klattenhoff C., Bratu D. P., McGinnis-Schultz N., Koppetsch B. S., Cook H. A., and Theurkauf W. E. (2007) Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev. Cell 12, 45–55 [DOI] [PubMed] [Google Scholar]
- 39. Schmid A. B., Lagleder S., Gräwert M. A., Röhl A., Hagn F., Wandinger S. K., Cox M. B., Demmer O., Richter K., Groll M., Kessler H., and Buchner J. (2012) The architecture of functional modules in the Hsp90 co-chaperone Sti1/Hop. EMBO J. 31, 1506–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yamamoto S., Subedi G. P., Hanashima S., Satoh T., Otaka M., Wakui H., Sawada K., Yokota S., Yamaguchi Y., Kubota H., and Itoh H. (2014) ATPase activity and ATP-dependent conformational change in the co-chaperone HSP70/HSP90-organizing protein (HOP). J. Biol. Chem. 289, 9880–9886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ni J. Q., Markstein M., Binari R., Pfeiffer B., Liu L. P., Villalta C., Booker M., Perkins L., and Perrimon N. (2008) Vector and parameters for targeted transgenic RNA interference in Drosophila melanogaster. Nat. Methods 5, 49–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Perkins L. A., Holderbaum L., Tao R., Hu Y., Sopko R., McCall K., Yang-Zhou D., Flockhart I., Binari R., Shim H. S., Miller A., Housden A., Foos M., Randkelv S., Kelley C., et al. (2015) The Transgenic RNAi Project at Harvard Medical School: Resources and Validation. Genetics 201, 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hu Y., Roesel C., Flockhart I., Perkins L., Perrimon N., and Mohr S. E. (2013) UP-TORR: online tool for accurate and up-to-date annotation of RNAi Reagents. Genetics 195, 37–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang Z., Xu J., Koppetsch B. S., Wang J., Tipping C., Ma S., Weng Z., Theurkauf W. E., and Zamore P. D. (2011) Heterotypic piRNA Ping-Pong requires Qin, a protein with both E3 ligase and Tudor domains. Mol. Cell 44, 572–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li B., and Dewey C. N. (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Leng N., Dawson J. A., Thomson J. A., Ruotti V., Rissman A. I., Smits B. M., Haag J. D., Gould M. N., Stewart R. M., and Kendziorski C. (2013) EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 29, 1035–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Madigan J. P., Chotkowski H. L., and Glaser R. L. (2002) DNA double-strand break-induced phosphorylation of Drosophila histone variant H2Av helps prevent radiation-induced apoptosis. Nucleic Acids Res. 30, 3698–3705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rogakou E. P., Pilch D. R., Orr A. H., Ivanova V. S., and Bonner W. M. (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868 [DOI] [PubMed] [Google Scholar]
- 49. Modesti M., and Kanaar R. (2001) DNA repair: spot(light)s on chromatin. Curr. Biol. 11, R229–232 [DOI] [PubMed] [Google Scholar]
- 50. Redon C., Pilch D., Rogakou E., Sedelnikova O., Newrock K., and Bonner W. (2002) Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev. 12, 162–169 [DOI] [PubMed] [Google Scholar]
- 51. Cox D. N., Chao A., and Lin H. (2000) piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development 127, 503–514 [DOI] [PubMed] [Google Scholar]
- 52. Saito K., Nishida K. M., Mori T., Kawamura Y., Miyoshi K., Nagami T., Siomi H., and Siomi M. C. (2006) Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 20, 2214–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lim A. K., and Kai T. (2007) Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 104, 6714–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lin H., and Spradling A. C. (1993) Germline stem cell division and egg chamber development in transplanted Drosophila germaria. Dev. Biol. 159, 140–152 [DOI] [PubMed] [Google Scholar]
- 55. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., and Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wickersheim M. L., and Blumenstiel J. P. (2013) Terminator oligo blocking efficiently eliminates rRNA from Drosophila small RNA sequencing libraries. BioTechniques 55, 269–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Han B. W., Wang W., Zamore P. D., and Weng Z. (2015) piPipes: a set of pipelines for piRNA and transposon analysis via small RNA-seq, RNA-seq, degradome- and CAGE-seq, ChIP-seq and genomic DNA sequencing. Bioinformatics 31, 593–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Langmead B., and Salzberg S. L. (2012) Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Langmead B., Trapnell C., Pop M., and Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Karolchik D., Hinrichs A. S., Furey T. S., Roskin K. M., Sugnet C. W., Haussler D., and Kent W. J. (2004) The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32, D493–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rosenkranz D., Han C. T., Roovers E. F., Zischler H., and Ketting R. F. (2015) Piwi proteins and piRNAs in mammalian oocytes and early embryos: from sample to sequence. Genom. Data 5, 309–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Roovers E. F., Rosenkranz D., Mahdipour M., Han C. T., He N., Chuva de Sousa Lopes S. M., van der Westerlaken L. A., Zischler H., Butter F., Roelen B. A., and Ketting R. F. (2015) Piwi proteins and piRNAs in mammalian oocytes and early embryos. Cell Rep. 10, 2069–2082 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.