Figure 1. Subunit interactions within GluN1/2A/2B triheteromeric NMDARs.
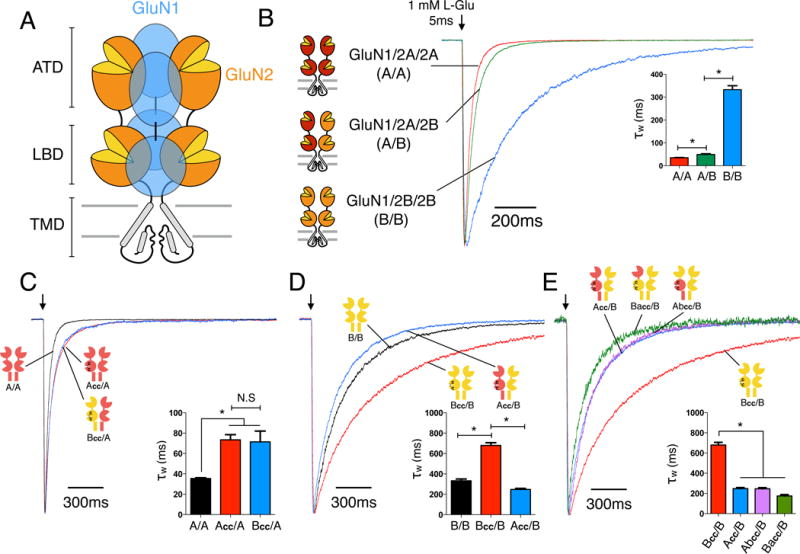
(A) Cartoon of the heterotetrameric NMDAR structure with three distinctive layers formed by the amino-terminal domains (ATDs), ligand binding domains (LBDs), and transmembrane domains (TMDs). The majority of NMDARs are assembled from two glycine-binding GluN1 and two glutamate-binding GluN2 subunits. Diheteromeric NMDARs are assembled from GluN1 and one type of GluN2 subunit, such as GluN1/2A (A/A) and GluN1/2B (B/B), whereas triheteromeric NMDARs are assembled from GluN1 and two different GluN2 subunits, such as GluN1/2A/2B (A/B). (B) Deactivation kinetics of diheteromeric A/A, B/B and triheteromeric A/B NMDARs in response to a 5ms pulse of 1mM L-glutamate pulse indicated by the arrow (* indicates significantly different, p<0.05, one-way ANOVA with Tukey posttest). (C) Deactivation kinetics of A/A receptors and NMDARs containing one wildtype GluN2A and one mutant subunit with crosslinked LBD (Acc/A or Bcc/A). Crosslinked LBDs are generated by introducing two cysteine mutations (GluN2A: K487C + N687C; GluN2B: K488C + N688C;) which are designed to mimic a “constitutively liganded” state. (D) Deactivation kinetics of B/B, Bcc/B and Acc/B receptors. (E) Deactivation kinetics of receptors with one wildtype GluN2B and one crosslinked chimeric subunit with the ATD interchanged between GluN2A and GluN2B (Abcc/B or Bacc/B), compared to those of Bcc/B and Acc/B. All current responses are normalized to their peaks. See also Figures S1–S4.
