Abstract
Personalized medicine offers the promise of better diagnoses, targeted therapies and individualized treatment plans. Pharmacogenomics is an integral component of personalized medicine; it aids in the prediction of an individual’s response to medications. Despite growing public acceptance and emerging clinical evidence, this rapidly expanding field of medicine is slow to be adopted and utilized by healthcare providers, although many believe that they should be knowledgeable and able to apply pharmacogenomics in clinical practice. Institutional infrastructure must be built to support pharmacogenomic implementation. Multidisciplinary education for healthcare providers is a critical component for pharmacogenomics to achieve its full potential to optimize patient care. We describe our recent experience at the Mayo Clinic implementing pharmacogenomics education in a large, academic healthcare system facilitated by the Mayo Clinic Center for Individualized Medicine.
Keywords: e-learning, genomic decision-making, healthcare systems, individualized medicine, multidisciplinary education, personalized medicine, pharmacogenetics, pharmacogenomics, point-of-care online resources
Personalized medicine offers the promise of better diagnoses, targeted therapies and individualized treatment plans [1]. Increasing evidence points toward the powerful influence of genetics on the current diagnosis and treatment paradigms used in many healthcare systems. Pharmacogenomics has been described as the study of drug response and genetic variation, and its importance stems from the ability to predict an individual’s response to medications [2]. Personalized drug therapy aims to optimize treatment outcomes while eliminating drugs that have no treatment effect on patients or, worse, put them at risk for serious adverse effects. This burgeoning field is expanding rapidly, moving from the laboratory bench to the patient bedside and leaving in its wake a genomic information revolution.
Growing access to direct-to-consumer testing has placed genetic test results in the hands of consumers and has moved genomic information away from the traditional healthcare provider infrastructure. Though limited evidence is available, initial investigations into patients’ attitudes toward pharmacogenomics have begun to yield supportive results [3,4]. Although the public has been shown to possess a basic knowledge of genetics [5], in one study, surveyed adults indicated interest in pharmacogenomic testing to assist with selection and dosing of drugs and to predict drug side effects [4]. Despite increasingly affordable and accessible genotyping [6] and the public’s growing interest in genetic testing, adoption of genomics into routine medical care remains a challenge. One recent study of primary care physicians reported that the majority believed direct-to-consumer genomic test results (including pharmacogenetic results) were understandable, and physicians would be open to discussing and using these results as part of patient care [7].
There is increasing evidence of pharmacogenomics’ clinical utility, but adoption by clinicians into practice has been slow [8,9]. Barriers to the translation and adoption into clinical practice have been described by the Clinical Pharmacogenetics Implementation Consortium (CPIC) of the Pharmacogenetics Research Network [10]. Operational, technical and human (cognitive/psychological) barriers include limitations of currently fragmented healthcare systems to manage patients’ genetic tests results over their lifetimes; limited use of electronic medical records; lack of reward for disease or adverse drug reaction prevention in the current healthcare environment; lack of clinician knowledge and readiness regarding genomics; lack of preemptive genetic testing results for point-of-care decision making; and privacy, ethical and legal concerns [8,10–12]. Despite these challenges, computerized decision support models are beginning to aid the incorporation of genetically guided personalized medicine and pharmacogenomics into patient care [13–18]. Evidence for clinical utility is growing as institutions are beginning to report positive outcomes of impact to patient care [9,19–21].
With increasing computer support guidance for pharmacogenomic decision making, education for healthcare providers delivering care at the patient bedside is the next logical step. Education for aiding clinicians to knowledgeably prescribe, dispense and administer drugs with pharmacogenomic biomarkers in drug labeling is critical [22–31]. There is an urgent need to educate clinicians in this emerging and rapidly expanding area of medicine to provide optimal patient care [9,32–35]. This paper describes the approach our institution is taking to implement a multidisciplinary pharmacogenomics education strategy.
Educational needs for health professionals
To foster a genomically-informed environment for multidisciplinary healthcare providers, an inter-professional approach to education may be considered for healthcare providers in-training including students, residents and fellows. General principles of inter-professional education (IPE) include interactive learning by healthcare professionals to improve collaborations and/or the health of patients through shared skills and knowledge [36,37]. The goal of IPE is to create teams that collaborate to provide patient-centered care. Several IPE models illustrate the interdependency between the health professionals’ education competency development and the system of collaborative healthcare and include four competency domains: values and ethics for inter-professional practice; roles and responsibilities for collaborative practice; inter-professional communication practices; and inter-professional teamwork and team-based practice [38,39]. Numerous studies have investigated the necessary elements for successful IPE implementation, process and outcomes [36,40]. Benefits of inter-professional education are increased quality, safety and outcomes of patient care [41]. While most IPE strategies focus on developing teamwork in undergraduate and graduate level healthcare students using simulation and field study, there is a growing emphasis on providing IPE in situ as a part of continuous professional development [42,43].
Key stakeholder groups for dissemination of pharmacogenomics education include point-of-care clinicians. This encompasses physicians, pharmacists, genetic counselors, nurses, nurse practitioners, nurse anesthetists, respiratory therapists and healthcare providers in training (residents/fellows). It is a challenging task to educate this diverse group of stakeholders within a real-life healthcare setting on a complex topic such as pharmacogenomics that continues to rapidly emerge and evolve. Healthcare providers in recent studies have been found to be uncomfortable ordering and interpreting pharmacogenomic tests. This is likely impacted by limited knowledge of pharmacogenomics by physicians, pharmacists, genetic counselors and other health practitioners [25,44,45]. Despite lacking knowledge of pharmacogenomics, practitioners have indicated that they believe that they should be knowledgeable, able to select appropriate tests, interpret test results and provide pharmacogenomic test results to patients [25,44,46]. As these tests continue to emerge and evolve, medical geneticists and genetic counselors will play an important role in providing consultation as part of multidisciplinary, team-based care [47,48].
From a federal public health policy perspective, it is critically important to develop a well-prepared and well-educated health professional workforce that can incorporate genomics into clinical practice. The Secretary’s Advisory Committee on Genetics, Health and Society (SACGHS) issued a report in 2011 that identified the genetics education and training efforts needed for point-of-care healthcare providers. Each professional group’s educational efforts should focus on undergraduate and graduate trainees and continuing education and development for practicing professionals [49]. In its report, SACGHS “…found evidence that suggests inadequate education of healthcare professionals is a significant factor limiting the appropriate integration of genetics into clinical care…,” and genetics education programs have been created in isolation without an inter-professional approach to patient care [49]. In order to lead this important educational effort, professional societies must collaborate and provide guidance for the successful translation, development and integration of personalized medicine into patient care [49].
To foster genomic literacy of physicians and other healthcare providers, the National Human Genome Research Institute (NHGRI) in 2013 took proactive steps and fostered the Inter-Society Coordinating Committee for Practitioner Education in Genomics (ISCC-PEG) whose charge is “…To improve genomic literacy of physicians and other practitioners and enhance the practice of genomic medicine…through sharing of educational approaches and joint identification of educational needs” [50]. The purpose of the ISCC is to facilitate inter-professional collaborations to apply the expanding knowledge and evidence of genomics to patient care. As part of this and other disciplinary-driven genomics education initiatives, some professional groups have taken important steps forward to create genomics education guidance documents for their respective health professional disciplines [24,51,52]. Yet, the number of groups that have developed guidelines is low and more work needs to be done [53]. The importance of an inter-professional approach cannot be understated; however, a discipline-tailored yet collaborative model is needed to address each discipline’s contributions at critical points in the healthcare continuum.
What does a multidisciplinary approach to genomics education look like? Our institution’s perspective is that it involves development of education customized to healthcare professionals based upon the individual discipline’s needs to perform patient care duties. For physicians and other prescribers, this may include evaluating genetic risks, ordering genetic testing and prescribing appropriate medications based on genetic information. For pharmacists, this may include providing pharmacogenomic testing support and guidance to prescribers, reviewing and interpreting test results and evaluating pharmacogenomic therapeutic results. For genetic counselors, this may include communicating and assisting clinician prescribers, pharmacists and patients to understand pharmacogenomic information [54,55]. Our institution’s pharmacogenomics education model has taken a laser-focused, common sense approach to provide critical education at the patient point-of-care as an adjunct to developing broad-based pharmacogenomics education that is well-received but which previous research at our institution has shown to have limited effect over time [55,56].
Challenges to delivering pharmacogenomics education
Wide dissemination of pharmacogenomics education to healthcare providers is challenging. Clinicians have difficulties in staying abreast of the rapidly emerging and changing science of genomic medicine. Approximately half of practicing physicians were trained before the completion of the Human Genome Project in 2003, and these practitioners did not have genomics as part of their educational curriculum [51,57]. Conversely, it is also a challenge for newly trained clinicians to stay current on genomic advances due to the rapidly emerging and changing aspects that outdate their genomics training by the time clinicians begin their practice. Evaluating and addressing knowledge gaps for both practicing clinicians and clinician trainees is critical [58]. The rapidly evolving nature of genomics is a barrier to the implementation of genomics into clinical practice. There is a relentless discovery and development of new genetic technologies, laboratory techniques, genomic variants and incorporation into Food and Drug Administration biomarker information in drug labeling [23,59,60]. Further, systematic methods and infrastructure are needed to translate and deliver this evolving information to diverse patient points-of-care across an academic healthcare continuum [61,62].
Several academic institutions have begun striving toward the goal of developing computer decision support (CDS) algorithms to assist healthcare providers with genotype-guided therapeutic decision making in drug prescribing systems [13–18]. Creating informatics infrastructure for genomically-guided therapeutic decision making is complex and includes consistent integration of genomic laboratory results into the electronic medical record; warehousing large amounts of genomic data; and creation of drug-gene algorithms and computer alerts for healthcare providers at the patient point-of-care [63]. Closing the pharmacogenomics knowledge gap of healthcare providers and building institutional infrastructures will complement and facilitate the integration of genomic medicine CDS algorithms and will begin to address challenges of pharmacogenomics implementation.
One institution’s experience: Mayo Clinic Center for Individualized Medicine
The central unifying infrastructure for pharmacogenomics at Mayo Clinic is the Center of Individualized Medicine (CIM) [64]. The mission of CIM is to solve clinical challenges by bringing the latest scientific discoveries to healthcare providers through cutting-edge genomics-based tests and treatments [64]. CIM brings key institutional groups together for pharmacogenomics clinical practice decisions, decision support algorithms and education. Multidisciplinary groups, including medicine, genetics, pharmacy, laboratory medicine, health informatics, statisticians, educators and scientists, work together on institutional pharmacogenomics program development, implementation and evaluation to create an overarching institutional pharmacogenomics paradigm for the large, multisite academic health system (Figure 1) to move pharmacogenomics to the point-of-care [11,65,66]. Subsequently, education for different groups is developed focusing on the knowledge required to facilitate pharmacogenomic therapeutic decision making at the point-of-care. This point-of-care education consists of electronic pharmacogenomic alerts that are linked to online knowledge resources and clinical experts. Rather than relying on individual pharmacogenomics knowledge, our institution’s integrated approach aims to serve our patients in a more focused and comprehensive manner.
Figure 1. The Mayo Clinic Center for Individualized Medicine provides overarching guidance for pharmacogenomics implementation through direct interactions with prescribers, pharmacists and laboratory medicine.
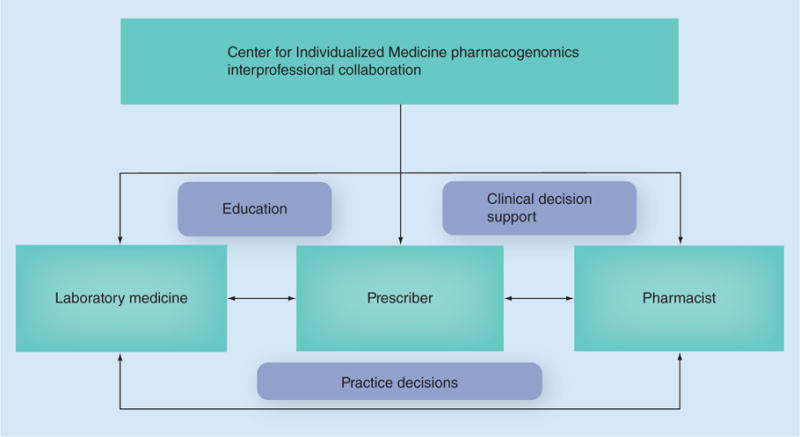
These individuals work as a team to support practice decisions, computerized decision support and education. Used with permission of Mayo Foundation for Medical Education and Research; all rights reserved.
Rapidly emerging and continuously evolving pharmacogenomics evidence and guidelines provide a compelling reason to create point-of-care education for large numbers of diverse disciplines. The immense challenge of delivering timely pharmacogenomics education for large numbers of diverse healthcare providers at points-of-care through the healthcare continuum should not be underestimated. From our institution’s perspective, the resources and infrastructure needed to support this effort are large and there must be institutional support and buy-in for this to be effective [11,65,66]. Other considerations that have impacted our institution’s ability to quickly deliver pharmacogenomics education to the point-of-care include multiple computer systems within a large healthcare system, multiple electronic medical records, poor trackability of lifetime genetic test results and diverse roles of healthcare professionals throughout the hospitals and clinics providing patient care. Understanding how to maximize point-of-care education effectiveness for patient care is key to its successful implementation [67,68]. In the initial rollout phase, the paradigm of education delivery into daily workflow at our institution was through provision of pharmacogenomics education to healthcare providers at the time of prescribing medications at points-of-care. This has been accomplished in part by using preemptive genotype results (when available) and clinical decision support linked to the electronic medical record [65]. Once development and implementation of clinical decision support for genomically-guided therapeutic decision making was completed, the next step was to develop formal pharmacogenomics education for multidisciplinary healthcare providers through an inter-professional approach.
To begin planning for implementation of pharmacogenomics education at our large, academic multicampus institution, members of the Center for Individualized Medicine Pharmacogenomics Task Force considered it as a practice issue: which professionals would serve the most logical role for drug-gene interaction management? With this question in mind, the first phase of educating healthcare providers focused on competency-based, online modules targeting approximately 500 pharmacists (inpatient and outpatient) across the large academic health system. Using this strategy, pharmacists serve as the first line of pharmacogenomics support in a manner that is similar to the current manner that pharmacists support prescribers. In a step-wise appoach, if the first line pharmacist is unable to answer a pharmacogenomics question, then the pharmacist escalates the question to one of the established institutional pharmacogenomics pharmacy specialists who have clinical pharmacy practice and personalized medicine expertise. Focused infrastructure has been implemented to support healthcare providers at points-of-care across the healthcare continuum.
Pharmacogenomics education competencies for pharmacists were piloted and tested at the main campus for quality assurance purposes before releasing the educational program to the entire health system. Results of the pilot have been positive. Preliminary pharmacogenomics education results showed that of 284 pharmacists, the Introduction module was completed by 232 pharmacists (82% completion) and the Hypersensitivity module was completed by 217 pharmacists (76% completion) [69]. The Introduction module showed an average Pretest score of 47.9, an average Posttest score of 93 and an average improvement of 45.1 points [69]. The Hypersensitivity module demonstrated an average Pretest score of 51.4, an average Posttest score of 96.6 and an average improvement of 45.2 points [69]. Feedback from formative evaluations on usability and effectiveness provided information for module improvement. Preliminary evaluations indicate the modules have been enthusiastically embraced by pharmacists and there is strong support for expansion of education to other campuses. Future summative evaluation studies are planned to evaluate the long-term effects of education on pharmacogenomics competency.
To prepare the online modules for broader dissemination to the academic health system, adaptations were made to include relevant site-specific information, including computer screen shots for the different electronic medical records systems. In addition, plans are underway to tailor the education for other groups including, medical trainees, nurses, and physicians. Modifications will be designed to support the different roles in the clinical care teams and develop understanding of how other team members contribute to the delivery of patient-centered pharmacogenomic care. For example, rarely have genetic counselors partnered with pharmacists in coordinated care, but it has been suggested [54] that this may be a model to consider. Additional inter-professional education strategies are necessary to support new paradigms of care resulting from integration of pharmacogenomics in practice.
To maximize educational effectiveness and meet the diverse pharmacogenomics education needs of busy practitioners, other educational approaches are also being employed in addition to e-learning (Table 1). Pharmacogenomics is taught by pharmacists and physicians via traditional methods such as lectures, courses, grand rounds and journal clubs, and via less formal methods such as the Genomic Tumor Board and Genomic Odyssey Board. A face-to-face annual conference and pre-/post-conference learning activities promote teaching and learning pharmacogenomics team-based care. In addition, the preconference course uses a blended model of delivery which includes online preparatory materials and a face-to-face, case-based session.
Table 1.
Pharmacogenomics educational courses developed for healthcare providers at our institution.
| Lecture | Format | Target audience |
|---|---|---|
| Clinical Pharmacogenomics: From Base Pairs to Bedside | Face-to-face lecture, online | Pharmacists Rochester campus |
| Critical Care Pharmacogenomics | Face-to-face lecture, online | Pharmacists Rochester campus |
| Applying Pharmacogenomics to the Management of the Patient with HIV | Face-to-face lecture, online | Pharmacists Rochester campus |
| Pharmacogenomic Considerations in Anesthesia and Analgesia | Face-to-face lecture, online | Pharmacists Rochester campus |
| Clinical Pharmacogenomics: Focus on Cardiovascular Drugs | Online | Pharmacists Rochester campus |
| Pharmacogenomics in Psychiatry: Moving towards Individualized Medication Therapy | Face-to-face lecture, online | Pharmacists Rochester campus |
| Can Pharmacogenomics Predict Efficacy or Toxicity of Chemotherapy? | Face-to-Face lecture, online | Pharmacists Rochester campus |
| Module 1: Pharmacogenomics 101: Moving Science to the Bedside | Online competency | Pharmacists Health system |
| Module 2: Pharmacogenomic Considerations for Hypersensitivity with Abacavir and Carbamazepine | Online competency | Pharmacists Health system |
| Module 3: Pharmacogenomic Considerations for CYP2D6 with Codeine, Tramadol and Tamoxifen | Online competency | Pharmacists Health system |
| Module 4: Pharmacogenomic Considerations for TPMT | Online competency | Pharmacists Health system |
| Pharmacogenomics Grand Rounds | Face-to-face lecture, videoconference to 3 academic sites | Multidisciplinary clinical and research |
| Pharmacogenomics Journal Club | Face-to-face lecture | Multidisciplinary clinical and research |
| Genomic Tumor Board | Videoconference with teaching cases to 3 academic sites | Multidisciplinary clinical and research |
| Genomic Odyssey Board | Videoconference to 3 academic sites | Multidisciplinary clinical and research |
| Mayo Clinic Individualizing Medicine Conference (annual meeting) | Face-to-face lectures | Multidisciplinary clinical and research |
| 2014 Mayo Clinic Individualizing Medicine Conference: Preconference Workshop | Blended model; online and face-to-face lectures | Multidisciplinary clinical and research |
| 2014 Mayo Clinic Individualizing Medicine Conference: Postconference Workshop | Face-to-face lectures | Multidisciplinary clinical and research |
The three largest academic sites include the Mayo Clinic located in Rochester, Minnesota; Scottsdale and Phoenix, Arizona; and Jacksonville, Florida.
Tamoxifen example of multidisciplinary pharmacogenomics
Tamoxifen is a prodrug used in the treatment of breast cancer. It is metabolized by hepatic Cytochrome P450 2D6 (CYP2D6) to the active metabolite endoxifen. Since most individuals are treated with tamoxifen therapy for multiple years, the known efficiency of the CYP2D6 enzyme may be advantageous to patient therapy [70]. Consider the situation when a patient is tested for the CYP2D6 genotype prior to beginning tamoxifen therapy, and the lab test results return as ‘CYP2D6*1/*4 ‘; this likely has little clinical utility for the typical prescriber. Laboratory medicine is essential in not only test development but also in providing phenotypic interpretations for a genotypic report in a manner that the clinical care team can understand and utilize. However, in some cases, a lab report providing information that the individual is a ‘CYP2D6 intermediate metabolizer’ phenotype provides additional information, but since this is a new science, it still might create some confusion for many healthcare providers on the clinical care team. Here, the pharmacy or medical laboratory team members may be in the best position to describe the metabolic outcome of this phenotype to the prescriber. Often, national guidelines such as the Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines will provide assistance in clinical application; however, in the specific case of tamoxifen, an institution may have its own internal recommendations for medication use.
Let us consider a clinical care scenario that is based on the available evidence, and the prescriber decides to proceed with tamoxifen medication therapy. However, at a later time, a different healthcare provider now decides to initiate therapy with fluoxetine for depression for this patient who currently is taking tamoxifen. Of clinical concern, fluoxetine is a strong enzyme inhibitor of CYP2D6 and using it concomitantly with tamoxifen will likely cause this patient with CYP2D6 intermediate metabolism appear much more like a CYP2D6 poor metabolizer in metabolic function, resulting in little to no conversion of tamoxifen into endoxifen. Pharmacy is in the best position to detect this drug-gene interaction. By incorporating pharmacogenomics knowledge into clinical practice, the pharmacist can now discuss the increased risk in a patient with CYP2D6 intermediate metabolism, thus, allowing for better alternative drug selection by healthcare providers. It is clear from this common clinical care scenario that multidisciplinary efforts are required in the clinical application of point-of-care pharmacogenomics.
Limitations
Though our model does not describe an ideal or universal education approach, early evaluations are positive. Because of the vast resources needed to develop education to support implementation of pharmacogenomics, efforts have been limited to large, academic medical centers [71].
Conclusion & future perspective
Despite lack of pharmacogenomics knowledge, healthcare providers have indicated that they believe that they should be knowledgeable and be able to apply pharmacogenomics information to their patients’ care needs. The overall plan of providing education for large numbers of diverse disciplines on rapidly emerging and evolving pharmacogenomics evidence and guidelines should be considered within a multidisciplinary framework. This information can be further passed down in individual departments (e.g., medicine, pharmacy, laboratory medicine) and tailored to the needs of the individual specialty. One of the challenges to overcome is that many healthcare providers were trained before the advent of genomic medicine and these practitioners did not have genomics as part of their educational curriculum. In addition, the needs of clinicians-in-training must also be addressed. Currently, the resources and infrastructure needed to support this educational effort are large, and there must be institutional support and buy-in for this to be effective. National genomics guidelines are a great assistance to aid implementation; however, institutional, multidisciplinary educational efforts at the local institutional level will be required to achieve pharmacogenomics’ full potential in clinical practice. The most successful healthcare systems in the next five years will be those that take up this challenge and translate evidence-based guidelines to the patient point-of-care in a new healthcare paradigm.
In terms of extrapolating our experience to other academic institutions, it is our observation that the use of online education for healthcare providers has proven to be critical to timely and effective delivery of pharmacogenomics education across multiple campuses and healthcare provider groups. Other institutions may benefit from developing and piloting local education for a targeted group of healthcare providers (pharmacists in our institution). Once the local education program has been piloted successfully, the education can be shared with sister academic institutions and extended to other healthcare provider groups.
Executive Summary.
Personalized medicine
Personalized medicine offers the promise of earlier diagnoses, targeted therapies, and individualized treatment plans.
Integration into practice
There is increasing evidence of pharmacogenomics’ clinical utility, but adoption by clinicians into practice has been slow.
In recent studies, healthcare providers have been found to be uncomfortable ordering and interpreting pharmacogenomics tests.
Translation requires education
Translation and implementation of pharmacogenomics at the patient bedside is the next logical step. Education for aiding clinicians to knowledgeably prescribe, dispense, and administer drugs with pharmacogenomic biomarkers in drug labeling is critical.
Educational challenges
Many healthcare providers were trained before the advent of genomic medicine and these practitioners did not have genomics as part of their educational curriculum.
Conversely, it is also a challenge for newly trained clinicians due to the rapidly emerging and changing aspects of their genomics training that may outdate by the time they begin their clinical practice.
Multidisciplinary institutional approach
Pharmacogenomics education has been developed by a multidisciplinary team at our institution. Education delivery into daily workflow at our institution has been accomplished through provision of pharmacogenomics education to prescribers at the time of prescribing medications.
Conclusion
Healthcare providers have indicated that they believe that they should be knowledgeable and be able to apply pharmacogenomics information.
Evidence and guidelines should be considered within a multidisciplinary framework (e.g., medicine, pharmacy, laboratory medicine) and tailored to the needs of the individual discipline.
While pharmacogenomics guidelines are of great assistance, multidisciplinary educational efforts at the level of local institutions will be required to achieve the full potential of pharmacogenomics.
Footnotes
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
•• of considerable interest
- 1.Green ED, Guyer MS. National Human Genome Research Institute. Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470(7333):204–213. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- 2.Wang L, McLeod HL, Weinshilboum RM. Genomics and drug response. N Engl J Med. 2011;364(12):1144–1153. doi: 10.1056/NEJMra1010600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Madadi P, Joly Y, Avard D, et al. Communicating pharmacogenetic research results to breastfeeding mothers taking codeine: a pilot study of perceptions and benefits. Clin Pharmacol Ther. 2010;88(6):792–795. doi: 10.1038/clpt.2010.125. [DOI] [PubMed] [Google Scholar]
- 4••.Haga SB, O’Daniel JM, Tindall GM, Lipkus IR, Agans R. Survey of U.S. public attitudes towards pharmacogenetic testing. Pharmacogenomics J. 2012;12(3):197–204. doi: 10.1038/tpj.2011.1. Study involving lay opinions of pharmacogenomic testing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lanie AD, Jayaratne TE, Sheldon JP, et al. Exploring the public understanding of basic genetic concepts. J Genet Couns. 2004;13(4):305–320. doi: 10.1023/b:jogc.0000035524.66944.6d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wetterstrand KA. DNA Sequencing Costs. Data from the NHGRI Genome Sequencing Program (GSP) 2014 www.genome.gov/sequencingcosts.
- 7.Bernhardt BA, Zayac C, Gordon ES, Wawak L, Pyeritz RE, Gollust SE. Incorporating direct-to-consumer genomic information into patient care: attitudes and experiences of primary care physicians. Per Med. 2012;9(7):683–692. doi: 10.2217/pme.12.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Squassina A, Manchia M, Manolopoulos VG, et al. Realities and expectations of pharmacogenomics and personalized medicine: impact of translating genetic knowledge into clinical practice. Pharmacogenomics. 2010;11(8):1149–1167. doi: 10.2217/pgs.10.97. [DOI] [PubMed] [Google Scholar]
- 9.Feero WG, Green ED. Genomics education for health care professionals in the 21st century. JAMA. 2011;306(9):989–990. doi: 10.1001/jama.2011.1245. [DOI] [PubMed] [Google Scholar]
- 10.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther. 2011;89(3):464–467. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11••.Farrugia G, Weinshilboum RM. Challenges in implementing genomic medicine: the Mayo Clinic Center for Individualized Medicine. Clin Pharmacol Ther. 2013;94(2):204–206. doi: 10.1038/clpt.2013.52. Addresses challenges of pharmacogenomics implementation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burton H, Cole T, Lucassen AM. Genomic medicine: challenges and opportunities for physicians. Clin Med. 2012;12(5):416–419. doi: 10.7861/clinmedicine.12-5-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crews KR, Hicks JK, Pui CH, Relling MV, Evans WE. Pharmacogenomics and individualized medicine: translating science into practice. Clin Pharmacol Ther. 2012;92(4):467–475. doi: 10.1038/clpt.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peterson JF, Bowton E, Field JR, et al. Electronic health record design and implementation for pharmacogenomics: a local perspective. Genet Med. 2013;15:833–841. doi: 10.1038/gim.2013.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bell GC, Crews KR, Wilkinson MR, et al. Development and use of active clinical decision support for preemptive pharmacogenomics. J Am Med Inform Assoc. 2014;21(e1):e93–e99. doi: 10.1136/amiajnl-2013-001993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Overby CL, Tarczy-Hornoch P, Kalet IJ, et al. Developing a prototype system for integrating pharmacogenomics findings into clinical practice. J Pers Med. 2012;2(4):241–256. doi: 10.3390/jpm2040241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weitzel KW, Elsey AR, Langaee TY, et al. Clinical pharmacogenetics implementation: approaches, successes, and challenges. Am J Med Genet C Semin Med Genet. 2014;166(1):56–67. doi: 10.1002/ajmg.c.31390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welch BM, Kawamoto K. Clinical decision support for genetically guided personalized medicine: a systematic review. J Am Med Inform Assoc. 2013;20(2):388–400. doi: 10.1136/amiajnl-2012-000892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19••.Schildcrout JS, Denny JC, Bowton E, et al. Optimizing drug outcomes through pharmacogenetics: a case for preemptive genotyping. Clin Pharmacol Ther. 2012;92(2):235–242. doi: 10.1038/clpt.2012.66. Early study of impact of implementing pharmacogenomics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffman JM, Haidar CE, Wilkinson MR, et al. PG4KDS: A model for the clinical implementation of pre-emptive pharmacogenetics. Am J Med Genet C Semin Med Genet. 2014;166(1):45–55. doi: 10.1002/ajmg.c.31391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Donnell PH, Danahey K, Jacobs M, et al. Adoption of a clinical pharmacogenomics implementation program during outpatient care–initial results of the University of Chicago “1,200 Patients Project”. Am J Med Genet C Semin Med Genet. 2014;166(1):68–75. doi: 10.1002/ajmg.c.31385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.US Food and Drug Administration (FDA) www.fda.gov.
- 23.Frueh FW, Amur S, Mummaneni P, et al. Pharmacogenomic biomarker information in drug labels approved by the United States food and drug administration: prevalence of related drug use. Pharmacotherapy. 2008;28(8):992–998. doi: 10.1592/phco.28.8.992. [DOI] [PubMed] [Google Scholar]
- 24.Rackover M, Goldgar C, Wolpert C, Healy K, Feiger J, Jenkins J. Establishing essential physician assistant clinical competencies guidelines for genetics and genomics. J Physician Assist Educ. 2007;18(2):47–48. [Google Scholar]
- 25.McCullough KB, Formea CM, Berg KD, et al. Assessment of the pharmacogenomics educational needs of pharmacists. Am J Pharm Educ. 2011;75(3):51. doi: 10.5688/ajpe75351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benzeroual KE, Bupendra S, Shinde S. Pharmacogenomics: assessing educational exposure, confidence in knowledge and training elements of pharmacists. Per Med. 2012;9(4):387–393. doi: 10.2217/pme.12.44. [DOI] [PubMed] [Google Scholar]
- 27.Sansgiry SS, Kulkarni AS. The human genome project: assessing confidence in knowledge and training requirements for community pharmacists. Am J Pharm Educ. 2003;67(2):39. [Google Scholar]
- 28.Roederer MW, Van Riper M, Valgus J, Knafl G, McLeod H. Knowledge, attitudes and education of pharmacists regarding pharmacogenetic testing. Per Med. 2012;9(1):19–27. doi: 10.2217/pme.11.87. [DOI] [PubMed] [Google Scholar]
- 29.Lesko LJ, Johnson JA. Academia at the crossroads: education and training in pharmacogenomics. Per Med. 2012;9(5):497–506. doi: 10.2217/pme.12.54. [DOI] [PubMed] [Google Scholar]
- 30.Dodson C. Knowledge and attitudes concerning pharmacogenomics among health care professionals. Per Med. 2011;8(4):421–428. doi: 10.2217/pme.11.28. [DOI] [PubMed] [Google Scholar]
- 31.Callier SL, Toma I, McCaffrey T, Harralson AF, O’Brien TJ. Engaging the next generation of healthcare professionals in genomics: planning for the future. Per Med. 2014;11(1):89–98. doi: 10.2217/pme.13.99. [DOI] [PubMed] [Google Scholar]
- 32.Carlberg C. The need for education in personalized medicine. Per Med. 2012;9(2):147–150. doi: 10.2217/pme.11.96. [DOI] [PubMed] [Google Scholar]
- 33.Frueh FW, Gurwitz D. From pharmacogenetics to personalized medicine: a vital need for educating health professionals and the community. Pharmacogenomics. 2004;5(5):571–579. doi: 10.1517/14622416.5.5.571. [DOI] [PubMed] [Google Scholar]
- 34.Hedgecoe A. Education, ethics and knowledge deficits in clinical pharmacogenetics. Pharmacogenomics. 2007;8(3):267–270. doi: 10.2217/14622416.8.3.267. [DOI] [PubMed] [Google Scholar]
- 35.Passamani E. Educational challenges in implementing genomic medicine. Clin Pharmacol Ther. 2013;94(2):192–195. doi: 10.1038/clpt.2013.38. [DOI] [PubMed] [Google Scholar]
- 36.Reeves S, Perrier L, Goldman J, Freeth D, Zwarenstein M. Interprofessional education: effects on professional practice and healthcare outcomes (update) Cochrane Database Syst Rev. 2013;28(3):CD002213. doi: 10.1002/14651858.CD002213.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bridges DR, Davidson RA, Odegard PS, Maki IV, Tomkowiak J. Interprofessional collaboration: three best practice models of interprofessional education. Med Educ Online. 2011;16 doi: 10.3402/meo.v16i0.6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Interprofessional Education Collaborative Expert Panel. Core competencies for interprofessional collaborative practice: report of an expert panel. Interprofessional Education Collaborative; Washington, DC, USA: 2011. [Google Scholar]
- 39.Martinez IL, Pfeifle AL, Ballard JA. Framing competency-based assessment for interprofessional education. Med Sci Educ. 2013;23(3S):562–565. [Google Scholar]
- 40.Hammick M, Freeth D, Koppel I, Reeves S, Barr H. A best evidence systematic review of interprofessional education: BEME Guide no. 9. Med Teach. 2007;29(8):735–751. doi: 10.1080/01421590701682576. [DOI] [PubMed] [Google Scholar]
- 41.WHO study group on Interprofessional Education and Collaborative Practice. Baker PG. Framework for Action on Interprofessional Education and Collaborative Practice. Vol. 64. World Health Organisation Press; Geneva, Switzerland: 2010. [Google Scholar]
- 42.Institute of Medicine. Interprofessional education for collaboration: learning how to improve health from interprofessional models across the continuum of education to practice: workshop summary. The National Academies Press; Washington, DC, USA: 2013. [PubMed] [Google Scholar]
- 43.Gunaldo TP, Mangum SW. Providing interprofessional continuing education promotes continuing competence in healthcare providers. Med Sci Educ. 2013;23(3S):559–561. [Google Scholar]
- 44.Haga SB, Burke W, Ginsburg GS, Mills R, Agans R. Primary care physicians’ knowledge of and experience with pharmacogenomic testing. Clin Genet. 2012;82(4):388–394. doi: 10.1111/j.1399-0004.2012.01908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haga SB, O’Daniel JM, Tindall GM, Mills R, Lipkus IM, Agans R. Survey of genetic counselors and clinical geneticists’ use and attitudes towards pharmacogenetic testing. Clin Genet. 2012;82(2):115–120. doi: 10.1111/j.1399-0004.2012.01848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46••.Stanek EJ, Sanders CL, JohansenTaber KA, et al. Adoption of pharmacogenomic testing by US physicians: results of a nationwide survey. Clin Pharmacol Ther. 2012;91(3):450–458. doi: 10.1038/clpt.2011.306. Study of US physicians’ attitudes and beliefs. [DOI] [PubMed] [Google Scholar]
- 47.Ormond KE. From genetic counseling to “genomic counseling”. Mol Genet Genomic Med. 2013;1(4):189–193. doi: 10.1002/mgg3.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Newman WG, Murphy BF, Callard A, Payne K. A role for genetic counsellors and clinical geneticists in pharmacogenetics? Clin Genet. 2012;82(2):201–202. doi: 10.1111/j.1399-0004.2012.01872.x. [DOI] [PubMed] [Google Scholar]
- 49.Secretary’s Advisory Committee on Genetics, Health, and Society. Genetics education and training: report of the Secretary’s Advisory Committee on Genetics, Health, and Society. 2011 http://oba.od.nih.gov.
- 50.National Human Genome Research Institute. Inter-Society Coordinating Committee for Practitioner Education in Genomics (ISCC-PEG): proposed charge, goals, activities, and metrics. 2013 www.genome.gov.
- 51.Korf BR, Berry AB, Limson M, et al. Framework for development of physician competencies in genomic medicine: report of the Competencies Working Group of the Inter-Society Coordinating Committee for Physician Education in Genomics. Genet Med. 2014 doi: 10.1038/gim.2014.35. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 52.Consensus Panel on Genetic/Genomic Nursing Competencies. Essentials of Genetic and Genomic Nursing: Competencies, Curricula Guidelines, and Outcome Indicator. 2nd. American Nurses Association; MD, USA: 2009. [Google Scholar]
- 53.McInerney JD, Edelman E, Nissen T, Reed K, Scott JA. Preparing health professionals for individualized medicine. Per Med. 2012;9(5):529–537. doi: 10.2217/pme.12.46. [DOI] [PubMed] [Google Scholar]
- 54.Mills R, Haga SB. Clinical delivery of pharmacogenetic testing services: a proposed partnership between genetic counselors and pharmacists. Pharmacogenomics. 2013;14(8):957–968. doi: 10.2217/pgs.13.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pestka EL, Brown JK. Genomics education for nurses in practice. J Nurses Staff Dev. 2004;20(3):145–149. doi: 10.1097/00124645-200405000-00008. [DOI] [PubMed] [Google Scholar]
- 56.Formea CM, Nicholson WT, McCullough KB, et al. Development and evaluation of a pharmacogenomics educational program for pharmacists. Am J Pharm Educ. 2013;77(1):10. doi: 10.5688/ajpe77110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Young A, Chaudhry HJ, Thomas JV, Dugan M. A census of actively licensed physicians in the United States, 2012. J Med Reg. 2013;99(2):11–24. [Google Scholar]
- 58.Demmer LA, Waggoner DJ. Professional medical education and genomics. Annu Rev Genomics Hum Genet. 2014;1510(3):1–3. doi: 10.1146/annurev-genom-090413-025522. [DOI] [PubMed] [Google Scholar]
- 59.Surh LC, Pacanowski MA, Haga SB, et al. Learning from product labels and label changes: how to build pharmacogenomics into drug-development programs. Pharmacogenomics. 2010;11(12):1637–1647. doi: 10.2217/pgs.10.138. [DOI] [PubMed] [Google Scholar]
- 60.United States Food and Drug Administration. Table of pharmacogenomic biomarkers in drug labeling. www.fda.gov.
- 61.Roederer MW. NAVAGATE: a rubric to move from pharmacogenomics science to pharmacogenomics practice. Pharmacogenomics. 2012;13(11):1307–1313. doi: 10.2217/pgs.12.110. [DOI] [PubMed] [Google Scholar]
- 62.Mills R, Voora D, Peyser B, Haga SB. Delivering pharmacogenetic testing in a primary care setting. Pharmgenomics Pers Med. 2013;6:105–112. doi: 10.2147/PGPM.S50598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63••.Manolio TA, Chisholm RX, Ozenberger B, et al. Implementing genomic medicine in the clinic: the future is here. Genet Med. 2013;15(4):258–267. doi: 10.1038/gim.2012.157. Addresses challenges to implementation of genomic medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Center for Individualized Medicine. Pharmacogenomics program. Mayo Clinic; http://mayoresearch.mayo.edu. [Google Scholar]
- 65.Bielinski SJ, Olson JE, Pathak J, et al. Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time-using genomic data to individualize treatment protocol. Mayo Clin Proc. 2014;89(1):25–33. doi: 10.1016/j.mayocp.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lazaridis KN, McAllister TM, Babovic-Vuksanovic D, et al. Implementing individualized medicine into the medical practice. Am J Genet C Semin Med Genet. 2014;166(1):15–23. doi: 10.1002/ajmg.c.31387. [DOI] [PubMed] [Google Scholar]
- 67.Cook DA, Sorensen KJ, Hersh W, Berger RA, Wilkinson JM. Features of effective medical knowledge resources to support point of care learning: a focus group study. PLoS ONE. 2013;8(11):e80318. doi: 10.1371/journal.pone.0080318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cook DA, Sorensen KJ, Nishimura RA, Ommen SR, Lloyd FJ. A comprehensive system to support physician learning at the point of care. Acad Med. 2015;90(1):33–39. doi: 10.1097/ACM.0000000000000551. [DOI] [PubMed] [Google Scholar]
- 69.Formea CM, Nicholson WT, McCullough KB, et al. Development, pilot, and quality assessment of a pharmacogenomics education program for pharmacists. Presented at: American College of Clinical Pharmacists 2014 Annual Meeting; Austin, TX, USA. 12–15 October 2014. [Google Scholar]
- 70.Goetz MP, Rae JM, Suman VJ, et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol. 2005;23(36):9312–9318. doi: 10.1200/JCO.2005.03.3266. [DOI] [PubMed] [Google Scholar]
- 71.Scott SA. Clinical pharmacogenomics: opportunities and challenges at point-of-care. Clin Pharmacol Ther. 2013;93(1):33–35. doi: 10.1038/clpt.2012.196. [DOI] [PMC free article] [PubMed] [Google Scholar]


