Abstract
The discovery and application of CRISPR/Cas9 technology for genome editing has greatly accelerated targeted mutagenesis in a variety of organisms. CRISPR/Cas9-mediated site-specific cleavage is typically exploited for the generation of insertions or deletions (indels) after aberrant dsDNA repair via the endogenous non-homology end-joining (NHEJ) pathway or, alternatively, for enhancing homology-directed repair to facilitate the generation of a specific mutation (or “knock-in”). However, there is a need for efficient cellular assays that can measure Cas9/guide RNA activity. Reliable methods for enriching and identifying desired mutants are also lacking. Here we describe a method using the Piggybac transposon for stable genomic integration of an H2B-GFP reporter or a hygromycin resistance gene for assaying Cas9 target cleavage and homology-directed repair. The H2B-GFP fusion protein provides increased stability and an obvious pattern of nuclear localization. This method, called SRIRACCHA (i.e. a stable, but reversible, integrated reporter for assaying CRISPR/Cas-stimulated HDR activity), enables the enrichment of mutants via selection of GFP-positive or hygromycin-resistant mammalian cells (immortalized or non-immortalized) as a surrogate for the modification of the endogenous target site. Currently available hyperactive Piggybac transposase mutants allow both delivery and removal of the surrogate reporters, with minimal risk of generating undesirable mutations. This assay permits rapid screening for efficient guide RNAs and the accelerated identification of mutant clones and is applicable to many cell types. We foresee the utility of this approach in contexts in which the maintenance of genomic integrity is essential, for example, when engineering cells for therapeutic purposes.
Keywords: CRISPR/Cas, DNA endonuclease, gene knockout, homologous recombination, transposable element (TE), HDR, Piggybac, genome editing, surrogate reporter
Introduction
CRISPR/Cas9 is an RNA-guided endonuclease (RGEN)2 that was discovered as part of an immune defense mechanism in bacteria whereby foreign DNA, e.g. from pathogenic phage, is cleaved by a specialized nuclease using RNA molecules (gRNAs) to dictate sequence specificity, thus preventing horizontal gene transfer (1). The adoption of the CRISPR/Cas9 system from Staphylococcus pyogenes, typically employing a single protein SpCas9 and a single synthetic gRNA, has brought about widespread implementation of this powerful site-specific RGEN for generating knockouts or precise gene editing in a wide variety of eukaryotic cell types, e.g. in plants, invertebrates, and mammals, including human cells.
Since the description of the CRISPR/Cas9 technology in eukaryotes, it has quickly gained popularity over other similar approaches such as the transcription activator-like effector nuclease (TALEN). The popularity of RGENs is due to their simplicity of design and high efficiency in a variety of mammalian cell types and embryos for both somatic and germ line mutagenesis. Specificity of this RGEN system is conferred by a 20-nucleotide region of the gRNA, which targets the Cas9 nuclease to complementary dsDNA sequences (i.e. protospacers) immediately followed by a protospacer adjacent motif (PAM), which for SpCas9 systems is NGG, and is required for Cas9 recognition and cleavage. Although both RGEN and TALEN (transcription activator-like effector nuclease) techniques harness nonspecific DNA endonucleases to generate double-stranded breaks (DSBs) (with both systems able to achieve high efficiency), RGEN use has accelerated because of the ease of generating gRNAs using standard cloning techniques and in vitro transcription or RNA polymerase III promoters. However, CRISPR/Cas9 RGENs have the potential for off-target effects (2–5), and therefore, one must typically screen many gRNAs to select for both high efficiency and high specificity. Alternatively, higher specificity can be achieved through modified RGEN approaches, such as the paired nickase approach with the Cas9D10A mutant and two gRNAs, which has greatly improved fidelity of RGENs for the generation of DSBs without a substantial loss of efficiency (3, 5); however, the paired nickase approach still requires the preselection of highly efficient pairs of gRNAs.
The simple, flexible, and highly efficient nature of CRISPR/Cas9 propels this RGEN system to become the most widely adopted technique for gene editing. The potential of the CRISPR/Cas9 system has been demonstrated through successful reverse genetic approaches in cells and whole organisms (6–9), multiplexed gene editing for simultaneously targeting multiple genes (7, 8, 10, 11), genome-wide genetic screens (12–20), and also for gene therapy in human cells (21–24). However, the practical application of CRISPR/Cas9 for generating knockouts (through the generation of DSBs and deletions) or precise gene editing (for knock-ins via homology directed repair with a donor template) still requires assaying multiple gRNAs for optimal activity/specificity and screening many cellular clones to identify the desired mutation.
The Piggybac (PB) transposon is a cut-and-paste mobile DNA element originally isolated from Trichoplusia ni and has undergone successive modifications through both codon optimization and directed evolution to generate one of the most highly active transposons for use in mammalian cells (25, 26). Typical applications exploit PB for stable integration of foreign DNA into the genome as a safer and easier alternative to retroviral vectors in cell types that can be transfected with low or modest efficiency. The PB transposon acts by precise mobilization into TTAA sequences, with a modest preference for transcriptional units (27). PB can mobilize multiple copies of a transposon into the genome with high efficiency (28) and can also mobilize very large pieces of DNA (up to 100 kb) (29). Another highly unique and practical property of the PB DNA transposon is the ability to both mobilize and remove integrated transposons using the PB transposase (PBase). With the development of an excision-only (Exc+Int−) mutant PBase (30), transposons can now be removed without the risk of reintegration. Lastly, one key advantage of PB is its “footprint-free” mobilization whereby sequence integrity around the TTAA integration site is preserved after excision of the PB transposon (28, 31–34).
Despite the success of RGENs for gene knock-out or editing, their performance is largely determined by the quality and specificity of guide RNAs. In the nucleus, chromatin accessibility, nuclease cleavage efficiency, and successful repair through NHEJ or HDR likely affect the successful generation of desired mutations. Examining site-specific nuclease activity in cells after the introduction of RGEN components is possible using Surveyor® or T7 endonuclease 1 (T7E1). These mismatch detection enzymes cleave bulges in DNA heteroduplexes consisting of wild type and mutated DNA strands (35, 36). However, these assays are not very sensitive or easily quantifiable, whereas more quantitative methods, such as high throughput targeted deep sequencing, can be prohibitively expensive, especially when assaying large numbers of gRNAs. Our goal here is to exploit the PB DNA transposon to deliver into the mammalian genome of cultured cells a reporter construct that provides a read-out of site-specific nuclease cleavage. Through transfection of a repair “donor” template, reporter expression (H2B-GFP or a hygromycin resistance gene) is restored through HDR after the generation of a DSB. Perhaps most useful of all, purification of H2B-GFP cells and/or selection of hygromycin-resistant cells enables the enrichment of mutant clones. Furthermore, the properties of the PB transposon enable the seamless removal of integrated transposons after the selection and characterization of desired mutations at endogenous target loci. This approach both accelerates gRNA validation and the generation of desired mutations.
Results
A transposable GFP reporter to assay CRISPR activity
The transposable SRIRACCHA GFP reporter consists of a CMV-driven puromycin resistance gene followed by a single cloning site for inserting endonuclease target sequences, a short polyadenylation signal, and an out-of-frame H2B-tagged GFP, which lacks a start codon followed by two chicken hypersensitive site IV (HSIV) insulator sequences to prevent silencing by heterochromatin. A schematic for a paired nickase (Cas9D10A) target, with two SpCas9 protospacers and PAMs, is shown (Fig. 1A). A donor plasmid is co-transfected with RGEN components (Cas9 and gRNAs) to promote repair and gene conversion via HDR. The donor plasmid contains the puromycin resistance gene linked to the H2B-GFP open reading frame via a T2A peptide coding sequence. A typical SRIRACCHA assay with this GFP reporter entails co-transfection of all components, including a plasmid encoding the hyperactive PB transposase (CMV-hyPBase) and co-transfection with an RFP expression plasmid (CMV-tdT) for normalization. Cutting and HDR of the transposon target yields expression of GFP. Quantification of GFP-positive cells relative to RFP-positive cells thus provides a read-out of nuclease activity. To optimize signal to noise, we titrated transposon, PBase, donor, and gRNA (Fig. 1, B and C). Ratios of 3:1 for transposon:hyPBase and 3:2 for donor:gRNA were optimal, with an approximate 4-fold induction in RGEN activity with relatively low background with this gRNA. These ratios were adopted for subsequent experiments.
Figure 1.
Optimization of the H2B-GFP SRIRACCHA reporter and use with an inducible Cas9D10A nuclease. A, constituents of the Piggybac target transposon with an H2B-GFP reporter and schematic of the SRIRACCHA process. The target transposon consists of a CMV-driven puromycin resistance gene, the target sequence (cloned into a BstEII site), a polyadenylation (pA) signal, an out-of-frame H2B-GFP open reading frame, an additional SV40 pA signal, and two HSIV core insulator sequences at the 3′ end. The Piggybac transposon sequences, including inverted terminal repeats, are represented by two outward facing arrows that flank the CMV-driven expression cassette. The Let-7 protospacer sequences are highlighted in red, and the PAM is in blue. After transposition into the genome by hyPBase and nuclease cutting, the donor plasmid supplies the template for HDR, which restores expression of H2B-GFP via a T2A cleavage sequence. B, the effects of different ratios of transposon and transposase plasmids (by mass) in HEK293T cells. Cells were transfected with plasmids driving constitutive expression of the transposase, wild type Cas9, gRNAs, and pCMV-tdT along with the target transposon and donor plasmid. The target and gRNA sequences are from a site in mouse Mirlet7g. Puromycin was added 48 h after transfection, and expression of GFP and RFP was analyzed 2 days after puromycin selection. Signals from nonspecific (NS) gRNAs or targeting gRNAs were also compared. C, the effects of different ratios of donor plasmid and gRNA plasmid (by mass) in HEK293T cells. Cells were transfected as in B. D, schematic of plasmids used for Cas9D10A induction (iCas9N) with dox and 4-OH-T. E, inducible Cas9D10A SRIRACCHA assay in the presence and absence of the Piggybac transposase. HeLa cells were transfected as above using a 3:1 transposon to transposase ratio and a 3:2 ratio of donor to gRNA but substituting hCas9 with plasmids in D. At 24 h, 500 ng/ml dox and 0.5 μm 4-OH-T was added to the transfected cells to induce Cas9D10A expression for 48 h. Fluorescence was examined 72 h after withdrawing dox and 4-OH-T. F, inducibility of Cas9D10A was compared with dox and/or 4-OH-T treatment, as transfected in E, with fluorescence examined 48 h after drug addition. G and H, SRIRACCHA Cas9D10A assay in HeLa (G) and HEK293T (H) cells. HeLa or HEK293T cells were transfected and induced as in E, with fluorescence examined 24 h (for HeLa) or 48 h (for HEK293T) after drug addition. Error bars represent S.E. of a representative experiment performed in triplicate (and replicated in a total of 2–3 experiments), where Student's t test was performed to determine significance relative to vehicle in E–H, with p < 0.05 (*) or p < 0.01 (**).
Because genome engineering should ideally exhibit high fidelity, we tested a strategy using an inducible Cas9 nuclease to limit the temporal window of nuclease activity. In conjunction with this, we used the Cas9 nickase (Cas9D10A) for paired nicking of target sites, which has been demonstrated to significantly improve fidelity (3, 5). The dual nickase approach also generates larger deletions after NHEJ (37, 38) and may be more useful for generating null alleles. In our approach 4-hydroxytamoxifen (4-OH-T) and doxycycline (dox) regulate Cas9D10A expression and nuclear localization (Fig. 1D). As shown in Fig. 1E, there is very little nuclease activity in the absence of 4-OH-T and dox compared with a nonspecific (NS) gRNA using this inducible SRIRACCHA (iSRIRACCHA) method. Dual control with both 4-OH-T and dox further reduced leaky expression and nuclease activity compared with either 4-OH-T or dox alone (Fig. 1F). We also used iSRIRACCHA to measure HDR with and without the transposase (Fig. 1E). Without the transposase, the signal was significantly reduced due to the transient nature of the episomal plasmid. The use of PB, therefore, amplifies and extends the lifetime of the signal via stable integration of the GFP reporter into the genome. The iSRIRACCHA method generated robust inducible signal in both HeLa and HEK293T cells, but for unknown reasons the background GFP signal was lower in HeLa cells (Fig. 1, G and H), and this cell line was used for subsequent assays.
gRNAs with the highest efficiency were predicted by SRIRACCHA using Cas9D10A
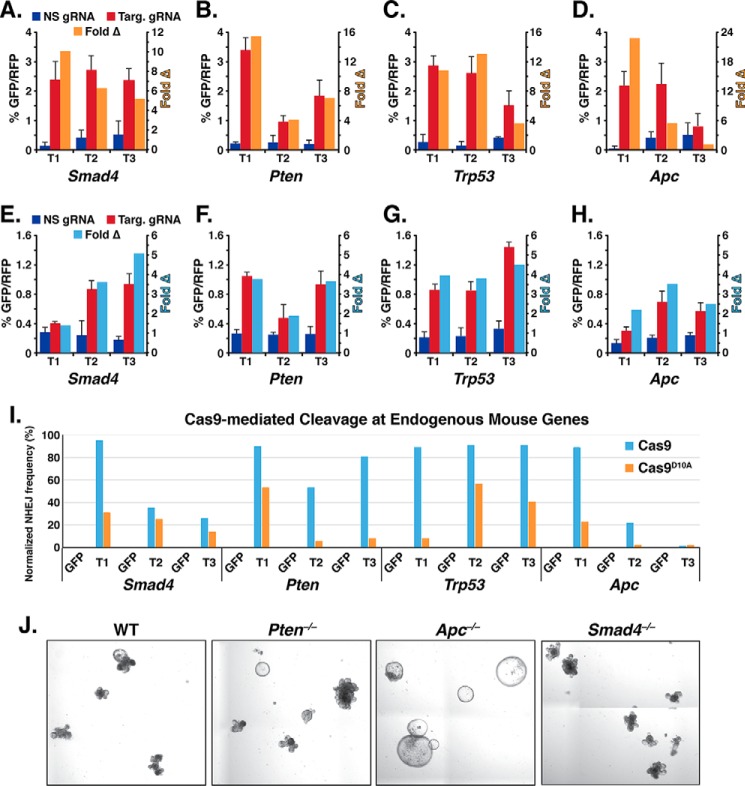
We next sought to determine whether SRIRACCHA could distinguish different activities of gRNAs in their ability to generate insertions or deletions (indels) at endogenous target loci. We designed gRNAs corresponding to three targets sites in mouse Smad4, Pten, Trp53, and Apc and assayed activity using the paired nickase approach and two gRNAs per target (Fig. 2, A–D) or the wild type Cas9 and a single gRNA per target (Fig. 2, E–H). One gRNA of each gRNA pair (used for the paired nickase assay) was used for the wild type Cas9 assay. As expected, varying CRISPR activity was observed with different gRNAs that target the same gene. The gRNA pairs targeting Apc exhibited the most variation using the paired nickase with varying signal to noise ratios, whereas Smad4 exhibited the greatest variability using the wild type Cas9.
Figure 2.
Using SRIRACCHA to test the targeting efficiency of gRNAs. Pairs of gRNAs targeting three unique sites in each of mouse Smad4 (A), Pten (B), Trp53 (C), and Apc (D) genes were assayed by a single transfection of SRIRACCHA H2B-GFP components with the inducible Cas9D10A in HeLa cells. Sequences for variable regions of each gRNA are shown in Table 1. HeLa cells were transfected and induced as described in Fig. 1E. Fluorescence was quantified 3 days after drug addition. SRIRACCHA assay with wild type Cas9 was performed to test the efficiency of individual gRNAs against Smad4 (E), Pten (F), Trp53 (G), and Apc (H) in HeLa cells. One gRNA from each gRNA pair (used in A–D) was assayed with wild type Cas9. I, targeted deep sequencing to test the efficiency of gRNA pairs or individual gRNAs leading to indels at endogenous target sites. Cells were either transfected with a GFP plasmid, as a negative control, or CRISPR/Cas9 plus gRNAs. Indels were quantified as representative of NHEJ frequency. Error bars represent S.E. of a representative experiment, performed in triplicate (and replicated in a total of three experiments). Deep sequencing was performed once for each gRNA. J, bright-field images of wild type mouse enteroids and CRISPR/CasD10A-mutated enteroids harboring loss-of-function mutations in Pten, Apc, or Smad4 genes.
To determine whether the CRISPR/Cas9 nuclease activity measured by SRIRACCHA corresponds to the cutting frequency of endogenous sites in the genome, we subjected single gRNAs or gRNA pairs (using wild type Cas9 or the Cas9 nickase) for testing by targeted deep sequencing of nuclease target sites in the mouse Smad4, Pten, Trp53, and Apc genes after transfection in mouse N2A cells. This enabled the quantification of indels after NHEJ, which is proportional to the cutting frequency by Cas9. Murine N2A cells were nucleofected with gRNAs for RGEN cleavage by the wild type or nickase Cas9 nuclease. Nuclease target sites in Smad4, Pten, Trp53, or Apc were PCR-amplified and subsequently analyzed by Illumina deep sequencing. As shown in Fig. 2I, when Cas9D10A was used, the relative NHEJ activity of gRNA pairs targeting each gene was consistent with the relative -fold change of activity reflected by our CRISPR reporter assay, except for two gRNAs specific for target sites in Trp53. However, in this nickase experiment, gRNA pairs specific for the Trp53 target site T2 exhibited the highest signal to noise ratio both by deep sequencing assay and SRIRACCHA. Both assays yielded a similar trend for Pten gRNAs and Trp53 gRNAs with wild type Cas9. This was not the case for targeting of Smad4 and Apc using wild type Cas9. We consistently see greater sensitivity and signal using Cas9D10A in the SRIRACCHA assay compared with wild type Cas9. This could possibly reflect enhanced HDR using the paired nickase.
We then used the T1 gRNA pairs and the Cas9D10A nickase to introduce indels into the Pten, Apc, or Smad4 genes in wild type C57BL/6 (B6) mouse small intestine enteroids. The hCas9D10A and gRNA constructs were transfected into stem cell-enriched enteroids as previously described (39). Apc-inactivated enteroids were selected with medium lacking R-spondin. Pten-inactivated enteroids were selected with medium lacking EGF but containing the epidermal GFR/HER2 inhibitor lapatinib. And Smad4-inactivated enteroids were selected with medium lacking noggin. Mutations of Apc and Smad4 in enteroids were confirmed by targeted deep sequencing, whereas Pten mutations were quantified using the T7E1 assay (data not shown). Notably, these Apc−/− and Smad4−/− mouse enteroids exhibited similar morphological changes with the reported APC−/− and SMAD4−/− human intestinal enteroids (40) (Fig. 2J).
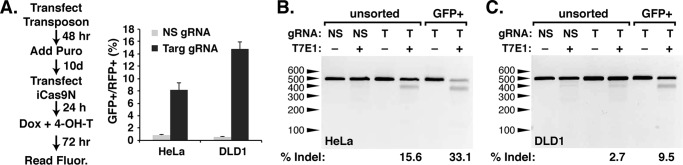
SRIRACCHA enabled the enrichment of mutant cells
The stable property of the SRIRACCHA reporter allows the identification of cells demonstrating CRISPR/Cas-stimulated HDR of the H2B-GFP reporter. We suspected that enrichment of GFP+ cells, which have undergone HDR after cleavage at the surrogate/synthetic site, would also enrich for cells containing indels at the endogenous target site. To confirm this we generated HeLa and DLD1 cell lines with a stably integrated GFP Piggybac transposon reporter containing a target site for NFE2L2 (Fig. 3A, left panel). The puromycin-selected cells were then transfected with the inducible RGEN nickase (iCas9N) and gRNA components. Twenty-four hours after transfection, dox and 4-OH-T were added to induce nuclease expression. When assayed 3 days after induction, SRIRACCHA reporter activity was induced >8-fold in HeLa cells and >20-fold in DLD1 cells (Fig. 3A, right panel). H2B-GFP-positive cells were also assayed for the presence of indels at endogenous NFE2L2 target sites, revealing a 2–4-fold increase in mutation frequency in H2B-GFP-positive cells relative to unsorted cells (Fig. 3, B and C).
Figure 3.
Sorting of GFP-positive cells enriches for mutations of targeted genes. A, left panel, workflow of two-step transfection procedure for targeted mutagenesis using the SRIRACCHA GFP reporter for analysis and cell sorting. Fluorescence read-out (A, right panel) in HeLa and DLD1 cells following this procedure. The NFE2L2 gene was targeted in these experiments. Cells were treated with dox and 4-OH-T for 72 h, expanded, and subjected to FACS. Mutagenesis was determined by T7E1 assays of genomic DNA from unsorted or GFP-sorted HeLa (B) or DLD1 (C) cells. NS denotes nonspecific gRNA, and T denotes gRNAs specific for NFE2L2. Indel frequency was quantified by densitometry of each band and is indicated below each gel. Results are shown from two experiments in HeLa and DLD1 cells, each performed in triplicate.
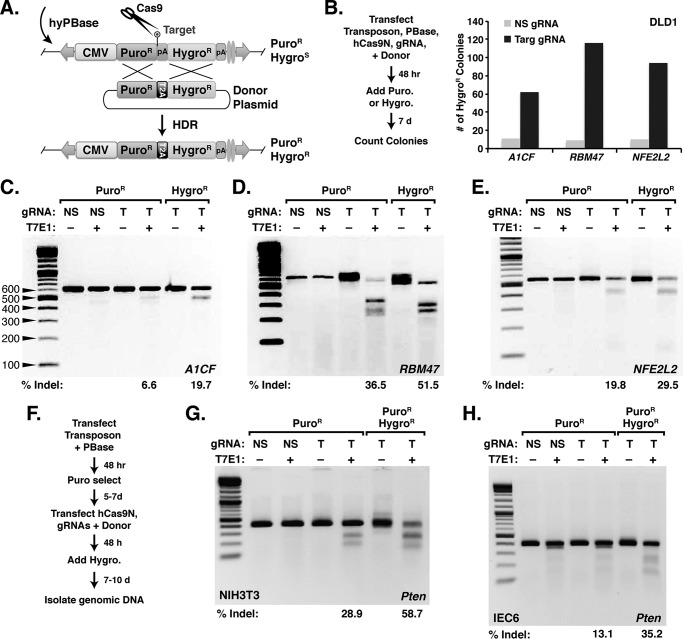
A SRIRACCHA hygromycin reporter for enrichment of mutant clones
Because FACS sorting proved to be a reliable method for enriching mutant cells, we sought to streamline the selection process by substituting the H2B-GFP gene with a hygromycin resistance cassette that lacks a start codon (Fig. 4A). To test the hygromycin reporter assay, DLD1 cells were transfected with transposon and CRISPR/Cas9 nickase components. Then, 48 h after transfection, DLD1 cells were subjected to puromycin selection or hygromycin selection (Fig. 4B, left panel). After selection, surviving colonies were counted then lysed for gDNA isolation and analysis of indel frequency using the T7E1 assay. Three SRIRACCHA hygromycin reporters corresponding to the A1CF, RBM47, and NFE2L2 genes were analyzed. After selection of transfected DLD1 cells, targeting gRNAs yielded 6–12 times as many colonies as non-targeting gRNAs (Fig. 4B, right panel). By T7E1 assay, the hygromycin-resistant clones exhibited a higher mutation frequency than puromycin-resistant colonies (Fig. 4, C–E).
Figure 4.
Enrichment of mutant cells with a target transposon conferring hygromycin resistance. A, schematic of HDR of an integrated Piggybac target transposon with a hygromycin resistance cassette. B, using the hygromycin reporter for SRIRACCHA with Cas9D10A in DLD1 cells against targets in A1CF, RBM47, and NFE2L2. The assay workflow is shown on the left. The bar graph on the right shows the number of surviving colonies after 7 days of hygromycin selection for targeting gRNAs and a nonspecific (NS) gRNA. C–E, T7E1 assay to analyze mutation rate in DLD1 cells after puromycin or hygromycin selection reveals that hygromycin enriches for mutations in the A1CF (C), RBM47 (D), and NFE2L2 (E) genes. F, two-step transfection approach using the BII-C3H transposon containing the Pten-T1 target site for Pten mutagenesis in NIH3T3 and IEC6 cells. G, T7E1 assay for mutations in the mouse Pten gene after selection of transfected NIH3T3 with hygromycin and/or puromycin after mutagenesis with Pten-T1 gRNAs (or NS gRNAs) using the paired nickase approach and Cas9D10A. H, T7E1 assay for mutations in the rat Pten gene following selection of IEC6, as described in G. Indel frequency (% Indel) is calculated from band intensity of fragments generated from T7E1 cleavage.
SRIRACCHA enriches mutations in non-transformed and non-immortalized cells
Using gRNAs specific for mouse/rat Pten (Pten-T1; Fig. 2, F and I), we used a two-step transfection approach, entailing selection for stable integration of the transposon reporter (Fig. 4A) before transfection of CRISPR/Cas9 components and donor plasmid (Fig. 4F) in mouse NIH3T3 (non-transformed) and rat IEC6 (non-immortalized) cells. After selection, indel frequency was measured by T7E1 assay. In these cell lines we saw a 2–3-fold increase of indel mutations after hygromycin selection (Fig. 4, G and H). This demonstrates that SRIRACCHA can be exploited for enriching mutants in multiple cell types, both non-transformed and non-immortalized.
Enriching for null alleles with SRIRACCHA
To generate knock-out clones in DLD1 and HepG2 cells, we used a two-step transfection method (Fig. 5A) that yielded results similar to the two-step transfection approach using the H2B-GFP reporter (Figs. 3A and 5B). Comparing each transfection approach, the two-step method yielded more colonies, whereas relative gRNA efficiencies were similar (Figs. 4B versus 5B). Twelve hygromycin-resistant DLD1 colonies were evaluated for loss of protein after RBM47 targeting. In Western blots one of these clones lacked detectable expression of RBM47 (Fig. 5C). In a similar experiment with HepG2 cells, targeting A1CF, among seven clones, five appeared to lack A1CF protein (Fig. 5C).
Figure 5.
Enriching for null and knock-in alleles with SRIRACCHA. A, modified workflow for 2-step transfection and hygromycin SRIRACCHA assay. B, images of hygromycin-resistant DLD1 colonies in 6-well plates for targeting gRNAs and a nonspecific (NS) gRNA. C, upper blot, Western blots to detect RBM47 expression in parental (P, untreated) and hygromycin-resistant DLD1 clones after undergoing SRIRACCHA with the hygromycin vector. C, lower blot, Western blots to detect A1CF expression in 7 HepG2 clones, also after undergoing SRIRACCHA with the hygromycin vector. A total of 20 HepG2 clones were assessed for A1CF expression by Western blot, with 12 clones showing loss of A1CF (not shown). In both cases DLD1 and HepG2 cells were subjected to the two-step transfection method (as in A). Results are from three experiments with the two-step method (B and C). D, T7E1 assay to analyze mutation rate of NFE2L2 in DLD1 clones subjected to two-step hygromycin SRIRACCHA with gRNAs targeting NFE2L2. Controls NS1, NS2, and positive control (Pos.) are pools of cells transfected with the indicated target transposons or gRNAs. The majority (84%) of clones are mutated at the endogenous NFE2L2 target site. E, schematic for targeting AKT with Cas9N and a ssODN for generating an AKT allele with a myristoylation signal at the amino terminus. Successful targeting was quantified by Taqman QPCR using primers flanking the target site and a fluorescent probe nested within the myristoylation sequence. F, Taqman results for QPCR of the myristoylation sequence in AKT after a one-step SRIRACCHA transfection using the paired nickase approach in HeLa cells. G, Taqman QPCR results for the myristoylation sequence following a two-step SRIRACCHA transfection using the paired nickase approach in DLD1 cells. Taqman QPCR levels were normalized to totalAKT signal from a separate EvaGreen QPCR reaction using the indicated primers in E. Error bars represent S.E. of an experiment performed in triplicate, where Student's t test was performed to evaluate significance, with p values indicated.
One advantage of the two-step method is that the surrogate and endogenous targets can be subjected to repeat CRISPR/Cas9 mutagenesis via subsequent additional transfections of RGEN components. Puromycin-resistant DLD1 cells containing the NFE2L2 transposon were transfected twice with RGEN nickase components specific for this NFE2L2 target site and then selected with hygromycin. Among 51 hygromycin-resistant clones, 43 were identified as mutant by T7E1 assay (Fig. 5D). Thus, with the hygromycin SRIRACCHA reporter, a very high frequency of mutagenesis can be attained.
SRIRACCHA enables the enrichment of knock-in alleles with a single-stranded oligodeoxynucleotide (ssODN) donor
Next we determined the enhancement of HDR using SRIRACCHA and an ssODN donor for insertion of a myristoylation signal at the AKT locus (Fig. 5E). In HeLa cells we pursued a one-step transfection of all components as we found that this cell line transfects with high efficiency. In this approach we used a target transposon that confers blasticidin resistance and, after donor repair, hygromycin resistance. In HeLa cells, hygromycin selection enriches for the myrAKT knock-in allele ∼3-fold (Fig. 5F), as assayed by Taqman QPCR. In a two-step approach in DLD1 cells, hygromycin selection enriches for the myrAKT knock-in allele >300-fold (Fig. 5G). Although transfection efficiency will invariably affect mutagenesis, SRIRACCHA consistently enriches for indels (through NHEJ) or knock-ins (through HDR) as desired.
Removal of the integrated target transposon
The Piggybac transposon reporter permits removal of the reporter from the genome using the “excision-only” mutant transposase PBaseExc+ Int− (or PBex) (Fig. 6A). FACS sorted GFP+ HeLa or DLD1 cells were transfected with the PBex plasmid or a control plasmid. Subsequently, GFP and RFP intensity was analyzed every 24 h starting 2 days after transfection. Over the 10 days after transfection of PBex, GFP intensity in transfected HeLa and DLD1 cells gradually decreased to 40–50% relative to that of control-transfected cells (Fig. 6, B and C), indicating that PBex triggered the successful removal of the transposon reporter from the genome.
Figure 6.
The target transposon is removable with the excision-only PB transposase. A, schematic showing mobilization of the H2B-GFP target transposon (after HDR) by the excision-only Piggybac transposase (PBaseExc+Int−) and subsequent degradation to yield GFP-negative cells. B, mean GFP intensity per RFP+ cell at 10 days post-transfection in GFP-sorted HeLa or GFP-sorted DLD1 cells transfected with pCMV-PBaseExc+Int− or a negative control vector (pCMV-Luc) together with an RFP vector (pCMV-tdT) for normalization. C, mean GFP intensity of cells transfected with pCMV-PBaseExc+Int− relative to GFP intensity of cells transfected with a negative control vector (pCMV-Luc) starting 2 days after transfection. GFP signal was only quantified in cells that are RFP-positive. Error bars represent S.E. of an experiment performed in triplicate, where Student's t test was performed to evaluate significance, with p < 0.05 (*).
Discussion
We have established a new approach for gene editing using the Piggybac DNA transposon as a surrogate RGEN reporter in our SRIRACCHA method, which permits the efficient measurement of nuclease activity and long term enrichment of targeted cells. The robust and novel nature of SRIRACCHA stems from stable expression conferred after the highly efficient mobilization and integration of the Piggybac reporter into the host genome. For single transfection approaches to assay RGEN nuclease activity, the use of Piggybac greatly amplifies and sustains the signal obtained from the H2B-GFP reporter. Our staggered (two-step) transfection procedure (in which the Piggybac reporter is first mobilized into the genome before introduction of RGENs and donor plasmids) permits the measurement of nuclease activity at the surrogate target site exclusively in the context of the host genome. This is likely more representative of native genomic targets, in contrast to episomal reporters (in which chromatin proteins are likely absent), and we speculate that SRIRACCHA may yield a more accurate characterization of RGEN activity in vivo.
Multiple examples of episomal surrogate reporters have been previously described for mutation enrichment after genome manipulation with site-specific nucleases (41–44, 46, 47). These reporters provide a read-out of DNA repair via the NHEJ or single-strand annealing pathways. In the case of NHEJ reporters, a deletion will yield expression of a fluorescent protein if the deletion restores the reading frame. This low level yield per deletion could account for the relatively poor sensitivity of the NHEJ-based reporters (47). Of note, all of the episomal reporters share one limitation: they rely on plasmid DNA transfected into cells where it exists transiently as an extra-chromosomal fragment. The use of transient episomal DNA likely limits the utility of such surrogate reporter assays to cell lines that can be transfected with high efficiency, such as HEK293 cells, a cell type frequently used for demonstrating the efficacy of episomal reporters (41–44). Furthermore, transient episomal reporters that confer resistance to antibiotics such as hygromycin or puromycin are cumbersome for achieving optimal enrichment of modified cells before loss of the episomal vector; the timing and efficacy of such selection is dubious because the precise moment of episomal loss is unpredictable, which complicates antibiotic treatments that require >3–4 days of selection. Long term and complete antibiotic selection with episomal reporters is thus unreliable and simply not tenable for most cell lines because of the limitations of transfection efficiency and the temporary nature of episomal expression.
In addition to other advantages, SRIRACCHA provides an unmet need for an HDR reporter. The use of an HDR reporter also ensures that mutagenesis occurs in dividing cells, considering that HDR occurs only in cells undergoing DNA synthesis (48). Thus, for the enrichment and propagation of mutants, selecting against non-dividing cells with an HDR reporter may help select clones capable of expansion, which is necessary for genotyping and phenotypic characterization. Although NHEJ reporters may provide greater signal (despite missing deletions that do not restore the reading frame of the reporter), our HDR reporter provides a robust read-out with the key advantage that SRIRACCHA enriches for both NHEJ and HDR events. This is not surprising as NHEJ is the dominant repair pathway of double-strand breaks (49) and is likely frequent in cells undergoing HDR.
We envision the greatest utility of our Piggybac-based SRIRACCHA method lies in the ability to enrich mutated cells. Stable expression enables long term enrichment of clones expressing H2B-GFP or the hygromycin resistance gene. This selection in turn enriches for clones with mutations at the endogenous target sites in the host genome. Although the level of enrichment varies (from 1.5–3.5-fold by T7E1 assay), this differential may be linked to the efficiency of targeting gRNAs; i.e. poorer gRNAs benefit more than those of higher efficiency (Fig. 4, B–E). T7E1 assays of hygromycin-selected cells indicate that A1CF targeting benefits the most from enrichment with the surrogate reporter, whereas RBM47 appears to benefit the least (Fig. 4, C–E). This phenomenon may indicate that the high level enrichment (e.g. up to 35-fold) of mutated cells seen with Cas9 episomal surrogate reporters (42, 43) could be predicated on lower gRNA/RGEN efficiency as previously noted (42). On the other hand, studies using episomal surrogates of NHEJ and single-strand annealing also report modest enrichment (2–3-fold, by T7E1) (47). In the former studies (42, 43) assays were performed in HEK293 and HeLa cells, whereas assays in the latter used porcine embryonic fibroblasts (47). These different cell types may also account for differences in enrichment. However, some studies in HEK293 also report lower enrichment (1.9–3-fold) using NHEJ reporters (44), so nuclease efficiency may play a greater role in dictating -fold enhancement.
In contrast to previous descriptions of episomal surrogate reporters for site-specific nucleases (41–44, 46, 47), our gRNAs were either preselected for optimal activity (against NFE2L2 and Pten) before assays of mutation enrichment or we compared enrichment after adjusting for transfection efficiency (e.g. Hygro-resistant versus Puro-resistant cells in Fig. 4, C–E). Thus, our “baseline” may be higher because of this prescreening or normalization. Consistent with this, we always detect mutations by T7E1 assays in mutated cells, whereas previous descriptions of episomal reporters often could not detect mutations in unselected cells (41, 43, 46). In such instances where the mutation load in unselected cells is undetectable by T7E1 assay (41, 43, 46), the -fold changes were calculated to be quite high, based on a denominator that is estimated (i.e. a mutation frequency of 0.5–1%). However, recent deep sequencing and T7E1 assays we performed using wild type Cas9 indicated that T7E1 does not detect a mutation frequency below 3% (data not shown), a level similar to previous reports (44). Thus, underestimated mutation frequencies in controls may underlie the high levels of mutation enrichment seen with other surrogate reporters.
Regardless, selection and enrichment using SRIRACCHA may be particularly valuable in situations where a precise location of nuclease cleavage is needed (to mutate a particular codon, e.g.), in which case only one (or a few) target site(s) may be available, such as the case where we knock-in a myristoylation signal at the start codon of AKT. For achieving knockouts, our final numbers are also encouraging; we achieved a very high mutation rate of NFE2L2 in DLD1 cells, where we found that the majority (84%) of clones were mutant by T7E1 assay (Fig. 4H). For targeted mutagenesis of A1CF and RBM47, we also obtained 13 apparent knockouts among 32 clones examined by Western blot. Thus, high mutation rates can be readily achieved with SRIRACCHA. After the characterization of individual mutant clones using SRIRACCHA, the Piggybac transposon reporter can be removed with the integration-defective PB transposase (PBaseExc+Int−). Sorting for H2B-GFP-negative cells enables selection of cells that have lost the transposon reporter after mobilization by the PB transposase. Alternatively, we have also developed a negative selection vector using HSV-TK (used in Fig. 5, F and G) to select cells resistant to the anti-viral drug ganciclovir. Removal of the Piggybac reporter may be critical for reversing any mutagenic effects of the integrated transposon or for restoring puromycin/hygromycin sensitivity or eliminating H2B-GFP fluorescence if desired. Previous studies have documented that the removal of the Piggybac DNA transposon results in clean, scar-less restoration of the genomic integration site in >99% of cases (28, 31–34). This footprint-free quality of the Piggybac transposon coupled with the unique availability of both hyperactive (25) and integration defective PB transposases (30) clearly makes the Piggybac transposon a superior tool for implementation of SRIRACCHA. Alternatively, retroviral vectors are commonly used for CRISPR/Cas delivery (14, 50–53) but are highly mutagenic because of their preferential insertion in transcriptional units (54–58) and cannot be easily removed from the genome. Thus, the use of high fidelity CRISPR systems, such as the Cas9 paired nickase (3, 5) or enhanced-specificity Cas9 mutants (59, 60), in conjunction with the Piggybac DNA transposon for surrogate-based enrichment is optimal for minimizing unintended mutations.
Using the highly efficient Piggybac transposase, the SRIRACCHA method works well in many cell types, including seven transformed, non-transformed, and non-immortalized cell lines used here: HeLa, HEK293T, DLD1, SW480, HepG2, NIH3T3, and IEC6 cells. This was accomplished using commercial reagents without extensive optimization of transfection conditions (see “Experimental procedures”). Transfection efficiency of cell lines is undoubtedly a limitation of any in vitro mutagenesis approach, but here we demonstrate that SRIRACCHA is able to enrich for mutations in cell lines that are easily transfected (such as HeLa) and in cell lines that are inefficiently transfected (such as IEC6). Many cultured cell lines should be amenable to the Piggybac-based SRIRACCHA method, provided that cells possess a functional HDR repair pathway. This method should prove valuable for manipulation of many cell lines and potentially aid in the development of ex vivo manipulated primary cells for therapeutic purposes. In sum, SRIRACCHA provides a robust and powerful approach for enhancing and enriching nuclease-mediated mutagenesis events in cultured cells.
Experimental procedures
Plasmid constructs
The pBS-U6-gRNA plasmid was constructed by PCR-amplifying the U6-gRNA fragment from the pX335 plasmid and cloning it into the EcoRV site of the pBluescript II KS(−) vector (pBSII) (Stratagene). Subsequent cloning into this vector was accomplished via ligation of double-stranded oligos into the BbsI sites by standard molecular cloning techniques. The pX335(D10A) plasmid was a gift from Dr. Feng Zhang (Addgene plasmid #42335) (7). Oligos for gRNA cloning are listed in Table 1. Vectors generated and described here specifically for SRIRACCHA (PB transposon target vectors, gRNA vectors, and donor templates) are schematically depicted in Fig. 7.
Table 1.
Oligonucleotides for cloning gRNAs
| Oligo name | Sense oligo sequence |
|---|---|
| Apc_t1_LA | CACCGTAAAGTATTTGTCTGGCTC |
| Apc_t1_RA | CACCGCATTATTGAAAGTGGAGGT |
| Apc_t2_LA | CACCGATACCGTAGTATTTAAATA |
| Apc_t2_RA | CACCGCCCAGCTCTTCTTCCTCAA |
| Apc_t3_LA | CACCGCGCAGCTGATTCTGCCTTT |
| Apc_t3_RA | CACCGCAAGTTCAGCACAAAGAAA |
| Pten_t1_LA | CACCGCTAACGATCTCTTTGATGA |
| Pten_t1_RA | CACCGAAAAGGAGATATCAAGAGGA |
| Pten_t2_LA | CACCGTGATAAGTTCTAGCTGTGGT |
| Pten_t2_RA | CACCGTCTGTGAAGATCTTGACCAA |
| Pten_t3_LA | CACCGTGTCATCTTCACTTAGCCAT |
| Pten_t3_RA | CACCGCAGCAATTCACTGTAAAGC |
| Smad4_t1_LA | CACCGACTATGTACAATGCTCAGAC |
| Smad4_t1_RA | CACCGTTGATGTGTCATAGACAAGG |
| Smad4_t2_LA | CACCGCGCGATTACTTGGCGGGTGT |
| Smad4_t2_RA | CACCGAACGGAGACGTACAGCGCCC |
| Smad4_t3_LA | CACCGATGGCTGCCCGCCAGAATAC |
| Smad4_t3_RA | CACCGGACTGTTGCAGATAGCTTC |
| P53_t1_LA | CACCGCAGGAGCTCCTGACACTCGG |
| P53_t1_RA | CACCGACCCTGTCACCGAGACCCC |
| P53_t2_LA | CACCGTTCCACCCGGATAAGATGCT |
| P53_t2_RA | CACCGAAATTTGTATCCCGAGTATC |
| P53_t3_LA | CACCGTGCCTGTCTTCCAGATACTC |
| P53_t3_RA | CACCGACTTTTCGCCACAGCGTGG |
| NFE2L2_T1_PSL | CACCGGGATCTGCCAACTACTCCC |
| NFE2L2_T1_PSR | CACCGGGTTTCTGACTGGATGTGC |
| mLet7g_T2_PSL | CACCGAGCGCTGGAGCAGCTCTGC |
| mLet7g_T2_PSR | CACCGTTCTCTTTTGCCTGATTCC |
| AKT_PSR | CACCGAGGGTTGGCTGCACAAACG |
| AKT_PSL | ACACCGACAATAGCCACGTCGCTCAG |
| Non-specific (NS) | CACCGGGTCTTCGAGAAGACCT |
Figure 7.
Vectors for SRIRACCHA. A–C, PB transposon vectors for insertion of synthetic target sequences containing puromycin resistance cassettes and H2B-GFP reporters A, puromycin and hygromycin resistance cassettes (B) and blasticidin and hygromycin resistance cassettes (C). Each target has a unique BstEII site for cloning target sequences. Vectors are driven by either the CMV promoter or the CMVe/EF1α hybrid promoter (C-hEF1a) devoid of CpG dinucleotides. Some vectors also contain an IRES-TK module for negative selection. D and E, vectors for gRNA expression, with a single gRNA expression cassette (D) or dual gRNA expression cassettes (E) with BpiI sites, as indicated, for cloning gRNA sequences. F, lastly, three donor vectors provide a template for HDR after nuclease cleavage at surrogate target sites.
To construct the BII-CPspaH2B (GFP reporter), BII-CPspaH (hygromycin reporter), BII-CPspaHTK (hygromycin IRES thymidine kinase reporter), and BII-B3HTK (blasticidin-pA-hygromycin IRES thymidine kinase reporter) target vectors, two copies of the 240-bp core insulator from the chicken β-globin HSIV were cloned between the XbaI and EcoRI sites of the pXL-BacII vector to generate the pXL-BII-2xInsul vector. An expression cassette consisting of the CMV promoter, the puromycin resistance gene, a 48-bp HSV thymidine kinase polyadenylation (TKpA) signal, the H2B-GFP open reading frame (lacking an ATG), and an SV40 polyadenylation signal were cloned between the EcoRV and XbaI sites of the pXL-BII-2xInsul vector to generate the BII-CPspaH2B vector. The H2B-GFP sequence in the BII-CPspaH2B vector was replaced with the hygromycin resistance gene (lacking an ATG) to create the BII-CPspaH vector. To generate the BII-CPspaHTK vector, an IRES-HSV-TK (i.e. the thymidine kinase gene from the Herpes simplex virus) sequence was inserted between the hygromycin resistance gene and the SV40 polyadenylation signal in the BII-CPspaH vector. The BII-B3HTK vector is similar to the BII-CPspaHTK vector but confers blasticidin resistance and is driven by the CMV enhancer and hEF1α promoter from pCpGfree (Invivogen Inc.). Target sequences (generated as double stranded oligonucleotides, listed in Table 2) were cloned into a unique BstEII site located between the puromycin (or blasticidin) resistance gene and the TKpA in all of these target vectors (BII-CPspaH2B, BII-CPspaH, or BII-CPspaHTK).
Table 2.
Oligos for generating paired nickase target sites in target transposon vectors
The sense strand of the right target site is shown in bold. The antisense strand of the left target site is underlined.
| Target | Target sequence (cloned into BstEII site of BII-CPspaH2BG/CPspaH/C3H/B3HTK) |
|---|---|
| Apc_t1 | CCGGAGCCAGACAAATACTTTAGCCATTATTGAAAGTGGAGGTGGG |
| Apc_t2 | CCATATTTAAATACTACGGTATTGCCCAGCTCTTCTTCCTCAAGGG |
| Apc_t3 | CCAAAAGGCAGAATCAGCTGCGTCCAAGTTCAGCACAAAGAAATGG |
| Pten_t1 | CCATCATCAAAGAGATCGTTAGCAGAAACAAAAGGAGATATCAAGAGGATGG |
| Pten_t2 | CCCACCACAGCTAGAACTTATCAAACCCTTCTGTGAAGATCTTGACCAATGG |
| Pten_t3 | CCAATGGCTAAGTGAAGATGACAATCATGTTGCAGCAATTCACTGTAAAGCTGG |
| Smad4_t1 | CCTGTCTGAGCATTGTACATAGTTTGATGTGTCATAGACAAGGTGG |
| Smad4_t2 | CCAACACCCGCCAAGTAATCGCGCATCAACGGAGACGTACAGCGCCCCGG |
| Smad4_t3 | CCAGTATTCTGGCGGGCAGCCATAGTGAAGGACTGTTGCAGATAGCTTCAGG |
| P53_t1 | CCTCCGAGTGTCAGGAGCTCCTGCAGCACAGGACCCTGTCACCGAGACCCCTGG |
| P53_t2 | CCCAGCATCTTATCCGGGTGGAAGGAAATTTGTATCCCGAGTATCTGG |
| P53_t3 | CCCGAGTATCTGGAAGACAGGCAGACTTTTCGCCACAGCGTGGTGG |
| NFE2L2_T1 | CCAGCACATCCAGTCAGAAACCAGTGGATCTGCCAACTACTCCCAGG |
| mLet7g_T2 | CCTGCAGAGCTGCTCCAGCGCTCCGTTCTCTTTTGCCTGATTCCAGG |
| AKT | CCATGAGCGACGTGGCTATTGTGAAGGAGGGTTGGCTGCACAAACGAGG |
The donor vectors were constructed by first cloning a cassette consisting of the puromycin resistance gene, a T2A peptide sequence, followed by the H2B-GFP sequence between the HindIII and SpeI sites in the pBSII vector (Stratagene) to create the pBS-PtH2B vector. The pBS-PtH vector was generated by replacing the H2B-GFP sequence in pBS-PtH2B with the hygromycin resistance gene. The pBS-PtH2B donor vector is used in conjunction with the BII-CPspaH2B vector, whereas the pBS-PtH donor is used in conjunction with vectors containing puromycin and hygromycin.
For conditional expression of the Cas9 nickase (Cas9D10A), a lox-stop-lox sequence (LSL) was cloned downstream of the TRE3G promoter (Clontech) located in the pGIT3G-insul vector, which was described previously (61). The LSL sequence consists of loxP sites flanking a splice acceptor from adenovirus 2 (Ad2SA) and four tandem arrayed polyadenylation signals from mouse Csf3, mouse Prm1, bovine GH1, and rabbit HBB2 genes, in this order. An Ad2SA-TKpA sequence was inserted upstream of the TRE3G promoter to reduce read-through transcripts and leaky expression. The Cas9D10A sequence from pX335 was cloned 3′ of the LSL sequence to generate the pGIT3G-LSL-Cas9N vector.
The pCMV-rtTA and pCMV-tdTomato vectors were constructed by replacing the β-galactosidase sequence in pCMV-β-gal (Stratagene) with the rtTAM2 or tdTomato coding sequences. The rtTAM2 and tdTomato sequences were PCR-amplified from the iiLet7 and Vil-Lin28b-itdT vectors, respectively, which were both described previously (61).
The hyperactive PB transposase (CMV-hyPBase) expression plasmid was obtained from System Biosciences Inc. (catalog #PB210PA-1). The expression plasmid (CMV-PBex) encoding the mutant integration-defective PB transposase (Exc+Int−) was also obtained from System Biosciences Inc. (catalog #PB220PA-1). The pCAG-ERT2CreERT2 vector was a gift from Connie Cepko (Addgene plasmid #13777) (62). The expression plasmids hCas9 and hCas9-D10A (hCas9N) were gifts from George Church (Addgene plasmids #41815 and #41816) (8). All DNA oligos were ordered from IDT. All vectors were verified by Sanger dideoxy sequencing after cloning with double-stranded oligos or other double-stranded DNA fragments.
T7E1 assays
Cells from 1 well of a 6-well plate were washed with PBS and lysed by adding 0.8 ml TNESII buffer (50 mm Tris-HCl, pH 8.0, 100 mm EDTA, 100 mm NaCl, and 1% SDS) and 20 μl of 10 mg/ml proteinase K (Roche Applied Science) and incubated at 55 °C overnight. Then, 0.28 ml of saturated NaCl was added to the lysate and mixed thoroughly on a shaker for 5 min. The sample was centrifuged at a maximum speed for 10 min, and 0.75 ml of supernatant was then transferred into a new tube. Genomic DNA was precipitated with 0.5 ml isopropyl alcohol and washed with 70% ethanol. The DNA pellet was dissolved in polyamine buffer (10 mm Tris-HCl, pH 8.0, 30 μm spermine, and 70 μm spermidine) in the presence of 0.1 mg/ml RNaseA and incubated at 37 °C for 15 min.
To perform T7E1 assays, gene fragments encompassing the nuclease target sites were PCR-amplified from genomic DNA using OneTaq DNA polymerase (New England BioLabs) or OneTaq Hot Start DNA Polymerase (New England BioLabs) for 30–35 cycles with 60 s per Kb extension time. Primers used for PCR are listed in Table 3. Then 5 μl of the PCR reaction was annealed and digested with 2 units of T7 endonuclease I (New England BioLabs) at 37 °C for 15 min. Digested DNA fragments were then separated and analyzed on a 2.3% agarose gel. Mutation frequency from T7E1 assays was quantified by measuring the total fluorescence of cut fragments relative to uncut band fluorescence using a Bio-Rad Chemidoc XRS Imaging System.
Table 3.
Primers used for target amplification and T7E1 assays
| Primer name | Sequence |
|---|---|
| RBM47_204F | TTTCCAGAACGCCTCCATC |
| RBM47_288R | TGTGGCAGTACATGACGAAG |
| AICF_82F | CCCACAGCCATACATTTTCAAC |
| AICF_419R | GTTCTGGGTTTCTTTAGTGCATG |
| NFE2L2_4500F | ATTTGTAATCTCCCCACTTCCC |
| NFE2L2_4935R | GGCAAAGCTGGAACTCAAATC |
| Apc_T1_F | AACCTGTCTGCACACTGCAC |
| Apc_T1_R | ATATACCTGTGGTCTTCGTTTGTAG |
| Smad4_T1_F | ACTCCAGAAATTGGAGAGTTGGA |
| Smad4_T1_R | TTGCTAGGATGAGCTCCATTTGTA |
| Pten_T1_F | ATCCTGCAGAAGAAGCCTCG |
| Pten_T1_R | AAAGAGGAACAGCCGCAGAA |
| Pten_T1_R2 | CGAACACTCCCTAGGTTGAATAC |
| P53_T2_F | ATGGTAAGCCCTCAACACCG |
| P53_T2_R | AATTACAGACCTCGGGTGGC |
Enteroid culture and transfection
Mouse enteroid/tumoroid culture and transfection protocols were adapted from methods as previously reported (39). For regular maintenance, mouse enteroids were grown in 100% Geltrex matrix (Thermo-Fisher Scientific) or Matrigel (Corning Inc.) in growth medium: advanced DMEM medium supplemented with 2 mm glutamax (Thermo Fisher Scientific), 10 mm HEPES, 1× penicillin-streptomycin, 50 ng/ml mEGF (R&D Systems Inc.), 5% Noggin- and Rspo1-conditioned medium, 1 mm N-acetylcysteine, 1× N2 supplement (Thermo Fisher Scientific), and 1× B27 supplement (Thermo Fisher Scientific). Noggin- and Rspo1-conditioned medium was obtained from HEK293T cells transduced with a vector containing the mouse Noggin and Rspo1 open reading frames (ORFs). Similarly, Wnt3a-conditioned medium was obtained from HEK293T cells transduced with a vector containing the human Wnt3a ORF.
To prepare cells for transfection and expand stem cell numbers, mouse enteroids were cultured for 3 days in expansion medium (growth medium plus 10 mm nicotinamide and 50% Wnt-conditioned medium). Before transfection, stem cell-enriched enteroids were mechanically dissociated and incubated with TrypLE Express (Thermo Fisher Scientific) for 5 min with periodic vortexing (every minute). Dissociated cells were spun at 100 × g for 2 min and resuspended in growth medium plus 10 mm nicotinamide, 30% Wnt-conditioned medium, and 10 μm Rho kinase inhibitor (Y-27632) (Santa Cruz Biotechnology Inc.). Then, 450 μl of resuspended cells were plated in 1 well of a 24-well plate before transfection with Lipofectamine 2000 (Thermo Fisher Scientific). A typical transfection reaction was prepared by mixing 1 μg of DNA and 3 μl of Lipofectamine 2000 each in 25 μl of DMEM, combining these mixtures, and incubating for 5 min at room temperature. After the transfection mixture was added to cells, the plate was centrifuged at 600 × g for 1 h at 32 °C followed by a 4-h incubation at 37 °C. Cells were then collected and cultured in 100% Geltrex matrix (or Matrigel) in the presence of 10 μm Y-27632.
Transfection with the SRIRACCHA H2B-GFP reporter
HEK293T, HeLa, and DLD1 cells were cultured in DMEM, 10% FBS, and 1× penicillin-streptomycin. Unless noted otherwise in individual experiments, cells were transfected as follows. HEK293T cells were seeded at a density of 30,000 cells/well of a 24-well plate, and HeLa cells were seeded at a density of 35,000–50,000 cells/well of 48-well plate. The next day cells were transfected with 250 ng of total DNA of a plasmid mix consisting of 10 ng of pCMV-tdTomato, 65 ng of pBII-CPspaH2B, 20 ng of CMV-hyPBase, 39 ng of pBS-PtH2B, 25 ng of pCMV-rtTA, 40 ng of pGIT3G-LSL-Cas9N, 25 ng of CAGG-ERT2-CRE-ERT2, and 26 ng of pBS-U6-gRNA using Lipofectamine 2000 (Thermo Fisher Scientific) with a Lipofectamine:DNA ratio of 3:1. Twenty-four hours after transfection, medium was replaced with fresh medium containing 0.5 μg/ml dox (Sigma) and 1 μm 4-OH-T (Sigma). GFP fluorescence was analyzed 72 h after the addition of doxycycline and 4-OH-T. For transfection of wild-type Cas9 in HeLa cells, 225 ng of total DNA consisting of 10 ng of pCMV-tdTomato, 39 ng of pBS-PtH2B, 65 ng of pBII-CPspaH2B, 20 ng of CMV-hyPBase, 65 ng of hCas9, and 26 ng of pBS-U6-gRNA were transfected into each well using JetPrime transfection reagent (Polyplus) according to manufacturer's instruction at a 2:1 ratio (JetPrime:DNA). GFP fluorescence was then examined 72 h after transfection. GFP and RFP fluorescence was analyzed on a Cytation3 (Biotek) and quantified using Gen5 software. The number of GFP+ cells was normalized to the number of RFP+ cells.
Suitable gRNAs against NFE2L2 were first selected among four gRNA pairs after screening for double-strand breaks induced by Cas9D10A in HEK293T cells (not shown). To obtain DLD1 or HeLa cells with stable integration of the BII-CPspaH2B Piggybac reporter for the selected NFE2L2 target site, DLD1 or HeLa cells were seeded at a density of 4 × 105 cells/well of a 6-well plate 18 h before transfection. Then, 1.5 μg of the BII-CPspaH2Btarget construct and 0.5 μg CMV-hyPBase were transfected into each well using 8 μl of Lipofectamine 2000. To select for transposon reporter integration, 2 μg/ml or 5 μg/ml puromycin were added to the transfected HeLa or DLD1 cells, respectively, 48 h post-transfection. The resulting stable DLD1 and HeLa cell lines (containing the BII-CPspaH2B reporter with the NFE2L2 target site) were used for subsequent CRISPR assays. In these H2B-GFP SRIRACCHA assays each cell line was transfected as described above for HeLa cells except that the BII-CPspaH2B and CMV-hyPBase plasmids were excluded from the transfection mixture. HeLa or DLD1 cells (containing the reporter) from 1 well of a 48-well plate were transferred to one well of a 6-well plate and cultured until confluent. Cells were then sorted to purify H2B-GFP-positive cells using a FACSAria II cell sorter (BD Biosciences). Sorted GFP+ HeLa or DLD1 cells were cultured, and their genotype was then analyzed by T7E1 assay. In experiments testing for transposon removal with the integration defective PB transposase (Exc+Int−), H2B-GFP-positive DLD1 or HeLa cells were expanded and plated at a density of 50,000 cells per well of a 24-well plate 18 h before transfection. Then, 450 ng of pCMV-Luc or pCMV-PBex along with 50 ng pCMV-tdTomato were co-transfected into each well using 2 μl of Lipofectamine 2000. Using the Cytation3 and Gen5 Software (Biotek), GFP and RFP fluorescence were quantified every 24 h starting at 48 h post-transfection. To evaluate the efficiency of transposon removal using the integration defective PB transposase (Exc+Int−), the GFP intensity of cells that were successfully transfected (and RFP-positive) was quantified.
Comparison of Cas9 WT and Cas9D10A NHEJ frequencies by deep sequencing
Mouse N2A cells were nucleofected with equal amounts of GFP or a site-specific gRNA and Cas9/Cas9D10A expression plasmid using the Lonza 4D nucleofector with 16-well Nucleocuvette Strips and Lonza published parameters. Three days post nucleofection, N2A cells were harvested, crude lysates were prepared, and PCRs were performed for the NHEJ analysis. PCRs were performed using Illumina protocols and primer sequences to add a barcode and adapters to PCR amplicons. Amplicons were sequenced on an Illumina MiSeq. Indel frequencies were calculated by comparison of MiSeq sample reads to the corresponding NCBI reference sequence. Samples were normalized to sequence reads from GFP transfected controls to correct for sequencing errors.
Transfection with the SRIRACCHA hygromycin reporter
For one-step transfection DLD1 cells were plated at a density of 5 × 105 cells per well of a 6-well plate for assaying the Piggybac hygromycin reporter (BII-CPspaH) containing target sites for A1CF, RBM47, or NFE2L2. Cells were transfected 18 h later with 290 ng of hCas9N, 290 ng of BII-CPspaH, 170 ng of pBS-PtH, 120 ng of pBS-gRNA (targeting or nonspecific), 90 ng of pCMV-hyPBase, and 40 ng of pCMV-tdTomato using 4 μl of Lipofectamine 2000 (Life Technologies). After 48 h cells were split 1:2 into an identical 6-well plate, and either puromycin (5 μg/ml) or hygromycin (600 μg/ml) was added to each plate. After 7 days of selection, hygromycin-resistant colonies were quantified. Twelve hygromycin-resistant colonies targeted at RBM47 were picked, expanded, and characterized for RBM47 protein expression by Western blot. For the remaining colonies, after 12 days of selection both hygromycin- and puromycin-resistant cells were pooled and harvested for isolating genomic DNA for T7E1 assays.
For two-step transfection of DLD1, cells were plated as above, then transfected with 0.5 μg pCMV-hyPBase and 1.5 μg of the Piggybac hygromycin reporter (BII-CPspaH) containing target sites for either A1CF, RBM47, or NFE2L2 genes using 8 μl of Lipofectamine 2000. After 24 h, cells were split 1:2 into 2 wells of a 6-well plate. After 48 h, 5 μg/ml puromycin was added, and cells were selected for 72 h. Cells were then transfected with 1 μg of hCas9N, 0.6 μg of pBS-PtH, and 0.4 μg of pBS-gRNA (targeting or nonspecific). Cells were selected for 7 days, stained with methylene blue, and photographed.
For two-step transfection of NIH3T3 cells and IEC6 cells 50,000 cells were seeded into 1 well of a 12-well plate and then transfected the next day with 200 ng of pCMV-hyPBase and 800 ng of BII-C3H-PtenT1 vector. NIH3T3 were transfected using 3 μl of JetPrime (Polyplus), whereas IEC6 were transfected with 3 μl of Lipofectamine 2000 per the manufacturer's instructions. After 48 h cells were selected with puromycin for 5 days (NIH3T3, 2 μg/ml) or for 7 days (IEC6, 4 μg/ml). These selected cells were then seeded in 6-well plates at a density of 3 × 105 cells per well and transfected the following day with 1.0 μg of hCas9N, 600 ng of pBS-PtH, and 400 ng of pBS-U6gRNAx2 (PtenT1 or nonspecific gRNAs) with the transfection reagents above. After 48 h, hygromycin was added, and cells were selected for 3 days (NIH3T3, 400 μg/ml) or 7 days (IEC6, 100 μg/ml), then expanded for 7 days before isolation of gDNA. PCR and T7E1 assays were performed as described above, using Pten_T1_F and Pten_T1_R2 primers (see Table 3).
For characterization of individual NFE2L2-targeted DLD1 clones, 4 × 105 cells were plated and transfected 18 h later using 7.5 μl of Lipofectamine 2000 with 0.5 μg of pCMV-hyPBase and 2 μg of the Piggybac hygromycin-TK reporter (BII-CPspaHTK) containing no target site or a target site for NFE2L2. After 48 h 5 μg/ml puromycin was added, and cells were selected for 5 days. Cells were split between 2 wells of a 6-well plate and transfected after 18 h with 1 μg of hCas9N, 0.6 μg of pBS-PtH, and 0.4 μg of pBS-gRNA (targeting or nonspecific) using 4 μl of JetPrime reagent (Polyplus). After 48 h the transfection of CRISPR components and the donor vector was repeated. After 48 h of this second transfection, one-half of the cells were split into one 10-cm plate and selected with 600 μg/ml hygromycin for 10 days. Individual hygromycin-resistant colonies were then picked and expanded for isolating genomic DNA for T7E1 assays.
For targeting AKT in HeLa cells for knock-in of a myristoylation signal, 7.5 × 105 cells were seeded and transfected the next day with 30 pmol of myrAKT ssODN, 500 ng of hCas9N, 450 ng of BII-B3HTK-AKT, 250 ng of pBS-BtH, 170 ng of pBS-U6gRNAx2, and 130 ng of pCMV-hyPBase using 5 μl of JetPrime (Polyplus) per the manufacturer's instructions. A nonspecific ssODN (MTOR_S2215Y) was used as a control. Oligo donors and QPCR primers are listed in Table 4. 48 h after transfection cells were selected with either blasticidin (8 μg/ml) or hygromycin (400 μg/ml) for 9 days, and then gDNA was isolated for Taqman QPCR using 50 ng of DNA per 20-μl reaction using the iTaq Universal Probes Mastermix (Bio-Rad). Levels of myrAKT were normalized to endogenous AKT levels with AKT_990(F) and AKT_1146(R) primers (Table 3) using the SsoFast EvaGreen Supermix (Bio-Rad).
Table 4.
Oligos and primers used for targeting AKT
*, phosphorothioate linkage.
| Primer/oligo name | Sequence | Notes |
|---|---|---|
| myrAKT ssODN | G*A*GAGGAGCGCGTGAGCGTCGCGGGAGCCTCGGGCACCATGGGGAGCAGCAAGAGCAAGCCCAAGTCTAGAATGAACGACGTGGCTATTGTGAAGGAGGGTTGGCTGCACAAACGAGGTTAGTA*C*C | Donor oligo for knock-in |
| PAGE-purified | ||
| MTOR_S2215Y ssODN | C*T*TCGGCCTGGTTAACACCCTTCTGGCCAATGACCCAACATATCTTCGGAAAAACCTCAGGTATTCAGAAACCCTGAAATGGCA*A*C | Negative control (non-specific) donor oligo |
| PAGE-purified | ||
| AKT_990 (F) | CTTCCTTTGCTTCTCCCAGAG | QPCR (SYBR or Taqman) with AKT_1146(R) |
| AKT_1146 (R) | CTCGTTTGTGCAGCCAAC | QPCR (SYBR or Taqman) with AKT_990(R) |
| AKT_probe | GAGCAGCAAGAGCAAGCCCAAG | Taqman probe for QPCR with AKT_990(F) and AKT_1146(R) |
For targeting AKT in DLD1, cells were first transfected with pCMV-hyPBase and BII-B3HTK-AKT as described above for the first part of the two-step transfection method. After 5 days of selection with 5 μg/ml blasticidin, 7.5 × 105 cells were seeded and transfected the next day with 30 pmol of myrAKT ssODN, 500 ng of hCas9N, 300 ng of pBS-BtH, and 200 ng of pBS-U6gRNAx2 using 4 μl of JetPrime (Polyplus) per the manufacturer's instructions. 48 h following transfection cells were selected with hygromycin (600 μg/ml) for 12 days and expanded for 3 additional days, and then gDNA was isolated for QPCR as described above for HeLa cells.
The human hepatocellular carcinoma cell line, HepG2, was transfected with a total of 1 μg of plasmid mix containing 750 ng of BII-CPspaH-A1CF target vector and 250 ng of pCMV-hyPBase using 3 μl of X-tremeGENE 9 DNA transfection (Roche Applied Science). Two days after transfection cells were placed under puromycin selection (5 μg/ml) for 1 week. Surviving cells were transfected with 1 μg of plasmid mix containing 500 ng of hCas9N, 200 ng of pBS-A1CF gRNA, and 300 ng of pBS-PtH. After 48 h, cells were expanded in a 100-mm dish. After five rounds of transfection of CRISPR/Cas9 components, cells were placed under hygromycin selection (600 μg/ml). Individual resistant colonies were picked and expanded for protein expression analysis. Of the 20 clones analyzed 12 showed a >97% decreased A1CF expression.
Author contributions
B. B. M. conceived and designed all vectors and all experiments with assistance from G. L. and T. P. B. B. M. and Y. W. wrote the paper. N. O. D. and S. M. P.-M. provided advice and comments. B. B. M. and G. L. performed experiments in Fig. 1. Y. W. performed SRIRACCHA and enteroid manipulations in Fig. 2. S. M. P.-M. and J. P. C. performed the nucleofections of N2A cells and deep sequencing in Fig 2. Y. W. performed the experiments in Fig. 3, T7E1 assays in Fig. 4, and PBex experiments in Fig. 5. B. B. M. carried out SRIRACCHA in DLD1 and HeLa cells and quantified the colonies in Fig. 4. V. B. manipulated and characterized DLD1 and HepG2 mutations for A1CF and RBM47. B. B. M. characterized NFE2L2-targeted hygromycin-resistant clones. TZ/B. B. M. performed experiments on NIH3T3 and IEC6 in Fig. 4.
This work was supported by National Institutes of Health Grants K01 DK093885, R03 DK108764, and P30 DK052574 Pilot (to B. B. M.) and HL-38180, DK-56260, and DK-52574 (to N. O. D.). This work was also supported by a Cancer Research Foundation Young Investigator Award and the Siteman Investment Program at the Siteman Cancer Center (to B. B. M.) and by the Digestive Disease Research Core Center (ARAC and Murine Cores) (to N. O. D.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- RGEN
- RNA-guided endonuclease
- gRNA
- guide RNA
- NHEJ
- non-homology end joining
- HDR
- homology directed repair
- SRIRACCHA
- stable but reversible integrated reporter for assaying CRISPR/Cas9-stimulated HDR activity
- iSRIRACCHA
- inducible SRIRACCHA
- PBase
- Piggybac transposase
- hyPBase
- hyperactive PBase
- PBex
- excision-only PBase
- DSB
- double strand break
- indels
- insertions or deletions
- PAM
- protospacer adjacent motif
- PB
- transposase
- T7E1
- T7 endonuclease 1
- HSIV
- hypersensitive site IV
- RFP
- red fluorescent protein
- 4-OH-T
- 4-hydroxytamoxifen
- dox
- doxycycline
- NS
- nonspecific
- ssODN
- single-stranded oligodeoxynucleotide
- QPCR
- quantitative PCR
- TKpA
- thymidine kinase polyadenylation
- IRES
- internal ribosome entry site
- LSL sequence
- lox-stop-lox sequence.
References
- 1. Marraffini L. A., and Sontheimer E. J. (2008) CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322, 1843–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fu Y., Foden J. A., Khayter C., Maeder M. L., Reyon D., Joung J. K., and Sander J. D. (2013) High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31, 822–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ran F. A., Hsu P. D., Lin C. Y., Gootenberg J. S., Konermann S., Trevino A. E., Scott D. A., Inoue A., Matoba S., Zhang Y., and Zhang F. (2013) Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cradick T. J., Fine E. J., Antico C. J., and Bao G. (2013) CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 41, 9584–9592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cho S. W., Kim S., Kim Y., Kweon J., Kim H. S., Bae S., and Kim J. S. (2014) Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res 24, 132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang H., Yang H., Shivalila C. S., Dawlaty M. M., Cheng A. W., Zhang F., and Jaenisch R. (2013) One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A., and Zhang F. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mali P., Yang L., Esvelt K. M., Aach J., Guell M., DiCarlo J. E., Norville J. E., and Church G. M. (2013) RNA-guided human genome engineering via Cas9. Science 339, 823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jinek M., East A., Cheng A., Lin S., Ma E., and Doudna J. (2013) RNA-programmed genome editing in human cells. eLife 2, e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Findlay G. M., Boyle E. A., Hause R. J., Klein J. C., and Shendure J. (2014) Saturation editing of genomic regions by multiplex homology-directed repair. Nature 513, 120–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mans R., van Rossum H. M., Wijsman M., Backx A., Kuijpers N. G., van den Broek M., Daran-Lapujade P., Pronk J. T., van Maris A. J., and Daran J. M. (2015) CRISPR/Cas9: a molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae. FEMS Yeast Res. 15, fov004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Doench J. G., Fusi N., Sullender M., Hegde M., Vaimberg E. W., Donovan K. F., Smith I., Tothova Z., Wilen C., Orchard R., Virgin H. W., Listgarten J., and Root D. E. (2016) Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ma H., Dang Y., Wu Y., Jia G., Anaya E., Zhang J., Abraham S., Choi J. G., Shi G., Qi L., Manjunath N., and Wu H. (2015) A CRISPR-based screen identifies genes essential for west-nile-virus-induced cell death. Cell Rep. 12, 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hart T., Chandrashekhar M., Aregger M., Steinhart Z., Brown K. R., MacLeod G., Mis M., Zimmermann M., Fradet-Turcotte A., Sun S., Mero P., Dirks P., Sidhu S., Roth F. P., Rissland O. S., Durocher D., Angers S., and Moffat J. (2015) High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163, 1515–1526 [DOI] [PubMed] [Google Scholar]
- 15. Gilbert L. A., Horlbeck M. A., Adamson B., Villalta J. E., Chen Y., Whitehead E. H., Guimaraes C., Panning B., Ploegh H. L., Bassik M. C., Qi L. S., Kampmann M., and Weissman J. S. (2014) Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159, 647–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Konermann S., Brigham M. D., Trevino A. E., Joung J., Abudayyeh O. O., Barcena C., Hsu P. D., Habib N., Gootenberg J. S., Nishimasu H., Nureki O., and Zhang F. (2015) Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang T., Birsoy K., Hughes N. W., Krupczak K. M., Post Y., Wei J. J., Lander E. S., and Sabatini D. M. (2015) Identification and characterization of essential genes in the human genome. Science 350, 1096–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang T., Wei J. J., Sabatini D. M., and Lander E. S. (2014) Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koike-Yusa H., Li Y., Tan E. P., Velasco-Herrera Mdel C., and Yusa K. (2014) Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat. Biotechnol. 32, 267–273 [DOI] [PubMed] [Google Scholar]
- 20. Shalem O., Sanjana N. E., Hartenian E., Shi X., Scott D. A., Mikkelsen T. S., Heckl D., Ebert B. L., Root D. E., Doench J. G., and Zhang F. (2014) Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ebina H., Misawa N., Kanemura Y., and Koyanagi Y. (2013) Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci. Rep. 3, 2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang W., Ye C., Liu J., Zhang D., Kimata J. T., and Zhou P. (2014) CCR5 gene disruption via lentiviral vectors expressing Cas9 and single guided RNA renders cells resistant to HIV-1 infection. PLoS ONE 9, e115987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu Y., Liang D., Wang Y., Bai M., Tang W., Bao S., Yan Z., Li D., and Li J. (2013) Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell 13, 659–662 [DOI] [PubMed] [Google Scholar]
- 24. Schwank G., Koo B. K., Sasselli V., Dekkers J. F., Heo I., Demircan T., Sasaki N., Boymans S., Cuppen E., van der Ent C. K., Nieuwenhuis E. E., Beekman J. M., and Clevers H. (2013) Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 13, 653–658 [DOI] [PubMed] [Google Scholar]
- 25. Yusa K., Zhou L., Li M. A., Bradley A., and Craig N. L. (2011) A hyperactive piggyBac transposase for mammalian applications. Proc. Natl. Acad. Sci. U.S.A. 108, 1531–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cadiñanos J., and Bradley A. (2007) Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res. 35, e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liang Q., Kong J., Stalker J., and Bradley A. (2009) Chromosomal mobilization and reintegration of Sleeping Beauty and PiggyBac transposons. Genesis 47, 404–408 [DOI] [PubMed] [Google Scholar]
- 28. Ding S., Wu X., Li G., Han M., Zhuang Y., and Xu T. (2005) Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122, 473–483 [DOI] [PubMed] [Google Scholar]
- 29. Li M. A., Turner D. J., Ning Z., Yusa K., Liang Q., Eckert S., Rad L., Fitzgerald T. W., Craig N. L., and Bradley A. (2011) Mobilization of giant piggyBac transposons in the mouse genome. Nucleic Acids Res. 39, e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li X., Burnight E. R., Cooney A. L., Malani N., Brady T., Sander J. D., Staber J., Wheelan S. J., Joung J. K., McCray P. B. Jr., Bushman F. D., Sinn P. L., and Craig N. L. (2013) piggyBac transposase tools for genome engineering. Proc. Natl. Acad. Sci. U.S.A. 110, E2279–E2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang W., Lin C., Lu D., Ning Z., Cox T., Melvin D., Wang X., Bradley A., and Liu P. (2008) Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 105, 9290–9295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mitra R., Fain-Thornton J., and Craig N. L. (2008) piggyBac can bypass DNA synthesis during cut and paste transposition. EMBO J. 27, 1097–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fraser M. J., Ciszczon T., Elick T., and Bauser C. (1996) Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect Mol. Biol. 5, 141–151 [DOI] [PubMed] [Google Scholar]
- 34. Elick T. A., Bauser C. A., and Fraser M. J. (1996) Excision of the piggyBac transposable element in vitro is a precise event that is enhanced by the expression of its encoded transposase. Genetica 98, 33–41 [DOI] [PubMed] [Google Scholar]
- 35. Mashal R. D., Koontz J., and Sklar J. (1995) Detection of mutations by cleavage of DNA heteroduplexes with bacteriophage resolvases. Nat. Genet. 9, 177–183 [DOI] [PubMed] [Google Scholar]
- 36. Qiu P., Shandilya H., D'Alessio J. M., O'Connor K., Durocher J., and Gerard G. F. (2004) Mutation detection using Surveyor nuclease. Biotechniques 36, 702–707 [DOI] [PubMed] [Google Scholar]
- 37. Zhou J., Wang J., Shen B., Chen L., Su Y., Yang J., Zhang W., Tian X., and Huang X. (2014) Dual sgRNAs facilitate CRISPR/Cas9-mediated mouse genome targeting. FEBS J. 281, 1717–1725 [DOI] [PubMed] [Google Scholar]
- 38. Dow L. E., Fisher J., O'Rourke K. P., Muley A., Kastenhuber E. R., Livshits G., Tschaharganeh D. F., Socci N. D., and Lowe S. W. (2015) Inducible in vivo genome editing with CRISPR-Cas9. Nat. Biotechnol. 33, 390–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schwank G., Andersson-Rolf A., Koo B. K., Sasaki N., and Clevers H. (2013) Generation of BAC transgenic epithelial organoids. PLoS ONE 8, e76871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Matano M., Date S., Shimokawa M., Takano A., Fujii M., Ohta Y., Watanabe T., Kanai T., and Sato T. (2015) Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med. 21, 256–262 [DOI] [PubMed] [Google Scholar]
- 41. Kim H., Um E., Cho S. R., Jung C., Kim H., and Kim J. S. (2011) Surrogate reporters for enrichment of cells with nuclease-induced mutations. Nat. Methods 8, 941–943 [DOI] [PubMed] [Google Scholar]
- 42. Ramakrishna S., Cho S. W., Kim S., Song M., Gopalappa R., Kim J. S., and Kim H. (2014) Surrogate reporter-based enrichment of cells containing RNA-guided Cas9 nuclease-induced mutations. Nat. Commun. 5, 3378. [DOI] [PubMed] [Google Scholar]
- 43. Ren C., Xu K., Liu Z., Shen J., Han F., Chen Z., and Zhang Z. (2015) Dual-reporter surrogate systems for efficient enrichment of genetically modified cells. Cell. Mol. Life Sci. 72, 2763–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim Y. H., Ramakrishna S., Kim H., and Kim J. S. (2014) Enrichment of cells with TALEN-induced mutations using surrogate reporters. Methods 69, 108–117 [DOI] [PubMed] [Google Scholar]
- 45. Deleted in proof
- 46. Kim H., Kim M. S., Wee G., Lee C. I., Kim H., and Kim J. S. (2013) Magnetic separation and antibiotics selection enable enrichment of cells with ZFN/TALEN-induced mutations. PLoS ONE 8, e56476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. He Z., Shi X., Liu M., Sun G., Proudfoot C., Whitelaw C. B., Lillico S. G., and Chen Y. (2016) Comparison of surrogate reporter systems for enrichment of cells with mutations induced by genome editors. J. Biotechnol. 221, 49–54 [DOI] [PubMed] [Google Scholar]
- 48. Prado F., Cortés-Ledesma F., Huertas P., and Aguilera A. (2003) Mitotic recombination in Saccharomyces cerevisiae. Curr. Genet. 42, 185–198 [DOI] [PubMed] [Google Scholar]
- 49. Rulten S. L., and Grundy G. J. (2017) Non-homologous end joining: Common interaction sites and exchange of multiple factors in the DNA repair process. Bioessays 10.1002/bies.201600209 [DOI] [PubMed] [Google Scholar]
- 50. Malina A., Mills J. R., Cencic R., Yan Y., Fraser J., Schippers L. M., Paquet M., Dostie J., and Pelletier J. (2013) Repurposing CRISPR/Cas9 for in situ functional assays. Genes Dev. 27, 2602–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sanjana N. E., Shalem O., and Zhang F. (2014) Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shi J., Wang E., Milazzo J. P., Wang Z., Kinney J. B., and Vakoc C. R. (2015) Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat. Biotechnol. 33, 661–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen S., Sanjana N. E., Zheng K., Shalem O., Lee K., Shi X., Scott D. A., Song J., Pan J. Q., Weissleder R., Lee H., Zhang F., and Sharp P. A. (2015) Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell 160, 1246–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schröder A. R., Shinn P., Chen H., Berry C., Ecker J. R., and Bushman F. (2002) HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110, 521–529 [DOI] [PubMed] [Google Scholar]
- 55. Wu X., Li Y., Crise B., and Burgess S. M. (2003) Transcription start regions in the human genome are favored targets for MLV integration. Science 300, 1749–1751 [DOI] [PubMed] [Google Scholar]
- 56. Mitchell R. S., Beitzel B. F., Schroder A. R., Shinn P., Chen H., Berry C. C., Ecker J. R., and Bushman F. D. (2004) Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLos Biol. 2, E234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lewinski M. K., Yamashita M., Emerman M., Ciuffi A., Marshall H., Crawford G., Collins F., Shinn P., Leipzig J., Hannenhalli S., Berry C. C., Ecker J. R., and Bushman F. D. (2006) Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog. 2, e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Derse D., Crise B., Li Y., Princler G., Lum N., Stewart C., McGrath C. F., Hughes S. H., Munroe D. J., and Wu X. (2007) Human T-cell leukemia virus type 1 integration target sites in the human genome: comparison with those of other retroviruses. J. Virol. 81, 6731–6741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Slaymaker I. M., Gao L., Zetsche B., Scott D. A., Yan W. X., and Zhang F. (2016) Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kleinstiver B. P., Pattanayak V., Prew M. S., Tsai S. Q., Nguyen N. T., Zheng Z., and Joung J. K. (2016) High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Madison B. B., Liu Q., Zhong X., Hahn C. M., Lin N., Emmett M. J., Stanger B. Z., Lee J. S., and Rustgi A. K. (2013) LIN28B promotes growth and tumorigenesis of the intestinal epithelium via Let-7. Genes Dev. 27, 2233–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Matsuda T., and Cepko C. L. (2007) Controlled expression of transgenes introduced by in vivo electroporation. Proc. Natl. Acad. Sci. U.S.A. 104, 1027–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]