Abstract
In preclinical studies, heregulin (HRG) expression was shown to be the most relevant predictive biomarker for response to patritumab, a fully human anti–epidermal growth factor receptor 3 monoclonal antibody. In support of a phase 2 study of erlotinib ± patritumab in non–small cell lung cancer (NSCLC), a reverse-transcription quantitative polymerase chain reaction (RT-qPCR) assay for relative quantification of HRG expression from formalin-fixed paraffin-embedded (FFPE) NSCLC tissue samples was developed and validated and described herein. Test specimens included matched FFPE normal lung and NSCLC and frozen NSCLC tissue, and HRG-positive and HRG-negative cell lines. Formalin-fixed paraffin-embedded tissue was examined for functional performance. Heregulin distribution was also analyzed across 200 NSCLC commercial samples. Applied Biosystems TaqMan Gene Expression Assays were run on the Bio-Rad CFX96 real-time PCR platform. Heregulin RT-qPCR assay specificity, PCR efficiency, PCR linearity, and reproducibility were demonstrated. The final assay parameters included the Qiagen FFPE RNA Extraction Kit for RNA extraction from FFPE NSCLC tissue, 50 ng of RNA input, and 3 reference (housekeeping) genes (HMBS, IPO8, and EIF2B1), which had expression levels similar to HRG expression levels and were stable among FFPE NSCLC samples. Using the validated assay, unimodal HRG distribution was confirmed across 185 evaluable FFPE NSCLC commercial samples. Feasibility of an RT-qPCR assay for the quantification of HRG expression in FFPE NSCLC specimens was demonstrated.
Keywords: Real-time reverse-transcription polymerase chain reaction (RT-qPCR), heregulin, non–small cell lung cancer, validation
Introduction
Lung cancer is currently the leading cause of cancer-related death in the United States,1 and non–small cell lung cancer (NSCLC) accounts for more than 83% of all primary lung cancers.2 Treatment options may include tumor resection (for resectable disease), radiation therapy, and/or chemotherapy or targeted and immune therapy (ie, immunotherapy).2 Although immunotherapy offers the promise of durable responses for a subset of patients,3-5 there are still unmet needs for patients who either do not respond to immunotherapy or who eventually relapse. Furthermore, data show that disease recurrence occurs in 18% to over 70% of all cases in which patients were treated with modalities such as tumor resection, curative-intent radiotherapy, and chemotherapy.6-9 Treatment efficacy and risk of developing toxicities to various cancer treatments also vary among patients and may be related to individual genetics.10 It is therefore crucial to establish reliable tests for predictive biomarkers that can help match patient tumor characteristics with appropriate drugs (ie, personalized medicine)10 to maximize the benefit to risk ratio, as well as guide clinical decision making.
Elevated expression of human epidermal growth factor receptor 3 (HER3) and its ligand heregulin (HRG) has been identified in various solid tumors, including NSCLC,11-15 and in NSCLC, HER3 and HRG may play a role in the mediation of resistance to anti–epidermal growth factor receptor (anti-EGFR) treatment.16,17 Although HER3 upregulation and reactivation may play a role in resistance to EGFR tyrosine kinase inhibitors,16-19 in preclinical models, HRG has been shown to reverse EGFR sensitivity,20 suggesting possible synergism between HER3 and EGFR inhibition to more completely block HER signaling. Given that checkpoint inhibitors do not significantly increase survival in patients with EGFR-mutant NSCLC, combining an HER3 inhibitor with anti-EGFR treatment may provide another therapeutic option.
Patritumab, a fully human anti-HER3 monoclonal antibody, was studied in the phase 2 HERALD (HER family Antagonism in Lung cancer, Daiichi-sankyo) trial in combination with erlotinib in EGFR treatment–naive patients with advanced NSCLC who had failed at least 2 prior chemotherapies (NCT02350712).21 Patritumab has been shown to inhibit HRG-mediated signaling through the HER3 pathway.22-24 Preclinical studies showed that HRG expression, compared with HER3 and pHER3 expression, was the most relevant predictive biomarker for patritumab response.25,26 It was hypothesized that high messenger RNA (mRNA) levels of HRG in tissue would correspond to greater clinical benefit to patritumab treatment.
Therefore, a sensitive and specific real-time reverse-transcription quantitative polymerase chain reaction (RT-qPCR) assay for use in the HERALD study was developed for the relative quantification of HRG expression in formalin-fixed paraffin-embedded (FFPE) NSCLC tissue samples. Herein, the development and validation of the HRG RT-qPCR assay in NSCLC are described.
Materials and Methods
Feasibility and assay development studies were performed to determine an optimal RNA extraction method (kit) for FFPE NSCLC tissues, optimal RNA input for RT-qPCR, primer/probe selection, selection of optimal reference (housekeeping) genes, and HRG expression levels in FFPE NSCLC specimens. The final expression assays selected for HRG RT-qPCR analysis were then validated for specificity, PCR efficiency, PCR linearity, and reproducibility. The validation was intended to demonstrate the performance and define the parameters of TaqMan Gene Expression Assays for HRG and reference genes to be used for RT-qPCR analysis of FFPE NSCLC samples.
Cell lines and tissue samples
Cell line cryovial stocks T47D (HRG negative, from ductal carcinoma) and A549 (HRG positive, from lung cancer) were cultured in 1:1 media (F12-K:Dulbecco Modified Eagle Medium). Cells were processed as either fresh (RNA extracted from frozen cell pellet) or to create a simulated FFPE sample. Cells were grown, pelleted, fixed, embedded, and sectioned, and sections were used for RNA extraction.
Matched frozen NSCLC, FFPE normal lung, and FFPE NSCLC tissue specimens were purchased from Asterand Bioscience (Detroit, MI, USA) and BioServe (Beltsville, MD, USA). Non–small cell lung cancer specimens (≥60% tumor cell content) were sectioned to a 5-µm thickness and were used for hematoxylin-eosin (H&E) staining and RNA extraction. Only those FFPE tissues that yielded RNA of acceptable purity (1.5-2.2 using a spectrophotometric absorbance ratio of A260/280) and acceptable functional performance (cycle threshold [Ct] < 35) via reference gene assay analysis were used.
Identification of an optimal RNA extraction method
RNA yields and quality from FFPE NSCLC samples were compared using the Qiagen FFPE RNA Extraction Kit (Qiagen, Germantown, MD, USA), the Ambion PureLink FFPE RNA Isolation Kit (purchased from Life Technologies, Carlsbad, CA), and a modified method using the BIOstic FFPE Extraction Kit developed by MO BIO (Carlsbad, CA, USA). Deoxyribonuclease treatment was not included in any of these methods. RNA yields were determined by A260/A280 ratios.
RNA extraction during assay validation
Histopathology and percent of tumor content were reviewed and confirmed by a pathologist from the H&E-stained tissue sections; macrodissection was not necessary due to a high level of tumor cellularity (ie, ≥60%). The Qiagen FFPE RNA Extraction Kit was used for RNA extraction.
Identification of optimal RNA input for RT-qPCR
To identify the optimal RNA input for analysis of HRG expression levels in FFPE NSCLC patient samples, 3 different amounts of RNA (20, 50, and 100 ng) were tested. The 20-ng quantity was chosen as the starting level as this is the lowest RNA input amount recommended by the manufacturer for the complementary DNA (cDNA) synthesis kit. The 100-ng quantity was chosen as the final amount as this is the upper limit of cDNA reaction mix input recommended by the manufacturer for RT-qPCR reactions. Input amounts of cDNA generated from the Qiagen and MO BIO RNA extraction kits were tested using 20, 50, or 100 ng cDNA volumes in a 20-µL reaction mix. All RT-qPCR assays were performed in triplicate.
HRG primer/probe selection
For assay optimization experiments, 3 HRG primer/probe sets with amplicon sizes of 93 base pairs (bp) (Hs00247620_m1 [primer/probe “A”]), 90 bp (Hs01108479_m1 [primer/probe “B”]), and 72 bp (Hs01101537_m1 [primer/probe “C”]) were analyzed. The 3 assays (primer/probe sets) were chosen due to their detection of HRG-α and HRG-β isoforms, which are important for the HER3 pathway, as well as for their small amplicon sizes. Because RNA in FFPE specimens is heavily fragmented, small amplicons are important for successful qPCR.27
Identification of optimal reference genes for HRG data normalization
Preliminary reference gene screening
To select reference genes, preliminary screening was conducted across 32 potential genes using RNA extracted from fresh frozen A549 cells.
Final reference gene selection
The purpose of the final gene selection was to identify reference genes that are appropriate to be used for normalization of HRG expression levels in FFPE NSCLC tissue samples. Optimal reference genes should be expressed in the range of HRG levels found in HRG-positive FFPE NSCLC samples and should not be affected by sample matrix or tissue type (ie, normal versus tumor).
Matched frozen NSCLC versus FFPE NSCLC tissue
The expression levels of 8 reference genes (from the original 32 genes) and HRG were measured in frozen NSCLC tissue samples to confirm that their expression levels were close to HRG expression levels. The Ct value shift between matched frozen NSCLC and FFPE NSCLC tissue was expected to be similar among HRG and housekeeping genes.
Matched FFPE NSCLC versus FFPE normal lung tissue
To show that the reference gene expression was not biased between FFPE normal lung and FFPE NSCLC tissue, RNA was extracted from FFPE normal lung and FFPE NSCLC samples using the Qiagen FFPE RNA Extraction Kit. A preliminary assessment of HRG expression levels in FFPE normal lung tissue was also performed using 5 samples of FFPE normal lung tissues matched to the FFPE NSCLC cases and 3 nonmatched (random) FFPE normal samples.
HRG RT-qPCR assay validation
The purpose of the validation was to verify that the required performance characteristics of the RT-qPCR assays were met and determine whether the assays were suitable for relative quantification of HRG expression in FFPE NSCLC tissue samples.
Reverse-transcription quantitative polymerase chain reaction
RNA was extracted from FFPE normal lung and FFPE NSCLC samples using the Qiagen FFPE RNA Extraction Kit. Reverse transcription was completed with 1000 ng of RNA input in 40 µL total volume. First strand cDNA synthesis was performed using the Applied Biosystems High Capacity cDNA Synthesis Kit (Thermo Fisher Scientific, Carlsbad, CA, USA) according to manufacturer’s instructions. Complementary DNA reaction was further diluted to 80 µL to create a 12.5 ng/µL working stock. The RT-qPCR analysis was completed using 4 µL (50 ng cDNA) input in 20 µL total volume. All qPCR reactions were run in triplicate. The qPCR analysis was performed using Applied Biosystems predesigned TaqMan Gene Expression assays and TaqMan Gene Expression Master Mix on the Bio-Rad CFX96 real-time PCR machine. Cycling was per manufacturer’s instructions. Data analysis was performed using the Bio-Rad CFX96 version 1.5 software set with a threshold of 100 and an auto baseline.
Target specificity
Bioinformatic analysis was performed by Applied Biosystems, using proprietary primers and probes, to show that HRG assays specifically targeted HRG. Functional testing of each assay was performed using genomic DNA (gDNA) and cDNA from A549 cells (gDNA and cDNA derived from fresh cell pellets and FFPE samples, respectively), and target specificity was confirmed by visualization of PCR amplicons from HRG and the 3 selected reference gene assays on a 2% agarose ethidium bromide gel. The gene expression assays were run in triplicate using gDNA and cDNA templates.
PCR efficiency and linearity
To determine whether the target and reference gene assays could be used for relative quantification experiments, PCR efficiency and linearity were assessed to verify similarity across a range of template input amounts (0.01, 0.1, 1, 5, 10, 25, 50 and 100 ng) using Universal Human Reference RNA from Stratagene (La Jolla, CA, USA). Acceptance criteria for PCR efficiency were 90% to 110% for each assay, corresponding to a slope of −3.6 to −3.1. The coefficient of determination (R2) was calculated to determine the correlation between RNA input and Ct value for each assay. Each individual assay, HRG, HMBS, EIF2B1, and IPO8, must have an R2 ≥ 0.99 to distinguish a 2-fold difference in template concentration. For each assay, 6 curves were run, in duplicate, over 3 days. The RT-qPCR analysis was completed with the gene expression assays HRG (primer/probe C), HMBS (Hs00609297_m1), EIF2B1 (Hs00426752_m1), and IPO8 (Hs00183533_m1) using the Bio-Rad CFX96 instrument platform.
Intra-run and inter-run reproducibility
For intra-run reproducibility, 6 RNA extractions were performed within 1 day for each FFPE sample. The number of tissue sections equivalent to 1.5 cm2 tissue area from each case was used for each RNA extraction. The RT-qPCR analysis was completed within a single day. The mean ΔCt and standard deviation were calculated across intra-run replicates.
For inter-run reproducibility, the same set of FFPE NSCLC samples was subject to 5 separate RNA extractions (each set comprised 6 FFPE NSCLC samples) that were completed over 3 days. RNA extraction and RT-qPCR analysis were completed by 2 operators.
The Universal Human Reference RNA was run as a positive control. No template controls were composed of qPCR master mix and water for confirmation of contaminant-free reagents.
Data analysis
Heregulin expression levels were normalized according to the following formula: ΔCt = Mean Ct HRG − Mean Ct (reference genes).
To further evaluate the precision of intra- and inter-run reproducibility, the fold difference in ΔCt values was obtained from the set of 5 or 6 extractions per sample by the following formula (lower values represent higher expression):
HRG distribution in NSCLC tumor specimens
In total, 200 NSCLC commercial tissue blocks were purchased from multiple commercial tissue depository vendors, and newly sectioned slides were used to test for HRG gene expression using the validated RT-qPCR assay. The distribution of NSCLC subtypes in the commercial samples was matched to the distribution of NSCLC subtypes observed in the HERALD study. Biopsies were performed between December 2009 and August 2013, and specimens were analyzed between 1 and 46 months post biopsy (time between biopsy and analysis was staggered to detect any possible variation in the integrity of the stored specimens over approximately 4 years).
HRG mRNA distribution
Shames et al28 showed a bimodal distribution of HRG gene expression in squamous cell carcinoma of the head and neck tissue samples. In the HERALD study, HRG gene distribution was shown as a unimodal distribution among 103 samples.21,29 To confirm the unimodal distribution observed in a larger sample set, the NSCLC FFPE commercial tissue blocks were analyzed to show the distribution of HRG expression.
Results
Identification of an optimal RNA extraction method
When the Qiagen FFPE RNA and Ambion PureLink extraction kits were compared, better RNA yield and purity were obtained using the Qiagen kit (Supplemental Table S1).
Subsequently, 7 FFPE NSCLC tissue samples and 2 FFPE cell line samples (A549 and T47D) were used in the comparison between the Qiagen FFPE RNA Extraction Kit and MO BIO–purified RNA kit. RNA yield and purity of RNA extractions were compared (Table 1). In all, 6 out of 7 NSCLC samples had higher RNA yield and better RNA purity with the Qiagen kit. The Qiagen FFPE RNA Extraction Kit was selected for use in the final assay validation studies.
Table 1.
Final RNA extraction comparison.
| RNA origin | RNA concentration, ng/µL |
Total RNA yield, ng |
A260/A280 |
|||
|---|---|---|---|---|---|---|
| Qiagen | MO BIO | Qiagen | MO BIO | Qiagen | MO BIO | |
| FFPE NSCLC | ||||||
| 1173692B | 119.7 | 74.6 | 3590 | 2239 | 1.78 | 1.84 |
| 1173127B | 82.3 | 69.2 | 2470 | 2076 | 1.80 | 1.89 |
| 1173740B | 154.7 | 191.2 | 4640 | 5735 | 1.88 | 1.95 |
| 2YHODAZN | 104.1 | 80.3 | 3122 | 2409 | 1.90 | 2.05 |
| AF8ALAN9 | 45.5 | 35.7 | 1364 | 1070 | 1.81 | 2.03 |
| QISIZA1M | 88.9 | 69.7 | 2666 | 2090 | 1.91 | 1.99 |
| TRQR7A81 | 258.1 | 167.5 | 7742 | 5025 | 1.94 | 1.76 |
| Cell linesa | ||||||
| FFPE HRG+ | 65.4 | 91.2 | 1963 | 2735 | 1.90 | 2.06 |
| FFPE HRG(−) | 215.9 | 103.6 | 6476 | 3108 | 1.94 | 1.95 |
Abbreviations: FFPE, formalin-fixed paraffin-embedded; HRG, heregulin; NSCLC, non–small cell lung cancer.
HRG-positive cell line = A549 and HRG-negative cell line = T47D.
Identification of optimal RNA input for RT-qPCR and HRG primer/probe selection
Replicate and mean Ct values were obtained for HRG assay performed with 20, 50, and 100 ng of RNA (Table 2). Among the 3 tested primer/probe sets (A, B, and C), C generated the smallest amplicon (72 bp) and had the best performance (per detection of HRG transcripts) and was thus selected for further validation studies.
Table 2.
Selection of HRG primer and amount of RNA input for the Qiagen kit.a
| RNA origin |
Ct values |
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
HRG primer/probe Bb |
HRG primer/probe Ab |
HRG primer probe Cb |
|||||||
| 20 ng | 50 ng | 100 ng | 20 ng | 50 ng | 100 ng | 20 ng | 50 ng | 100 ng | |
| FFPE NSCLC | |||||||||
| 1173692B | N/A | N/A | N/A | N/A | 36.7 | 36.6 | 38.2 | 37.3 | 35.4 |
| 1173127B | N/A | N/A | N/A | N/A | 38.6 | 37.7 | 38.0 | 36.5 | 37.0 |
| 1173740B | N/A | N/A | N/A | N/A | N/A | 38.7 | 39.0 | 38.2 | 37.3 |
| 2YHODAZN | 39.8 | N/A | 39.4 | 39.8 | 37.2 | 35.2 | 36.9 | 35.6 | 35.1 |
| AF8ALAN9 | 39.8 | 38.8 | 38.2 | 39.8 | 34.5 | 33.3 | 34.8 | 33.7 | 32.5 |
| QISIZA1M | 36.5 | 35.0 | 34.3 | 36.5 | 32.6 | 31.3 | 34.4 | 33.4 | 31.8 |
| TRQR7A81 | 32.1 | 31.5 | 30.0 | 32.1 | 28.4 | 27.3 | 29.4 | 28.3 | 26.8 |
| Cell linesc | |||||||||
| Fresh HRG+ | 29.1 | 29.0 | 27.2 | 27.3 | 26.9 | 25.4 | 26.9 | 26.4 | 24.5 |
| FFPE HRG+ | 36.7 | 35.4 | 34.3 | 36.7 | 31.2 | 30.3 | 32.1 | 30.9 | 29.3 |
| Fresh HRG(−) | N/A | N/A | N/A | N/A | N/A | 39.4 | 38.8 | 37.3 | 38.2 |
| FFPE HRG(−) | N/A | N/A | N/A | N/A | N/A | 38.8 | N/A | 38.8 | 37.1 |
Abbreviations: Ct, cycle threshold; FFPE, formalin-fixed paraffin-embedded; HRG, heregulin; N/A, not amplifiable; NSCLC, non–small cell lung cancer.
Mean Ct values for each assay for the 3 input amounts using Qiagen kit–extracted RNA.
Primer/probe A = Hs00247620_m1, B = Hs01108479_m1, and C = Hs01101537_m1.
HRG-positive cell line = A549 and HRG-negative cell line = T47D.
Identification of optimal reference genes for HRG data normalization
Preliminary reference gene screening
During preliminary screening, most reference genes had higher expression levels than HRG expression in frozen A549 cells (data not shown). Seven reference genes (HMBS, PUM1, ABL1, IPO8, EIF2B1, GADD45A, and TBP) that demonstrated similar expression levels to HRG in frozen A549 cells were selected for further testing; 1 additional reference gene with higher levels of expression (UBC) was also included, for a total of 8 reference genes.
For the primer/probe set C in frozen NSCLC samples, mean HRG Ct values ranged from 25 to 32 (mean 28.2) (Table 3). When RNA from frozen NSCLC tumor samples was analyzed, the mean Ct values for HMBS (24.5), GADD45A (25.1), and TBP (25.3) were closest to levels of HRG expression in frozen NSCLC samples (Table 4) and were therefore selected as the initial reference genes for subsequent analysis. When the mean difference in Ct between matched frozen NSCLC and FFPE NSCLC samples for HRG and the 3 reference genes (HMBS, GADD45A, and TBP) was compared using 7 paired samples, the data indicated that the Ct value shift of TBP and GADD45A was significantly higher than the Ct value shift of HRG (data not shown). Therefore, HMBS, EIF2B1, IPO8, PUM1, and ABL1 were selected for further consideration.
Table 3.
HRG gene expression assay analysis in frozen NSCLC samples.a
| RNA origin |
Ct values for HRG primer/probe Cb |
|
|---|---|---|
| Mean | SD | |
| Frozen NSCLC | ||
| 1173692F-30 | 28.2 | 0.05 |
| 1173127F-30 | 29.8 | 0.05 |
| 1173740F-30 | 32.0 | 0.19 |
| 2YHOD | 28.6 | 0.02 |
| AF8AL | 27.8 | 0.06 |
| Q151Z | 25.4 | 0.02 |
| TRQR7 | 25.6 | 0.09 |
| Mean | 28.2 | |
| SD | 2.29 | |
| Fresh cell linesc | ||
| HRG+ | 25.5 | 0.05 |
| HRG(−) | 38.8 | NC |
Abbreviations: Ct, cycle threshold; HRG, heregulin; NC, could not be calculated; NSCLC, non–small cell lung cancer.
Three HRG gene expression assays (only primer/probe C is shown) were tested in triplicate on 7 frozen NSCLC samples and 2 cell line RNA controls. Cell line RNA controls were included to correlate with previous work and not used in the final calculations for the mean and SD.
Primer/probe C = Hs01101537_m1.
HRG-positive cell line = A549 and HRG-negative cell line = T47D.
Table 4.
Reference gene expression analysis in frozen NSCLC tumor samples.a
| RNA origin |
Ct values |
|||||||
|---|---|---|---|---|---|---|---|---|
|
UBC
|
TBP
|
GADD45A
|
ABL1
|
|||||
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| Frozen NSCLC | ||||||||
| 1173692F-30 | 19.5 | 0.06 | 25.1 | 0.15 | 25.4 | 0.19 | 23.8 | 0.04 |
| 1173127F-30 | 20.2 | 0.03 | 26.1 | 0.12 | 24.6 | 0.13 | 24.3 | 0.07 |
| 1173740F-30 | 20.9 | 0.19 | 24.0 | 0.10 | 26.0 | 0.02 | 24.2 | 0.17 |
| 2YHOD | 19.9 | 0.14 | 25.1 | 0.04 | 25.8 | 0.04 | 23.7 | 0.08 |
| AF8AL | 19.4 | 0.19 | 25.8 | 0.19 | 24.0 | 0.08 | 23.2 | 0.09 |
| Q151Z | 19.8 | 0.09 | 25.1 | 0.12 | 24.8 | 0.13 | 22.6 | 0.08 |
| TRQR7 | 20.6 | 0.09 | 26.1 | 0.07 | 24.9 | 0.08 | 23.9 | 0.11 |
| Mean | 20.0 | 25.3 | 25.1 | 23.7 | ||||
| SD | 0.57 | 0.76 | 0.71 | 0.58 | ||||
| Fresh cell linesb | ||||||||
| HRG+ | 21.3 | 0.07 | 27.6 | 0.14 | 27.1 | 0.14 | 25.9 | 0.15 |
| HRG(−) | 21.7 | 0.12 | 28.6 | 0.11 | 29.2 | 0.12 | 26.1 | 0.10 |
| RNA origin | Ct values | |||||||
| EIF2B1 | PUM1 | HMBS | IP08 | |||||
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| Frozen NSCLC | ||||||||
| 1173692F-30 | 23.2 | 0.16 | 21.5 | 0.08 | 24.2 | 0.07 | 23.9 | 0.16 |
| 1173127F-30 | 25.0 | 0.21 | 22.2 | 0.06 | 25.0 | 0.16 | 24.7 | 0.09 |
| 1173740F-30 | 23.3 | 0.10 | 21.6 | 0.10 | 24.4 | 0.09 | 24.1 | 0.10 |
| 2YHOD | 23.5 | 0.05 | 20.9 | 0.07 | 24.2 | 0.05 | 24.4 | 0.02 |
| AF8AL | 23.2 | 0.16 | 21.1 | 0.09 | 23.8 | 0.12 | 23.5 | 0.12 |
| Q151Z | 23.1 | 0.09 | 21.4 | 0.08 | 24.4 | 0.09 | 23.6 | 0.17 |
| TRQR7 | 24.4 | 0.02 | 22.5 | 0.03 | 25.3 | 0.08 | 24.6 | 0.18 |
| Mean | 23.7 | 21.6 | 24.5 | 24.1 | ||||
| SD | 0.72 | 0.58 | 0.53 | 0.48 | ||||
| Fresh cell linesb | ||||||||
| HRG+ | 25.7 | 0.25 | 24.7 | 0.11 | 24.6 | 0.01 | 25.9 | 0.17 |
| HRG(−) | 25.5 | 0.04 | 24.7 | 0.12 | 24.5 | 0.03 | 25.6 | 0.12 |
Abbreviations: HRG, heregulin; NSCLC, non–small cell lung cancer.
Eight reference gene assays were tested in triplicate on 7 extracted RNA samples and 2 cell line RNA control samples. Cell line RNA controls were included as controls for comparison across experiments. The cell line values were not used in the final calculations for the mean and SD.
HRG-positive cell line = A549 and HRG-negative cell line = T47D.
Final reference gene selection
Matched frozen NSCLC versus FFPE NSCLC tissue
The mean difference in Ct between matched frozen NSCLC and FFPE NSCLC samples for HRG and the 5 reference genes (HMBS, EIF2B1, IPO8, PUM1, and ABL1) ranged from 6.7 to 9.2 (Table 5). The 3 reference genes with a difference closest to that observed for HRG (6.7) were IPO8 (6.7), EIF2B1 (7.7), and HMBS (7.9). These assays also demonstrated the lowest standard deviation (1.06, 1.31, and 1.32, respectively) across samples.
Table 5.
Comparison of Ct values in matched frozen to FFPE NSCLC tissue with HRG plus 5 reference genes.
| Mean Ct value |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
|
HRG (72 bp) |
HMBS (64 bp) |
EIF2B1 (75 bp) |
|||||||
| Frozen NSCLC | FFPE NSCLC | ΔCt FFPE vs frozen NSCLC | Frozen NSCLC | FFPE NSCLC | ΔCt FFPE vs frozen NSCLC | Frozen NSCLC | FFPE NSCLC | ΔCt FFPE vs frozen NSCLC | |
| FFPE NSCLC | |||||||||
| TRQR7A81 | 25.6 | 28.3 | 2.7 | 25.3 | 30.9 | 5.6 | 24.4 | 29.6 | 5.2 |
| 2YHODAZN | 28.6 | 36.8 | 8.2 | 24.2 | 31.8 | 7.6 | 23.5 | 31.1 | 7.6 |
| QISIZA1M | 25.4 | 33.8 | 8.3 | 24.4 | 32.0 | 7.7 | 23.1 | 31.5 | 8.3 |
| AF8ALAN9 | 27.8 | 33.4 | 5.6 | 23.8 | 31.2 | 7.4 | 23.2 | 30.0 | 6.8 |
| 1173692B | 28.2 | 36.5 | 8.4 | 24.2 | 34.0 | 9.8 | 23.2 | 32.1 | 9.0 |
| 1173127B | 29.8 | 37.5 | 7.8 | 25.0 | 33.9 | 8.9 | 25.0 | 33.2 | 8.2 |
| 1173740B | 32.0 | 37.9 | 5.9 | 24.4 | 32.4 | 8.0 | 23.3 | 31.9 | 8.5 |
| Mean | 28.2 | 34.9 | 6.7 | 24.4 | 32.3 | 7.9 | 23.7 | 31.3 | 7.7 |
| SD | 2.29 | 3.38 | 2.10 | 0.53 | 1.21 | 1.32 | 0.72 | 1.25 | 1.31 |
| Mean Ct value | |||||||||
| IPO8 (71 bp) | ABL1 (91 bp) | PUM1 (89 bp) | |||||||
| Frozen NSCLC | FFPE NSCLC | ΔCt FFPE vs frozen NSCLC | Frozen NSCLC | FFPE NSCLC | ΔCt FFPE vs frozen NSCLC | Frozen NSCLC | FFPE NSCLC | ΔCt FFPE vs frozen NSCLC | |
| FFPE NSCLC | |||||||||
| TRQR7A81 | 24.6 | 29.3 | 4.6 | 23.9 | 28.9 | 5.1 | 22.5 | 28.3 | 5.8 |
| 2YHODAZN | 24.4 | 31.5 | 7.0 | 23.7 | 32.4 | 8.7 | 20.9 | 30.1 | 9.1 |
| QISIZA1M | 23.6 | 31.1 | 7.5 | 22.6 | 31.7 | 9.1 | 21.4 | 31.2 | 9.8 |
| AF8ALAN9 | 23.5 | 29.5 | 6.1 | 23.2 | 29.9 | 6.7 | 21.1 | 29.3 | 8.2 |
| 1173692B | 23.9 | 31.6 | 7.7 | 23.8 | 33.1 | 9.3 | 21.5 | 32.0 | 10.5 |
| 1173127B | 24.7 | 31.9 | 7.2 | 24.3 | 32.3 | 8.1 | 22.2 | 32.5 | 10.2 |
| 1173740B | 24.1 | 30.9 | 6.8 | 24.2 | 33.7 | 9.5 | 21.6 | 32.1 | 10.4 |
| Mean | 24.1 | 30.8 | 6.7 | 23.7 | 31.7 | 8.1 | 21.6 | 30.8 | 9.2 |
| SD | 0.48 | 1.03 | 1.06 | 0.58 | 1.70 | 1.63 | 0.58 | 1.60 | 1.70 |
Abbreviations: Ct, cycle threshold; FFPE, formalin-fixed paraffin-embedded; NSCLC, non–small cell lung cancer.
Matched FFPE NSCLC versus FFPE normal lung tissue
The mean difference in Ct between matched FFPE NSCLC and FFPE normal lung tissue for the 5 reference gene (HBMS, EIF2B1, IPO8, PUM1, and ABL1) ranged from −1.5 (PUM1) to −0.5 (IPO8) (Table 6). There did not appear to be bias in the expression level of the 5 reference genes due to tissue type (ie, normal versus tumor). The 3 genes with the smallest mean difference in Ct values were EIF2B1 (−1.2), ABL1 (−0.8), and IPO8 (−0.5). The lowest standard deviation across samples was observed for IPO8 (0.68), HMBS (0.72), EIF2B1 (0.75), and ABL1 (0.72).
Table 6.
Comparison of Ct values in FFPE NSCLC tissues and FFPE normal lung tissue for HRG plus 5 reference genes.
| Mean Ct values |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
|
HRG (72 bp) |
HMBS (64 bp) |
EIF2B1 (75 bp) |
|||||||
| FFPE NSCLC | FFPE normal lung | ΔCt FFPE NSCLC vs normal lung | FFPE NSCLC | FFPE normal lung | ΔCt FFPE NSCLC vs normal lung | FFPE NSCLC | FFPE normal lung | ΔCt FFPE NSCLC vs normal lung | |
| FFPE NSCLC | |||||||||
| TRQR7A81 | 28.3 | 34.3 | −6.0 | 30.9 | 32.9 | −2.0 | 29.6 | 31.0 | −1.4 |
| 2YHODAZN | 36.8 | 36.1 | 0.7 | 31.8 | 33.4 | −1.6 | 31.1 | 32.9 | −1.8 |
| QISIZA1M | 33.8 | 34.3 | −0.6 | 32.0 | 32.2 | −0.1 | 31.5 | 31.4 | 0.1 |
| AF8ALAN9 | 33.4 | 34.9 | −1.5 | 31.2 | 32.4 | −1.3 | 30.0 | 31.3 | −1.3 |
| 1173692B | 36.5 | 37.6 | −1.0 | 34.0 | 35.6 | −1.7 | 32.1 | 33.7 | −1.6 |
| FFPE normal lung | |||||||||
| 1171535B | NA | 35.1 | NA | NA | 35.5 | NA | NA | 32.6 | NA |
| 1173394B | NA | 29.0 | NA | NA | 29.8 | NA | NA | 27.6 | NA |
| 1173400B | NA | 29.2 | NA | NA | 29.7 | NA | NA | 27.6 | NA |
| Mean | 33.77 | 33.8 | −1.7 | 32.0 | 32.7 | −1.3 | 30.9 | 31.0 | −1.2 |
| SD | 3.41 | 3.10 | 2.55 | 1.21 | 2.21 | 0.72 | 1.06 | 2.29 | 0.75 |
| IPO8 (71 bp) | ABL1 (91 bp) | PUM1 (89 bp) | |||||||
| FFPE NSCLC | |||||||||
| TRQR7A81 | 29.3 | 30.2 | −2.0 | 28.9 | 30.5 | −1.6 | 28.3 | 30.4 | −2.1 |
| 2YHODAZN | 31.5 | 31.7 | −0.3 | 32.4 | 33.4 | −1.0 | 30.1 | 32.4 | −2.4 |
| IPO8 (71 bp) | ABL1 (91 bp) | PUM1 (89 bp) | |||||||
| QISIZA1M | 31.1 | 30.5 | 0.6 | 31.7 | 31.3 | 0.4 | 31.2 | 31.2 | 0.0 |
| AF8ALAN9 | 29.5 | 30.5 | −1.0 | 29.9 | 30.9 | −0.9 | 29.3 | 30.8 | −1.5 |
| 1173692B | 31.6 | 32.4 | −0.8 | 33.1 | 33.7 | −0.7 | 32.0 | 33.7 | −1.7 |
| FFPE Normal lung | |||||||||
| 1171535B | NA | 31.5 | NA | NA | 32.7 | NA | NA | 32.3 | NA |
| 1173394B | NA | 27.1 | NA | NA | 27.0 | NA | NA | 25.9 | NA |
| 1173400B | NA | 27.2 | NA | NA | 26.6 | NA | NA | 25.9 | NA |
| Mean | 30.6 | 30.2 | −0.5 | 31.2 | 30.8 | −0.8 | 30.2 | 30.3 | −1.5 |
| SD | 1.12 | 1.97 | 0.68 | 1.72 | 2.71 | 0.72 | 1.49 | 2.94 | 0.94 |
Abbreviations: Ct, cycle threshold; FFPE, formalin-fixed paraffin-embedded; HRG, heregulin; NA, not applicable (no matched tissue available); NSCLC, non–small cell lung cancer.
Overall, IPO8, EIF2B1, and HMBS generated the lowest difference in standard deviation values for matched frozen NSCLC, FFPE normal lung, and FFPE NSCLC tissue, across all FFPE NSCLC samples (suggesting that they can perform well across varying RNA quality and are suitable for normalization of HRG Ct values), and were thus selected as the final 3 reference genes.
HRG RT-qPCR assay validation
Target specificity
Polymerase chain reactions (HRG, HMBS, EIF2B1, and IPO8) produced amplification from a cDNA template, and no amplification was detected from the gDNA template. When PCR products were run on a 2% agarose ethidium bromide gel, a single expected band for HRG, HMBS, EIF2B1, or IPO8 was observed from HRG, HMBS, EIF2B1, or IPO8 PCR reactions, respectively, when cDNA was used as template; no band was observed from PCR reactions when gDNA was used as template (expected, because of the use of primers that are designed to span exon-exon boundaries; data not shown). These data demonstrated specificity of the primer/probes for HRG and the 3 reference gene assays.
PCR efficiency and linearity
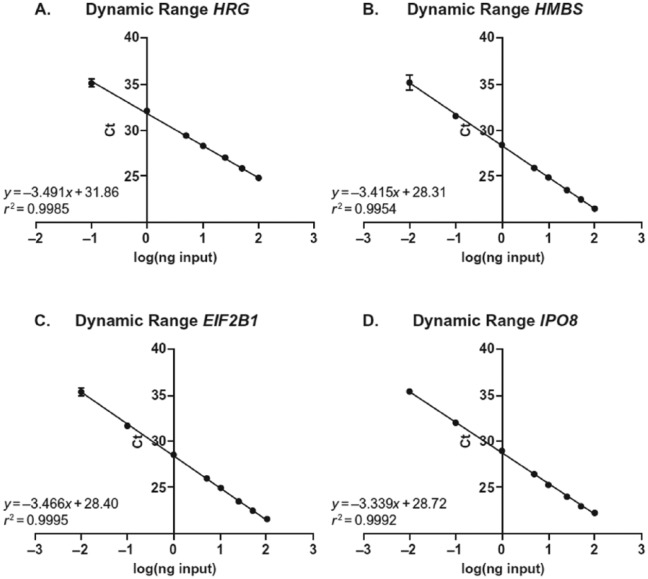
For HRG and the 3 housekeeping genes (HMBS, EIF2B1, and IPO8), based on the mean values for slope and R2 across 6 runs, the average PCR efficiency and linearity across 6 runs met the predefined PCR efficiency (90%-110%; slope −3.6 to −3.1) and coefficient of determination (R2 ≥ 0.99) requirements (Table 7 and representative graphs in Figure 1).
Table 7.
PCR efficiency and linearity for HRG and the 3 housekeeping genes (HMBS, EIF2B1, and IPO8).
| Gene target | Run 1 | Run 2 | Run 3 | Run 4 | Run 5 | Run 6 | Mean |
|---|---|---|---|---|---|---|---|
| PCR efficiency, % | |||||||
| HRG | 93.3a | 97.6a | 92.6a | 91.0a | 98.1a | 88.5a | 93.5a |
| HMBS | 94.8 | 93.3 | 91.2 | 97.8 | 99.0 | 102.3 | 96.4 |
| EIF2B1 | 93.3 | 93.6 | 94.7 | 92.5 | 93.8 | 98.4 | 94.4 |
| IPO8 | 100.0 | 101.0 | 99.1 | 99.8 | 96.1 | 99.9 | 99.3 |
| R2 | |||||||
| HRG | 0.9966a | 0.9922a | 0.9986a | 0.9954a | 0.9958a | 0.9972a | 0.9960a |
| HMBS | 0.9988 | 0.9925 | 0.9925 | 0.9987 | 0.9986 | 0.9977 | 0.9965 |
| EIF2B1 | 0.9987 | 0.9994 | 0.9988 | 0.9986 | 0.9982 | 0.9979 | 0.9986 |
| IPO8 | 0.9992 | 0.9986 | 0.9989 | 0.9991 | 0.9989 | 0.9984 | 0.9989 |
Abbreviation: HRG, heregulin.
No amplification was seen in the HRG assay at 0.01 ng complementary DNA. The calculation was based on 7 data points instead of 8 data points.
Figure 1.
PCR efficiency and linearity for HRG and 3 housekeeping genes: (A) HRGa,b (B) HMBSb, (C) EIF2B1b, and (D) IPO8b. Ct values as a function of input template. Ct indicates cycle threshold.
aThe HRG figure represents a 7-point curve analysis due to no amplification seen in lowest point of 0.01 ng RNA.
bEach point represents the mean Ct across 6 runs.
Intra-run and inter-run reproducibility
The intra-run reproducibility experiments demonstrated that normalized HRG expression levels could vary from 1.1-fold to 5.4-fold between independent extractions (Table 8). In FFPE specimens, inter-run reproducibility experiments demonstrated that normalized HRG expression levels could vary from 1.1-fold to 2.5-fold between independent extractions, independent RT-qPCR runs, and 2 operators (Table 9). The fold difference in normalized expression levels for the positive control sample was 1.1. The results also support data from the PCR efficiency and linearity experiments which demonstrated that mean Ct values greater than 35 for either HRG or reference gene expression assays trend toward a higher SD. Variability in HRG ΔCt values can also be attributed to the use of serial tissue sections for each independent extraction due to tumor heterogeneity.
Table 8.
Intra-run reproducibility: normalized Ct values for HRG (ΔCt) for each RNA extraction and FFPE sample and mean ΔCt deviation comparison.
| Sample ID | Intra-run ΔCt |
Intra-run mean ΔCt deviation comparisona |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Run 1 | Run 2 | Run 3 | Run 4 | Run 5 | Run 6 | Mean | SD | ΔCt highest | ΔCt lowest | X = highest − lowest | ΔCt deviation (2x) | |
| FFPE NSCLC | ||||||||||||
| 1173692B | 3.6 | N/A | 3.3 | 3.0 | 3.0 | 2.4 | 3.0 | 0.43 | 3.6 | 2.4 | 1.2 | 2.3 |
| 1173127B | 5.5 | 4.3 | 4.7 | 3.6 | 4.2 | 4.1 | 4.4 | 0.65 | 5.5 | 3.6 | 2.0 | 3.8 |
| 1173740B | 7.4 | 7.0 | N/A | 7.2 | 6.6 | 7.0 | 7.0 | 0.27 | 7.4 | 6.6 | 0.7 | 1.7 |
| 2YHODAZN | 5.9 | 5.8 | 5.4 | 5.3 | 7.7 | 5.7 | 6.0 | 0.89 | 7.7 | 5.3 | 2.4 | 5.4 |
| TRQR7A81 | −1.6 | −1.7 | −1.5 | −1.6 | −1.4 | −1.7 | −1.6 | 0.11 | −1.4 | −1.7 | 0.3 | 1.2 |
| FFPE normal lungb | 3.3 | 3.0 | 3.1 | 3.0 | 3.4 | 3.1 | 3.2 | 0.16 | 3.4 | 3.0 | 0.4 | 1.3 |
| Positive controlc | 3.7 | 3.7 | 3.6 | 3.6 | 3.6 | 3.6 | 3.6 | 0.04 | 3.7 | 3.6 | 0.1 | 1.1 |
Abbreviations: Ct, cycle threshold; Ext., extraction (RNA); HRG, heregulin.
ΔCt deviation calculations using highest and lowest HRG expression values for each patient extraction set.
TRQR7N58.
Universal Human Reference RNA.
Table 9.
Inter-run reproducibility: normalized Ct values for HRG (ΔCt) for each run and FFPE sample and mean ΔCt deviation comparison.
| Sample ID | Inter-run ΔCt |
Inter-run mean ΔCt deviation comparisona |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Run 1 | Run 2 | Run 3 | Run 4 | Run 5 | Mean | SD | ΔCt highest | ΔCt lowest | X = highest − lowest | ΔCt deviation (2x) | |
| FFPE NSCLC | |||||||||||
| 1173692B | 3.5 | 3.8 | 3.7 | 4.2 | 3.6 | 3.7 | 0.27 | 4.2 | 3.5 | 0.7 | 1.7 |
| 1173127B | 5.3 | 4.7 | 5.4 | 4.1 | 5.4 | 5.0 | 0.58 | 5.4 | 4.1 | 1.3 | 2.5 |
| 1173740B | 6.6 | 7.9 | 7.1 | 6.9 | 6.6 | 7.0 | 0.53 | 7.9 | 6.6 | 1.3 | 2.5 |
| 2YHODAZN | 5.4 | 5.5 | 5.2 | 6.0 | 6.2 | 5.7 | 0.43 | 6.2 | 5.2 | 1.0 | 2.0 |
| TRQR7A81 | −1.2 | −1.2 | −1.2 | −1.3 | −1.1 | −1.2 | 0.06 | −1.1 | −1.3 | 0.2 | 1.1 |
| FFPE normal lungb | 3.0 | 2.9 | 3.0 | 2.8 | 3.1 | 3.0 | 0.09 | 3.1 | 2.8 | 0.2 | 1.2 |
| Positive controlc | 3.5 | 3.6 | 3.6 | 3.6 | 3.6 | 3.6 | 0.04 | 3.6 | 3.5 | 0.1 | 1.1 |
Abbreviations: Ct, cycle threshold; HRG, heregulin.
ΔCt deviation calculations using highest and lowest HRG expression values for each patient extraction set.
TRQR7N58.
Universal Human Reference RNA.
Results of the HRG gene expression assay validation study are summarized in Table 10; the performance of the validation met predefined acceptance criteria and the assay was deemed to be validated and suitable for use for analysis of clinical samples.
Table 10.
Summary of HRG gene expression assay validation and final assay conditions for HRG gene expression assay analysis.
| Measure or requirement | Final result |
|---|---|
| Assay validation | |
| Tissue samples used | FFPE NSCLC samples FFPE normal lung tissue |
| RNA sample used | Universal Human Reference RNA |
| Cell lines used | T47D (fresh and FFPE): HRG negative A549 (fresh and FFPE): HRG positive |
| Specificity | Single amplicon generated with cDNA template No amplification with gDNA template |
| PCR efficiency and linearity | Target and reference gene expression assays amplify with an efficiency of 100 ± 10% and a coefficient of determination (R2) ≥0.99 across 7 or 8 RNA input amounts, respectively |
| Reproducibility | Complete intra-run extractions and analysis within 1 day and inter-run extractions and analysis over 3 days |
| Intraplate | Fold difference between highest and lowest ΔCt values ≤5.4 |
| Interplate | Fold difference between highest and lowest ΔCt values ≤2.5 |
Abbreviations: cDNA, complementary DNA; FFPE, formalin-fixed paraffin-embedded; gDNA, genomic DNA; HRG, heregulin; NSCLC, non–small cell lung cancer; PCR, polymerase chain reaction.
HRG Distribution in NSCLC Tumor Specimens
Of the 200 NSCLC commercial tissue blocks freshly sectioned and analyzed, most (59.0%) were adenocarcinoma subtypes (Table 11). The distribution of NSCLC subtypes in 200 NSCLC commercial samples was matched to the distribution of NSCLC subtypes observed in the HERALD study. Tumor stage was known for 197 of the 200 matched HERALD specimens in patients with histologically confirmed stage I (26.8%), stage II (37.1%), stage III (30.4%), and stage IV (5.7%) NSCLC; most of the specimens (52.5%) had ≥85% tumor content (91.5% had ≥65% tumor content).
Table 11.
Characteristics of NSCLC commercial specimens used for RT-qPCR assay validation (N = 200).
| Characteristic | n | % | ΔCt (N = 185)a,b |
|
|---|---|---|---|---|
| Median | Range | |||
| NSCLC tumor subtype | N = 200 | |||
| Adenocarcinoma | 118 | 59.0 | 4.9 | 1.7–9.4 |
| Squamous cell carcinoma | 62 | 31.0 | 2.9a | 0.4–8.1a |
| Large cell carcinoma | 18 | 9.0 | 6.2 | 1.1–9.2 |
| Adenosquamous carcinoma | 2 | 1.0 | [4.3]f | [4.3]f |
| Tumor stage c | N = 194d | |||
| I | 52 | 26.8 | 4.3 | 1.2–8.5 |
| II | 72 | 37.1 | 4.6 | 0.4–9.2 |
| III | 59 | 30.4 | 4.2a | 1.1–9.4a |
| IV | 11 | 5.7 | 3.6 | 0.6–8.1 |
| Tumor content, % | N = 200 | |||
| 50–60 | 17 | 8.5 | 4.3 | 2.6–7.9 |
| 65–70 | 36 | 18.0 | 3.9 | 0.4–8.1 |
| 75–80 | 42 | 21.0 | 4.6 | 0.6–8.1 |
| 85–90 | 73 | 36.5 | 4.4e | 0.5–9.4a |
| ≥95 | 32 | 16.0 | 5.9 | 1.3–8.9 |
| Time from biopsy to analysis, mo | N = 197e | |||
| 1–6 | 31 | 15.7 | 4.1 | 2.0–7.4 |
| 7–12 | 55 | 27.9 | 4.1 | 0.5–7.1 |
| 13–18 | 7 | 3.6 | 2.9 | 1.3–6.9 |
| 19–24 | 31 | 15.7 | 3.97 | 1.7–8.1 |
| 25–30 | 16 | 8.1 | 4.4a | 0.4–8.9a |
| 31–36 | 31 | 15.7 | 4.8 | 0.6–9.4 |
| 37–42 | 15 | 7.6 | 5.8 | 1.4–7.8 |
| 43–46 | 11 | 5.6 | 5.6 | 2.8–5.9 |
Abbreviations: Ct, cycle threshold; NSCLC, non–small cell lung cancer.
One specimen (squamous cell carcinoma, stage IIIA) was excluded from the analysis due to an outlier ΔCt value of −7.2.
ΔCt results missing for 10 adenocarcinoma, 2 squamous cell carcinoma, 1 large-cell carcinoma, and 1 adenosquamous carcinoma specimen.
Stage I includes stage IA, stage II includes stages IIA and IIB, and stage III includes stage IIIA.
N = 194: tumor stage was unknown for 6 specimens.
N = 197: biopsy dates were missing for 3 specimens.
ΔCt value available from only 1 patient with adenosquamous carcinoma.
HRG mRNA distribution
Figure 2 illustrates a unimodal distribution of HRG expression levels across 185 evaluable samples, with a median ΔCt of 4.6. Approximately, 43% to 44% of NSCLC samples could be classified as HRG “high expressors” ([HRG-high] high and low HRG expression was defined by the median ΔCt of blinded data from the RT-qPCR assay in the HERALD study).29
Figure 2.
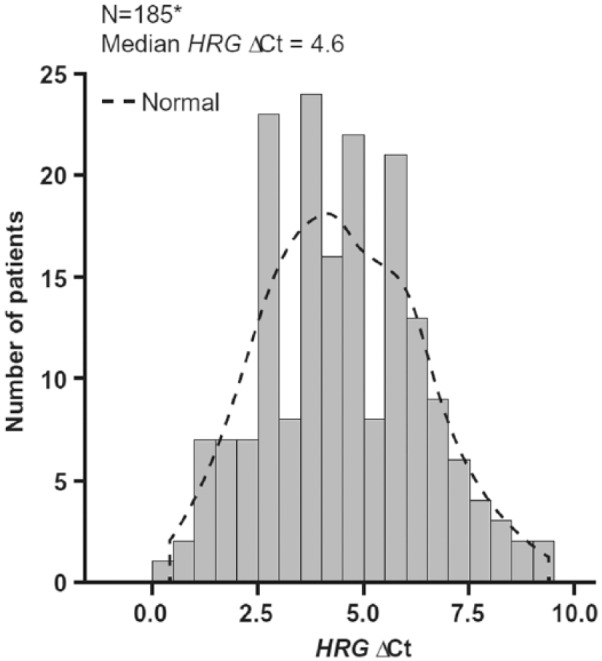
Unimodal HRG distribution in NSCLC commercial samples using the validated RT-qPCR assay. Ct indicates cycle threshold; HRG, heregulin.
*One sample with ΔCt = −7.2 excluded from the analysis.
Discussion
An RT-qPCR assay was developed and validated for the relative quantification of HRG expression in FFPE NSCLC tissue samples. Assay feasibility was demonstrated by comparison of data from fresh and FFPE cell line pellets, matched frozen NSCLC and FFPE NSCLC samples, and FFPE normal lung and FFPE NSCLC. Performance was demonstrated and parameters were defined for the Applied Biosystems TaqMan Gene Expression assays for the target gene, HRG, and 3 reference genes (HMBS, IPO8, and E12FB1). This validated RT-qPCR assay was subsequently used to measure HRG mRNA from FFPE tissue in the phase 2 HERALD study and in commercial samples, in patients with NSCLC.
The RT-qPCR assay was used to analyze 2 sets of samples. The first set included 186 FFPE NSCLC tissue samples procured from commercial vendors. When plotted, the distribution of expression levels of 185 evaluable samples was representative of a unimodal distribution (unimodal HRG distribution was confirmed in a separate analysis; data not shown). This finding is in contrast to a study by Shames et al,28 in which data showed a bimodal distribution of HRG (on a log10 scale) in squamous cell carcinoma of the head and neck. This may be due to an insufficient number of samples used to define the bimodal distribution, different assay parameters, different tumor samples (primary vs metastasis), or different tumor histology. In addition, these results were comparable with the distribution observed in the samples from the phase 2 HERALD trial.29
Acceptable RT-qPCR results were obtained from tissue samples ranging in age from 1 month to approximately 4 years from the time of collection. A limitation to the RT-qPCR assay is that, compared with immunohistochemistry (IHC), multiple tissue sections are required for RNA extraction. The availability of sufficient tissue sections (1000 ng RNA needed for a final 50-ng yield for assay input) may pose a challenge in the clinical trial setting, in which biopsy tissue, which may be used for multiple purposes, including morphological interpretation and IHC, may be inadequate for molecular studies.30 Another limitation is that, compared with IHC or RNAscope in situ hybridization (a commercially available branched in situ hybridization method31), higher tumor content is required for RT-qPCR to limit the inference of RNA extracted from normal cells. Immunohistochemistry can also show variations between individual cells in expression level, whereas RT-qPCR gives an average value across the sample. However, an overall advantage of RT-qPCR is an objective data output, in contrast to the subjective nature of IHC or RNAscope in situ hybridization data interpretation.
Data from preclinical studies (mouse xenograft model) have demonstrated that HRG mRNA is the best biomarker for correlation of in vivo response to treatment with single-agent patritumab.25,26 A secondary objective of HERALD was to identify which patients would most likely benefit from the addition of patritumab to erlotinib treatment by defining a primary biomarker hypothesis prior to data unblinding and then testing.32 In HERALD, patients were randomized to treatment with erlotinib plus either placebo or patritumab (high or low dose), and levels of mRNA from HRG and the 3 reference genes (HMBS, EIF2B1, and IPO8) identified herein were measured by RT-qPCR. In 2 separate analyses (including a simulation analysis to account for any possible imbalances between arms in patients with sensitizing EGFR mutations [6.6%]),29 data indicated significant clinical benefits for patritumab-treated patients with high HRG mRNA levels.21,29 Subgroup analysis by HRG mRNA expression levels demonstrated clinical benefit as judged by hazard ratio (HR) for progression-free survival from patritumab in patients with high HRG mRNA levels, including those patients in the patritumab high-dose (HR: 0.37; P = .0283) and low-dose (HR: 0.29; P = .0027) treatment arms.21,29
Overall, data from this study demonstrated the feasibility of a RT-qPCR assay for the quantification of HRG expression in RNA-extracted FFPE NSCLC specimens. Data from the HRG distribution study in 200 NSCLC FFPE commercial tumor specimens further indicated a unimodal distribution of HRG levels (median ΔCt: 4.6). Previously published data (median ΔCt: 4.1) included the 186 FFPE samples from the current analysis plus 93 fresh frozen tissue samples.29 The slight difference between median ΔCt values is caused by different sample sets. The HRG RT-qPCR assay was analytically validated prior to clinical sample analysis in the HERALD phase 1/2b study in patients with advanced NSCLC, and, based on the results of HERALD, may have identified a subgroup of patients who will receive clinical benefits from patritumab therapy.
Supplementary Material
Acknowledgments
Third-party writing assistance was provided by BlueMomentum, an Ashfield Company, part of UDG Healthcare plc, and supported by Daiichi Sankyo, Inc.
Footnotes
Peer review:Four peer reviewers contributed to the peer review report. Reviewers’ reports totaled 533 words, excluding any confidential comments to the academic editor.
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Study design, collection, assays, analysis, interpretation of data, and writing of the manuscript were funded by Daiichi Sankyo, Inc. Daiichi Sankyo provided the decision to submit.
Declaration of conflicting interests:The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: JK and KS are former employees of MolecularMD. XJ, RAB, and DF are former employees of Daiichi Sankyo Pharma Development. WF is an employee of Daiichi Sankyo Pharma Development. KN is an employee of Daiichi Sankyo Co., Ltd. MS was an employee of U3 Pharma GmbH. CS is an employee of MolecularMD.
Author Contributions: KN, MS, CS, and WF designed the study. JK and KS collected and assembled the data for the study. XJ, RAB, DF, CS, and WF performed data analysis and interpreted the data. All authors wrote the report and approved the final version of the manuscript.
Statement of Ethical Assurance: WF is the guarantor of this work and, as such, had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. [DOI] [PubMed] [Google Scholar]
- 2. National Comprehensive Cancer Network (NCCN). Clinical Practice Guidelines in Oncology (NCCN Guidelines) Non-small cell lung cancer. Version 4. http://www.nccn.org. Published 2016. Accessed September 1, 2016.
- 3. Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus docetaxel in advanced nonsquamous non–small-cell lung cancer. N Engl J Med. 2015;373:1627–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus docetaxel in advanced squamous-cell non–small-cell lung cancer. N Engl J Med. 2015;373:123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Garon EB, Rizvi NA, Hui R, et al. ; KEYNOTE-001 Investigators. Pembrolizumab for the treatment of non–small-cell lung cancer. N Engl J Med. 2015;372:2018–2028. [DOI] [PubMed] [Google Scholar]
- 6. Hung JJ, Jeng WJ, Hsu WH, et al. Prognostic factors of postrecurrence survival in completely resected stage I non-small cell lung cancer with distant metastasis. Thorax. 2010;65:241–245. [DOI] [PubMed] [Google Scholar]
- 7. Sugimura H, Nichols FC, Yang P, et al. Survival after recurrent non–small-cell lung cancer after complete pulmonary resection. Ann Thorac Surg. 2007;83:409–417. [DOI] [PubMed] [Google Scholar]
- 8. Milano MT, Katz AW, Okunieff P. Patterns of recurrence after curative-intent radiation for oligometastases confined to one organ. Am J Clin Oncol. 2010;33:157–163. [DOI] [PubMed] [Google Scholar]
- 9. Consonni D, Pierobon M, Gail MH, et al. Lung cancer prognosis before and after recurrence in a population-based setting. J Natl Cancer Inst. 2015;107:djv059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rodríguez-Antona C, Taron M. Pharmacogenomic biomarkers for personalized cancer treatment. J Intern Med. 2015;277:201–217. [DOI] [PubMed] [Google Scholar]
- 11. al Moustafa AE, Alaoui-Jamali M, Paterson J, O’Connor-McCourt M. Expression of P185erbB-2, P160erbB-3, P180erbB-4, and heregulin alpha in human normal bronchial epithelial and lung cancer cell lines. Anticancer Res. 1999;19:481–486. [PubMed] [Google Scholar]
- 12. Bièche I, Onody P, Tozlu S, et al. Prognostic value of ERBB family mRNA expression in breast carcinomas. Int J Cancer. 2003;106:758–765. [DOI] [PubMed] [Google Scholar]
- 13. Müller-Tidow C, Diederichs S, Bulk E, et al. Identification of metastasis-associated receptor tyrosine kinases in non-small cell lung cancer. Cancer Res. 2005;65:1778–1782. [DOI] [PubMed] [Google Scholar]
- 14. Tanner B, Hasenclever D, Stern K, et al. ErbB-3 predicts survival in ovarian cancer. J Clin Oncol. 2006;24:4317–4323. [DOI] [PubMed] [Google Scholar]
- 15. Yi ES, Harclerode D, Gondo M, et al. High c-erbB-3 protein expression is associated with shorter survival in advanced non-small cell lung carcinomas. Mod Pathol. 1997;10:142–148. [PubMed] [Google Scholar]
- 16. Xia W, Gerard CM, Liu L, Baudson NM, Ory TL, Spector NL. Combining lapatinib (GW572016), a small molecule inhibitor of ErbB1 and ErbB2 tyrosine kinases, with therapeutic anti-ErbB2 antibodies enhances apoptosis of ErbB2-overexpressing breast cancer cells. Oncogene. 2005;24:6213–6221. [DOI] [PubMed] [Google Scholar]
- 17. Sergina NV, Rausch M, Wang D, et al. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature. 2007;445:437–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Baselga J, Swain SM. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer. 2009;9:463–475. [DOI] [PubMed] [Google Scholar]
- 19. Garrett JT, Olivares MG, Rinehart C, et al. Transcriptional and posttranslational up-regulation of HER3 (ErbB3) compensates for inhibition of the HER2 tyrosine kinase. Proc Natl Acad Sci U S A. 2011;108:5021–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Motoyama AB, Hynes NE, Lane HA. The efficacy of ErbB receptor-targeted anticancer therapeutics is influenced by the availability of epidermal growth factor-related peptides. Cancer Res. 2002;62:3151–3158. [PubMed] [Google Scholar]
- 21. von Pawel J, Tseng J, Dediu M, et al. Patritumab with erlotinib in advanced, EGFR treatment-naive non–small cell lung cancer: phase Ib/II HERALD trial. J Clin Oncol. 2014;32:8045. [Google Scholar]
- 22. Freeman D, Ogbagabriel S, Rothe M, Radinsky R, Treder M. Fully human anti-HER3 monoclonal antibodies (mAbs) have unique in vitro and in vivo functional and antitumor activities versus other HER family inhibitors. Cancer Res. 2008;68:LB-21. [Google Scholar]
- 23. Treder M, Hartmann S, Ogbagabriel S, et al. Fully human anti-HER3 monoclonal antibodies (mAbs) inhibit oncogenic signaling and tumor cell growth in vitro and in vivo. Cancer Res. 2008;68:LB-20. [Google Scholar]
- 24. Treder M, Ogbagabriel S, Moor R, et al. Fully human anti-HER3 mAb U3-1287 (AMG 888) demonstrates unique in vitro and in vivo activities versus other HER family inhibitors in NSCLC models. EJC Suppl. 2008;12:99. [Google Scholar]
- 25. Schneider M, Blum S, Wenzl C, et al. Heregulin expression level as a predictive biomarker for patritumab efficacy. J Clin Oncol. 2014;32:2618. [Google Scholar]
- 26. Yonesaka K, Kawakami H, Kaneda H, et al. The expression level of HER3 ligand heregulin mRNA as a predictive biomarker for anti-HER3 antibody patritumab combined with erlotinib in non-small cell lung cancer. J Clin Oncol. 2014;32:e19082. [Google Scholar]
- 27. Dietrich D, Uhl B, Sailer V, et al. Improved PCR performance using template DNA from formalin-fixed and paraffin-embedded tissues by overcoming PCR inhibition. PLoS ONE. 2013;8:e77771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shames DS, Carbon J, Walter K, et al. High heregulin expression is associated with activated HER3 and may define an actionable biomarker in patients with squamous cell carcinomas of the head and neck. PLoS ONE. 2013;8:e56765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mendell J, Freeman DJ, Feng W, et al. Clinical translation and validation of a predictive biomarker for patritumab, an anti-human epidermal growth factor receptor 3 (HER3) monoclonal antibody, in patients with advanced non-small cell lung cancer. eBioMedicine. 2015;2:264–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Travis WD, Rekhtman N. Pathological diagnosis and classification of lung cancer in small biopsies and cytology: strategic management of tissue for molecular testing. Semin Respir Crit Care Med. 2011;32:22–31. [DOI] [PubMed] [Google Scholar]
- 31. Wang F, Flanagan J, Su N, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14:22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Beckman RA, Clark J, Chen C. Integrating predictive biomarkers and classifiers into oncology clinical development programmes. Nat Rev Drug Discov. 2011;10:735–748. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.