Abstract
The molecular basis of the pathophysiological role of oxidative stress in autism is understudied. Herein, we used polymerase chain reaction (PCR) array to analyze transcriptional pattern of 84 oxidative stress genes in peripheral blood mononuclear cell pools isolated from 32 autistic patients (16 mild/moderate and 16 severe) and 16 healthy subjects (each sample is a pool from 4 autistic patients or 4 controls). The PCR array data were further validated by quantitative real-time PCR in 80 autistic children (55 mild/moderate and 25 severe) and 60 healthy subjects. Our data revealed downregulation in GCLM, SOD2, NCF2, PRNP, and PTGS2 transcripts (1.5, 3.8, 1.2, 1.7, and 2.2, respectively;P < .05 for all) in autistic group compared with controls. In addition, TXN and FTH1 exhibited 1.4- and 1.7-fold downregulation, respectively, in severe autistic patients when compared with mild/moderate group (P = .005 and .0008, respectively). This study helps in a better understanding of the underlying biology and related genetic factors of autism, and most importantly, it presents suggested candidate biomarkers for diagnosis and prognosis purposes as well as targets for therapeutic intervention.
Keywords: Autism, gene expression, oxidative stress, neurodevelopmental disorders, PCR array, ROS
Introduction
Autism is a set of childhood neurodevelopmental disorders that affect young children (<3 years old) with severe manifestations, such as impaired social skills, abnormal verbal and nonverbal communication, and restricted-repetitive behaviors.1 The fifth edition of Diagnostic and Statistical Manual of Mental Disorders (DSM-V) considered the autistic disorder, pervasive developmental disorders not otherwise stated, and the Asperger syndrome as a single disorder with variable severity in the communication impairment and/or the repetitive behaviors domain.2 Autism prevalence is on rise over the recent few years (current global incidence rate is 0.15% versus 0.03% prior to 1990).3 Several causes contribute to the development of autism, including immunological (immune dysregulation),4 neurological (neuroinflammation and altered neurotransmission),5 and environmental factors,6 in addition to the genetic background of the host.7 Gene variants associated with autism are myriad and regional specific8; they can even be perpetuated by some factors such as consanguineous marriage which is highly prevalent in Egypt.9 For instance, a study found a specific BCKDK gene variant mutation among autistic offspring in some consanguineous families where certain amino acids may relieve symptoms of this rare form of autism.10 Oxidative stress is the underlying mechanism that links different causes and is the main determinant of autism development and progression.11–13
Oxidative stress refers to a pathologic state arisen from the imbalance between the cellular reactive oxygen species (ROS) and the ability of the cell to detoxify them with the resulting severe damage of all macromolecules (protein, lipid, and DNA) and disruption in several signaling pathways.14 Endogenous and dietary antioxidants combat oxidative stress through controlling the levels of ROS produced, scavenging excess ROS, and repairing the oxidative damaged biomolecules.15 The brain is highly exposed to increased oxidative stress due to the presence of excitatory amino acids whose catabolism ends with the production of ROS causing neuronal damage.14 Thus, increased oxidative stress is a primary risk factor for the pathophysiology of many neuropsychiatric disorders such as the Parkinson disease, the Alzheimer disease, the Huntington disease, and multiple sclerosis.16,17 Numerous oxidative stress markers (antioxidant enzymes, lipid peroxidation, and protein/DNA oxidation) were detected in abnormal levels in autistic children, such as protein dityrosine.18,19 Several studies have documented reduced levels of glutathione, glutathione peroxidase, methionine, and cysteine besides elevated levels of oxidized glutathione in children with autism.12,20 The excreted antioxidants are lower in autistic patients compared with healthy age-matching subjects and upon correlation with severity of the disease, such as superoxide dismutase.21,22 Also, ceruloplasmin and transferrin antioxidants show suboptimal levels in serum of autistic children, leading to abnormal metabolism of toxic and oxidative stress–mediating metal ions.23 Recent genetic studies have identified variants of some antioxidant enzyme-coding genes that increase the susceptibility to autism. For example, the interaction between glutathione S-transferase P1 and glutathione S-transferase M1 mutated genes contributed to autism risk.24
Autism is complicated by the absence of both specific medical diagnostic tests and definitive drug therapy. Its diagnosis is based mainly on the presence of abnormal behaviors associated with the disease,5 and the current therapeutic approaches aim to ameliorate those behavioral deficits.25 Although autism is associated with a high degree of heritability, there is a big research gap in studying the autism-driving genes. Identifying those panels of genes will help in developing reliable biomarkers for early diagnostic and therapeutic purposes. Transcriptome analysis identified new messenger RNA (mRNA) putative markers; for instance, a study developed a panel of 66 genes involved in neurological processes that showed significant dysregulation in autism.26 Similarly, proteome analysis revealed some autism-associated biomarkers, such as urinary kininogen 1.27
However, the transcriptional profile of most antioxidant enzyme families remains understudied in autism. In this study, we sought to uncover the molecular basis of the pathophysiologic role of oxidative stress in autism. We investigated the mRNA expression of several oxidative stress–related transcripts, noninvasively, in peripheral blood mononuclear cells (PBMCs) derived from children having autism with varying degrees using pathway-focused polymerase chain reaction (PCR) array. Transcriptional profile of several genes was altered in autistic patients compared with healthy controls. We then validated the altered mRNA abundance of 8 key signaling molecules in a larger number of patients by quantitative real-time polymerase chain reaction (qRT-PCR).
Materials and Methods
Ethical statement
All experiments were approved by the institution ethical review board (medical research ethics committee at National Research Centre, Cairo, Egypt) according to Helsinki Declaration 1975 revised in 2008. Written informed consent was obtained from the caregiver of each child before collecting blood samples. All studied cases (55 mild/moderate and 25 severe autistic patients in addition to 60 controls) were recruited from Autism Disorders Clinic, Medical Research Center of Excellence, and National Research Centre. Exclusion criteria included all subjects with other causes of mental subnormality, delayed language, and all patients receiving antioxidants. The typically developing (TD) controls included 60 age-matched children (3-6 years) sampled from 1 common kindergarten with no abnormal histories of motor, language, or social developmental disorders, as determined according to reports of parents and teachers. The 80 autistic cases included 60 boys (75%) and 20 girls (25%) along with frequency-matched controls. In addition, 22 autistic children (27.5%) had consanguineous parents. All patients were subjected to the following. (1) Detailed history taking including 3-generation pedigree analysis, pregnancy history, perinatal history, developmental history, similarly affected family members, and age of onset of presenting manifestation. (2) Clinical diagnosis was based on the criteria for autistic disorder as defined in the DSM-V2 and Autism Diagnostic Interview–Revised.28 The Childhood Autism Rating Scale is used for assessing autism severity,29 and it was completed by the caregivers of children with autism. Total scores were calculated for each child with autism (Tables 1 and 2). Criteria of selection for study were defined as Comprehensive Diagnostic Evaluation. This included looking at the child’s behavior and development and parents’ interview. It also included a hearing screening and neurological examination.
Table 1.
Classification of the autistic children according to the Childhood Autism Rating Scale score.
| Degree of autism | No. of cases | % |
|---|---|---|
| Mild to moderate | 55 | 68.75 |
| Severe cases | 25 | 31.25 |
Table 2.
Comparison between the ADI-R domains according to the severity of autism among the autistic cases.
| Degree of CARS |
P value | ||
|---|---|---|---|
| Mean ± SD |
|||
| Mild to moderate | Severe | ||
| Social domain | 19.5 ± 4.7 | 21.8 ± 2.2 | .05 |
| Verbal domain | 11.2 ± 3 | 9.3 ± 4.5 | .4 |
| Nonverbal domain | 8.5 ± 2.4 | 8.5 ± 3.8 | .6 |
| Repetitive domain | 5.8 ± 2.7 | 4.8 ± 1.7 | .1 |
Abbreviations: ADI-R, Autism Diagnostic Interview–Revised; CARS, Childhood Autism Rating Scale.
RNA extraction
RNA isolation was performed on freshly collected blood samples (3 mL) following the steps of the single-step method,30 in which we lysed and discarded red blood cells and then mixed the formed pellets with fixed ratios of guanidinium thiocyanate, sodium citrate, 2-mercaptoethanol, chloroform isoamyl alcohol, phenol, and sodium acetate; then, we precipitated, washed, and dissolved the RNA using isopropanol, 70% ethanol, and nuclease-free water, respectively. This was followed by quantification of the recovered RNA using a Thermo Scientific NanoDrop spectrophotometer.
PCR array
Eight hundred nanograms of RNA was reverse transcribed into complementary DNA (cDNA) using the RT2 PCR Array First Strand Kit (SABiosciences, Valencia, CA, USA); 102 µL of the total 111 µL synthesized cDNA was mixed with 1150 µL RT2 SYBR Green/ROX qPCR master mix (SABiosciences), and nuclease-free water was added to adjust the final volume to 2300 µL; 20 µL of the PCR mix was added to each well of the 100-well rounded plate. The PCR array used in this study was the human oxidative stress RT2 Profiler PCR Array (SABiosciences). The thermal profile was as follows: initial incubation for 10 minutes at 95°C (AmpliTaq Gold pre-activation) and then at 95°C for 15 seconds and 60°C for 1 minute repeated for 40 cycles. The PCR run was performed on Rotor Gene Real-Time PCR System (Qiagen, Santa Clarita, CA, USA).
Similar threshold was set for all qRT-PCR runs, and the cycle threshold (CT) values were exported to a blank data analysis excel sheet to proceed with statistical analysis. The RPLP0 housekeeping gene (HKG) was selected for normalization of data because it showed the smallest CT value across different samples. The statistical analysis of PCR array data was performed using the SABiosciences web-based software: (http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php).
We entered the CT data and the software automatically quantified the mRNA abundance using 2(−ΔΔCT) method in which the CT of the HKG is subtracted from each gene of interest to calculate ΔCT; then the mean ΔCT of all samples in control group was calculated and subtracted from ΔCT of each experimental sample (mild or severe autism) to calculate the ΔΔCT. The fold change is calculated as 2(−ΔΔCT).
Real-time polymerase chain reaction
Two hundred nanograms of RNA was used in the reverse transcription step. A master mix (25 µL) containing 1 µL of the synthesized cDNA, 1 µL of gene-specific PCR primer for human GCLM, SOD2, TXN, NCF2, PRNP, PTGS2, GPX7, and FTH1 (10 µM) (SABiosciences), 12.5 µL of RT2 SYBR Green/ROX qPCR master mix (SABiosciences), and 10.5 µL of H2O was incubated for 10 minutes at 95°C followed by 40 cycles at 95°C for 15 seconds and 60°C for 1 minute in rotor gene Real-Time PCR System (Qiagen). The same threshold was set for all the individual primer-based real-time polymerase chain reaction (RT-PCR) runs, the HKG human RPLP0 (SABiosciences) was used for normalization, and the expression of each individual gene was calculated by the 2(−ΔΔCT) method and expressed as fold change over the mean of control group as described above. This was followed by statistical comparison.
Statistical analysis
The statistical comparison of the data was performed using Prism software version 5. The data were analyzed by the parametric unpaired t test or the nonparametric Mann-Whitney U test according to the normal distribution curve. Data were presented as the mean and standard error of the mean unless mentioned. Statistically significant differences between the groups were considered if P ⩽ .05.
Results
Transcriptional profiling of oxidative stress–related genes in PBMCs of autistic children
An initial explorative experiment to identify the most regulated ROS genes was performed using pathway-focused PCR array, including 84 oxidative stress–related genes. The relative gene expression of the 84 transcripts was measured in PBMC RNAs from 16 mild autistic children, 16 severe autistic children, and 16 age-matched healthy children (RNAs from each 4 synonymous subjects were pooled in 1 sample to perform the PCR array experiment; these 32 cases and 16 controls are subsets of the total 80 autistic and 60 normal children enrolled in the study). The selected array had primers for transcripts encoding peroxidases, peroxiredoxins, antioxidants, oxidative stress–responsive proteins, and oxygen transporters in addition to genes involved in superoxide and ROS metabolism (Table 3). Only 13% of the arrayed genes (n = 11) showed altered expression in mild/moderate or severe autistic children when compared with the control group (greater or less than 1.5-fold differential regulation associated with statistical cutoff at P ⩽ .05; Table 3). Three oxidative stress responsive genes (CCL5, DHCR24, and GCLC) besides glutathione peroxidase (GPX7) showed dysregulation in mild autistic group compared with controls. CCL5 and GCLC were downregulated (1.7- and 1.8-fold downregulation, respectively; Table 4), whereas DHCR24 and GPX7 were upregulated (2.5- and 1.5-fold upregulation, respectively; Table 4). We observed a trend of downregulation in most of the differentially regulated transcripts of severe autistic group when compared with controls (GCLM, SOD2, TXN, NCF2, PRNP, PTGS2, and FTH1; the mean fold regulation ranges from −1.5 to −4.3; Table 4) and upregulation of a single transcript (GPX7; 1.6-fold regulation; Table 4). However, none of the 84 genes showed more than 1.5-fold regulation plus statistical significance at P ⩽ .05 between mild and severe autism. Together, these results suggested that, regardless of the severity of the disease, autism amended the antioxidative state in PBMCs.
Table 3.
Differential gene expression of oxidative stress genes by RT-PCR array.
| Gene ID | Fold regulation (mild and moderate)a | Fold regulation (severe)a | Gene ID | Fold regulation (mild and moderate)a | Fold regulation (severe)a |
|---|---|---|---|---|---|
| Glutathione peroxidases | BNIP3 | 1.0619 | 1.0473 | ||
| GPX1 | −1.2687 | −1.0497 | MPV17 | −1.0668 | −1.0668 |
| GPX2 | −1.1355 | −1.8618 | Oxidative stress–responsive genes | ||
| GPX3 | −1.162 | −1.5619 | TXN | −1.47 | −2.1735 |
| GPX4 | −1.4224 | −1.0817 | KRT1 | −1.6424 | −1.4373 |
| GPX5 | 1.0041 | 1.3379 | LPO | −2.2076 | −2.2605 |
| GPX6 | 1.633 | −1.1674 | TPO | −1.0122 | −2.7895 |
| GPX7 | 1.4515 | 1.5511 | MPO | −2.2423 | −1.3947 |
| GSTP1 | −1.2709 | −1.0093 | GCLM | −1.5324 | −1.651 |
| GSTZ1 | −1.3021 | −1.2255 | MSRA | −1.4658 | −1.632 |
| Peroxiredoxins | CAT | −1.5692 | −1.4012 | ||
| PRDX1 | 1.1467 | −2.555 | CCL5 | −1.7112 | −1.5087 |
| PRDX2 | 1.0749 | 1.2924 | DUSP1 | −1.5404 | −1.8361 |
| PRDX3 | −1.2491 | −1.8965 | HMOX1 | 1.0151 | −1.0595 |
| PRDX4 | 1.3195 | −1.2628 | TXNRD2 | 1.3558 | −2.7959 |
| PRDX5 | −1.2262 | −1.217 | ATOX1 | 1.0347 | 1.014 |
| PRDX6 | −1.4389 | −1.3629 | HSPA1A | 1.1076 | −1.8877 |
| Other peroxidases and antioxidants | TXNRD1 | −1.2333 | −2.2089 | ||
| CYBB | −1.4273 | −1.5227 | DHCR24 | 2.4566 | 1.1355 |
| MGST3 | −1.3457 | −1.2658 | NQO1 | 1.065 | −1.4743 |
| PTGS1 | −1.1534 | −1.162 | NUDT1 | −1.1348 | 1.007 |
| PTGS2 | −2.0837 | −4.2673 | TTN | 1.127 | −1.1728 |
| PXDN | 11.0171 | 5.5277 | GCLC | −1.7994 | −2.2191 |
| ALB | 2.9862 | 3.6723 | GSS | −1.0187 | 1.2058 |
| Superoxide dismutases | APOE | 2.3349 | 2.2089 | ||
| SOD1 | −1.276 | −1.1121 | OXR1 | −1.1218 | −2.0046 |
| SOD2 | −1.3964 | −2.4852 | OXSR1 | −1.3605 | −1.6702 |
| SOD3 | −2.7023 | −1.6663 | FOXM1 | 2.0777 | 2.3403 |
| Other genes involved in superoxide metabolism | PDLIM1 | 1.2226 | 1.0968 | ||
| NCF1 | −1.5819 | −2.1287 | PNKP | −1.0246 | −2.0753 |
| NCF2 | −1.8758 | −1.8704 | EPX | 1.2072 | −1.2198 |
| NOS2 | 1.0975 | −2.9214 | PRNP | −1.7151 | −1.9816 |
| NOX4 | 4.542 | 1.3379 | FTH1 | −1.5538 | −1.454 |
| NOX5 | 1.7573 | 1.3379 | RNF7 | −1.2782 | −1.3348 |
| PREX1 | −1.4931 | −1.6702 | SCARA3 | −1.8628 | −3.539 |
| UCP2 | 1.2433 | 1.1892 | VIMP | −1.1264 | −1.5192 |
| DUOX1 | 1.7901 | −1.1303 | SEPP1 | −1.2376 | −1.6058 |
| DUOX2 | 1.0662 | −1.2658 | GSR | −1.0413 | −1.4473 |
| MT3 | 1.6606 | 1.9274 | SIRT2 | −1.0081 | −1.3787 |
| CCS | −7.8173 | 1.146 | SQSTM1 | −1.5755 | −1.6857 |
| ALOX12 | 1.2205 | 1.2775 | SRXN1 | −1.6663 | −3.0596 |
| GTF2I | −1.5764 | −1.6283 | STK25 | −1.0041 | −1.2058 |
| Other genes involved in ROS metabolism | MBL2 | −1.2534 | −1.2255 | ||
| AOX1 | −2.6558 | −1.4175 | Oxygen transporters | ||
| SFTPD | −1.5502 | −1.7818 | CYGB | −1.6682 | −3.8906 |
| EPHX2 | 1.0099 | −1.0595 | MB | 2.5257 | 1.1355 |
Abbreviations: PBMCs: peripheral blood mononuclear cells; ROS, reactive oxygen species; RT-PCR, real-time polymerase chain reaction.
A negative sign indicates reduced expression in the autistic patients relative to the normal ones.
Fold regulation was calculated as the ratio of expression of each gene measured in RNA isolated from PBMCs of severe and mild/moderate autistic children relative to nonautistic healthy children (n = 4 per group, and each RNA sample is a pool of 4 samples).
Table 4.
Differential regulation of oxidative stress–related genes exhibiting statistically significant differences in autistic patients compared with healthy controls.
| Gene symbol | Gene description | Unigene | Refseq | Differential expression mild and moderate/C | P value |
|---|---|---|---|---|---|
| CCL5 | Chemokine (C-C motif) ligand 5 | Hs.514821 | NM_002985 | −1.7112 | .02 |
| GCLC | Glutamate-cysteine ligase, catalytic subunit | Hs.654465 | NM_001498 | −1.7994 | .03 |
| DHCR24 | 24-dehydrocholesterol reductase | Hs.498727 | NM_014762 | 2.4566 | .04 |
| GPX7 | Glutathione peroxidase 7 | Hs.43728 | NM_015696 | 1.4515 | .05 |
| Gene symbol | Gene description | Unigene | Refseq | Differential expression severe/C | P value |
| GCLM | Glutamate-cysteine ligase, modifier subunit | Hs.315562 | NM_002061 | −1.651 | .01 |
| GPX7 | Glutathione peroxidase 7 | Hs.43728 | NM_015696 | 1.5511 | .01 |
| SOD2 | Superoxide dismutase 2, mitochondrial | Hs.487046 | NM_000636 | −2.4852 | .01 |
| TXN | Thioredoxin | Hs.435136 | NM_003329 | −2.1735 | .03 |
| NCF2 | Neutrophil cytosolic factor 2 | Hs.587558 | NM_000433 | −1.8704 | .04 |
| PRNP | Prion protein | Hs.610285 | NM_183079 | −1.9816 | .04 |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | Hs.196384 | NM_000963 | −4.2673 | .05 |
| FTH1 | Ferritin, heavy polypeptide 1 | Hs.712676 | NM_002032 | −1.454 | .05 |
Differential expression of the most regulated oxidative stress–related genes (fold regulation ⩾ 1.5 [upregulation/downregulation] with P ≤ .05) in mild/moderate (Table 4) or severe (Table 4) autistic patients compared with healthy controls (C). A negative sign refers to downregulation in patients relative to the healthy ones. Genes are arranged in a descending order according to their significant P value.
Quantitative RT-PCR of dysregulated ROS transcripts confirms the data of explorative PCR array
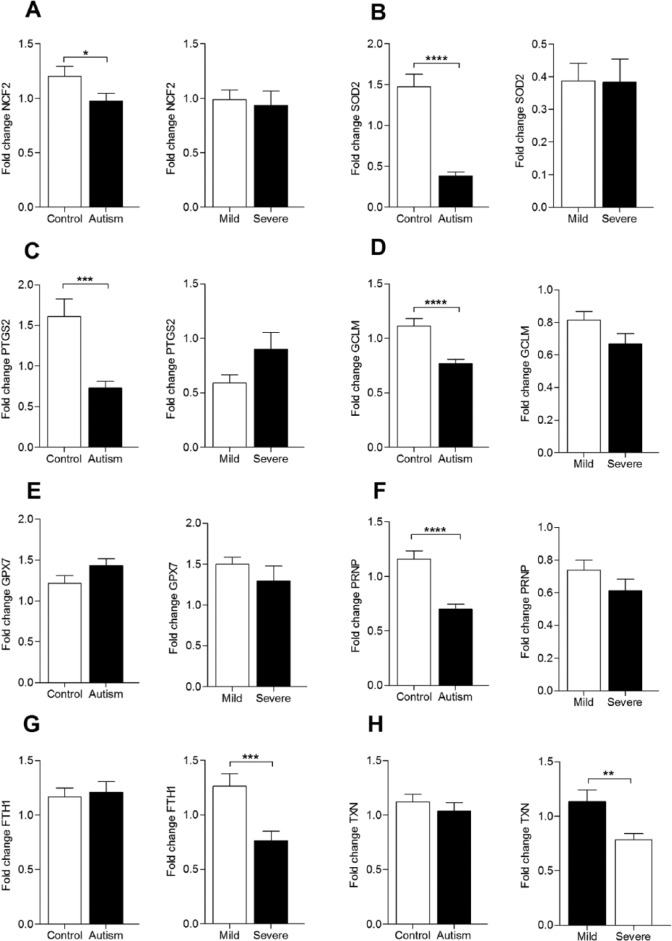
We further validated the PCR array data on a larger cohort size (60 healthy subjects and 80 autistic patients [55 mild cases and 25 severe cases]) using gene-specific qRT-PCR assay. We selected the 8 genes that showed differential regulation in severe autistic patients compared with controls (GCLM, SOD2, TXN, NCF2, PRNP, PTGS2, GPX7, and FTH1) for the validation experiment. First, we compared the transcriptional profile of the above 8 genes between autistic patients and controls. The data presented in Figure 1 showed downregulation in 5 transcripts (GCLM, SOD2, NCF2, PRNP, and PTGS2 [1.5, 3.8, 1.2, 1.7, and 2.2, respectively; P < .05 for all]) and similar regulation of 3 transcripts (FTH1, TXN, and GPX7) in autistic patients relative to healthy counterparts. Second, to test for a biomarker for severe autistic presentation, we compared the transcriptional profile of the 8 genes between mild/moderate and severe autistic patients. We noticed a dramatic downregulation of FTH1 (1.7-fold, P = .0008; Figure 1) and TXN (1.4-fold, P = .005; Figure 1) in severe autistic patients. However, mRNA abundance of the other transcripts was comparable between the 2 autistic groups.
Figure 1.
The expression pattern of 8 oxidative stress–related transcripts in autism patients and healthy controls. Quantitative real-time polymerase chain reaction was performed to measure the messenger RNA expression of (A) NCF2, (B) SOD2, (C) PTGS2, (D) GCLM, (E) GPX7, (F) PRNP, (G) FTH1, and (H) TXN in peripheral blood mononuclear cells of healthy controls (n = 60) and patients with mild/moderate autism (n = 55) and with severe autism(n = 25). The housekeeping gene RPLP0 was used for data normalization. Data are presented as fold change relative to the average of controls (*P < .05; **P < .01; ***P < .001; ****P < .0001).
Discussion
Oxidative stress is implicated in the pathophysiology of many neurological disorders. However, little is known about the linkage between oxidative stress and the occurrence or the progress of autism. The present data showed that autistic patients exhibited transcriptional dysregulation of 5 oxidative stress–regulating genes (GCLM, SOD2, NCF2, PRNP, and PTGS2) when compared with healthy subjects. Moreover, FTH1 and TXN genes showed altered regulation in different stages of autism; their mRNA abundance was less in severe autistic cases than in mild ones.
Studying the transcriptional profile of brain is limited by the highly invasive brain tissues and the poor quality of RNA recovered from neurological tissues. Therefore, there is a great demand to use alternate noninvasive tissue such as peripheral blood to characterize the neural transcriptome.31 The transcriptional profile of PBMCs has been widely studied in several neurological disorders, such as neurofibromatosis type I, tuberous sclerosis type II, and Down syndrome.32,33 X-box–binding protein 1 was initially recognized as a genetic risk factor for bipolar disorder based on some gene expression studies performed on lymphoblastoid cell lines derived from 2 discordant twins.34 In autism, studies performed on cell lines derived from autistic children showed that monozygotic twins with autism have differential gene expression in correlation with the disease severity and language impairment.35 The role of peripheral blood cells in understanding transcriptional dysregulation in autism was further confirmed in TD children who showed altered expression of dopamine- and serotonin-related genes in lymphoblastic cells.36
We focused on investigating the antioxidative milieu in autistic children. Brain tissues are rich in redox-active metals and fatty unsaturated lipids that may serve as substrates for lipid peroxidation37; in addition, they show high metabolic rate and oxygen consumption through the mitochondrial respiratory chain38; moreover, they have deficit in antioxidative stress with the consequent altered gene expression and cell apoptosis.13 In this study, the elaboration of redox strategies in peripheral cells may have significant relevance in this neuropsychiatric disease.
GPX7 is a member of glutathione peroxidase family that can neutralize H2O2 in the absence of glutathione.39 Several studies showed reduced blood levels of glutathione peroxidase in autistic children,40 but, however, some studies reported augmented activity level in autism.41 Our initial explorative experiment showed significant upregulation of GPX7 gene expression in both mild and severe autistic cases relative to TD controls, which may represent an adaptive mechanism to a probable glutathione deficiency that may ensue from GCLM gene downregulation. Although experimental42 and clinical43 studies suggested a protective role of GPX7 against oxidative stress, the data of the validation experiment showed similar GPX7 expression in the recruited cohort of autistic children and controls—a finding that requires further confirmation on larger number of cases. Generally, the discrepancies in results obtained from different studies may be attributed to the highly heterogeneous cause of autism. Such heterogeneity is most evident in SOD activities in autism.
The blood level of SOD in autistic patients did not reach consensus. Some studies showed increased SOD activities in erythrocytes,41,44 whereas other studies reported unchanged activities of erythrocyte SOD.45 The currently observed downregulation of SOD2 transcript in PBMCs of autistic children agrees with the previous literatures that reported reduced levels of SOD2 protein in the temporal lobe of brain46 and plasma45 as well as reduced SOD activity in erythrocytes of autistic children.47 Unlike SOD1 (CuZnSOD) which is mainly located in cytoplasm, SOD2 is the mitochondrial isoform of SOD which uses manganese to scavenge the superoxide radicals produced by mitochondria—the main source of cellular ROS. The downregulation of SOD2 may explain the mitochondrial dysfunction implicated in autism.48
Ferritin is the main iron-storing protein that protects the cell from the toxic effect of iron and from the free radicals leading to oxidative stress.49 Ferritin consists of multisubunits of heavy and light chains. FTH1 gene encodes the heavy chain of ferritin. Because variation in the composition of ferritin subunit highly affects the rates of iron storage and release, our finding of the low mRNA expression of FTH1 in severe autistic patients may present a possible explanation for the reduced levels of serum ferritin and iron in autistic children that was reported in the literature.50–52 FTH1 also delivers iron to many organs including brain,53 so the reported FTH1 downregulation may imply iron deficiency in brains of the studied cases. Because iron is essential for brain development,54 the current finding suggests a possible role of FTH1 gene downregulation in pathophysiology of autism. On clinical basis, severe autistic children in our study experienced a range of sleep fragmentation, where the exact mechanism could be due to low serum ferritin. This agreed with a previous study by Youssef et al.55
PRNP gene codes for the cellular isoform of the prion protein (PrPC). PrPC is expressed by neurons along with other cells where it plays a prominent antioxidant role.56 Experimental animal studies revealed that PrPC knockout mice are highly vulnerable to the toxic effect of oxidative stress agents.57 So, the reported downregulation of PRNP gene may explain the defective antioxidant status in autism. Because the prion protein plays vital roles in central nervous system development, neuroprotection, and synaptic plasticity,58,59 we suggest that the current PRNP gene downregulation may underlie the incidence or progression of autism. However, reports describing the regulation of PrPC in autism are scarce or even absent, and our study may be the first to depict the downregulation of PrPC-encoding gene (PRNP) in autism.
Similarly, our study is the first to report the dysregulation in NCF2 gene transcription. NCF2 gene encodes neutrophil cytosolic factor 2, a subunit of nicotinamide adenine dinucleotide phosphate (NADPH) oxidase—the enzyme that produces a burst of superoxide in neutrophil phagosome using NADPH and oxygen. Autistic children exhibited mitochondrial dysfunction associated with low oxidative burst and reduced NADH oxidase level60 and enzymatic activity.61 The present findings confirm the reduced NCF2 expression in autism, thus highlighting the mitochondrial dysfunction in this neuropsychiatric disease. The combined downregulation of SOD2 and NCF2 transcripts may cause mitochondrial dysfunction in our cohort of autistic children.
GCLM gene encodes the modulatory subunit of glutamate-cysteine ligase (GCL)—the enzyme that catalyzes the first rate-limiting step of glutathione synthesis.62 The present finding displays a downregulation of GCLM transcript in the autistic children and supports the findings of Gu et al63 who noticed reduced GCL activity in autistic patients and attributed this to the reduced GCLM protein expression in their cerebellum tissues. Both findings may explain the glutathione deficiency in children with autism. We also suppose that GCLM gene downregulation and the probable subsequent diminished abundance of glutathione may underlie the development of autism as they may weaken the detoxification of xenobiotics that interfere with early postnatal neurodevelopment.64,65
PTGS2 gene encodes cyclooxygenase 2 (COX-2).66 is a key enzyme in the pathway of prostaglandin synthesis with 2 existing isoforms COX-1 and COX-2. Cyclooxygenase 2 and oxidative stress play in a vicious cycle where the COX-2 expression is induced under oxidative stress condition67 with the consequent production of prostaglandin E2 that in turn leads to ultimate production of ROS.68 Several studies demonstrated elevated levels of plasma prostaglandin E2 in autistic patients,68,69 which in excess leads to neuroinflammation.70,71 Unexpectedly, we found diminished mRNA expression of PTGS2 in autistic children. Regulation of gene expression at the transcriptional level is not always associated with similar pattern of protein expression.72,73 Accordingly, PTGS2 protein might be regulated in autism at either posttranscriptional or posttranslational levels. An alternative explanation is that PBMCs may not properly reflect the PTGS2 regulation in nervous tissues. Therefore, tissue-specific studies at the protein level are warranted to confirm the current downregulation of PTGS2 expression in autism.
TXN gene encodes thioredoxin (TRX) which functions as an antioxidant with a redox-regulating activity.74 Children with autism exhibited elevated levels of serum TRX in correlation with the disease severity.75,76 Our data of the similar mRNA abundance of TXN gene between autistic children and controls as well as reduced levels of TXN transcript in severe autistic children suggest that the elevated protein level of TRX observed by the other studies is perhaps due to regulation at posttranscriptional level. In addition, we reported for the first time that the level of TXN varies with the severity of autism.
In summary, our data possibly expose the link between oxidative stress and autism at the molecular level and present possible novel noninvasive biomarker candidates—a finding that is believed to be so important for autism where definitive diagnostic tests and intervention monitoring tools are not available. Further studies are required on a large sample size to validate the current findings at the protein level to investigate the consequence of the altered transcriptional profile.
Footnotes
Peer review:Seven peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1266 words, excluding any confidential comments to the academic editor.
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Science and Technology Development Fund in Egypt (STDF) to Dr Nagwa A Meguid (grant number 5541).
Declaration of Conflicting Interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: NAM diagnosed the enrolled autistic children, designed the study and revised the paper; SASG and MFM critically revised the paper; MKI, RMD, NGBED, THA and MH performed the molecular biology experiments; MKI analyzed the data and wrote the paper; and finally MKEA designed the molecular experiments, analyzed the data and revised the paper.
References
- 1. Loucas T, Charman T, Pickles A, et al. Autistic symptomatology and language ability in autism spectrum disorder and specific language impairment. J Child Psychol Psychiatry. 2008;49:1184–1192. [DOI] [PubMed] [Google Scholar]
- 2. American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 5th ed. Washington, DC: American Psychiatric Association; 2013. [Google Scholar]
- 3. Ospina MB, Krebs Seida J, Clark B, et al. Behavioural and developmental interventions for autism spectrum disorder: a clinical systematic review. PLoS ONE. 2008;3:e3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ashwood P, Wills S, Van de Water J. The immune response in autism: a new frontier for autism research. J Leukoc Biol. 2006;80:1–15. [DOI] [PubMed] [Google Scholar]
- 5. Kumar B, Prakash A, Sewal RK, Medhi B, Modi M. Drug therapy in autism: a present and future perspective. Pharmacol Rep. 2012;64:1291–1304. [DOI] [PubMed] [Google Scholar]
- 6. London EA. The environment as an etiologic factor in autism: a new direction for research. Environ Health Perspect. 2000;108:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hertz-Picciotto I, Croen LA, Hansen R, Jones CR, van de Water J, Pessah IN. The CHARGE study: an epidemiologic investigation of genetic and environmental factors contributing to autism. Environ Health Perspect. 2006;114:1119–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dufault R, Lukiw WJ, Crider R, Schnoll R, Wallinga D, Deth R. A macroepigenetic approach to identify factors responsible for the autism epidemic in the United States. Clin Epigenetics. 2012;4:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yamamah G, Abdel-Raouf E, Talaat A, Saad-Hussein A, Hamamy H, Meguid NA. Prevalence of consanguineous marriages in South Sinai, Egypt. J Biosoc Sci. 2013;45:31–39. [DOI] [PubMed] [Google Scholar]
- 10. Novarino G, El-Fishawy P, Kayserili H, et al. Mutations in BCKD-kinase lead to a potentially treatable form of autism with epilepsy. Science. 2012;338:394–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chauhan A, Chauhan V. Oxidative stress in autism. Pathophysiology. 2006;13:171–181. [DOI] [PubMed] [Google Scholar]
- 12. Frustaci A, Neri M, Cesario A, et al. Oxidative stress-related biomarkers in autism: systematic review and meta-analyses. Free Radic Biol Med. 2012;52:2128-2141. [DOI] [PubMed] [Google Scholar]
- 13. Melnyk S, Fuchs GJ, Schulz E, et al. Metabolic imbalance associated with methylation dysregulation and oxidative damage in children with autism. J Autism Dev Disord. 2012;42:367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Uttara B, Singh AV, Zamboni P, Mahajan RT. Oxidative stress and neurodegenerative diseases: a review of upstream and downstream antioxidant therapeutic options. Current Neuropharmacol. 2009;7:65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kunwar A, Priyadarsini IK. Free radicals, oxidative stress and importance of antioxidants in human health. J Med Allied Sci. 2011;1:53–60. [Google Scholar]
- 16. Hwang O. Role of oxidative stress in Parkinson’s disease. Exp Neurobiol. 2013;22:11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Patel VP, Chu CT. Nuclear transport, oxidative stress, and neurodegeneration. Int J Clin Exp Pathol. 2011;4:215–229. [PMC free article] [PubMed] [Google Scholar]
- 18. Frye RE, Delatorre R, Taylor H, et al. Redox metabolism abnormalities in autistic children associated with mitochondrial disease. Transl Psychiatry. 2013;3:e273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Anwar A, Marini M, Abruzzo PM, et al. Quantitation of plasma thiamine, related metabolites and plasma protein oxidative damage markers in children with autism spectrum disorder and healthy controls. Free Radic Res. 2016;50:S85-S90. [DOI] [PubMed] [Google Scholar]
- 20. James SJ, Cutler P, Melnyk S, et al. Metabolic biomarkers of increased oxidative stress and impaired methylation capacity in children with autism. Am J Clin Nutr. 2004;80:1611–1617. [DOI] [PubMed] [Google Scholar]
- 21. Damodaran LP, Arumugam G. Urinary oxidative stress markers in children with autism. Redox Rep. 2011;16:216–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yui K, Tanuma N, Yamada H, Kawasaki Y. Reduced endogenous urinary total antioxidant power and its relation of plasma antioxidant activity of superoxide dismutase in individuals with autism spectrum disorder [published online ahead of print August 21, 2016]. Int J Dev Neurosci. [DOI] [PubMed] [Google Scholar]
- 23. Vellingiri B, Subramaniam M, Meyyazhagan A, Krishnan P, Iyer M. Peripheral blood markers of homocysteine, paraoxonase1 (PON1) activity and oxidative stress in autism. Int J Dev Neurosci. 2015;47:82–83. [Google Scholar]
- 24. Rahbar MH, Samms-Vaughan M, Ma J, et al. Interaction between GSTT1 and GSTP1 allele variants as a risk modulating-factor for autism spectrum disorders. Res Autism Spectr Disord. 2015;12:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karande S. Autism: a review for family physicians. Indian J Med Sci. 2006;60:205–215. [PubMed] [Google Scholar]
- 26. Diaz-Beltran L, Esteban FJ, Wall DP. A common molecular signature in ASD gene expression: following Root 66 to autism. Transl Psychiatry. 2016;6:e705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Suganya V, Geetha A, Sujatha S. Urine proteome analysis to evaluate protein biomarkers in children with autism. Clin Chim Acta. 2015;450:210–219. [DOI] [PubMed] [Google Scholar]
- 28. Rutter M, Le Couteur A, Lord C. Autism Diagnostic Interview™ Revised (ADI™ R). Los Angeles, CA: Western Psychological Services; 2003. [Google Scholar]
- 29. Schopler E, Reichler RJ, Renner BR. The Childhood Autism Rating Scale (CARS). Los Angeles, CA: Western Psychological Services; 1988. [Google Scholar]
- 30. Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. [DOI] [PubMed] [Google Scholar]
- 31. Gregg JP, Lit L, Baron CA, et al. Gene expression changes in children with autism. Genomics. 2008;91:22–29. [DOI] [PubMed] [Google Scholar]
- 32. Tang Y, Lu A, Ran R, et al. Human blood genomics: distinct profiles for gender, age and neurofibromatosis type 1. Brain Res Mol Brain Res. 2004;132:155–167. [DOI] [PubMed] [Google Scholar]
- 33. Tang Y, Schapiro MB, Franz DN, et al. Blood expression profiles for tuberous sclerosis complex 2, neurofibromatosis type 1, and Down’s syndrome. Ann Neurol. 2004;56:808–814. [DOI] [PubMed] [Google Scholar]
- 34. Kakiuchi C, Iwamoto K, Ishiwata M, et al. Impaired feedback regulation of XBP1 as a genetic risk factor for bipolar disorder. Nat Genet. 2003;35:171–175. [DOI] [PubMed] [Google Scholar]
- 35. Hu VW, Frank BC, Heine S, Lee NH, Quackenbush J. Gene expression profiling of lymphoblastoid cell lines from monozygotic twins discordant in severity of autism reveals differential regulation of neurologically relevant genes. BMC Genomics. 2006;7:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Baron CA, Liu SY, Hicks C, Gregg JP. Utilization of lymphoblastoid cell lines as a system for the molecular modeling of autism. J Autism Dev Disord. 2006; 36:973–982. [DOI] [PubMed] [Google Scholar]
- 37. Schipper HM. Redox neurology: visions of an emerging subspecialty. Ann N Y Acad Sci. 2004;1012:342–355. [DOI] [PubMed] [Google Scholar]
- 38. Jain V, Langham MC, Wehrli FW. MRI estimation of global brain oxygen consumption rate. J Cereb Blood Flow Metab. 2010;30:1598–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peng DF, Belkhiri A, Hu TL, et al. Glutathione peroxidase 7 protects against oxidative DNA damage in oesophageal cells. Gut. 2012;61:1250–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Laszlo A, Novak Z, Szollosi-Varga I, Hai du Q, Vetro A, Kovacs A. Blood lipid peroxidation, antioxidant enzyme activities and hemorheological changes in autistic children. Ideggyogy Sz. 2013;66:23–28. [PubMed] [Google Scholar]
- 41. Al-Gadani Y, El-Ansary A, Attas O, Al-Ayadhi L. Metabolic biomarkers related to oxidative stress and antioxidant status in Saudi autistic children. Clin Biochem. 2009;42:1032–1040. [DOI] [PubMed] [Google Scholar]
- 42. Wei PC, Hsieh YH, Su MI, et al. Loss of the oxidative stress sensor NPGPx compromises GRP78 chaperone activity and induces systemic disease. Mol Cell. 2012;48:747–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Utomo A, Jiang X, Furuta S, et al. Identification of a novel putative non-selenocysteine containing phospholipid hydroperoxide glutathione peroxidase (NPGPx) essential for alleviating oxidative stress generated from polyunsaturated fatty acids in breast cancer cells. J Biol Chem. 2004;279:43522–43529. [DOI] [PubMed] [Google Scholar]
- 44. Zoroglu SS, Armutcu F, Ozen S, et al. Increased oxidative stress and altered activities of erythrocyte free radical scavenging enzymes in autism. Eur Arch Psychiatry Clin Neurosci. 2004;254:143–147. [DOI] [PubMed] [Google Scholar]
- 45. Meguid NA, Dardir AA, Abdel-Raouf ER, Hashish A. Evaluation of oxidative stress in autism: defective antioxidant enzymes and increased lipid peroxidation. Biol Trace Elem Res. 2011;143:58–65. [DOI] [PubMed] [Google Scholar]
- 46. Tang G, Gutierrez Rios P, Kuo SH, et al. Mitochondrial abnormalities in temporal lobe of autistic brain. Neurobiol Dis. 2013;54:349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yorbik O, Sayal A, Akay C, Akbiyik DI, Sohmen T. Investigation of antioxidant enzymes in children with autistic disorder. Prostaglandins Leukot Essent Fatty Acids. 2002;67:341–343. [DOI] [PubMed] [Google Scholar]
- 48. Rossignol DA, Frye RE. Mitochondrial dysfunction in autism spectrum disorders: a systematic review and meta-analysis. Mol Psychiatry. 2012;17:290–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Eid R, Boucher E, Gharib N, et al. Identification of human ferritin, heavy polypeptide 1 (FTH1) and yeast RGI1 (YER067W) as pro-survival sequences that counteract the effects of Bax and copper in Saccharomyces cerevisiae. Exp Cell Res. 2016;342:52–61. [DOI] [PubMed] [Google Scholar]
- 50. Reynolds A, Krebs NF, Stewart PA, et al. Iron status in children with autism spectrum disorder. Pediatrics. 2012;130:S154–S159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dosman CF, Drmic IE, Brian JA, et al. Ferritin as an indicator of suspected iron deficiency in children with autism spectrum disorder: prevalence of low serum ferritin concentration. Dev Med Child Neurol. 2006;48:1008–1009. [DOI] [PubMed] [Google Scholar]
- 52. Latif A, Heinz P, Cook R. Iron deficiency in autism and Asperger syndrome. Autism. 2002;6:103–114. [DOI] [PubMed] [Google Scholar]
- 53. Fisher J, Devraj K, Ingram J, et al. Ferritin: a novel mechanism for delivery of iron to the brain and other organs. Am J Physiol Cell physiol. 2007;293:C641–C649. [DOI] [PubMed] [Google Scholar]
- 54. Georgieff MK. The role of iron in neurodevelopment: fetal iron deficiency and the developing hippocampus. Biochem Soc Trans. 2008;36:1267–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Youssef J, Singh K, Huntington N, Becker R, Kothare SV. Relationship of serum ferritin levels to sleep fragmentation and periodic limb movements of sleep on polysomnography in autism spectrum disorders. Pediatr Neurol. 2013;49:274–278. [DOI] [PubMed] [Google Scholar]
- 56. Zeng L, Zou W, Wang G. Cellular prion protein (PrPC) and its role in stress responses. Int J Clin Exp Med. 2015;8:8042–8050. [PMC free article] [PubMed] [Google Scholar]
- 57. Doeppner TR, Kaltwasser B, Schlechter J, et al. Cellular prion protein promotes post-ischemic neuronal survival, angioneurogenesis and enhances neural progenitor cell homing via proteasome inhibition. Cell Death Dis. 2015;6:e2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kaiser DM, Acharya M, Leighton PLA, et al. Amyloid beta precursor protein and prion protein have a conserved interaction affecting cell adhesion and CNS development. PLoS ONE. 2012;7:e51305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Llorens F, del Rio JA. Unraveling the neuroprotective mechanisms of PrP (C) in excitotoxicity. Prion. 2012;6:245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Napoli E, Wong S, Hertz-Picciotto I, Giulivi C. Deficits in bioenergetics and impaired immune response in granulocytes from children with autism. Pediatrics. 2014;133:e1405–e1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Giulivi C, Zhang YF, Omanska-Klusek A, et al. Mitochondrial dysfunction in autism. JAMA. 2010;304:2389–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gipp JJ, Bailey HH, Mulcahy RT. Cloning and sequencing of the cDNA for the light subunit of human liver gamma-glutamylcysteine synthetase and relative mRNA levels for heavy and light subunits in human normal tissues. Biochem Biophys Res Commun. 1995;206:584–589. [DOI] [PubMed] [Google Scholar]
- 63. Gu F, Chauhan V, Chauhan A. Impaired synthesis and antioxidant defense of glutathione in the cerebellum of autistic subjects: alterations in the activities and protein expression of glutathione-related enzymes. Free Radic Biol Med. 2013;65:488–496. [DOI] [PubMed] [Google Scholar]
- 64. Denes A, Miyan JA. Brain-immune interactions in health and disease. Front Neurosci. 2014;8:382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Main PAE, Angley MT, O’Doherty CE, Thomas P, Fenech M. The potential role of the antioxidant and detoxification properties of glutathione in autism spectrum disorders: a systematic review and meta-analysis. Nutr Metab (Lond). 2012;9:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hla T, Neilson K. Human cyclooxygenase-2 cDNA. Proc Natl Acad Sci U S A. 1992;89:7384–7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chung HY, Lee EK, Choi YJ, et al. Molecular inflammation as an underlying mechanism of the aging process and age-related diseases. J Dent Res. 2011;90:830–840. [DOI] [PubMed] [Google Scholar]
- 68. Brigandi SA, Shao H, Qian SY, Shen Y, Wu BL, Kang JX. Autistic children exhibit decreased levels of essential fatty acids in red blood cells. Int J Mol Sci. 2015;16:10061–10076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. El-Ansary A, Al-Ayadhi L. Lipid mediators in plasma of autism spectrum disorders. Lipids Health Dis. 2012;11:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tassoni D, Kaur G, Weisinger RS, Sinclair AJ. The role of eicosanoids in the brain. Asia Pac J Clin Nutr. 2008;17:220–228. [PubMed] [Google Scholar]
- 71. Tamiji J, Crawford DA. The neurobiology of lipid metabolism in autism spectrum disorders. Neurosignals. 2010;18:98–112. [DOI] [PubMed] [Google Scholar]
- 72. Mostoller K, Norbury CC, Jain P, Wigdahl B. Human T-cell leukemia virus type I Tax induces the expression of dendritic cell markers associated with maturation and activation. J Neurovirol. 2004;10:358–371. [DOI] [PubMed] [Google Scholar]
- 73. Jorgensen TN, Haase C, Michelsen BK. Treatment of an immortalized APC cell line with both cytokines and LPS ensures effective T-cell activation in vitro. Scand J Immunol. 2002;56:492–503. [DOI] [PubMed] [Google Scholar]
- 74. Jee C, Vanoaica L, Lee J, Park BJ, Ahnn J. Thioredoxin is related to life span regulation and oxidative stress response in Caenorhabditis elegans. Genes Cells. 2005;10:1203–1210. [DOI] [PubMed] [Google Scholar]
- 75. Al-Yafee YA, Al-Ayadhi LY, Haq SH, El-Ansary AK. Novel metabolic biomarkers related to sulfur-dependent detoxification pathways in autistic patients of Saudi Arabia. BMC Neurol. 2011;11:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhang QB, Gao SJ, Zhao HX. Thioredoxin: a novel, independent diagnosis marker in children with autism. Int J Dev Neurosci. 2015;40:92–96. [DOI] [PubMed] [Google Scholar]