Abstract
The ligase Itch plays major roles in signaling pathways by inducing ubiquitylation-dependent degradation of several substrates. Substrate recognition and binding are critical for the regulation of this reaction. Like closely related ligases, Itch can interact with proteins containing a PPXY motif via its WW domains. In addition to these WW domains, Itch possesses a proline-rich region (PRR) that has been shown to interact with several Src homology 3 (SH3) domain-containing proteins. We have previously established that despite the apparent surface uniformity and conserved fold of SH3 domains, they display different binding mechanisms and affinities for their interaction with the PRR of Itch. Here, we attempt to determine the molecular bases underlying the wide range of binding properties of the Itch PRR. Using pulldown assays combined with mass spectrometry analysis, we show that the Itch PRR preferentially forms complexes with endophilins, amphyphisins, and pacsins but can also target a variety of other SH3 domain-containing proteins. In addition, we map the binding sites of these proteins using a combination of PRR sub-sequences and mutants. We find that different SH3 domains target distinct proline-rich sequences overlapping significantly. We also structurally analyze these protein complexes using crystallography and molecular modeling. These structures depict the position of Itch PRR engaged in a 1:2 protein complex with β-PIX and a 1:1 complex with the other SH3 domain-containing proteins. Taken together, these results reveal the binding preferences of the Itch PRR toward its most common SH3 domain-containing partners and demonstrate that the PRR region is sufficient for binding.
Keywords: crystal structure, isothermal titration calorimetry (ITC), molecular modeling, protein-protein interaction, Src homology 3 domain (SH3 domain), amphiphysin, endophilin, Itch, pacsin, proline-rich region (PRR)
Introduction
The C2-WW-HECT (CWH)3 family ubiquitin ligases are involved in major signaling pathways that regulate cell growth and proliferation (1). Nedd4 and Itch are well-known members of the family that regulate T-cell activation and effector differentiation (2). To ubiquitylate their substrates, CWH ligases must first establish a direct contact with them. This is done via the interaction of their WW domains with short PPXY motifs present in the substrate (3, 4). Unlike other members of the CWH family, Itch possesses a conserved proline-rich region (PRR) in a non-conserved linker region located between the N-terminal C2 domain and the WW domains common to the other ligases of the family. This PRR is composed of 20 amino acids and is essential for Itch interaction with SH3 domain-containing proteins (5, 6).
SH3 domains are well-characterized protein-interaction modules composed of roughly 60 amino acids with a conserved fold that are often found in proteins involved in signaling, endocytosis, and trafficking. The Itch PRR is targeted by several of these SH3 domain-containing proteins, and this typically induces ubiquitylation of these binding partners by Itch (5, 7–10). A notable exception to this rule is β-PIX, which was not readily ubiquitylated by Itch (8).
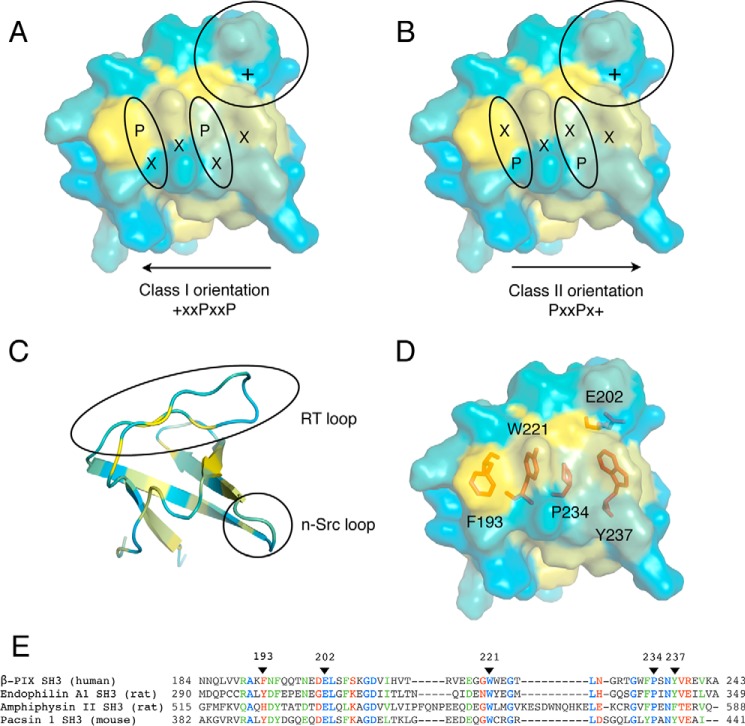
Despite the functional characterization of interactions involving the Itch PRR and several SH3 domain-containing proteins, very little is known about the selectivity and strength of these interactions. The conserved fold of SH3 domains generally exposes key aromatic residues and a proline on the surface, which form hydrophobic pockets required for interactions with the core PXXP sequence (Fig. 1). An additional specificity pocket, defined by negatively charged residues in the RT loop of the SH3 domain, completes the binding motif by forming interactions with a positively charged residue outside the proline core. The relative position of this positively charged residue defines the orientation of the typical class I (+XXPXXP) and class II (PXXPX+) ligands. Itch PRR consists of a compact succession of three class II binding motifs and one class I binding motif that are partially overlapping (8). Isothermal titration calorimetry (ITC) studies showed that the Itch PRR forms classical complexes with a 1:1 protein ratio with several SH3 domain-containing proteins. In the same set of experiments, it was shown that the Itch PRR can also interact simultaneously with two SH3 domains either from two different molecules in the case of β-PIX (8) or from a single protein in the case of Grb2.4 Thus, the PRR of Itch is employing at least two different mechanisms of binding with distinct stoichiometries to recognize SH3 domains. Although the overall structure and amino acid composition of SH3 domains are highly conserved, subtle variations can lead to drastic changes in their binding properties (11). Previous studies have also demonstrated that the affinity of the Itch PRR toward SH3 domains varies greatly and that it has a clear preference for endophilin. Interestingly, the dissociation constant reported for the endophilin·Itch complex places it among the strongest SH3-PRR affinities reported to date. However, the specific residues and the molecular basis of Itch PRR targeting these different SH3 domains remain to be elucidated. Here, we map the binding sites on the SH3 domain-containing proteins most frequently found in complex with the Itch PRR and find that they recognize distinct yet overlapping sites within the Itch PRR. We also solve the structure of the simultaneous interaction between Itch PRR and two β-PIX SH3s. This structure was then used as a template for the molecular modeling of SH3-binding interfaces between Itch PRR and other SH3 domain-containing proteins. Together, these results reveal the binding preferences of the Itch PRR toward several of its SH3 domain-containing partners.
Figure 1.
Representation of a typical SH3 domain interacting with canonical class I and II peptides. A–D, the SH3 domain of β-PIX (SH3″ of the present study) is represented in the same orientation and colored according to a hydrophobicity scale (yellow for hydrophobic). The surface representation reveals the position of the specificity pocket (circle) and hydrophobic pockets (oval shape) accommodating the proline residues of a peptide binding either in a class I (A) or class II orientation (B). C, the SH3″ domain of β-PIX is shown in a schematic representation to indicate the position of the n-Src and RT loops. D, the SH3″ domain of β-PIX is represented as in A with key residues at the binding interface represented as red sticks. E, alignment of a subset of SH3 domains known to interact with the PRR of Itch performed with T-Coffee (8, 45). Residues are numbered according to the full-length human protein sequences. Fully conserved residues, residues with strongly similar properties, and residues with weakly similar properties are colored blue, green, and red, respectively. Arrowheads indicate the position of the residues represented as sticks in D.
Results
SH3 domain-containing proteins binding to Itch PRR
The Itch PRR contains a variety of potential binding sites for SH3 domains, and it has been shown to interact with several different proteins (7–10). To further evaluate the capacity of the Itch PRR to interact with SH3 domain-containing proteins, we performed a pulldown assay with GST-fused PRR peptides on rat brain extracts and analyzed the bound proteins using mass spectrometry. All of the SH3 domain-containing proteins that bound to the Itch PRR are compiled in Table 1. Most of the previously known binding partners of the Itch PRR were identified, except for β-PIX, CIN85, and STAM-1 (9, 10). In addition, several peptide fragments from spectrin α, endophilin A1–3, pacsin 1, and amphiphysin I/II were found in the bound fraction. In agreement with its high affinity for the Itch PRR, a vast majority of spectra identified by MS/MS were matched to endophilin A1. Most of the other proteins detected by mass spectrometry were only represented by a limited number of peptide fragments. Nevertheless, among these proteins, we find several previously reported binding partners for Itch PRR as well as newly identified binding proteins including spectrin α, Src, cortactin, LASP1, DBNL, CD2AP, and STAC. Interestingly, all of these potential new partners of Itch are linked to the organization of the cytoskeleton, pointing to a possible new and unexplored role for Itch.
Table 1.
SH3 domain-containing proteins interacting with Itch PRR identified by mass spectrometry
| Identity | Ensembl | Total spectra | Unique peptide | %/%a | Mass |
|---|---|---|---|---|---|
| GST-Itch PRR (residues 249–269), upper gelb | kDa | ||||
| Endophilin A1 | ENSP00000369981 | 560 | 46 | 64/100 | 39.9 |
| Spectrin α | ENSP00000361824 | 46 | 36 | 18/22 | 284.9 |
| Endophilin A2 | ENSP00000269886 | 45 | 13 | 36/51 | 41.5 |
| Endophilin A3 | ENSP00000391372 | 18 | 6 | 10/14 | 39.3 |
| Amphiphysin II | ENSP00000365281 | 7 | 5 | 19/26 | 47.5 |
| Pacsin 1 | ENSP00000244458 | 2 | 2 | 5.9/8 | 50.9 |
| Endophilin B2 | ENSP00000361645 | 1 | 1 | 3.0/5 | 43.9 |
| SNX18 | ENSP00000317332 | 1 | 1 | 1.8/3 | 68.9 |
| v-Src | ENSP00000362680 | 1 | 1 | 1.3/2 | 59.8 |
| GST-Itch PRR (residues 249–269), lower gelb | |||||
| Endophilin A1 | ENSP00000369981 | 138 | 18 | 45/72 | 39.9 |
| Endophilin A2 | ENSP00000269886 | 18 | 5 | 20/28 | 41.5 |
| Grb2 | ENSP00000376347 | 12 | 5 | 29/32 | 25.2 |
| GST-Itch PRR (residues 224–276), upper gelb | |||||
| Endophilin A1 | ENSP00000369981 | 348 | 33 | 60/96 | 39.9 |
| Endophilin A2 | ENSP00000269886 | 44 | 17 | 36/52 | 41.5 |
| Spectrin α | ENSP00000361824 | 41 | 34 | 19/24 | 284.9 |
| Amphiphysin I | ENSP00000317441 | 18 | 11 | 22/39 | 71.9 |
| Endophilin A3 | ENSP00000391372 | 17 | 9 | 24/33 | 39.3 |
| Pacsin 1 | ENSP00000244458 | 6 | 6 | 19/26 | 50.9 |
| Cortactin | ENSP00000317189 | 5 | 3 | 9.4/17 | 57.4 |
| v-Src | ENSP00000362680 | 3 | 2 | 5.4/8 | 59.8 |
| LASP1 | ENSP00000325240 | 3 | 2 | 11/15 | 29.7 |
| DBNL | ENSP00000411701 | 2 | 2 | 10/16 | 48.2 |
| Pacsin 2 | ENSP00000263246 | 2 | 2 | 5.1/9 | 55.7 |
| SNX18 | ENSP00000317332 | 2 | 1 | 1.4/2 | 68.9 |
| CD2AP | ENSP00000352264 | 1 | 1 | 3.1/4 | 71.4 |
| SNX9 | ENSP00000376024 | 1 | 1 | 2.2/4 | 66.5 |
| STAC | ENSP00000273183 | 1 | 1 | 4.0/5 | 44.5 |
a The amino acid coverage of the protein in this assignment/the coverage corrected for peptide sequences that are unlikely to be observed using normal proteomics methods.
b Data were analyzed with the human genome using the Global Proteome Machine Organization (28).
Molecular dissection of Itch PRR-mediated interactions
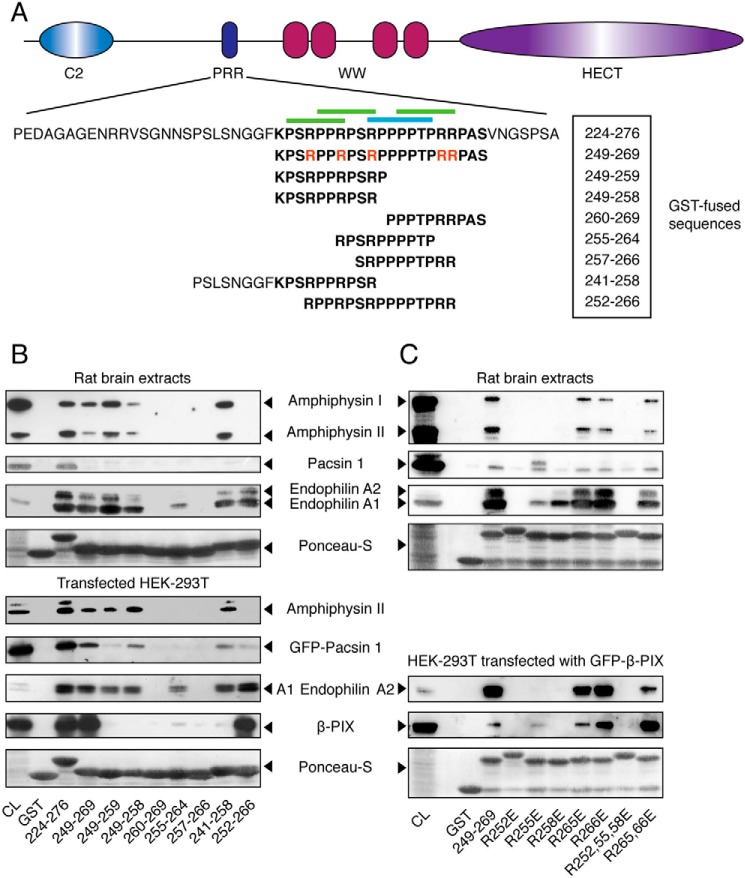
To determine the critical motifs within the Itch PRR required for the binding to the different SH3 domain-containing proteins, we constructed a series of overlapping peptides covering the Itch PRR in fusion with GST (Fig. 2A). These fusion proteins were used in pulldown assays with rat brain extracts (Fig. 2B, top) or with extracts from transfected HEK-293T cells (Fig. 2B, bottom) to determine the interaction with endophilin, pacsin, amphiphysin, and β-PIX. In these experiments, the anti-endophilin immunoblots revealed that this protein interacts with the first half of the Itch PRR, which contains two overlapping class II SH3-binding sites (peptide 249–259). endophilin was found to bind peptides minimally containing a RPPRPSR motif (residues 252–258). Removing the first residues of this class II motif (peptide 255–264) greatly impaired binding to endophilin. Interestingly, the class I and the most C-terminal class II motifs alone (peptide 257–266) did not bind to endophilin. In these experiments, the antibody used to detect endophilin recognizes endophilin A1, A2, and A3. Endophilin A1 and A3 have the same molecular mass (40 kDa) and cannot be distinguished. Because endophilin A1 is much more abundant, it is probably responsible for most of the signal observed on the blots. Only endophilin A2 is expressed in HEK-293T cells, and its higher molecular mass (45 kDa) also enables its detection in rat brain extracts. Both endophilin A1 and A2 have the same binding preferences toward GST-Itch PRR constructs.
Figure 2.
Binding preferences of the SH3 domains of amphiphysin, pacsin, endophilin, and β-PIX toward the PRR of Itch. A, schematic representation of the Itch protein highlighting the sequence of the full-length PRR (boldface type) as well as the specific sub-sequences used in these studies. Arginine residues mutated to glutamic acid in the context of the 249–269 peptide are indicated in red. Canonical class I and II binding sites are indicated by blue and green overbars, respectively. B, rat brain extracts were incubated with the indicated GST-PRR fusions bound to GSH resin. Immunoblots of bound proteins eluted from the resin were performed with anti-amphiphysin, anti-pan-endophilin, and anti-pacsin 1 antibodies to show their content, and 5% of the cell lysates served as a control (CL) (top panels). Extracts from HEK-293T cells transfected with the indicated constructs were incubated with the same GST fusions bound to GSH resin. The immunoblots of bound proteins eluted from the resin were performed with anti-amphiphysin, anti-GFP (GFP-pacsin 1 and GFP-endophilin A1), or anti-FLAG (FLAG-β-PIX). Immunoblots were performed on 5% of cell lysates (CL) to determine protein overexpression levels (bottom panels). C, rat brain extracts were incubated with the indicated GST-PRR mutant bound to GSH resin. The immunoblots of the bound proteins eluted from the resin were performed as in B to determine endogenous protein expression in 10% of cell lysates (CL) and their recovery in the pulldown assay (top panels). Extracts from HEK-293T cells overexpressing GFP-β-PIX were incubated with the GST-PRR mutants bound to GSH resin. The immunoblots of bound proteins eluted from the resin were performed with anti-pan-endophilin or anti-GFP to measure the quantities of endogenous endophilin and transfected GFP-β-PIX recovered from the same cell lysate (bottom panels).
Using a similar pulldown assay, we next evaluated amphiphysin binding to the Itch PRR. The anti-amphiphysin antibody used in these assays recognized both amphiphysin I and II and revealed that amphiphysin II was pulled down more efficiently than amphiphysin I relative to their respective expression levels in rat brain tissues. Despite this difference, both isoforms showed similar binding preferences. Like endophilin, the amphiphysins bind exclusively to sequences encompassing the first half of the Itch PRR containing the two overlapping class II motifs (peptide 249–259). In contrast to endophilin, amphiphysin binding does not tolerate any truncation of these class II motifs because binding was completely lost when peptides were truncated at both the N and C terminus (peptides 255–264) (Fig. 2B). Thus, both motifs were required for the interaction with amphiphysin, whereas a single intact class II motif could partially pull down endophilin.
The determination of the pacsin-binding site on the Itch PRR was more difficult to define due to its lower recovery in the pulldown assay (Fig. 2B). When overexpressed in HEK-293T cells, pacsin binding was readily detected, and it displays a binding pattern very similar to endophilin and amphiphysin, because the three proteins bind to constructs containing the first two class II motifs (peptide 249–259) of the Itch PRR (Fig. 2B). Like amphiphysin, pacsin binding was disrupted by truncation of either of the two class II motifs, but it still bound to a peptide containing the second class II motif, indicating that this motif contributes more to the interaction.
In contrast, the binding site for β-PIX is more extended than the other proteins tested, because none of the shorter Itch PRR constructs bind efficiently to β-PIX. The immunoblot performed with resin incubated with overexpressed β-PIX indicates that the only significant recovery occurred with the peptide 252–266, suggesting that this SH3 domain recognizes the class I motif and possibly part of the class II motifs flanking it in the Itch PRR (Fig. 2B). This result is consistent with previous experiments and demonstrates that the β-PIX interaction with Itch involves a different binding mode (9).
To further evaluate the binding preferences of the SH3 domain-containing proteins, we mutated several arginine residues to glutamic acid in the GST-FLAG-fused PRR of Itch (peptide 249–269) and performed pulldown studies with rat brain extracts (Fig. 2C). In agreement with results obtained with the PRR sub-sequences, the mutation analysis indicates that the first three arginine residues contained in the two overlapping class II motifs of the PRR play a key role in the binding to the four SH3 domains tested here. Interestingly, there were differences in the contribution of each of the first three arginines to the binding of the different SH3s. Whereas the R252E mutant abolishes the interaction with all of the SH3 domains tested under our experimental conditions, the R255E mutant retains partial binding toward endophilin A1 and β-PIX and appears to bind pacsin almost as efficiently as the WT sequence. In contrast, Arg-255 was required for the interaction of the Itch PRR with the SH3 domains of amphiphysin and endophilin A2. In turn, the R258E mutation only partially impairs binding of endophilin A1 but completely abolishes the interaction with endophilin A2, pacsin, β-PIX, and amphiphysin I and II. The R265E or R266E mutants, located exclusively within the last class II SH3-binding site, had no significant impact on its interaction with the SH3 domain-containing proteins. However, the combined R265E/R266E mutation slightly impairs binding to endophilin A1 and A2. Taken together, these results indicate that despite the fact that the preferred binding sites of these SH3 domain-containing proteins are overlapping, they target distinct residues within the PRR of Itch.
Residues located outside the proline-rich core do not contribute to the binding of SH3 domain-containing proteins
The pulldown experiments shown in Fig. 2B indicate that the SH3 domain of pacsin does not fully recognize the 249–269 peptide, suggesting that residues flanking the PRR contribute to the binding of some SH3 domains to Itch. We have previously reported that pacsin binding to the Itch PRR displayed a binding affinity comparable with that of β-PIX but much lower than that of endophilin. To determine whether residues outside the PRR contributed to the binding, we repeated the ITC experiments with the shorter sequence (peptide 249–269) and compared the binding affinity with that of a longer sequence (peptide 224–276) used previously (8). Thermograms for binding to the SH3 domain of endophilin, pacsin, β-PIX, and amphiphysin (Fig. 3A) indicate similar binding affinities for the 249–269 peptide and the longer 224–276 peptide (Fig. 3B). The binding region for the SH3 domains thus lies entirely within the core PRR sequence for endophilin, pacsin, and β-PIX. As reported previously, it was not possible to fit a saturation curve on thermograms obtained with amphiphysin, although significant heat dispersion was observed.
Figure 3.
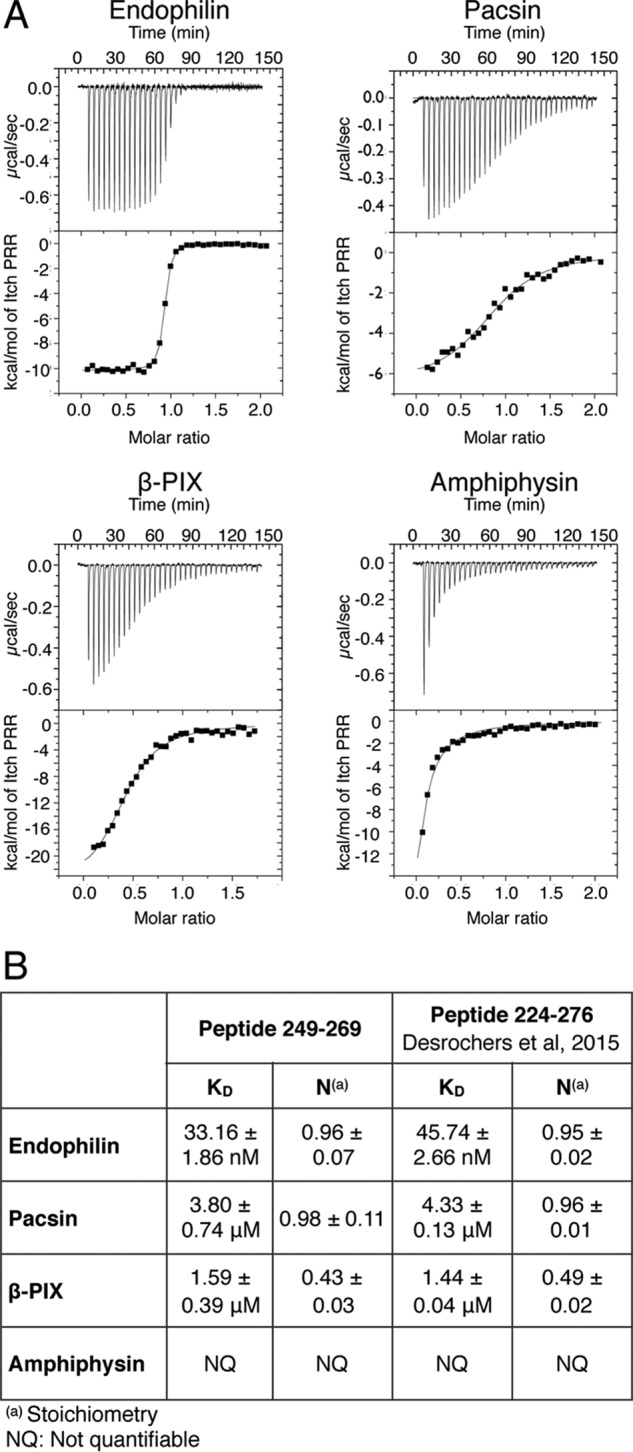
Representative ITC thermograms obtained by the successive addition of Itch PRR to a subset of SH3 domains. A, representative results obtained by ITC for the binding of the 249–269 peptide to the SH3 domain of endophilin, pacsin, β-PIX, and amphiphysin. B, the calculated dissociation constant (KD) and stoichiometries (N) are compiled with results obtained previously using a longer 224–276 peptide (8).
Crystal structure of the complex formed between β-PIX and Itch PRR reveals a super-SH3
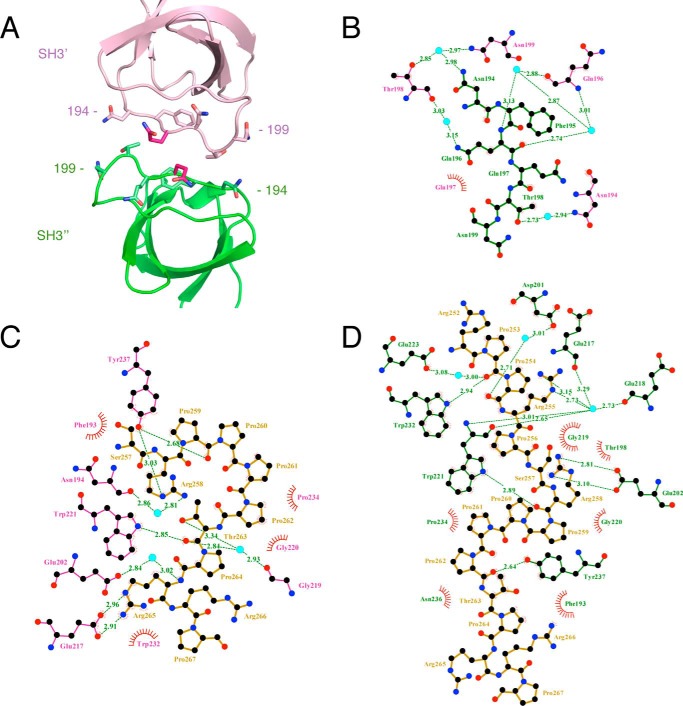
We have shown that different SH3 domain-containing proteins target overlapping sequences within the Itch PRR. To further understand the difference in target recognition at the molecular level, we attempted to crystallize complexes formed by the Itch PRR (peptide 249–269) and the SH3 domains of endophilin, amphiphysin, pacsin, or β-PIX, but we only succeeded in obtaining crystals of the Itch PRR and β-PIX SH3 complex. These crystals belong to the P1 space group, diffract to 1.65 Å resolution (Table 2), and contain four copies of the β-PIX SH3 domain and two copies of the Itch PRR in their unit cell (Fig. 4A). The proteins arrange to form two “super-SH3” domains, which are atypical SH3-PRR associations in which a single PRR simultaneously engages two SH3 domains (12). Each of these super-SH3 complexes thus contains two molecules of β-PIX (hereafter referred to as SH3′ and SH3″; Figs. 4–10) and one Itch PRR. The two super-SH3 domains are very similar to each other, as evidenced by an RMSD of 0.7 Å for 138 matched Cα, and are related to each other by a 2-fold screw axis non-crystallographic symmetry (Fig. 4B). Within each super-SH3, the PRR peptide is sandwiched between two β-PIX SH3 domains that are related to each other by 2-fold non-crystallographic symmetry. This super-SH3 arrangement is closely related to the one observed for cortactin in complex with an AMAP1 peptide (13) that also exhibits a 2-fold non-crystallographic symmetry between the cortactin SH3 domains (Fig. 5). The super-SH3 arrangement of β-PIX is stabilized by hydrophobic contacts and a rich network of water-mediated bonds contributed by the RT loops of the symmetry-related SH3 domains (Fig. 6, A and B). This water network is stabilized at one end by Arg-258 from the Itch PRR, which also interacts with SH3″ Glu-202 and SH3′ Tyr-237 (Figs. 6 (C and D) and 7).
Table 2.
Crystallographic data collection and refinement statistics
Values in parentheses are for the highest-resolution shell.
| Data collection | |
| Beamline | X25, NSLS |
| Wavelength (Å) | 1.1 |
| Space group | P1 |
| Unit-cell parameters | |
| a, b, c (Å) | 28.85, 43.45, 61.48, |
| α,β,γ (degrees) | 90.37, 101.01, 105.25 |
| Resolution (Å) | 30.12–1.55 (1.605–1.55) |
| No. of unique reflections | 32,525 (4639) |
| Multiplicity | 1.8 (1.8) |
| Completeness (%) | 95.7 (93.2) |
| Rmerge | 0.021 (0.069) |
| I/σ(I) | 18.2 (6.9) |
| Refinement statistics | |
| Resolution (Å) | 50–1.65 (1.71–1.65) |
| Reflections (total/test)a | 32,519/1390 |
| Rwork/Rfree (%) | 13.89/16.13 (14.15/18.16) |
| No. of atoms (excluding hydrogens) | |
| Protein | 2321 |
| Water | 383 |
| B factors | 19.10 |
| Protein | 17.94 |
| Water | 26.11 |
| RMSDs | |
| Bond lengths (Å) | 0.004 |
| Bond angles (degrees) | 0.71 |
| Ramachandranb | |
| Favored (%) | 98.9 |
| Allowed (%) | 1.1 |
| Outliers (%) | 0 |
| Clashscore | 2.20 |
a Reflections with F0 > 0.
b MolProbity analysis.
Figure 4.
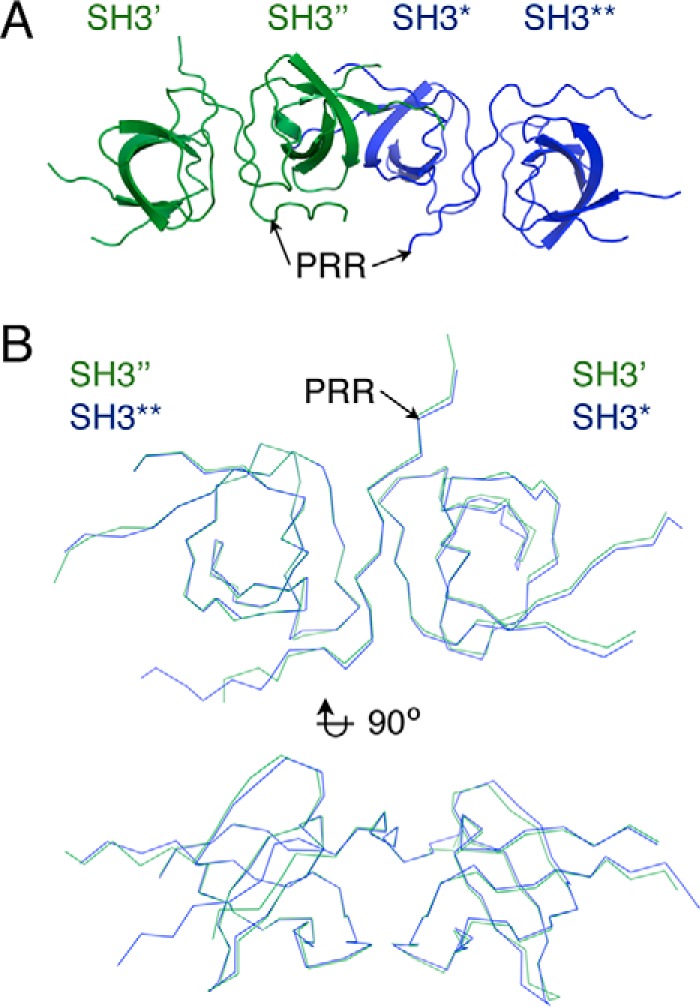
Representation and comparison of the two β-PIX·Itch PRR super-SH3 complexes found in the crystal lattice (PDB code 5SXP). A, schematic representation of the two super-SH3 complexes in the crystal lattice. B, superposition of the two β-PIX·Itch PRR complexes (ribbon).
Figure 5.
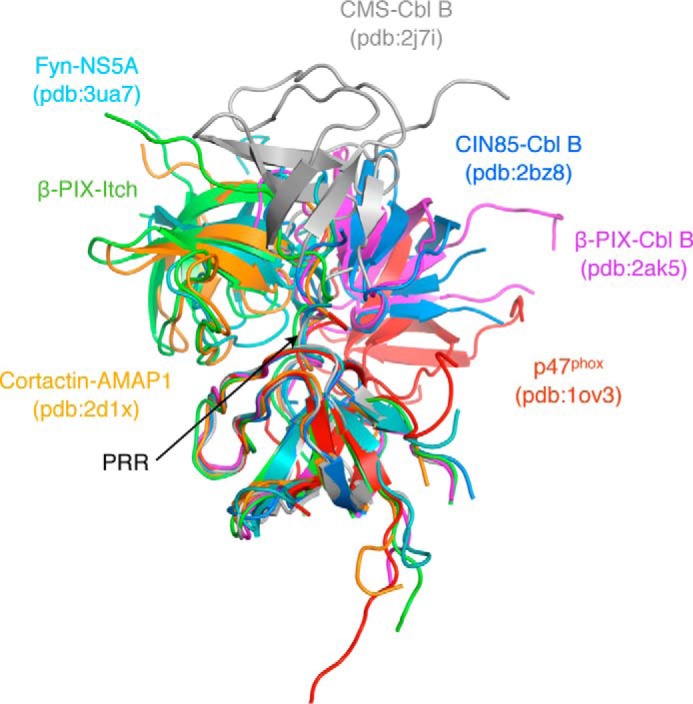
Comparison of the β-PIX·Itch PRR super-SH3 complex with other known complexes. Various SH3 structures were aligned to the β-PIX SH3′ domain to reveal the position of the second SH3 domain according to other known complexes. The structural alignment and schematic representation were done using PyMOL (Schrödinger) (12, 13, 21, 22, 46).
Figure 6.
LigPlot+ representation of the SH3-SH3 domain contacts and PRR-SH3 interactions within the super-SH3 complex β-PIX·Itch PRR (PDB code 5SXP). A, schematic representation of the β-PIX SH3 domains facing each other as in the super-SH3 structure with residues 194–199 of the SH3′ represented as purple sticks and their corresponding SH3″ residues in green. B, LigPlot+ analysis of the SH3′-SH3″ interactions (35, 36). Hydrophobic contacts are represented by red half-circles. Cyan spheres represent water molecules establishing a water-mediated bond between both SH3 domains. Hydrogen bonds and their length are displayed in green. Residues of the SH3′ and SH3″ domains are labeled in pink and green, respectively. Interactions between the Itch PRR (dark yellow) and β-PIX SH3′ (C) or SH3″ (D) were generated by Ligplot+ as in B.
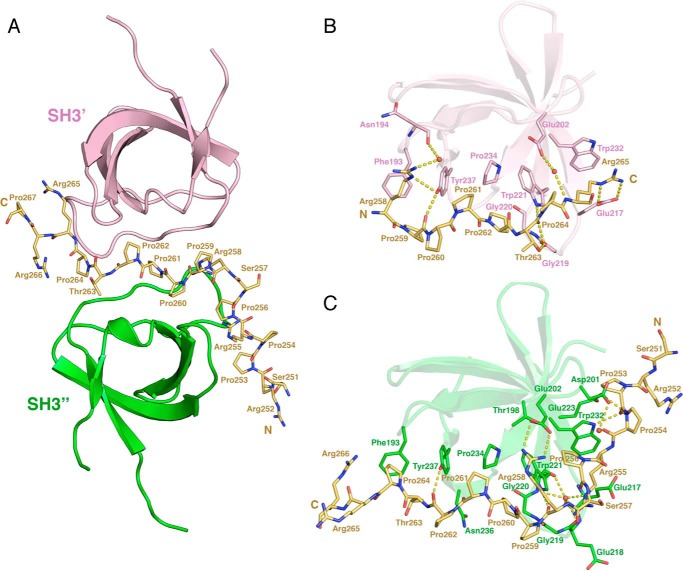
Figure 7.
Crystal structure of the super-SH3 interaction between the PRR of Itch and two β-PIX SH3 domains (PDB code 5SXP). A, schematic representation of the β-PIX SH3 domains. SH3′ and SH3″ correspond to chain D and B, respectively, present in the crystal structure. The PRR-containing peptide is represented as yellow sticks. B and C, detailed view of each SH3 domain interacting with the Itch PRR. SH3 residues contributing to the binding or Itch PRR were identified with LigPlot+ and are represented as sticks (35, 36). Water molecules contributing to the binding of the Itch PRR to the SH3 domains of β-PIX are represented as red spheres, and hydrogen bonds are shown as a dashed yellow line.
Figure 8.
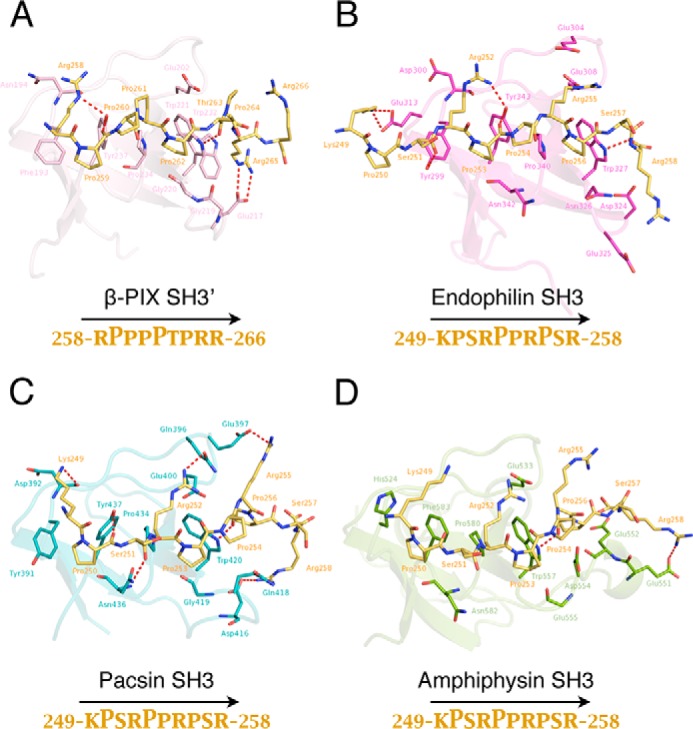
Modeling of the interactions of Itch PRR with different SH3 domains. The FlexPepDock protocol was performed using the structure of the β-PIX SH3′ in complex with Itch PRR (PDB code 5SXP) (A) (14, 15). In this complex, the prolines are oriented as a class II ligand and were used as a template to align the various binding sequences determined in our pulldown assays. The binding interface between Itch PRR and these SH3 domains was modeled as detailed under “Experimental procedures.” The complex formed between Itch PRR and β-PIX SH3′ (A) is shown here as a comparison with the models obtained (B–D). The sequence KPSRPPRPSR (peptide 249–258) of the ubiquitin ligase Itch (green sticks, darker arginines) interacts with the SH3 domains of endophilin (B), pacsin (C), and amphiphysin (D). The structures depict a schematic representation of the SH3 domains, with residues establishing either hydrophobic contacts or hydrogen bonds (dashed red line) displayed as sticks. Yellow sticks represent the Itch PRR. The interacting residues were defined using LigPlot+ (35, 36). The peptide orientation as well as prolines forming the core PXXP motifs are identified below the model.
Figure 9.
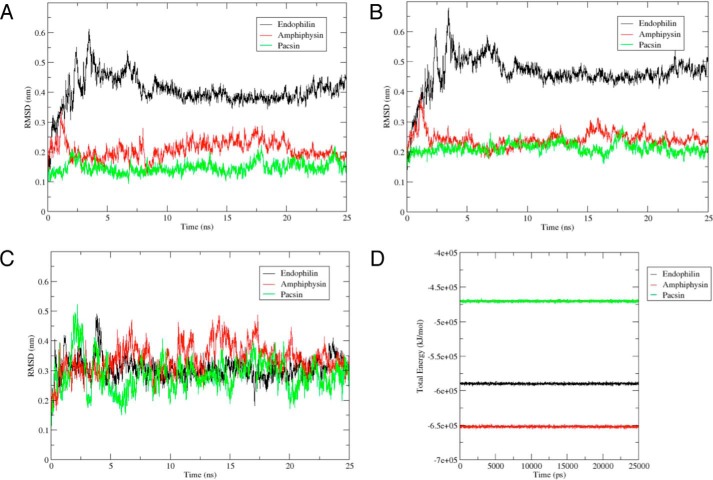
A–C, RMSDs as a function of simulation time for the three complexes (A), the three SH3 domains (B), and the three PRRs (C). The duration of each production run was 25 ns. The backbone of the complex was selected for both least squares fit and the group for RMSD calculations. RMSD shown is relative to the initial complex present in the equilibrated and minimized system. Represented in black, green, and red are the RMSD calculations for the PRR·endophilin complex, PRR·pacsin complex, and PRR·amphiphysin complex, respectively. D, total energy of the three complexes during the duration of the MD runs. The duration of each production run was 25 ns. Represented in black, green, and red are the total energy calculations for the PRR·endophilin complex, PRR·pacsin complex, and PRR·amphiphysin complex, respectively.
Figure 10.
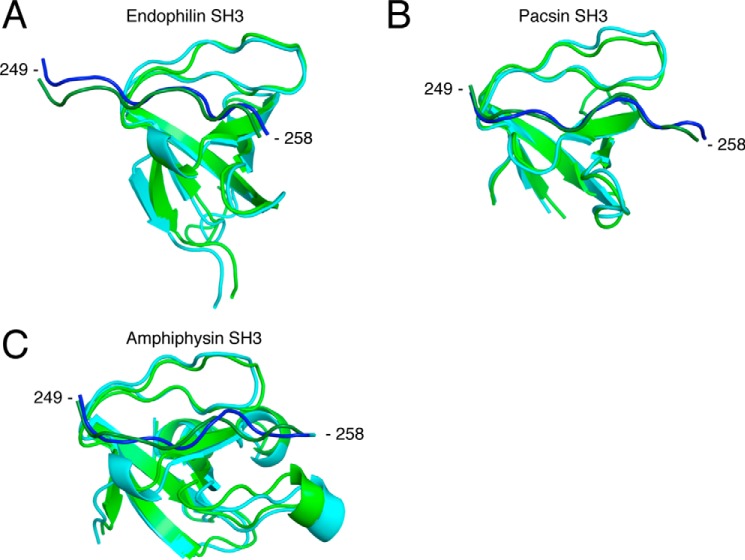
Structural alignment between the initial complex and the frame corresponding to the centroid from clustered frames post-MD simulations for the endophilin complex (A), for the pacsin complex (B), and for the amphiphysin complex (C). The RMSD cut-off for two structures to be considered neighbors in the GROMOS clustering algorithm was 0.20, 0.17, and 0.13 nm, respectively, for each complex. Shown is a representation of the initial complexes modeled from the Itch PRR (dark green) in complex with β-PIX-SH3′ (green). Blue, frame corresponding to the cluster centroid. Structural alignment and schematic representation were done using PyMOL (Schrödinger) (12, 13, 21, 22, 46).
The Itch PRR interacts as an atypical class II ligand with SH3′ by burying 374–384 Å2 of accessible surface area, whereas it interacts as a class I ligand with SH3″ by burying 615–654 Å2 of accessible surface area. This particular arrangement is enabled by a pseudo-palindrome in the recognition sequence: 258RPPP*PTPR265, where the asterisk denotes the center of the palindrome that coincides with the 2-fold symmetry center of the two SH3 domains. The conformation of the two Itch PRR and SH3″ complexes present in the unit cell is very similar to a previously reported 1:1 complex (9) with RMSDs of 0.4 and 0.7 Å for 66 matched Cα. As observed previously, the face of SH3″ containing the key aromatic/hydrophobic residues binds the RXXPXXP sequence from residue 258 to 264, and an additional face of the SH3″ extends the binding interface to accommodate the 252–257 sequence N-terminal to the class I ligand (9). Strikingly, despite also having reported a 1:2 Itch PRR/β-PIX stoichiometry in vitro, the structure presented by Janz et al. (9) does not adopt a super-SH3 arrangement, and it notably lacks the interaction between PRR and SH3′. In the structure presented here, the face of SH3′ containing the key aromatic/hydrophobic residues binds a PXXPXXR sequence (residues 259–265). The prolines of this motif are exposed and oriented as a class II ligand. However, Arg-265 is positioned one amino acid farther than the canonical motif and interacts with a residue of the n-Src loop (Glu-217) instead of a classical interaction established with the RT loop (Figs. 6 (C and D) and 7). In contrast to what is observed with the SH3″ domain, the second face of the SH3′ is not involved in the binding of Itch PRR. The absence of an extended binding interface in the case of SH3′ is the main reason for the difference in buried accessible area between the SH3′ and SH3″ domains upon binding to the Itch PRR. Taken together, these results suggest that a pseudo-palindromic sequence in Itch PRR allows for the recognition of two β-PIX molecules in a symmetric super-SH3 configuration.
Modeling of Itch PRR interaction with SH3 domains
Because we were unable to grow crystals with the other SH3 domains, we used the structure of the β-PIX complex to model their binding to the Itch PRR. For those modeling studies, we used the FlexPepDock protocol implemented with Rosetta (14, 15). The models obtained depict a potential structure for the Itch PRR in complex with each of these SH3 domains according to the binding preferences determined in the pulldown experiments (Fig. 8). The SH3 domain of endophilin would interact with the prolines 253 and 256, whereas pacsin and amphiphysin would target prolines 250 and 253. These complexes all require the participation of the first three arginines of the Itch PRR contacting the main interface of the SH3 domain. According to these models and the pulldown assays, the SH3 domain of endophilin was the only one that targets the first canonical class II binding site, whereas pacsin and amphiphysin bind to the second site (Fig. 8). Depending on its position and orientation, the PRR of Itch can thus accommodate SH3 domains in a variety of conformations, where it forms similar yet distinct molecular complexes.
The three complexes modeled from the Itch PRR in complex with β-PIX-SH3′ were each submitted to 25-ns production runs. All three systems stabilized after 5 ns (Fig. 9A). The complex with endophilin reached a higher RMSD of 4.03 Å, amphiphysin reached an RMSD of 2.08 Å, and pacsin reached a lower RMSD of 1.49 Å. The high RMSD observed in the endophilin complex can be explained by the fluctuations of the Itch peptide bound to the protein during the simulations (Fig. 9C) and movements in the C terminus end of the protein (Figs. 9B and 10A), which is displaced compared with its initial coordinates. The lowest RMSD complex, pacsin, has a relatively close alignment with its pre-MD structure (Fig. 10B). Frames from each production run were clustered with the GROMOS method. The centroid of the most populated cluster was used as the representative structure. Total energy analysis of the three complexes shows high energetic stability throughout the duration of the simulations, each of the run reaching an energetic plateau and maintaining it. The stability observed in both the RMSD calculations and the total energy analysis reveal that the three complexes formed by Itch PRR and its respective protein model are stable.
Discussion
Proline-rich regions often consist of a succession of potential SH3-binding sites within a sequence that can cover >100 amino acids. These highly flexible binding modules enable the interaction with multiple SH3 domain-containing proteins, which are often found in large protein complexes established during endocytosis and cell signaling. The endocytic proteins dynamin and synaptojanin are classic examples of proteins that contain a complex PRR. Mapping their SH3-binding sequences revealed that some binding partners interact with overlapping motifs, whereas others were found to bind distinct sites separated by several residues. The ubiquitin ligase Itch shares some of the SH3-binding partners identified for dynamin and synaptojanin, including endophilin, pacsin, amphiphysin, and Grb2 (16–19), but it forms an interaction using a single compact PRR that contains three overlapping class II motifs and one class I motif. A similar PRR organization can be found in the srGAP3 protein, which contains a short peptide encoding an overlapping class I and II motif, and a single PXXP motif within the peptide was found to be sufficient for its interaction with several SH3 domain-containing proteins (20). Interestingly, the key binding partners of srGAP3 identified by mass spectrometry were the same as those found here for the Itch PRR, although their PRRs differ considerably in terms of potential binding sites. ITC analysis further revealed that these common binding partners display weaker affinities toward srGAP3 than for the Itch PRR (8, 20). In addition to residues forming the canonical SH3-binding sites, adjacent amino acids are important for additional interactions with the surface of these SH3 domains and thereby confer binding specificity among PRRs and their ability to form high-affinity complexes.
We have mapped the sequence preferences of several binding partners of Itch by pulldown using PRR peptides in combination with mutational analysis. The results demonstrate that these SH3 domain-containing partners bind to distinct yet overlapping sequences within the PRR of Itch. This is consistent with previous results demonstrating that endophilin and β-PIX compete for the binding of the Itch PRR. Interestingly, β-PIX requires a more extended sequence compared with other binding partners. In conjunction with the 1:2 stoichiometry of this complex, this could reflect the need of docking two SH3 domains to the PRR to allow for the formation of a super-SH3 structure.
The structure of the super-SH3 complex formed by the simultaneous binding of two molecules of the SH3 domain of β-PIX to the PRR of Itch reveals interactions with a core symmetrical RPXPPXPR motif (residues 258–265). Residues preceding or following this motif also stabilized the complex by forming contacts with an additional face of these SH3 domains. CIN85 and β-PIX have also been previously shown to form a 2:1 protein complex with a similar pseudo-symmetrical proline-arginine motif within the PRR of Cbl (21, 22). These proteins and others shared a common binding mechanism, which involves a central positively charged residue simultaneously contacting both SH3 domains (21, 22).
This positively charged residue contacting both SH3 domains is absent from the complex between the Itch PRR and β-PIX. The molecular determinants that drive the formation of super-SH3 complexes remain poorly understood, but it appears that the exact sequence of the PRR contributes to these unusual stoichiometries as well as specific residues from the SH3 domains. It has been postulated that a GWW motif within the n-Src loop of the SH3 domain was crucial to form the binding interface for super-SH3 complexes. This motif is found in β-PIX and other SH3 domain-containing proteins that form super-SH3 complexes. However, this motif does not always form SH3-SH3 interactions (12, 13, 21, 22). β-PIX was also found to interact at a 2:1 stoichiometry with the PRR of Cbl and Itch, but it forms a 1:1 complex with the PRR of PAK (8, 9, 21, 23). Thus, it is clear that super-SH3 interactions are complex and rely on a combination of specific residues from both the SH3 domain and the PRR.
The crystal structure presented here depicts contacts between the two SH3 domains that occur exclusively between the RT loops (Fig. 6, A and B). This conformation is similar to what was observed in the AMAP1·cortactin structure (Fig. 5). In both complexes, there is a similar SH3 dimer interface formed by a key glutamine residue (Gln-196 of β-PIX and Gln-504 of cortactin SH3) (Fig. 6, A and B). In addition to cortactin, DBNL also has a glutamine at the same position in its RT loop and was found to interact with Itch in our mass spectrometry analysis (Table 1). CIN85 is another known binding partner for the Itch PRR that contains this glutamine residue (9). Although the stoichiometry for these specific complexes remains to be determined, we can speculate that these proteins could form similar super-SH3 complexes with the PRR of Itch. However, there is diversity within super-SH3 structures, and we cannot exclude a possible dimer formation involving the participation of novel SH3 residues or resembling other known super-SH3 structures (Fig. 5).
The interaction between the Itch PRR and β-PIX SH3″ also reveals similarities to the interaction between the E. coli secreted protein EspFu and the SH3 domain of the insulin receptor tyrosine kinase substrate (IRTKS) (24). In the EspFu·IRTKS complex, recognition of the PRR by the SH3 domain of IRTKS requires the binding of two PXXP motifs in tandem, which results in formation of an extensive binding surface. The PRR of Itch exhibits a similar L shape exposing tandem PXXP motifs on two faces of the SH3 domain. Unlike IRTKS, the β-PIX SH3″ lacks the hydrophobic pocket to accommodate the fourth proline. This would account for the great discrepancy observed in the respective affinity of these complexes, because an additional IP motif within EspFu was found to mediate high-affinity binding toward this pocket with a reported KD of 500 nm. In comparison, the PRR of Itch interacts as an extended class I ligand with a KD of 1.6 μm toward β-PIX.
Despite the fact that Itch PRR makes extensive hydrophobic contacts, salt bridges, and hydrogen bonds with both SH3 domains of β-PIX, its affinity is still similar to most of the other SH3 domains tested. Among the SH3 domain-containing proteins tested, only endophilin was able to establish a high affinity interaction with the PRR of Itch. This is consistent with our findings indicating that endophilin is a major binding partner of Itch (Table 1). Itch was found to ubiquitylate endophilin and target it for proteasomal degradation, thereby impacting signaling by the EGF receptor (5, 8, 25).
Intriguingly, endophilin was the only SH3 domain-containing protein targeting the first PXXP motif of the Itch PRR. Assuming that the PRR adopts the same conformation in complex with endophilin as it does with β-PIX, this would place the turn of the L-shaped peptide in a proper conformation to allow another face of the SH3 domain to contact additional motifs in the peptide. In the absence of crystallographic data for the complex formed between Itch and endophilin, we can only speculate that extensive rearrangements of a longer PRR sequence and/or the SH3 domain could favor a more ideal fit with endophilin. Thus, the specific architecture of the Itch PRR can accommodate the formation of a variety of protein complexes. In addition to β-PIX, other SH3 domain-containing proteins may also dimerize on the Itch PRR. We indeed isolated the Grb2 protein in our proteomics studies. Grb2 contains two distinct SH3 domains that could bind in tandem to the PRR of Itch, and this tandem binding would resemble a super-SH3-like complex.
Interestingly, in contrast to the characterized SH3 domain-containing partners of Itch, there is still no evidence that β-PIX is ubiquitylated (8). This suggests that super-SH3 complexes with Itch serve an alternative biological function that might be independent of a classical ubiquitylation process.
Experimental procedures
Cell culture and transfection
HEK-293T cells were obtained from the ATCC and were maintained at 37 °C with 5% CO2 in high-glucose DMEM (Gibco) supplemented with 10% cosmic calf serum (HyClone), penicillin (Invitrogen; 100 units/ml), and streptomycin (Invitrogen; 100 mg/ml). Cells were transfected with the indicated plasmids using calcium/phosphate precipitates (26). Typically, cells were transfected using 5 μg of plasmid/55 cm2.
Plasmids
The plasmids encoding amphiphysin II, GFP-pacsin 1, and GFP-endophilin A1 were a kind gift from P. S. McPherson (Montreal Neurological Institute, McGill University). GFP and FLAG-tagged β-PIX were generously provided by A. Weiss (Howard Hughes Medical Institute, University of California) and J. P. Fawcett (Dalhousie University). We have produced the various PRR sub-sequences by annealing oligonucleotides (Thermo Fisher) coding the indicated amino acids with an additional tyrosine at the C terminus of the peptide to facilitate protein quantification by UV spectroscopy (absorbance at 280 nm) and introduced these sequences in the pGEX-4T1 vector. Another GST-PRR (peptide 249–269) construct was created by the insertion of the PRR coding sequence preceded by three glycines in a modified pGEX-4T1 vector, where the thrombin cutting site was replaced with a TEV cutting site. This construct was used for large-scale protein purification (for ITC and crystallography assays) along with GST-fused constructs expressing endophilin, pacsin, amphiphysin, or β-PIX SH3s. These plasmids were obtained by PCR amplification and subcloning into the pGEX-4T1 TEV modified vector as described previously (8). We have further included an annealed sequence coding three copies of the FLAG epitope in the BamHI restriction site of the TEV modified GST-PRR (peptide 249–269). The FLAG-tagged construct behaved similarly in pulldown experiments and was used to introduce point mutations to change arginine to glutamic acids in Itch PRR (27).
Antibodies
Polyclonal antibodies against pacsin 1 were purchased from Genetex (GTX103078). Polyclonal antibodies against pan-endophilin and the GFP tag were bought from Thermo Fisher (36-3400, A-11122). Anti-amphiphysin I/II monoclonal antibodies were purchased from Santa Cruz Biotechnology, Inc. (SC-58227). Monoclonal antibodies against the FLAG epitope were purchased from Sigma-Aldrich (F3165). Goat anti-rabbit-HRP and goat anti-mouse HRP IgG were from Jackson ImmunoResearch Laboratories (111-035-003 and 115-035-003).
Pulldown experiments
Transfected HEK-293T cells were washed in PBS and resuspended in buffer A (20 mm Hepes, pH 7.4, 150 mm NaCl) containing protease inhibitors. The cells were lysed by sonication, and Triton X-100 was added to a final concentration of 1%. For rat brain extracts (Pel-Freez Biologicals), tissue was homogenized in buffer A and centrifuged at 1000 rpm for 10 min before Triton X-100 was added to the resulting supernatant. Extracts were incubated for 20 min at 4 °C and centrifuged at 45,000 rpm at 4 °C. Extracts were incubated with 10 μg of the appropriate GST-fusion protein coupled to GSH Sepharose 4B (Bio-World) for 16 h at 4 °C. Beads were washed extensively in the same buffer and prepared for Western blotting analysis. To normalize the quantity of GST-fusion proteins used in each assay, purified beads were run on 10% SDS-PAGE along with a standard curve of BSA. The gel was stained with Coomassie, and densitometry analysis allowed the determination of the volume of beads needed to obtain the desired amount of GST-fusion protein.
Western blotting analysis
Protein extracts and purified proteins obtained by pulldown assays were separated by SDS-PAGE. Proteins were then transferred to nitrocellulose for blotting with the appropriate primary and secondary antibodies. 0.1 μg/ml of goat anti-rabbit-HRP or goat anti-mouse-HRP conjugated IgG were used (Jackson ImmunoResearch Laboratories). Antibody incubation and membrane washings were performed in PBS supplemented with 5% dry milk and 0.05% Tween 20. Immunoreactivity was detected by chemiluminescence using West-Pico SuperSignal (Thermo Fisher Scientific).
Expression and purification of proteins
The SH3 domains and PRR peptides were expressed as GST-fusion proteins in E. coli host strain TOPP2 (Stratagene). The cells were grown at 37°C in Luria Broth medium, and protein expression was induced for 4 h at 30 °C with 0.7 mm isopropyl-β-d-thiogalactopyranoside (Inalco). The cells were harvested by centrifugation, resuspended in lysis buffer (20 mm Tris-HCl, pH 7.4, 1 m NaCl, 0.2 mm EDTA, and 1 mm DTT), lysed by passage through a French press, and centrifuged at 105,000 × g for 1 h at 4°C. The supernatant was then collected and incubated for 1 h with GSH Sepharose 4B resin (GE Healthcare) at 4 °C. Following incubation, the resin was collected by centrifugation and washed with lysis buffer and TEV buffer (25 mm Na2HPO4, 125 mm NaCl, and 5 mm DTT). The GST tag was cleaved by incubating the resin for 2 h with 100 units of TEV protease. The proteins were eluted by extensive washes in TEV buffer. The SH3 domains were further purified using Q-Sepharose High Performance (GE Healthcare). PRR peptides were further purified over a C4-reverse phase HPLC column (Vydac). Proteins and peptides were desalted, quantified by absorbance at 280 nm, flash-frozen, lyophilized, and kept at −80°C until they were processed for ITC experiments and crystallography.
Isothermal titration calorimetry studies
ITC titrations were performed at 25 °C in 20 mm phosphate buffer at pH 7.4 using a MicroCal VP-ITC system. Concentrations of injected PRR peptides in the syringe and SH3 domains in the cell varied from 150 to 350 μm and from 15 to 35 μm, respectively, keeping a molar ratio of about 10:1 between the syringe and the cell. Data were analyzed using MicroCal Origin Software, and all experiments fit the single binding site model with a 1:1 stoichiometry. Errors in KD values were estimated from duplicate measurements.
Mass spectrometry
GST pulldowns with rat brain extracts (Pel-Freez Biologicals) were performed using GST alone, GST-PRR (residues 224–276), or GST-FLAG-PRR (residues 249–269). Fusion proteins alone were loaded as a control, and proteins were separated by SDS-PAGE. The resulting gels were stained with Imperial protein stain (Thermo Fisher), and gel sections corresponding to proteins above or below GST-fusion proteins (upper or lower gels) were processed with in-gel tryptic digest and subsequent nano-LC-MS/MS analysis by the proteomics platform of the Institute for Research in Immunology and Cancer (Montreal, Canada). Data analysis was performed using the Global Proteome Machine to display SH3 domain-containing proteins identified in the screening (28, 29).
Crystallography
Lyophilized PRR peptide and SH3 domains were suspended in water at a final concentration of 4.8 and 4 mm and mixed to a final 1.7:1 molar ratio. Crystals were obtained for β-PIX SH3 at 20 °C using the vapor diffusion method with a hanging drop containing an equal volume of protein complex and well solution (100 mm MIB buffer, pH 5.0, and 25% PEG 1500). Crystals were mounted in a loop and flash-cooled in a stream of nitrogen gas at 100 K. Diffraction data were collected using a Pilatus 6M detector at beamline X25 at the National Synchrotron Light Source of the Brookhaven National Laboratory. The data set was indexed and integrated using XDS and scaled with XSCALE and the Diffraction Anisotropy Server (UCLA) (30, 31). Due to diffraction anisotropy, the resolution was cut to 1.55 Å to maintain decent completeness in the high-resolution bins. Molecular replacement was performed with Phenix using the crystal structure of AIP4 and β-PIX (PDB code 2P4R) as a search template. Model building was performed in Coot, refined with Phenix, and validated using MolProbity (32–34).
Modeling
The structure of the Itch PRR in complex with β-PIX-SH3′ obtained in the crystallography study above served as an initial template. This structure depicts an atypical class II interaction that involves the participation of prolines 259 and 262. A copy of the PRR peptide was then oriented to this complex, and prolines 253 and 256 were aligned to create the class II motif in the PRR·β-PIX complex. We then replaced the SH3 domain of β-PIX by the SH3 domain of endophilin (PDB code 3IQL) and removed the original PRR·β-PIX to obtain a chimeric PRR·endophilin complex. The same approach was used for pacsin (PDB code 2X3X) and amphiphysin (PDB code 1BB9) as well, except that the prolines 250 and 253 were instead aligned to the class II motif of the PRR·β-PIX complex in agreement with the results obtained in the pulldown studies. The resulting chimeric structures were then used as a template for the high-resolution modeling protocol FlexPepDock, implemented with the Rosetta framework (14, 15). FlexPepDock produced 300 low-resolution and 300 high-resolution structures. The resulting models had a peptide backbone RMSD that ranged between 2.5 and 4 Å when overlapped with the initial chimeric structure, showing that the flexible docking procedure was not only exploring local minima. The representation of the best model according to the FlexPepDock score was performed with PyMOL for the endophilin SH3 in complex with Itch PRR (Schrödinger). For pacsin and amphiphysin, we selected the third models for representation because they displayed a combination of properly docked prolines, optimized interactions, and best protein geometry according to MolProbity analyses (33). Residues establishing a hydrogen bond or a hydrophobic contact were identified using LigPlot+ (35, 36). The surface hydrophobicity color script used for representation was generously provided by H. A. Steinberg (Artforscience).
MD simulations
MD simulations and system equilibration were performed with the GROMACS version 5.0.5 molecular simulation package (37), using the GROMOS96 54a7 force field (38). The models of the SH3 domain·PRR peptide complexes obtained from FlexPepDock were centered in a cubic box. The periodic water box edges were extended at least 20 Å away from the complex to allow it to move freely during the production runs. The SPC216 water model (39) was used to solvate the system and add water molecules to the box. Chloride ions were added to neutralize the net charge of the solvated system. All three systems were submitted to 5000 steps of energy minimization using a steepest descent method to remove initial steric clashes and ensure appropriate geometry. The particle-mesh Ewald algorithm (40) was used in all calculations to consider electrostatic interactions with grid spacing around 1 Å, and van der Waals forces were considered with a cut-off distance of 1.4 Å. Each system was subjected to two rounds of equilibration after minimization. They were first gradually heated in a NVT ensemble from 0 to 300 K over 100 ps using the V-rescale coupling algorithm (41) with position restrains on the protein and the ligand. This was followed by NPT equilibration of 100 ps to reach the reference pressure of 1 atm by using the Parrinello-Rahman coupling algorithm (42) with isotropic coupling and with position restrains on the protein and the ligand. The LINCS algorithm was used to constrain all bonds involving hydrogen atoms (43). Each system ran for a 25-ns MD simulation under the same conditions as the equilibration procedures with a time step of 2 fs but without any position restrains. Computations were made on the supercomputer Guillimin from McGill University, managed by Calcul Québec and Compute Canada. Production runs requested 36 cores. The analysis for the trajectories was carried out using the standard software tools provided by the GROMACS package, and visualization was performed with VMD (44) and PyMOL (Schrödinger).
Author contributions
A. A. conceived all experiments, except for ITC and crystallography, which were conceived by J. G. O.; G. D. collected the data presented in this paper; L. C. analyzed the crystal structure; M. L.-P. gave extensive technical assistance for the large-scale protein purifications and ITC experiments; MD experiments were performed and analyzed by A.-T. T. and A. S.; R. A. contributed to molecular cloning; G. D., L. C., J. G. O., and A. A. collectively analyzed the data and wrote the paper.
This work was supported by Natural Sciences and Engineering Research Council of Canada (NSERC) Grant 288238 (to A. A.). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (code 5SXP) have been deposited in the Protein Data Bank (http://wwpdb.org/).
G. Desrochers, M. Lussier-Price, J. G. Omichinski, and A. Angers, unpublished observations.
- CWH
- C2-WW-HECT
- IRTKS
- insulin receptor tyrosine kinase substrate
- ITC
- isothermal titration calorimetry
- PRR
- proline-rich region
- SH3
- Src homology 3
- RMSD
- root mean square deviation
- MD
- molecular dynamics
- TEV
- tobacco etch virus
- PDB
- Protein Data Bank.
References
- 1. Rotin D., and Kumar S. (2009) Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 10, 398–409 [DOI] [PubMed] [Google Scholar]
- 2. Gay D. L., Ramón H., and Oliver P. M. (2008) Cbl- and Nedd4-family ubiquitin ligases: balancing tolerance and immunity. Immunol. Res. 42, 51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kay B. K., Williamson M. P., and Sudol M. (2000) The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 14, 231–241 [PubMed] [Google Scholar]
- 4. Sudol M., and Hunter T. (2000) New wrinkles for an old domain. Cell 103, 1001–1004 [DOI] [PubMed] [Google Scholar]
- 5. Angers A., Ramjaun A. R., and McPherson P. S. (2004) The HECT domain ligase itch ubiquitinates endophilin and localizes to the trans-Golgi network and endosomal system. J. Biol. Chem. 279, 11471–11479 [DOI] [PubMed] [Google Scholar]
- 6. Desrochers G., Corbeil L., and Angers A. (2013) From a conserved structure to regulation: CWH ubiquitin ligases tightly regulate key cellular events. in Advances in Medicine and Biology, Vol. 62 (Berhardt L. V., ed) pp. 177–215, Nova Science Publishers, Hauppauge, NY [Google Scholar]
- 7. Baumann C., Lindholm C. K., Rimoldi D., and Lévy F. (2010) The E3 ubiquitin ligase itch regulates sorting nexin 9 through an unconventional substrate recognition domain. FEBS J. 277, 2803–2814 [DOI] [PubMed] [Google Scholar]
- 8. Desrochers G., Lussier-Price M., Omichinski J. G., and Angers A. (2015) Multiple src homology 3 binding to the ubiquitin ligase itch conserved proline-rich region. Biochemistry 54, 7345–7354 [DOI] [PubMed] [Google Scholar]
- 9. Janz J. M., Sakmar T. P., and Min K. C. (2007) A novel interaction between atrophin-interacting protein 4 and β-p21-activated kinase-interactive exchange factor is mediated by an SH3 domain. J. Biol. Chem. 282, 28893–28903 [DOI] [PubMed] [Google Scholar]
- 10. Malik R., Soh U. J., Trejo J., and Marchese A. (2012) Novel roles for the E3 ubiquitin ligase atrophin-interacting protein 4 and signal transduction adaptor molecule 1 in G protein-coupled receptor signaling. J. Biol. Chem. 287, 9013–9027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kelil A., Levy E. D., and Michnick S. W. (2016) Evolution of domain-peptide interactions to coadapt specificity and affinity to functional diversity. Proc. Natl. Acad. Sci. U.S.A. 113, E3862–E3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Groemping Y., Lapouge K., Smerdon S. J., and Rittinger K. (2003) Molecular basis of phosphorylation-induced activation of the NADPH oxidase. Cell 113, 343–355 [DOI] [PubMed] [Google Scholar]
- 13. Hashimoto S., Hirose M., Hashimoto A., Morishige M., Yamada A., Hosaka H., Akagi K., Ogawa E., Oneyama C., Agatsuma T., Okada M., Kobayashi H., Wada H., Nakano H., Ikegami T., et al. (2006) Targeting AMAP1 and cortactin binding bearing an atypical src homology 3/proline interface for prevention of breast cancer invasion and metastasis. Proc. Natl. Acad. Sci. U.S.A. 103, 7036–7041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. London N., Raveh B., Cohen E., Fathi G., and Schueler-Furman O. (2011) Rosetta flexpepdock web server: high resolution modeling of peptide-protein interactions. Nucleic Acids Res. 39, W249–W253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Raveh B., London N., and Schueler-Furman O. (2010) Sub-angstrom modeling of complexes between flexible peptides and globular proteins. Proteins 78, 2029–2040 [DOI] [PubMed] [Google Scholar]
- 16. Anggono V., and Robinson P. J. (2007) Syndapin I and endophilin I bind overlapping proline-rich regions of dynamin I: role in synaptic vesicle endocytosis. J. Neurochem. 102, 931–943 [DOI] [PubMed] [Google Scholar]
- 17. Cestra G., Castagnoli L., Dente L., Minenkova O., Petrelli A., Migone N., Hoffmüller U., Schneider-Mergener J., and Cesareni G. (1999) The SH3 domains of endophilin and amphiphysin bind to the proline-rich region of synaptojanin 1 at distinct sites that display an unconventional binding specificity. J. Biol. Chem. 274, 32001–32007 [DOI] [PubMed] [Google Scholar]
- 18. Solomaha E., Szeto F. L., Yousef M. A., and Palfrey H. C. (2005) Kinetics of Src homology 3 domain association with the proline-rich domain of dynamins: specificity, occlusion, and the effects of phosphorylation. J. Biol. Chem. 280, 23147–23156 [DOI] [PubMed] [Google Scholar]
- 19. Zucconi A., Dente L., Santonico E., Castagnoli L., and Cesareni G. (2001) Selection of ligands by panning of domain libraries displayed on phage λ reveals new potential partners of synaptojanin 1. J. Mol. Biol. 307, 1329–1339 [DOI] [PubMed] [Google Scholar]
- 20. Wuertenberger S., and Groemping Y. (2015) A single PXXP motif in the C-terminal region of srGAP3 mediates binding to multiple SH3 domains. FEBS Lett. 589, 1156–1163 [DOI] [PubMed] [Google Scholar]
- 21. Jozic D., Cárdenes N., Deribe Y. L., Moncalián G., Hoeller D., Groemping Y., Dikic I., Rittinger K., and Bravo J. (2005) Cbl promotes clustering of endocytic adaptor proteins. Nat. Struct. Mol. Biol. 12, 972–979 [DOI] [PubMed] [Google Scholar]
- 22. Moncalián G., Cárdenes N., Deribe Y. L., Spínola-Amilibia M., Dikic I., and Bravo J. (2006) Atypical polyproline recognition by the CMS N-terminal Src homology 3 domain. J. Biol. Chem. 281, 38845–38853 [DOI] [PubMed] [Google Scholar]
- 23. Hoelz A., Janz J. M., Lawrie S. D., Corwin B., Lee A., and Sakmar T. P. (2006) Crystal structure of the SH3 domain of betapix in complex with a high affinity peptide from pak2. J. Mol. Biol. 358, 509–522 [DOI] [PubMed] [Google Scholar]
- 24. Aitio O., Hellman M., Kazlauskas A., Vingadassalom D. F., Leong J. M., Saksela K., and Permi P. (2010) Recognition of tandem PxxP motifs as a unique Src homology 3-binding mode triggers pathogen-driven actin assembly. Proc. Natl. Acad. Sci. U.S.A. 107, 21743–21748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Azakir B. A., Desrochers G., and Angers A. (2010) The ubiquitin ligase Itch mediates the antiapoptotic activity of epidermal growth factor by promoting the ubiquitylation and degradation of the truncated C-terminal portion of Bid. FEBS J 277, 1319–1330 [DOI] [PubMed] [Google Scholar]
- 26. Kingston R. E., Chen C. A., and Rose J. K. (2003) Calcium phosphate transfection. Curr. Protoc. Mol. Biol. 10.1002/0471142727.mb0901s63 [DOI] [PubMed] [Google Scholar]
- 27. Blott E. J., Bossi G., Clark R., Zvelebil M., and Griffiths G. M. (2001) Fas ligand is targeted to secretory lysosomes via a proline-rich domain in its cytoplasmic tail. J. Cell Sci. 114, 2405–2416 [DOI] [PubMed] [Google Scholar]
- 28. Craig R., and Beavis R. C. (2004) Tandem: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 29. Fenyö D., Eriksson J., and Beavis R. (2010) Mass spectrometric protein identification using the global proteome machine. Methods Mol. Biol. 673, 189–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Strong M., Sawaya M. R., Wang S., Phillips M., Cascio D., and Eisenberg D. (2006) Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 103, 8060–8065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) Phenix: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Laskowski R. A., and Swindells M. B. (2011) Ligplot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 51, 2778–2786 [DOI] [PubMed] [Google Scholar]
- 36. Wallace A. C., Laskowski R. A., and Thornton J. M. (1995) Ligplot: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 8, 127–134 [DOI] [PubMed] [Google Scholar]
- 37. Abraham M. J., Murtola T., Schulz R., Páll S., Smith J. C., Hess B., and Lindahl E. (2015) Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 [Google Scholar]
- 38. Schmid N., Eichenberger A. P., Choutko A., Riniker S., Winger M., Mark A. E., and van Gunsteren W. F. (2011) Definition and testing of the GROMOS force-field versions 54a7 and 54b7. Eur. Biophys. J. 40, 843–856 [DOI] [PubMed] [Google Scholar]
- 39. Berendsen H. J. C., Grigera J. R., and Straatsma T. P. (1987) The missing term in effective pair potentials. J. Phys. Chem. 91, 6269–6271 [Google Scholar]
- 40. Essmann U., Perera L., Berkowitz M. L., Darden T., Lee H., and Pedersen L. G. (1995) A smooth particle mesh ewald method. J. Chem. Phys. 103, 8577–8593 [Google Scholar]
- 41. Bussi G., Donadio D., and Parrinello M. (2007) Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 [DOI] [PubMed] [Google Scholar]
- 42. Parrinello M., and Rahman A. (1981) Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 [Google Scholar]
- 43. Hess B., Bekker H., Berendsen H. J. C., Fraaije J. G. E. M. (1997) Lincs: A linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 [Google Scholar]
- 44. Humphrey W., Dalke A., and Schulten K. (1996) Vmd: visual molecular dynamics. J. Mol. Graph. 14, 33–38, 27–28 [DOI] [PubMed] [Google Scholar]
- 45. Di Tommaso P., Moretti S., Xenarios I., Orobitg M., Montanyola A., Chang J. M., Taly J. F., and Notredame C. (2011) T-coffee: a web server for the multiple sequence alignment of protein and rna sequences using structural information and homology extension. Nucleic Acids Res. 39, W13–W17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Martin-Garcia J. M., Luque I., Ruiz-Sanz J., and Camara-Artigas A. (2012) The promiscuous binding of the Fyn SH3 domain to a peptide from the ns5a protein. Acta Crystallogr. D Biol. Crystallogr. 68, 1030–1040 [DOI] [PubMed] [Google Scholar]