Abstract
Protein O-glycosylation is an important post-translational modification in all organisms, but deciphering the specific functions of these glycans is difficult due to their structural complexity. Understanding the glycosylation of mucin-like proteins presents a particular challenge as they are modified numerous times with both the enzymes involved and the glycosylation patterns being poorly understood. Here we systematically explored the O-glycosylation pathway of a mucin-like serine-rich repeat protein PsrP from the human pathogen Streptococcus pneumoniae TIGR4. Previous works have assigned the function of 3 of the 10 glycosyltransferases thought to modify PsrP, GtfA/B, and Gtf3 as catalyzing the first two reactions to form a unified disaccharide core structure. We now use in vivo and in vitro glycosylation assays combined with hydrolytic activity assays to identify the glycosyltransferases capable of decorating this core structure in the third and fourth steps of glycosylation. Specifically, the full-length GlyE and GlyG proteins and the GlyD DUF1792 domain participate in both steps, whereas full-length GlyA and the GlyD GT8 domain catalyze only the fourth step. Incorporation of different sugars to the disaccharide core structure at multiple sites along the serine-rich repeats results in a highly polymorphic product. Furthermore, crystal structures of apo- and UDP-complexed GlyE combined with structural analyses reveal a novel Rossmann-fold “add-on” domain that we speculate to function as a universal module shared by GlyD, GlyE, and GlyA to forward the peptide acceptor from one enzyme to another. These findings define the complete glycosylation pathway of a bacterial glycoprotein and offer a testable hypothesis of how glycosyltransferase coordination facilitates glycan assembly.
Keywords: bacterial pathogenesis, crystal structure, enzyme, glycosylation, glycosyltransferase, Streptococcus pneumoniae, polymorphism, serine-rich repeat protein
Introduction
Protein glycosylation, catalyzed by glycosyltransferases, is an important protein posttranslational modification widespread in both prokaryotes (1) and eukaryotes (2). More than two-thirds of eukaryotic proteins are subjected to glycosylation (3) for executing diverse cellular functions (4–6). Most glycosylated proteins are exposed to the cell surface, thus usually participating in cell-cell recognition, signaling transduction, and immune modulation (7). Aberrant protein glycosylations are correlated with many serious human diseases (5), including cancer, neurological disorder, tissue dysfunction, and bone disease. For instance, the most abundant human glycoprotein mucin, which modulates cell-cell recognition and adhesion as lubricants and chemical barriers (8, 9), is an important tumor-associated antigen (10). Nascent mucin are initially modified with O-linked N-acetylgalactosamine (GalNAc) at numerous Ser and Thr residues (11) and then sequentially glycosylated with more saccharide residues in a stepwise manner (12), resulting in varying types of core structure of two-three residues in different tissues (13). Moreover, in some specialized tissues or developmental stages, these core structures are further elongated and modified by N-acetylglucosamine (GlcNAc), galactose (Gal) and fucose and usually terminated with sialylation or sometimes sulfation, leading to an extended linear or branched glycan structure (13). However, the fine structure of mucin glycan and the glycosylation pathway remain poorly understood.
It has been recognized that O-glycosylation is also a common modification in prokaryotes (1, 14), which are involved in pathogenesis and/or immune modulation/escape (15). For example, O-glycosylated flagellar proteins are important adhesins in Gram-negative bacteria (16). Glycosylation of flagellin contributes to the recognition of Burkholderia cenocepacia toward human receptors, leading to a reduced inflammatory response in vitro (17). More interestingly, the Gram-positive bacteria streptococci, staphylococci, and lactobacilli encode a family of mucin-like proteins, the serine-rich repeat proteins, termed SRRPs.4 Previous reports indicated that SRRPs participate in bacterial adhesion, immune evasion, colonization, and biofilm formation (18–23) and thus contribute to bacterial infections that cause infective endocarditis, pneumococcal pneumonia, neonatal sepsis, and meningitis (23). SRRPs usually harbor two serine-rich repeat regions (SRR1 and SRR2), which are subjected to heavy O-glycosylation (23, 24), a key modification that contributes to the biogenesis and pathogenesis (24–28). For example, disruption of gtfA or gtfB results in the formation of intracellular aggregates of Streptococcus gordonii SRRP protein GspB, which in turn blocks the transportation of GspB to the bacterial surface (29, 30). Therefore, SRRPs and biogenesis pathways are potential targets for developing novel vaccines or antibacterial agents (23).
Streptococcus pneumoniae TIGR4 encodes an SRRP termed PsrP that promotes biofilm formation through interaction with extracellular DNA in the biofilm matrix and adhering to keratin 10 expressing lung epithelial cells (20, 26, 31, 32). The glycosylation and secretion of PsrP are controlled by a downstream gene cluster, which encodes 10 putative glycosyltransferases and 2 general secretory pathway proteins in addition to 5 accessory secretion components (33). Gene synteny analyses suggest that the psrP locus and counterpart loci share a conserved core region of seven genes: secY2, asp1-3, secA2, gftA, and gtfB (Fig. 1A). Beyond this core region, the gene cluster harbors diverse insertions in different species that encode extra putative glycosyltransferases. The conserved core region may provide bacteria as a common mechanism for the biosynthesis of SRRPs, whereas the diversity of extra glycosyltransferases, responsible for the heavy O-glycosylation, might enable bacteria to adapt to changing ecological niches mediated by SRRPs (24). Previous structural and biochemical studies have demonstrated that the first two steps of streptococcal SRRP glycosylation are sequentially catalyzed by an O-GlcNAc transferase complex GtfA/B (34–37) and Gtf3 (38, 39). Recent reports on Streptococcus parasanguinis SRRP, termed Fap1, revealed that the third and fourth steps of glycosylation are respectively catalyzed by the DUF1792 domain and the GT2 domain of a dual-functional glycosyltransferase dGT1 (40, 41). Remarkably, S. pneumoniae psrP gene locus encodes the most diverse glycosyltransferases (Fig. 1A), strongly suggesting that PsrP is most likely subjected to a more diverse and complex modification. Thus, PsrP might be an ideal model to comprehensively illustrate this heavy O-glycosylation pathway. However, the subsequent steps of PsrP glycosylation remain unclear except for the first and second steps.
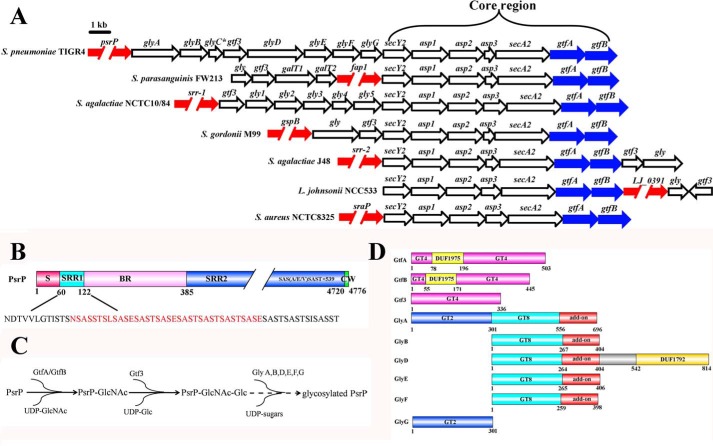
Figure 1.
The SRRP loci. A, SRRP genes are colored in red, whereas gtfA and gtfB are colored in blue. Arrows indicate the direction of transcription. B, domain organization of PsrP. The BR represents the ligand-binding region of PsrP. The sequence of SRR1 is shown, and the residues for glycosylation assays in this study are colored in red. C, a scheme for the previously identified glycosylation pathway of PsrP. D, domain organizations of the glycosyltransferases encoded by the psrP gene cluster.
Here we performed systematic enzymatic activity assays on the nine glycosyltransferases within the psrP locus, except for the pseudogene glyC. After adding the first two sugar residues by GtfA/B and Gtf3, the third step of glycosylation is catalyzed by GlyD, GlyE, or GlyG using different sugar donors, whereas the fourth sugar residue could be added by GlyD, GlyE, GlyA, or GlyG. As a result, the glycosylation of PsrP exhibits a very high polymorphism. Furthermore, we revealed a novel add-on domain of a Rossmann fold shared by GlyD, GlyE, and GlyA that might function as a universal module to forward the peptide acceptor from one enzyme to another. Our findings not only provide the catalytic mechanism of SRRPs but also reveal the molecular basis for the polymorphism of O-glycosylation of surface adhering proteins.
Results
Organization of S. pneumoniae TIGR4 psrP locus
The open reading frame of psrP gene is of 14,331 bp in length which encodes a 4776-residue protein PsrP with a theoretical molecular mass of 412 kDa. PsrP consists of a signal peptide, a short serine-rich repeat region SRR1, and a ligand-binding region BR followed by a second extremely large serine-rich repeat region SRR2 and a C-terminal cell-wall anchor domain (Fig. 1B). The glycosylation and secretion pathway of PsrP contain nine putative glycosyltransferases (GtfA/B, Gtf3, GlyA-G) and two general secretory pathway proteins (SecY2 and SecA2) in addition to five accessory secretion components Asp1–5 (33). It has been reported that GtfA/B and Gtf3 catalyze the first and second steps of PsrP glycosylation (Fig. 1C), and all these three proteins share a GT-B fold and belong to the GT4 family (36, 37, 39). Bioinformatic analyses reveal a pairwise identity of 33–38% along the GT8 domain of putative glycosyltransferases GlyA, GlyB, GlyD, GlyE, and GlyF. In addition, GlyG and the N-terminal domain of GlyA share a GT2 family domain with a sequence identity of 36% (Fig. 1D).
Hydrolytic activity assays toward various sugar donors
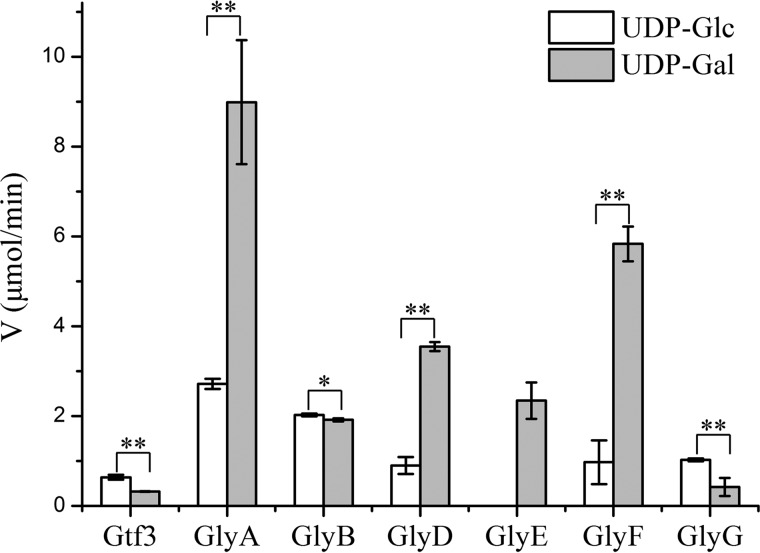
Previous reports showed that GtfA/B catalyzes the first step of PsrP glycosylation by transferring GlcNAc to multiple serine residues of PsrP (36), in which GtfA harbors the active site, whereas GtfB provides the primary binding site for the acceptor (37). To identify the sugar donors of the remaining glycosyltransferases, we performed a series of hydrolytic assays using the common sugar donors UDP-Glc, UDP-Gal, UDP-GlcNAc, ADP-Glc, GDP-Glc, and GDP-Man, respectively. The results showed that only two sugar donors, UDP-Glc and UDP-Gal, could be hydrolyzed by these glycosyltransferases. Similar to the previous report (39), Gtf3 shows a higher hydrolytic activity toward UDP-Glc compared with UDP-Gal. GlyG also has a significantly higher activity toward UDP-Glc, whereas GlyA, GlyD, GlyE, and GlyF are more active toward UDP-Gal (Fig. 2). Meanwhile, GlyB shows a comparable activity toward both UDP-Glc and UDP-Gal (Fig. 2). It demonstrated that all these seven enzymes indeed possess hydrolytic activity toward a given sugar donor. Moreover, it suggested that the GT8 domain might favor UDP-Gal, whereas the GT2 domain prefers UDP-Glc (Figs. 1D and 2).
Figure 2.
The hydrolytic activities toward UDP-Glc and UDP-Gal of putative glycosyltransferases encoded by the psrP gene cluster. The reaction lasted for 60 min at 37 °C in the presence of 10 μm enzyme and 1 mm UDP-Glc or UDP-Gal. The velocities were calculated by determining the production of UDP (μm) per minute. Data are presented as the means ± S.D. from three independent assays. Two-tailed Student's t test was used for the comparison of statistical significance. p values of <0.05 and <0.01 are indicated with *, and **, respectively.
The third step of PsrP glycosylation: GlyD or GlyE
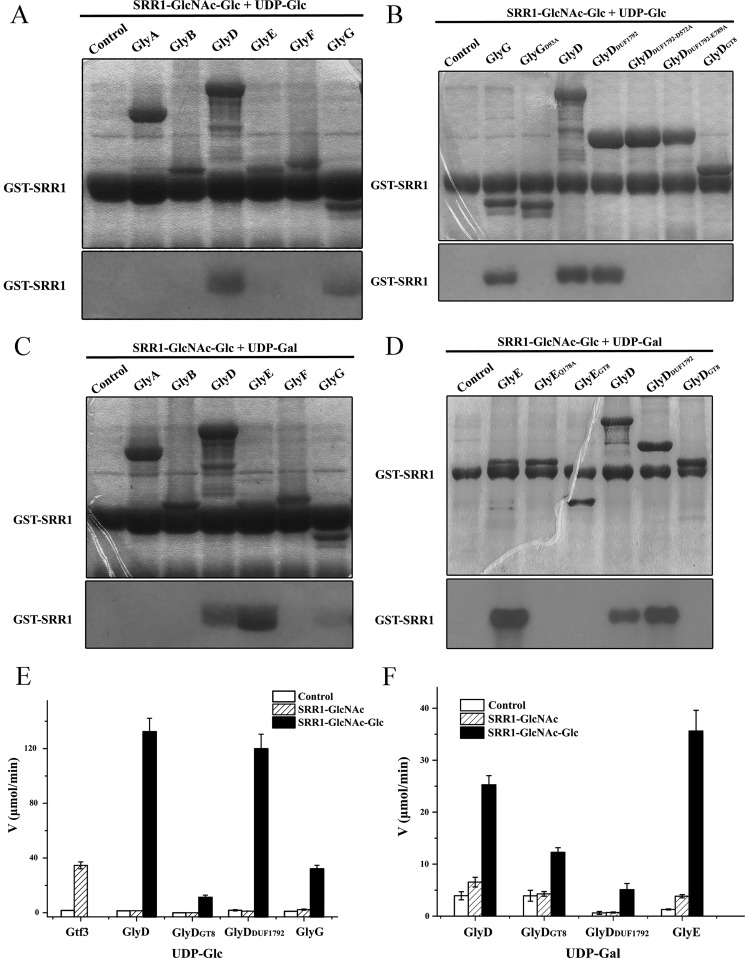
The DUF1792 domain of dGT1 from S. parasanguinis has been identified to catalyze the third step of Fap1 glycosylation (40). Sequence comparison indicated that the C terminus of GlyD in S. pneumoniae TIGR4 also has a DUF1792 domain that shares a sequence identity of 56% with the N-terminal DUF1792 domain of S. parasanguinis dGT1. Beyond the shared DUF1792 domain, GlyD possesses an N-terminal GT8 domain, whereas dGT1 has a C-terminal GT2 domain. To identify which glycosyltransferase catalyzes the third step of PsrP glycosylation, we applied in vitro assays to detect the glycosylation activity using the 3H-labeled sugar donor UDP-Gal or UDP-Glc. The acceptor SRR1-GlcNAc-Glc was prepared by in vivo co-expression of GST-SRR1, GtfA/B, and Gtf3 in Escherichia coli. A glycosylated GST-SRR1 could be visualized as a single band using electrophoresis followed by autoradiography.
Using UDP-Glc as the sugar donor, the two enzymes GlyD and GlyG possess the glucosyltransferase activity, with a GlyD of 2-fold activity to that of GlyG, suggesting GlyD plays a primary role in the third step of SRR1 glycosylation (Fig. 3A). As GlyD possesses an N-terminal GT8 domain (residues 1–404, termed GlyDGT8) and a C-terminal DUF1792 domain (residues 542–814, termed GlyDDUF1792) (Fig. 1D), we further purified the two distinct domains applied to activity assays. Similar to S. parasanguinis dGT1 (40), GlyDDUF1792, but not GlyDGT8, is responsible for the third-step glycosylation (Fig. 3B). It has been reported that Asp-31 in the metal-binding motif of dGT1 and the catalytic residue Glu-248 are critical for the glycosyltransferase activity (40). As predicted, mutation of the counterpart residues Asp-572 and Glu-789 of GlyDDUF1792 completely abolished the glycosyltransferase activity (Fig. 3B). In addition, S. pneumoniae GlyG shares a sequence homology of 33% to the C-terminal GT2 domain of dGT1, which participates in the fourth-step glycosylation of Fap1. Moreover, mutation of residue Asp-93 of GlyG that is a counterpart to a conserved metal-binding residue in dGT1 resulted in the loss of glycosyltransferase activity (Fig. 3B).
Figure 3.
The third-step glycosylation: glycosylation of SRR1-GlcNAc-Glc. The glycosyltransferase activity assays of putative enzymes and mutants using UDP-[3H]glucose or UDP-[3H]galactose as the sugar donor are shown in A–D, respectively. Separation of the reaction mixture was performed by SDS-PAGE (upper column), which was further detected by 3H autoradiography (lower column). GST-SRR1 was labeled, whereas the other bonds in the SDS-PAGE correspond to different glycosyltransferases. Augmentation of hydrolytic activities in the presence of SRR1-GlcNAc or SRR1-GlcNAc-Glc toward UDP-Glc (E) or UDP-Gal (F).
Alternatively, when taking UDP-Gal as the sugar donor, we found that the two enzymes, GlyE and GlyD, have galactosyltransferase activity, with GlyE of 2-fold activity to that of GlyD (Fig. 3C). Further analysis suggested that GlyDDUF1792, but not GlyDGT8, is responsible for the galactosyltransferase activity of GlyD (Fig. 3D). GlyDDUF1972 is capable of utilizing both UDP-Glc and UDP-Gal as the sugar donors, maybe due to its unique GT-D fold that has a novel Rossmann-like nucleotide-binding fold (40). Analysis of the active-site pocket reveals a plasticity of the UDP-sugar binding loops, which might accommodate different sugar donors. To further identify which sugar donor is preferred by GlyDDUF1972, we compared its hydrolytic activity toward the two sugar donors in the presence of the acceptor SRR1-GlcNAc-Glc and revealed a much higher augmentation of activity toward UDP-Glc (Fig. 3, E and F). It is also in agreement with the results that GlyDDUF1972 plays a primary role in the third-step glycosylation using UDP-Glc as donor and a secondary role when using UDP-Gal as donor.
In fact, in the presence of the acceptor SRR1-GlcNAc, the hydrolytic activity of Gtf3 toward UDP-Glc is increased by 21-fold (Fig. 3E) in agreement with the previous proposal that the activity of a glycosyltransferase could be dramatically increased by >100-fold in the presence of an optimal acceptor (42). As expected, upon the addition of SRR1-GlcNAc-Glc, the hydrolytic activity toward UDP-Glc of either the full-length GlyD or GlyDDUF1792 was increased to ∼100-fold (Fig. 3E). Moreover, in the presence of SRR1-GlcNAc-Glc, GlyG showed a 30-fold higher activity toward UDP-Glc. Similarly, the addition of SRR1-GlcNAc-Glc resulted in ∼7- and 28-fold increase of hydrolytic activity toward UDP-Gal for GlyD and GlyE, respectively (Fig. 3F). These results further proved that Gtf3 is the only enzyme responsible for adding second sugar, whereas GlyDDUF1792, GlyE, and GlyG are the enzymes that catalyze the third step of PsrP glycosylation.
The fourth step: diverse and heterogeneous
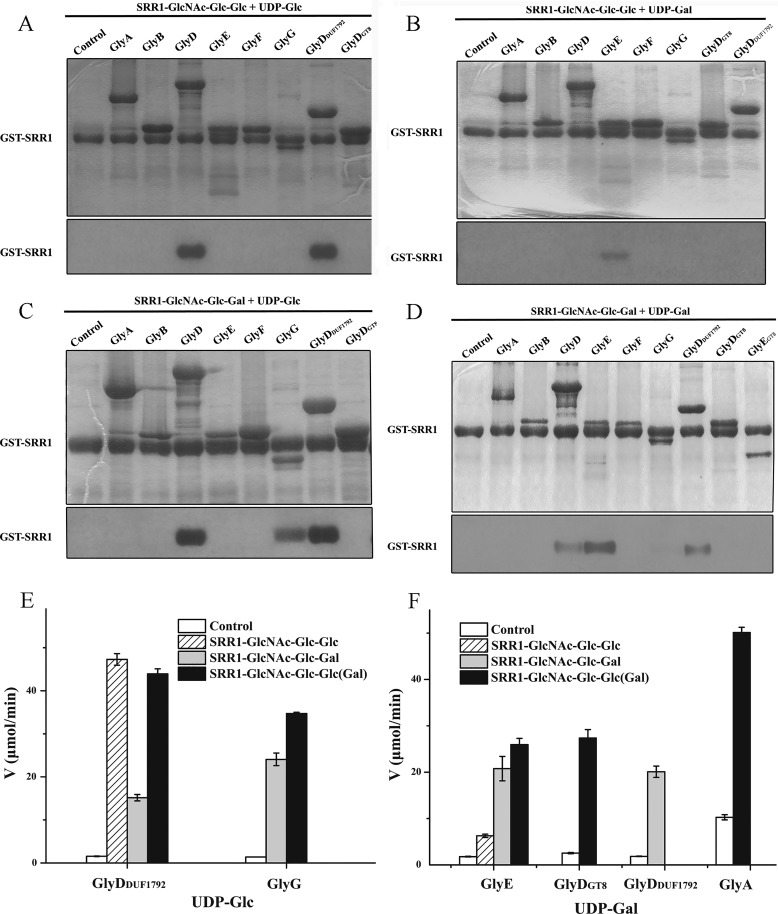
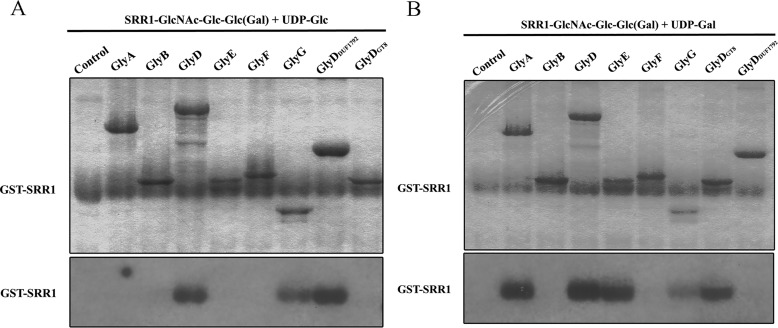
To further explore the subsequent glycosylation of PsrP, we purified trisaccharide modified acceptors SRR1-GlcNAc-Glc-Glc by co-expression of GST-SRR1, GtfA/B, Gtf3, and GlyG, and SRR1-GlcNAc-Glc-Gal by co-expression of GST-SRR1, GtfA/B, Gtf3, and GlyE in E. coli, respectively. The two glycosylated acceptors were subjected to in vitro glycosylation assays. With UDP-Glc as the donor, SRR1-GlcNAc-Glc-Glc could be further modified by GlyDDUF1792 as well as the full-length GlyD to produce SRR1-GlcNAc-Glc-Glc-Glc (Fig. 4A). Alternatively, taking UDP-Gal as the donor, SRR1-GlcNAc-Glc-Glc could be further glycosylated by GlyE to produce SRR1-GlcNAc-Glc-Glc-Gal (Fig. 4B). Glycosylation activity assays of another trisaccharide-modified acceptor, SRR1-GlcNAc-Glc-Gal, indicated that GlyD, more precisely GlyDDUF1792, is the primary enzyme to produce SRR1-GlcNAc-Glc-Gal-Glc (Fig. 4C), whereas both GlyDDUF1792 and GlyE contribute to the production of SRR1-GlcNAc-Glc-Gal-Gal (Fig. 4D). Besides, GlyG also exhibits a relatively lower fourth-step glycosylation activity using UDP-Glc as the donor (Fig. 4C). Hydrolytic activity assays in the presence of corresponding acceptor also revealed a significantly activity increase for GlyE, GlyDDUF1792, and GlyG (Fig. 4, E and F), further proving that these three enzymes catalyze the fourth-step glycosylation.
Figure 4.
The fourth-step glycosylation of PsrP. The glycosyltransferase activities were performed using the substrates SRR1-GlcNAc-Glc-Glc and UDP-[3H]glucose (A) or UDP-[3H]galactose (B) or SRR1-GlcNAc-Glc-Gal, and UDP-[3H]glucose (C) or UDP-[3H]galactose (D). Augmentation of hydrolytic activities in the presence of trisaccharide-modified SRR1 toward UDP-Glc (E) or UDP-Gal (F).
As GlyDDUF1792 is capable of adding the third sugar using either UDP-Glc or UDP-Gal as the donor, co-expression of GST-SRR1, GtfA/B, Gtf3, and GlyDDUF1792 in E. coli was supposed to produce a chimeric trisaccharide-modified SRR1 ending with a Glc or Gal residue. Assays using this chimeric acceptor revealed that two more enzymes, GlyG and GlyA, in addition to GlyDGT8 are also capable of catalyzing the fourth-step glycosylation (Fig. 5, A and B) beyond the two primary enzymes GlyDDUF1792 and GlyE. In fact, upon the addition of the hypothetical chimeric acceptor, the hydrolytic activity of GlyG toward UDP-Glc as well as GlyA and GlyDGT8 toward UDP-Gal is significantly augmented (Fig. 4, E and F).
Figure 5.
The fourth-step glycosylation of PsrP using the mixed acceptors of trisaccharide-modified SRR1 (SRR1-GlcNAc-Glc-Glc and SRR1-GlcNAc-Glc-Gal) in the presence of UDP-[3H]glucose (A) or UDP-[3H]galactose (B).
Overall structure and substrate-binding site of GlyE
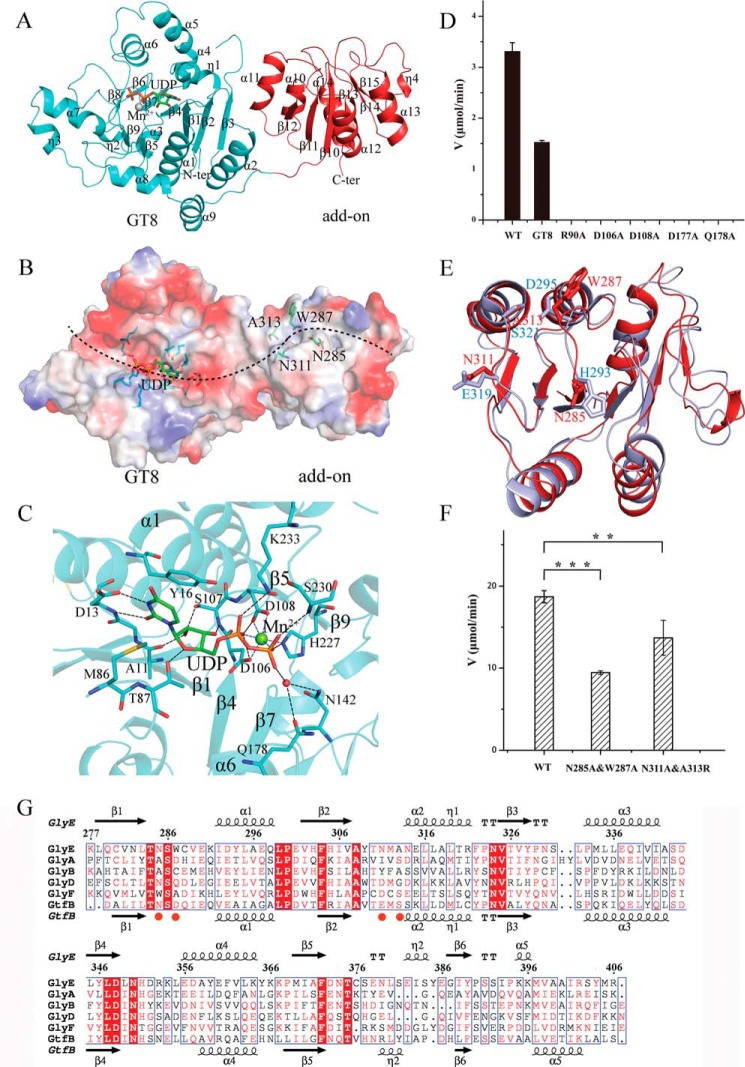
As GlyE possesses a typical GT8 domain that is shared by most enzymes participating in the third- and fourth-step glycosylations of PsrP (Fig. 1D), we solved the apo-form and UDP-complexed structures of GlyE to better understand the structural insights. In the complex structure, a manganese ion and a UDP molecule at the active site could be well defined (Fig. 6A). Atomic absorption spectrum also confirmed the presence of manganese in GlyE at a molar ratio of ∼1:1.
Figure 6.
Overall structure and active-site pocket of GlyE. A, schematic representation of GlyE with the secondary structural elements labeled sequentially. The UDP molecule is shown as sticks and Mn2+ is presented as a sphere. The GT8 domain is colored in cyan, whereas the add-on domain is colored in red. B, the substrate-binding pocket. The UDP-binding residues are shown as sticks, and the putative acceptor-binding groove is indicated as a dotted black line. C, the binding site of UDP. The UDP molecule and UDP-binding residues are shown as sticks, whereas the Mn2+ is shown as a sphere. The polar interactions are indicated as dashed lines. D, the hydrolytic activities of the wild-type GlyE and mutants from the UDP-binding pocket. E, structural comparison of the add-on domain of GlyE (red) against the C-terminal Rossmann-fold domain of GtfB (light blue). The putative acceptor-binding residues of GlyE and GtfB are shown as sticks. F, the glycosyltransferase activities of the wild-type GlyE and mutants of acceptor-binding residues in the presence of SRR1-GlcNAc-Glc. The p values of < 0.01 and 0.001 are indicated with ** and ***, respectively. G, structure-based sequence alignment of the shared add-on domains within the GT8 glycosyltransferases and GtfB. The secondary structural elements of GlyE and Gtf3 are labeled on the top and at the bottom, respectively. The putative acceptor-binding residues are marked with red spheres.
The overall structure of GlyE is composed of two distinct domains connected by a linker (residues Ser-266–Lys-277). The N-terminal domain (residues Asn-3–Lys-265, termed GT8) adopts a canonical glycosyltransferase GT-A fold that contains two abutting Rossmann-like folds (Fig. 6A). Beyond the GT8 domain, GlyE has a C-terminal domain of a Rossmann-like fold (termed “add-on” domain) that consists of a central six-stranded parallel β-sheet sandwiched by two helices on one side and three helices on the other. Structural comparison of the apo- and UDP-bound GlyE structures yields a root mean square deviation (r.m.s.d.) of 0.57 Å >390 Cα atoms, indicating very slight conformational changes of the overall structure upon binding to UDP. The most obvious differences come from the variations of the loop between β3 and η1 and helices α4-α6. In the apo-form GlyE, the active-site pocket is open and surface-exposed. Binding of UDP makes the active-site pocket undergo an induced fit, resulting in a compact active-site pocket to perfectly accommodate UDP. Structural homology search using DALI server (43) revealed a top structural homolog, Neisseria meningitidis galactosyltransferase LgtC (N. meningitidis galactosyltransferase) (44). In the structure of GlyE-UDP, the UDP molecule binds at the cleft formed by the central β-sheet and is almost surface-exposed to solvent (Fig. 6B). In details, the uracil base of UDP is stabilized by Asp-13, Tyr-16, and Met-86, whereas the ribose binds to Ala-11 and Ser-107. In addition, the two phosphate groups form hydrogen bonds with Asp-106, Asn-142, Gln-178, His-227, Ser-230, and Lys-233 (Fig. 6C).
In the GlyE-UDP complex structure, a single well ordered Mn2+ is coordinated in an octahedral fashion by the two phosphate oxygens of UDP as well as by His-227, Asp-106, and Asp-108 (Fig. 6C) in which Asp-106 and Asp-108 comprise the typical DXD sequence motif required for the coordination of a divalent cation in the binding of the nucleotide sugar (45). Indeed, mutation of either Asp-106 or Asp-108 completely abolished the hydrolytic activity against UDP-Gal (Fig. 6D), consistent with their important role in catalysis (46–48). In addition, structural superposition against LgtC (N. meningitidis galactosyltransferase) in complex with the sugar donor enabled us to assign the key residues Arg-90, Asn-142, Asp-177, and Gln-178 binding to the sugar moiety. As predicted, mutation of the key residues, for instance Gln-178, Arg-90, and Asp-177, also completely abolished the hydrolytic activity (Fig. 6D). The individual GT8 domain of GlyE remains ∼40% hydrolytic activity toward UDP-Gal compared with the full-length GlyE (Fig. 6D); however, deletion of the add-on domain of GlyE resulted in the complete loss of glycosyltransferase activity (Figs. 3D and 4D), indicating an essential role of the add-on domain in glycosyltransfer reaction.
Discussion
The add-on domain might be involved in forwarding the acceptor
Previous structural and biochemical studies demonstrated that the first step of SRRP glycosylation is catalyzed by an O-GlcNAc transferase complex GtfA and GtfB in a nonprocessive manner (34–37). GtfA harbors the catalytic pocket, whereas GtfB possesses the primary binding site of acceptor (37). Interestingly, the add-on domain of GlyE shares a Rossmann-fold similar to the C-terminal domain of GtfB that contains the putative binding residues of His-293, Asp-295, Glu-319, and Ser-321 (Fig. 6E). Electrostatic surface potential reveals a continuous groove on GlyE that extends from the UDP-binding site to the add-on domain (Fig. 6B). Notably, residues Asn-285, Trp-287, Asn-311, and Ala-313 in the add-on domain of GlyE that correspond to the putative acceptor-binding residues of GtfB are evenly distributed along this long groove. Either the mutant N285A/W287A or N311A/A313R has a significantly decreased glycosyltransferase activity in the presence of sugar acceptor SRR1-GlcNAc-Glc (Fig. 6F). Thus we speculated that the glycosylated peptide acceptor slides along this groove to make the serine residues subject to further glycosylation. As predicted, deletion of the add-on domain of GlyE resulted in the complete loss of glycosyltransferase activity (Figs. 3D and 4D). Moreover, this surface-exposed groove could accommodate the polypeptide acceptor at varying degrees of glycosylation.
Different from the five previous structure-known GT8 glycosyltransferases (37, 44, 49–51), GlyE represents the first structure that possesses a GT8 domain and an add-on domain, which is most likely involved in recruiting the substrate to the catalytic domain. Moreover, GlyA, GlyB, GlyD, and GlyF also contain a GT8 domain followed by a similar Rossmann-fold add-on domain (Fig. 1D). Structure-based sequence alignment revealed that these add-on domains are highly conserved (Fig. 6G). As GlyA, GlyD, and GlyE participate in different steps of PsrP glycosylation, these add-on domains might assist to forward the glycosylated acceptor en route from one enzyme to another using a similar binding pattern. Notably, despite possessing the hydrolytic activity toward both UDP-Glc and UDP-Gal (Fig. 2), GlyB and GlyF did not show any glycosyltransferase activity in our in vitro glycosylation assays, probably due to variations at the acceptor-binding site (Fig. 6G).
A putative pathway for the heavy O-glycosylation of PsrP
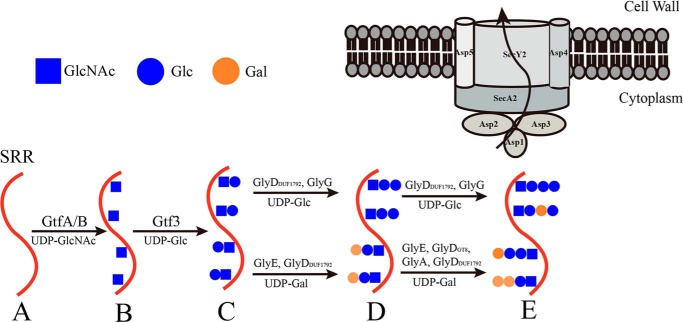
In the sequential transfer model, glycosyltransferases add the sugar residues one by one to a peptide acceptor using the nucleotide-activated sugar donor. However, the fine glycosylation pathway and mechanism are largely unknown. Moreover, the glycan modification at multiple sites of a polypeptide acceptor remains a mystery. Here we have systematically analyzed and demonstrated the heavy O-glycosylation of PsrP, an ideal model for the sequential O-glycosylation of a bacterial adhesin.
Based on previous reports (37, 39, 40) and our glycosylation assays, we propose a pathway for the polymorphic glycosylation of PsrP (Fig. 7). The nascent SRR (Fig. 7A) is first subjected to O-glycosylation catalyzed by a GtfA/B complex to add the GlcNAc residue in a cooperative mechanism (36, 37), which is highly conserved in all Gram-positive pathogens that possess SRRPs. Afterward, Gtf3 catalyzes the second step of glycosylation that adds a Glc residue to the GlcNAc-modified SRR (Fig. 7B), which is accommodated in an open active-site pocket (39). These two initial steps are specifically catalyzed by a given enzyme/complex, forming the unified disaccharide core structure of the glycan (Fig. 7C). Along with the extension of glycan chains at the third step, the disaccharide-modified SRR could be recognized by a couple of glycosyltransferases, including GlyG, GlyE, and GlyDDUF1792, using either UDP-Glc or UDP-Gal as the sugar donor. Thus two types of sugar residues could be randomly incorporated at the third step, resulting in a chimeric glycosylation pattern (Fig. 7D). Notably, the three glycosyltransferases working at this step differ a lot with each other. GlyE consists of a GT8 domain followed by a Rossmann-fold add-on domain, whereas GlyG and GlyDDUF1792 are composed of a single GT2 domain and GT-D fold, respectively. These varying enzymes produce a chimeric SRR acceptor that harbors different non-reducing sugars at multiple sites subjected to further glycosylation. As predicted, the fourth step could be catalyzed by as more as five different glycosyltransferases using two types of sugar donor. In consequence, the produced glycosylated SRR contains four types of tetrasaccharide chains that decorate the serine residues (Fig. 7E). It is worth noting that both the GT8 and DUF1792 domains of GlyD, which are structurally distinct from each other, are capable of incorporating a Gal residue at the fourth step. In addition, GlyA was identified as participating in the fourth-step glycosylation, most likely using its GT8 and add-on domains, as UDP-Gal is the favorable sugar donor of the GT8 domain. Moreover, GlyDDUF1792, GlyE, and GlyG participate in both the third and fourth steps of glycosylation, indicating their broad substrate spectrum. All together, our results indicated that the glycosylation of the SRR domains of PsrP exhibits a very high polymorphism, leading to highly diverse mature-form PsrP proteins.
Figure 7.
A proposed pathway for the heavy O-glycosylation of PsrP.
Furthermore, as all serine residues along the serine-rich repeat regions are randomly subjected to glycosylation at various degrees, the glycosylated PsrP should be heterogeneous that contains diverse O-linked glycans of different lengths. This phenomenon has also been found in human mucin, which undergoes a very complex O-glycosylation involved in a variety of biological processes (52). Here we have identified a unified disaccharide core structure and highly polymorphic extensions of PsrP glycan, providing insightful hints to the mechanism of heavy O-glycosylation. More investigations of pneumococcal pathogenesis mediated by precisely controlled glycosylation of PsrP will help to correlate the physiological functions with the polymorphic glycans.
Experimental procedures
Cloning, expression, and purification of glycosyltransferases and mutants
The coding regions of glycosyltransferases were amplified from the genomic DNA of S. pneumoniae TIGR4 and cloned into a 2B-T vector with an N-terminal hexahistidine tag using ligation-independent cloning system. The E. coli BL21 (DE3) strain was used for the expression of recombinant proteins. The transformed cells were grown at 37 °C in LB culture medium (10 g of NaCl, 10 g of Bacto-Tryptone, and 5 g of yeast extract per liter) containing appropriate antibiotics until the A600 nm reached ∼0.6. Protein expression was then induced with 0.2 mm isopropyl 1-thio-β-d-galactopyranoside (IPTG) by another 20 h at 16 °C. Cells were harvested by centrifugation (6000 × g, 4 °C, 10 min) and resuspended in 40 ml of lysis buffer (20 mm Tris-Cl, pH 8.0, 100 mm NaCl). After 5 min of sonication and centrifugation at 12,000 × g for 30 min, the supernatant containing the soluble target protein was collected and loaded onto a nickel-nitrilotriacetic acid column (Qiagen, Mississauga ON) equilibrated with the binding buffer (20 mm Tris-Cl, pH 8.0, 100 mm NaCl). The target protein was eluted with 300 mm imidazole and further loaded onto a Superdex 75 column (GE Healthcare) equilibrated with 20 mm Tris-Cl, pH 7.0, 100 mm NaCl. The target protein samples at the peak were pooled, and protein purity was evaluated by electrophoresis and samples were stored at −80 °C.
The selenomethionine (Se-Met)-labeled GlyE protein was expressed in E. coli strain B834 (DE3) (Novagen, Madison, WI). Transformed cells were inoculated into LB medium at 37 °C overnight. The cells were harvested and washed twice with the M9 medium. Then the cells were cultured in Se-Met medium (M9 medium with 50 mg/liter Se-Met and other essential amino acids at 50 mg/liter) to an A600 nm of ∼0.6. Protein expression and purification steps were carried out as described above for the native protein.
Site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) with the plasmid encoding the wild-type glycosyltransferases as the template. The mutant proteins were expressed, purified, and stored in the same manner as the wild-type protein.
Atomic absorption spectroscopy (Atomscan Advantage, Thermo Ash Jarrell Corp.) was performed to determine the metal content of GlyE. Before analysis, purified GlyE protein in 20 mm Tris-Cl, pH 7.0, and 100 mm NaCl was concentrated to ∼1 mg/ml with the total volume of 10 ml.
Crystallization, data collection, and processing
Before crystallization, the protein sample was concentrated to 10 mg/ml by ultrafiltration (Millipore Amicon). Crystallization trials of GlyE were done using a Mosquito robot (TTP Labtech) in 96-well plates (Greiner) at 16 °C. The UDP-bound crystals were obtained using the hanging drop vapor-diffusion method with the initial condition of equilibrating 0.1 μl of 10 mg/ml Se-Met-substituted protein (mixed with UDP to the final concentration of 5 mm) with and equal volume of the reservoir solution (0.2 m MgCl2, 0.1 m HEPES, pH 7.5, 25% polyethylene glycol 3350). After exhaustive optimization trials by microseeding, the crystals of a square shape were grown to the optimal size with the addition of 5 mm DTT. The apo-form crystals were obtained in the same condition as the UDP-bound crystals using the native GlyE protein at 10 mg/ml. All the crystals were transferred to cryoprotectant (reservoir solution supplemented with 30% ethylene glycol) and flash-cooled with liquid nitrogen. The data were collected at 100 K in a liquid nitrogen stream using beamline 17U with a Q315r CCD (ADSC, MARresearch, Germany) at the Shanghai Synchrotron Radiation Facility (SSRF).
Structure determination and refinement
All diffraction data were integrated and scaled with the program HKL2000 (53). The GlyE proteins in the presence of UDP were crystallized in the space group of P212121. The crystal structure of GlyE in complex with UDP was determined using single-wavelength anomalous dispersion (SAD) phasing (54) method from a single Se-Met-substituted protein crystal to a highest resolution of 1.95 Å. The AutoSol program (55) implemented in PHENIX (56) was used to locate the selenium atoms, and the initial phase was calculated by Resolve (57). Electron density maps showed clear features of secondary structural elements. Automatic model building was carried out using Autobuild in PHENIX. The resultant model was refined using the maximum likelihood method implemented in REFMAC5 (58) as part of the CCP4i (59) program suite and rebuilt interactively using the program COOT (60). The apo-form structure of GlyE was determined by the Molecular Replacement method (64) using the GlyE-UDP structure as the search model. The model was refined using the same method as the GlyE-UDP structure. The final structures were evaluated with the programs MOLPROBITY (61) and PROCHECK (62). Crystallographic parameters are listed in Table 1. All structure figures were prepared with PyMOL (63).
Table 1.
Crystal parameters, data collection, and structure refinement
| GlyE-UDP | ApoGlyE | |
|---|---|---|
| Data collection | ||
| Space group | P212121 | P21 |
| Unit cell | 75.170 88.210 128.360 | 89.119, 84.211, 130.113 |
| a, b, c (Å) | 90.00 | 90.00, 89.97, 90.00 |
| α, β, γ (°) | ||
| Resolution range (Å) | 48.81-1.95 (2.06-1.95)a | 50.00-2.40 (2.49-2.40) |
| Unique reflections | 61,392 (8,485) | 74,936 (7,360) |
| Completeness (%) | 97.7 (93.9) | 99.2 (98.1) |
| 〈I/σ(I)〉 | 7.3 (3.0) | 11.2 (2.2) |
| Rmergeb (%) | 13.3 (39.4) | 9.4 (45.0) |
| Average redundancy | 4.9 (4.9) | 3.1 (3.0) |
| Structure refinement | ||
| Resolution range (Å) | 48.81 − 1.95 | 50.00 − 2.40 |
| R-factorc/R-freed (%) | 20.4/25.7 | 19.8/25.5 |
| Number of protein atoms | 6,390 | 12,808 |
| Number of water atoms | 553 | 487 |
| r.m.s.d. bond lengths (Å) | 0.005 | 0.012 |
| r.m.s.d. bond angles (°) | 0.942 | 1.541 |
| Mean B factors (Å2) | 16.6 | 50.0 |
| Ramachandran plote (residues, %) | ||
| Most favored (%) | 97.5 | 94.7 |
| Additional allowed (%) | 2.2 | 4.0 |
| Outliers (%) | 0.3 | 1.3 |
| PDB entry | 5GVV | 5GVW |
a The values in parentheses refer to statistics in the highest bin.
b Rmerge = ∑hkl∑i Ii(hkl) − 〈I(hkl)〉 /∑hkl∑iIi(hkl), where Ii(hkl) is the intensity of an observation, and 〈I(hkl)〉 is the mean value for its unique reflection. Summations are over all reflections.
c Rwork = ∑i Fo(h) − Fc(h) /∑iFo(h), where Fo and Fc are the observed and calculated structure factor amplitudes, respectively.
d Rfree was calculated with 5% of the data excluded from the refinement.
e The categories were defined by Molprobity.
Hydrolytic activity assays
The hydrolytic activities of the glycosyltransferases were assayed by high performance liquid chromatography (HPLC). All assays were performed at 37 °C in buffer containing 20 mm Tris-Cl, pH 7.0, 100 mm NaCl, 1 mm MnCl2, 1 mm MgCl2, with 1 mm UDP-Gal or UDP-Glc (sigma) as the sugar donor. The donors were diluted to a series of concentrations from 100 mm stock solution. The reaction in the 10-μl system was triggered by adding the purified enzyme solution at a final concentration of 10 μm. The reaction lasted for 60 min and was terminated by heating at 100 °C for 10 min. For the glycosyltransferase activity, the acceptor SRR1 of different modifications was also added in the solution at a final concentration of 0.25 mm. For different enzymes and acceptors, the reaction period (2–60 min) was screened to ensure the production of UDP is proportional to the time. All samples were centrifuged at 10,000 × g for 10 min. The supernatant in a volume of 10 μl was subjected to the HPLC system (Agilent 1200 Series). A buffer of 100 mm KH2PO4/K2HPO4 pH 6.5, 10 mm tetrabutylammonium bromide was used for equilibration of the column (Zorbax 300SB-C18 column, 4.6 × 150 mm, Agilent), and separation of the components was at a flow rate of 1 ml/min. The product UDP was used as the standard and quantified by the absorption at 254 nm. The enzymatic reaction velocities were calculated by determining the generation of product per minute. Three independent assays were performed to calculate the means and S.D.
Co-expression studies
Glutathione S-transferase (GST)-tagged SRR1 (GST-SRR1) was cloned within the first multiple-cloning site of pETDuet. DNA encoding GtfA and GtfB was amplified as a single DNA fragment from the genomic DNA from S. pneumoniae TIGR4 and cloned within the second multiple-cloning site of pETDuet. DNA encoding Gtf3 was cloned as an N-terminal His tag within the plasmid pET28a, whereas other glycosyltransferases were cloned into the plasmid pCDFDuet-1. Co-expression of GST-SRR1 with defined glycosyltransferases was carried out as described previously (36). The disaccharide modified SRR1 was obtained by co-expression of GST-SRR1, GtfA/B, and Gtf3, whereas the trisaccharide-modified SRR1 was obtained by co-expression of GST-SRR1, GtfA/B, Gtf3, and GlyG, GST-SRR1, GtfA/B, Gtf3, and GlyE, or GST-SRR1, GtfA/B, Gtf3, and GlyDDUF1792, respectively. E. coli BL21 (DE3) cells were simultaneously transformed with the designated plasmid sets, and recombinant colonies were selected on plates containing the appropriate antibiotics. The GST-SRR1 was purified using the GSH resin followed by the size-exclusion chromatography.
In vitro glycosylation assays
The PsrP substrates with different modifications were obtained from E. coli by co-expression of GST-SRR1 with different glycosyltransferases. The in vitro glycosylation assays were performed as described above, with the addition of 5 μg of GST-SRR1 and 0.4 μCi of UDP-[3H]glucose or UDP-[3H]galactose (15–30 Ci/mmol; American Radiolabeled Chemicals, Inc). Enzyme of 10 μm was added to the final 10 μl system. The reaction lasted for 2 h at 37 °C and was terminated by heating at 100 °C for 10 min. The reaction mixtures were then separated on a 12% SDS-PAGE gel followed by Coomassie Blue staining. Incorporation of UDP-[3H]glucose or UDP-[3H]galactose was visualized by 3H autoradiography. The intensity of the bands was scaled and integrated by the software ImageJ. The assays were performed in at least three independent experiments.
Author contributions
C.-Z. Z. and Y. C. conceived, designed, and supervised the project. C.-Z. Z., Y. C., and Y.-L. J. analyzed the data and wrote the manuscript. Y.-L. J., H. J., and S. W. performed the protein purification and glycosylation assays. Y.-L. J., H. J., R.-L. Z., and H. B. Y. performed crystal screening and optimization, data collection, and structural determination. All authors discussed the data and read the manuscript.
Acknowledgments
We thank Prof. Hui Wu at the University of Alabama at Birmingham for the assistance in experimental design. We thank the staff at the Shanghai Synchrotron Radiation Facility and the Core Facility Center for Life Sciences at University of Science and Technology of China for technical assistance.
This work was supported by Ministry of Science and Technology of China Grants 2015CB910100, 2013CB835300, and 2014CB910100, National Natural Science Foundation of China Grants 31270781, 31570728, and 31400628, and by the Innovation Center for Cell Signaling Network. The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (codes 5GVV and 5GVW) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- SRRP
- serine-rich repeat (SRR) proteins
- SRR1
- a short serine-rich repeat region
- SRR2
- a large serine-rich repeat region
- Se-Met
- selenomethionine
- r.m.s.d.
- root mean square deviation
- dGT1
- S. parasanguinis glycosyltransferases.
References
- 1. Nothaft H., and Szymanski C. M. (2010) Protein glycosylation in bacteria: sweeter than ever. Nat. Rev. Microbiol. 8, 765–778 [DOI] [PubMed] [Google Scholar]
- 2. Schwarz F., and Aebi M. (2011) Mechanisms and principles of N-linked protein glycosylation. Curr. Opin Struct. Biol. 21, 576–582 [DOI] [PubMed] [Google Scholar]
- 3. Apweiler R., Hermjakob H., and Sharon N. (1999) On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta 1473, 4–8 [DOI] [PubMed] [Google Scholar]
- 4. Fletcher C. M., Coyne M. J., Villa O. F., Chatzidaki-Livanis M., and Comstock L. E. (2009) A general O-glycosylation system important to the physiology of a major human intestinal symbiont. Cell 137, 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ohtsubo K., and Marth J. D. (2006) Glycosylation in cellular mechanisms of health and disease. Cell 126, 855–867 [DOI] [PubMed] [Google Scholar]
- 6. Varki A. (2006) Nothing in glycobiology makes sense, except in the light of evolution. Cell 126, 841–845 [DOI] [PubMed] [Google Scholar]
- 7. Lu Q., Li S., and Shao F. (2015) Sweet talk: protein glycosylation in bacterial interaction with the host. Trends Microbiol. 23, 630–641 [DOI] [PubMed] [Google Scholar]
- 8. Corfield A. P. (2015) Mucins: a biologically relevant glycan barrier in mucosal protection. Biochim. Biophys. Acta 1850, 236–252 [DOI] [PubMed] [Google Scholar]
- 9. Fukuda M. (2002) Roles of mucin-type O-glycans in cell adhesion. Biochim. Biophys. Acta 1573, 394–405 [DOI] [PubMed] [Google Scholar]
- 10. Saeland E., van Vliet S. J., Bäckström M., van den Berg V. C., Geijtenbeek T. B., Meijer G. A., and van Kooyk Y. (2007) The C-type lectin MGL expressed by dendritic cells detects glycan changes on MUC1 in colon carcinoma. Cancer Immunol. Immunother. 56, 1225–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tian E., and Ten Hagen K. G. (2009) Recent insights into the biological roles of mucin-type O-glycosylation. Glycoconj. J. 26, 325–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ju T., Cummings R. D., and Canfield W. M. (2002) Purification, characterization, and subunit structure of rat core 1 β1,3-galactosyltransferase. J. Biol. Chem. 277, 169–177 [DOI] [PubMed] [Google Scholar]
- 13. Hang H. C., and Bertozzi C. R. (2005) The chemistry and biology of mucin-type O-linked glycosylation. Bioorg Med. Chem. 13, 5021–5034 [DOI] [PubMed] [Google Scholar]
- 14. Iwashkiw J. A., Vozza N. F., Kinsella R. L., and Feldman M. F. (2013) Pour some sugar on it: the expanding world of bacterial protein O-linked glycosylation. Mol. Microbiol. 89, 14–28 [DOI] [PubMed] [Google Scholar]
- 15. Tan F. Y., Tang C. M., and Exley R. M. (2015) Sugar coating: bacterial protein glycosylation and host-microbe interactions. Trends Biochem. Sci. 40, 342–350 [DOI] [PubMed] [Google Scholar]
- 16. Logan S. M. (2006) Flagellar glycosylation: a new component of the motility repertoire? Microbiology 152, 1249–1262 [DOI] [PubMed] [Google Scholar]
- 17. Hanuszkiewicz A., Pittock P., Humphries F., Moll H., Rosales A. R., Molinaro A., Moynagh P. N., Lajoie G. A., and Valvano M. A. (2014) Identification of the flagellin glycosylation system in Burkholderia cenocepacia and the contribution of glycosylated flagellin to evasion of human innate immune responses. J. Biol. Chem. 289, 19231–19244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kukita K., Kawada-Matsuo M., Oho T., Nagatomo M., Oogai Y., Hashimoto M., Suda Y., Tanaka T., and Komatsuzawa H. (2013) Staphylococcus aureus SasA is responsible for binding to the salivary agglutinin gp340, derived from human saliva. Infect. Immun. 81, 1870–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sheen T. R., Jimenez A., Wang N. Y., Banerjee A., van Sorge N. M., and Doran K. S. (2011) Serine-rich repeat proteins and pili promote Streptococcus agalactiae colonization of the vaginal tract. J. Bacteriol. 193, 6834–6842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sanchez C. J., Shivshankar P., Stol K., Trakhtenbroit S., Sullam P. M., Sauer K., Hermans P. W., and Orihuela C. J. (2010) The pneumococcal serine-rich repeat protein is an intra-species bacterial adhesin that promotes bacterial aggregation in vivo and in biofilms. PLoS Pathog. 6, e1001044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pyburn T. M., Bensing B. A., Xiong Y. Q., Melancon B. J., Tomasiak T. M., Ward N. J., Yankovskaya V., Oliver K. M., Cecchini G., Sulikowski G. A., Tyska M. J., Sullam P. M., and Iverson T. M. (2011) A structural model for binding of the serine-rich repeat adhesin GspB to host carbohydrate receptors. PLoS Pathog. 7, e1002112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. King N. P., Beatson S. A., Totsika M., Ulett G. C., Alm R. A., Manning P. A., and Schembri M. A. (2011) UafB is a serine-rich repeat adhesin of Staphylococcus saprophyticus that mediates binding to fibronectin, fibrinogen, and human uroepithelial cells. Microbiology 157, 1161–1175 [DOI] [PubMed] [Google Scholar]
- 23. Lizcano A., Sanchez C. J., and Orihuela C. J. (2012) A role for glycosylated serine-rich repeat proteins in gram-positive bacterial pathogenesis. Mol. Oral Microbiol. 27, 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhou M., and Wu H. (2009) Glycosylation and biogenesis of a family of serine-rich bacterial adhesins. Microbiology 155, 317–327 [DOI] [PubMed] [Google Scholar]
- 25. Takamatsu D., Bensing B. A., and Sullam P. M. (2004) Four proteins encoded in the gspB-secY2A2 operon of Streptococcus gordonii mediate the intracellular glycosylation of the platelet-binding protein GspB. J. Bacteriol. 186, 7100–7111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shivshankar P., Sanchez C., Rose L. F., and Orihuela C. J. (2009) The Streptococcus pneumoniae adhesin PsrP binds to Keratin 10 on lung cells. Mol. Microbiol. 73, 663–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hegge F. T., Hitchen P. G., Aas F. E., Kristiansen H., Løvold C., Egge-Jacobsen W., Panico M., Leong W. Y., Bull V., Virji M., Morris H. R., Dell A., and Koomey M. (2004) Unique modifications with phosphocholine and phosphoethanolamine define alternate antigenic forms of Neisseria gonorrhoeae type IV pili. Proc. Natl. Acad. Sci. U.S.A. 101, 10798–10803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Seepersaud R., Bensing B. A., Yen Y. T., and Sullam P. M. (2012) The accessory Sec protein Asp2 modulates GlcNAc deposition onto the serine-rich repeat glycoprotein GspB. J. Bacteriol. 194, 5564–5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Takamatsu D., Bensing B. A., and Sullam P. M. (2004) Genes in the accessory sec locus of Streptococcus gordonii have three functionally distinct effects on the expression of the platelet-binding protein GspB. Mol. Microbiol. 52, 189–203 [DOI] [PubMed] [Google Scholar]
- 30. Bensing B. A., Takamatsu D., and Sullam P. M. (2005) Determinants of the streptococcal surface glycoprotein GspB that facilitate export by the accessory Sec system. Mol. Microbiol. 58, 1468–1481 [DOI] [PubMed] [Google Scholar]
- 31. Muñoz-Almagro C., Selva L., Sanchez C. J., Esteva C., de Sevilla M. F., Pallares R., and Orihuela C. J. (2010) PsrP, a protective pneumococcal antigen, is highly prevalent in children with pneumonia and is strongly associated with clonal type. Clin Vaccine Immunol. 17, 1672–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schulte T., Mikaelsson C., Beaussart A., Kikhney A., Deshmukh M., Wolniak S., Pathak A., Ebel C., Löfling J., Fogolari F., Henriques-Normark B., Dufrêne Y. F., Svergun D., Nygren P. Å., and Achour A. (2016) The BR domain of PsrP interacts with extracellular DNA to promote bacterial aggregation; structural insights into pneumococcal biofilm formation. Sci. Rep. 6, 32371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Obert C., Sublett J., Kaushal D., Hinojosa E., Barton T., Tuomanen E. I., and Orihuela C. J. (2006) Identification of a candidate Streptococcus pneumoniae core genome and regions of diversity correlated with invasive pneumococcal disease. Infect. Immun. 74, 4766–4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu R., Zhou M., and Wu H. (2010) Purification and characterization of an active N-acetylglucosaminyltransferase enzyme complex from streptococci. Appl. Environ. Microbiol. 76, 7966–7971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu R., and Wu H. (2011) A molecular chaperone mediates a two-protein enzyme complex and glycosylation of serine-rich streptococcal adhesins. J. Biol. Chem. 286, 34923–34931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shi W. W., Jiang Y. L., Zhu F., Yang Y. H., Shao Q. Y., Yang H. B., Ren Y. M., Wu H., Chen Y., and Zhou C. Z. (2014) Structure of a novel O-linked N-acetyl-d-glucosamine (O-GlcNAc) transferase, GtfA, reveals insights into the glycosylation of pneumococcal serine-rich repeat adhesins. J. Biol. Chem. 289, 20898–20907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen Y., Seepersaud R., Bensing B. A., Sullam P. M., and Rapoport T. A. (2016) Mechanism of a cytosolic O-glycosyltransferase essential for the synthesis of a bacterial adhesion protein. Proc. Natl. Acad. Sci. U.S.A. 113, E1190–E1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou M., Zhu F., Dong S., Pritchard D. G., and Wu H. (2010) A novel glucosyltransferase is required for glycosylation of a serine-rich adhesin and biofilm formation by Streptococcus parasanguinis. J. Biol. Chem. 285, 12140–12148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhu F., Erlandsen H., Ding L., Li J., Huang Y., Zhou M., Liang X., Ma J., and Wu H. (2011) Structural and functional analysis of a new subfamily of glycosyltransferases required for glycosylation of serine-rich streptococcal adhesins. J. Biol. Chem. 286, 27048–27057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang H., Zhu F., Yang T., Ding L., Zhou M., Li J., Haslam S. M., Dell A., Erlandsen H., and Wu H. (2014) The highly conserved domain of unknown function 1792 has a distinct glycosyltransferase fold. Nat. Commun. 5, 4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang H., Zhou M., Yang T., Haslam S. M., Dell A., and Wu H. (2016) A new helical binding domain mediates a unique glycosyltransferase activity of a bifunctional protein. J. Biol. Chem. 291, 22106–22117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sindhuwinata N., Munoz E., Munoz F. J., Palcic M. M., Peters H., and Peters T. (2010) Binding of an acceptor substrate analog enhances the enzymatic activity of human blood group B galactosyltransferase. Glycobiology 20, 718–723 [DOI] [PubMed] [Google Scholar]
- 43. Holm L., and Sander C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233, 123–138 [DOI] [PubMed] [Google Scholar]
- 44. Persson K., Ly H. D., Dieckelmann M., Wakarchuk W. W., Withers S. G., and Strynadka N. C. (2001) Crystal structure of the retaining galactosyltransferase LgtC from Neisseria meningitidis in complex with donor and acceptor sugar analogs. Nat. Struct. Biol. 8, 166–175 [DOI] [PubMed] [Google Scholar]
- 45. Busch C., Hofmann F., Selzer J., Munro S., Jeckel D., and Aktories K. (1998) A common motif of eukaryotic glycosyltransferases is essential for the enzyme activity of large clostridial cytotoxins. J. Biol. Chem. 273, 19566–19572 [DOI] [PubMed] [Google Scholar]
- 46. Shibayama K., Ohsuka S., Tanaka T., Arakawa Y., and Ohta M. (1998) Conserved structural regions involved in the catalytic mechanism of Escherichia coli K-12 WaaO (RfaI). J. Bacteriol 180, 5313–5318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wiggins C. A., and Munro S. (1998) Activity of the yeast MNN1 α-1,3-mannosyltransferase requires a motif conserved in many other families of glycosyltransferases. Proc. Natl. Acad. Sci. U.S.A. 95, 7945–7950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hagen F. K., Hazes B., Raffo R., deSa D., and Tabak L. A. (1999) Structure-function analysis of the UDP-N-acetyl-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase. Essential residues lie in a predicted active site cleft resembling a lactose repressor fold. J. Biol. Chem. 274, 6797–6803 [DOI] [PubMed] [Google Scholar]
- 49. Yu H., Takeuchi M., LeBarron J., Kantharia J., London E., Bakker H., Haltiwanger R. S., Li H., and Takeuchi H. (2015) Notch-modifying xylosyltransferase structures support an SNi-like retaining mechanism. Nat. Chem. Biol. 11, 847–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zeqiraj E., Tang X., Hunter R. W., García-Rocha M., Judd A., Deak M., von Wilamowitz-Moellendorff A., Kurinov I., Guinovart J. J., Tyers M., Sakamoto K., and Sicheri F. (2014) Structural basis for the recruitment of glycogen synthase by glycogenin. Proc. Natl. Acad. Sci. U.S.A. 111, E2831–E2840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chaikuad A., Froese D. S., Berridge G., von Delft F., Oppermann U., and Yue W. W. (2011) Conformational plasticity of glycogenin and its maltosaccharide substrate during glycogen biogenesis. Proc. Natl. Acad. Sci. U.S.A. 108, 21028–21033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hanisch F. G. (2001) O-Glycosylation of the mucin type. Biol. Chem. 382, 143–149 [DOI] [PubMed] [Google Scholar]
- 53. Otwinowski Z., and Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 54. Brodersen D. E., de La Fortelle E., Vonrhein C., Bricogne G., Nyborg J., and Kjeldgaard M. (2000) Applications of single-wavelength anomalous dispersion at high and atomic resolution. Acta Crystallogr. D. Biol. Crystallogr. 56, 431–441 [DOI] [PubMed] [Google Scholar]
- 55. Sheldrick G. M. (2008) A short history of SHELX. Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 56. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., and Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hao Q., Gu Y. X., Zheng C. D., and Fan H. F. (2000) OASIS: a computer program for breaking phase ambiguity in one-wavelength anomalous scattering or single isomorphous substitution (replacement) data. J. Appl. Cryst. 33, 980–981 [Google Scholar]
- 58. Murshudov G. N., Vagin A. A, and Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 59. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 60. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 61. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., and Arendall W. B. 3rd, Snoeyink J., Richardson J. S., and Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Laskowski R., Macarthur M., Moss D., and Thornton J. (1993) Procheck: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26, 283–291 [Google Scholar]
- 63. DeLano W. L. (2002) The PyMOL Molecular Graphics System, De Lano Scientific, San Carlos, CA [Google Scholar]
- 64. Evans P., and McCoy A. (2008) An introduction to molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 64, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]