Abstract
TATA box-binding protein (TBP)-associated factors (TAFs), evolutionarily conserved from yeast to humans, play a central role during transcription initiation. A subset of TAF proteins is shared in transcription factor II D (TFIID) and SAGA transcription regulatory complexes. Although higher eukaryotes contain multiple TAF variants that specify tissue- and developmental stage-specific organization of TFIID or SAGA complexes, in unicellular genomes, however, each TAF is encoded by a single gene. Surprisingly, we found that the genome of Candida albicans, the predominant human fungal pathogen, contains two paralogous TAF12 genes, CaTAF12L and CaTAF12, encoding H2B-like histone-fold domain-containing variants. Of the available fungal genome sequences, only seven other closely related diploid pathogenic Candida genomes encode the two TAF12 paralogs. Using affinity purifications from C. albicans cell extracts, we demonstrate that CaTAF12L uniquely associates with the SAGA complex and CaTAF12 associates with the TFIID complex. We further show that CaTAF12, but not CaTAF12L, is essential for C. albicans growth. Conditional depletion of the two TAF12 variant proteins caused distinct cellular and colony phenotypes. Together our results define a specialized organization of the TAF12 variants and non-redundant roles for the two TAF12 variants in the unicellular C. albicans genome.
Keywords: Candida albicans, fungi, gene regulation, transcription regulation, transcriptional coactivator, SAGA, TAF12, TFIID
Introduction
Initiation of transcription leading to the formation of a preinitiation complex at the core promoter is a critical control point in gene regulation (1). TFIID2 is a multisubunit complex that functions as a general transcription factor and contains the TBP and ∼14 TBP-associated factors (TAFs) (1, 2). All TAFs are evolutionarily conserved from yeast to humans (3, 4). Barring TAF14, all other TAFs are essential for yeast cell growth; moreover, ∼84% of the total yeast genes are dependent on one or more TAFs for their expression in nutrient-rich medium (5, 6). Nine of the TAFs including TAF4 and TAF12 contain a histone-fold domain essential for heterodimerization (reviewed in Refs. 2 and 7). Moreover, TAF5, TAF6, TAF9, TAF10, and TAF12 are also integral components of the yeast SAGA complex (8). The SAGA complex composed of the Spt-Ada-Gcn5-Acetyltranferase subunits is a histone-modifying complex involved in acetylation of histone H3-lysine 9/14 and TBP delivery to facilitate activated transcription, therefore serving a coactivator function (9).
Mechanistically, TAFs have multiple roles in transcription, with their primary role being to deliver TBP to target promoters as part of the TFIID (1). They also function as promoter selectivity factors as part of the TFIID (reviewed in Ref. 10), and as coactivators by mediating interactions between the activators located upstream of the transcription start site and the components of the core transcriptional machinery (11–14). Variants of TBP and several TAFs differentially regulate transcription by virtue of their cell type-, tissue-, or developmental stage-specific expression in higher eukaryotes. In Drosophila melanogaster, five TAF paralogs are exclusively expressed in the testis and associate with core TFIID to form testis-specific TFIID (15). In human cells, TAF1L and TAF7L that are specifically expressed during male germ cell differentiation form alternate TFIID complexes to regulate spermatogenesis (16, 17). TAF4b, a paralog of TAF4, is also expressed in a cell type-specific manner (18, 19), and is a part of an alternate TFIID complex that regulates specific sets of genes (20). On the other hand, TAF9 and its paralog TAF9b, although both expressed in multiple cell lines, have limited overlapping functions (21). Indeed, the diverse spectrum of TBP- and TAF-like factors allows the higher eukaryotic cells to form multiple TFIID complexes that differ in their subunit composition, binding specificity, and function (22).
In multicellular organisms, certain TAF variants, such as TAF6L in human SAGA/STAGA (23–25) and the SAF6, which heterodimerizes with TAF9 in Drosophila (26), also specifically associate with SAGA complex to carry out tissue-specific functions. TBP or TAF isoforms are not known in unicellular organisms, with the exception of two TAF5-like genes SpTAF5 (originally called taf72+) and SpTAF5b (originally called taf73+) in Schizosaccharomyces pombe (27, 28). Although both SpTAF5 and SpTAF5b associate with the TFIID, only the SpTAF5b protein is associated with Gcn5 in S. pombe (27, 28).
In this study, we report that two novel paralogous TAF12 genes, CaTAF12L/orf19.470 and CaTAF12/orf19.6820, have non-redundant functions in Candida albicans, the most predominant fungal pathogen causing invasive candidiasis in humans (29). Using coimmunoprecipitation assays, we show that CaTAF12L is specifically associated with SAGA, and CaTAF12 is specifically associated with the TFIID complex. Although CaTAF12L deletion was not lethal, the deletion of CaTAF12 was lethal in C. albicans. Depletion of CaTAF12L and CaTAF12 protein levels caused distinct cellular and colony morphologies. We further show that CaTAF12L is required for oxidative stress resistance of C. albicans.
Results
C. albicans genome encodes two TAF12 variants
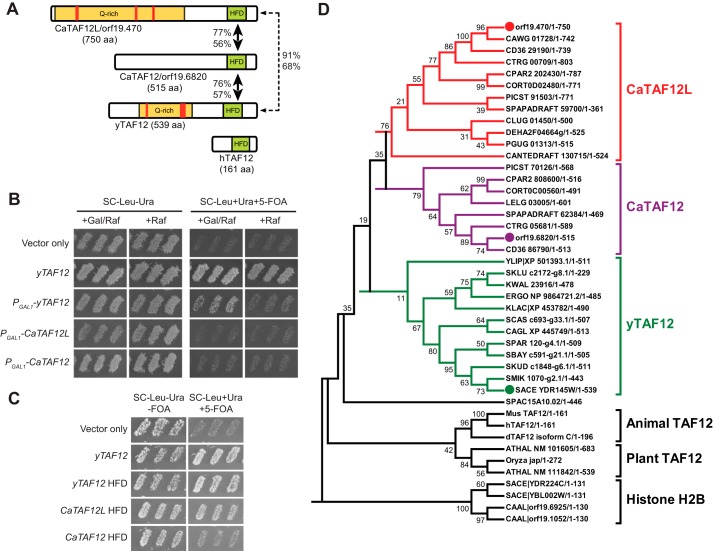
C. albicans genome surprisingly has two ORFs orf19.470 (named here as CaTAF12L) and orf19.6820 (CaTAF12) encoding histone H2B-like histone-fold domain (HF domain) (Fig. 1A). The rest of the 13 TAF genes are encoded as single ORFs. Each of the two C. albicans TAF12 protein sequences have high sequence similarity (identity >56%) with the yTAF12 only in their HF domains. Although both CaTAF12L and CaTAF12 have extended amino-terminal regions, only CaTAF12L contains the Gln-rich region found in yTAF12 (Fig. 1A). To test whether the two C. albicans TAF12 genes could function in the budding yeast Saccharomyces cerevisiae, we expressed the CaTAF12L and CaTAF12 genes from the yeast GAL1 promoter. The phenotype data showed that overexpression of neither TAF12L nor TAF12 whole ORFs could complement the taf12Δ deletion (Fig. 1B). However, overexpression of TAF12L was dominant negative over yTAF12 and resulted in slow growth (SC−Leu−Ura+Gal/Raf) (Fig. 1B). Previous studies showed that the TAF12 HF domain alone was sufficient for S. cerevisiae viability (30) and coactivator function in S. cerevisiae (31). Therefore we cloned and expressed CaTAF12L and CaTAF12 fragments encoding the HF domain alone from high copy plasmids in the S. cerevisiae taf12Δ strain. Although the yTAF12 histone-fold domain (HFD) fully complemented taf12Δ deletion, only CaTAF12L HFD, but not that of CaTAF12, provided at least partial complementation (+5-FOA; Fig. 1C). Thus the HF domains of CaTAF12L and CaTAF12 also seem to have functionally diverged between each other.
Figure 1.
Diploid Candida clade genomes encode two TAF12 proteins bearing H2B-like histone-fold domain. A, the schematic diagram showing the two C. albicans TAF12-like sequences (orf19.470 and orf19.6820), the S. cerevisiae TAF12 (yTAF12), and the human TAF12. The yellow-shaded region contains abundant Gln residues, and the red-filled boxes indicate tandem Gln repeats. The extent of homology (percentage of similarity and identity) of each HFD sequence with respect to the yTAF12 HFD is shown. aa, amino acids. B and C, genetic complementation analysis of S. cerevisiae taf12Δ strain by CaTAF12L and CaTAF12. Shown is the growth phenotype of S. cerevisiae strains bearing yTAF12 (ISC21), CaTAF12L (ISC22), or CaTAF12 (ISC23) under the control of PGAL1 promoter in Gal/Raf (activating conditions) and Raf (basal expression) conditions. Yeast strains bearing plasmid-borne yTAF12 (ISC25) and vector-only (ISC20) were used as controls. C, growth phenotype of S. cerevisiae strains bearing TAF12L or TAF12 HFD expressed from yTAF12 promoter. S. cerevisiae strains bearing yTAF12 (ISC25), yTAF12 HFD (ISC26), CaTAF12L HFD (ISC27), or CaTAF12 HFD (ISC28) or vector alone (ISC24) were tested by replica printing for growth upon eviction of a plasmid bearing yTAF12 on SC−Leu−Ura and SC−Leu+Ura+5-FOA plates, respectively, at 25 °C. D, phylogenetic tree of the TAF12 sequences. A multiple sequence alignment (total taxa = 43) was constructed using TAF12 amino acid sequences from Saccharomycotina species and from higher eukaryotes, as well as the histone-fold domain of histone H2B from S. cerevisiae and C. albicans. A phylogenetic tree was constructed using the Neighbor-Joining method in the MEGA6 software (56). All default parameters were chosen except that bootstrap test (1000 replicates) was used for the test of phylogeny. The percentage of replicate trees in which the associated taxa clustered together is shown next to the branches. All positions containing alignment gaps and missing data in pairwise sequence comparisons were eliminated. The clusters CaTAF12L, CaTAF12, yTAF12, and animal and plant TAF12 are indicated on the right. The histone H2B sequences formed an outgroup. The abbreviated species names in the tree are expanded in supplemental Table S1.
BLAST search of the publicly available fungal genome sequences identified two TAF12-like ORFs from eight asexually reproducing Candida clade species. All other fungal genomes examined, including the sexually reproducing Candida clade species, contained a single TAF12 gene. Among the higher eukaryotic genomes searched, only the Arabidopsis thaliana genome contains two TAF12 genes (32), but their distinct functional roles are not understood. (Fig. 1D) To understand the evolutionary history of the two CaTAF12 sequences, the TAF12 amino acid sequences from several genomes were aligned and a phylogenetic tree was constructed. The different TAF12 proteins formed five major clusters with the two TAF12 sequences from C. albicans clustering into separate branches occupied by the orf19.470-like (CaTAF12L) and orf19.6820-like (CaTAF12) sequences (Fig. 1D). These analyses showed that although CaTAF12L-like sequences bearing a glutamine-rich amino-terminal region were found in all fungal species examined, the CaTAF12-like sequences were found only in the eight Candida clade species (Fig. 1D) that are diploid fungal pathogens (33, 34). Moreover, interrogation of the Candida Gene Order Database showed that both CaTAF12L and CaTAF12 genes seem to have lost synteny in all Candida clade genomes with reference to the S. cerevisiae TAF12 gene (35, 36). Therefore we suggest that the two TAF12 ORFs may have originated within the fungal kingdom during the course of the evolution but perhaps were retained only in the diploid Candida clade species in the absence of meiosis.
CaTAF12L associates with SAGA, whereas CaTAF12 associates with TFIID
The architecture of TFIID and the SAGA is governed by multiple TAF-TAF HFD interactions. Although TAF4 and TAF12 form a histone H2A-and H2B-like dimer in TFIID (37), TAF4 is absent in the SAGA complex. In its wake, the TAF12 protein heterodimerizes with Ada1, an H2A-like HFD-containing protein in the SAGA (38). Thus there is a distinct specificity in the interaction of TAF12 with its partner in TFIID and SAGA complexes. Moreover, the observations that TFIID and SAGA complexes each contain two molecules of TAF12 (39–41) suggest that both CaTAF12 homologs may associate with either TFIID or SAGA in two copies. Therefore we tested whether one or both of the two TAF12 variants could associate with the TFIID and/or the SAGA complex in C. albicans.
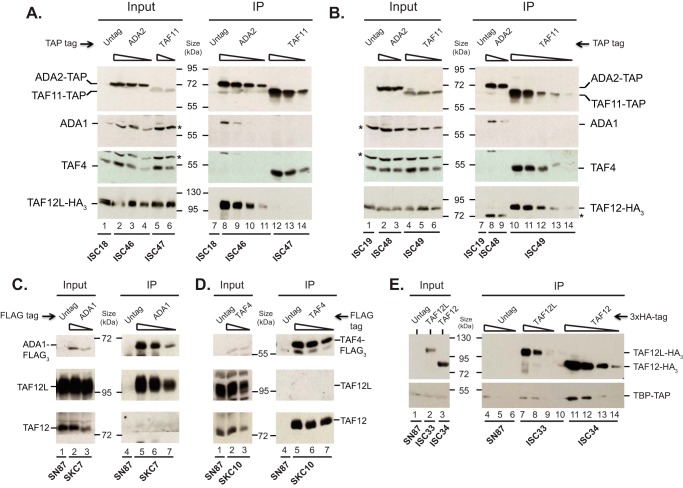
Western blotting analysis of C. albicans cell extracts showed that both CaTAF12L and CaTAF12 are expressed constitutively in rich medium (YPD) in wild-type cells (Figs. 2, A and B, and 3B). Therefore homozygous strains bearing either TAP-tagged TAF11, a component of the TFIID complex, or TAP-tagged Ada2, a component of the SAGA complex, in the background of either 3×HA-tagged CaTAF12L or 3×HA-tagged CaTAF12 expressed from their native loci were constructed. Total cell extracts from these epitope-tagged strains were used for coimmunoprecipitation assays. Both Ada2-TAP and TAF11-TAP were efficiently pulled down from the respective cell extracts (Fig. 2, A and B). Interestingly, CaTAF12L-HA3 was detected only in the Ada2-TAP pulldown, but not in the TAF11-TAP pulldown (Fig. 2A). In contrast, CaTAF12-HA3 was detected only in the TAF11-TAP pulldown but not in the Ada2-TAP pulldown (Fig. 2B). The blots were also probed with polyclonal antibodies against Ada1 and TAF4, the TAF12 dimerization partners in SAGA and TFIID, respectively. We found that Ada1, but not TAF4, was coimmunoprecipitated with Ada2-TAP (Fig. 2A), and that TAF4, but not Ada1, was coimmunoprecipitated with TAF11-TAP (Fig. 2B).
Figure 2.
Coimmunoprecipitation of CaTAF12L with SAGA and CaTAF12 with TFIID. A, CaTAF12L and Ada1 coimmunoprecipitate with Ada2-TAP but not with TAF11-TAP. B, CaTAF12 and TAF4 coimmunoprecipitate with TAF11-TAP but not with Ada2-TAP. Whole-cell protein extracts from control (ISC18 or ISC19) or the various TAP-tagged strains (ISC46-ISC49) were used for immunoprecipitation with IgG-Sepharose beads. 2-fold serial dilutions of the input cell extracts (starting at 100 μg) or the IP eluates (starting at 50% eluate) were blotted, excised into appropriate strips, and probed directly or stripped and reprobed as follows. The TAP-tagged proteins were detected with rabbit IgG Fc fragment, the 3×HA-tagged TAF12L and TAF12 proteins were detected with anti-HA (12CA5, Roche Applied Science) antibody, and the ADA1 and TAF4 proteins were detected with respective mouse polyclonal antibodies in the immunoblot. A nonspecific band detected by the anti-ADA1 antibody is indicated by an asterisk; this band was also found in the TAF4 input panels due to incomplete stripping of the blot, which was first probed with anti-Ada1 antibody prior to the anti-TAF4 antibody probe. Similarly, the residual Ada2-TAP band was seen in the IP lanes due to incomplete stripping prior to anti-ADA1, anti-TAF4, and anti-HA probes (asterisk; lanes 8 and 9). C and D, coimmunoprecipitation of CaTAF12L with Ada1-FLAG3 (C) and CaTAF12 with TAF4-FLAG3 (D). Whole-cell protein extracts from control (SN87) or the 3×FLAG-tagged strains SKC7 and SKC10 were used for immunoprecipitation with FLAG-M2 affinity agarose beads (Sigma-Aldrich), and eluates and input whole-cell extracts were analyzed by Western blotting. Anti-FLAG antibody was used to detect Ada1-FLAG3, and mouse polyclonal anti-TAF4 antibody was used to detect TAF4-FLAG3. TAF12L and TAF12 were detected in the immunoblots with respective rabbit polyclonal antibodies. E, TBP coimmunoprecipitates with CaTAF12. Whole-cell protein extracts from untagged (SN87) or TBP-TAP strains containing either CaTAF12L-HA3 (ISC33) or CaTAF12-HA3 (ISC34) were used for immunoprecipitation with anti-HA antibody, and the input samples and the IP eluates were blotted as above and probed with rabbit IgG Fc fragment or anti-HA (12CA5) antibodies.
Figure 3.
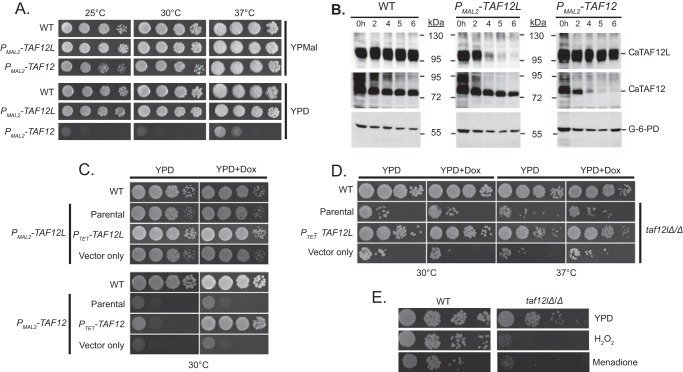
CaTAF12 but not CaTAF12L is essential for growth of C. albicans. A–C, conditional depletion of CaTAF12L and CaTAF12. A, the strains bearing glucose-repressible CaTAF12L (ISC11) or CaTAF12 (ISC12) alleles or the wild type (SN95) as control were cultured in YP maltose (promoter-on), serial dilutions were spotted on YPMal or YPD (promoter-off) plates, and growth was monitored for 2 days. B, CaTAF12L and CaTAF12 protein levels were depleted upon promoter shut-off. Wild type, PMAL2-CaTAF12L, and PMAL2-CaTAF12 strains were pre-grown in YPM and cultured in YPD, and cells were harvested. Whole-cell extracts from indicated time points were immunoblotted and probed with polyclonal rabbit anti-CaTAF12L, anti-CaTAF12, or the control anti-glucose 6-phosphate dehydrogenase (G-6-PD) antibodies. The positions of molecular mass markers are indicated in kDa. C, growth defects of CaTAF12L- and CaTAF12-depleted strains were rescued by CaTAF12L and CaTAF12, respectively. The strains WT (SN95), the parental strains bearing PMAL2-CaTAF12L (ISC11) or PMAL2-CaTAF12 (ISC12) alleles, and the complemented strains PTET-CaTAF12L (SDC13) and PTET-CaTAF12 (SDC16) or the vector-only control strains (SDC3 and SDC6) were grown overnight in the YPMal medium, and 10-fold serial dilutions were spotted on either YPD or YPD+doxycycline (YPD+Dox) plates, and then plates were incubated at 30 °C for 2 days. D, C. albicans taf12lΔ/Δ null mutant is viable. The WT (SN95), taf12lΔ/Δ mutant (parental; ISC36), and isogenic derivative strains bearing either PTET-CaTAF12L (SDC17) or vector alone (SDC12) were cultured in YPD, serial dilutions were spotted onto YPD or YPD+Dox plates, and growth was monitored at 30 and 37 °C. E, CaTAF12L is required for oxidative stress resistance. C. albicans WT strain SN95 and taf12lΔ/Δ null mutant (ISC36) were cultured in YPD, serial dilutions were spotted onto YPD or YPD plus 15 mm H2O2 or 60 μm menadione, and growth was monitored at 30 °C.
To independently establish the association of CaTAF12L and CaTAF12 with SAGA and TFIID, respectively, we carried out immunoprecipitation using cell extracts from strains expressing 3×FLAG-tagged Ada1 or TAF4 and probed the Western blots with polyclonal CaTAF12L and CaTAF12 antibodies. Consistent with the TAP immunoprecipitation assays, the CaTAF12L protein exclusively coimmunoprecipitated with Ada1-FLAG3 protein (Fig. 2C), whereas the CaTAF12 protein exclusively coimmunoprecipitated with TAF4-FLAG3 protein (Fig. 2D). These immunoprecipitation assays established that CaTAF12L and CaTAF12 associate differentially with the two transcriptional regulatory complexes SAGA and TFIID in C. albicans.
Next we tested whether one or both of the TAF12-like proteins could interact with TBP. Coimmunoprecipitation assays were conducted using cell extracts from strains expressing either CaTAF12L-HA3 or CaTAF12-HA3 in the background of TAP-tagged TBP. Indeed, TBP was coimmunoprecipitated with CaTAF12-HA3 (ISC34; Fig. 2E). However, in the CaTAF12L-HA3 immunoprecipitation, the amount of TBP recovered was at or close to the background level obtained in the untagged control SN87 cell extract (ISC33; Fig. 2E). Previous studies in yeast showed that although TBP can genetically and physically interact with SAGA subunits (42), TBP is not purified as an integral component of SAGA complex (8, 43, 44). Together these biochemical data established an in vivo association of CaTAF12L with SAGA complex and CaTAF12with TFIID complex in C. albicans.
CaTAF12 but not CaTAF12L is essential for viability of C. albicans
To investigate the in vivo functions of CaTAF12L and CaTAF12 genes in C. albicans, we constructed C. albicans strains bearing glucose-repressible CaTAF12L and CaTAF12 genes, viz. PMAL2-CaTAF12L and PMAL2-CaTAF12, and examined cell growth upon promoter shut-off in YPD medium. Although we did not see any measurable deleterious effect of overexpression of CaTAF12L from the PMAL2 promoter, overexpression of CaTAF12 caused slight growth defect at 25 °C (YPMal, Fig. 3A). Upon promoter shut-off (YPD), the PMAL2-CaTAF12 strain showed extreme slow growth as compared with the promoter-on condition (YPMal; Fig. 3A). In contrast, the PMAL2-CaTAF12L strain showed only a slight growth defect upon promoter shut-off (Fig. 3A). Western blotting analysis confirmed the depletion of the CaTAF12L and CaTAF12 proteins, respectively, in the PMAL2-CaTAF12L and PMAL2-CaTAF12 strains within 5–6 h of promoter shut-off (Fig. 3B). Moreover, depletion of either of the TAF12 variants did not significantly alter the steady-state level of the other TAF12 variant.
To complement the mutant phenotypes of PMAL2-CaTAF12L and PMAL2-CaTAF12 strains in glucose medium, CaTAF12L or CaTAF12, respectively, was reintroduced from the PTET constructs. Indeed, the PTET-CaTAF12L and PTET-CaTAF12 constructs yielded robust complementation of the defective growth phenotypes of Cataf12l (PMAL2-CaTAF12L) and Cataf12 (PMAL2-CaTAF12) mutants specifically in doxycycline-containing medium (Fig. 3C). The control vector-only transformants did not alter the growth defect (Fig. 3C). We also attempted to construct homozygous null mutant strains using different deletion strategies but were unable to obtain taf12Δ/Δ mutant strain (data not shown), indicating that CaTAF12 is essential for C. albicans growth. Although the taf12lΔ/Δ mutant was viable, the strain exhibited severe slow-growth phenotype (Fig. 3D). Expression of CaTAF12L from the TET promoter complemented the growth defect of the Cataf12lΔ/Δ mutant at both 30 °C and 37 °C (Fig. 3D). Examination of growth under oxidative stress conditions imposed by H2O2 and menadione caused severe growth impairment of the mutant (Fig. 3E). Thus CaTAF12L is non-essential for vegetative growth but required for oxidative stress resistance in C. albicans.
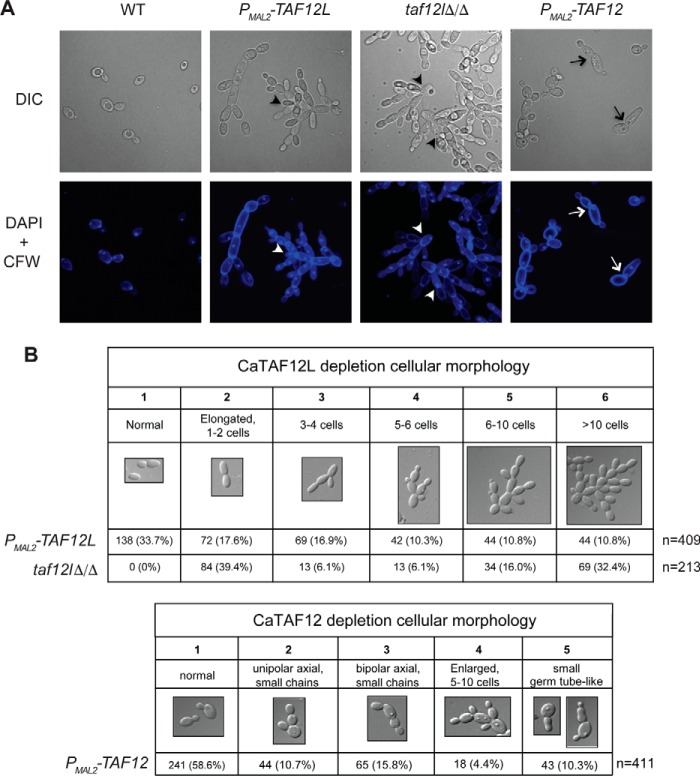
The CaTAF12L- and CaTAF12-depleted strains showed distinct cellular morphologies (Fig. 4). Depletion of CaTAF12L, both in the promoter shut-off mutant ISC11 (PMAL2-CaTAF12L) as well as in the taf12lΔ/Δ null mutant ISC36, led to pseudohyphal morphology comprising long, extensive chains of cells, each bearing a nucleus, indicating a cell separation defect (Fig. 4, A and B). CaTAF12 depletion, however, led to cells exhibiting unipolar buds or bipolar axial buds, or with small germ tube-like extensions beyond which the cells do not grow further (Fig. 4, A and B). Moreover, the CaTAF12L-depleted mutant showed reduced colony filamentation in different pseudohyphae-inducing solid media, viz. yeast carbon base (YCB) and synthetic low-ammonia dextrose media (SLAD), as well as attenuated hyphal colonies in serum-containing medium (supplemental Fig. S1). The C. albicans taf12lΔ/Δ null mutant, like the CaTAF12L-depleted mutant, formed rough colony morphology in all media tested, as well as deficient filamentation and agar invasion (supplemental Fig. S1). Together these results indicate that the CaTAF12L and CaTAF12 genes make distinct contributions to C. albicans growth.
Figure 4.
The taf12l and taf12 mutants have distinct cellular morphologies. A, strains SN95 (WT), ISC11 (PMAL2-CaTAF12L), ISC36 (taf12lΔ/Δ), or ISC12 (PMAL2-CaTAF12) were cultured overnight in YPM, diluted to fresh YPD, cultured for 6 h at 30 °C, and then fixed and stained with DAPI and CFW. Top row images were acquired by differential interference contrast (DIC) microscopy, and the bottom DAPI+CFW images were acquired by fluorescence microscopy, and images were digitally zoomed (2×). The altered cellular morphologies showing chains of cells are indicated by arrowheads in taf12lΔ/Δ and PMAL2-CaTAF12L, and the bipolar axial budding or short evaginations in PMAL2-CaTAF12 are indicated by arrows. B, summary of the cellular morphology of C. albicans depleted for TAF12L or TAF12. Top, cell counts from the different categories of cellular morphologies of C. albicans strains. Bottom, summary of cell counts from different cellular morphologies upon TAF12 depletion.
Discussion
TAFs are required for transcription of a wide array of genes. In this study, we investigated the role of two TAF12 variants encoded by the paralogous genes orf19.470 (CaTAF12L) and orf19.6820 (CaTAF12) in C. albicans. Both the TAF12 variants contain the histone H2B-like histone-fold domain, shown previously to be essential for TAF12 function in yeast (30). Among all the fungal genome sequences examined, only eight diploid Candida clade genomes encoded the two TAF12 variants (Fig. 1D). Although orf19.6820/CaTAF12 coimmunoprecipitated with TBP, we found no specific enrichment of orf19.470/CaTAF12L with TBP, and they were accordingly named TAF12 and TAF12L as per the convention for TAF nomenclature (4). Our biochemical and genetic analyses revealed that CaTAF12L and CaTAF12 are physically and functionally specialized to regulate gene expression by uniquely associating with the SAGA and the TFIID complexes, respectively.
In multicellular organisms, several paralogous TAF genes are tissue-restricted in their expression. Consequently, alternative TFIID and SAGA complexes are formed that function in a tissue- and cell type-specific manner (45, 46). In this respect, we found that the C. albicans TAF12L and CaTAF12 variants are constitutively expressed in vegetatively growing C. albicans cells (Fig. 2). Moreover, in yeast cells, TAF12 can directly heterodimerize with either TAF4 (47) or Ada1 (38). Therefore it is conceivable that CaTAF12L could associate with the SAGA complex through heterodimerization with Ada1 and CaTAF12 with the TFIID complex by heterodimerization with TAF4. Moreover, our S. cerevisiae complementation experiments indicated that CaTAF12L and CaTAF12 histone-fold domains are functionally distinct because only CaTAF12L HFD provided partial complementation in yeast.
How might the unique specialization of the two TAF12 variants for TFIID and SAGA influence cellular function? Past studies have shown that TAF12 is involved in interactions of the SAGA complex with the yeast transcriptional activators Gal4 and Gcn4 (31, 48–50). Interestingly, Gal4 activation domain cross-links to TAF12 only in the context of SAGA and not TFIID, indicating a complex-specific preferential role for TAF12 in activator interactions. Furthermore, the Gcn4 activation domain specifically cross-links to the amino-terminal 1–277 residues in the yeast TAF12 (50). However, C. albicans CaTAF12L and CaTAF12 are completely unrelated outside the HF domain, suggesting that a putative transcriptional activator is likely to be highly selective for CaTAF12L and CaTAF12 in C. albicans. Moreover, the Drosophila TAF12 along with TAF4, TAF5, TAF6, and TAF9 constitute a stable core of the Drosophila TFIID complex to which other TAFs and TBP associate (51). Our finding that CaTAF12 interacts with TFIID subunits and is essential for C. albicans growth is supportive of its central role in stabilizing the TFIID.
Although CaTAF12L and CaTAF12 associate specifically with separate complexes, our microscopy data showed that both the genes are required for optimal cell cycle progression but that they act in distinct steps. Although CaTAF12L mutation led to chains of cells that failed to undergo cell separation, CaTAF12 mutation caused arrest of cells either with small bud or with a slight evagination. Our finding that CaTAF12L is essential for oxidative stress resistance is supported by a previous report that the SAGA subunit Ada2 is required for oxidative stress resistance in C. albicans (52). Thus CaTAF12L, being non-essential for vegetative growth, paradoxically has an essential role in modulating stress resistance in C. albicans. The evolution of two TAF12 variants in C. albicans and the closely related seven diploid pathogenic Candida genomes, although this could be a relic of speciation, nevertheless seems to provide this group of fungal pathogens a means to regulate cell growth in a distinct manner. We anticipate that our findings would lead to future studies to dissect the non-essential TAF12L to better understand the role of shared TAFs in supporting genome-wide transcription.
Experimental procedures
Growth conditions, strains, and plasmids
C. albicans strains SN87, SN95, and SN152 (53) were used as parental strains for all strains derived in this study. S. cerevisiae strains were derived from BY4743. Both S. cerevisiae and C. albicans strains were cultured in yeast extract-peptone (YP)-rich medium (54) for genetic analyses and protein extraction. All media were supplemented with 20 g/liter of either glucose or maltose as indicated.
The genome sequences and coordinates of genes of our interest were obtained from the Candida Genome Database (CGD). C. albicans SC5314 genomic DNA was used as template for all plasmid constructs, unless indicated otherwise, and all constructs were verified by restriction digestion and sequencing. Details of strain and plasmid constructions are described in the supplemental text. A list of Candida albicans genes reported in this study is provided in supplemental Table S1. Plasmids and strains used in this study are listed in supplemental Tables S2 and S3, and the list of oligonucleotides is provided in supplemental Table S4.
Immunoblotting
C. albicans strains were grown overnight (∼18–20 h) in YP medium containing maltose and inoculated to a starting A600 of 0.15 (SN95 and PMAL2-CaTAF12L) or 0.25 (PMAL2-CaTAF12) into fresh YP medium containing glucose. Cultures were grown at 30 °C, and cells were harvested at different time points by centrifugation at 2450 × g for 5 min. Protein extracts were prepared and quantitated as described (55). About 50 μg of protein extracts were resolved on 8% SDS-PAGE gel and immunoblotted as described (55). The blots were probed with anti-CaTAF12L (1:1000) and anti-CaTAF12 (1:1000) rabbit polyclonal antibodies, with glucose 6-phosphate antibody (1:1000) as a control (Sigma). All blots were visualized using ECL Plus chemiluminescent system (GE Healthcare). The details of construction and purification of recombinant CaTAF12L and CaTAF12, as well as rabbit anti-CaTAF12L and anti-CaTAF12 polyclonal antibody production (see supplemental text).
Coimmunoprecipitation
The indicated C. albicans strains were cultured in YP medium containing glucose at 30 °C for 5 h, and whole-cell extracts were prepared as described previously (55). About 1 mg each of the whole-cell extracts was mixed with either 5 μl of IgG-Sepharose beads for TAP IPs or 10 μl of anti-FLAG M2-agarose beads (Sigma) for FLAG IPs and then incubated on a rotating platform at 4 °C for 2 h. After IP, the beads were collected, washed, and boiled in 1× SDS-PAGE sample buffer. 2-fold serial dilutions of the immunoprecipitated samples and 100 μg of input cell extracts were separated by 8% SDS-PAGE, and then immunoblotted as described (55). The following antibodies were used at 1:1000 dilution to probe the immunoblots: rabbit IgG Fc fragment (Jackson ImmunoResearch Laboratories), anti-HA (12CA5, Roche Applied Science), mouse polyclonal anti-TAF4 and anti-Ada1, and rabbit polyclonal anti-CaTAF12L and anti-CaTAF12 antibodies. Anti-FLAG antibody was used at 1:2500. All blots were visualized using ECL Plus chemiluminescent system (GE Healthcare).
Microscopy
C. albicans strains were cultured in the indicated medium, and then cells were harvested, washed with sterile 1× PBS, and fixed in 4% formaldehyde for 1–2 h at room temperature on a rotator. The fixed cells were again washed several times with 1× PBS. For cell imaging, an 0.1–0.2 A600 equivalent of the fixed cells was stained with a 1:20 dilution of 0.1 mg/ml DAPI for 20–30 min in the dark. Cells were pelleted, and a 100-fold dilution of 1 mg/ml calcofluor white (CFW) was added, incubated for 10 min in the dark, washed four times with 1× PBS, and mounted on slides, and images were acquired in a confocal microscope (Olympus FluoView FV1000) using 100× objective (oil immersion).
Author contributions
I. S., P. P., S. S., and K. N. designed research; I. S., P. P., and S. S. performed experiments; I. S., P. P., and K. N. analyzed data; I. S. drafted the manuscript, and K. N. edited and completed the manuscript. All authors read and approved the manuscript.
Supplementary Material
Acknowledgments
We are grateful to K. Ganesan for providing the promoter-replacement constructs prior to their publication. We also thank Suzanne Noble, Alexander Johnson, and Joachim Morschhauser for other plasmids and strains, and Shambhu Kumar for strain construction of FLAG-tagged strains. We thank Jerry Workman, Swaminathan Venkatesh, and members of the Workman laboratory for helpful discussions, and members of the Natarajan laboratory for discussions and comments on the manuscript. We acknowledge equipment use at the Central Instrumentation Facility (CIF), School of Life Sciences and the Confocal Microscopy Facility at Advanced Instrumentation Research Facility (AIRF), Jawaharlal Nehru University (JNU).
This work was partially supported by a grant from the Department of Biotechnology and through the DST-PURSE and UPE2 grants, and departmental funding under DST-FIST and UGC-NRC grants. This work was also supported by junior and senior research fellowships from the Council of Scientific and Industrial Research, New Delhi (to I. S. and P. P.) and by a senior research fellowship from the Indian Council for Medical Research, New Delhi (to S. S.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental text, Fig. S1, and Tables S1–S5.
- TFIID
- transcription factor II D
- TAF
- TBP-associated factor
- TBP
- TATA box-binding protein
- HFD
- histone-fold domain
- TAP
- tandem affinity purification
- IP
- immunoprecipitation
- YPD
- yeast extract peptone dextrose
- YPMal
- yeast extract peptone maltose
- CFW
- calcofluor white
- 5-FOA
- 5-fluoroorotic acid
- Ca
- Candida albicans.
References
- 1. Hahn S. (2004) Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Biol. 11, 394–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Burley S. K., and Roeder R. G. (1996) Biochemistry and structural biology of transcription factor IID (TFIID). Annu. Rev. Biochem. 65, 769–799 [DOI] [PubMed] [Google Scholar]
- 3. Albright S. R., and Tjian R. (2000) TAFs revisited: more data reveal new twists and confirm old ideas. Gene 242, 1–13 [DOI] [PubMed] [Google Scholar]
- 4. Tora L. (2002) A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev. 16, 673–675 [DOI] [PubMed] [Google Scholar]
- 5. Shen W. C., Bhaumik S. R., Causton H. C., Simon I., Zhu X., Jennings E. G., Wang T. H., Young R. A., and Green M. R. (2003) Systematic analysis of essential yeast TAFs in genome-wide transcription and preinitiation complex assembly. EMBO J. 22, 3395–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Apone L. M., and Green M. R. (1998) Transcription sans TBP. Nature 393, 114–115 [DOI] [PubMed] [Google Scholar]
- 7. Gangloff Y. G., Romier C., Thuault S., Werten S., and Davidson I. (2001) The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem. Sci. 26, 250–257 [DOI] [PubMed] [Google Scholar]
- 8. Grant P. A., Schieltz D., Pray-Grant M. G., Steger D. J., Reese J. C., Yates J. R. 3rd, and Workman J. L. (1998) A subset of TAFIIs are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94, 45–53 [DOI] [PubMed] [Google Scholar]
- 9. Weake V. M., and Workman J. L. (2012) SAGA function in tissue-specific gene expression. Trends Cell Biol. 22, 177–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Juven-Gershon T., and Kadonaga J. T. (2010) Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev. Biol. 339, 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Garbett K. A., Tripathi M. K., Cencki B., Layer J. H., and Weil P. A. (2007) Yeast TFIID serves as a coactivator for Rap1p by direct protein-protein interaction. Mol. Cell. Biol. 27, 297–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu W. L., Coleman R. A., Ma E., Grob P., Yang J. L., Zhang Y., Dailey G., Nogales E., and Tjian R. (2009) Structures of three distinct activator-TFIID complexes. Genes Dev. 23, 1510–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stringer K. F., Ingles C. J., and Greenblatt J. (1990) Direct and selective binding of an acidic transcription factor domain to the TATA-box factor TFIID. Nature 345, 783–786 [DOI] [PubMed] [Google Scholar]
- 14. Mencía M., Moqtaderi Z., Geisberg J. V., Kuras L., and Struhl K. (2002) Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol. Cell 9, 823–833 [DOI] [PubMed] [Google Scholar]
- 15. Hiller M., Chen X., Pringle M. J., Suchorolski M., Sancak Y., Viswanathan S., Bolival B., Lin T. Y., Marino S., and Fuller M. T. (2004) Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development 131, 5297–5308 [DOI] [PubMed] [Google Scholar]
- 16. Wang P. J., and Page D. C. (2002) Functional substitution for TAFII250 by a retroposed homolog that is expressed in human spermatogenesis. Hum. Mol. Genet. 11, 2341–2346 [DOI] [PubMed] [Google Scholar]
- 17. Pointud J. C., Mengus G., Brancorsini S., Monaco L., Parvinen M., Sassone-Corsi P., and Davidson I. (2003) The intracellular localisation of TAF7L, a paralogue of transcription factor TFIID subunit TAF7, is developmentally regulated during male germ-cell differentiation. J. Cell Sci. 116, 1847–1858 [DOI] [PubMed] [Google Scholar]
- 18. Freiman R. N., Albright S. R., Zheng S., Sha W. C., Hammer R. E., and Tjian R. (2001) Requirement of tissue-selective TBP-associated factor TAFII105 in ovarian development. Science 293, 2084–2087 [DOI] [PubMed] [Google Scholar]
- 19. Dikstein R., Zhou S., and Tjian R. (1996) Human TAFII 105 is a cell type-specific TFIID subunit related to hTAFII130. Cell 87, 137–146 [DOI] [PubMed] [Google Scholar]
- 20. Liu W. L., Coleman R. A., Grob P., King D. S., Florens L., Washburn M. P., Geles K. G., Yang J. L., Ramey V., Nogales E., and Tjian R. (2008) Structural changes in TAF4b-TFIID correlate with promoter selectivity. Mol. Cell 29, 81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Frontini M., Soutoglou E., Argentini M., Bole-Feysot C., Jost B., Scheer E., and Tora L. (2005) TAF9b (formerly TAF9L) is a bona fide TAF that has unique and overlapping roles with TAF9. Mol. Cell. Biol. 25, 4638–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Müller F., and Tora L. (2004) The multicoloured world of promoter recognition complexes. EMBO J. 23, 2–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ogryzko V. V., Kotani T., Zhang X., Schiltz R. L., Howard T., Yang X. J., Howard B. H., Qin J., and Nakatani Y. (1998) Histone-like TAFs within the PCAF histone acetylase complex. Cell 94, 35–44 [DOI] [PubMed] [Google Scholar]
- 24. Martinez E., Palhan V. B., Tjernberg A., Lymar E. S., Gamper A. M., Kundu T. K., Chait B. T., and Roeder R. G. (2001) Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol. 21, 6782–6795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nagy Z., Riss A., Fujiyama S., Krebs A., Orpinell M., Jansen P., Cohen A., Stunnenberg H. G., Kato S., and Tora L. (2010) The metazoan ATAC and SAGA coactivator HAT complexes regulate different sets of inducible target genes. Cell. Mol. Life Sci. 67, 611–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Weake V. M., Swanson S. K., Mushegian A., Florens L., Washburn M. P., Abmayr S. M., and Workman J. L. (2009) A novel histone fold domain-containing protein that replaces TAF6 in Drosophila SAGA is required for SAGA-dependent gene expression. Genes Dev. 23, 2818–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mitsuzawa H., Seino H., Yamao F., and Ishihama A. (2001) Two WD repeat-containing TATA-binding protein-associated factors in fission yeast that suppress defects in the anaphase-promoting complex. J. Biol. Chem. 276, 17117–17124 [DOI] [PubMed] [Google Scholar]
- 28. Mitsuzawa H., and Ishihama A. (2002) Identification of histone H4-like TAF in Schizosaccharomyces pombe as a protein that interacts with WD repeat-containing TAF. Nucleic Acids Res. 30, 1952–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pfaller M. A., and Diekema D. J. (2007) Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20, 133–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moqtaderi Z., Yale J. D., Struhl K., and Buratowski S. (1996) Yeast homologues of higher eukaryotic TFIID subunits. Proc. Natl. Acad. Sci. U.S.A. 93, 14654–14658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Natarajan K., Jackson B. M., Rhee E., and Hinnebusch A. G. (1998) yTAFII61 has a general role in RNA polymerase II transcription and is required by Gcn4p to recruit the SAGA coactivator complex. Mol. Cell 2, 683–692 [DOI] [PubMed] [Google Scholar]
- 32. Lago C., Clerici E., Mizzi L., Colombo L., and Kater M. M. (2004) TBP-associated factors in Arabidopsis. Gene 342, 231–241 [DOI] [PubMed] [Google Scholar]
- 33. Dujon B., Sherman D., Fischer G., Durrens P., Casaregola S., Lafontaine I., De Montigny J., Marck C., Neuvéglise C., Talla E., Goffard N., Frangeul L., Aigle M., Anthouard V., Babour A., et al. (2004) Genome evolution in yeasts. Nature 430, 35–44 [DOI] [PubMed] [Google Scholar]
- 34. Butler G., Rasmussen M. D., Lin M. F., Santos M. A., Sakthikumar S., Munro C. A., Rheinbay E., Grabherr M., Forche A., Reedy J. L., Agrafioti I., Arnaud M. B., Bates S., Brown A. J., Brunke S., et al. (2009) Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459, 657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fitzpatrick D. A., O'Gaora P., Byrne K. P., and Butler G. (2010) Analysis of gene evolution and metabolic pathways using the Candida Gene Order Browser. BMC Genomics 11, 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maguire S. L., ÓhÉigeartaigh S. S., Byrne K. P., Schröder M. S., O'Gaora P., Wolfe K. H., and Butler G. (2013) Comparative genome analysis and gene finding in Candida species using CGOB. Mol. Biol. Evol. 30, 1281–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Werten S., Mitschler A., Romier C., Gangloff Y.-G., Thuault S., Davidson I., and Moras D. (2002) Crystal structure of a subcomplex of human transcription factor TFIID formed by TATA binding protein-associated factors hTAF4 (hTAFII135) and hTAF12 (hTAFII20). J. Biol. Chem. 277, 45502–45509 [DOI] [PubMed] [Google Scholar]
- 38. Gangloff Y. G., Werten S., Romier C., Carré L., Poch O., Moras D., and Davidson I. (2000) The human TFIID components TAFII135 and TAFII20 and the yeast SAGA components ADA1 and TAFII68 heterodimerize to form histone-like pairs. Mol. Cell. Biol. 20, 340–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sanders S. L., Garbett K. A., and Weil P. A. (2002) Molecular characterization of Saccharomyces cerevisiae TFIID. Mol. Cell. Biol. 22, 6000–6013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Leurent C., Sanders S., Ruhlmann C., Mallouh V., Weil P. A., Kirschner D. B., Tora L., and Schultz P. (2002) Mapping histone fold TAFs within yeast TFIID. EMBO J. 21, 3424–3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu P. Y., Ruhlmann C., Winston F., and Schultz P. (2004) Molecular architecture of the S. cerevisiae SAGA complex. Molecular cell 15, 199–208 [DOI] [PubMed] [Google Scholar]
- 42. Roberts S. M., and Winston F. (1997) Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics 147, 451–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Drysdale C. M., Jackson B. M., McVeigh R., Klebanow E. R., Bai Y., Kokubo T., Swanson M., Nakatani Y., Weil P. A., and Hinnebusch A. G. (1998) The Gcn4p activation domain interacts specifically in vitro with RNA polymerase II holoenzyme, TFIID, and the Adap-Gcn5p coactivator complex. Mol. Cell. Biol. 18, 1711–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Han Y., Luo J., Ranish J., and Hahn S. (2014) Architecture of the Saccharomyces cerevisiae SAGA transcription coactivator complex. EMBO J. 33, 2534–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Müller F., Zaucker A., and Tora L. (2010) Developmental regulation of transcription initiation: more than just changing the actors. Curr. Opin. Genet. Dev. 20, 533–540 [DOI] [PubMed] [Google Scholar]
- 46. Spedale G., Timmers H. T., and Pijnappel W. W. (2012) ATAC-king the complexity of SAGA during evolution. Genes Dev. 26, 527–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Thuault S., Gangloff Y.-G., Kirchner J., Sanders S., Werten S., Romier C., Weil P. A., and Davidson I. (2002) Functional analysis of the TFIID-specific yeast TAF4 (yTAFII48) reveals an unexpected organization of its histone-fold domain. J. Biol. Chem. 277, 45510–45517 [DOI] [PubMed] [Google Scholar]
- 48. Reeves W. M., and Hahn S. (2005) Targets of the Gal4 transcription activator in functional transcription complexes. Mol. Cell. Biol. 25, 9092–9102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fishburn J., Mohibullah N., and Hahn S. (2005) Function of a eukaryotic transcription activator during the transcription cycle. Mol. Cell 18, 369–378 [DOI] [PubMed] [Google Scholar]
- 50. Herbig E., Warfield L., Fish L., Fishburn J., Knutson B. A., Moorefield B., Pacheco D., and Hahn S. (2010) Mechanism of Mediator recruitment by tandem Gcn4 activation domains and three Gal11 activator-binding domains. Mol. Cell. Biol. 30, 2376–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wright K. J., Marr M. T. 2nd, and Tjian R. (2006) TAF4 nucleates a core subcomplex of TFIID and mediates activated transcription from a TATA-less promoter. Proc. Natl. Acad. Sci. U.S.A. 103, 12347–12352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sellam A., Askew C., Epp E., Lavoie H., Whiteway M., and Nantel A. (2009) Genome-wide mapping of the coactivator Ada2p yields insight into the functional roles of SAGA/ADA complex in Candida albicans. Mol. Biol. Cell 20, 2389–2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hromatka B. S., Noble S. M., and Johnson A. D. (2005) Transcriptional response of Candida albicans to nitric oxide and the role of the YHB1 gene in nitrosative stress and virulence. Mol. Biol. Cell 16, 4814–4826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sherman F. (1991) Getting started with yeast. Methods Enzymol. 194, 3–21 [DOI] [PubMed] [Google Scholar]
- 55. Singh R. P., Prasad H. K., Sinha I., Agarwal N., and Natarajan K. (2011) Cap2-HAP complex is a critical transcriptional regulator that has dual but contrasting roles in regulation of iron homeostasis in Candida albicans. J. Biol. Chem. 286, 25154–25170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tamura K., Stecher G., Peterson D., Filipski A., and Kumar S. (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.