Abstract
In Drosophila, the final immune deficiency (IMD) pathway-dependent signal is transmitted through proteolytic conversion of the nuclear factor-κB (NF-κB)-like transcription factor Relish to the active N-terminal fragment Relish-N. Relish-N is then translocated from the cytosol into the nucleus for the expression of IMD-controlled genes. We previously demonstrated that transglutaminase (TG) suppresses the IMD pathway by polymerizing Relish-N to inhibit its nuclear translocation. Conversely, we also demonstrated that orally ingested synthetic amines, such as monodansylcadaverine (DCA) and biotin-labeled pentylamine, are TG-dependently incorporated into Relish-N, causing the nuclear translocation of modified Relish-N in gut epithelial cells. It remains unclear, however, whether polyamine-containing Relish-N retains transcriptional activity. Here, we used mass spectrometry analysis of a recombinant Relish-N modified with DCA by TG activity after proteolytic digestion and show that the DCA-modified Gln residues are located in the DNA-binding region of Relish-N. TG-catalyzed DCA incorporation inhibited binding of Relish-N to the Rel-responsive element in the NF-κB-binding DNA sequence. Subcellular fractionation of TG-expressing Drosophila S2 cells indicated that TG was localized in both the cytosol and nucleus. Of note, natural polyamines, including spermidine and spermine, competitively inhibited TG-dependent DCA incorporation into Relish-N. Moreover, in vivo experiments demonstrated that Relish-N was modified by spermine and that this modification reduced transcription of IMD pathway-controlled cecropin A1 and diptericin genes. These findings suggest that intracellular TG regulates Relish-N-mediated transcriptional activity by incorporating polyamines into Relish-N and via protein-protein cross-linking.
Keywords: DNA-binding protein, Drosophila, enzyme, innate immunity, NF-κB transcription factor, polyamine, protein cross-linking, transglutaminase
Introduction
Protein-protein cross-linking catalyzed by transglutaminase (TG)3 plays important and diverse roles in various physiologic phenomena, such as blood coagulation, skin barrier formation, and apoptosis (1). TG catalyzes the isopeptide formation of ϵ-(γ-glutamyl) lysine bonds in a Ca2+-dependent manner (2). TGs function both intracellularly and extracellularly, and many TG substrates have been identified in metazoans (3–5). The mammalian genome contains eight encoded isoenzymes with each member exhibiting diverse functions: factor XIIIa is essential for blood coagulation, and TG2 is involved in apoptosis, cell adhesion, and inflammatory responses (1). In invertebrates, such as the horseshoe crab Tachypleus tridentatus, the crayfish Pacifastacus leniusculus, and the fruit fly Drosophila melanogaster, TGs also exhibit pleiotropic functions, including hemolymph coagulation, cuticle formation, and immobilization of invading pathogens, that depend on TG-mediated cross-linking of specific proteins (6–9). Genome analysis of Drosophila identified a single TG gene encoding an 87-kDa protein that functions in both intracellular and extracellular spaces, although there is no authentic secretion signal sequence in the N terminus. Drosophila TG forms clots and traps invading pathogenic microbes (10, 11) and cross-links drosocrystallin on the peritrophic matrix, a semipermeable barrier structure in insects, to form a stabilized fiber structure against toxic proteases released by orally infected pathogenic bacteria (12). Moreover, RNAi directed against the TG gene in Drosophila leads to a pupal semilethal phenotype, abnormal morphology, and shorter lifespan compared with non-RNAi flies (8, 13).
Drosophila has two major immune signal pathways, the Toll and immune deficiency (IMD) pathways (14, 15). The Toll pathway is driven by yeast and Gram-positive bacteria, and the IMD pathway is driven by diaminopimelic acid-type peptidoglycans of Gram-negative bacteria or some strains of Gram-positive bacteria. The Toll and IMD pathways control the production of antimicrobial peptides by the activation of nuclear factor-κB (NF-κB)-like nuclear factors Dorsal/Dif and Relish (16–20), respectively. The transcriptional factor Relish, which is regulated by the IMD pathway, is endoproteolytically activated by Dredd, a homolog of mammalian caspase-8 (21). Upon stimulation with diaminopimelic acid-type peptidoglycans, Relish is converted to the N-terminal fragment (Relish-N) containing the Rel homology domain with transcriptional activity and the C-terminal fragment (Relish-C) containing an ankyrin-repeat domain that inhibits the nuclear translocation of Relish-N (Fig. 1A). We previously reported that the translocation of Relish-N from the cytosol into the nucleus is inhibited by TG-catalyzed protein-protein cross-linking in gut epithelial cells to promote immune tolerance against commensal bacteria, that full-length Relish must be proteolytically cleaved by Dredd before being cross-linked, and that the resulting Relish-N must be exposed by proteolytic conversion to become a suitable substrate for TG (13). Conversely, synthetic amines, such as monodansylcadaverine (DCA) and biotin-labeled pentylamine, ingested by flies are TG-dependently incorporated into Relish-N, which inhibits protein-protein cross-linking of Relish-N and leads to the nuclear translocation of Relish-N modified with the amines as well as native Relish-N (13). It remains unclear, however, whether the amine-incorporated Relish-N retains transcriptional activity in the nucleus. Natural polyamines, such as spermidine and spermine, are ubiquitously present in organisms and are involved in critically important intracellular functions, such as gene transcription, cellular growth, and differentiation (22–24). In the present study, we examined the effects of these polyamines on TG-dependent regulation of the transcriptional activity of a recombinant Relish-N (rRelish-N) expressed in Escherichia coli. Moreover, TG-dependent polyamine incorporation into native Relish-N and the effects of the modification on the transcription of antimicrobial peptide genes controlled by the IMD pathway were also examined in vivo.
Figure 1.
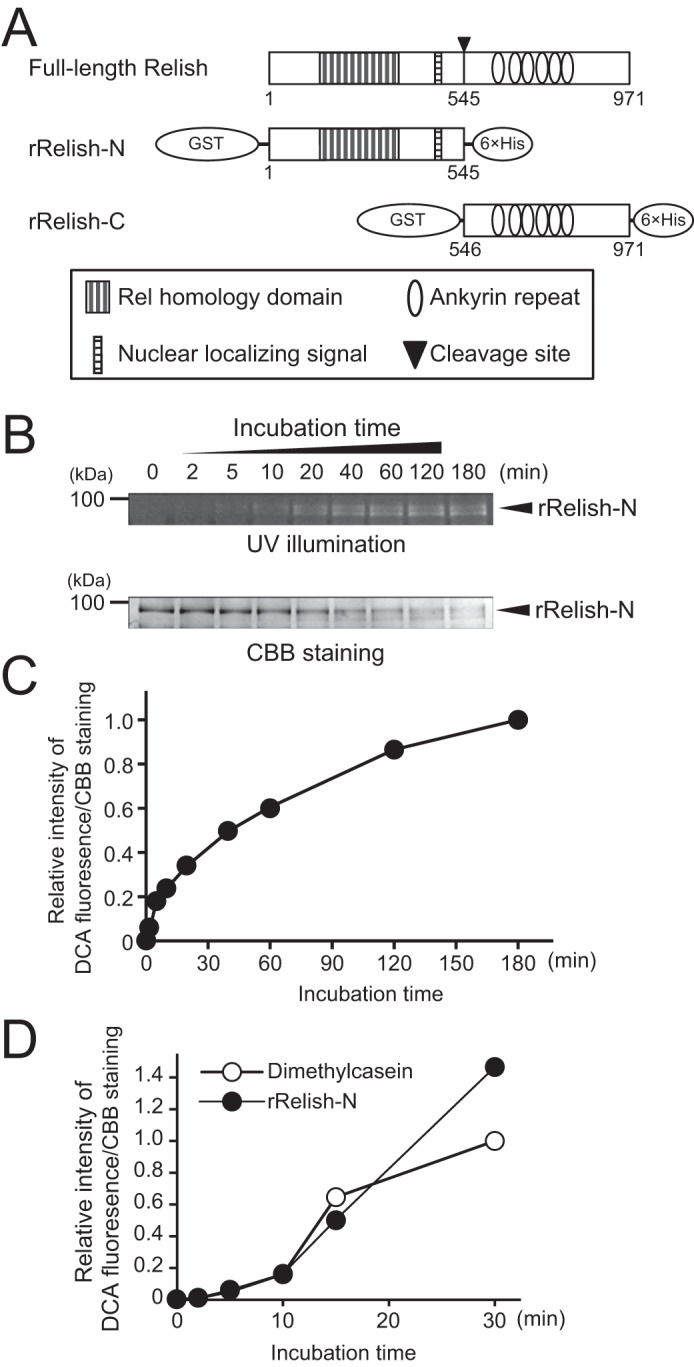
TG-dependent incorporation of DCA into rRelish-N. A, schematic domain structures of full-length Relish, rRelish-N, and rRelish-C. B, rRelish-N (50 nm) was incubated with rTG in the presence of DCA (1 mm) for various incubation times. Treated proteins were separated by SDS-PAGE and detected by UV illumination and CBB staining. C, density of bands stained with CBB or modified with DCA fluorescence was quantitated using ImageJ software (54), and the quantity of DCA incorporation was calculated based on CBB staining. Relative intensity of DCA incorporation at 180 min was defined as 1.0. D, rTG was incubated with N,N′-dimethylcasein or rRelish-N in the presence of DCA (1 mm) for various incubation times. Treated proteins were separated by SDS-PAGE and detected by UV illumination and CBB staining. Relative intensity of DCA incorporation was quantified as described in C. Data for B, C, and D are representative of three independent experiments.
Results
TG-dependent DCA incorporation into rRelish-N
We previously demonstrated that Drosophila TG catalyzes the cross-linking of Relish-N to suppress antimicrobial production (13). To confirm whether rRelish-N, as shown in Fig. 1A, functions as a substrate for TG, rRelish-N was incubated with a recombinant TG (rTG) in the presence of DCA, and the fluorescence of the DCA-incorporated rRelish was detected by UV illumination. DCA was incorporated into rRelish-N in a time-dependent manner (Fig. 1, B and C). In Coomassie Brilliant Blue (CBB)-stained gels, the amount of rRelish-N protein decreased depending on the incubation time because a part of rRelish-N was transformed into high-molecular-weight polymers through TG-dependent protein-protein cross-linking as shown in the following experiments. In addition, the incorporation of DCA into rRelish-N was equivalent to that into N,N′-dimethylcasein, which is a standard TG substrate, indicating that rRelish-N is a proper substrate for TG and equivalent to N,N′-dimethylcasein (Fig. 1D). In the present study, rRelish-N was tagged with GST derived from Schistosoma japonicum at its N terminus because human GST is a known substrate of TG2 (25). To confirm that the GST tag in rRelish-N is not a target of Drosophila TG, rRelish-N was cleaved by PreScission Protease (GE Healthcare), and the resulting proteins were incubated with rTG in the presence of DCA. As a result, DCA was only incorporated into rRelish-N, clearly indicating that rRelish-N, but not the GST tag, is a substrate for TG (Fig. 2, A and B).
Figure 2.
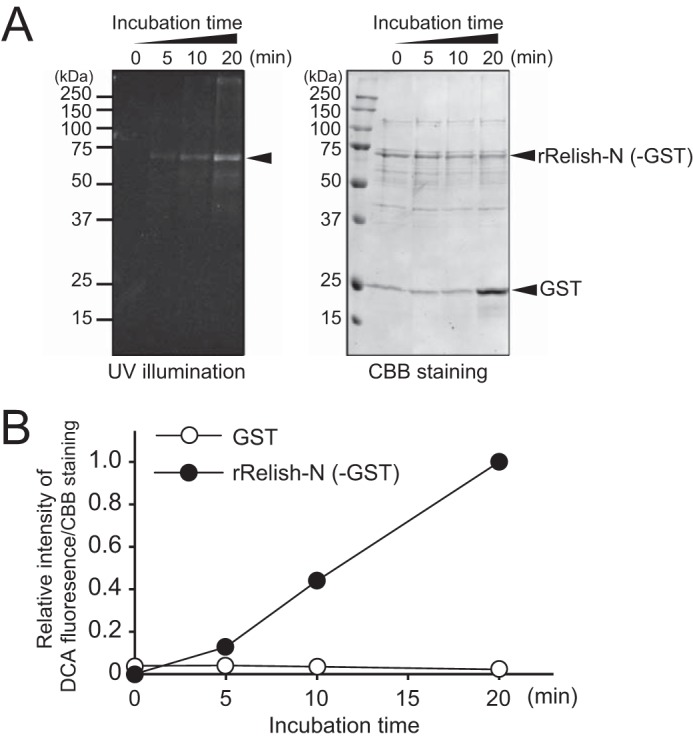
TG-dependent incorporation of DCA into rRelish-N without the GST tag. A, rRelish-N digested with PreScission Protease (2 units) was incubated with rTG in the presence of DCA (1 mm) for different incubation times. Treated proteins were separated by SDS-PAGE and detected by UV illumination (left) and CBB staining (right). B, CBB-stained bands and the DCA fluorescence were quantitated using ImageJ software. Data for A and B are representative of three independent experiments.
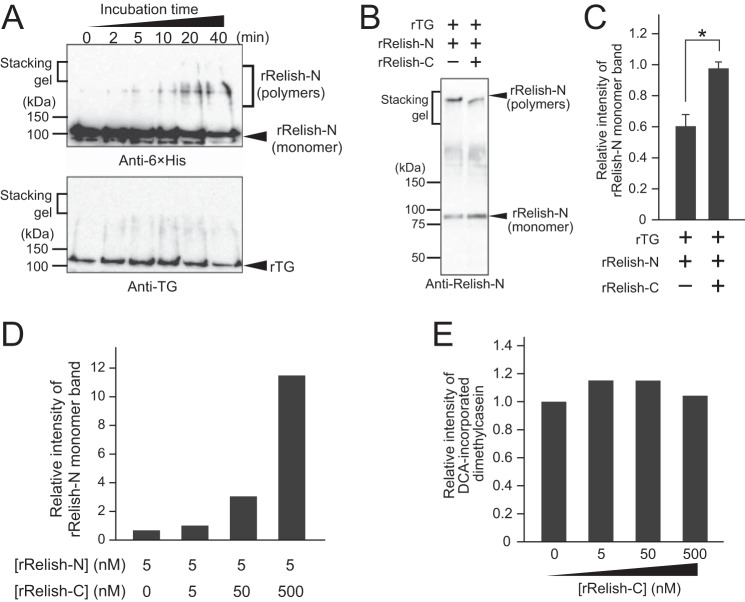
We previously reported that rRelish-N expressed in Drosophila S2 cells is cross-linked by TG to form a high-molecular-weight polymer (13). To examine whether rRelish-N can form a homopolymer in vitro, rRelish-N and rTG were incubated and subjected to SDS-PAGE. The high-molecular-weight bands were clearly detected by anti-His tag antibody depending on the incubation time (Fig. 3A, upper panel) but not by anti-TG antibody (Fig. 3A, lower panel), indicating that rRelish-N forms TG-dependent covalent homopolymers. We also previously reported that the full-length Relish must be proteolytically cleaved by Dredd to release Relish-N, leading to the exposure of Gln or Lys residues on Relish-N for TG-dependent cross-linking (13). In contrast, the C-terminal portion of Relish, Relish-C, contains an ankyrin-repeat domain that inhibits the dimerization of Relish-N (16–20). To examine the inhibitory effects of Relish-C on the protein-protein cross-linking of Relish-N, rRelish-N was incubated with rTG in the presence of rRelish-C. In Western blotting, the intensity of the monomer band of rRelish-N in the absence of rRelish-C was increased ∼65% by the addition of an equimolar concentration of rRelish-C (Fig. 3, B and C) and increased dramatically depending on the dose increase of rRelish-C (Fig. 3D), indicating that rRelish-C interacts with rRelish-N through masking TG-dependent cross-linking sites on rRelish-N. To determine whether rRelish-C itself has an inhibitory effect on the cross-linking activity of rTG, TG-dependent DCA incorporation into dimethylcasein was measured in the presence of rRelish-C (Fig. 3E). The band intensity of DCA-incorporated dimethylcasein on SDS-PAGE was not affected by increasing concentrations of rRelish-C, indicating that the blocking mechanism of TG-dependent rRelish-N polymerization is due to the presence of rRelish-C at the rRelish-N cross-linking sites.
Figure 3.
Polymerization of rRelish-N by TG. A, rRelish-N was incubated with rTG. Treated proteins were separated by SDS-PAGE and detected by Western blotting. B, rRelish-N (5 nm) was incubated with rTG in the presence of rRelish-C (5 nm). Treated proteins were separated by SDS-PAGE and detected by Western blotting. C, the intensity of rRelish-N (monomer) was analyzed by UltraQuant ID Gel Analysis Software (Aplegen). Error bars represent standard errors of mean values (n = 3). *, p < 0.05. D, rRelish-N (5 nm) was incubated with rTG in the presence of different concentrations of rRelish-C (0, 5, 50, and 500 nm). The intensity of the rRelish-N (monomer) band was analyzed by ImageJ software. E, dimethylcasein (5 nm) was incubated with rTG in the presence of different concentrations of rRelish-C (0, 5, 50, and 500 nm). The intensity of the DCA-incorporated dimethylcasein band was analyzed by ImageJ software. Data for A, B, D, and E are representative of three independent experiments.
Determination of amine-incorporation sites in Relish-N
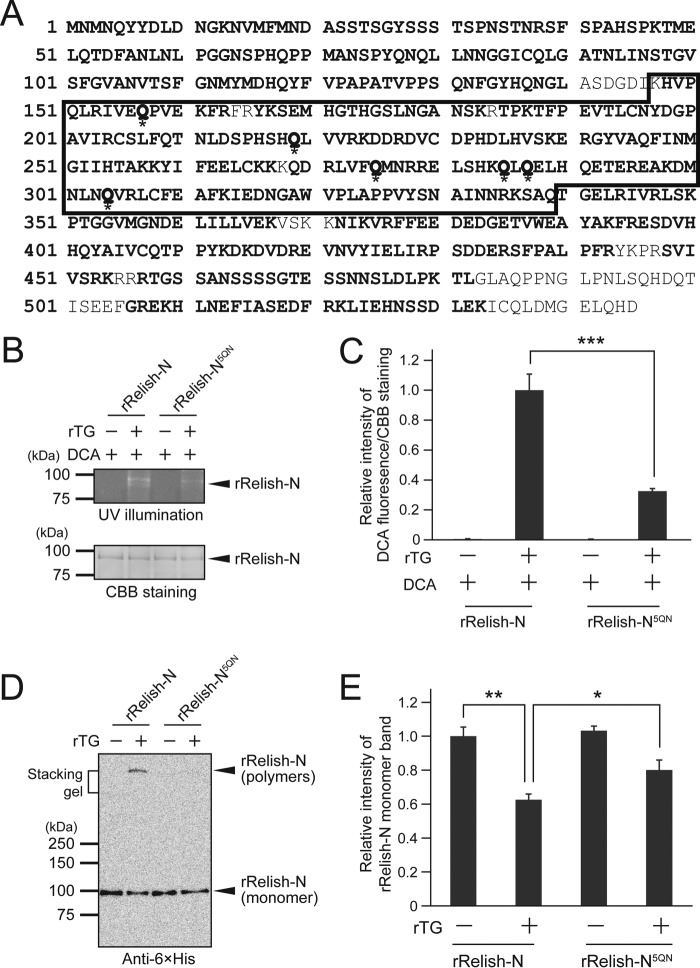
In mammals, inhibitor of NF-κB (I-κB), a mammalian homolog of Relish-C, is a substrate of TG2, and the TG2-catalyzed cross-linking sites have been identified by mass spectrometry (26). To identify the amine-incorporation sites of Relish-N, rRelish-N was incubated with rTG in the presence of DCA, and DCA-incorporated Gln residues were determined by mass spectrometry after proteolytic digestion (Tables 1–3). In total, 58 peptides were identified by LC-MS/MS analysis of the tryptic or chymotryptic digestion of DCA-modified rRelish-N, covering 90% of the whole sequence of Relish-N (Tables 1–3). Six Gln residues in rRelish-N were identified as DCA incorporation sites, including Gln157, Gln219, Gln275, Gln285, Gln287, and Gln304 (Table 3). Interestingly, the modified Gln residues were all located in the Rel homology domain of Relish-N, functioning as the binding site for the κB site of target genes (Fig. 4A).
Table 1.
Mass spectrometry of trypsin-digested peptides
| Mass |
ΔMass | Amino acids | Expected peptide | |
|---|---|---|---|---|
| Measured | Calculated | |||
| Da | Da | |||
| 2641.098 | 2641.102 | −0.004 | 14–38 | K↓NVMFMNDASSTSGYSSSTSPNSTNR↓S |
| 956.471 | 956.472 | −0.001 | 39–47 | R↓SFSPAHSPK↓T |
| 748.434 | 748.434 | 0.000 | 148–153 | K↓HVPQLR↓I |
| 1243.692 | 1243.693 | −0.001 | 154–163 | R↓IVEQPVEKFR↓F |
| 1916.878 | 1916.880 | −0.002 | 166–183 | R↓YKSEMHGTHGSLNGANSK↓R |
| 1950.948 | 1950.951 | −0.003 | 188–204 | K↓TFPEVTLCNYDGPAVIR↓C |
| 2237.097 | 2237.101 | −0.004 | 205–223 | R↓CSLFQTNLDSPHSHQLVVR↓K |
| 1806.794 | 1806.796 | −0.002 | 225–239 | K↓DDRDVCDPHDLHVSK↓E |
| 1761.921 | 1761.924 | −0.003 | 242–257 | R↓GYVAQFINMGIIHTAK↓K |
| 1228.616 | 1228.616 | −0.001 | 258–266 | K↓KYIFEELCK↓K |
| 1305.659 | 1305.661 | −0.002 | 269–278 | K↓QDRLVFQMNR↓R |
| 1409.688 | 1409.690 | −0.002 | 285–295 | K↓QLQELHQETER↓E |
| 988.476 | 988.476 | 0.000 | 299–306 | K↓DMNLNQVR↓L |
| 913.437 | 913.437 | 0.000 | 307–313 | R↓LCFEAFK↓I |
| 2537.302 | 2537.303 | −0.001 | 314–336 | K↓IEDNGAWVPLAPPVYSNAINNRK↓S |
| 1228.688 | 1228.689 | −0.001 | 337–347 | K↓SAQTGELRIVR↓L |
| 2112.147 | 2112.150 | −0.003 | 348–367 | R↓LSKPTGGVMGNDELILLVEK↓V |
| 2319.028 | 2319.033 | −0.005 | 375–393 | K↓VRFFEEDEDGETVWEAYAK↓F |
| 2171.009 | 2171.011 | −0.002 | 396–413 | R↓ESDVHHQYAIVCQTPPYK↓D |
| 1945.973 | 1945.975 | −0.002 | 420–435 | R↓EVNVYIELIRPSDDER↓S |
| 933.507 | 933.507 | 0.000 | 436–443 | R↓SFPALPFR↓Y |
| 659.397 | 659.397 | 0.000 | 448–453 | R↓SVIVSR↓K |
| 2225.986 | 2225.988 | −0.003 | 458–480 | R↓TGSSANSSSSGTESSNNSLDLPK↓T |
| 1733.835 | 1733.837 | −0.002 | 508–521 | R↓EKHLNEFIASEDFR↓K |
| 1411.729 | 1411.731 | −0.002 | 522–533 | R↓KLIEHNSSDLEK↓I |
Table 2.
Mass spectrometry of chymotrypsin-digested peptides
| Mass |
ΔMass | Amino acids | Expected peptide | |
|---|---|---|---|---|
| Measured | Calculated | |||
| Da | Da | |||
| 1222.474 | 1222.463 | 0.011 | 1–9 | -↓MNMNQYYDL↓D + 2 oxidation (Met) |
| 1151.527 | 1151.528 | −0.001 | 8–17 | Y↓DLDNGKNVMF↓M |
| 1031.387 | 1031.387 | 0.000 | 18–27 | F↓MNDASSTSGY↓S |
| 1370.606 | 1370.606 | 0.000 | 28–40 | Y↓SSSTSPNSTNRSF↓S |
| 1687.788 | 1687.788 | 0.000 | 41–55 | F↓SPAHSPKTMELQTDF↓A |
| 2175.012 | 2175.017 | −0.005 | 56–76 | F↓ANLNLPGGNSPHQPPMANSPY↓Q |
| 1470.727 | 1470.725 | 0.002 | 77–89 | Y↓QNQLLNNGGICQL↓G |
| 1279.638 | 1279.641 | −0.002 | 90–102 | L↓GATNLINSTGVSF↓G |
| 1258.564 | 1258.565 | −0.001 | 103–114 | F↓GVANVTSFGNMY↓M |
| 1157.427 | 1157.427 | 0.000 | 111–119 | F↓GNMYMDHQY↓F |
| 1470.750 | 1470.751 | −0.001 | 120–133 | Y↓FVPAPATVPPSQNF↓G |
| 787.361 | 787.361 | 0.000 | 134–140 | F↓GYHQNGL↓A |
| 1182.545 | 1182.545 | −0.001 | 167–177 | Y↓KSEMHGTHGSL↓N |
| 1670.753 | 1670.755 | −0.002 | 195–208 | L↓CNYDGPAVIRCSLF↓Q |
| 1837.871 | 1837.874 | −0.003 | 221–235 | L↓VVRKDDRDVCDPHDL↓H |
| 974.493 | 974.493 | 0.000 | 236–243 | L↓HVSKERGY↓V |
| 1387.763 | 1387.765 | −0.001 | 248–259 | F↓INMGIIHTAKKY↓I |
| 981.523 | 981.524 | −0.001 | 282–289 | L↓SHKQLQEL↓H |
| 628.366 | 628.366 | 0.000 | 303–307 | L↓NQVRL↓C |
| 1767.918 | 1767.920 | −0.002 | 313–328 | F↓KIEDNGAWVPLAPPVY↓S |
| 1601.812 | 1601.812 | 0.000 | 329–343 | Y↓SNAINNRKSAQTGEL↓R |
| 1658.854 | 1658.855 | −0.001 | 349–364 | L↓SKPTGGVMGNDELILL↓V + oxidation (Met) |
| 1515.722 | 1515.722 | 0.000 | 392–403 | Y↓AKFRESDVHHQY↓A |
| 1047.505 | 1047.506 | −0.001 | 404–412 | Y↓AIVCQTPPY↓K |
| 1478.736 | 1478.737 | 0.000 | 413–424 | Y↓KDKDVDREVNVY↓I |
| 685.401 | 685.401 | 0.000 | 477–482 | L↓DLPKTL↓G |
| 1189.645 | 1189.646 | 0.000 | 483–494 | L↓GLAQPPNGLPNL↓S |
| 1077.581 | 1077.582 | −0.001 | 515–523 | F↓IASEDFRKL↓I |
Table 3.
Dansylated peptides
| Mass |
ΔMass | Amino acids | Expected peptide | |
|---|---|---|---|---|
| Measured | Calculated | |||
| Da | Da | |||
| 1989.088 | 1989.087 | 0.001 | 148–161 | K↓HVPQLRIVEQPVEK↓F + DCA (Gln) |
| 1693.785 | 1693.788 | −0.003 | 209–220 | F↓QTNLDSPHSHQL↓V + DCA (Gln) |
| 1240.607 | 1240.610 | −0.003 | 272–278 | R↓LVFQMNR↓R + DCA (Gln), oxidation (Met) |
| 2045.978 | 2045.970 | 0.008 | 285–295 | K↓QLQELHQETER↓E + 2 DCA (Gln) |
| 1306.615 | 1306.616 | −0.002 | 299–306 | K↓DMNLNQVR↓L + DCA (Gln) |
Figure 4.
TG-dependent DCA incorporation into rRelish-N and rRelish-N5QN. A, amino acids detected by mass spectrometry are shown in bold letters. Asterisks indicate the DCA-incorporated Gln residues. A box indicates the Rel homology domain. B, rRelish-N or rRelish-N5QN was incubated with rTG in the presence of DCA. Treated proteins were separated by SDS-PAGE and detected by UV illumination and CBB staining. C, the bands were analyzed by ImageJ software. D, rRelish-N or rRelish-N5QN was incubated with rTG. Treated proteins were separated by SDS-PAGE and determined by Western blotting. E, the intensity of the rRelish-N monomer band was analyzed by ImageJ software. Data for B and D are representative of three independent experiments. Error bars for C and E represent standard errors of mean values (n = 3). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
To assess the mechanism underlying the specificity of the identified Gln residues for TG-dependent cross-linking of rRelish-N, all six Gln residues were mutated to Asn residues (rRelish-N6QN). The expression of rRelish-N6QN, however, was too low to be used for the experiments. Fortunately, rRelish-N5QN with five Gln-to-Asn replacements at Gln157, Gln275, Gln285, Gln287, and Gln304 was successfully prepared. rRelish-N5QN was incubated with rTG in the presence of DCA and subjected to SDS-PAGE. The DCA incorporation into rRelish-N5QN was strongly inhibited by the mutations, and the fluorescence intensity of the band of DCA-incorporated rRelish-N5QN was ∼30% that of wild-type rRelish-N (Fig. 4, B and C). Moreover, the amount of homopolymers of rRelish-N5QN cross-linked by rTG was reduced to an undetectable level (Fig. 4D), and the ratio of Relish-N monomers to Relish-N polymers in Western blotting was significantly higher in rRelish-N5QN than in rRelish-N (Fig. 4E), indicating a loss of the reactivity of rRelish-N5QN against the TG-dependent protein-protein cross-linking. These results demonstrated that the Gln residues identified by LC-MS/MS analysis are fairly specific for TG-dependent cross-linking in rRelish-N.
Polyamine incorporation of rRelish-N inhibits its ability to bind to the κB site
Mammalian TG2 functions not only in the cytosol but also in the nucleus (27–31). To confirm the subcellular localization of TG in Drosophila, rTG was expressed in S2 cells with no detectable endogenous TG antigen, and cytoplasmic and nuclear fractions were prepared. Analysis of the relative intensity of the rTG band by Western blotting indicated that the amount of rTG in the nucleus was approximately half that in the cytoplasm (Fig. 5, A and B), suggesting that Drosophila TG works not only in the cytosol but also in the nucleus.
Figure 5.
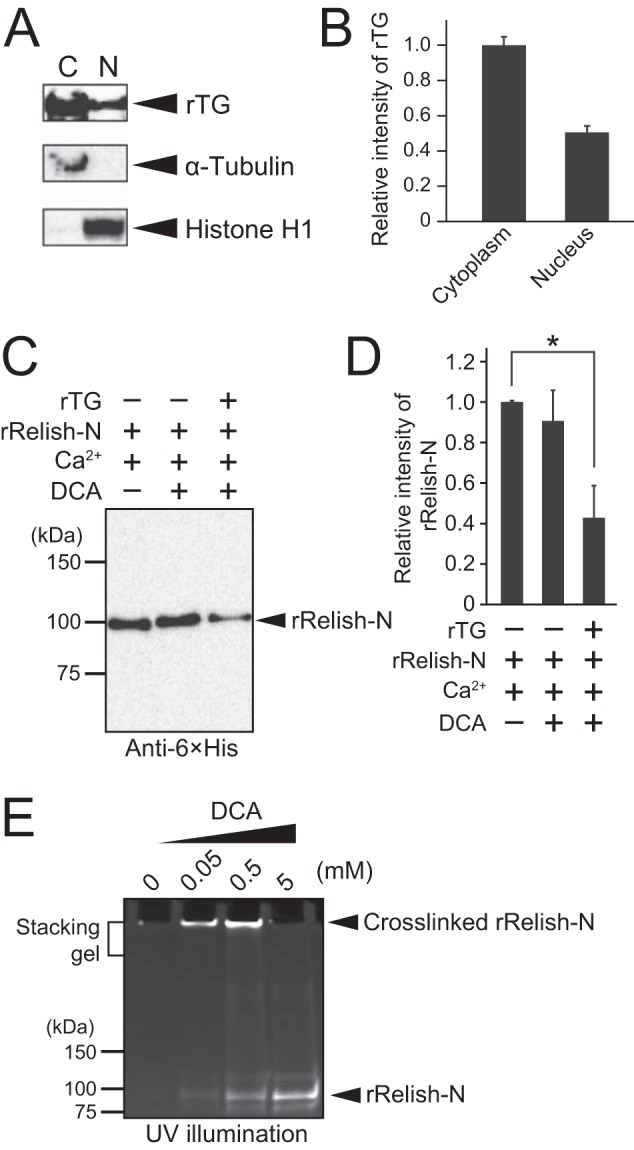
TG localization in S2 cells and the inhibition of the κB-binding ability of rRelish-N by TG-dependent DCA incorporation. A, TG was expressed in S2 cells, and the cytoplasmic (C) and nuclear (N) fractions were prepared. V5-tagged rTG in the subcellular fractions was detected by anti-V5 antibody, and anti-α-tubulin and anti-histone H1 antibodies were used to detect the reference proteins of the cytoplasmic and nuclear fractions. B, the intensity of the rTG band was analyzed by ImageJ software. C, the DNA-binding ability of rRelish-N was analyzed by pulldown assay. rRelish-N was incubated with rTG in the presence of DCA (5 mm) with TG buffer at 37 °C for 1 h. The resulting DCA-incorporated rRelish-N was incubated with 50 nm biotinylated κB oligo at 4 °C overnight. Streptavidin-immobilized agarose was added to the sample, and the resulting biotin-κB-bound rRelish-N was obtained by centrifugation. Proteins were separated by SDS-PAGE and detected by Western blotting using anti-histidine tag antibody. D, the intensity of the Relish-N monomer band was analyzed by ImageJ software. E, rRelish-N was incubated with rTG in different concentrations of DCA (0, 0.05, 0.5, and 5 mm). Treated proteins were separated by SDS-PAGE and analyzed by UV illumination. Data for A, C, and E are representative of three independent experiments. Error bars for B and D represent standard errors of mean values (n = 3). *, p < 0.05.
DCA- or biotin-pentylamine-incorporated Relish-N is translocated into the nucleus in gut epithelial cells, but whether the amine-incorporated Relish-N retains transcriptional activity in the nucleus is unknown (13). To evaluate the DNA-binding ability of rRelish-N cross-linked with DCA, rRelish-N treated with 5 mm DCA was incubated with the biotinylated κB oligonucleotide, and the protein-oligonucleotide complex was collected using streptavidin-immobilized agarose. The amount of κB-bound rRelish-N was significantly decreased only when incubated with rTG and DCA (Fig. 5, C and D). To confirm that the decrease in the binding activity of DCA-treated rRelish-N to the κB oligonucleotide was not caused by TG-dependent protein-protein cross-linking of rRelish-N, rRelish-N was incubated with rTG in different concentrations of DCA and subjected to SDS-PAGE. The polymer band of rRelish-N on SDS-PAGE disappeared at 5 mm DCA (Fig. 5E), indicating that the protein-protein cross-linking of rRelish-N by rTG was almost completely inhibited under these conditions.
Polyamines, such as putrescine, spermidine, and spermine, are natural substrates for TG (32), and intracellular polyamines are present in millimolar concentrations (33, 34). To confirm whether these polyamines are TG-dependently incorporated into rRelish-N, DCA was incubated with rRelish-N in the presence of different concentrations of spermidine or spermine. The incorporation of DCA into rRelish-N was inhibited in the presence of the polyamines in a concentration-dependent manner, suggesting that these polyamines function as substrates for TG in the cytosol or nucleus (Fig. 6, A and B). The incorporation of spermine into rRelish-N was also confirmed by Western blotting using anti-spermine antibody in the presence of DCA (Fig. 6, C and D). DCA incorporation into Relish-N was strongly inhibited by increasing concentrations of spermine, consistent with the incorporation of spermine in a concentration-dependent manner, indicating that spermine and DCA were competitively cross-linked with Relish-N by rTG (Fig. 6, E and F).
Figure 6.
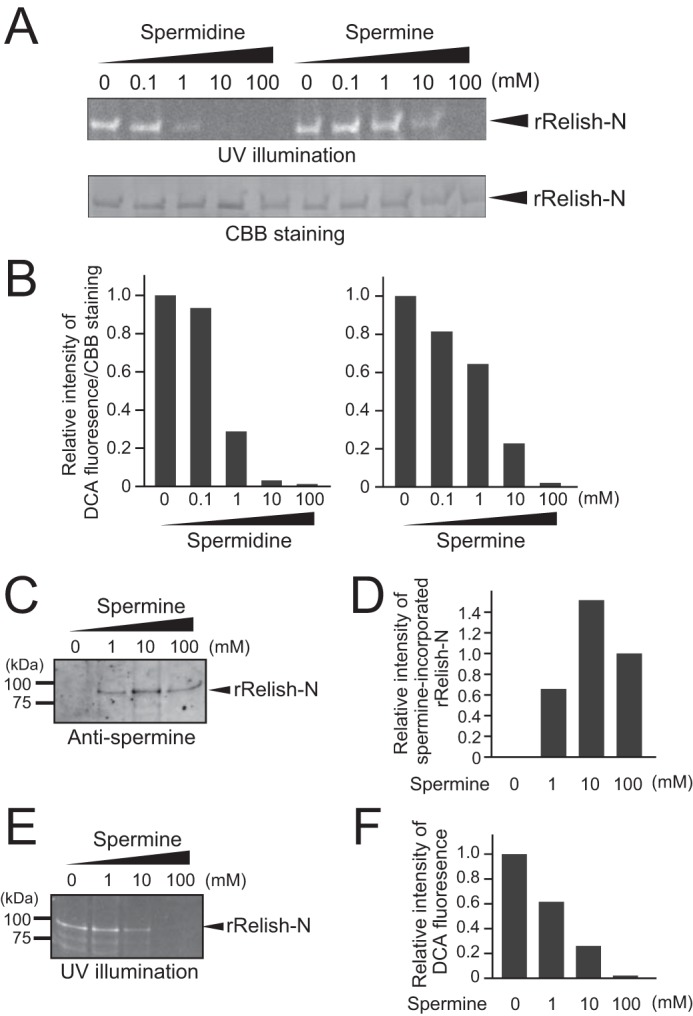
Polyamines and DCA were competitively incorporated into rRelish-N. A, rRelish-N was incubated with rTG in the presence of DCA (1 mm) and different concentrations of spermidine or spermine. Treated proteins were separated by SDS-PAGE and detected by UV illumination and CBB staining. B, the density of bands stained with CBB or modified with DCA fluorescence was quantitated using ImageJ software, and the quantity of DCA incorporation was calculated based on CBB staining. Relative intensity of DCA incorporation at 0 mm spermidine or spermine was defined as 1.0. C, rRelish-N was incubated with rTG in the presence of DCA (1 mm) and different concentrations of the spermine. Treated proteins were separated by SDS-PAGE and detected by Western blotting using anti-spermine antibody. D, the bands of spermine-incorporated rRelish-N were quantified by ImageJ software. Relative intensity of spermine-incorporated rRelish-N at 100 mm spermine was defined as 1.0. E, rRelish-N was incubated with rTG in the presence of DCA (1 mm) and different concentrations of spermine. Treated proteins were separated by SDS-PAGE and detected by UV illumination. F, the quantity of DCA incorporation was measured by ImageJ software. Relative intensity of DCA incorporation at 0 mm spermine was defined as 1.0. Data for A–F are representative of three independent experiments.
To examine polyamine modification of Relish-N in vivo, intestinal proteins extracted from flies fed a spermine-containing diet were analyzed by Western blotting using anti-spermine and anti-Relish-N antibodies. Protein bands of ∼68 kDa corresponding to native Relish-N in wild-type flies not fed a spermine-containing diet were detected by anti-spermine antibody (Fig. 7A), and endogenous spermine was incorporated into Relish-N in the absence of exogenous spermine (Fig. 7A, left panel). The relative intensity of the ∼68-kDa bands increased 2-fold depending on the amount of spermine ingested (Fig. 7, A and B). In contrast, the incorporation of spermine into the ∼68-kDa bands was not detectable in Relish-null mutant flies (Fig. 7, A and B). These findings indicate that polyamines are incorporated into Relish-N in vivo. To investigate the effect of polyamine incorporation on the transcriptional activity of Relish-N, mRNA expression of the genes encoding the antimicrobial peptides cecropin A1 and diptericin, which are controlled by the IMD pathway, was measured by quantitative PCR. S2 cells with no TG activity exhibited the same expression level of cecropin A1 both in the presence and absence of spermine (Fig. 7C). In contrast, in cells expressing TG, the transcription of cecropin A1 was decreased significantly in the presence of spermine (Fig. 7C). Moreover, the transcription of diptericin was significantly reduced in wild-type flies fed a spermine-containing diet (Fig. 7D). In TG-RNAi flies, the transcriptional level of diptericin remained constant regardless of the presence of spermine. These findings suggest that the transcriptional activity of Relish-N is controlled by polyamines in a TG-dependent manner. Fig. 7D shows that diptericin expression was not significantly enhanced in TG-RNAi flies because 3-day-old flies were used for the experiments. We recently found that the overexpression of antimicrobial peptide is not induced in young TG-RNAi flies (35).
Figure 7.
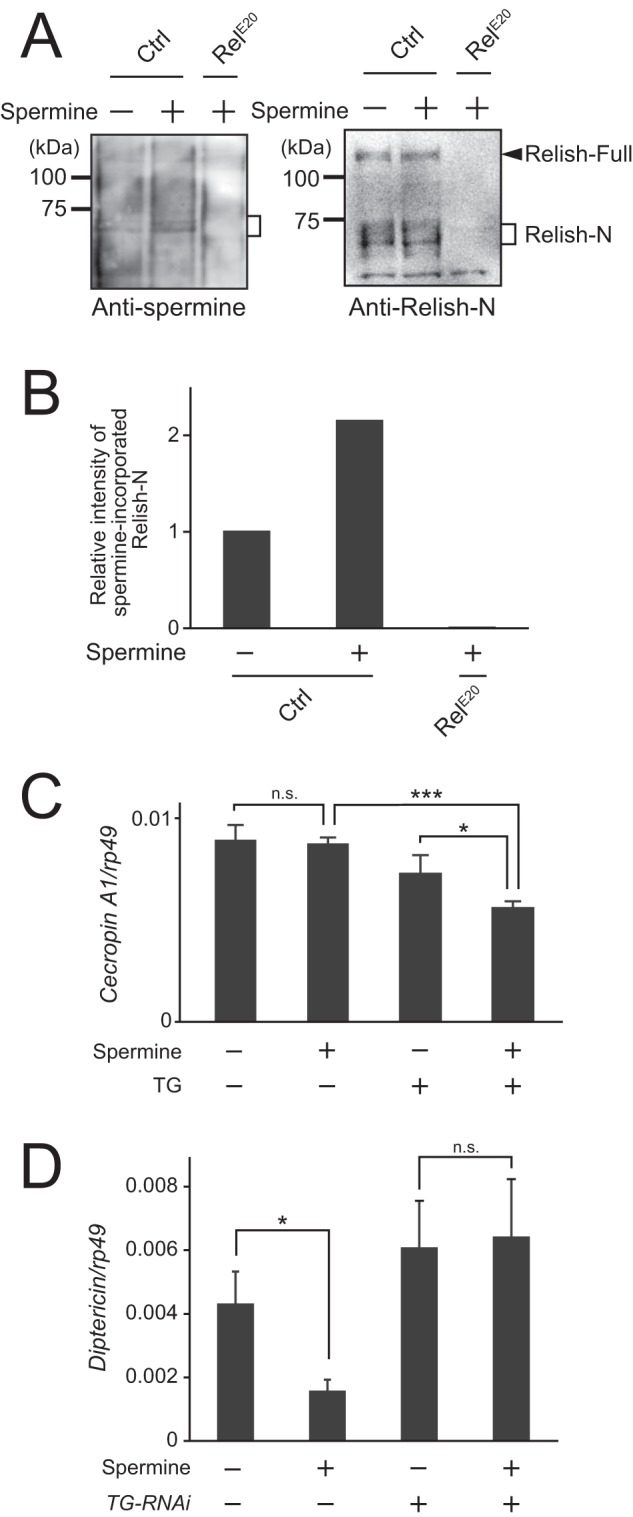
Polyamine incorporation into native Relish in vivo. A, flies were fed 5 mm spermine for 48 h and then dissected, and midguts of the flies were examined by Western blotting. RelE20 indicates Relish-null mutant flies. B, the density of the bands of spermine-incorporated Relish-N was quantitated using ImageJ software. C, TG was expressed in S2 cells with medium containing 5 mm spermine. The expression level of cecropin A1 was analyzed by quantitative PCR. D, TG-RNAi flies were fed 5 mm spermine for 48 h. The expression level of diptericin was analyzed by quantitative PCR. Data for A and B are representative of three independent experiments. Error bars for C and D represent standard errors of mean values (n = 3). *, p < 0.05; ***, p < 0.001; n.s., not significant. Ctrl, control.
Discussion
Many factors are involved in regulating the IMD pathway in Drosophila (36, 37): peptidoglycan-recognition protein (PGRP)-LB and PGRP-SC (amidases against peptidoglycans) digest peptidoglycans to non-stimulatory fragments (38–40), Casper represses the expression of IMD pathway-controlled antimicrobial peptide genes (41), PIMS/Rudra/Pirk interacts with the complex of PGRP-LC (a membrane-bound receptor for peptidoglycans) and the IMD protein to reduce IMD pathway immune signaling (42–44), the Pvr receptor tyrosine kinase attenuates the JNK and NF-κB arms of the IMD pathway (45), PGRP-LF forms a non-signaling complex with PGRP-LC (46), Drosophila Ring and YY1-binding protein promotes the proteasomal degradation of Relish (47), and defense repressor 1 regulates the activation of the IMD pathway in the brain (48, 49). Chronic activation of the IMD pathway leads to inflammatory gut diseases and reduces lifespan (50, 51), clearly indicating that distinct proteins and enzymes have evolved into essential regulatory factors to evade uncontrollable conditions for the IMD pathway.
We previously reported that TG-catalyzed Relish-N cross-linking suppresses the IMD pathway to maintain the threshold required for immune tolerance against microbes in the gut and that the regulated amounts of antimicrobial peptides may function to maintain gut microbiota (13). In mammals, specific transcription factors involved in intracellular signaling pathways are regulated through TG2-dependent protein-protein cross-linking in the cytosol or the nucleus. Park et al. (26) reported an I-κB kinase-independent activation mechanism of NF-κB in a lipopolysaccharide-treated microglial cell line: the TG2-dependent polymer formation of I-κB in the cytosol results in the translocation of NF-κB into the nucleus and the subsequent up-regulation of inflammatory genes. In contrast, Tatsukawa et al. (30) reported hepatocyte apoptosis by TG2-dependent cross-linking of the transcription factor Sp1 in the nucleus: the cross-linking of Sp1 down-regulates expression of the c-Met gene, which encodes a receptor for hepatocyte growth factor required for hepatocyte viability. Therefore, the regulation of signaling pathways through TG-dependent protein-protein cross-linking of specific transcription factors or regulatory components appears to be highly conserved between mammals and insects.
Conversely, our previous study revealed that synthetic amines, such as DCA and biotin-labeled pentylamine, ingested by flies are incorporated into Relish-N by TG activity, leading to nuclear translocation of the modified Relish-N into gut epithelial cells (13). Here we found that the incorporation sites for DCA identified in rRelish-N are all localized in the DNA-binding domain of Relish-N and that the DCA modification of rRelish-N reduces the binding of the κB oligonucleotide (Fig. 5, C and D). We previously reported that ingested or injected DCA triggers the activation of the IMD pathway in the fly gut, although the expression of diptericin in DCA-treated flies was not as high as that in TG-RNAi flies (13). The present study did not investigate the transcriptional activity of rRelish-N partially modified with DCA except that DCA incorporation of rRelish-N5QN was reduced to ∼30% of that in wild-type rRelish-N (Fig. 4C). Some Relish-N partially modified with DCA that is translocated into the nucleus may retain a lower level of transcriptional activity. Natural polyamines, including spermidine and spermine, are also incorporated into Relish-N (Fig. 6). These findings suggest that the polyamine-incorporated Relish-N in the nucleus has reduced transcriptional activity in the nucleus. These polyamines are ubiquitously located in the cytosol and nucleus and play critical roles in gene regulation, cell growth, and wound healing after injury (22–24). Their intracellular concentrations are strictly regulated through metabolism and cellular import and export. We found that TG localizes not only in the cytosol but also in the nucleus in Drosophila S2 cells (Fig. 5, A and B). Therefore, these polyamines may be TG-dependently incorporated into Relish-N to reduce its transcriptional activity under physiologic or pathologic conditions.
TG requires Ca2+ for its activity (1). In addition to the production of antimicrobial peptides by the IMD pathway, reactive oxygen species-dependent innate immunity mediated by the dual-oxidase pathway in the gut is very important for host survival (52). Lee et al. (53) reported that uracil secreted from non-resident microbes is recognized by a G protein-coupled receptor(s), triggering the activation of phospholipase Cβ to increase intracellular Ca2+ sufficient for spontaneous activation of the dual-oxidase pathway. We speculate that this basal concentration of Ca2+ in gut epithelial cells is both required and sufficient to maintain TG activity for the cross-linking of Relish-N to suppress its nuclear translocation or for polyamine incorporation into Relish to reduce its transcriptional activity (Fig. 8). We conclude that TG regulates the transcriptional activity of Relish-N through the incorporation of polyamines into Relish-N as well as through the protein-protein cross-linking of Relish-N in Drosophila.
Figure 8.
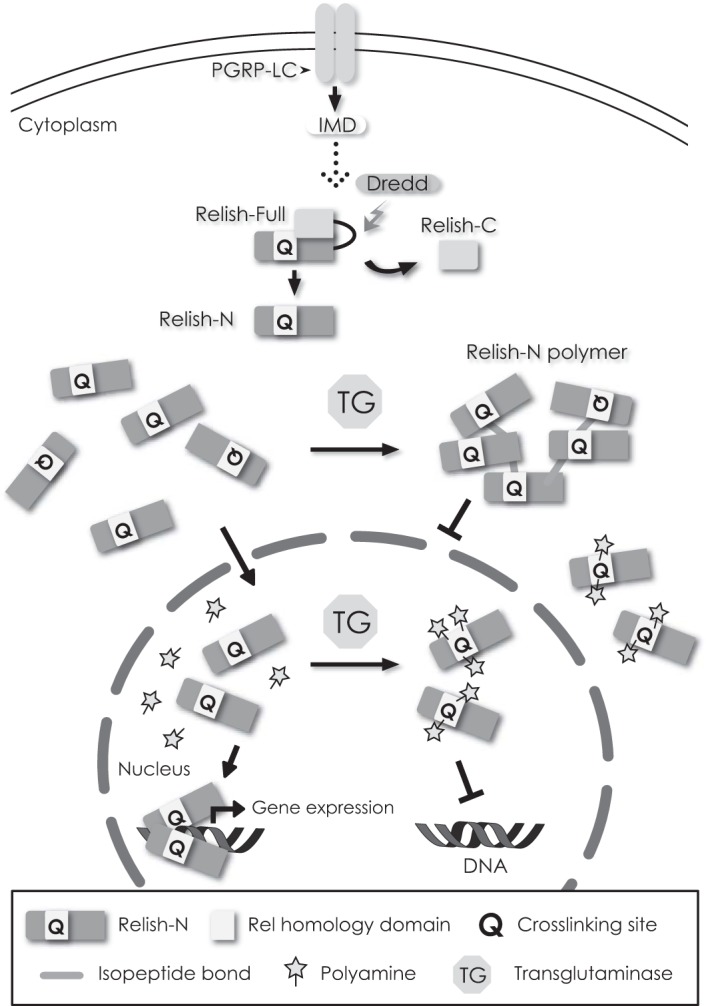
TG functions as a suppressor of Relish. Intracellular TG catalyzes the polymerization of Relish-N to inhibit its translocation into the nucleus. Conversely, polyamines are incorporated into Relish-N to reduce the DNA-binding or transcriptional ability of Relish-N.
Experimental procedures
Materials
All chemicals used in this study were of analytical grade unless otherwise stated. DCA, N,N′-dimethylcasein, and streptavidin immobilized on agarose CL-4B were purchased from Sigma-Aldrich. Drosophila S2 cells were purchased from Thermo Fisher Scientific (Waltham, MA). The biotinylated κB oligonucleotide was obtained from Hokkaido System Science (Sapporo, Japan).
Fly stocks
Da-Gal4 and w1118 were obtained from Bloomington Stock Center; UAS-TG-RNAi was obtained from National Institute of Genetics; and RelE20 was obtained from Prof. D. Hultmark, Umeå University. All flies were maintained in standard medium at 25 °C.
Preparation of primers for PCR
The sequence of Relish (CG11992) or the TG (CG7356) gene was amplified by PCR. For the genetic template, the complimentary DNA library of Drosophila was used. A mutant of Relish-N was generated using a site-directed mutagenesis method. The primers used in this study were as follows: Relish-N (residues 1–545), 5′-CGGGATCCGATGAACATGAATCAGTACTACGAC-3′ (forward) and 5′-TAGTTTAGCGGCCGCATCGTGCTGCAACTC-3′ (reverse); Relish-C (residues 546-971), 5′-CGCGGATCCGGGTCATAACCGGGCGGAA-3′ (forward) and 5′-ATAGTTTAGCGGCCGCAGTTGGGTTAACCAGTAG-3′ (reverse); TG, 5′-GGCGGATCCATGAGTTATTGGTATCGG-3′ (forward) and 5′-CCGCCCGGGAGCTATTACATCGGTGCG-3′ (reverse); Relish-N (Q157N), 5′-AACCCGGTGGAGAAGTTCCGC-3′ (forward) and 5′-CTCAACGATCCGCAGCTG-3′ (reverse); Relish-N (Q275N), 5′-GTCTTTAACATGAACCGCCGCGAG-3′ (forward) and 5′-CAGGCGATCCTGCTTCTTC-3′ (reverse); Relish-N (Q285N,Q287N), 5′-CTAAACGAACTGCATCAGGAGACAGAG-3′ (forward) and 5′-GTTTTTGTGGGACAACTCGCGGC-3′ (reverse); and Relish-N (Q304N), 5′-AACGTGCGGCTCTGCTTTGAGGCC-3′ (forward) and 5′-GTTCAAGTTCATGTCCTTGGC-3′ (reverse). The constructed plasmids were confirmed by DNA sequencing.
Western blotting
Proteins were separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane. The membrane was blocked with 5% skim milk in Tris-buffered saline (20 mm Tris-HCl, pH 7.5, containing 150 mm NaCl). The blocked membrane was incubated with each antibody at room temperature for 1 h, including anti-His tag mAb-HRP-DirecT (catalogue number D291-7, MBL, Nagoya, Japan), anti-TG polyclonal antibody (8), anti-Relish-N polyclonal antibody (13), anti-V5 tag-HRP-DirecT (catalogue number PM003-7, MBL), anti-α-tubulin-HRP-DirecT (catalogue number PM054-7, MBL), anti-spermine antibody (catalogue number ab26975, Abcam, Cambridge, UK), and anti-histone H1 antibody (catalogue number 39575, Active Motif, Carlsbad, CA). Protein bands were visualized by WesternBright Quantum (Advansta, Menlo Park, CA). Chemiluminescence was detected using an Omega Lum G fluorescence imager (Aplegen, Pleasanton, CA).
Preparation of recombinant proteins
The nucleotide sequences of Relish-N and Relish-C were inserted into the pET-22b vector (Novagen, Madison, WI) containing GST and His6 tags (Fig. 1A). For wild-type TG (rTG), the entire TG sequence was inserted into the pGEX-4T-2 vector (GE Healthcare). Transformed colonies were picked up and grown in Luria-Bertani medium to reach an optical density at 600-nm wavelength of ∼0.6. At this stage, to induce protein expression, 0.05 mm isopropyl β-d-1-thiogalactopyranoside was added and incubated at 18 °C for 24 h with shaking at 180 rpm. After induction, the collected bacteria were sonicated in lysis buffer (50 mm Tris-HCl, pH 8.0, containing 200 mm NaCl, 1% Triton X-100, 10% glycerol, 1 mm DTT, 1 mm EDTA, and 1 mm PMSF) by a Branson Sonifier 250 for 1 min with an output control of 2.5 and a duty cycle of 50%. The supernatant containing soluble recombinant proteins was separated by centrifugation (13,000 × g, 4 °C, 20 min) and filtered. GST-tagged recombinant proteins were loaded on a glutathione-Sepharose 4B column (GE Healthcare). The column was washed with washing buffer (50 mm Tris-HCl, pH 8.0, containing 200 mm NaCl) and eluted with elution buffer (50 mm Tris-HCl, pH 8.0, containing 200 mm NaCl and 20 mm reduced glutathione).
TG-dependent DCA incorporation into rRelish-N
rRelish-N was incubated with rTG in TG buffer (50 mm Tris-HCl, pH 8.5, containing 10 mm CaCl2, 10 mm DTT, and 1 mm DCA) at 37 °C. The resulting rRelish-N modified with DCA was precipitated with TCA and separated by SDS-PAGE. The fluorescence of DCA was detected by UV illumination (wavelength, 365 nm), and loaded proteins were also stained with CBB.
Mass spectrometry
DCA-incorporated rRelish-N separated by SDS-PAGE was detected by silver staining or CBB staining, and the protein band was excised, digested with trypsin or chymotrypsin, and subjected to LC-MS/MS analysis. Peak lists obtained from the mass spectra were used to identify fragments using the Mascot search engine (Matrix Science, Boston, MA). The analysis was carried out at the Proteomics center, Laboratory for Technical Support, Medical Institute for Bioregulation, Kyushu University.
Pulldown assay
rRelish-N was incubated with rTG in the presence of DCA with TG buffer at 37 °C for 1 h. The resulting DCA-incorporated rRelish-N was incubated with 50 nm biotinylated κB oligo (biotin-5′-CATCGGGGATTCCTTTT-3′) in the κB-binding buffer (10 mm Tris-HCl, pH 7.5, containing 50 mm KCl, 1 mm DTT, and 1 mg/ml BSA) in the presence of 2.5% glycerol, 0.02% Nonidet P-40, and 5 mm MgCl2 at 4 °C overnight. Streptavidin immobilized on agarose was added to the sample, and the resulting biotin-κB-bound rRelish-N was obtained by centrifugation. The proteins were separated by SDS-PAGE and detected by Western blotting.
Subcellular fractionation of the nucleus and cytosol
Drosophila S2 cells expressing rTG were prepared as described previously (13). The NE-PER Nuclear and Cytoplasmic Extraction Reagents kit (Thermo Fisher Scientific) was used to prepare the nuclear and cytoplasmic fractions according to the manufacturer's instructions.
Total RNA extraction and reverse transcription
Drosophila S2 cells were maintained at 27 °C in Insect-Xpress protein-free medium (Lonza Japan, Tokyo, Japan) with 1% penicillin-streptomycin and 50 μg/ml gentamicin. The cells were transfected with FuGENE HD (Promega, Madison, WI). A pIB vector (Thermo Fisher Scientific) was used for expression of the C-terminal FLAG-tagged TG. Transfected cells were incubated for 18 h with 1 μm 20-hydroxyecdysone (Sigma). After the incubation, the cells were treated with or without 0.5 mm spermine for 1 h and then stimulated with 1 μg/ml peptidoglycan from E. coli O111:B4 (InvivoGen, San Diego, CA) for 8 h. The cells were washed with PBS, and total RNA was extracted with RNAiso Plus (Takara, Shiga, Japan) and treated with deoxyribonuclease I. Total RNA (500 ng) was used for reverse transcription with PrimeScript RT Master Mix (Perfect Real Time, Takara) according to the manufacturer's instructions. Three-day-old adult female flies were fed 5 mm spermine for 48 h. Spermine was dissolved in 5% sucrose. The reagent was added to a vial containing standard medium covered with glass microfiber filters GF/A (diameter, 21 mm; GE Healthcare). After feeding, flies' midguts (three replicates of three intestines) were dissected in PBS. Total RNA (300 ng) extracted from midguts was used for reverse transcription.
Quantitative PCR
Quantitative PCR was performed using FastStart Essential DNA Green Master (Roche Applied Science) on a LightCycler Nano (Roche Applied Science). Copy number was determined by the relative standard curve method. Target gene inserted into pMD19 (Takara) was used as the standard. Ribosomal protein 49 (rp49) was used as a housekeeping gene. The following primers were used: rp49, 5′-AGATCGTGAAGAAGCGCACCAAG-3′ (forward) and 5′-CACCAGGAACTTCTTGAATCCGG-3′ (reverse); diptericin, 5′-GGCTTATCCGATGCCCGACG-3′ (forward) and 5′-TCTGTAGGTGTAGGTGCTTCCC-3′ (reverse); and cecropin A1, 5′-CATCTTCGTTTTCGTCGCTC-3′ (forward) and 5′-CGACATTGGCGGCTTGTTGA-3′ (reverse).
Spermine incorporation into rRelish-N
rRelish-N was incubated with rTG in the presence of DCA (1 mm) and different concentrations of the polyamines. Treated proteins were separated by SDS-PAGE and detected by UV illumination and Western blotting with anti-spermine antibody. To detect spermine incorporated into Relish, flies were fed 5 mm spermine for 48 h. Spermine-fed flies were dissected, and midgut proteins were extracted by lysis buffer. The proteins were separated by SDS-PAGE and confirmed by Western blotting.
Statistical analysis
Student's t test was used to calculate p values. A p value of less than 0.05 was considered significant.
Author contributions
K. M. and T. S. performed the experiments. K. M., T. S., and S. K. analyzed the data and interpreted the experimental results. K. M., T. S., and S. K. designed the experiments and wrote the paper.
Acknowledgments
We thank D. Hultmark for gifting us the flies (Umeå University, Umeå, Sweden), R. Ueda for providing fly strains (National Institute of Genetics, Mishima, Japan), M. Matsumoto and M. Oda for the mass spectrometry analysis (Kyushu University, Fukuoka, Japan), Y. Fukae for preparing rTG (Kyushu University, Fukuoka, Japan), and Y. Ikeda and Y. Fukuda for technical assistance with the Western blotting analysis (Kyushu University, Fukuoka, Japan).
This work was supported in part by Grants-in-aid for Scientific Research (B) 15H04353 (to S. K.), for Young Scientists (B) 26860333 (to T. S.), and for Japan Society for the Promotion of Science Fellows 16J02938 (to K. M.) and the Ministry of Education, Culture, Sports, Science, and Technology of Japan and the Kyushu University Interdisciplinary Programs in Education and Projects in Research Development Fund Grant 26701 (Tenure track type; to T. S.). The authors declare that they have no conflicts of interest with the contents of this article.
- TG
- transglutaminase
- IMD
- immune deficiency
- NF-κB
- nuclear factor-κB
- DCA
- monodansylcadaverine
- CBB
- Coomassie Brilliant Blue
- I-κB
- inhibitor of NF-κB
- PGRP
- peptidoglycan-recognition protein
- rRelish
- recombinant Relish
- rTG
- recombinant TG
- dansyl
- 5-dimethylaminonaphthalene-1-sulfonyl.
References
- 1. Eckert R. L., Kaartinen M. T., Nurminskaya M., Belkin A. M., Colak G., Johnson G. V., and Mehta K. (2014) Transglutaminase regulation of cell function. Physiol. Rev. 94, 383–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lorand L., and Graham R. M. (2003) Transglutaminases: crosslinking enzymes with pleiotropic functions. Nat. Rev. Mol. Cell Biol. 4, 140–156 [DOI] [PubMed] [Google Scholar]
- 3. Klöck C., and Khosla C. (2012) Regulation of the activities of the mammalian transglutaminase family of enzymes. Protein Sci. 21, 1781–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Király R., Demény M., and Fésüs L. (2011) Protein transamidation by transglutaminase 2 in cells: a disputed Ca2+-dependent action of a multifunctional protein. FEBS J. 278, 4717–4739 [DOI] [PubMed] [Google Scholar]
- 5. Dickneite G., Herwald H., Korte W., Allanore Y., Denton C. P., and Matucci Cerinic M. (2015) Coagulation factor XIII: a multifunctional transglutaminase with clinical potential in a range of conditions. Thromb. Haemost. 113, 686–697 [DOI] [PubMed] [Google Scholar]
- 6. Matsuda Y., Koshiba T., Osaki T., Suyama H., Arisaka F., Toh Y., and Kawabata S. (2007) An arthropod cuticular chitin-binding protein endows injured sites with transglutaminase-dependent mesh. J. Biol. Chem. 282, 37316–37324 [DOI] [PubMed] [Google Scholar]
- 7. Wang R., Liang Z., Hal M., and Söderhall K. (2001) A transglutaminase involved in the coagulation system of the freshwater crayfish, Pacifastacus leniusculus. Tissue localisation and cDNA cloning. Fish Shellfish Immunol. 11, 623–637 [DOI] [PubMed] [Google Scholar]
- 8. Shibata T., Ariki S., Shinzawa N., Miyaji R., Suyama H., Sako M., Inomata N., Koshiba T., Kanuka H., and Kawabata S. (2010) Protein crosslinking by transglutaminase controls cuticle morphogenesis in. Drosophila. PLoS One 5, e13477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shibata T., and Kawabata S. (2015) Transglutaminase in invertebrates, in Transglutaminases (Hitomi K., Kojima S., and Fesus L., eds) pp. 117–127, Springer, New York [Google Scholar]
- 10. Lindgren M., Riazi R., Lesch C., Wilhelmsson C., Theopold U., and Dushay M. S. (2008) Fondue and transglutaminase in the Drosophila larval clot. J. Insect Physiol. 54, 586–592 [DOI] [PubMed] [Google Scholar]
- 11. Wang Z., Wilhelmsson C., Hyrsl P., Loof T. G., Dobes P., Klupp M., Loseva O., Mörgelin M., Iklé J., Cripps R. M., Herwald H., and Theopold U. (2010) Pathogen entrapment by transglutaminase—a conserved early innate immune mechanism. PLoS Pathog. 6, e1000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shibata T., Maki K., Hadano J., Fujikawa T., Kitazaki K., Koshiba T., and Kawabata S. (2015) Crosslinking of a peritrophic matrix protein protects gut epithelia from bacterial exotoxins. PLoS Pathog. 11, e1005244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shibata T., Sekihara S., Fujikawa T., Miyaji R., Maki K., Ishihara T., Koshiba T., and Kawabata S. (2013) Transglutaminase-catalyzed protein-protein cross-linking suppresses the activity of the NF-κB-like transcription factor relish. Sci. Signal. 6, ra61. [DOI] [PubMed] [Google Scholar]
- 14. Lemaitre B., and Hoffmann J. (2007) The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743 [DOI] [PubMed] [Google Scholar]
- 15. Ferrandon D. (2013) The complementary facets of epithelial host defenses in the genetic model organism Drosophila melanogaster: from resistance to resilience. Curr. Opin. Immunol. 25, 59–70 [DOI] [PubMed] [Google Scholar]
- 16. Hetru C., and Hoffmann J. A. (2009) NF-κB in the immune response of Drosophila. Cold Spring Harb. Perspect. Biol. 1, a000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dushay M. S., Asling B., and Hultmark D. (1996) Origins of immunity: Relish, a compound Rel-like gene in the antibacterial defense of Drosophila. Proc. Natl. Acad. Sci. U.S.A. 93, 10343–10347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Petersen U. M., Björklund G., Ip Y. T., and Engström Y. (1995) The dorsal-related immunity factor, Dif, is a sequence-specific trans-activator of Drosophila Cecropin gene expression. EMBO J. 14, 3146–3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gross I., Georgel P., Kappler C., Reichhart J. M., and Hoffmann J. A. (1996) Drosophila immunity: a comparative analysis of the Rel proteins dorsal and Dif in the induction of the genes encoding diptericin and. cecropin. Nucleic Acids Res. 24, 1238–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Manfruelli P., Reichhart J. M., Steward R., Hoffmann J. A., and Lemaitre B. (1999) A mosaic analysis in Drosophila fat body cells of the control of antimicrobial peptide genes by the Rel proteins Dorsal and DIF. EMBO J. 18, 3380–3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stoven S., Silverman N., Junell A., Hedengren-Olcott M., Erturk D., Engstrom Y., Maniatis T., and Hultmark D. (2003) Caspase-mediated processing of the Drosophila NF-κB factor Relish. Proc. Natl. Acad. Sci. U.S.A. 100, 5991–5996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pegg A. E. (2009) Mammalian polyamine metabolism and function. IUBMB Life 61, 880–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gao J. H., Guo L. J., Huang Z. Y., Rao J. N., and Tang C. W. (2013) Roles of cellular polyamines in mucosal healing in the gastrointestinal tract. J. Physiol. Pharmacol. 64, 681–693 [PubMed] [Google Scholar]
- 24. Djouhri-Bouktab L., Rolain J. M., and Brunel J. M. (2014) Mini-review: polyamines metabolism, toxicity and potent therapeutical use. Antiinfect. Agents 12, 95–103 [Google Scholar]
- 25. Piredda L., Farrace M. G., Lo Bello M., Malorni W., Melino G., Petruzzelli R., and Piacentini M. (1999) Identification of 'tissue' transglutaminase binding proteins in neural cells committed to apoptosis. FASEB J. 13, 355–364 [DOI] [PubMed] [Google Scholar]
- 26. Park S. S., Kim J. M., Kim D. S., Kim I. H., and Kim S. Y. (2006) Transglutaminase 2 mediates polymer formation of I-κBα through C-terminal glutamine cluster. J. Biol. Chem. 281, 34965–34972 [DOI] [PubMed] [Google Scholar]
- 27. Takeuchi Y., Ohashi H., Birckbichler P. J., and Ikejima T. (1998) Nuclear translocation of tissue type transglutaminase during sphingosine-induced cell death: a novel aspect of the enzyme with DNA hydrolytic activity. Z. Naturforsch. C 53, 352–358 [DOI] [PubMed] [Google Scholar]
- 28. Lesort M., Attanavanich K., Zhang J., and Johnson G. V. (1998) Distinct nuclear localization and activity of tissue transglutaminase. J. Biol. Chem. 273, 11991–11994 [DOI] [PubMed] [Google Scholar]
- 29. Peng X., Zhang Y., Zhang H., Graner S., Williams J. F., Levitt M. L., and Lokshin A. (1999) Interaction of tissue transglutaminase with nuclear transport protein importin-α3. FEBS Lett. 446, 35–39 [DOI] [PubMed] [Google Scholar]
- 30. Tatsukawa H., Fukaya Y., Frampton G., Martinez-Fuentes A., Suzuki K., Kuo T. F., Nagatsuma K., Shimokado K., Okuno M., Wu J., Iismaa S., Matsuura T., Tsukamoto H., Zern M. A., Graham R. M., et al. (2009) Role of transglutaminase 2 in liver injury via cross-linking and silencing of transcription factor Sp1. Gastroenterology 136, 1783.e10–1795.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shrestha R., Tatsukawa H., Shrestha R., Ishibashi N., Matsuura T., Kagechika H., Kose S., Hitomi K., Imamoto N., and Kojima S. (2015) Molecular mechanism by which acyclic retinoid induces nuclear localization of transglutaminase 2 in human hepatocellular carcinoma cells. Cell. Death Dis. 6, e2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Folk J. E., Park M. H., Chung S. I., Schrode J., Lester E. P., and Cooper H. L. (1980) Polyamines as physiological substrates for transglutaminases. J. Biol. Chem. 255, 3695–3700 [PubMed] [Google Scholar]
- 33. Watanabe S., Kusama-Eguchi K., Kobayashi H., and Igarashi K. (1991) Estimation of polyamine binding to macromolecules and ATP in bovine lymphocytes and rat liver. J. Biol. Chem. 266, 20803–20809 [PubMed] [Google Scholar]
- 34. Igarashi K., and Kashiwagi K. (2010) Modulation of cellular function by polyamines. Int. J. Biochem. Cell Biol. 42, 39–51 [DOI] [PubMed] [Google Scholar]
- 35. Sekihara S., Shibata T., Hyakkendani M., and Kawabata S. (2016) RNA interference directed against the transglutaminase gene triggers dysbiosis of gut microbiota in Drosophila. J. Biol. Chem. 291, 25077–25087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kuraishi T., Hori A., and Kurata S. (2013) Host-microbe interactions in the gut of Drosophila melanogaster. Front. Physiol. 4, 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee K. Z., and Ferrandon D. (2011) Negative regulation of immune responses on the fly. EMBO J. 30, 988–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zaidman-Rémy A., Hervé M., Poidevin M., Pili-Floury S., Kim M. S., Blanot D., Oh B. H., Ueda R., Mengin-Lecreulx D., and Lemaitre B. (2006) The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity 24, 463–473 [DOI] [PubMed] [Google Scholar]
- 39. Paredes J. C., Welchman D. P., Poidevin M., and Lemaitre B. (2011) Negative regulation by amidase PGRPs shapes the Drosophila antibacterial response and protects the fly from innocuous infection. Immunity 35, 770–779 [DOI] [PubMed] [Google Scholar]
- 40. Bischoff V., Vignal C., Duvic B., Boneca I. G., Hoffmann J. A., and Royet J. (2006) Downregulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. PLoS Pathog. 2, e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim M., Lee J. H., Lee S. Y., Kim E., and Chung J. (2006) Caspar, a suppressor of antibacterial immunity in. Drosophila. Proc. Natl. Acad. Sci. U.S.A. 103, 16358–16363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kleino A., Myllymäki H., Kallio J., Vanha-aho L. M, Oksanen K., Ulvila J., Hultmark D., Valanne S., and Rämet M. (2008) Pirk is a negative regulator of the Drosophila Imd pathway. J. Immunol. 180, 5413–5422 [DOI] [PubMed] [Google Scholar]
- 43. Lhocine N., Ribeiro P. S., Buchon N., Wepf A., Wilson R., Tenev T., Lemaitre B., Gstaiger M., Meier P., and Leulier F. (2008) PIMS modulates immune tolerance by negatively regulating Drosophila innate immune signaling. Cell Host Microbe 4, 147–158 [DOI] [PubMed] [Google Scholar]
- 44. Aggarwal K., Rus F., Vriesema-Magnuson C., Ertürk-Hasdemir D., Paquette N., and Silverman N. (2008) Rudra interrupts receptor signaling complexes to negatively regulate the IMD pathway. PLoS Pathog. 4, e1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bond D., and Foley E. (2009) A quantitative RNAi screen for JNK modifiers identifies Pvr as a novel regulator of Drosophila immune signaling. PLoS Pathog. 5, e1000655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Basbous N., Coste F., Leone P., Vincentelli R., Royet J., Kellenberger C., and Roussel A. (2011) The Drosophila peptidoglycan-recognition protein LF interacts with peptidoglycan-recognition protein LC to downregulate the Imd pathway. EMBO Rep. 12, 327–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Aparicio R., Neyen C., Lemaitre B., and Busturia A. (2013) dRYBP contributes to the negative regulation of the Drosophila Imd pathway. PLoS One 8, e62052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Foley E., and O'Farrell P. H. (2004) Functional dissection of an innate immune response by a genome-wide RNAi screen. PLoS Biol. 2, E203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cao Y., Chtarbanova S., Petersen A. J., and Ganetzky B. (2013) Dnr1 mutations cause neurodegeneration in Drosophila by activating the innate immune response in the brain. Proc. Natl. Acad. Sci. U.S.A. 110, E1752–E1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ryu J. H., Kim S. H., Lee H. Y., Bai J. Y., Nam Y. D., Bae J. W., Lee D. G., Shin S. C., Ha E. M., and Lee W. J. (2008) Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in. Drosophila. Science 319, 777–782 [DOI] [PubMed] [Google Scholar]
- 51. Bonnay F., Cohen-Berros E., Hoffmann M., Kim S. Y., Boulianne G. L., Hoffmann J. A., Matt N., and Reichhart J. (2013) big bang gene modulates gut immune tolerance in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 110, 2957–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kim S. H., and Lee W. J. (2014) Role of DUOX in gut inflammation: lessons from Drosophila model of gut-microbiota interactions. Front. Cell. Infect. Microbiol. 3, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lee K. A., Kim S. H., Kim E. K., Ha E. M., You H., Kim B., Kim M. J., Kwon Y., Ryu J. H., and Lee W. J. (2013) Bacterial-derived uracil as a modulator of mucosal immunity and gut-microbe homeostasis in. Drosophila. Cell 153, 797–811 [DOI] [PubMed] [Google Scholar]
- 54. Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]