Abstract
The concept of protein intrinsic disorder has taken the driving seat to understand regulatory proteins in general. Reports suggest that in mammals nearly 75% of signalling proteins contain long disordered regions with greater than 30 amino acid residues. Therefore, intrinsically disordered proteins (IDPs) have been implicated in several human diseases and should be considered as potential novel drug targets. Moreover, intrinsic disorder provides a huge multifunctional capability to hub proteins such as c-Myc and p53. c-Myc is the hot spot for understanding and developing therapeutics against cancers and cancer stem cells. Our past understanding is mainly based on in vitro and in vivo experiments conducted using c-Myc as whole protein. Using the reductionist approach, c-Myc oncoprotein has been divided into structured and disordered domains. A wealth of data is available dealing with the structured perspectives of c-Myc, but understanding c-Myc in terms of disordered domains has just begun. Disorderness provides enormous flexibility to proteins in general for binding to numerous partners. Here, we have reviewed the current progress on understanding c-Myc using the emerging concept of IDPs.
Keywords: Intrinsically disordered proteins, transcription factors, conformational ensembles, therapeutics and drug development, and molecular recognition elements
Introduction
Proteins are the vital effectors of biological system whose functional diversity is responsible for their multifaceted role in different biological processes. Versatility of function arises from heterogeneous structure.1 Conventionally, it is a well-established paradigm that a protein function is dependent on folded 3-dimensional (3D) structure of its polypeptide chain.2,3 However, recent research has evidenced that protein function is not solely dependent on folded conformation, but there are regions of amino acid sequences or patches of sequences which are unable to fold into tertiary structures.4 These regions are called intrinsically disordered regions (IDRs), and proteins are named intrinsically disordered proteins (IDPs). The term ‘disordered’ has been used by Jirgensons5 to describe poorly structured regions while classifying proteins on the basis of conformations.4 Several studies have predicted the abundance of IDRs or IDPs in all forms of life, from viruses to bacteria to higher eukaryotes.6 In a recent study, we have also predicted the prevalence of intrinsic disorder in complete proteome of Zika virus.7
Several computational and experimental approaches have been used to predict the disorder in proteins, and it has been estimated that most of the regulatory and signalling proteins contain a larger fraction of disorder regions.4,8 It is reported that more than one-third of human proteome is disordered with nearly 75% of regulatory proteins falling under this category.4,9 Intrinsically disordered proteins have been differentiated from structured proteins in several aspects, such as amino acid sequence composition, charge, hydrophobicity, flexibility, and aromaticity.10 The IDPs/IDRs mainly contain abundant charged amino acids that show disorder-promoting propensity.10,11 This sequence biasness has been used to develop several disorder predictors, such as PONDER (predictor of naturally disordered region), IUPred, GlobPlot, SPRITZ, DisoPred, and DisEMBL.12-15 These predictors have been used to analyse the frequency of intrinsic disorder in 3 kingdoms of life.14 It has been observed that eukaryotic proteome has high prevalence of intrinsic disorder, relative to bacteria and archaea. Furthermore, analysis of eukaryotic proteome has shown that most of the disordered proteins are located in nucleus and belong to the family of transcription factors (TFs) and cell-signalling proteins.14 Similarly, a study has been conducted to find the pattern of conserved protein disorder among different protein families in all kingdoms of life. Interestingly, it has been observed that viruses and eukaryotes have the highest prevalence of long regions of conserved protein disorder than bacteria and archaea.16 Similarly, in humans, a study has been conducted in which all disease-associated proteins were retrieved and analysed for the prevalence of intrinsic disorder.17 It has been found that nearly 70% of signalling and disease-associated proteins show abundance of intrinsic disorder with long disordered regions. Furthermore, imbalance in the regulation of IDPs has been shown to associate with the onset of several diseases such as cancer, cardiovascular disease, amyloidosis, neurodegenerative diseases, and diabetes.17
Intrinsic disorder provides structural plasticity and functional diversity to perform multiple interactions simultaneously. Under physiological conditions, IDPs lack 3D structures.18 These IDPs show peculiar folding behaviour than ordered proteins, which is not yet fully understood. However, some IDPs can undergo disorder-to-order transitions after binding to a partner, and interestingly, it has been reported that still there is significant amount of disorder preserved by these proteins in the bound state.19 It is believed that these disorder-to-order transitions or coupled folding and binding mechanisms are responsible for providing structural plasticity to IDPs.20,21 Free-energy landscape studies have clearly demonstrated the conformational plasticity of IDPs.22 The comparison of energy landscapes has shown rugged funnel-like model for IDPs with respect to ordered proteins.23 This also signifies the presence of large number of isoenergetic conformations in IDPs.22 Conformational heterogeneity of IDPs has been studied by several techniques, such as nuclear magnetic resonance (NMR), fluorescence anisotropy, tryptophan quenching, and fluorescence correlation spectroscopy.24 However, NMR techniques have been mostly used and are well suited for studying IDP structural dynamics.25 Several new developments have been made in NMR methods, such as paramagnetic relaxation enhancements (PREs) or residual dipolar couplings (RDCs), and 13C direct experiments. Data obtained from all these NMR methods have been further processed through computational tools to produce ensemble conformations of IDPs under different functional conditons.25
There are several IDP examples that use the structural plasticity provided by intrinsic disorder to perform multiple interactions. For example, adenoviral early region protein E1A has been seen to form ternary complex such as E1A-CBP-pRb with the help of intrinsic disorder character possessed by E1A.26 This multicomplex formation has also been found to show allosteric modulation. In case of activator for thyroid hormone and retinoid receptors (ACTR) and nuclear co-activator binding domain (NCBD) interaction model, a complex mechanism of binding has been seen in which conformational selection and subsequent induced folding upon binding were observed.27 There is another example of structural plasticity named ‘fly-casting’ mechanism that was used to describe fast kinetics of coupled binding and folding of IDPs. This mechanism was well documented by studying binding kinetics between phosphorylated kinase–inducible domain and kinase-inducible interacting domain (KIX) of c-AMP response element binding proteins.28 In another study, the mechanism of IDP folding upon binding was elucidated where c-Myb transactivation–disordered domain has been seen to acquire alpha-helical structure upon binding to KIX partner.21,29,30
There are several examples in the literature that have shown that unlimited structural flexibilities of IDPs correlated with multiple protein-protein interactions. Transcription factors are one of the types of proteins in eukaryotic proteome that act as central hub to regulate gene expression.31 In further part of the review, we shed light on IDP perspective of TFs and role of c-Myc as disordered protein TF.
TFs as IDPs
Transcriptional regulation is an important biological function to perform controlled gene expression by regulatory proteins named TFs. Transcription factors regulate the transcription of genes (positively or negatively) by binding to the target sequence on DNA and can also bind to other co-activator proteins. These multiple interactions of TFs have been associated with their IDRs.32 Disorder prediction studies have shown that there are more than 49% of intrinsic disorders in human TFs.32 Eukaryotic TFs have shown greater disordered character over prokaryotic TFs. Several human diseases, such as cancer, diabetes, and autoimmune, have been found to associate with deregulation of TFs.33 Deregulation of multiple TFs has been reported in cancer progression. Extensively studied TFs which have shown a major role in progression of cancer phenotypes are p53 and c-Myc proteins.32,33 In this review, we discuss how c-Myc disordered regions play a crucial role in controlling gene expression, disease development, and several therapeutic strategies available at present.
Myc proteins are TFs that serve as central regulators of several physiological processes, such as apoptosis, cell proliferation, differentiation, metabolism, and biosynthesis of proteins. The Myc protein genes are classified as c-myc, N-myc, and L-myc protooncogenes. Among all, c-myc has been shown to involve in different pathways significantly. Due to their involvement in several human and animal cancers, these genes have gained a greater consideration by the researchers.34,35 Myc proteins induce a cellular response after binding to the DNA of target genes. c-Myc expression is tightly regulated by various ligand-associated receptor signalling under normal conditions. Almost 50% of human cancers show deregulation and activation of c-Myc, and it has been observed that a 2-fold rise in c-Myc expression may affect the cell cycle progression that ultimately leads to cancer.36 In case of the Burkitt lymphoma, translocation of immunoglobulin gene with c-myc gene causes overexpression of c-Myc, and amplification of gene leads to development of hepatocellular carcinoma in humans.36
A remarkable function of c-Myc protein has been identified where overexpression leads to the reprogramming of somatic cell into pluripotent stem cell.37 Hence, these proteins are documented as an important factor for maintenance of undifferentiated state of stem cells. It has been shown that in normal cells inadequate expression of c-Myc induces apoptosis.
It has been reported that protein disorder patterns are conserved in Myc proteins during evolution instead of amino acid sequences.38 Interestingly, these findings become more significant as studies have shown that disordered regions in proteins are more prone to amino acid substitutions which are correlated with Darwinian adaptation under positive selection.39 Disorder prediction studies show that the C-terminal region (410-490) of c-Myc is the most conserved region in protein disorder pattern.40 Similarly, the N-terminal region is unstructured in the absence of binding partner, and ANCHOR prediction algorithm has mapped several eukaryotic linear motifs (ELM) on this region.38 These ELMs represent short amino acid sequences that provide an interaction site for other proteins which are involved in several posttranslational modifications.41 Together with computational and experimental investigations, it is clear that c-Myc extensively uses its disorder regions to perform diverse interactions.
In this mini-review, we try to summarize the importance of protein intrinsic disorder in c-Myc functioning and also emphasize the concept of disorder-based drug designing as a new strategy that could greatly benefit current cancer therapeutics.
C-Myc Domain Architecture: Role in Folding and Binding
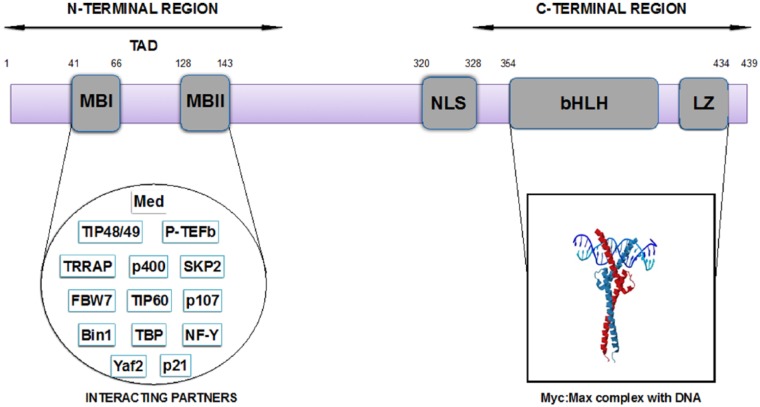
These diverse functions of c-Myc proteins are attributed to their heterogeneous structure (Figure 1). It contains a C-terminal domain which accounts for dimerization during DNA binding and an N-terminal transactivation domain (TAD) which empowers critical transcriptional and cellular transforming functions.38,42 C-terminal being an important domain for its DNA binding regulatory activity contains a basic helix-loop-helix-leucine-zipper (bHLH-ZIP) motif (residues 354-454) that helps in the formation of heterodimer along with Myc-associated protein X (MAX). This domain is partially folded which gains full structure when interacting with the bHLH-ZIP region of the MAX protein. c-Myc homodimers have not been reported in the literature. However, c-Myc and MAX heterodimers have shown a critical interaction which is required for DNA binding activity.38 This heterodimer binds to the target DNA at E-box (enhancer box). It has been investigated that DNA binding of c-Myc:Max heterodimer is through DNA looping mechanism in which dimers of heterodimers are bounded to sequentially arranged E-boxes.43
Figure 1.
Domain architecture of c-Myc protein and the interacting partners’ network. This figure shows that c-Myc protein residues have different functional characteristics: N-terminal region serves as transactivation domain (TAD) by performing multiple interactions with several interacting partners and C-terminal region forms heterodimer with MAX protein that ultimately performs DNA binding activities. These 2 regions are further divided into small motifs: MB1 (Myc Box 1) and MB2 (Myc Box 2) present in TAD domain (N-terminal) and BHLH (basic helix-loop-helix) and LZ (leucine zipper) present at C-terminal. NLS, Nuclear localization Signal or Sequence.
There are about 15% of genes coded by RNA pol 2 which are directly regulated by c-Myc.44 Diverse interactions of c-Myc are contributed to its N-terminal TAD.45 This domain is responsible for tight regulation of c-Myc levels inside the cell. It has Myc Box 1 (MB1) which contains site for proteasome-mediated degradation, and this region acts as hot spot for different mutations, which results in the development of cancer phenotypes.
c-Myc is an IDP which attains ordered structure only after binding to its disordered partner MAX protein. Bioinformatics and experimental approaches have been used to study the disordered regions of Myc proteins, and it has been predicted that c-Myc protein has less tendency to form structures. According to disorder prediction algorithm, it has been estimated that c-Myc contains more than 45% of residues which has propensity for disordered structure formation.9 Nuclear magnetic resonance studies of c-Myc disordered region (1-88) have attributed to its functional plasticity and multiprotein complex formation ability.46 The N-terminal domain spans residues 1 to 167 and links to the C-terminal region by a flexible linker. This TAD region is the central hub of all transactivation activities regulated through binding to several proteins.47,48 It contains 2 highly conserved regions spanning residues 41 to 66 and 28 to 143; these regions are named MB1 and Myc box 2 (MB2), respectively. It has been identified that these Myc boxes are important sites for recognizing c-Myc by several regulatory proteins. Importance of Myc boxes in transforming and proliferation activities have been proved by site directed mutagenesis experiments, though the actual biophysical mechanism of c-Myc-mediated transactivation is still not clear because of little information about structure of TAD region. However, it has been established that TAD region spanning residues 1 to 143 is needed to perform neoplastic transformations, differentiation, and apoptosis activities.43,46 Since c-Myc is a central hub protein in several cancers, therefore investigation of biophysical aspects would provide significant understanding towards diverse protein interactions.29 To understand this aspect, a recent NMR study has been done to map the structural dynamics of c-Myc in the presence of bin-1 protein. It was observed that unlike most of the disordered proteins, c-Myc has not shown folding after binding mechanisms; instead, it acquired disordered state throughout with little transient structured region.46
Mutational Aspects of Intrinsic Disorder in c-Myc
Over the past decades, only structured proteins were considered to show the impact of disease-associated mutations.49 Several structure-based methods have been well established to characterize single-nucleotide polymorphism (SNPs) associated with diseases. Similarly, in previous studies, mutations in disordered regions were considered to be nondamaging or neutral. However, with the advent of intrinsic disorder concept and its relation to disease, studies have emphasized the damaging consequences of mutations in disordered regions.49 c-Myc protein level is tightly regulated inside the cells. The N-terminal region is responsible for its degradation through proteasome-mediated pathways. However, mutations in the N-terminal region hamper its degradation and increased level associated with the development of lymphomas.50 Deletion-mapping experiments have indicated varying transforming potentials of N-terminal mutants. In more than 60% of tumours, most of the mutations are found in the N-terminal region. Several studies have reported the presence of 2 predominant (P57S and T58I) naturally occurring mutations in most of the tumours. Proline-rich region spanning residues 57 to 64 contains high mutational frequency than rest of the protein. This region is essential for phosphorylation, and the mutations in this region hamper the phosphorylation and dephosphorylation processes. Thr58 phosphorylation site is considered to be a hotspot mutation and is found to enhance the transforming ability of c-Myc. However, the exact mechanism of how phosphorylation regulates the transforming activity of c-Myc is not yet known, but it has been found that T58 mutants have reduced degradation through proteasome-mediated pathways.50,51
Therapeutic Target and Drug Development
Traditional drug design approaches are mainly focused on targeting structured active site region of the proteins. Several successful drugs have been developed by targeting active sites of enzymes.52 Most of the inhibitor molecules modulate the enzyme function by competitive or noncompetitive inhibition. A report suggests that 65% of marketed drugs follow the mechanism of inhibition either by targeting enzyme active site or by showing structural similarity with the substrate.53 In addition, recent studies have shown a significant understanding towards targeting protein-protein interactions for inhibition. The main rationale behind targeting protein interactions is related to the energy of protein-protein interaction that concentrates in smaller regions called hot spots, and these regions could be blocked by small molecules effectively.54 However, there are several challenges in targeting protein-protein interactions that limit the application of this approach, but still this approach may provide a new insight into the direction of developing a more specific drug against target.55 Similar challenges are further posed by evolving concept of IDPs because these IPDs are ‘protein clouds’ that lack 3D structure and show conformational heterogeneity where rational drug design approaches seem to fail.52 Therefore, IDPs demand a novel disorder-based approach for drug development.
In recent years, a significant understanding has been developed for the mechanism of IDP-mediated protein-protein interactions. It has been evidenced that small molecules play a major role in modulating protein-protein interactions, as, in case, one of the protein regions is disordered which becomes ordered upon binding to structured partner.55 Because several proteins that play a critical role in the disease process are IDPs, targeting their IDRs that facilitate their myriad interactions may represent a novel strategy to develop new therapeutics.
To target IDRs, a strategy has been developed in which small molecules are being used which can bind to the molecular recognition elements or features (MoREs/MoRFs) of IDP and thus can stabilize its disordered state which ultimately leads to the inhibition of protein interaction with structured partner.55 Molecular recognition elements are the sequences that are involved in the interaction of IDPs with other partners, and these are characterized by analysing the type of conformation adopted by the complex during binding. These MoRFs have been classified on the basis of acquired structure after interaction with the binding partner as follows: α-MoRFs, β-MoRFs, and ι-MoRFs.56 These MoRFs have been used to screen binding partners which can be used as inhibitors in drug designing.54,56 In case of c-Myc, 3 binding sites have been characterized within the 85-amino-acid–disordered bHLH-ZIP domain.57 It has been observed that first binding site was located between amino acid residues 402 and 409, and this site was predicted to contain disorder-promoting amino acid residues. Similarly, other 2 binding sites correspond to amino acid residues 366 to 375 and 375 to 385, respectively, and have been reported to show considerable structural pasticity.57
Reports suggest that c-Myc:MAX interactions are important targets for anticancer therapy. Studies on animal mouse models have shown that c-Myc inhibition can completely stop tumour growth and can also inhibit cancer stem cell progression.31 However, it is very difficult to target IDPs. It was suggested that there are 2 possible binding modes of inhibition to c-Myc:Max. Inhibitor can bind to the nonfunctional conformation of c-Myc (370-409), preventing Myc:Max interaction, whereas in the other mode, the inhibitor will mimic the Max structure and bind to the c-Myc competitively.59 A mass spectrometry–based method was developed to characterize different binding modes of synthetic inhibitors, and it was observed that most of the promising inhibitors were following first mode of binding to the c-Myc, which was named trap mode.31 Peptidomimetic screening of compound libraries has yielded 2 molecules, IIA6B17 and IIA4B20, which were found to disrupt initial formation of Myc:MAX dimers. Subsequently, other molecules, mycmycin-1 and mycmycin-2, were derived from previous molecules to improve the specificity of the inhibition.33
Using high-throughput assays, several small-molecule inhibitors have been reported previously which have shown to inhibit Myc:Max dimerization (Figure 3). However, recent studies have developed small molecules (10058-F4 and 10074-G5) which have shown binding affinities to the disordered region of c-Myc. Interestingly, it was investigated that those molecules binding to the c-Myc have maintained the disordered state in its monomeric form.59 As described in Figure 2C, 10074-A4 inhibitor has been shown to bind to c-Myc370-409 at different sites by stabilizing the disordered state of c-Myc as dynamic ensemble.60
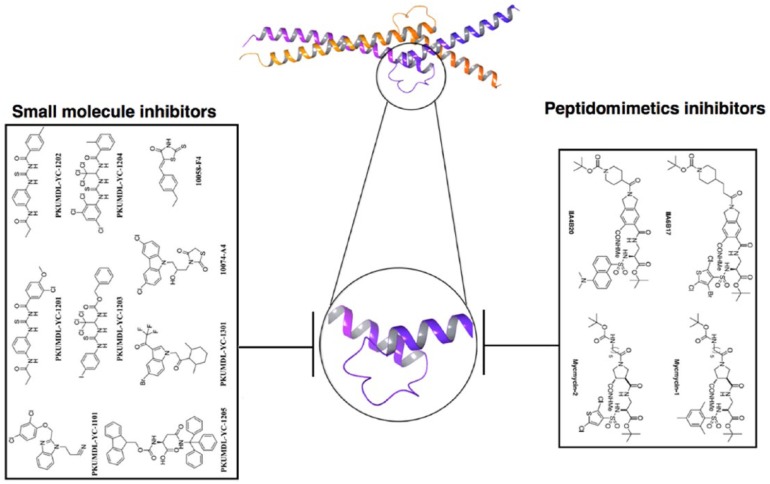
Figure 3.
Myc:Max dimer–associated peptidomimetic and small-molecule inhibitors. This figure represents the C-terminal disordered loop (purple enlarged in circle) in leucine zipper of c-Myc that has been targeted by peptidomimetic compound inhibitors, such as IIA6B17 and IIA4B20, and their analogues, mycmycin-1 and mycmycin-2. In addition, further effective small-molecule inhibitors, such as 10058-F4, 10074-G5, PKUMDL-YC-1101, PKUMDL-YC-1201, PKUMDL-YC-1202, PKUMDL-YC-1203, PKUMDL-YC-1204, PKUMDL-YC-1205, and PKUMDL-YC-1301, were also developed to inhibit Myc:Max dimer formation.
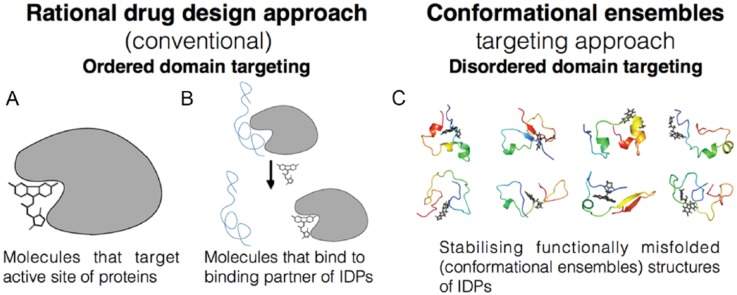
Figure 2.
Various drug development strategies showing the origin of disorder-based drug targeting. (A) This cartoon represents the conventional method of structure-based drug design and targeting catalytic active site of structured proteins using small inhibitor molecules (black inhibitor molecule shown here is just symbolic). (B) This shows the strategy to inhibit protein-protein interactions between disordered protein (blue) and its interacting partner (grey) by the inhibitor molecule (black symbolic) that can bind to the ordered structure of interacting partner. (C) This figure shows the stabilizing function of small-molecule inhibitor which binds to ensemble states of disordered proteins in different conformations and inhibits the functional ability. IDPs indicate intrinsically disordered proteins.
Reproduced with permission from Jin et al (2013)58, representing the complex formation between conformational ensembles of c-Myc370-409/10074-A4 inhibitor molecule using molecular dynamic simulations).
Seven compounds have been discovered recently using computational virtual screening based on small-molecule inhibitor approach to bind multiple conformations of IDPs. The screening was done on the basis of previous history of small-molecule binding pockets between residues 370 and 409 of c-Myc C-terminal domain. Seven Myc-Max–specific low-molecular-weight inhibitors (PKUMDL-YC-1101, PKUMDL-YC-1201, PKUMDL-YC-1202, PKUMDL-YC-1203, PKUMDL-YC-1204, PKUMDL-YC-1205, and PKUMDL-YC-1301) were found to be directly targeting the disordered bHLH-LZ domain of c-Myc and have shown good binding strength.61 Four compounds (as shown in Figure 3) have shown inhibitory action on HL-60 cell growth in cancer cell–based assays.
Recently, a new class of direct Myc:MAX inhibitors have been reported where the inhibitor mimicks the alpha helix of c-Myc and shows inhibition by disrupting the binding of Myc:MAX complex to E-box without disturbing the association of dimers.62
Taken together, all these studies provide a significant understanding towards disorder-based drug targeting. However, the field is challenging and still needs to be explored extensively. Despite above challenges, c-Myc:MAX system appears to be promising for potential inhibitor discovery and further development of disorder-based drug targeting approach.
Conclusions
Till date, most of the inhibitors have been designed to inhibit Myc:Max interaction which leads to complete blockage of signalling pathway. However, c-Myc is required for normal somatic cell growth and differentiation, so its complete inhibition may lead to the death of normal cells along with cancer cells. Most of the available inhibitors of c-Myc target C-terminal region only. Therefore, to target c-Myc more specifically, it is needed to look upon its alternate interaction network which is governed by the TAD region. The TAD region is intrinsically disordered which interacts with several regulatory proteins which are responsible for modulating c-Myc activity. However, it could be a safe and specific therapeutic target to modulate c-Myc function. To accomplish this task, it is needed to characterize biophysical and structural aspects of c-Myc TAD region which are not yet fully discovered.
Footnotes
Peer review:Five peer reviewers contributed to the peer review report. Reviewers’ reports totalled 1075 words, excluding any confidential comments to the academic editor.
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was partially supported by DST grant, India (YSS/2015/000613), to RG and IIT Mandi, India, to RG. DK is supported by ICMR fellowship and NS is supported by MHRD fellowship, both in India.
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: Conception and design, writing and review of the manuscript and study supervision: RG. Writing of the manuscript: DK and NS.
References
- 1. Uversky VN. Functional roles of transiently and intrinsically disordered regions within proteins. FEBS J. 2015;282:1182–1189. [DOI] [PubMed] [Google Scholar]
- 2. Van Der Lee R, Buljan M, Lang B, et al. Classification of intrinsically disordered regions and proteins, Chem Rev. 2014;114:6589–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chothia C, Lesk AM. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986;5:823–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Oldfield CJ, Dunker AK. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem. 2014;83:553–584. [DOI] [PubMed] [Google Scholar]
- 5. Jirgensons B. Classification of proteins according to conformation: Die Makromol. Chemie 1966;91:74–86. [Google Scholar]
- 6. Forman-Kay JD, Mittag T. From sequence and forces to structure, function, and evolution of intrinsically disordered proteins. Structure. 2013;21:1492–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Giri R, Kumar D, Sharma N, Uversky VN. Intrinsically disordered side of the Zika virus proteome. Front Cell Infect Microbiol. 2016;6:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dunker AK, Oldfield CJ, Meng J, et al. The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genomics. 2008;9:S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008;18:756–764. [DOI] [PubMed] [Google Scholar]
- 10. Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. [DOI] [PubMed] [Google Scholar]
- 11. Habchi J, Tompa P, Longhi S, Uversky VN. Introducing protein intrinsic disorder. Chem Rev. 2014;114:6561–6588. [DOI] [PubMed] [Google Scholar]
- 12. Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim Biophys Acta. 2010;1804:996–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Linding R, Russell RB, Neduva V, Gibson TJ. GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31:3701–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. [DOI] [PubMed] [Google Scholar]
- 15. Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11:1453–1459. [DOI] [PubMed] [Google Scholar]
- 16. Chen JW, Romero P, Uversky VN, Dunker AK. Conservation of intrinsic disorder in protein domains and families: I. A database of conserved predicted disordered regions. J Proteome Res. 2006;5:879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cheng Y, LeGall T, Oldfield CJ, et al. Rational drug design via intrinsically disordered protein. Trends Biotechnol. 2006;24:435–442. [DOI] [PubMed] [Google Scholar]
- 18. Uversky VN, Dunker AK. Understanding protein non-folding. Biochim Biophys Acta. 2010;1804:1231–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci. 2008;33:2–8. [DOI] [PubMed] [Google Scholar]
- 20. Dogan J, Mu X, Engström Å, et al. The transition state structure for coupled binding and folding of disordered protein domains. Sci Rep. 2013;3:6573–6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Toto A, Camilloni C, Giri R, et al. Molecular recognition by templated folding of an intrinsically disordered protein. Sci Rep. 2016;6:21994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Uversky VN. Unusual biophysics of intrinsically disordered proteins. Biochim Biophys Acta. 2013;1834:932–951. [DOI] [PubMed] [Google Scholar]
- 23. Brunori M, Gianni S, Giri R, Morrone A, Travaglini-Allocatelli C. Morphogenesis of a protein: folding pathways and the energy landscape. Biochem Soc Trans. 2012;40:429–432. [DOI] [PubMed] [Google Scholar]
- 24. Gibbs EB, Showalter SA. Quantitative biophysical characterization of intrinsically disordered proteins. Biochemistry. 2015;54:1314–1326. [DOI] [PubMed] [Google Scholar]
- 25. Kosol S, Contreras-Martos S, Cedeño C, Tompa P. Structural characterization of intrinsically disordered proteins by NMR spectroscopy. Molecules. 2013;18:10802–10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ferreon ACM, Ferreon JC, Wright PE, Deniz AA. Modulation of allostery by protein intrinsic disorder. Nature. 2013;498:390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jemth P, Mu X, Engström Å, Dogan J. A frustrated binding interface for intrinsically disordered proteins. J Biol Chem. 2014;289:5528–5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang Y, Liu Z. Kinetic advantage of intrinsically disordered proteins in coupled folding-binding process: a critical assessment of the ‘fly-casting’ mechanism. J Mol Biol. 2009;393:1143–1159. [DOI] [PubMed] [Google Scholar]
- 29. Giri R, Morrone A, Toto A, Brunori M, Gianni S. Structure of the transition state for the binding of c-Myb and KIX highlights an unexpected order for a disordered system. Proc Natl Acad Sci U S A. 2013;110:14942–14947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Toto A, Giri R, Brunori M, Gianni S. The mechanism of binding of the KIX domain to the mixed lineage leukemia protein and its allosteric role in the recognition of c-Myb. Protein Sci. 2014;23:962–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sammak S, Zinzalla G. Targeting protein-protein interactions (PPIs) of transcription factors: challenges of intrinsically disordered proteins (IDPs) and regions (IDRs). Prog Biophys Mol Biol. 2015;119:41–46. [DOI] [PubMed] [Google Scholar]
- 32. Dunker AK, Uversky VN. Drugs for ‘protein clouds’: targeting intrinsically disordered transcription factors. Curr Opin Pharmacol. 2010;10:782–788. [DOI] [PubMed] [Google Scholar]
- 33. Uversky VN, Davé V, Iakoucheva LM, et al. Pathological unfoldomics of uncontrolled chaos: intrinsically disordered proteins and human diseases. Chem Rev. 2014;114:6844–6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Eilers M, Eisenman RN. Myc’s broad reach. Genes Dev. 2008;22:2755–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lüscher B, Vervoorts J. Regulation of gene transcription by the oncoprotein Myc. Gene. 2012;494:145–160. [DOI] [PubMed] [Google Scholar]
- 36. Yan C, Higgins PJ. Drugging the undruggable: transcription therapy for cancer. Biochim Biophys Acta. 2013;1835:76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Buganim Y, Faddah DA, Jaenisch R. Mechanisms and models of somatic cell reprogramming. Nat Rev Genet. 2013;14:427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mahani A, Henriksson J, Wright APH. Origins of Myc proteins – using intrinsic protein disorder to trace distant relatives. PLoS ONE. 2013;8:e75057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nilsson J, Grahn M, Wright AP, et al. Proteome-wide evidence for enhanced positive Darwinian selection within intrinsically disordered regions in proteins. Genome Biol. 2011;12:R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xue B, Oldfield CJ, Van Y-Y, Dunker AK, Uversky VN. Protein intrinsic disorder and induced pluripotent stem cells. Mol Biosyst. 2012;8:134–150. [DOI] [PubMed] [Google Scholar]
- 41. Dinkel H, Michael S, Weatheritt RJ, et al. ELM – the database of eukaryotic linear motifs. Nucleic Acids Res. 2012;40:D242–D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cole MD, McMahon SB. The Myc oncoprotein: a critical evaluation of transactivation and target gene regulation. Oncogene. 1999;18:2916–2924. [DOI] [PubMed] [Google Scholar]
- 43. Fladvad M, Zhou K, Moshref A, Pursglove S, Säfsten P, Sunnerhagen M. N and C-terminal sub-regions in the c-Myc transactivation region and their joint role in creating versatility in folding and binding. J Mol Biol. 2005;346:175–189. [DOI] [PubMed] [Google Scholar]
- 44. Iakoucheva LM, Brown CJ, Lawson JD, Obradović Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323:573–584. [DOI] [PubMed] [Google Scholar]
- 45. Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–645. [DOI] [PubMed] [Google Scholar]
- 46. Andresen C, Helander S, Lemak A, et al. Transient structure and dynamics in the disordered c-Myc transactivation domain affect Bin1 binding. Nucleic Acids Res. 2012;40:6353–6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sakamuro D, Prendergast GC. New Myc-interacting proteins: a second Myc network emerges. Oncogene. 1999;18:2942–2954. [DOI] [PubMed] [Google Scholar]
- 48. Gianni S, Morrone A, Giri R, Brunori M. A folding-after-binding mechanism describes the recognition between the transactivation domain of c-Myb and the KIX domain of the CREB-binding protein. Biochem Biophys Res Commun. 2012;428:205–209. [DOI] [PubMed] [Google Scholar]
- 49. Vacic V, Iakoucheva LM. Disease mutations in disordered regions – exception to the rule? Mol Biosyst. 2012;8:27–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bahram F, von der Lehr N, Cetinkaya C, Larsson LG. c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood. 2000;95:2104–2110. [PubMed] [Google Scholar]
- 51. Chang DW, Claassen GF, Hann SR, Cole MD. The c-Myc transactivation domain is a direct modulator of apoptotic versus proliferative signals. Mol Cell Biol. 2000;20:4309–4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Uversky VN. Intrinsically disordered proteins and novel strategies for drug discovery. Expert Opin Drug Discov. 2012;7:475–488. [DOI] [PubMed] [Google Scholar]
- 53. Robertson JG. Mechanistic basis of enzyme-targeted drugs. Biochemistry. 2005;44:5561–5571. [DOI] [PubMed] [Google Scholar]
- 54. Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. [DOI] [PubMed] [Google Scholar]
- 55. Metallo SJ. Intrinsically disordered proteins are potential drug targets. Curr Opin Chem Biol. 2010;14:481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vacic V, Oldfield CJ, Mohan A, et al. Characterization of molecular recognition features, MoRFs, and their binding partners. J Proteome Res. 2007;6:2351–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hammoudeh DI, Follis AV, Prochownik EV, Metallo SJ. Multiple independent binding sites for small-molecule inhibitors on the oncoprotein c-Myc. J Am Chem Soc. 2009;131:7390–7401. [DOI] [PubMed] [Google Scholar]
- 58. Jin F, et al. Ligand Clouds around Protein Clouds: A Scenario of Ligand Binding with Intrinsically Disordered Proteins. PLoS Comput. Biol. 2013;9:e1003249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Follis AV, Hammoudeh DI, Wang H, Prochownik EV, Metallo SJ. Structural rationale for the coupled binding and unfolding of the c-Myc oncoprotein by small molecules. Chem Biol. 2008;15:1149–1155. [DOI] [PubMed] [Google Scholar]
- 60. Jin F, Yu C, Lai L, et al. Ligand clouds around protein clouds: a scenario of ligand binding with intrinsically disordered proteins. PLoS Comput Biol. 2013;9:e1003249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yu C, Niu X, Jin F, Liu Z, Jin C, Lai L. Structure-based inhibitor design for the intrinsically disordered protein c-Myc. Sci Rep. 2016;6:22298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jung K-Y, Wang H, Teriete P, et al. Perturbation of the c-Myc-Max protein-protein interaction via synthetic α-helix mimetics. J Med Chem. 2015;58:3002–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]