Abstract
The propeptides of subtilisin-like serine proteinases (subtilases, SBTs) serve dual functions as intramolecular chaperones that are required for enzyme folding and as inhibitors of the mature proteases. SBT propeptides are homologous to the I9 family of protease inhibitors that have only been described in fungi. Here we report the identification and characterization of subtilisin propeptide-like inhibitor 1 (SPI-1) from Arabidopsis thaliana. Sequence similarity and the shared β-α-β-β-α-β core structure identified SPI-1 as a member of the I9 inhibitor family and as the first independent I9 inhibitor in higher eukaryotes. SPI-1 was characterized as a high-affinity, tight-binding inhibitor of Arabidopsis subtilase SBT4.13 with Kd and Ki values in the picomolar range. SPI-1 acted as a stable inhibitor of SBT4.13 over the physiologically relevant range of pH, and its inhibitory profile included many other SBTs from plants but not bovine chymotrypsin or bacterial subtilisin A. Upon binding to SBT4.13, the C-terminal extension of SPI-1 was proteolytically cleaved. The last four amino acids at the newly formed C terminus of SPI-1 matched both the cleavage specificity of SBT4.13 and the consensus sequence of Arabidopsis SBTs at the junction of the propeptide with the catalytic domain. The data suggest that the C terminus of SPI-1 acts as a competitive inhibitor of target proteases as it remains bound to the active site in a product-like manner. SPI-1 thus resembles SBT propeptides with respect to its mode of protease inhibition. However, in contrast to SBT propeptides, SPI-1 could not substitute as a folding assistant for SBT4.13.
Keywords: molecular chaperone, plant biochemistry, protease inhibitor, protein processing, proteolysis, serine protease, intramolecular chaperone, prodomain, subtilase, subtilisin
Introduction
Proteolysis is essential for life. Proteolytic enzymes are required in all organisms for protein turnover by non-selective protein degradation. Proteolysis also serves important regulatory roles as many aspects of growth and development are controlled by selective degradation of regulatory proteins. In addition to protein degradation, the specific processing of precursor proteins controls the activity of enzymes, regulatory proteins, and peptide hormones. Post-translational processing of proteins may also be required for protein assembly or subcellular targeting. As the process of proteolysis is essentially irreversible under physiological conditions, it obviously needs tight regulation. Proteases are indeed regulated at the transcript level but most importantly by post-translational mechanisms. To prevent unwanted protein degradation and to enable spatial and temporal control of proteolytic activity, most proteases are produced as inactive precursors, zymogens, that rely on specific activation processes for enzyme maturation (1). Finally, proteolytic activity of the mature enzyme can be regulated by substrate availability, cofactors, pH, the assembly of oligomeric complexes, and, last but not least, a plethora of proteinase inhibitors (PIs)2 (2–5).
Based on their amino acid sequences, the MEROPS database currently distinguishes 79 families of peptidase inhibitors that are assigned to 38 different clans according to tertiary structure (release 10.0) (6, 7). The family of I9 inhibitors belongs to clan JC exemplified by the propeptide (PP) of subtilisin BPN′ from Bacillus amyloliquefaciens. The subtilisin PP features an α-β sandwich formed by a four-stranded antiparallel β-sheet and two three-turn α-helices as the characteristic I9 fold (8). I9 family members exclusively inhibit subtilases (SBTs), i.e. the S8 family of serine peptidases (MEROPS), including subtilisins in bacteria and plants as well as kexin in yeast and the related mammalian proprotein convertases (PCs) (9).
Most members of the I9 family are not independent proteins but are rather found as N-terminal domain in SBTs, i.e. the prodomain, or propeptide. As PPs, these I9 inhibitors are responsible for the latency of the zymogens (1, 4, 5), and, on top of their inhibitor function, they are also required for protein folding. The dual function of PPs as intramolecular chaperones and enzyme inhibitors was first described for subtilisin E of Bacillus subtilis (10) and was later reported for a wide range of other proteases as well (11–14). When expressed without their respective PPs, subtilisins accumulate in an inactive, semifolded, molten globule-like state. By co-expression of their respective PPs or by stoichiometric addition of the PP to the unfolded protein, the intermediate acquires the native state, and active protein is regained (11, 12). The chaperoning function of PPs was confirmed for SBTs in mammals (14, 15) and, most recently, in plants (16). A PP deletion mutant of tomato SBT3 fails to fold correctly and accumulates intracellularly, whereas co-expression in trans of its cognate prodomain restores secretion of the active protease to the apoplast (16). Upon completion of folding and autocatalytic cleavage, the PP binds to the subtilisin domain in an autoinhibited complex of exceptional stability, thus switching from an intramolecular chaperone to an inhibitor of the enzyme (16–19).
Crystal structure analysis of PP·SBT complexes revealed a common mechanism of autoinhibition for bacterial subtilisins (8, 20), mammalian PCSK9 (21), and cucumisin, an abundant SBT in melon fruits (22). In each case, the β-sheet of the propeptide packs against two parallel surface helices of the respective protease, whereas its C terminus binds to the nonprime subsites of the catalytic center in a product-like manner (subsites up- and downstream of the cleavage site are called nonprime and prime subsites, respectively, according to Schechter and Berger (65)). Release from autoinhibition requires a second cleavage of the prodomain. For several mammalian PCs as well as tomato SBT3, secondary cleavage was found to be pH-dependent, and therefore it occurs in a compartment-specific manner as the pH drops along the secretory pathway (15, 16, 23).
Despite their prevalence, the PPs of subtilases are not the only members of the I9 family. Two I9 inhibitors have been described in fungi that are independent proteins and not part of a protease precursor, namely Saccharomyces cerevisiae proteinase B inhibitor 2 (ScIB2) and Pleurotus ostreatus proteinase A inhibitor 1 (PoIA1) (24, 25). In line with the mechanism of PP-mediated SBT inhibition, the inhibitory activity of ScIB2 and PoIA1 against vacuolar proteinase B and proteinase ProA of P. ostreatus also was found to depend on the extreme C termini of the two inhibitor proteins (24, 26, 27). PoIA1 was confirmed as a member of the I9 family by protein NMR (28). Comparing the relative accessible surface area between PoIA1 and the PP of subtilisin in its free and protease-bound forms, the mode of binding and protease inhibition by PoIA1 was shown to resemble that of subtilisin PP (28). The similarity of PoIA1 and PPs also includes the ability to assist in protein folding. When fused to the catalytic domain of subtilisin BPN′, PoIA1 substitutes for its PP as an intramolecular chaperone (29). Interestingly, yeast ScIB2 has additional functions that are independent from its interaction with proteinase B. In a heterodimeric complex with thioredoxin, ScIB2 binds to the SNARE complex at yeast vacuolar membranes, stabilizes the target membrane-associated SNARE Vamp3P, and facilitates vacuole fusion (30, 31).
Higher eukaryotes appear to lack independent I9 inhibitors (I9s) (5). However, in our continuing effort to functionally characterize the SBT gene family in plants (9, 32), we noticed that plant genomes comprise genes that may be wrongly annotated as SBTs. Their open reading frames comprise the N-terminal part of SBTs, including a signal peptide for secretion, the PP, and a short C-terminal extension, but they do not contain the catalytic domain. Hypothesizing that the predicted proteins may actually represent independent I9s, we focused on the gene At1g71950 in Arabidopsis. We show here that At1g71950 codes for the first subtilisin propeptide-like inhibitor (SPI-1) in plants. The relationship between SPI-1 and the PPs of plant SBTs was addressed in phylogenetic analyses and by functional comparison of their inhibitory and chaperoning activities.
Results
Phylogeny of I9 inhibitors and primary structure of SPI-1
The Arabidopsis genome comprises 56 SBT genes (36). In contrast, there are only two genes for potential I9s, At1g71950 (SPI-1; the one characterized here) and At2g39851 (SPI-2). To address the phylogenetic relationship of SPIs, the PPs of SBTs, and I9s in fungi, we first performed database searches using the Basic Local Alignment Search Tool for proteins (blastP) to identify SPI-1 homologs in other species. SPI-1 homologs were found across the plant kingdom, including monocots, dicots, and mosses, but not in algae. Sequence identity with SPI-1 ranged from 30.4% in Physcomitrella patens (Pp00121G00530) to 60.3% in tomato, Solanum lycopersicum (Sl06g065370). SPI-2 shares only 22% sequence identity with SPI-1, and SPI-2 homologs were found only in the Brassicales. For sequence comparison with SPIs, we chose the well characterized PPs of subtilisin A from Bacillus licheniformis and the PPs of all Arabidopsis SBTs in clades 1–5 in addition to the fungal I9 inhibitors PoIA1 and ScIB2. The signal peptides predicted by SignalP 3.0 for all proteins except PoIA1 and ScIB2 were omitted during sequence comparison. Because sequence identity between SPI-1 and the fungal I9s is less than 10%, PROMALS3D was used to calculate the sequence alignment taking additional information from secondary structure predictions and available 3D structures into account (33). Based on the sequence alignment, a phylogenetic tree was generated using the Genome-to-Genome Distance Calculator web server (34). Phylogenetic analysis clearly separated SPI-1 homologs from SPI-2, SBT PPs, and fungal inhibitors (Fig. 1). SPI-1 homologs clustered in a monophyletic clade consisting of two subclades with the P. patens inhibitor as an outgroup. Among the SPI-1 homologs, two I9 inhibitors from tomato were found to be most closely related to SPI-1 (Fig. 1). The results suggest that SPI-1 and related I9s diverged early in the plant lineage and evolved independently from SBT PPs and SPI-2. They also indicate that plant I9s are only distantly related to PoIA1 and ScIB2 in fungi, which cluster together with bacterial PPs.
Figure 1.
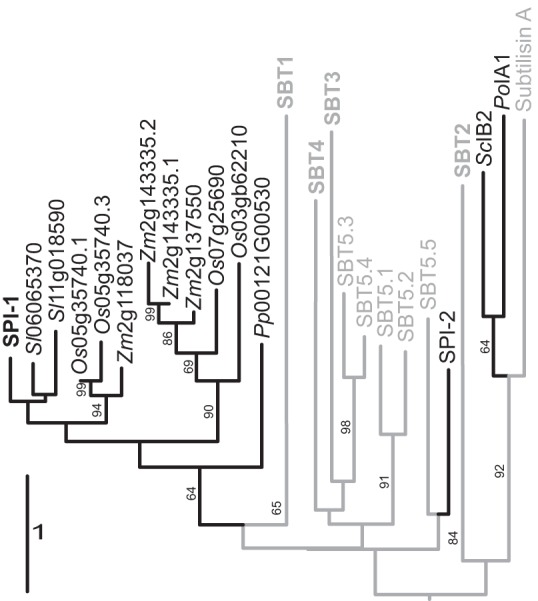
Phylogenetic relationship of propeptides and I9 inhibitors. Maximum likelihood tree was calculated on the Genome-to-Genome Distance Calculator web server and rooted by midpoint rooting of SPI-1 with plant homologs (black) from tomato (Sl); Sl replaces Solyc in tomato gene identifiers, rice (Oryza sativa (Os)), and maize (Zea mays (Zm); Zm replaces GRMZM in maize gene identifiers), also including propeptides of subtilases (gray) from Arabidopsis SBT subfamilies 1–5, subtilisin A from B. licheniformis, and the fungal I9 inhibitors and Arabidopsis SPI-2 (black). The PPs of Arabidopsis SBT subfamilies 1–4 clustered in four distinct clades; these clades were collapsed and are labeled SBT1, -2, -3, and -4, respectively. The branches are scaled in terms of the expected number of substitutions per site. Maximum likelihood bootstrapping values are provided on the left of the branches when larger than 60%.
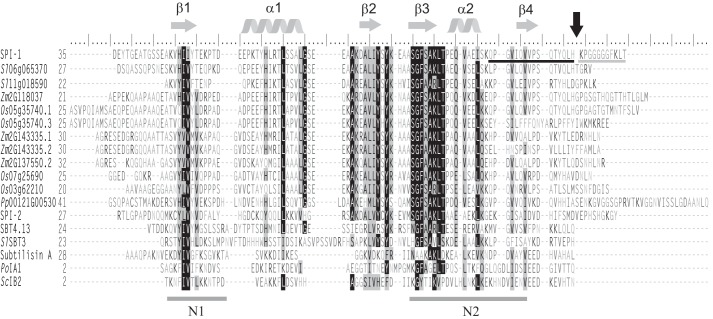
Taking a closer look at the protein sequences, the structural scaffold of two β-α-β motifs was found to be conserved in the PPs of bacterial subtilisins, mammalian PCs, and plant SBTs alike (Fig. 2). This core structure, which is indispensable for the interaction of PPs with their cognate proteases (8, 14, 16, 35), was also predicted in SPI-1 and its plant homologs (Fig. 2). Sequence variability between PPs and I9s was greatest at their N and C termini. As judged from the crystal structure of the cucumisin·prodomain complex, the N terminus of the PP does not seem to be important for binding (22). Therefore, it may not be subject to evolutionary selection, possibly explaining N-terminal variability in length and sequence. The C-terminal extensions also varied considerably, even within the group of I9s that are otherwise well conserved in their β-α-β-β-α-β core. In the group of SPI homologs, C-terminal variability was most obvious for two splicing variants in maize and rice, which are identical in sequence except for their C termini.
Figure 2.
Amino acid sequence alignment of SPI-1, its plant homologues, SPI-2, the propeptides of AtSBT4.13, SlSBT3, subtilisin A, and two fungal I9 inhibitors after removal of the predicted N-terminal signal peptides (SignalP 3.0; Ref. 64). The sequence alignment with secondary structure prediction was generated using PROMALS3D (33). Highly conserved residues (identical in >70% of the sequences) and partially conserved residues are shaded in black and gray, respectively. Conserved α-helices and β-strands are indicated as gray helices or arrows above the alignment. Conserved hydrophobic regions N1 and N2 are underlined. The C-terminal cleavage site of SPI-1 is marked by a black arrow. ESI-MS- and MALDI-TOF MS-identified peptides flanking the cleavage site are underlined in black and gray, respectively. Residues are numbered from the first Met. Os, O. sativa; Zm, Z. mays.
Considering on the one hand the early separation and independent evolution of plant SPI-1 homologs and on the other hand their conserved core structure, we investigated to what extent Arabidopsis SPI-1 shares the chaperoning and inhibitor functions that are well described for subtilase PPs and I9s in fungi. The functional significance of C-terminal variability was also addressed.
Proteinase inhibitor activity and specificity of SPI-1
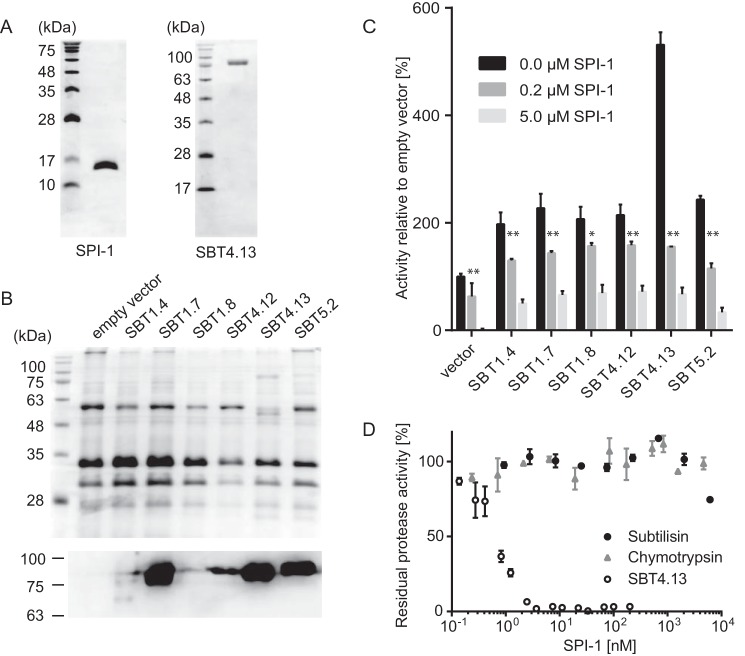
To assess the inhibitory potential of SPI-1 and its specificity for individual SBTs, the protein was expressed in Escherichia coli with an N-terminal His6 tag replacing the signal peptide and purified to homogeneity (Fig. 3A). Unlike the PPs of plant SBTs that accumulate as insoluble inclusion bodies when expressed in E. coli (16, 19), SPI-1 could be purified from the soluble protein fraction by metal-chelate affinity chromatography. Arabidopsis SBTs were expressed in Nicotiana benthamiana by agroinfiltration. SBTs were chosen that could be detected in cell wall extracts after transient expression and assayed in apoplastic washes using FITC-casein as a universal proteinase substrate. Expression of these SBTs was confirmed by Coomassie Brilliant Blue staining for the most highly expressed SBTs and by immunoblotting analysis (Fig. 3B). SBTs shown in Fig. 3C were found to cleave FITC-casein resulting in an increase in relative fluorescence as compared with the control (extracts from empty vector-infiltrated plants). The activity of all these SBTs was increasingly inhibited by addition of 0.2 and 5 μm SPI1–1, respectively. Interestingly, the background activity observed for the empty vector control also was significantly inhibited, suggesting that SPI-1 is rather non-selective, inhibiting a variety of SBTs from Arabidopsis as well as unidentified extracellular proteases in N. benthamiana (Fig. 3C). Further supporting this notion, SPI-1 was also found to be active against SBTs from tomato.3
Figure 3.
Expression of Arabidopsis SBTs and inhibition by SPI-1. A, purification of SPI-1 and SBT4.13. Two micrograms of recombinant SPI-1 (left) and SBT4.13 (right) purified form E. coli extracts (SPI-1) and apoplastic washes of agroinfiltrated N. benthamiana plants (SBT4.13) were separated by 15 or 12% SDS-PAGE, respectively. Gels were stained with Coomassie Brilliant Blue R-250. The molecular mass of the marker proteins is indicated. B, SDS-PAGE and Western blotting analysis of transiently expressed Arabidopsis subtilases. The Coomassie-stained gel is shown (top) as a control for protein loading. Only the most highly expressed SBTs (SBT1.7, -4.13, and -5.2) are visualized by Coomassie staining. To detect SBTs on the immunoblot, a mixture of polyclonal antisera directed against tomato SBTs 1–4 (1:10,000) was used in combination with a peroxidase-conjugated secondary antibody (1:10,000; Calbiochem) with enhanced chemiluminescence detection. C, activity of Arabidopsis SBTs and inhibition by SPI-1. SBT activity was assayed with fluorescence-labeled casein as a universal substrate (Pierce) in the presence of 0, 0.2, or 5 μm SPI-1. Activity was normalized against protein concentration assayed using the Bradford procedure, and it is shown relative to background activity in extracts of empty vector-infiltrated control plants (background in the presence of 5 μg of SPI-1 was set at 0). Data represent the mean of three experiments using independent inhibitor preparations; error bars indicate standard deviation. Two-way analysis of variance in combination with Tukey's multiple-comparisons test (GraphPad Prism 6) was performed, and significant differences are shown between samples without and with 0.2 μm SPI-1 at p ≤ 0.001 (*) and p ≤ 0.0001 (**), respectively. D, activity of SPI-1 against SBT4.13, α-chymotrypsin, and subtilisin A. Protease activity was assayed with 1 mm N-succinyl-Ala-Ala-Pro-Phe p-nitroanilide as the substrate for chymotrypsin and subtilisin A and with an internally quenched fluorigenic peptide substrate (25 μm) for SBT4.13. Assays were performed at room temperature with increasing concentrations of SPI-1. Activity is expressed as percentage of the respective protease activity without inhibitor. Error bars indicate standard deviation.
We further tested whether the inhibitory spectrum of SPI-1 also includes bacterial subtilisins and other serine proteinases. In contrast to Arabidopsis SBT4.13 purified to homogeneity from cell wall extracts of agroinfiltrated N. benthamiana plants (Fig. 3A) that was inhibited by SPI-1 in a dose-dependent manner, no inhibition was observed for subtilisin A and chymotrypsin (Fig. 3D). Therefore, SPI-1 is not a general inhibitor of subtilases. Unlike the Kazal-like extracellular PIs (EPI1 and EPI10) of Phytophthora infestans (37, 38) that are active against both plant and bacterial SBTs (32), SPI-1 appears to be a specific inhibitor of only plant SBTs. Its inhibitory profile is thus more similar to that of (at least some) plant PPs that show no activity against subtilisin but inhibit several plant SBTs in addition to their own catalytic domain (19).
Kinetic analysis of SPI-1 binding and inhibition of SBT4.13
SBT4.13 showed highest expression levels of all tested SBTs and was strongly inhibited by SPI-1 (Fig. 3C). SBT4.13 was thus chosen to further characterize enzyme-inhibitor interaction. The enzyme was purified to homogeneity (Fig. 3A), and its inhibition by SPI-1 was analyzed under steady-state conditions using an internally quenched fluorigenic peptide substrate. Activity assays required a minimum SBT4.13 concentration of 1.5 nm. SPI-1 acted as a tight-binding inhibitor under these conditions (Ki ≪ [E]0), and therefore conventional kinetics could not be applied. We therefore fitted the untransformed velocity data to the Morrison equation that applies to tight-binding conditions (39, 40). The apparent Ki for the inhibition of SBT4.13 by SPI-1 was indeed very low (Ki(app) < 10 × [E]0) at 123 ± 48 pm (Fig. 4A). To verify the apparent high-affinity interaction, we determined the dissociation constant (Kd) of the SBT4.13·SPI-1 complex by microscale thermophoresis. Fluorescently labeled SBT4.13 was titrated with increasing concentrations of unlabeled SPI-1. Complex formation was recorded as the ligand-dependent change in thermophoretic behavior and plotted against SPI-1 concentration (Fig. 4B). A Kd of 546 ± 155 pm was derived from the saturation curve, which is in good agreement with the apparent Ki value, indicating that SPI-1 is a high-affinity, tight-binding inhibitor of SBT4.13.
Figure 4.
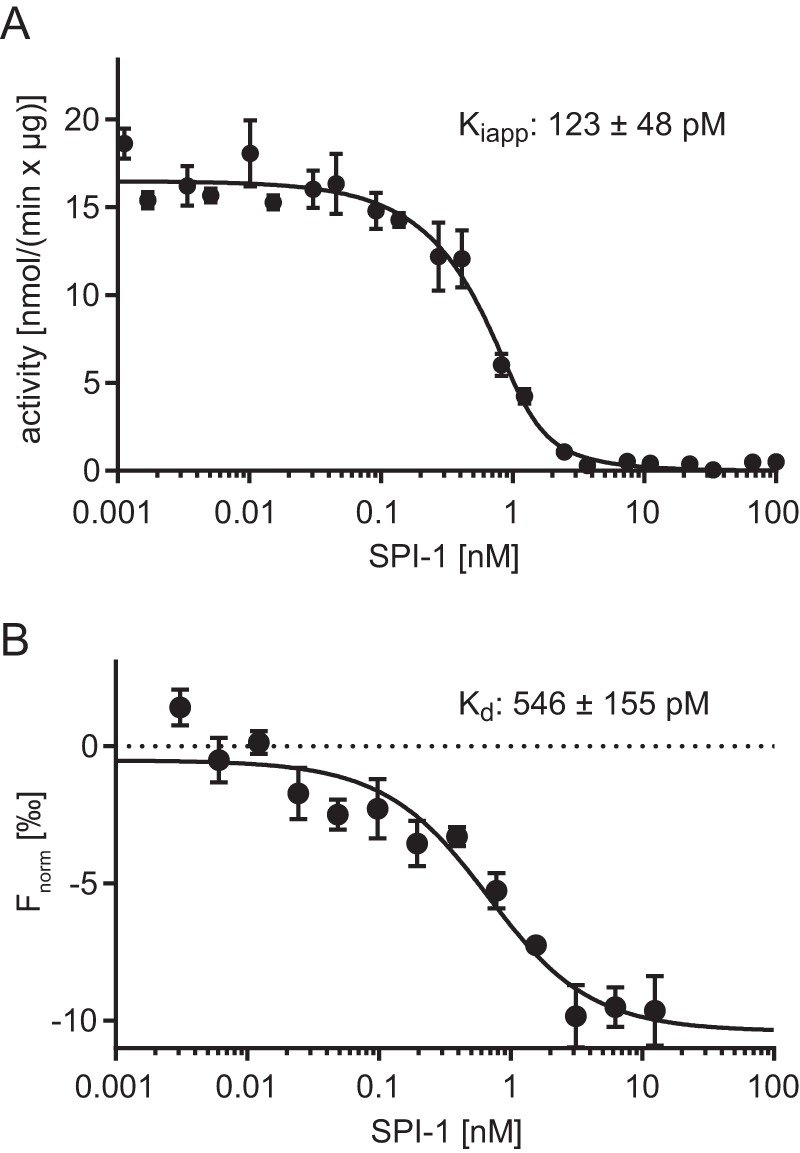
Interaction of SBT4.13 with SPI-1. A, apparent inhibition constant. Purified SBT4.13 (1.5 nm) was incubated with two serial dilutions (1:2) of SPI-1 starting at 100 and 200 nm, respectively. Activity was recorded over 10 min at room temperature using a fluorigenic peptide substrate at 25 μm. An apparent inhibition constant (Ki(app)) of 123 ± 48 pm was calculated using the Morrison equation (39, 40) in GraphPad Prism 6 (R2 = 0.979). Results represent the mean of three biological replicates with independent SPI-1 preparations. B, dissociation constant analyzed by MST. Labeled SBT4.13 (250 pm) was titrated against a serial dilution (1:1) of unlabeled SPI-1 starting at 12.5 nm. The baseline-corrected normalized fluorescence (Fnorm) is plotted against the SPI-1 concentration. A Kd of 546 ± 155 pm was calculated with GraphPad Prism 6 (R2 = 0.911). Results represent the mean ± S.E. (error bars) of three biological replicates using independent SPI-1 preparations, all run in technical triplicates.
Characterization of the SPI-1·SBT4.13 complex
To further investigate the interaction of SPI-1 and SBT4.13, the two proteins were mixed at 4 °C using the inhibitor at 3-fold molar excess followed by gel filtration to separate the complex from unbound SPI-1 (Fig. 5). SDS-PAGE analysis of peak fractions confirmed the formation of a stable and inactive complex as SPI-1 co-eluted with SBT4.13, and proteolytic activity was inhibited as compared with the free protease (Fig. 5, A and B). To assess the stability of SPI-1 during its interaction with SBT4.13, the two proteins were incubated at room temperature at a molar protease-to-inhibitor ratio of 1:1.5 to allow for complex formation and analyzed by SDS-PAGE after different time intervals (Fig. 6A). Intriguingly, SPI-1 exhibited a small shift in size already at the earliest time point (after 5 min). The resulting smaller protein was stable for at least 4 h. Only after overnight incubation was band intensity somewhat reduced (Fig. 6A), indicating that under these conditions SPI-1 is a rather stable inhibitor of SBT4.13.
Figure 5.
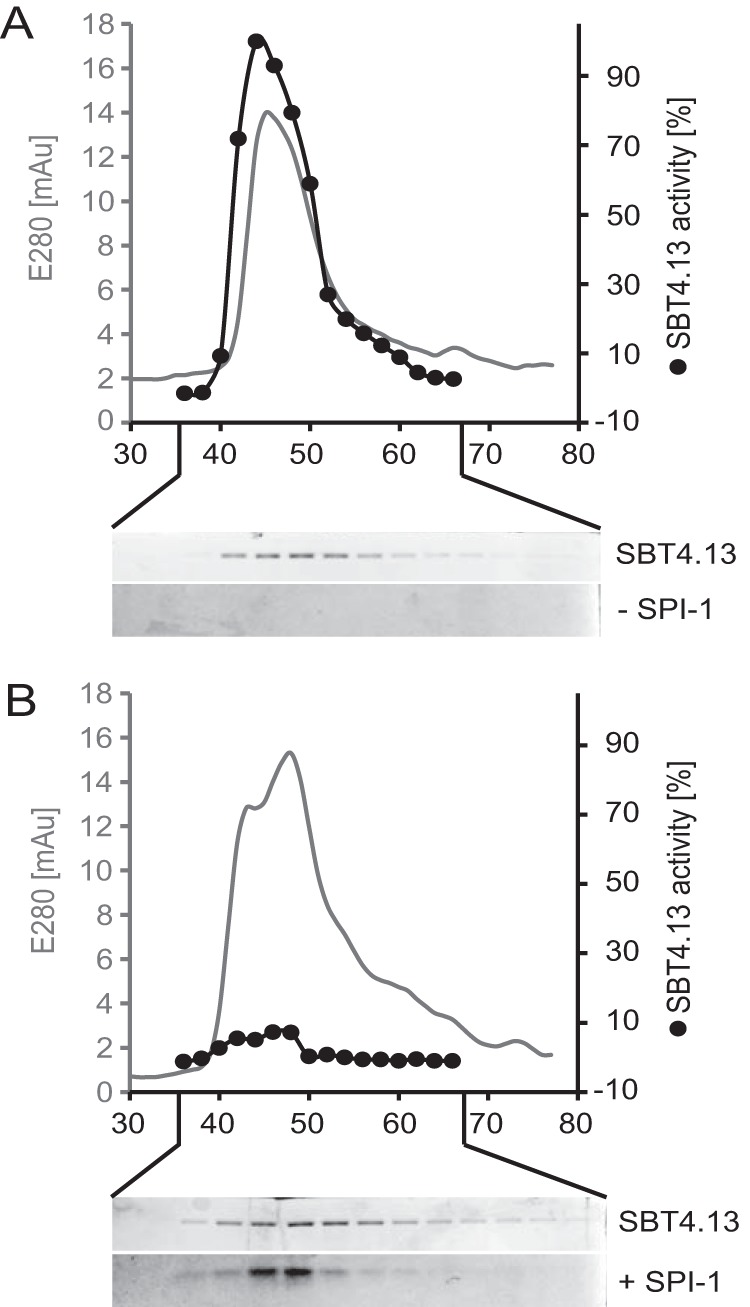
Purification of the SBT4.13·SPI-1 complex. SBT4.13 and the SBT4.13·SPI-1 complex were separated by gel filtration. Fractions were analyzed by SDS-PAGE, and SBT4.13 activity was assayed using a fluorigenic peptide substrate at 25 μm. A, gel filtration of SBT4.13 (100 μg) monitored at 280 nm (gray line) and collected in 200-μl fractions. SBT4.13 activity was assayed in 0.5-μl aliquots (black dots) in two technical replicates. Fifteen-microliter aliquots of the same fractions were separated by 15% SDS-PAGE and Coomassie-stained as shown below the chromatogram. B, SBT4.13 (100 μg) incubated for 10 min in the presence of a 3-fold molar excess of SPI-1 was separated by gel filtration, and fractions were analyzed as described for A. mAU, milliabsorbance units.
Figure 6.
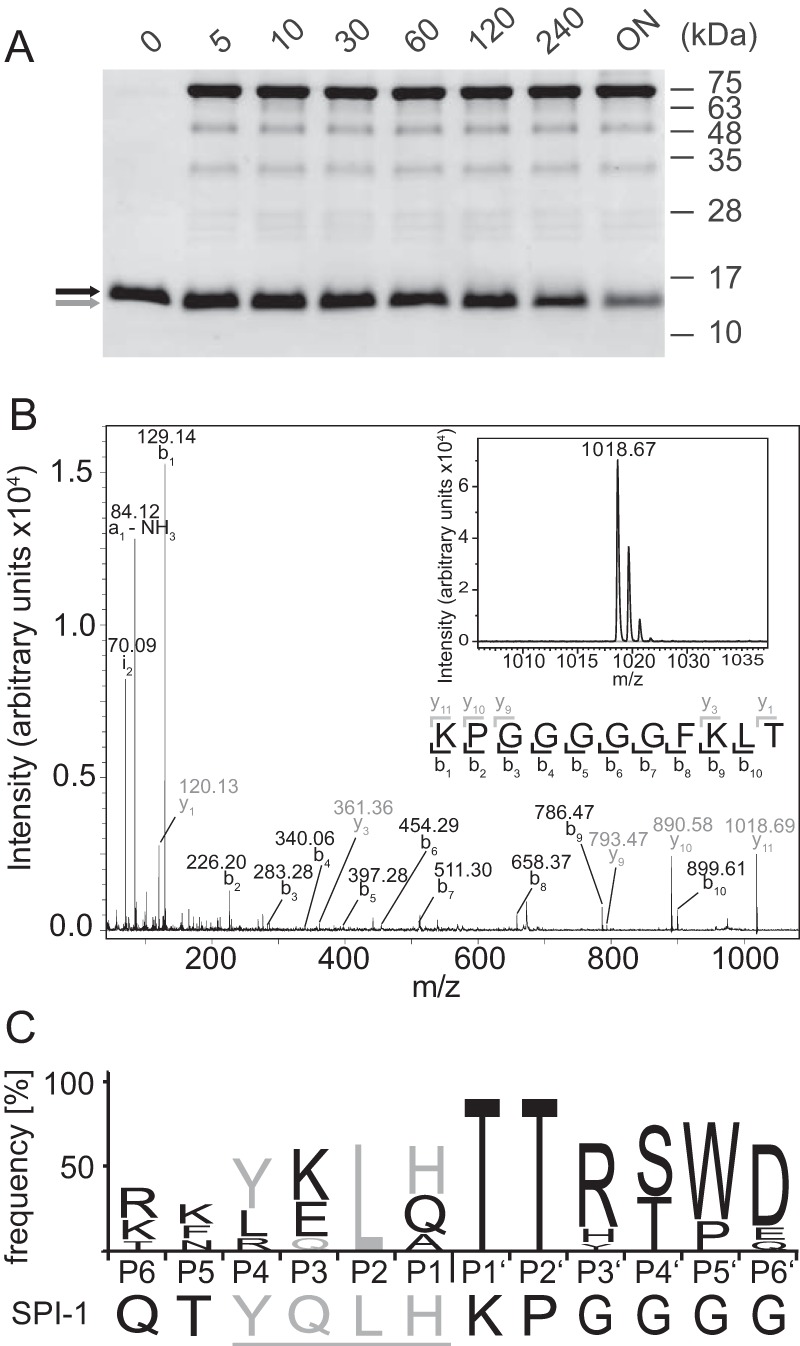
Complex stability over time and C-terminal cleavage of SPI-1. A, SBT4.13 (5 μm) was incubated with SPI-1 in 1.5-fold molar excess, and at the indicated time points aliquots containing 1.4 μg of SPI-1 and 5 μg of SBT4.13 were separated by 15% SDS-PAGE and Coomassie-stained. Full-length SPI-1 and its cleavage product are marked by black and gray arrows, respectively. B, identification of the C-terminal SPI-1 cleavage product generated after 10-min incubation with SBT4.13. The low-molecular-mass fraction (10-kDa cutoff) of the SBT4.13·SPI-1 digest and a control consisting of only SPI-1 were subjected to MALDI-TOF peptide mass fingerprinting to identify peptides that were released upon binding of the protease. The relevant part of the peptide mass fingerprints is shown in the inset, revealing a mass signal of 1018.67 Da in the SBT4.13-treated sample (black) that was not detected in SPI-1 control (gray baseline signal). The identity and sequence of the 11-mer peptide were confirmed by b- and y-ion series as indicated in black and gray, respectively. C, Icelogo showing sequence conservation in six positions upstream (P1–P6) and downstream (P1′–P6′) of the propeptide processing site of the 54 Arabidopsis SBTs in subfamilies 1–5 as compared with the C-terminal cleavage site in SPI-1. Amino acid distribution is expressed as percent difference (p = 0.05) over the representation in the Arabidopsis proteome (Swiss-Prot protein database for Arabidopsis thaliana). Amino acids of SPI-1 that are over-represented in SBTs are shown in gray and underlined. ON, overnight.
To analyze whether the shift in size observed after 5 min resulted from cleavage of the inhibitor, the SPI-1 band was cut out from the gel and digested with trypsin, and resulting peptides were analyzed by ESI-MS. In parallel, the complex was established and subjected to ultrafiltration. Potential cleavage products that would have been lost during SDS-PAGE were recovered in the filtrate and analyzed by MALDI-TOF MS. Both approaches identified a single cleavage site close to the C terminus of SPI-1 (indicated in Fig. 2B). The C-terminal cleavage product is shown in Fig. 6B. Comparing the amino acids flanking the cleavage site with the cleavage specificity of SBT4.13, the four residues on the nonprime side (upstream) of the scissile bond (YQLH) were found to match the substrate preference of SBT4.13 (32). The data suggest that SBT4.13 cleaves SPI-1 upon binding, resulting in the stable protease·inhibitor complex.
We further compared the sequence flanking the cleavage site of SPI-1 with the amino acid sequence at the junction between the PP and the catalytic domain of all Arabidopsis subtilases. The sequence logo in Fig. 6C shows the relative abundance of individual amino acids six positions upstream (P1–P6) and six positions downstream (P1′–P6′) of the propeptide processing site in Arabidopsis SBTs as compared with the SPI-1 sequence. Prime-side residues in positions P1′–P5′ represent the N terminus of the mature SBTs that was found to be highly conserved (TTX(S/T)(W/P)) as reported previously for SBTs in Arabidopsis and tomato (16, 36, 41). SPI-1 did not show any sequence similarity in this region. In contrast, SPI-1 residues on the non-prime side of the cleavage site, particularly the sequence YQLH in positions P4–P1, strongly resembled the C terminus of Arabidopsis PPs (Fig. 6C). These observations suggest that SPI-1 is recognized as a substrate and is efficiently cleaved by SBT4.13 at a position corresponding to the autocatalytic propeptide processing site. Resembling the residues upstream of the conserved processing site, the newly formed C terminus of SPI-1 is perfectly suited to occupy the S1–S4 substrate-binding pockets of SBT4.13 and other Arabidopsis SBTs, thus explaining its broad inhibitory activity (Fig. 3C). The data suggest that the neo-C terminus of SPI-1 remains bound and blocks the active site of the protease in a product-like manner as reported for PPs of bacterial and plant SBTs (20, 22) and inferred for fungal I9s (28).
pH stability of the proteinase·inhibitor complex
PPs are temporary inhibitors of their cognate proteases. As part of the maturation process, PPs need to be cleaved a second time, resulting in the release of the active protease. SPI-1 appears to be resistant to secondary cleavage, allowing for more permanent inhibition of SBT4.13 (Fig. 6A), at least under these experimental conditions. For the PPs of tomato SBT3 and some mammalian PCs, secondary cleavage was shown to be pH-dependent. Therefore, these SBTs are inhibited by their cognate PPs only until the pH drops as they transit the secretory pathway, and they are activated by autocatalytic processing in the acidic environment of the trans-Golgi network or dense-core secretory granules, respectively (15, 16, 23). To test whether SPI-1 also is cleaved conditionally and its inhibitory activity is therefore transient, we assessed the stability of the SBT4.13·SPI-1 complex under different pH conditions. His-tagged SPI-1 was bound to Ni-NTA-agarose and incubated with SBT4.13 at neutral pH to establish the complex on a solid matrix. Stepwise change of pH from pH 7 down to pH 5 or from pH 7 up to pH 10 did not release the protease from the matrix, indicating stability of the complex in this pH range. At pH 12 and pH 4, the interaction was lost as SBT4.13 was eluted, whereas most of the His-tagged inhibitor remained attached to the matrix (Fig. 7, A and B). Microscale thermophoresis was then used to determine the dissociation constant of the complex at a pH ranging from 7 to 4 (Fig. 7C). Consistent with the stability of the complex at pH 6 (Fig. 7B), there was no significant difference in Kd when the pH was lowered from 7 to 6 (Fig. 7C). At pH 5, however, the Kd increased to 30 nm, and at pH 4, no interaction was observed anymore. Because Kd values for protease-inhibitor interaction in the low nanomolar range are sufficient for effective protease inhibition in vivo (32), we expect that the Kd of 30 nm at pH 5 still allows for SBT inhibition by SPI-1 in planta. Considering the pH in different cellular compartments, ranging from pH 8.2 in peroxisomes to pH 5.2 in the vacuole (42), we conclude that SPI-1 is a stable inhibitor of SBTs in the physiologically relevant range of pH.
Figure 7.
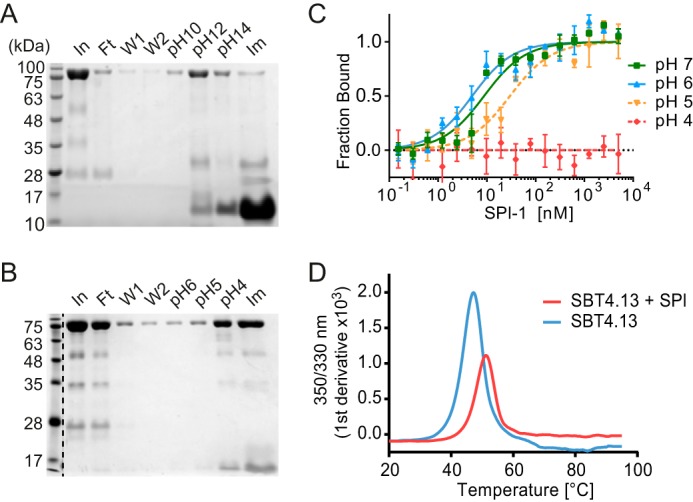
pH stability and thermal stability of the SBT4.13·SPI-1 complex. The SBT4.13·SPI-1 complex was established on Ni-NTA in sodium phosphate buffer, pH 7 (A), or formate/acetate/phosphate buffer, pH 7 (B). Lanes marked In, Ft, W1, and W2 show proteins in the input, flow-through, and wash fractions, respectively. Elution at different pH values was performed during 20 min at 4 °C in a batch procedure. Residual protein was eluted using 200 mm imidazole (Im). Samples were analyzed by 15% SDS-PAGE and Coomassie staining. C, pH dependence of binding affinities. Labeled SBT4.13 (8 nm) was titrated against unlabeled SPI-1 in formate/acetate/phosphate buffer at pH 7 (green squares), pH 6 (blue triangles), pH 5 (yellow triangles), and pH 4 (red diamonds). The fraction of SBT4.13 bound in the complex is plotted against the SPI-1 concentration. Data points represent the mean ± S.E. of three technical replicates. GraphPad Prism 6 was used for curve fitting (R2pH7 = 0.932, R2pH6 = 0.913, and R2pH5 = 0.912). D, thermal stability of SBT4.13·SPI-1 complex. Thermal unfolding was analyzed in a Prometheus NT.48 instrument. The change in tryptophan fluorescence with increasing temperature was recorded for SBT4.13 (blue line) and the SBT4.13·SPI-1 complex (red line) at 350 and 330 nm, and the first derivative of the 350/330 nm fluorescence ratio is plotted against the temperature.
Thermal stability of the proteinase·inhibitor complex
The stability of proteins is typically increased upon binding of a ligand (43). Likewise, thermal stability of tomato SBT3·PP complex is considerably increased as compared with the free protease (16). Subtilisin E, however, is destabilized in its PP-bound state (44). The effect of SPI-1 binding on thermal stability of SBT4.13 was analyzed by nano-differential scanning fluorometry (nanoDSF). NanoDSF monitors changes in intrinsic tryptophan fluorescence upon thermal unfolding and was used here to determine the melting point of SBT4.13 as compared with the SBT4.13·SPI-1 complex (Fig. 7D). Free SBT4.13 exhibited a melting point of 47.2 °C, whereas the thermal transition of the SBT4.13·SPI-1 complex was increased to 51.3 °C (Fig. 7D). The 4 °C increase in melting temperature, reflecting the free energy of inhibitor binding, indicated higher stability for the complex as compared with the free protease. Dissociation of the complex therefore is an unfavorable reaction, and inhibition by SPI-1 is likely to be irreversible unless the inhibitor is released by other factors, e.g. by proteolytic cleavage by other proteases.
Assessment of SPI-1 as folding assistant
The PPs of SBTs in bacteria, mammals, and plants are required for folding of their cognate protease domains. When expressed without their PPs, SBTs accumulate as partially folded, inactive intermediates that are kinetically trapped in a molten globule-like conformation. By lowering the free energy of the transition state, PPs allow SBTs to proceed from the molten globule to the native conformation. Therefore, the role of PPs during SBT folding is best described as that of a single-turnover catalyst (12, 45). The fungal I9 inhibitor PoIA1 is able to substitute for the PP of subtilisin BPN′ as folding assistant (29). The tertiary structure of PoIA1 and its ability to tightly bind subtilisin BPN′ were found to be critical for its role as a foldase (29). Considering the tight-binding inhibition of SBT4.13 by SPI-1, we wondered whether SPI-1 can also assist in SBT folding.
Because misfolded proteins fail to pass quality control in the endoplasmic reticulum, correct folding is a prerequisite for subcellular targeting and secretion (46, 47). Successful sorting along the secretory pathway is thus indicative of folding and has been used to assess foldase activity of PPs in both mammals and plants (15, 16). We recently showed that tomato SBT3 lacking its PP fails to be secreted and accumulates intracellularly when transiently expressed in N. benthamiana plants. Consistent with its role as folding assistant, secretion was restored when the PP of SBT3 was co-expressed as a separate polypeptide chain in trans (16). The same assay was used here to test whether SPI-1 can assist in folding of SBT4.13. A propeptide deletion mutant of SBT4.13 (SBT4.13ΔPP) failed to be secreted, indicating that also SBT4.13 depends on its PP to pass endoplasmic reticulum quality control (Fig. 8). Secretion was restored upon co-expression of the SBT4.13 propeptide (SBT4.13PP), confirming its role as folding assistant. SPI-1 could not substitute for SBT4.13PP, indicating that the plant I9 inhibitor, unlike fungal PoAI1, does not support SBT folding (Fig. 8).
Figure 8.
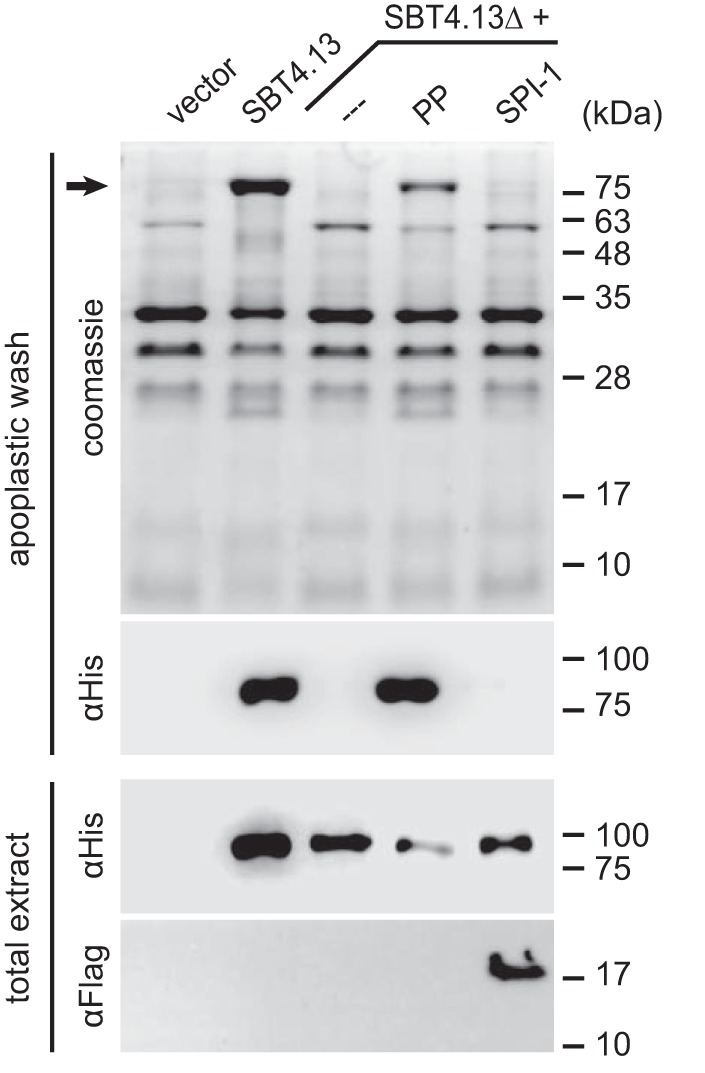
Chaperoning activity of SBT4. 13PP and SPI-1. A SBT4.13 propeptide deletion mutant (SBT4.13ΔPP) was expressed in N. benthamiana leaves with or without co-expression of the SBT4.13 propeptide (SBT4.13PP) or SPI-1. Twenty micrograms of total leaf extract or 4 μg of apoplastic proteins were separated by 15% SDS-PAGE. Proteins were visualized by Coomassie staining and Western blotting as indicated. SBT4.13 and SBT4.13ΔPP (PP) were detected with a monoclonal mouse anti-His6 antibody (1:5000; Dianova) in combination with a peroxidase-conjugated secondary antibody (1:10,000; Calbiochem) with enhanced chemiluminescence detection in apoplastic washes and total leaf extracts. Expression of SPI-1 was confirmed in total leaf extracts using a monoclonal FLAG M2-peroxidase conjugate (1:5000; Sigma-Aldrich). The molecular masses of marker proteins and that of SBT4.13 (black arrow) in the Coomassie-stained gel are indicated.
Discussion
SPI-1 was characterized as a high-affinity, tight-binding inhibitor of Arabidopsis subtilase SBT4.13 with Kd and Ki values in the picomolar range. SPI-1 acted as a stable inhibitor of SBT4.13 over the physiologically relevant pH range, and its inhibitory profile included many other SBTs from plants but not bacterial subtilisin A or bovine chymotrypsin. Sequence similarity and the shared β-α-β-β-α-β core structure indicated homology to the propeptides of SBTs, identifying SPI-1 as a member of the I9 inhibitor family and as the first independent I9 inhibitor of higher eukaryotes. Related inhibitor sequences were found in the genomes of other plants but not in algae, animals, or bacteria. Phylogenetic analysis indicated a clear separation of SPI-1-related inhibitors in plants from the I9 inhibitors in fungi, from the PPs of SBTs, and from SPI-2, suggesting that the SPI-1 family diverged very early during the evolution of land plants and, therefore, evolved independently of plant SBTs (Fig. 1). Consistent with this notion, the second activity of PPs, that of a chaperone assisting in SBT folding, has apparently been lost during the evolution of SPI-1.
The apparent evolutionary separation of the two activities is consistent with previous reports showing that the ability of PPs to bind and inhibit SBTs is not necessarily correlated to their chaperoning activity. The PP of Aqualysin I, for example, is 10-fold more potent as an inhibitor of subtilisin E than its own PP but shows only half the activity as a folding catalyst (48). In contrast, high chaperoning activity was observed for PPs that are poor SBT inhibitors (49). Furthermore, in the PP of tomato SBT3, inhibitor and chaperoning functions could be separated by site-directed mutagenesis. An 11-amino acid deletion (Ser-57 to Lys-70) resulted in the loss of chaperoning activity, whereas the inhibitory activity of the SBT3 PP toward mature SBT3 was only marginally impaired (16).
Most of the well characterized PIs of serine proteinases, including those of the Kazal, Kunitz, and Bowman-Birk families, bind the protease in a lock-and-key fashion using the standard (or Laskowski) mechanism of serine protease inhibition (3, 50, 51). The reactive site of these inhibitors comprises a peptide bond that binds to the active site of the cognate protease in a substrate-like manner but, unlike true substrates, is hydrolyzed very slowly (3, 50). The reactive site is typically encompassed in a disulfide loop, and the cleavage products thus remain physically attached (3). The stability of the enzyme·inhibitor complex and exclusion of water from the active site favor reformation of the peptide bond as compared with hydrolysis of the acyl-enzyme intermediate, thus explaining the slow rate of hydrolysis (52). SPI-1 is also bound in a substrate-like manner but, unlike standard inhibitors, it is readily cleaved, and because of the lack of disulfide bridges the C-terminal cleavage product is released. Two factors appear to contribute to the affinity and stability of the resulting enzyme·inhibitor complex: (i) the interaction of the newly formed SPI-1 C terminus with the active site of the protease and (ii) exosite interactions of the protease with the I9 core structure.
In regard to (i), the four C-terminal amino acids of cleaved SPI-1 (YQLH) match the substrate specificity of SBT4.13 on the nonprime side (upstream) of the scissile bond. Therefore, these amino acids are easily accommodated by the S1–S4 pockets of the protease's substrate binding site (see Ref. 51 for active site nomenclature). We propose that the C terminus of cleaved SPI-1 occupies the active site in a product-like manner, and by blocking substrate access it acts as a competitive inhibitor of the protease. This model is consistent with the PP mode of SBT inhibition. The autoinhibitory function of SBT PPs is known to involve their C termini (18, 19, 45), which were shown to interact directly with the active center of cognate SBTs (8, 20–22). Furthermore, the C-terminal amino acids of the fungal I9s ScIB2 and PoIA1 were shown to be critical for inhibitor activity (26, 27). According to this model, inhibitor specificity is determined (at least in part) by its C-terminal amino acids as only proteases that accommodate these residues in their active sites will be inhibited. Vice versa, all proteases targeted by SPI-1 would need to bind the same residues and, therefore, should resemble each other in their nonprime substrate binding sites (pockets S1–S4). Consistent with SPI-1 inhibiting all Arabidopsis SBTs that were tested, the sequences of these SBTs were conserved upstream of the site of autocatalytic prodomain processing, and the conserved sequence also resembled the C terminus of the inhibitor (Fig. 6C). However, the active-site interaction by itself cannot explain the high affinity of inhibitor binding.
In regard to (ii), exosite interactions are thus likely to contribute to the stability of the enzyme·inhibitor complex. Structural studies revealed exosite interactions of PPs and fungal I9s with cognate proteases involving the binding of the central β-sheet of the I9 fold to two parallel surface helices of the respective SBT (8, 20–22). As the I9 core structure and the subtilisin fold are conserved in SPI-1 and SBT4.13, respectively, similar exosite interactions are expected in the SPI-1·SBT4.13 complex. The relative contributions of exosite and active-site interactions to the overall free energy of binding, resulting in the 4 °C increase in melting temperature of the enzyme·inhibitor complex as compared with the free protease (Fig. 7D), remain to be determined.
In regard to the physiological role of SPI-1, we can only speculate at this time. PIs are widely distributed in plants, and they serve a multitude of different functions. High concentrations of PIs are often found in tissues that need to be protected from herbivory, such as reproductive and storage organs, including seed and tubers (53–55). Many PIs of different inhibitor families target trypsin or chymotrypsin-like serine proteases that are absent from plants but typical digestive enzymes of herbivores. First shown in tomato plants, PIs are known to accumulate systemically after local injury, thus taking an active role in plant defense (56). Induction of PI and other defense genes following herbivore attack is controlled by the octadecanoid pathway and jasmonate signaling (55, 57). Publicly accessible microarray data do not reveal any regulation of SPI-1 expression in response to biotic stress or jasmonates (58). This observation combined with the specificity of SPI-1 for plant SBTs rather than digestive serine proteinases makes a role in defense against pests or pathogens rather unlikely.
Endogenous SBTs are more likely physiological targets of SPI-1. Microarray data indicate constitutive expression of SPI-1 (58), which increases upon cold stress and during senescence (59). Consistent with a reduction of SBT activity in response to cold stress, the expression of several SBTs in clade 1 is reduced after 24 h of cold treatment, including those clade 1 SBTs that were shown here to be inhibited by SPI-1. The up-regulation of SPI-1 expression during senescence, however, points to a possible interaction with SBT1.4 (also known as senescence-associated subtilisin protease). Considering the broad specificity of SPI-1 against Arabidopsis SBTs, SPI-1 may thus contribute to the control of proteolytic activity in these processes. However, whether the inhibition observed for clade 1 SBTs in vitro is relevant for the regulation of SBT activity in vivo remains to be seen.
In addition to cold stress and senescence, SPI-1 expression levels are particularly high in the chalazal endosperm of developing seeds and in dry seeds where expression declines during imbibition (58). Interestingly, SBT4.13 shows a similar pattern of expression in the chalazal endosperm (60). It is thus tempting to propose that the strong interaction and in vitro inhibition of SBT4.13 is relevant in vivo for the control of SBT4.13 activity during seed development.
Experimental procedures
Cloning of SBTs
The open reading frames (ORFs) of SBT1.4 (At3g14067; ABRC clone U65235), SBT1.7 (At5g67360; ABRC clone U09277), SBT1.8 (At2g05920; ABRC clone U09230), SBT4.12 (At5g59090; ABRC clone U17589), SBT4.13 (At5g59120; ABRC clone U17749), and SBT5.2 (At1g20160; RAFL16-19-210) were amplified by PCR using the primer combinations given in Table 1.
Table 1.
Oligonucleotide primers used in this study
Sequences are given in 5′ to 3′ orientation; F (forward) and R (reverse) refer to the direction of priming. In some cases, additional bases were added at the 5′-end for increased efficiency of restriction. Restriction sites are underlined, and the restriction enzyme used is indicated in the primer name. All oligonucleotides were obtained from Eurofins Genomics GmbH (Ebersberg, Germany). The His6 tag (His), FLAG tag (FLAG) and deletions (Δ) are indicated in the primer name and the name of the product. In the case of primers binding inside the cDNA sequence, position is counted from the start codon.
| Primer sequence | Name | Product |
|---|---|---|
| CCATGGCTAAGCTCTCTCTTTCC | SBT1.4-F | SBT1.4 |
| CCTCAGAAGGACTGAACTGATCCCTG | SBT1.4-R | SBT1.4 |
| TCATGTCTTCTTCGTTTCTCTCCT | SBT1.7-F | SBT1.7 |
| GACTATGTCCAGCTAATCGCCAC | SBT1.7-R | SBT1.7 |
| ATGGCTTCTTCTTCTTCTTCTTCTT | SBT1.8-F | SBT1.8 |
| TATCAAAACCGGTTCCAAGAGAAT | SBT1.8-R | SBT1.8 |
| GGCTCGAGATGGCGAATCTAGCTGCGTC | SBT4.12-XhoI-F | SBT4.12-His |
| GGATCCTCAGTGATGGTGATGGTGATGTGCCTCATCAACAACCATTATG | SBT4.12-BamHI-His-R | SBT4.12-His |
| CGCTCGAGATGGCGACGCTAGCAG | SBT4.13-XhoI-F | SBT4.13-(His/PP) |
| CGCCCGGGTCAGTAATCACTAGTATAAACAACAATGGG | SBT4.13-SmaI-R | SBT4.13 |
| GGATCCTCAGTGATGGTGATGGTGATGGTAATCACTAGTATAAACAACA | SBT4.13-BamHI-His-R | SBT4.13-His |
| AAGTCCCAAGACGTCGTCGTAGCTGAAACTGAACTCAAGAACAAA | SBT4.13-SP-72 R | SBT4.13ΔPP |
| TTTGTTCTTGAGTTCAGTTTCAGCTACGACGACGTCTTGGGACTT | SBT4.13-327-F | SBT4.13ΔPP |
| CCGGCGGAATTCACAGTGAGAA | SBT4.13-894-R | SBT4.13ΔPP |
| GGCTCGAGATGAAAGGCATTACATTCTTCACA | SBT5.2-XhoI-F | SBT5.2-His |
| GGATCCTCAGTGATGGTGATGGTGATGGTTTGTGCGGCTACTCTCGCTAC | SBT5.2-His-BamHI-R | SBT5.2-His |
| CGCTCGAGATGCAAACATTTGCTCCTGGAAC | SPI-XhoI-F | SPI-FLAG |
| GGCCCGGGTCATTTGTCGTCATCATCCTTGTAGTCGGTCAACTTGAAACCACCACCA | SPI-FLAG-SmaI-R | SPI-FLAG |
| GTCCCGGGTCATTTGTCGTCATCATCCTTGTAGTCTTGAAGTTGTAACTTCTTGTTTGGG | SBT4.13PP-FLAG-SmaI-R | SBT4.13PP |
| CCCATATGCACCACCACCACCACCACGATGAATATACCGGAGAAGCAACGG | SPI-His-NdeI-104-F | SPI-His |
| CCGTCGACTTAGGTCAACTTGAAACCACCACC | SPI-SalI-R | SPI-His |
PCR products were cloned into pCR2.1-TOPO (Invitrogen) and amplified in E. coli DH10B. Inserts were mobilized with KpnI/XbaI for SBT1.4; EcoRI for SBT1.7; XhoI/HindIII for SBT1.8; XhoI/BamHI for SBT4.12-His, SBT4.13-His, and SBT5.2-His; and XhoI/SmaI for SBT4.13 and cloned into the respective restriction sites between the cauliflower mosaic virus 35S promoter and terminator in pART7. The expression cassettes were then transferred from pART7 into the binary vector pART27 using NotI. The expression plasmids were then introduced into Agrobacterium tumefaciens strain C58C1.
Cloning of the SBT4.13 propeptide deletion mutant, the PP of SBT4.13, and SPI-1 for expression in N. benthamiana
A SBT4.13 deletion mutant (SBT4.13ΔPP) lacking the entire prodomain (Ser-21 to Asn-109) was generated in two successive PCRs. The SBT4.13 ORFs corresponding to the signal peptide and to part of the catalytic domain were amplified from ABRC clone U17749 using the primer combinations SBT4.13-XhoI-F/SBT4.13-SP-72-R and SBT4.13-327-F/SBT4.13-894-R, respectively (Table 1). The two PCR products were joined in a second PCR with primers SBT4.13-XhoI-F and SBT4.13-894-R, thus linking the signal peptide directly to the catalytic domain. The 3′-primer included a natural EcoRI site that was used to replace the 5′-end of C-terminally His6-tagged wild-type SBT4.13 in pCR2.1-TOPO, resulting in SBT4.13ΔPP that was transferred into pART7/pART27 as described above.
SPI-1 and the propeptide of SBT4.13 were cloned by PCR from Arabidopsis cDNA and equipped with a C-terminal FLAG tag using the primer pairs SPI-XhoI-F/SPI-FLAG-SmaI-R and SBT4.13-XhoI-F/SBT4.13PP-FLAG-SmaI-R, respectively (Table 1). XhoI and SmaI restriction sites were used to clone the PCR products into pART7, and NotI was used for transfer into pART27.
Transient protein expression in N. benthamiana
For agroinfiltration, A. tumefaciens C58C1 strains containing the different expression constructs in pART27 and C58C1 containing the p19 suppressor of silencing were grown for 2 days on LB plates with appropriate antibiotics (rifampicin, tetracycline, and spectinomycin for C58C1/pART27; kanamycin instead of spectinomycin for C58C1/p19). Bacterial colonies were washed off the plates in 10 ml of 10 mm MES, pH 5.6, containing 10 mm MgCl2 and mixed at an A600 ratio of 0.7 (C58C1/pART27):1 (C58C1/p19). For purification of SBT4.13, the A600 ratio was adjusted to 0.35:0.5. A blunt syringe was used to infiltrate the bacterial suspension into leaves of 6-week-old N. benthamiana plants. Five days after agroinfiltration, the leaves were harvested, and apoplastic extracts were prepared as described (16).
Purification of SBT4.13
SBT4.13 was expressed in N. benthamiana and purified from cell wall extracts. Apoplastic washes were subjected to ammonium sulfate precipitation at 95% saturation. The precipitate was dissolved in one-tenth of the original volume and dialyzed against 25 mm sodium acetate, pH 5. Proteins were absorbed to a strong cation exchanger (SP Sepharose Fast Flow, GE Healthcare) and eluted with 200 mm NaCl in 25 mm sodium acetate, pH 5, in a batch procedure. The eluate was subjected to gel filtration over an Enrich SEC 650 size-exclusion column on a NGC Discover chromatography system (Bio-Rad) in 50 mm HEPES, pH 7.5, 200 mm NaCl. SBT4.13 was visualized as a single protein band after SDS-PAGE and Coomassie Brilliant Blue staining (Fig. 3A). One milligram of SBT4.13 was obtained from 40 infiltrated plants. The identity of the protein was confirmed by tryptic digest and MALDI-TOF MS, resulting in a sequence coverage of 50.6%. Peptides identified by MS included those corresponding to the very C terminus (SPIVVYTSDY) and the N terminus predicted for mature SBT4.13 (TTTSWDFMGLK), confirming the identity and the integrity of the purified protein.
Expression and purification of N-terminally His6-tagged SPI-1
The ORF of SPI-1 lacking the signal peptide (as predicted by SignalP 3.0) was amplified by PCR from Arabidopsis cDNA using oligonucleotide primers SPI-His-NdeI-103-F and SPI-SalI-R comprising NdeI and SalI restriction sites to facilitate subsequent cloning into pET21a. The expression vector was transfected into E. coli BL21-CodonPlus(DE3)-RIL (Agilent Technologies). Cells were grown in 500 ml of LB medium at 37 °C and 200 rpm until A600 reached 0.8. The culture was cooled down to 30 °C, and 1 mm isopropyl β-d-1-thiogalactopyranoside was added to induce protein expression. After 3 h at 30 °C, cells were harvested by centrifugation and lysed by sonication on ice in phosphate-buffered saline containing a spatula tip of DNase I (3200 units/mg; AppliChem, Darmstadt, Germany), 5 mm phenylmethylsulfonyl fluoride, 10 mm benzamidine hydrochloride, and 10 mm imidazole. Recombinant SPI-1 was purified from bacterial extracts on Ni-NTA-agarose (Qiagen, Hilden, Germany) used according to the manufacturer's instructions. The eluate was dialyzed (6-kDa-molecular-mass cutoff) against 50 mm HEPES, pH 7.5, and stored in aliquots at −20 °C.
Protease activity against FITC-casein and inhibition by SPI-1
The activity of Arabidopsis subtilases in apoplastic extracts of agroinfiltrated N. benthamiana plants was measured using the a Fluorescent Protease Assay kit (Pierce) with FITC-labeled casein as a universal protease substrate according to the manufacturer's instructions. In a total volume of 200 μl, reactions contained 50 μl of the apoplastic extract supplemented with 0, 0.2, or 5 μm recombinant SPI-1 using three independent preparations of the inhibitor. Specific activity was calculated as the increase in relative fluorescence during the linear phase in the first 10 min of the reaction per milligram of protein using an Infinite M200 Pro microplate reader (Tecan). The Bradford assay (Roth) was used for protein quantification with bovine serum albumin as the reference protein.
Inhibition of α-chymotrypsin and subtilisin A activity by recombinant SPI-1
The activity of 12 nm α-chymotrypsin (Sigma) or 6 nm subtilisin A (Sigma) was measured with N-succinyl-Ala-Ala-Pro-Phe p-nitroanilide (Sigma) as the substrate in 80 mm Tris/HCl, pH 7.8, 100 mm CaCl2 and 50 mm Tris/HCl, pH 8, 1 mm CaCl2, respectively. The assay contained 1 mm substrate and serial 1:3 dilutions of recombinant SPI-1. Assays were performed in duplicate using two independent preparations of SPI-1. Substrate cleavage was recorded in an Infinite M200 Pro microplate reader as the increase in absorbance at 412 nm (9-nm bandwidth). Protease activity was calculated from the linear phase in the first 3.5 (subtilisin A) or 7 min (α-chymotrypsin) of the reaction.
SBT4.13 enzyme kinetics
A fluorigenic peptide substrate, Abz-YLPA↓GVPI-Tyr(NO2) (PepMic, Suzhou, China), was used for kinetic measurements of SBT4.13 activity under steady-state conditions. The cleavage site of SBT4.13 is indicated by an arrow. Activity of 10 ng of SBT4.13 was measured against 25 μm substrate in 50 mm HEPES, pH 7.5, 0.05% Tween 20, 1 mg/ml PEG 8000. Recombinant SPI-1 was added in two serial 1:2 dilution series ranging from 100 nm to 0.56 pm and 200 nm to 1.12 pm, respectively. Assays were run at room temperature, and substrate cleavage was recorded in an Infinite M200 Pro microplate reader as the increase in relative fluorescence (λex = 320 nm, 9-nm bandwidth; λem = 420 nm, 20-nm bandwidth). SBT4.13 activity was calculated from the linear phase in the first 10 min of the reaction. Assays were performed in triplicate using three independent protein preparations of the inhibitor. Apparent Ki (Ki(app)) was calculated in GraphPad Prism 6 (GraphPad Software, San Diego, CA) using the Morrison equation (40).
Dissociation constants for the SPI-1·SBT4.13 complex
The Kd of the SPI-1·SBT4.13 complex was determined by microscale thermophoresis (MST). SBT4.13 (10 μm) was labeled using NT-647-N-hydroxysuccinimid (RED-NHS) (amine-reactive) dye at a 1:1 protein:dye ratio in a total volume of 200 μl according to the manufacturer's instructions (NanoTemper Technologies, Munich, Germany). Unreacted dye was removed using the size-exclusion columns provided with the kit, resulting in a stock solution of 1.66 μm labeled SBT4.13 in a total volume of 600 μl. Labeled SBT4.13 was adjusted to 200 pm in reaction buffer (50 mm HEPES, pH 7.5, 1 mg/ml PEG 8000, 0.05% (v/v) Tween 20) and titrated with serial (1:1) dilutions of SPI-1 (starting at 12.5 nm). The protease·inhibitor complex was allowed to establish for 5 min on ice. The samples were treated against bleaching (antibleaching kit, NanoTemper Technologies), loaded into standard MST capillaries, and incubated in the instrument for 10 min before measurement. MST was performed in a Monolith NT.115 Pico (NanoTemper Technologies) at 25 °C with 95% light-emitting diode and 40% MST power. A binding curve showing changes in the thermophoresis signal (Fnorm = Fhot/Fcold × 1000) depending on the concentration of SPI-1 (Fnorm(I)) was plotted using the NT Analysis software (NanoTemper Technologies). Binding constants were derived from three independent experiments (three independent preparations of SPI-1), each analyzed in three technical replicates.
Binding constants were calculated in GraphPad Prism 6 using the following equation (where B is response value of bound state, U is response value of unbound state, E is concentration of labeled SBT4.13, and I is concentration of SPI-1).
| (Eq. 1) |
To determine pH dependence of the SBT4.13-inhibitor interaction, 8 nm labeled SBT4.13 was titrated with serial (1:1) dilutions of 5 μm His6-tagged SPI-1 in 8.33 mm formate/formic acid, 16.66 mm acetate/acetic acid, 25 mm Na2HPO4/NaH2PO4, 0.05% Tween 20, 0.5 mg/ml BSA at varying pH values. MST was performed in a Monolith NT.115 instrument at 25 °C with 40% light-emitting diode and 60% MST power. The fraction of SBT4.13 complexed with the inhibitor (fraction bound) was calculated from the thermophoresis signal using the NT Analysis software.
Binding constants were derived from three technical replicates for each pH value using the following equation in GraphPad Prism 6 (where E is concentration of labeled SBT4.13 and I is concentration inhibitor).
| (Eq. 2) |
Determination of pH stability of the SBT4.13·SPI-1 complex
For the analysis of complex stability at acidic pH, 120 μg of recombinant His6-tagged SPI-1 was bound to 50 μl of Ni-NTA-agarose (50% slurry) and incubated with 100 μg of SBT4.13 in 8.33 mm formate/formic acid, 16.66 mm acetate/acetic acid, 25 mm Na2HPO4/NaH2PO4, pH 7, supplemented with 10 mm imidazole. For each elution step, samples were incubated for 20 min at 4 °C in 500 μl of the same buffer adjusted to the respective pH value, and proteins recovered in the supernatant were analyzed by SDS-PAGE. Residual protein was eluted in a final step using 200 mm imidazole.
For basic pH, 50 μl of Ni-NTA-agarose were saturated with His6-tagged SPI-1 in Na2HPO4/NaH2PO4, pH 7, containing 10 mm imidazole to allow binding of 50 μg of SBT4.13 to the matrix. Stepwise elution was done as described before for acidic pH values. To reach pH 12 and 14, NaOH was used for pH adjustment of the phosphate buffer.
Analysis of the melting temperature of SBT4.13 and SBT4.13·SPI-1 inhibitor complex
SBT4.13 (2 μm) and an equimolar mixture of SBT4.13 with His6-tagged SPI-1 in 50 mm HEPES, pH 7.5, were incubated on ice for 10 min. Thermal unfolding of SBT4.13 was analyzed by nanoDSF in a Prometheus NT.48 instrument (NanoTemper Technologies) with a temperature slope of 1 °C/min. Tryptophan fluorescence was recorded at 350 and 330 nm, and the 350:330 nm ratio was plotted against the temperature. The melting point (Tm) was determined for SBT4.13 and the SBT4.13·SPI-1 complex as the maximum of the first derivative of the fluorescence ratio plots (Fig. 7D). As SPI-1 does not contain any tryptophan residues, the method was not suitable to determine Tm of the free inhibitor.
Identification of the C-terminal cleavage site of SPI-1
His6-tagged SPI-1 (7.5 μm) was added to SBT4.13 (5 μm) at room temperature. The reaction was stopped after 5 min by addition of SDS-PAGE sample loading buffer and heat denaturation followed by SDS-PAGE and Coomassie Brilliant Blue staining. The protein band corresponding to SPI-1 and an untreated SPI-1 control were subjected to in-gel trypsin (Roche Applied Science) digestion. Tryptic peptides were analyzed by nano-LC-ESI-MS/MS on an ACQUITY nano-UPLC system (Waters) coupled to a LTQ-Orbitrap XL hybrid mass spectrometer (Thermo Fisher Scientific, Germany). Digests were concentrated and desalted on a precolumn (2 cm × 180 μm, Symmetry C18, 5-μm particle size; Waters) and separated on a 20-cm × 75-μm BEH 130 C18 reversed phase column (1.7-μm particle size; Waters). Gradient elution was performed from 1 to 40% acetonitrile (ACN) in 0.1% formic acid within 60 min. The LTQ-Orbitrap was operated under the control of XCalibur 2.1.0 software. Survey spectra (m/z = 250–1800) were recorded in the Orbitrap at a resolution of 60,000 at m/z = 400. Data-dependent tandem mass spectra were generated for the seven most abundant peptide precursors in the linear ion trap. For all measurements with the Orbitrap detector, internal calibration was performed using lock-mass ions from ambient air as described previously (61). MS data were analyzed using MaxQuant (62) with carbamidomethylation of cysteine (+H3C2NO) and oxidation of methionine (+O) as fixed and variable modifications of semitryptic peptides, respectively.
To identify the small C-terminal SPI-1 peptide that is released by SBT4.13, the two proteins were incubated as before for 10 min at room temperature and then filtered through a Vivaspin concentrator (10-kDa-molecular-mass cutoff; Sartorius) to recover small cleavage products. The filtrate was purified on C18 StageTips (63); resuspended in 50% ACN, 0.1% trifluoroacetic acid (TFA); and mixed with an equal volume of the crystallization matrix (5 mg/ml α-cyano-4-hydroxy-trans-cinnamic acid in 50% ACN, 0.1% TFA). Mass spectra were recorded with an AutofexIII mass spectrometer (Bruker Daltonics) in the reflector mode with external calibration (Peptide Calibration Standard II, Bruker Daltonics). Flex Analysis 3.0 was used for data analysis with a mass tolerance of 30 ppm for precursor ions. A single peptide with a mass of 1018.67 Da was detected in the protease-treated sample. MS/MS fragmentation spectra were recorded for this mass signal to obtain sequence information and confirm its identity.
Author contributions
A. St. and A. Sc. conceived the study. M. H. designed and performed the experiments. Data were analyzed and interpreted by M. H., A. St., and A. Sc. M. H. and A. Sc. wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Ursula Glück-Behrens and Denisa Heindel for excellent technical assistance. We also thank Jens Pfannstiel and Berit Würtz at the University's Service Unit Mass Spectrometry and Waltraud Schulze (Department of Plant Systems Biology, University of Hohenheim) for mass spectral analyses and Nina Schlinck and Amit Gupta from NanoTemper Technologies for help with microscale thermophoresis and melting curve analysis using the Monolith Pico and Prometheus instruments.
The authors declare that they have no conflicts of interest with the contents of this article.
M. Hohl, A. Stintzi, and A. Schaller, unpublished observation.
- PI
- proteinase inhibitor
- MST
- microscale thermophoresis
- PP
- propeptide
- SBT
- subtilase
- PC
- proprotein convertase
- SPI
- subtilisin propeptide-like inhibitor
- Sc
- S. cerevisiae
- Po
- P. ostreatus
- At
- A. thaliana
- I9s
- I9 inhibitors
- Pp
- P. patens
- Sl
- S. lycopersicum
- ESI
- electrospray ionization
- Ni-NTA
- nickel-nitrilotriacetic acid
- DSF
- differential scanning fluorometry
- ABRC
- Arabidopsis Biological Resource Center
- Abz
- aminobenzoic acid
- ACN
- acetonitrile.
References
- 1. Khan A. R., and James M. N. (1998) Molecular mechanisms for the conversion of zymogens to active proteolytic enzymes. Protein Sci. 7, 815–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bode W., and Huber R. (1992) Natural protein proteinase inhibitors and their interaction with proteinases. Eur. J. Biochem. 204, 433–451 [DOI] [PubMed] [Google Scholar]
- 3. Laskowski M. Jr., and Kato I. (1980) Protein inhibitors of proteinases. Annu. Rev. Biochem. 49, 593–626 [DOI] [PubMed] [Google Scholar]
- 4. Kantyka T., Rawlings N. D., and Potempa J. (2010) Prokaryote-derived protein inhibitors of peptidases: a sketchy occurrence and mostly unknown function. Biochimie 92, 1644–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Santamaría M. E., Diaz-Mendoza M., Diaz I., and Martinez M. (2014) Plant protein peptidase inhibitors: an evolutionary overview based on comparative genomics. BMC Genomics 15, 812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rawlings N. D., Barrett A. J., and Finn R. (2016) Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 44, D343–D350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rawlings N. D., Barrett A. J., and Bateman A. (2010) MEROPS: the peptidase database. Nucleic Acids Res. 38, D227–D233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gallagher T., Gilliland G., Wang L., and Bryan P. (1995) The prosegment-subtilisin BPN′ complex: crystal structure of a specific 'foldase'. Structure 3, 907–914 [DOI] [PubMed] [Google Scholar]
- 9. Schaller A., Stintzi A., and Graff L. (2012) Subtilases—versatile tools for protein turnover, plant development, and interactions with the environment. Physiol. Plant. 145, 52–66 [DOI] [PubMed] [Google Scholar]
- 10. Ikemura H., Takagi H., and Inouye M. (1987) Requirement of pro-sequence for the production of active subtilisin E in Escherichia coli. J. Biol. Chem. 262, 7859–7864 [PubMed] [Google Scholar]
- 11. Shinde U., and Inouye M. (1994) The structural and functional organization of intramolecular chaperones: the N-terminal propeptides which mediate protein folding. J. Biochem. 115, 629–636 [DOI] [PubMed] [Google Scholar]
- 12. Eder J., and Fersht A. R. (1995) Pro-sequence-assisted protein folding. Mol. Microbiol. 16, 609–614 [DOI] [PubMed] [Google Scholar]
- 13. Demidyuk I. V., Shubin A. V., Gasanov E. V., and Kostrov S. V. (2010) Propeptides as modulators of functional activity of proteases. Biomol. Concepts 1, 305–322 [DOI] [PubMed] [Google Scholar]
- 14. Shinde U., and Thomas G. (2011) Insights from bacterial subtilases into the mechanisms of intramolecular chaperone-mediated activation of furin. Methods Mol. Biol. 768, 59–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dillon S. L., Williamson D. M., Elferich J., Radler D., Joshi R., Thomas G., and Shinde U. (2012) Propeptides are sufficient to regulate organelle-specific pH-dependent activation of furin and proprotein convertase 1/3. J. Mol. Biol. 423, 47–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Meyer M., Leptihn S., Welz M., and Schaller A. (2016) Functional characterization of propeptides in plant subtilases as intramolecular chaperones and inhibitors of the mature protease. J. Biol. Chem. 291, 19449–19461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li Y., Hu Z., Jordan F., and Inouye M. (1995) Functional analysis of the propeptide of subtilisin E as an intramolecular chaperone for protein folding. Refolding and inhibitory abilities of propeptide mutants. J. Biol. Chem. 270, 25127–25132 [DOI] [PubMed] [Google Scholar]
- 18. Fugère M., Limperis P. C., Beaulieu-Audy V., Gagnon F., Lavigne P., Klarskov K., Leduc R., and Day R. (2002) Inhibitory potency and specificity of subtilase-like pro-protein convertase (SPC) prodomains. J. Biol. Chem. 277, 7648–7656 [DOI] [PubMed] [Google Scholar]
- 19. Nakagawa M., Ueyama M., Tsuruta H., Uno T., Kanamaru K., Mikami B., and Yamagata H. (2010) Functional analysis of the cucumisin propeptide as a potent inhibitor of its mature enzyme. J. Biol. Chem. 285, 29797–29807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jain S. C., Shinde U., Li Y., Inouye M., and Berman H. M. (1998) The crystal structure of an autoprocessed Ser221Cys-subtilisin E-propeptide complex at 2.0 Å resolution. J. Mol. Biol. 284, 137–144 [DOI] [PubMed] [Google Scholar]
- 21. Piper D. E., Jackson S., Liu Q., Romanow W. G., Shetterly S., Thibault S. T., Shan B., and Walker N. P. (2007) The crystal structure of PCSK9: a regulator of plasma LDL-cholesterol. Structure 15, 545–552 [DOI] [PubMed] [Google Scholar]
- 22. Sotokawauchi A., Kato-Murayama M., Murayama K., Hosaka T., Maeda I., Onjo M., Ohsawa N., Kato D.-I., Arima K., and Shirouzu M. (2017) Structural basis of cucumisin protease activity regulation by Its propeptide. J. Biochem. 161, 45–53 [DOI] [PubMed] [Google Scholar]
- 23. Anderson E. D., Molloy S. S., Jean F., Fei H., Shimamura S., and Thomas G. (2002) The ordered and compartment-specific autoproteolytic removal of the furin intramolecular chaperone is required for enzyme activation. J. Biol. Chem. 277, 12879–12890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maier K., Müller H., and Holzer H. (1979) Purification and molecular characterization of two inhibitors of yeast proteinase B. J. Biol. Chem. 254, 8491–8497 [PubMed] [Google Scholar]
- 25. Dohmae N., Takio K., Tsumuraya Y., and Hashimoto Y. (1995) The complete amino acid sequences of two serine proteinase inhibitors from the fruiting bodies of a basidiomycete, Pleurotus ostreatus. Arch. Biochem. Biophys. 316, 498–506 [DOI] [PubMed] [Google Scholar]
- 26. Kojima S., Deguchi M., and Miura K. (1999) Involvement of the C-terminal region of yeast proteinase B inhibitor 2 in its inhibitory action. J. Mol. Biol. 286, 775–785 [DOI] [PubMed] [Google Scholar]
- 27. Kojima S., Hisano Y., and Miura K. (2001) Alteration of inhibitory properties of Pleurotus ostreatus proteinase a inhibitor 1 by mutation of its C-terminal region. Biochem. Biophys. Res. Commun. 281, 1271–1276 [DOI] [PubMed] [Google Scholar]
- 28. Sasakawa H., Yoshinaga S., Kojima S., and Tamura A. (2002) Structure of POIA1, a homologous protein to the propeptide of subtilisin: implication for protein foldability and the function as an intramolecular chaperone. J. Mol. Biol. 317, 159–167 [DOI] [PubMed] [Google Scholar]
- 29. Kojima S., Iwahara A., and Yanai H. (2005) Inhibitor-assisted refolding of protease: a protease inhibitor as an intramolecular chaperone. FEBS Lett. 579, 4430–4436 [DOI] [PubMed] [Google Scholar]
- 30. Xu Z., Sato K., and Wickner W. (1998) LMA1 binds to vacuoles at Sec18p (NSF), transfers upon ATP hydrolysis to a t-SNARE (Vam3p) complex, and is released during fusion. Cell 93, 1125–1134 [DOI] [PubMed] [Google Scholar]
- 31. Slusarewicz P., Xu Z., Seefeld K., Haas A., and Wickner W. T. (1997) I2/(B) is a small cytosolic protein that participates in vacuole fusion. Proc. Natl. Acad. Sci. U.S.A. 94, 5582–5587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schardon K., Hohl M., Graff L., Pfannstiel J., Schulze W., Stintzi A., and Schaller A. (2016) Precursor processing for plant peptide hormone maturation by subtilisin-like serine proteinases. Science 354, 1594–1597 [DOI] [PubMed] [Google Scholar]
- 33. Pei J., Kim B.-H., and Grishin N. V. (2008) PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 36, 2295–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Meier-Kolthoff J. P., Auch A. F., Klenk H. P., and Göker M. (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tangrea M. A., Bryan P. N., Sari N., and Orban J. (2002) Solution structure of the pro-hormone convertase 1 pro-domain from Mus musculus. J. Mol. Biol. 320, 801–812 [DOI] [PubMed] [Google Scholar]
- 36. Rautengarten C., Steinhauser D., Büssis D., Stintzi A., Schaller A., Kopka J., and Altmann T. (2005) Inferring hypotheses on functional relationships of genes: analysis of the Arabidopsis thaliana subtilase gene family. PLoS Comput. Biol. 1, e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tian M., Huitema E., Da Cunha L., Torto-Alalibo T., and Kamoun S. (2004) A Kazal-like extracellular serine protease inhibitor from Phytophthora infestans targets the tomato pathogenesis-related protease P69B. J. Biol. Chem. 279, 26370–26377 [DOI] [PubMed] [Google Scholar]
- 38. Tian M., Benedetti B., and Kamoun S. (2005) A second Kazal-like protease inhibitor from Phytophthora infestans inhibits and interacts with the apoplastic pathogenesis-related protease P69B of tomato. Plant Physiol. 138, 1785–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Williams J. W., and Morrison J. F. (1979) The kinetics of reversible tight-binding inhibition. Methods Enzymol. 63, 437–467 [DOI] [PubMed] [Google Scholar]
- 40. Greco W. R., and Hakala M. T. (1979) Evaluation of methods for estimating the dissociation constant of tight binding enzyme inhibitors. J. Biol. Chem. 254, 12104–12109 [PubMed] [Google Scholar]
- 41. Meichtry J., Amrhein N., and Schaller A. (1999) Characterization of the subtilase gene family in tomato (Lycopersicon esculentum Mill.). Plant Mol. Biol. 39, 749–760 [DOI] [PubMed] [Google Scholar]
- 42. Shen J., Zeng Y., Zhuang X., Sun L., Yao X., Pimpl P., and Jiang L. (2013) Organelle pH in the Arabidopsis endomembrane system. Mol. Plant 6, 1419–1437 [DOI] [PubMed] [Google Scholar]
- 43. Layton C. J., and Hellinga H. W. (2011) Quantitation of protein-protein interactions by thermal stability shift analysis. Protein Sci. 20, 1439–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shinde U., Fu X., and Inouye M. (1999) A pathway for conformational diversity in proteins mediated by intramolecular chaperones. J. Biol. Chem. 274, 15615–15621 [DOI] [PubMed] [Google Scholar]
- 45. Bryan P. N. (2002) Prodomains and protein folding catalysis. Chemical Reviews 102, 4805–4816 [DOI] [PubMed] [Google Scholar]
- 46. Gething M. J., McCammon K., and Sambrook J. (1986) Expression of wild-type and mutant forms of influenza hemagglutinin: the role of folding in intracellular transport. Cell 46, 939–950 [DOI] [PubMed] [Google Scholar]
- 47. Brodsky J. L., and Skach W. R. (2011) Protein folding and quality control in the endoplasmic reticulum: recent lessons from yeast and mammalian cell systems. Curr. Opin. Cell Biol. 23, 464–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Marie-Claire C., Yabuta Y., Suefuji K., Matsuzawa H., and Shinde U. (2001) Folding pathway mediated by an intramolecular chaperone: the structural and functional characterization of the aqualysin I propeptide. J. Mol. Biol. 305, 151–165 [DOI] [PubMed] [Google Scholar]
- 49. Fu X., Inouye M., and Shinde U. (2000) Folding pathway mediated by an intramolecular chaperone. The inhibitory and chaperone functions of the subtilisin propeptide are not obligatorily linked. J. Biol. Chem. 275, 16871–16878 [DOI] [PubMed] [Google Scholar]
- 50. Birk Y. (2003) Plant Protease Inhibitors: Significance in Nutrition, Plant Protection, Cancer Prevention and Genetic Engineering, 1st Ed., pp. 5–70, Springer, Berlin, Germany [Google Scholar]
- 51. Farady C. J., and Craik C. S. (2010) Mechanisms of macromolecular protease inhibitors. ChemBioChem 11, 2341–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zakharova E., Horvath M. P., and Goldenberg D. P. (2009) Structure of a serine protease poised to resynthesize a peptide bond. Proc. Natl. Acad. Sci. U.S.A. 106, 11034–11039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ryan C. A. (1990) Protease inhibitors in plants: genes for improving defenses against insects and pathogens. Annu. Rev. Phytopathol. 28, 425–449 [Google Scholar]
- 54. Howe G. A., and Schaller A. (2008) Direct defenses in plants and their induction by wounding and insect herbivores, in Induced Plant Resistance against Herbivory (Schaller A., ed) pp. 7–29, Springer, Heidelberg, Germany [Google Scholar]
- 55. Koiwa H., Bressan R. A., and Hasegawa P. M. (1997) Regulation of protease inhibitors and plant defense. Trends Plant Sci. 2, 379–384 [Google Scholar]
- 56. Green T. R., and Ryan C. A. (1972) Wound-induced proteinase inhibitor in plant leaves: a possible defense mechanism against insects. Science 175, 776–777 [DOI] [PubMed] [Google Scholar]
- 57. Farmer E. E., and Ryan C. A. (1992) Octadecanoid-derived signals in plants. Trends Cell Biol. 2, 236–241 [DOI] [PubMed] [Google Scholar]
- 58. Winter D., Vinegar B., Nahal H., Ammar R., Wilson G. V., and Provart N. J. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2, e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gepstein S., Sabehi G., Carp M. J., Hajouj T., Nesher M. F., Yariv I., Dor C., and Bassani M. (2003) Large-scale identification of leaf senescence-associated genes. Plant J. 36, 629–642 [DOI] [PubMed] [Google Scholar]
- 60. Bleckmann A., and Dresselhaus T. (2016) Fluorescent whole-mount RNA in situ hybridization (F-WISH) in plant germ cells and the fertilized ovule. Methods 98, 66–73 [DOI] [PubMed] [Google Scholar]
- 61. Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., and Mann M. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 62. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 63. Rappsilber J., Ishihama Y., and Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 64. Petersen T. N., Brunak S., von Heijne G., and Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 65. Schechter I., and Berger A. (1967) On the size of the active site in proteases. I. papain. Biochem. Biophys. Res. Commun. 27, 157–162 [DOI] [PubMed] [Google Scholar]