Abstract
An improved understanding of the molecular mechanisms in synapse formation provides insight into both learning and memory and the etiology of neurodegenerative disorders. Coactivator-associated arginine methyltransferase 1 (CARM1) is a protein methyltransferase that negatively regulates synaptic gene expression and inhibits neuronal differentiation. Despite its regulatory function in neurons, little is known about the CARM1 cellular location and its role in dendritic maturation and synapse formation. Here, we examined the effects of CARM1 inhibition on dendritic spine and synapse morphology in the rat hippocampus. CARM1 was localized in hippocampal post-synapses, with immunocytochemistry and electron microscopy revealing co-localization of CARM1 with post-synaptic density (PSD)-95 protein, a post-synaptic marker. Specific siRNA-mediated suppression of CARM1 expression resulted in precocious dendritic maturation, with increased spine width and density at sites along dendrites and induction of mushroom-type spines. These changes were accompanied by a striking increase in the cluster size and number of key synaptic proteins, including N-methyl-d-aspartate receptor subunit 2B (NR2B) and PSD-95. Similarly, pharmacological inhibition of CARM1 activity with the CARM1-specific inhibitor AMI-1 significantly increased spine width and mushroom-type spines and also increased the cluster size and number of NR2B and cluster size of PSD-95. These results suggest that CARM1 is a post-synaptic protein that plays roles in dendritic maturation and synaptic formation and that spatiotemporal regulation of CARM1 activity modulates neuronal connectivity and improves synaptic dysfunction.
Keywords: dendritic spine, hippocampus, neuron, signal transduction, synapse, CARM1, PSD, dendritic maturation, hippocampal neurons, synaptic clustering
Introduction
Understanding the cellular and molecular mechanisms underlying synaptic formation will provide insight into not only learning and memory processes, but also the basis for neurodevelopmental and neurodegenerative disorders (1–5). Synapses, neuronal structures that form intercellular junctions between axon terminals and small protrusions called dendritic spines, accumulate information in the form of life-long alterations in their molecular and structural composition (6). Dendritic spines receive synaptic input from pre-synaptic axon terminals and regulate post-synaptic transmission (7). Spine morphology is highly correlated with synaptic function (8). Interestingly, spine structures are highly plastic, undergoing continuous change that modifies synaptic transmission and strength (9).
Dendritic spines contain a prominent thickening at the cytoplasmic surface of the post-synaptic membrane, called the post-synaptic density (PSD)2 (10, 11). The PSD is a dynamic region that consists of a variety of scaffolding proteins such as members of the membrane-associated guanylate kinase family including PSD-95; membrane receptors such as N-methyl-d-aspartate (NMDA) receptors, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors, and neurotrophic receptors; intracellular signaling molecules such as protein kinase C (PKC), calcium/calmodulin-dependent protein kinase 2 (CaMKII), and small GTPase family members; and structural proteins such as actin and microtubule components (11–14). The spatiotemporal regulation of the various synaptic proteins at the dendritic spine holds the key to understanding the mechanisms by which synaptic structures are remodeled and synaptic function is regulated.
Post-translational modification of proteins within signaling cascades, such as phosphorylation and methylation of synaptic proteins, is a fundamental and essential mechanism for regulating axonal elongation and dendrite branching (8, 15). For example, protein phosphorylation by PKCϵ regulates synaptogenic gene expression and rescues synaptic plasticity that is impaired in neurodegenerative diseases like Alzheimer's disease (16, 17). Activated PKCϵ modulates morphological changes in dendritic spines and enhances long-term memory by activating structural changes in the synaptic cytoskeleton (18–20) or by up-regulating HuD-mediated post-transcriptional controls (21, 22).
Arginine methylation of synaptic proteins, catalyzed by protein arginine methyltransferases (PRMTs), is another common post-translational modification that occurs in neuronal cells (23–25). Dysregulation and aberrant expression of PRMTs are associated with various disease states (24, 26, 27). The regulation and substrate specificity of PRMTs have been rapidly elucidated in the past few years, but the biological functions of PRMTs in the brain and the effects of arginine methylation of synaptic proteins by PRMTs remains unclear. Coactivator-associated arginine methyltransferase 1 (CARM1), also called PRMT4, modulates protein methylation in neurons (28, 29). CARM1 methylates HuD protein and plays an important role in mRNA processing (22, 30). It also inhibits neuronal differentiation in vitro (28). CARM1 activity is modulated upon phosphorylation by PKCϵ (22, 31). Taken together, these findings strongly suggest that regulation of CARM1 activity through PKCϵ may influence learning-specific synaptic formation in the brain. Despite data suggesting that CARM1 functions in neurons, no studies have further characterized the cellular localization and function of CARM1 on dendritic maturation and synaptic formation.
In the present study, we tested the hypothesis that CARM1 is expressed in the rat brain, particularly in the PSD fraction, where it regulates dendrite and synaptic morphology. We examined the effects of genetic and pharmacological inhibition of CARM1 on dendritic complexity, spine number and density, and arborization, and the accumulation of excitatory synaptic proteins at synaptic sites in primary cultures of differentiating rat hippocampal neurons.
Results
CARM1 is a post-synaptic protein that clusters at synapses in hippocampal neurons
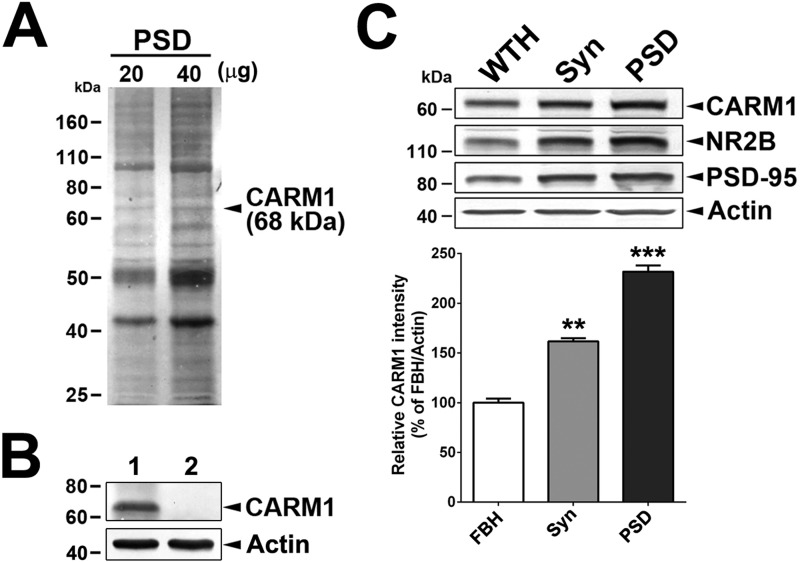
To understand CARM1 function at dendritic synapses in hippocampal neurons, we first examined CARM1 expression in brain tissue subcellular fractions. The One-Triton PSD fraction (50) was separated by SDS-PAGE, and a protein band of 68 kDa (Fig. 1A) was excised from the gel after Coomassie Brilliant Blue staining, extracted, separated by SDS-PAGE, and then immunoblotted using anti-CARM1 antibody (Fig. 1B), revealing strong CARM1 expression. To verify the specificity of the reaction, the anti-CARM1 antibody was pre-absorbed with CARM1 antigen. Pre-absorption of the CARM1 antibody completely abolished detection of CARM1 in the PSD fraction (Fig. 1B).
Figure 1.
CARM1 is a post-synaptic protein. A, a total of 20 or 40 μg PSD fraction isolated from rat hippocampi was separated in SDS-PAGE, and gels were stained with Coomassie Brilliant Blue solution. Arrow indicates CARM1 at 68 kDa. B, immunoblotting analysis to detect CARM1 protein levels in protein complexes extracted from PAGE gels in A using anti-CARM1 (lane 1) or anti-serum pre-absorbed with CARM1 antigen (lane 2). C, immunoblotting analysis to detect CARM1 protein levels in whole tissue homogenates (WTH), synaptosomes (Syn), or One-Triton PSD fraction (10 μg/each fraction). Mean ± S.E., three independent experiments. **, p < 0.01; ***, p < 0.001, compared with WTH).
We next tested whether CARM1 localizes to the PSD fraction in rat hippocampal neurons. The relative enrichment of CARM1 in whole hippocampal tissue homogenate, synaptosomes, and PSD fractions treated once with Triton X-100 was tested by immunoblotting analyses and compared with that of NR2B and PSD-95 proteins that are known to be specifically associated with the PSD fraction. Immunoblots revealed that the amount of CARM1 was enriched in the synaptosome and PSD fractions by 161.6 and 231.7%, respectively, compared with whole-tissue homogenate (Fig. 1C). These results confirm that CARM1 is enriched in the PSD fraction.
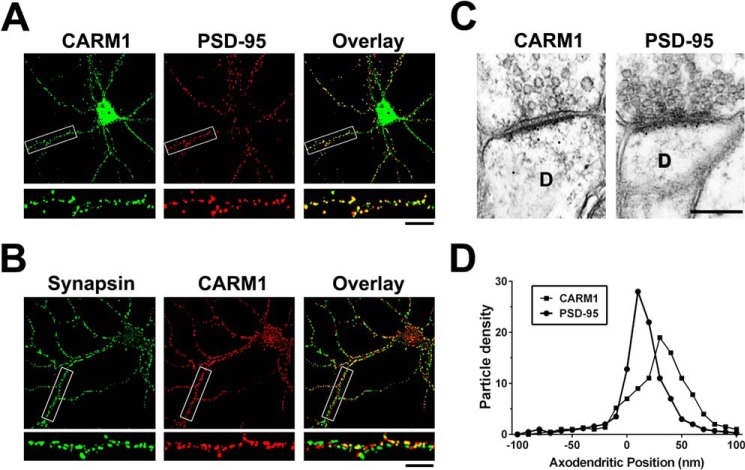
We examined the subcellular distribution of CARM1 by fluorescence immunocytochemistry of dissociated rat hippocampal neurons. Dissociated hippocampal neurons were grown for 14 days in vitro (DIV), fixed, and double-stained with antibodies against CARM1 and the pre-synaptic marker synapsin or the excitatory post-synaptic marker PSD-95. Percentage of co-localization was calculated from overlay images (Fig. 2). High-resolution confocal microscope images showed that CARM1 formed clusters at sites that also contain PSD-95 (Mander's coefficient = 0.726 ± 0.041) (Fig. 2A). However, CARM1 barely co-localized with synapsin (Mander's coefficient = 0.292 ± 0.034) (Fig. 2B).
Figure 2.
CARM1 co-localizes with PSD-95 in hippocampal neurons. A and B, cultured hippocampal neurons (14 DIV) were double-stained with anti-CARM1 and PSD-95 (a post-synaptic marker protein) or synapsin (a pre-synaptic marker), and then co-localization from overlay images was analyzed. Scale bar, 10 μm. C, rat brain sections were labeled with antibodies against CARM1 or PSD-95 and gold particle labelings in the CA1 stratum radiatum of the hippocampus were observed using electron microscopy to show post-embedded localization of CARM1 or PSD-95. Scale bar, 500 nm. D, each CARM1 or PSD-95 labeling was counted to show pre- or post-synaptic positions of gold particles and the axodendritic distribution was quantitatively expressed.
To verify that CARM1 is located at synapses in vivo and to determine whether CARM1 is predominantly located at the pre-synaptic or post-synaptic membrane, the distribution of CARM1 at synapses from the CA1 stratum radiatum of the hippocampus was examined with the use of immunoelectron microscopy. Brain sections were immunolabeled with an antibody against either CARM1 or PSD-95. The immunogold label in neurons for both antibodies was present primarily at excitatory synapses, identified by the presence of a Gray's Type I PSD, round pre-synaptic vesicles, and a distinct synaptic cleft (Fig. 2C). Additionally, no labeled synapses were detected in control experiments in which sections were stained with antibodies that were pre-absorbed with their antigen, respectively, or stained with secondary antibody alone (data not shown). Thus the labeling of excitatory synapses accurately reflects localization of CARM1. Then we quantitatively analyzed the axodendritic distribution of gold particles against CARM1 to determine whether the position of the label was over the pre-synaptic or post-synaptic compartment and was compared with that against PSD-95. The axodendritic distribution of the gold particles against CARM1 was analyzed by measuring the distance from the extracellular perimeter of the post-synaptic membrane to the center of the gold particle. The immunogold label against CARM1 was primarily over the post-synaptic membrane and compartment, confirming that over 87% of the gold particles were detected from the post-synaptic membrane and spine, with 15% of the label within 10 nm (the diameter of the gold particle) of the outer perimeter of the post-synaptic membrane and 81% within 50 nm (Fig. 2D). PSD-95 also preferentially labeled post-synaptic structures, with 89% of the gold particles over the post-synaptic compartment, with 41% of the label within 10 nm (the diameter of the gold particle) of the outer perimeter of the post-synaptic membrane and 83% within 50 nm (Fig. 2D). The prominent clustering of CARM1 at post-synaptic sites, revealed by higher co-localization with PSD-95, is consistent with its strong enrichment in the PSD fraction.
Knockdown of CARM1 expression increases the complexity of dendritic arborization of cultured hippocampal neurons
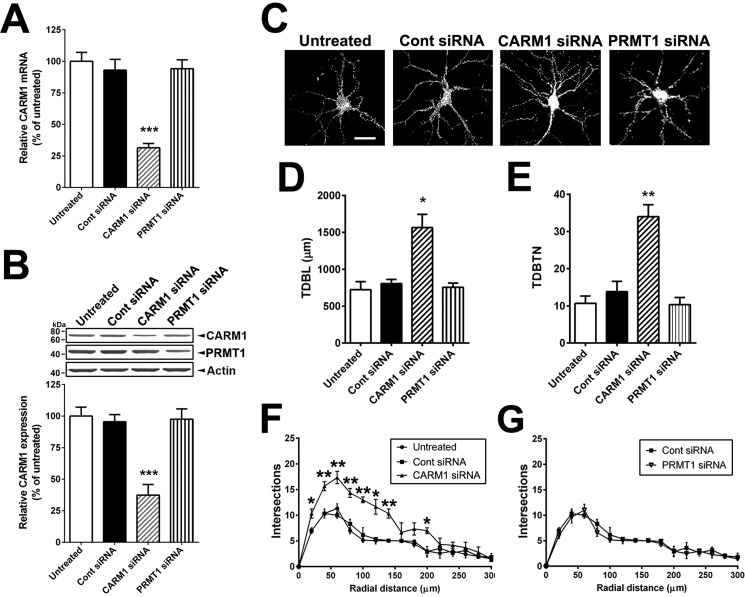
We showed previously that CARM1 inhibition by a specific CARM1 inhibitor or PKCϵ-mediated inhibition promotes the dendritic arborization of differentiating hippocampal neurons in vitro (22). To confirm whether CARM1 is important for dendritic maturation of dissociated hippocampal neurons, we suppressed CARM1 expression with specific siRNAs against CARM1 or PRMT1 (a predominant type I protein methyltransferase in mammalian cells, accounting for 85% of cellular PRMT activity) as negative control (32). Using RT-qPCR, we confirmed that transduction of CARM1-specific siRNA into neurons reduced CARM1 mRNA levels by 69% compared with untreated cells, and there was no difference in CARM1 mRNA levels in cells transduced with a scrambled control or PRMT1 siRNA compared with untreated cells (Fig. 3A). By immunoblotting analysis, neurons transduced with CARM1-specific siRNA showed a 62% decrease in CARM1 protein levels, compared with neurons transduced with scrambled control siRNA or untreated cells. Neurons transduced with PRMT1-specific siRNA showed no difference in CARM1 protein levels compared with untreated cells, even that reduced PRMT1 protein levels by 55%, compared with untreated cells (Fig. 3B).
Figure 3.
Reduction of CARM1 expression increases the complexity of dendritic arborization of cultured hippocampal neurons. A, cultured hippocampal neurons were untreated or transduced with control scrambled (Cont), CARM1-specific, or PRMT1-specific siRNA at 9 DIV for 5 days. At 14 DIV, neurons were lysed and used for total RNA isolation and RT-qPCR. Relative CARM1 mRNA level was analyzed after normalization with GAPDH mRNA level. B, immunoblotting results with anti-CARM1, anti-PRMT1, and anti-actin antibodies from hippocampal neurons treated as in (A). C, cultured hippocampal neurons treated as in (A) were fixed at 14 DIV and stained with MAP-2, a dendritic marker. Whole cell shape is shown. Scale bar, 30 μm. D and E, quantitative analysis of TDBL (μm) and TDBTN from tracing images of neurons treated as in (C). F and G, Sholl analysis of dendritic complexity in neurons treated as in (C). Mean ± S.E., n = 40 neurons/condition from three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001, compared with untreated condition.
To determine the effects of CARM1 silencing on dendritic maturation of hippocampal neurons, neurons transduced with control, CARM1-specific, or PRMT1-specific siRNA were fixed, stained with the dendritic marker microtubule-associated protein 2 (MAP-2), and imaged by confocal microscopy. Neurons transduced with CARM1-specific siRNA showed increased dendritic branching and an enhanced dendritic arbor, but not with PRMT1-specific siRNA (Fig. 3C). To quantify the dendritic structure response to CARM1 silencing, images of the neurons stained with MAP-2 were traced in Neurolucida software for analysis of total dendritic branch length (TDBL) and total dendritic branch tip number (TDBTN). Transduction of neurons with CARM1-specific siRNA caused a dramatic increase in TDBL (1565.0 ± 179.5 μm), compared with untreated (723.3 ± 110.4 μm), control siRNAs (806.7 ± 56.7 μm), or PRMT1 siRNAs (756.7 ± 57.6 μm). TDBTN was also increased in neurons transduced with CARM1-specific siRNA (34.00 ± 3.19), compared with untreated (10.67 ± 1.99), control siRNA (13.83 ± 2.75), or PRMT1 siRNAs (10.33 ± 1.91) (Fig. 3, D and E). To quantify dendritic complexity, we used Sholl analysis to measure the number of dendritic branches that intersect in concentric circles at 20-μm intervals starting 20 μm from the center of the soma. The number of intersections was significantly increased by CARM1-specific siRNA, as far as 200 μm from the soma. The most dramatic effects were seen in the first 100 μm of the dendrites, where transduction of CARM1-specific siRNAs induced an ∼1.7-fold increase in the number of intersections, compared with untreated or control siRNA (Fig. 3F). Transduction of PRMT1 siRNAs did not show any effect on dendritic complexity, compared with control siRNA (Fig. 3G). Together, these results demonstrate that CARM1 inhibition increases the total dendritic length and branching of cultured hippocampal neurons.
Treatment of cultured hippocampal neurons with CARM1 inhibitor affects spine morphology
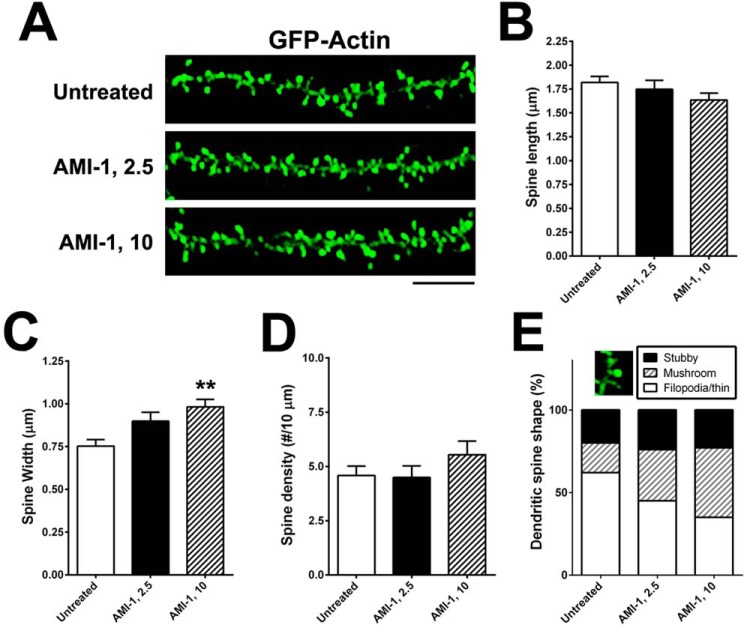
Because decreased expression of CARM1 led to an increase in dendritic complexity in cultured hippocampal neurons, we tested whether inhibition of CARM1 signaling would have a similar effect on dendritic spine morphology. Primary cultures of rat hippocampal neurons were transfected at 7 DIV with a GFP:actin plasmid to label the dendritic spines and then were untreated or treated with 2.5 or 10 μm of the CARM1-specific inhibitor arginine N-methyltransferase inhibitor-1 (AMI-1) for 4 days. Cultures were imaged by confocal microscopy (Fig. 4A) to evaluate spine length (from the base of the dendritic shaft to the tip of the spine) and width of each spine head (at its widest point). The length of the spines was slightly lower in cells treated with 10 μm AMI-1 (1.634 ± 0.072 μm), but the difference was not statistically significant compared with untreated controls (1.819 ± 0.064 μm) or cells treated with 2.5 μm (1.748 ± 0.093 μm) (Fig. 4B). The width of dendritic spines was slightly higher in cells treated with 2.5 μm AMI-1 (0.899 ± 0.052 μm) and significantly higher with 10 μm AMI-1 (0.982 ± 0.044 μm, p = 0.0085), compared with untreated control cells (0.753 ± 0.038 μm) (Fig. 4C). There was no significant difference in spine density (the number of spines per 10-μm dendrite) along the dendritic shaft: untreated (4.58 ± 0.43), 2.5 μm AMI-1 (4.49 ± 0.54), and 10 μm AMI-1 (5.54 ± 0.62) (Fig. 4D). We further investigated spine shapes using a morphometric analysis that takes into account the ratio of spine head and neck to the spine length to categorize the spines into three different types (stubby, mushroom, and filopodia, which includes long, thin-type spines). CARM1 inhibition with AMI-1 significantly increased the proportion of mushroom-type spines (p = 0019), and led to a dramatic decrease of the proportion of filopodia-type, including thin-type, spines (p = 0.0027) (Fig. 4E).
Figure 4.
Treatment of CARM1 inhibitor promotes dendritic spine maturation in cultured hippocampal neurons. A, cultured hippocampal neurons were transfected with GFP-actin at 7 DIV and untreated or treated at 10 DIV with control media (untreated) or AMI-1 (2.5 or 10 μm) for 4 days. At 14 DIV, neurons were fixed and dendritic spine morphology was visualized by confocal microscopy. Scale bar, 10 μm. B–D, spine length (B), spine head width (C), or spine density (number of spines/10 μm dendrite length) (D) was calculated and statistically analyzed. E, dendritic spine shape was counted and expressed as %. Mean ± S.E., n > 100 spines, from three independent experiments. **, p < 0.01, compared with untreated condition.
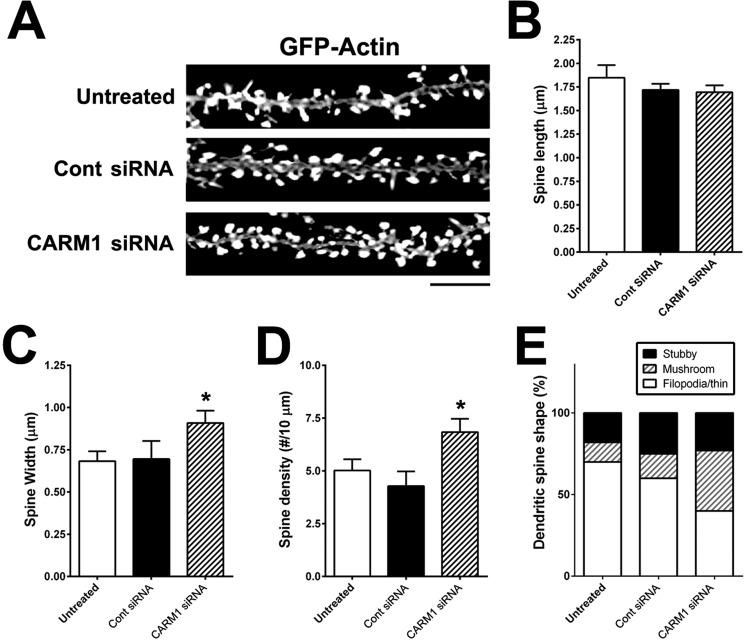
Knockdown of CARM1 expression alters spine morphology
We also examined the effects of CARM1-specific siRNA on dendritic spine morphology. Primary hippocampal neurons were transfected at 7 DIV with a GFP:actin plasmid to label dendritic spines and then were untreated or transduced with control siRNA or CARM1-specific siRNA for 5 days and imaged by confocal microscopy (Fig. 5A). Spine length was not significantly different among the three groups: untreated (1.848 ± 0.133 μm), control siRNA (1.719 ± 0.064 μm), or CARM1-specific siRNA (1.694 ± 0.0723 μm) (Fig. 5B). The width of the dendritic spines was significantly greater in cells transduced with CARM1-specific siRNA (0.909 ± 0.072 μm, p = 0.041), compared with untreated (0.683 ± 0.059 μm) or control siRNA (0.708 ± 0.107 μm) (Fig. 5C). In addition, dendritic spine density was statistically significantly higher in the CARM1-specific siRNA group (6.84 ± 0.62, p = 0.0294) compared with the untreated (5.02 ± 0.54) or control siRNA groups (4.28 ± 0.69) (Fig. 5D). Transduction of neurons with CARM1-specific siRNA also affected dendritic spine shape, causing a dramatic increase in the proportion of mushroom-type spines (∼37%, p = 0.0061), compared with untreated (12%) or control siRNA transduced (15%) cells. Conversely, the proportion of filopodia/thin-type spines was significantly lower in neurons transduced with CARM1-specific siRNA (∼40%, p = 0.0196), compared with untreated (70%) or control siRNA transduced (60%) cells (Fig. 5E). These results are consistent with the changes in dendritic spine morphology seen with AMI-1 inhibition of CARM1, further supporting a role for CARM1 signaling in spine maturation during hippocampal neuronal differentiation.
Figure 5.
Reduction of CARM1 expression modifies dendritic spine maturation in cultured hippocampal neurons. A, cultured hippocampal neurons were untreated or transfected with GFP-actin at 7 DIV. After 2 days, cells were transduced with control siRNA (Cont) or CARM1-specific siRNA and incubated for another 5 days. At 14 DIV, neurons were fixed and dendritic spine morphology was visualized by confocal microscopy. Scale bar, 10 μm. B–D, spine length (B), spine head width (C), or spine density (D) (number of spines/10-μm dendrite length) was calculated and statistically analyzed. E, dendritic spine shape was counted and expressed as %. Mean ± S.E., n > 100 spines, from three independent experiments. *, p < 0.05, compared with untreated condition).
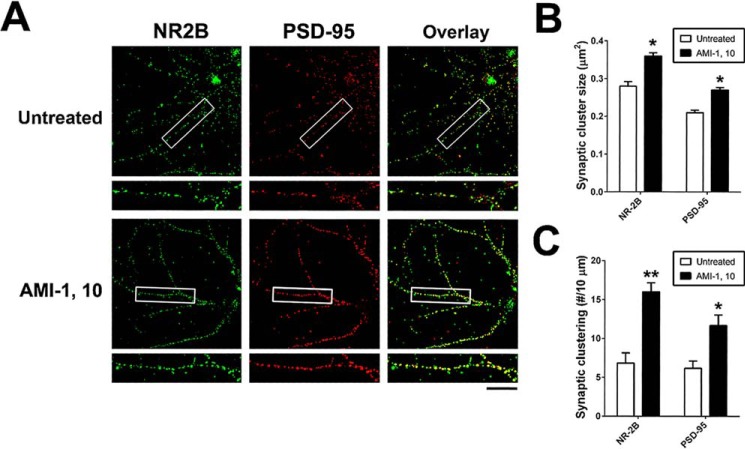
CARM1 inhibition increases post-synaptic targeting of NR2B and PSD-95
Spine morphological categories are highly correlated with synaptic function (33). We found that CARM1 inhibition promotes elaboration of dendrites. We therefore investigated whether CARM1 inhibition using the pharmacological inhibitor AMI-1 or genetic knockdown with specific siRNA affects clustering of synaptic proteins, including synapsin (pre-synaptic) or NR2B and PSD-95 (post-synaptic), that mediate signal transmission and information processing at synaptic sites (34, 35). Primary hippocampal neurons at 10 DIV were untreated or treated with 10 μm AMI-1. After a 4-day incubation, neurons were fixed and double-stained for NR2B and PSD-95, and the average cluster sizes and the number of synaptic clusters for each protein were evaluated (Fig. 6A). Inhibition of CARM1 signaling by AMI-1 treatment led to an increase in the synaptic cluster sizes (Fig. 6B) and the number of clusters (Fig. 6C) of NR2B and PSD-95 compared with untreated cells. There was no difference between AMI-1 treated and untreated cells in synaptic cluster size or number for the pre-synaptic synapsin protein (data not shown).
Figure 6.
CARM1 inhibition increases post-synaptic targeting of NR2B and PSD-95. A, cultured hippocampal neurons at 10 DIV were treated with control media (untreated) or AMI-1 (10 μm) and incubated for 4 days. At 14 DIV, neurons were fixed and used for double-staining with NR2B and PSD-95 antibodies. Scale bar, 10 μm. B and C, synaptic cluster size (μm2) and synaptic clustering (number per 10 μm) were calculated and statistically analyzed. Mean ± S.E., n > 100 spines, from three independent experiments. *, p < 0.05; **, p < 0.01, compared with untreated condition).
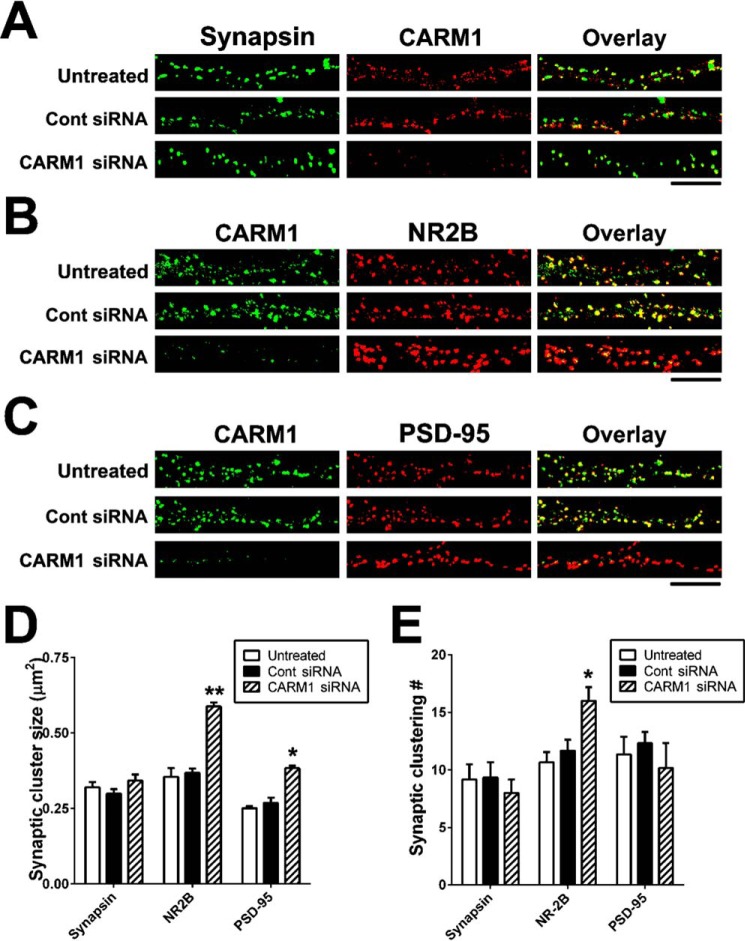
We confirmed these findings in neurons transduced with CARM1-specific siRNAs. Untreated hippocampal neurons or neurons transduced with control or CARM1-specific siRNA were double-stained for CARM1 and synapsin (Fig. 7A), CARM1 and NR2B (Fig. 7B), or CARM1 and PSD-95 (Fig. 7C). Immunocytochemical data clearly showed that transduction of CARM1-specific siRNA into hippocampal neurons decreased CARM1 expression (Fig. 7, A–C). Consistent with the results seen after CARM1 inhibitor treatment, there was no difference in either the synaptic cluster size or the cluster number of synapsin (Fig. 7, A, D, and E). By contrast, siRNA suppression of CARM1 expression led to a significant increase in NR2B cluster size and number compared with untreated or control siRNA-transduced cells (Fig. 7, B, D, and E). CARM1-specific siRNA transduction also led to a significant increase in the synaptic cluster size, not in cluster number, of PSD-95 compared with untreated or control siRNAs-transduced cells (Fig. 7, C–E). Taken together, these data imply that regulation of CARM1 signaling is necessary for clustering of post-synaptic proteins at synapses of hippocampal neurons.
Figure 7.
Knockdown of CARM1 protein promotes post-synaptic clustering of NR2B and PSD-95 proteins. A–C, cultured hippocampal neurons were untreated or transduced with control (Cont) or CARM1-specific siRNA at 9 DIV and incubated for 5 days. At 14 DIV, neurons were fixed and used for double-staining with CARM1 and synapsin (A), NR2B (B), or PSD-95 (C) antibodies. Scale bar, 10 μm. D and E, Synaptic cluster size (μm2) and synaptic clustering (number per 10 μm) were calculated and statistically analyzed. Mean ± S.E., three independent experiments. *, p < 0.05, **, p < 0.01, compared with untreated).
Discussion
In this study, we found that CARM1 is expressed in the PSD fraction of rat hippocampal neurons, confirmed by a series of SDS-PAGE and immunoblotting analyses. We also found that CARM1 is highly enriched in the PSD fraction and that it is present in clusters that co-localize with PSD-95, a post-synaptic marker, in dissociated hippocampal neurons and in dissected rat hippocampi, two main criteria for a protein to be considered a component of the PSD (10). We also demonstrated that inhibition of CARM1 activity, either pharmacologically or through siRNA knockdown, significantly increased dendritic complexity, with greater primary dendritic elongation and number of branches; induced dendritic maturation, as evidenced by an increase in mushroom-type spines; and enhanced clustering of the post-synaptic proteins NR2B and PSD-95 at synapses of differentiating hippocampal neurons in vitro.
CARM1 is a representative PRMT that inhibits neuronal differentiation in vitro (29). Despite data indicating that CARM1 functions in cellular systems, the role of CARM1 protein at the synapse in neurons has remained elusive. CARM1 may be involved in multiple steps during neuronal differentiation, and may be particularly important for the regulation of dendritic maturation and functional synapse formation. CARM1 has been shown previously to modulate mRNA stability and expression of synaptic genes through mRNA-binding proteins. It directly methylates HuD, an mRNA binding protein (30), and inhibits expression of neuronal genes including neurotrophic factors (NTF), such as brain-derived neurotrophic factor (BDNF), at the post-transcriptional level (22, 28, 29). NTF signaling, including that of mature BDNF, is important for dendritic spine formation and synaptogenesis (36–38). These findings suggest that CARM1 inhibition may be required for HuD-mRNA binding, stability, and expression of neuronal genes that are directly linked to dendritic synaptogenesis. Our previous study showed that treatment of a specific CARM1 inhibitor AMI-1 to hippocampal neurons in vitro increased HuD-mRNA binding and the stability and expression of NTFs, including BDNF, in hippocampal neurons (22). A growing body of evidence supports an important regulatory role for CARM1 in synapse function in the hippocampus, an area of the brain critical for learning, memory, and higher-level cognition.
CARM1 methylates diverse protein substrates and regulates intermolecular interactions and subcellular protein localization (39, 40), suggesting that it may affect not only gene expression but also protein function in the dendritic spines of hippocampal neurons, in turn affecting dendrite structure and dendritic spine morphology. Dendritic spines are structurally dynamic and undergo continuous remodeling even after complete maturation, reflecting modifications in synaptic strength and neural circuits (13, 41). During the fundamental processes of spine growth and shrinkage, polymerization and depolymerization of actin filament is required (42). A variety of small GTPases such as Rac1, Cdc42, and Ras are critical regulators of actin polymerization that influence spine number and morphology (43, 44). Cortactin directly binds to actin and phosphorylation-induced cortactin promotes the formation of dendritic protrusions (45). Therefore, future studies will investigate whether small GTPases and/or other actin-regulating factors lie downstream of CARM1 signaling, and how CARM1 may regulate these factors to modulate dendritic architecture and spine morphology and function.
In addition to regulating synaptic gene and protein expression, CARM1 may influence dendritic spine structure by regulating protein-protein interactions in the PSD. Interestingly, spine structures continuously change, reflecting the plasticity of synapses and the modification of synaptic transmission and strength. Changes in the molecular interactions among post-synaptic proteins in the PSD are required for morphological changes related to synaptic plasticity. PSD-95 is a MAGUK family member that interacts with NR2B. Upon neuronal activation, PSD-95 interacts with NR2B and helps to stabilize its localization at post-synaptic membranes, thereby strengthening post-synaptic transmission (46). We found that inhibition of CARM1 signaling increases the cluster sizes of NR2B and PSD-95 at synapses in cultured hippocampal neurons, suggesting that CARM1 inhibits synaptic clustering of PSD proteins and alters signal transmission in hippocampal neurons. This loss of PSD protein clustering has been strongly correlated with disrupted learning behavior in mice (47). CARM1 can methylate neuronal proteins that contain an RXR motif and inhibit specific protein-protein interactions (24, 48). Proteomic analyses to identify post-synaptic proteins containing the RXR motif as potential CARM1 substrates in the PSD may help reveal how CARM1 modulates dendritic morphogenesis and whether CARM1-induced methylation of post-synaptic proteins interferes with synapse maturation.
Post-translational modification of synaptic proteins is an important process underlying long-lasting memory (49). CARM1 activity is specifically modulated by protein phosphorylation (22, 31). PKC can directly phosphorylate CARM1 and abrogate CARM1 activity (30, 31), including decreasing HuD methylation (22). During neuronal differentiation or neural activation, activated PKCϵ enhances post-transcriptional activation of synaptic genes and promotes dendritic maturation, whereas CARM1 inhibits further activation or dendritic maturation. The spatiotemporal regulation of synaptic proteins by CARM1 activation or inhibition at the PSD level may be the key to elucidating the mechanism through which synaptic structures are remodeled. Taken together, our findings strongly suggest that inhibition of CARM1 activity, either directly with a specific inhibitor or indirectly via PKCϵ-mediated modification, may influence learning-specific synaptic formation at the post-translational level in the hippocampus. Further studies using specific CARM1 inhibitors are required to determine how CARM1 activity is regulated in vivo.
In conclusion, CARM1 is enriched in the PSD and co-localizes with post-synaptic proteins in differentiating hippocampal neurons. Our findings suggest that inhibition of CARM1 activity and down-regulation of CARM1 expression potentiates dendritic complexity and spine maturation during neuronal differentiation, which induces synapse formation and would affect the synaptic plasticity essential for learning and memory. Therefore, abnormal activation of CARM1 could contribute to neurological disorders, and inhibition of CARM1 may be a potential therapeutic approach to recover synaptic dysfunction in age-related or neurodegenerative diseases.
Experimental procedures
Ethics statement
Animals were carefully handled following the guidelines and all animal experiments were approved by the West Virginia University Institutional Animal Care and Use Committee.
Isolation of hippocampal subcellular protein fractions
Synaptic subcellular fractions were isolated from rat hippocampal tissue homogenates as described previously (50) with minor modifications. Six rat hippocampi were combined and homogenized in solution containing 0.32 m sucrose, 1 mm NaHCO3, 1 mm MgCl2, 0.5 mm CaCl2, 0.1 mm phenylmethylsulfonyl fluoride, and 1 mg/liter leupeptin. This suspension was placed on a discontinuous sucrose gradient composed of 0.85 m, 1.0 m, and 1.2 m sucrose fractions, and separated by centrifugation for 2 h at 82,500 × g in a TLA-120.1 rotor in a Beckman Coulter Optima Max-XP ultracentrifuge. The synaptosome fraction at the 1.2 m and 1.0 m sucrose interface was collected and extracted with 0.5% Triton X-100 for 15 min. This fraction was pelleted by centrifugation at 36,800 × g for 45 min in a TLA-120.1 rotor to yield the One-Triton PSD fraction and subjected to SDS-PAGE analyses, followed by Coomassie Blue staining (Bio-Rad). Protein concentrations of each fraction were determined by the bicinchoninic acid (BCA) (Thermo Scientific) method and subjected to immunoblotting analysis, as described below.
Hippocampal neuron culture and treatment
Cultures of dissociated hippocampal neurons from embryonic day 18 rats were prepared as described previously (51). Cultured neurons at 10 DIV were exposed to AMI-1 (2.5 or 10 μm) (Enzo Life Sciences) in plating media minus additional glutamate for the indicated times.
Immunoblotting analysis
Cultured neurons were lysed in cold lysis buffer (10 mm Tris-HCl, pH 7.4, 5 mm EDTA, 1% Triton X-100, 10% glycerol, 1 mm CaCl2, 1 mm MgCl2, 1× complete protease inhibitor mixture, and 1× phosphatase inhibitor mixture) (Thermo Scientific) for 1 h at 4 °C. After protein quantitation using the BCA method, equal quantities of cell lysates, tissue homogenates, or brain subcellular fractions were run on 8 or 10% SDS-PAGE gels and transferred to nitrocellulose membranes. Membranes were blocked for 1 h in 5% nonfat milk in TBST (10 mm Tris, pH 7.5, 200 mm NaCl, 0.2% Tween 20) and incubated with rabbit anti-CARM1 (1:1,000) (Cell Signaling Technology), rabbit anti-PRMT1 (1:500) (Fisher Scientific), rabbit anti-NR2B (1:2,000) (EMD Millipore), or rabbit anti-PSD-95 (1:1,000) (Cell Signaling Technology) antibodies. The membranes were washed in TBST and incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary IgG (1:10,000) (Jackson ImmunoResearch Laboratories). Bound antibodies were detected by enhanced chemiluminescence substrate and exposed to X-ray film (Thermo Scientific). As a control, membranes were stripped and re-probed with primary antibody against mouse anti-actin (1:5,000) (Sigma-Aldrich) and HRP-conjugated goat anti-mouse IgG (1:10,000) (Jackson ImmunoResearch Laboratories). Levels of immunoreactivity for each band were assessed by densitometric analysis of films using an HP Scanjet densitometer and ImageJ image analysis system software (1.44a) (NIH), and then normalized to actin levels.
Electron microscopy and quantification
Three-month-old rats were anesthetized with a mixture of ketamine HCl (100 mg/kg body weight) and xylazine (20 mg/kg body weight). The rats were transcardially perfused with saline followed by perfusion with 4% paraformaldehyde and 0.5% glutaraldehyde in 0.1 m phosphate buffer (PB), pH 7.4. After perfusion, the brains were removed and post-fixed in the same fixative overnight at 4 °C. The fixed brains were washed four times in PB and once in PB containing 4% glucose, then stored overnight in PB containing 4% glucose. The brain was cut into ∼350-μm sections on a vibratome and the sections immersed in PB containing 4% sucrose. The sections were then washed in 10 and 20% glycerol in PB for 1 h each, then in 30% glycerol in PB overnight. A portion of the CA1 region of the hippocampus was cut from the sections and embedded by freeze substitution. The tissue was plunge frozen in an AFS freeze substitution instrument (Leica, Vienna, Austria) by liquid nitrogen immersed in 1.5% uranyl acetate in methanol for 27 h at −90 °C, and then warmed to −45 °C, rinsed in anhydrous ethanol.
Sections were cut into ∼100-nm thickness and mounted on mesh grids. The sections were etched in 0.1% sodium borohydride, 50 mm glycine with TBS including Triton X-100 (TBS-T) (50 mm Tris-Cl, pH 7.4, 150 mm NaCl, 0.1% Triton X-100) for 10 min. The sections were washed in TBS-T twice for 5 min each and blocked with TBS-T including 2% bovine serum albumin for 30 min. The sections were incubated overnight with mouse anti-CARM1 (1:200) (Cell Signaling Technologies) or mouse anti-PSD-95 (1:100) (Cell Signaling Technologies). Control sections were incubated overnight either with antibodies that had been pre-absorbed with the specific blocking peptides or with the secondary antibodies. After rinsing in blocking buffer three times, incubation in primary antibody, the sections were then incubated with 10 nm gold-conjugated secondary antibodies (1:20 dilution in blocking buffer with 0.5% polyethylene glycol) (Jackson Immunoresearch Laboratories) for 2 h. After rinsing in distilled water and counterstaining in 2% uranyl acetate and in Reynolds lead citrate, the sections were observed on a Philips 300 transmission electron microscope. Excitatory synapses were identified by the presence of a distinct synaptic cleft, round pre-synaptic vesicles, and Gray's Type I PSD. All labeled synapses were photographed.
Quantification of the distribution of gold label was conducted on images of immunolabeled tissue derived from two rats. The digitized images were imported into NIH ImageJ software for further analysis by an investigator blind to the experimental conditions. To determine the axodendritic distribution of gold particles, the distance between the outer perimeter of the post-synaptic membrane and the center of each gold particle was measured. All gold particles within 100 nm of the post-synaptic membrane toward either the pre-synaptic or the post-synaptic side were measured. A total of 13 synapses and 54 gold particles were analyzed for synapses labeled with CARM1, and 17 synapses and 78 gold particles were analyzed for synapses labeled with the PSD-95. Distances to gold particles on the pre-synaptic side were given negative values, and distances to gold particles on the post-synaptic side were given positive values. To analyze the tangential distribution of labeling along the PSD, the distance between the gold particle and the midline of the PSD was measured. This distance was normalized as a percentage of the total distance between the midline and the lateral edge of the PSD.
Transfection and CARM1 gene silencing
To identify the effects of genetic knockdown of CARM1 on dendritic morphology and synaptic clustering of synapsin, NR2B, and PSD-95, hippocampal neurons were first transfected with the construct pCAG-mGFP-actin (a gift from Ryohei Yasuda (Addgene plasmid number 21948)) at 7 DIV with Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. After a 2-day incubation, neurons were untreated or transduced with rat CARM1- or PRMT1-specific siRNA or scrambled control siRNA lentiviral particles (5 × 103 viral particles/μl) (Santa Cruz Biotechnology) at a multiplicity of infection (m.o.i.) of 4 to ensure efficient infection following the manufacturer's protocol, followed by overnight incubation. After a media change with plating media minus glutamate, cells were incubated for an additional 4 days, and then a portion of the cells was either lysed and used for total RNA isolation and RT-qPCR experiments, lysed and used for immunoblotting analysis, or fixed for immunocytochemical analysis.
Real-time quantitative RT-PCR
Total RNA was isolated using the RNeasy Mini Kit (Qiagen) per the manufacturer's protocol. For the RT reaction, 500 ng of total RNA was reverse transcribed using oligo(dT) primer and Superscript III (Invitrogen) at 50 °C for 1 h. Real-time qPCR was performed for 40 cycles with SYBR Green 1 PCR Master Mix and processed on LightCycler 480 Instrument II (Roche) machine using specific primer sets against rat CARM1, PRMT1, or GAPDH (all from Qiagen). Reactions were run in triplicate for each sample and a dissociation curve was generated. Threshold cycles (Ct) for CARM1 amplification were normalized to the housekeeping gene GAPDH (ΔCt) and every experimental sample was referred to its control (ΔΔCt). Relative expression change values were expressed as 2−ΔΔCt.
Immunocytochemistry and quantification of dendritic branching
Cultured neurons were treated as described above, fixed, and then incubated with primary antibodies against mouse anti-MAP-2 (1:1,000) (Abcam) and Alexa 488-conjugated anti-mouse secondary IgG (1:1,000) (Invitrogen) as described previously (22). The immunostained neurons were viewed on a Zeiss LSM 710 laser-scanning confocal microscope. Neurons from blindly predetermined locations on the coverslip were imaged in Z-sections, and maximal projections of the images were used for analysis. To quantify changes in dendritic structure, images of neurons stained with MAP-2 were opened in Neurolucida software (MBF Bioscience) and the dendrites were traced and analyzed by a person blinded to the treatment condition. Tracings were then automatically analyzed in NeuroExplorer software (Nex Technologies) for TDBL and TDBTN. NeuroExplorer was also used for Sholl analysis to quantify the number of dendrite intersections in concentric rings centered about the soma beginning at a radius of 20 μm from the center of the soma and at 20-μm intervals thereafter. Forty neurons derived from three independent cultures were randomly selected, traced, and analyzed for each condition.
Immunocytochemistry and image acquirement
Culture neurons were treated as described above and then fixed in 2% paraformaldehyde, 4% sucrose, PBS for 10 min at room temperature followed by methanol for 20 min at −20 °C. The fixed neurons were incubated in pre-block buffer (20 mm phosphate buffer, pH 7.4, 5% normal goat serum, 0.05% Triton X-100, and 450 mm NaCl) at 4 °C overnight with the following primary antibodies: mouse anti-CARM1 (1:200) (Cell Signaling Technologies), rabbit anti-CARM1 (1:200) (Cell Signaling Technologies), rabbit anti-NR2B (1:200) (EMD Millipore), mouse anti-NR2B (1:150) (Thermo Fisher Scientific), mouse anti-PSD-95 (1:100) (Cell Signaling Technologies), mouse anti-synapsin (1:200) (Santa Cruz Biotechnology), or rabbit anti-synapsin (1:250) (EMD Millipore), followed by incubation with the appropriate secondary antibodies (Alexa 488 anti-rabbit or 546 anti-mouse; each at 1:1,000) (Invitrogen) in pre-block buffer at room temperature for 1 h. After immunostaining, the cells were mounted as described previously (50) and viewed on a Zeiss LSM 710 laser-scanning confocal microscope. Images were acquired under a 63× or 100× oil immersion objective lens and scanned for further analysis.
Image analysis
Confocal microscope images for each treatment from each of three independent sets of experiments were imported into Image J (NIH) software. To determine the proportion of CARM1 signal that co-localizes with PSD-95 or synapsin, Image J (NIH) software was used with Just Another Colocalization (JACoP) plugin set. A Mander's coefficient was calculated and the average value (n = 10) was further analyzed. Dendritic spine length (from dendritic shaft to the top of spine head) and dendritic spine width (diameter of the widest part of spine head) were measured and dendritic spine density (number of spines/10 μm of dendrite) was also counted. Dendritic spine shape (stubby, mushroom, or filopodium) was identified as described in previous research (5, 21). After measuring spine length (the distance from the base of the neck to the furthest point on the spine heads) and diameter of spine head and neck, the ratio from each spine was calculated and compared. Thin spines were judged if serial viewing revealed the length to be greater than the neck diameter, and the diameters of the head and neck to be similar. Mushroom spines were judged if the diameter of the head was three times larger than the diameter of their necks. Stubby spines were judged if the diameter of the neck was similar to the total length of the spine.
Quantification and statistical analysis
Quantitative data are expressed in arbitrary units (%) comparing untreated controls with cells treated with the indicated concentrations of inhibitors or transduced with siRNA. All data are presented as mean ± S.E. from three or more independent experiments unless otherwise indicated. Statistical comparisons between different treatment groups were conducted with Tukey's multiple comparison test after one-way analysis of variance (ANOVA) using GraphPad Prism 6 software (GraphPad Software Inc.). p values of less than 0.05 were considered to be statistically significant.
Author contributions
C. S. L. conceived the idea for the project, conducted most of the experiment, analyzed the results, and wrote most of the paper. D. L. A. helped to develop the idea for the project and wrote the paper with C. S. L.
The authors declare that they have no conflicts of interest with the contents of this article.
- PSD
- post-synaptic density
- CARM1
- coactivator-associated arginine methyltransferase 1
- DIV
- days in vitro
- NR2B
- N-methyl-d-aspartate subunit 2B
- PKC
- protein kinase C
- PRMTs
- protein arginine methyltransferases
- siRNA
- small interfering RNA
- RT-qPCR
- quantitative reverse transcription-polymerase chain reaction
- MAP-2
- microtubule-associated protein 2
- TDBL
- total dendritic branch length
- TDBTN
- total dendritic branch tip number
- AMI-1
- arginine N-methyltransferase inhibitor-1
- NTF
- neurotrophic factors
- BDNF
- brain-derived neurotrophic factor.
References
- 1. Kandel E. R., Dudai Y., and Mayford M. R. (2014) The molecular and systems biology of memory. Cell 157, 163–186 [DOI] [PubMed] [Google Scholar]
- 2. Sperling R. A., Dickerson B. C., Pihlajamaki M., Vannini P., LaViolette P. S., Vitolo O. V., Hedden T., Becker J. A., Rentz D. M., Selkoe D. J., and Johnson K. A. (2010) Functional alterations in memory networks in early Alzheimer's disease. Neuromolecular Med. 12, 27–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Walsh D. M., and Selkoe D. J. (2004) Deciphering the molecular basis of memory failure in Alzheimer's disease. Neuron 44, 181–193 [DOI] [PubMed] [Google Scholar]
- 4. Wondolowski J., and Dickman D. (2013) Emerging links between homeostatic synaptic plasticity and neurological disease. Front. Cell Neurosci. 7, 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hongpaisan J., Sun M. K., and Alkon D. L. (2011) PKCϵ activation prevents synaptic loss, Aβ elevation, and cognitive deficits in Alzheimer's disease transgenic mice. J. Neurosci. 31, 630–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harris K. M., and Weinberg R. J. (2012) Ultrastructure of synapses in the mammalian brain. Cold Spring Harb. Perspect. Biol. 4, a005587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rochefort N. L., and Konnerth A. (2012) Dendritic spines: from structure to in vivo function. EMBO Rep. 13, 699–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Amtul Z., and Atta-Ur-Rahman (2015) Neural plasticity and memory: molecular mechanism. Rev. Neurosci. 26, 253–268 [DOI] [PubMed] [Google Scholar]
- 9. Koleske A. J. (2013) Molecular mechanisms of dendrite stability. Nat. Rev. Neurosci. 14, 536–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kennedy M. B. (2000) Signal-processing machines at the postsynaptic density. Science 290, 750–754 [DOI] [PubMed] [Google Scholar]
- 11. Newpher T. M., and Ehlers M. D. (2009) Spine microdomains for postsynaptic signaling and plasticity. Trends Cell Biol. 19, 218–227 [DOI] [PubMed] [Google Scholar]
- 12. Kastellakis G., Cai D. J., Mednick S. C., Silva A. J., and Poirazi P. (2015) Synaptic clustering within dendrites: an emerging theory of memory formation. Prog. Neurobiol. 126, 19–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sala C., and Segal M. (2014) Dendritic spines: the locus of structural and functional plasticity. Physiol. Rev. 94, 141–188 [DOI] [PubMed] [Google Scholar]
- 14. Woolfrey K. M., and Srivastava D. P. (2016) Control of dendritic spine morphological and functional plasticity by small GTPases. Neural Plast. 2016, 3025948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Saneyoshi T., Fortin D. A., and Soderling T. R. (2010) Regulation of spine and synapse formation by activity-dependent intracellular signaling pathways. Curr. Opin. Neurobiol. 20, 108–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nelson T. J., Sun M. K., Hongpaisan J., and Alkon D. L. (2008) Insulin, PKC signaling pathways and synaptic remodeling during memory storage and neuronal repair. Eur. J. Pharmacol. 585, 76–87 [DOI] [PubMed] [Google Scholar]
- 17. Sun M. K., and Alkon D. L. (2010) Pharmacology of protein kinase C activators: cognition-enhancing and antidementic therapeutics. Pharmacol. Ther. 127, 66–77 [DOI] [PubMed] [Google Scholar]
- 18. Calabrese B., and Halpain S. (2005) Essential role for the PKC target MARCKS in maintaining dendritic spine morphology. Neuron 48, 77–90 [DOI] [PubMed] [Google Scholar]
- 19. Matsuoka Y., Li X., and Bennett V. (1998) Adducin is an in vivo substrate for protein kinase C: phosphorylation in the MARCKS-related domain inhibits activity in promoting spectrin-actin complexes and occurs in many cells, including dendritic spines of neurons. J. Cell Biol. 142, 485–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pascale A., Amadio M., Scapagnini G., Lanni C., Racchi M., Provenzani A., Govoni S., Alkon D. L., and Quattrone A. (2005) Neuronal ELAV proteins enhance mRNA stability by a PKCα-dependent pathway. Proc. Natl. Acad. Sci. U.S.A. 102, 12065–12070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hongpaisan J., and Alkon D. L. (2007) A structural basis for enhancement of long-term associative memory in single dendritic spines regulated by PKC. Proc. Natl. Acad. Sci. U.S.A. 104, 19571–19576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lim C. S., and Alkon D. L. (2012) Protein kinase C stimulates HuD-mediated mRNA stability and protein expression of neurotrophic factors and enhances dendritic maturation of hippocampal neurons in culture. Hippocampus 22, 2303–2319 [DOI] [PubMed] [Google Scholar]
- 23. Guo A., Gu H., Zhou J., Mulhern D., Wang Y., Lee K. A., Yang V., Aguiar M., Kornhauser J., Jia X., Ren J., Beausoleil S. A., Silva J. C., Vemulapalli V., Bedford M. T., and Comb M. J. (2014) Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol. Cell Proteomics 13, 372–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Morales Y., Cáceres T., May K., and Hevel J. M. (2016) Biochemistry and regulation of the protein arginine methyltransferases (PRMTs). Arch. Biochem. Biophys. 590, 138–152 [DOI] [PubMed] [Google Scholar]
- 25. Wolf S. S. (2009) The protein arginine methyltransferase family: an update about function, new perspectives and the physiological role in humans. Cell. Mol. Life Sci. 66, 2109–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hu H., Qian K., Ho M. C., and Zheng Y. G. (2016) Small molecule inhibitors of protein arginine methyltransferases. Expert Opin. Investig. Drugs 25, 335–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McBride A. E. (2006) 3 diverse roles of protein arginine methyltransferases. Enzymes 24, 51–103 [DOI] [PubMed] [Google Scholar]
- 28. Colombrita C., Silani V., and Ratti A. (2013) ELAV proteins along evolution: back to the nucleus? Mol. Cell. Neurosci. 56, 447–455 [DOI] [PubMed] [Google Scholar]
- 29. Fujiwara T., Mori Y., Chu D. L., Koyama Y., Miyata S., Tanaka H., Yachi K., Kubo T., Yoshikawa H., and Tohyama M. (2006) CARM1 regulates proliferation of PC12 cells by methylating HuD. Mol. Cell Biol. 26, 2273–2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Higashimoto K., Kuhn P., Desai D., Cheng X., and Xu W. (2007) Phosphorylation-mediated inactivation of coactivator-associated arginine methyltransferase 1. Proc. Natl. Acad. Sci. U.S.A. 104, 12318–12323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Feng Q., He B., Jung S. Y., Song Y., Qin J., Tsai S. Y., Tsai M. J., and O'Malley B. W. (2009) Biochemical control of CARM1 enzymatic activity by phosphorylation. J. Biol. Chem. 284, 36167–36174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang X., and Cheng X. (2003) Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides. Structure 11, 509–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nimchinsky E. A., Sabatini B. L., and Svoboda K. (2002) Structure and function of dendritic spines. Annu. Rev. Physiol. 64, 313–353 [DOI] [PubMed] [Google Scholar]
- 34. Hunt D. L., and Castillo P. E. (2012) Synaptic plasticity of NMDA receptors: mechanisms and functional implications. Curr. Opin. Neurobiol. 22, 496–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shepherd J. D., and Huganir R. L. (2007) The cell biology of synaptic plasticity: AMPA receptor trafficking. Annu. Rev. Cell Dev. Biol. 23, 613–643 [DOI] [PubMed] [Google Scholar]
- 36. Cunha C., Brambilla R., and Thomas K. L. (2010) A simple role for BDNF in learning and memory? Front. Mol. Neurosci. 3, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shen K., and Cowan C. W. (2010) Guidance molecules in synapse formation and plasticity. Cold Spring Harb. Perspect. Biol. 2, a001842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Waterhouse E. G., and Xu B. (2009) New insights into the role of brain-derived neurotrophic factor in synaptic plasticity. Mol. Cell. Neurosci. 42, 81–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bedford M. T. (2007) Arginine methylation at a glance. J. Cell Sci. 120, 4243–4246 [DOI] [PubMed] [Google Scholar]
- 40. Lee Y. H., and Stallcup M. R. (2009) Minireview: protein arginine methylation of nonhistone proteins in transcriptional regulation. Mol. Endocrinol. 23, 425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yuste R., and Bonhoeffer T. (2001) Morphological changes in dendritic spines associated with long-term synaptic plasticity. Annu. Rev. Neurosci. 24, 1071–1089 [DOI] [PubMed] [Google Scholar]
- 42. Schubert V., and Dotti C. G. (2007) Transmitting on actin: synaptic control of dendritic architecture. J. Cell Sci. 120, 205–212 [DOI] [PubMed] [Google Scholar]
- 43. Carlisle H. J., and Kennedy M. B. (2005) Spine architecture and synaptic plasticity. Trends Neurosci. 28, 182–187 [DOI] [PubMed] [Google Scholar]
- 44. Tashiro A., and Yuste R. (2004) Regulation of dendritic spine motility and stability by Rac1 and Rho kinase: evidence for two forms of spine motility. Mol. Cell. Neurosci. 26, 429–440 [DOI] [PubMed] [Google Scholar]
- 45. Pontrello C. G., and Ethell I. M. (2009) Accelerators, brakes, and gears of actin dynamics in dendritic spines. Open Neurosci. J. 3, 67–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhu J., Shang Y., and Zhang M. (2016) Mechanistic basis of MAGUK-organized complexes in synaptic development and signalling. Nat. Rev. Neurosci. 17, 209–223 [DOI] [PubMed] [Google Scholar]
- 47. Nagura H., Ishikawa Y., Kobayashi K., Takao K., Tanaka T., Nishikawa K., Tamura H., Shiosaka S., Suzuki H., Miyakawa T., Fujiyoshi Y., and Doi T. (2012) Impaired synaptic clustering of postsynaptic density proteins and altered signal transmission in hippocampal neurons, and disrupted learning behavior in PDZ1 and PDZ2 ligand binding-deficient PSD-95 knockin mice. Mol. Brain 5, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Basso M., and Pennuto M. (2015) Serine phosphorylation and arginine methylation at the crossroads to neurodegeneration. Exp. Neurol. 271, 77–83 [DOI] [PubMed] [Google Scholar]
- 49. Routtenberg A., and Rekart J. L. (2005) Post-translational protein modification as the substrate for long-lasting memory. Trends Neurosci. 28, 12–19 [DOI] [PubMed] [Google Scholar]
- 50. Walikonis R. S., Jensen O. N., Mann M., Provance D. W. Jr., Mercer J. A., and Kennedy M. B. (2000) Identification of proteins in the postsynaptic density fraction by mass spectrometry. J. Neurosci. 20, 4069–4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lim C. S., and Walikonis R. S. (2008) Hepatocyte growth factor and c-Met promote dendritic maturation during hippocampal neuron differentiation via the Akt pathway. Cell. Signal. 20, 825–835 [DOI] [PMC free article] [PubMed] [Google Scholar]