Abstract
Leading institutions throughout the country have established Precision Medicine programs to support personalized treatment of patients. A cornerstone for these programs is the establishment of enterprise-wide Clinical Data Warehouses. Working shoulder-to-shoulder, a team of physicians, systems biologists, engineers, and scientists at Rutgers Cancer Institute of New Jersey have designed, developed, and implemented the Warehouse with information originating from data sources, including Electronic Medical Records, Clinical Trial Management Systems, Tumor Registries, Biospecimen Repositories, Radiology and Pathology archives, and Next Generation Sequencing services. Innovative solutions were implemented to detect and extract unstructured clinical information that was embedded in paper/text documents, including synoptic pathology reports. Supporting important precision medicine use cases, the growing Warehouse enables physicians to systematically mine and review the molecular, genomic, image-based, and correlated clinical information of patient tumors individually or as part of large cohorts to identify changes and patterns that may influence treatment decisions and potential outcomes.
Keywords: Clinical data warehouse, precision medicine, semantic interoperability, synoptic pathology reports
Introduction
Leading health care centers across the country now recognize the paramount importance and utility of establishing a clinical data warehouse. In fact, some of the earliest pioneering projects have already crossed the 20-year mark.1–5 Primary benefits that have been realized as a result of these early efforts include cost containment and tracking patient outcomes, providing clinical decision support at point of care, improving prognostic accuracy, and facilitating research and clinical trials.6–8
Despite many positive reports from academic and clinical sites across the country regarding the development of data management and analytical tools, most centers report that the specific solutions that were implemented have notable limitations and flaws.9The most often cited challenges are related to the intrinsic complexity of the underlying biomedical and clinical data—the fact that information exists in both structured and unstructured formats, and the wide range of data ownership and regulatory issues associated with collecting and organizing the data.9 To address these issues, many clinical data warehouse projects required large up-front costs on Information Technology (IT) implementation, which relied, chiefly, on manual data entry. Once the data were migrated into the warehouse, there was often only limited support for performing ad hoc queries and only minimal bioinformatics and computational tools available to enable investigators and physicians to systematically mine and interrogate complex genomic signatures, or to detect and track subtle changes in patient response to therapy and treatment.
As part of our mission as a National Cancer Institute–designated Comprehensive Cancer Center, Rutgers Cancer Institute of New Jersey has made the goal of implementing a clinical data warehouse a high priority to facilitate improvements in prevention, detection, treatment, and care of cancer patients. Working shoulder-to-shoulder, a team of physicians, systems biologists, engineers, and scientists at our Center have designed, developed, and implemented the methods, protocols, and workflows to facilitate the extraction, standardization, and ongoing population of a Clinical Data Warehouse (Warehouse in subsequent text) with information originating from a full range of data sources, including not only the Electronic Medical Record (EMR) system which contains patient visit records, clinical history, physician order entries, and data originating from laboratory results, radiology reports, and pathology reports but also genomic sequencing studies and research-generated data records. Our overarching efforts focus on identifying and consolidating information most crucial to diagnosis, choices in treatment and therapy planning, as well as investigative research.
One of the great challenges when establishing a comprehensive Warehouse lies in the fact that a vast amount of clinical data are found in unstructured or semistructured format. Some of these reports are generated from outside consultations or referring laboratories. At many institutions, such documents are simply scanned into images or PDF and attached to the patient record. Others may arrive via structured vehicles such as HL7 format, while the clinical content of the message is lumped into a continuous ASCII (American Standard Code for Information Interchange) string. These solutions only satisfy rudimentary requirements of foundational interoperability by allowing the information to flow into another Healthcare Information Technology (HIT) system; however, the data cannot be easily interpreted in the destination database. To effectively incorporate this information into the Warehouse and achieve semantic interoperability,10,11 our team has contracted with Extract Systems (Madison, WI), a Wisconsin-based high-tech company, to develop and optimize software that semiautomatically extracts data that would otherwise remain locked in paper-based documents, so that the information can be reliably uploaded into discrete fields for integration with clinical data warehouse repository. Using this technology, our team is able to automatically capture more than 500 data elements, which had previously been trapped in pathology reports and historic gene sequencing reports. Even without human supervision, the software is able to extract, at an accuracy rate of 96.65%, not only the traditional pathological description of gross and microscopic pathologic findings in tumor specimen but also a full set of discrete elements as defined in individual cancer synoptic protocols from the College of American Pathologists,12 including the most critical pathology staging information, which is often missing from the EMR.
Whole exome sequencing and targeted exon sequencing have become the methods of choice for identifying actionable molecular traits of a given tumor. The capacity of a clinical data warehouse to support systematic queries facilitates the process of identifying candidates for new therapies and providing directions for future research. In this article, we demonstrate an automated work flow for incorporating sequencing results from both structured and unstructured sources.
During the course of this project our team has established productive collaborations with several vendors, which has led to the development of software that allows integration of radiology imaging studies and digitized pathology specimens within the Warehouse. The colocalization of correlated data elements, which represent the full spectrum of clinical information, imaging studies, and genomic information, coupled with our experience and expertise in advanced pattern recognition, high-performance computing, and data mining, has placed our team in an excellent position to optimize personalized treatment, refine best practices, and provide objective, reproducible insight as to the underlying mechanisms of disease onset and progression.
Methods
Governance
To support the planning and development of the Warehouse, a Data Governance Council has been established to provide clinical insight and guide the methods used for gathering, accessing, and sharing the data. The Council is led by a cross-section of physicians, scientists, administrators, and bioinformatics and IT staff members who work in concert in the planning, review, and decision-making activities of the Cancer Center. The overarching mission of the Data Governance Council is to provide policy and oversight for all key, data-centric, clinical, and research projects at Rutgers Cancer Institute of New Jersey. The Council is charged with establishing, reviewing, and optimizing all standing policies and standard operating procedures for evaluating requests from investigators for access and use of both de-identified and Protected Health Information (PHI) data sets.
As part of its charge, the Council is responsible for (1) providing support and training to the Rutgers Cancer community through a series of programs and workshops to support data literacy, (2) establishing a review process for evaluating investigator-initiated project proposals, (3) reviewing and providing storage infrastructure and security, (4) advocating for data quality and ease of access, (5) addressing data life cycle policy issues, (6) establishing standards for master reference data, and (7) staying abreast of the scientific, technological, legal, and ethical standards and guidelines related to data use and disclosure that form the basis of the Rutgers Cancer Institute of New Jersey policies and operational decisions in addition to State, Federal, and Institutional Review Board (IRB) protocols.
Design and components
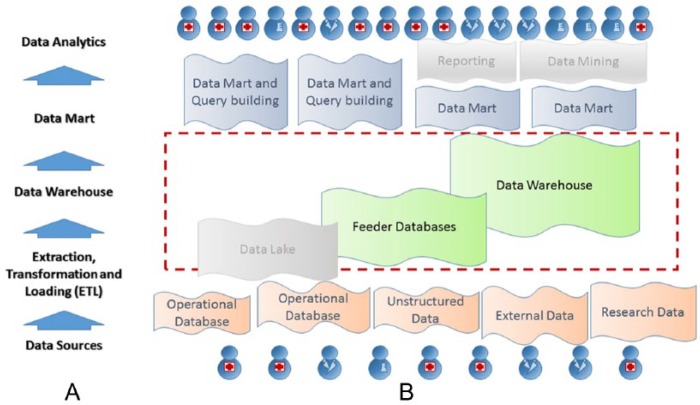
Figure 1 shows a high-level diagram which depicts the general work flow of the clinical data warehouse solution including: operational databases and external data sources, Extraction Transformation Loading (ETL) interfaces, data warehouse, data mart, and data analysis as shown in Figure 1A. Although the traditional approach for building such a system would begin with extracting data from the operational databases and external sources and then implementing an ETL to populate the warehouse, our team recognized the large number of failed attempts at other institutions to construct a functional system in this manner. Although the specific details may vary, the primary design flaw of many of those projects was that the primary focus was on the extraction of information from the data sources, before clearly identifying the use-case scenarios and clarifying the required data mapping. Being aware of the potential pitfalls of this approach, our team decided to implement a “backward-in” strategy for our project.
Figure 1.
Architecture of the clinical data warehouse project. (A) Key layers of the data warehouse layout. (B) Components in the implementation. The data lake component as well as further reporting and mining tools have not yet been implemented and are therefore rendered in gray.
The key feature of the “backward-in” strategy is to first establish a remotely hosted data mart which is then manually populated with data elements corresponding to a representative set of cases. As part of a pilot study, our staff entered approximately 1000 such cases and then began to exercise the query, visualization, and logical tools featured by each candidate data mart. A steering committee composed of surgical and medical oncologists worked closely with our biomedical informatics team and IT staff to determine whether the candidate data mart provided adequate support for the types of experiments and investigations which are underway. During the same time, the Committee made recommendations as to whether additional data modalities or elements should be included in the repository.
Once the Committee was satisfied with the data mart and the data elements that it housed, our development team turned attention toward building a fully automatic ETL. Having conducted the “backward-in” pilot study and having a solid understanding of how the data elements in the mart should be organized, the team knew exactly which data sources, mapping strategies, and interfaces would be required. The project then proceeded by completing the build and then taking advantage of the automated features of the ETL to allow prospective population of the clinical data warehouse and mart going forward.
Feeder databases and data lake
Development of a research-centric clinical data warehouse must offer the flexibility of accommodating new projects and protocols, which invariably presents new data requirements.
The industrial solution for such challenges has been to establish a Hadoop-based data lake,13 which can serve as a file reservoir to house both structured and unstructured data that have not yet been integrated into the warehouse schema. The data lake implementation uses a “schema-on-read” principle, which enables and requires data specialists and data scientists to process raw data on demand to satisfy analysis needs. As many of our clinical data warehouse end users are typically researchers and clinicians with limited IT experience, it was of vital importance for us to establish simple methods for managing data requirements and our staff’s efforts. We accomplished this by establishing feeder databases.
Feeder databases are efforts led by research scientists to establish prototypical relational databases of specific subject areas that can subsequently be forwarded to data marts for integration. Our current feeder databases include data elements derived from complete synoptic pathology reports as well as complete itemized variant report data from exon sequencing studies, whereas the Warehouse focuses on more refined and common data elements that apply to a wider number of projects and disease groups.
Moving data directly from feeder databases to data marts guarantees that the corresponding data tables have the same level of security as all the data in the Warehouse. More importantly, these data marts provide the opportunity for end users to consume and evaluate data from subspecialty areas ahead of Warehouse development, allowing feedback to the development team to facilitate the optimized design and ETL of the warehouse. The feeder databases allow continued execution of the “backward-in” strategy during ongoing development of the Warehouse.
Data mart and security
Among the software solutions offered by the company BioFortis, Inc. (Columbia, MD), is Labmatrix, which gained early adoption and recognition among prestigious institutions such as the National Human Genome Research Institute and the National Cancer Institute, and has since become a leading provider of information management solutions for the translational life science research community. The Qiagram software module supplements these capabilities with intuitive, user-friendly graphical query–building capabilities. We adopted a combination of these software models as our data mart solution, which established a unique, secure, and scalable scientific intelligent environment that enables our researchers to aggregate, explore, and interrogate information as it is collected. The BioFortis solution was designed to be subject-centric and has strong built-in capability to selectively display identified or de-identified query results to users according to their access privileges to specific IRB-approved research protocols. Users can either manipulate specific prebuilt reports for their desired output or work with honest brokers to build complicated ad hoc output of results. The graphical presentations of the queries clearly represent their logical formation and are exceptional tools which foster brainstorming among scientific and technical members of the team.
As approved by the IRB committee at Rutgers Cancer Institute of New Jersey, the precision medicine honest broker system14 is in place to help researchers use the infrastructure provided by the Warehouse (Rutgers Cancer Institute of New Jersey IRB protocols 0220100249, 2012002075, 0220090048, and 0220044862). The Data Governance Council has designated Honest Broker Administrators to review all data requests and associated IRB protocols prior to undertaking any new study. The retrieval can either exclude all patient identifying information or include limited data PHI elements as specified by the study IRB. Delivery of the information can be arranged in combination of several forms, including (1) 1-time data download, (2) limited-access data mart browsing capability including allowed data elements, (3) limited-time hypothesis-generating query capability over specified data points, and (4) live, online reports that can be filtered and aggregated by user.
Extracting discrete data from unstructured and paper forms
The necessity to integrate data originating from disparate EMR systems often makes it useful to store text or scanned paper reports in their entirety for subsequent access by the physicians and other clinical personnel. Although comprehensible by human readers, patients’ medical information embedded in these reports is not easily integrated into clinical databases, without enlisting advanced optical character recognition and text-based analytical technologies. Our team partnered with Extract Systems to implement the software and systems to automate the process of detecting and extracting unstructured clinical information that was embedded in paper or text documents such as pathology reports and genomic sequencing reports.
Synoptic cancer reporting12,15,16 and pathology staging17 are critical for producing accurate and complete cancer pathology reporting and supporting collaboration among clinicians for the optimal management of cancer patients. Although synoptic reporting has already been implemented in most Laboratory Information Systems (LIS), the EMR typically receives a full text report via version 2 HL7 protocol.18–20 By developing a set of universal rules based on CAP’s synoptic protocol with implementation details provided by the Department of Pathology and Laboratory Medicine, Robert Wood Johnson University Hospital, the approach that we implemented is able to reliably process the entire ensemble of information of each pathology report into discrete data elements and output them in a corresponding structured XML format that subsequently transformed into compatible data structure for the data warehouse. The Warehouse is therefore capable of supporting queries into every synoptic data element of the reports including but not limited to margin details, lymph node findings, and additional pathological findings. To date, our team has implemented rules for processing synoptic reports of breast, prostate, melanoma, colon, rectum, neuroendocrine tumor, and pancreas exocrine and pancreas endocrine tumors and plans to incorporate other tumor forms in near future. The rules can also be easily adapted to any future changes into the synoptic reporting scheme and be fully compatible with implementations in other hospitals and LIS systems.
Although genomic sequencing entities such as Foundation Medicine (Cambridge, MA) are capable of reporting in XML formats, many users still receive emailed or faxed reports from referring physicians and patients in PDF. The Extract Systems implementation is also proficient in extracting genomic alterations, significant negative mutations, and variants of unknown significance (VUS) from these reports into discrete form and allows cataloging of the information into databases.
Image archive
One of the data types most often overlooked when building a clinical data warehouse are medical images,21 which play a key role in cancer diagnosis and prognosis. As medical imaging data are heterogeneous and large, they take additional architectural layout, multiple software interfaces, and careful security planning to allow users to retrieve and display of this visual information.
Pathology specimens relevant to the precision medicine project as well as a range of other research protocols were imaged at the Imaging Shared Resource at Rutgers Cancer Institute of New Jersey. An Olympus VS120 scanner (Olympus Corporation, Center Valley, PA) is capable of digitizing slides with maximum magnification of 40× and throughput of 100 slides per load. The resulted images were transferred to an online database. Each whole slide image in itself does not contain patient identifier, and they require privileged information to be linked to research protocol and patient record. For example, pathology images were linked through pathology data types in the schema. Users can simply click on a URL from the data mart at which point a Web viewer interface provides access to the corresponding whole slide images for that case. The interface supports visualization and navigation about the imaged specimen. Radiology images were previously transmitted from the Picture Archiving and Communication Systems (PACS) located in the Department of Radiology at Robert Wood Johnson University Hospital using standard transfer protocols.
Each image record referenced in the clinical data warehouse is linked to the actual imaging data using URL into a Web image archive application, which supports user viewing and manipulation. The Web image archive applications supporting this access satisfy the following requirements:
The application should support proper visualization of the images via a Web browser. There are different requirements for displaying each type of medical images. Efficient traversing of whole slide images requires panning and zooming. Stack support is essential for browsing computed tomographic images.
Each image can be accessed via a direct URL. The application does not require user to search for a patient or time stamp to reach the desired image.
The URLs can be automatically retrieved from the application’s back-end database into the clinical database with complementary image metadata.
Image access is restricted based on user account and password. Therefore, if an URL is copied and sent to another physician, the recipient still needs his or her own proper privilege for access.
The application server is strategically configured for network security at the institution.
If some users do not have access to PHI, the image access is also stripped of protected information. This may demand PHI-free version of images to be served and/or a separate, PHI-free image server configuration to be established.
Document archive
The data warehouse ETL process closely integrates with the extraction workflows of unstructured information and tracks the storage of the source data files. The Warehouse maintains a reservoir of copies to the original pathologic report or genomic sequencing report so that user have the capability to easily call back, interrogate, and even reprocess these unstructured data, should more advanced processing algorithms are developed to understand such data. Similar to the images, these links can be invoked interactively through the BioFortis data mart, and through downstream applications.
Targeted exon sequencing
Rutgers Cancer Institute of New Jersey has arranged with Foundation Medicine and RUCDR Infinite Biologics (Piscataway, NJ) to receive a full complement of discrete data elements for targeted exon sequencing studies performed at their facilities. Annotated clinical summary reports in PDF as well as the corresponding data-rich XML reports are transferred, regularly, from these services through secure network connections and, with help of well-defined structure schema files, are extracted, transformed, and loaded into a prototype feeder database of the Warehouse with an interface developed using Informatica (Redwood City, CA) software. The resulting feeder database includes the variants with clinical significance, VUS, copy number gains and losses, gene rearrangement - all with clinical annotation as well as details on sequencing depth, allele frequency and sample purity measures. Gene names in the variants data set were mapped to latest Human Genome Organization (HUGO) gene names and indexes published by the HUGO Gene Nomenclature Committee (HGNC) (http://genenames.org).
A separately maintained data server is used to store exome sequencing data files transmitted from the sequencing services. The Warehouse maintains links to these files so that researchers, with proper permission, can access the server, directly, to retrieve these files in spite of their large size.
Results
The oncology clinical data warehouse project was built under the overall umbrella of the precision medicine initiative at the Rutgers Cancer Institute of New Jersey; therefore, the key use cases presented in this article are tightly tied to providing data support and propelling advances in clinical applications and investigative oncology research. The Molecular Tumor Board, among other programs under the initiative, as well as the Oncology Research Information Exchange Network (ORIEN) project provided much of the motivation that inspired and benchmarked our progress.
Institutional Molecular Tumor Board
The precision medicine programs and clinical services which are currently underway at Rutgers Cancer Institute of New Jersey required the establishment of a weekly Molecular Tumor Board to enable physicians to discuss challenging clinical cancer cases for which clinical-grade tumor sequencing data have been obtained. Patients presented at the Tumor Board have had clinical-grade sequencing of their tumor specimens obtained either in the context of a tumor sequencing protocol at Rutgers Cancer Institute of New Jersey or have had tumor sequencing performed at as part of their routine clinical care. Patients with rare histology cancers or with tumor refractory to standard treatment are prioritized for review. The Board is composed of a full range of expert clinicians, including medical oncologists, radiation therapists, surgeons, pathologists as well as basic scientists, systems biologists and bioinformatics scientists, clinical geneticists, representatives from our early phase clinical trials group, and experts in biomedical ethics.
At presentation, the sequencing data are reviewed in depth by a team of physician-scientists with experience in genomic analysis and cancer biology. The biological and clinical relevance of potential driver mutations are identified, and potential targeted treatment strategies are raised. A discussion regarding targeted therapies is held for each case, and a consensus reached regarding possible therapeutic options. Recommendations often include referral to appropriate clinical trials, potential use of off-label therapies when clinical trials are not available, and guidance regarding further diagnostic interventions. A letter summarizing the discussion is sent to the referring clinician for each case. Follow-up clinical information and outcome data are gathered on each case where available. The Rutgers Cancer Institute Tumor Board is routinely broadcast as interactive sessions with investigators located at collaborating Institutions using Health Insurance Portability and Accountability Act–approved procedures and protocols.22–24 The Warehouse has been designed to allow capture of the clinically salient data elements generated during the course of this process, including but not limited to patient demographics, family history, diagnosis, treatment, pathology report and staging, lab tests results, as well as genomic variations generated from paneled gene sequencing, to allow subsequent viewing, mining, and analysis by the clinical and research community.
Oncology Research Information Exchange Network
As a logical extension of the Warehouse and precision medicine activities which are being conducted, our institution has recently become a participating member of the Oncology Research Information Exchange Network (ORIEN). ORIEN is a national alliance, which was formed to enable investigators from leading institutions across the country to share “big data” for cancer research. The Network was established in 2014 by founding members Moffitt Cancer Center, Ohio State University Comprehensive Cancer Center, Arthur G. James Cancer Hospital, and Richard J. Solove Research Institute in Columbus. ORIEN members use a standardized protocol to follow patients throughout their lifetime (ie, Total Cancer Care Protocol) and share both clinical and molecular data from patients enrolled in the protocol to enhance discovery and create evidence of what is the most effective therapy for individual patients. In addition to the founding institutions, ORIEN members as of today include City of Hope Cancer Center, University of Virginia Cancer Center, University of Colorado Cancer Center, University of New Mexico Cancer Center, Morehouse School of Medicine, Rutgers Cancer Institute of New Jersey, University of Southern California Norris Cancer Center, and many more (http://oriencancer.org/#members-list). Our team at Rutgers will leverage the data collected through ORIEN to facilitate systematic investigations, which are focused on rare malignances for which any one institution is unable to acquire sufficient number of cases for rigorous statistical analysis.
The IT challenges of generating cohort patient data for the ORIEN project were in perfect alignment with our efforts directed toward developing the clinical data warehouse, with many of the data elements of interest overlapping, especially as they relate to patient diagnosis, mortality, treatment, and biospecimens. In this use case, information from source systems—EMR, cancer registry, biospecimen banking system, and molecular testing results—was integrated, de-identified, and transformed in accordance to the Observational Medical Outcomes Partnership (OMOP) data model25 so they could be transmitted and cataloged in the ORIEN central reservoir.
Example precision medicine use case
With the help of BioFortis technology, the data marts can host project-specific patient data to suit various needs of data access with user-based access control. Sophisticated queries can be built to execute complicated scientific logic and retrieve specific data sets.
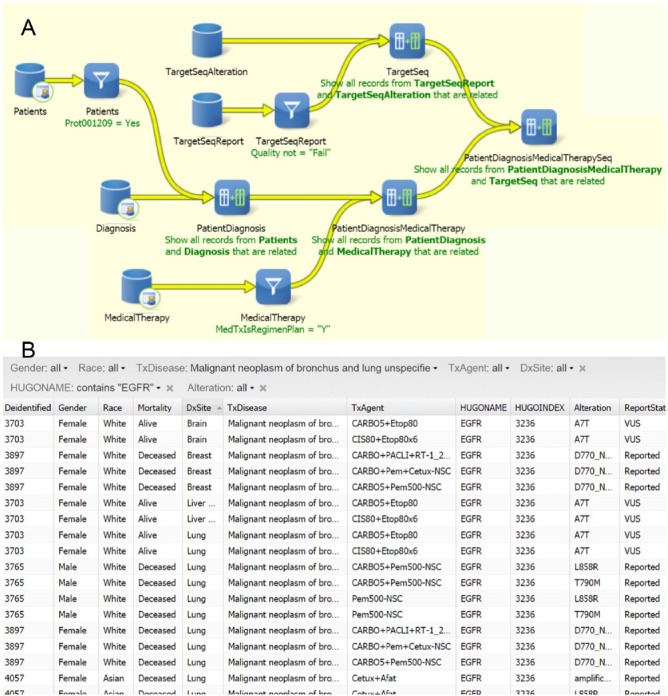
Although BioFortis queries can be constructed to suit a wide variety of specific data requirement, user can also leverage on BioFortis’s reporting capability to access data without immediate programming help. These data reports are based on prebuilt queries to suit general needs of the users. The data reports can easily incorporate filters to help user tailor the retrieval list. Figure 2 depicts an example where patient cohort was assembled with specific gene mutation and tumor diagnosis and treated with therapeutic agents. Users with appropriate IRB clearance are able to access additional information for a given data set, including the capacity to access and interrogate imaged pathology specimens. More advanced users can receive training to build ad hoc queries into their data mart or work closely with honest brokers to bring scientific logic to actual data.
Figure 2.
Example of using BioFortis Qiagram interface to formulate and execute precision medicine queries. (A) Query building diagram using Qiagram (simplified for display purposes). (B) The result report can be published for general user access. The report form allows drop-down menu selection for close examination according to individual interests. The example shows a cohort of lung cancer patients presenting with EGFR (Epidermal Growth Factor Receptor) mutation who have been treated with therapeutic agents.
Tissue microarray use case
Tissue microarray (TMA) technology26,27 is a cohort research tool for assessing tumor morphology and protein marker expression. Researchers often make discoveries by correlating TMA results with clinical information. Although some information was collected when each TMA was constructed, it was usually limited in comparison with what a comprehensive clinical data warehouse could offer. Therefore, as one of the first use cases to exercise our clinical data warehouse, as well as the first requests into the honest broker system, a research prostate cancer TMA, which was previously constructed by our cancer center’s BioSpecimen Repository Service and Histopathology Shared Resources, with no clinical data other than diagnosis of record (prostate adenocarcinoma) and Gleason score, was revisited by the team.
Adhering to policies of the honest broker system, researchers obtained new IRB approvals to collect further information on the TMA from the Office of Human Research Services before submitting data collection requests to honest broker administrators, which clearly described the project as well as the clinical information that was desired. Clinical members of the original research protocol inspected and quality-assured a subset (10%) of the clinical information before the data were made available in the Warehouse. The honest broker administrator mediated further communications between the user and the honest broker so that the honest broker can work with the data warehouse administrator to reidentify the patients included in the TMA, build specific queries into the data warehouse, and produce data worksheets as requested. After being inspected by the honest broker administrator, the final data set released to researchers included, with reference to location on the TMA block, de-identified clinical information, including demographics (without PHI), biopsy pathology result, prostatectomy pathology result, Gleason grading, pathology and clinical staging, patient visit information, medical treatment, selected pre- and postsurgical lab tests, as well as links to each patient’s tumor whole slide images. In addition, the honest broker built query to identify patients with biochemical recurrence defined as 1) two consecutive prostate-specific antigen tests after surgery with increasing result levels and 2) the final result being over 0.2 ng/mL.
Integrating pathology reports and scanned sequencing reports
Each pathology report enters in our EMR system in HL7 format and is subsequently incorporated into EMRs of corresponding patients in textual/ASCII format. When displayed to EMR users, these reports show proper alignment for optimal human comprehension. Feeding these reports into data warehouse, however, requires well-defined data elements to be properly recognized and extracted.
Our team worked closely with pathologists at Rutgers Robert Wood John University Hospital and data expert at Extract Systems to define data elements to be extracted from pathology reports and synoptic cancer reports. In general, surgical pathology reports include a final diagnosis text, entered as sentences or paragraphs of free texts with lack of standardized vocabularies among pathologists; gross and microscopic description of the specimen received and final pathologic diagnosis on each part of submitted surgical specimen; immunohistochemistry and special stains results; along with addendums and amendments. A surgical pathology report may also include 1 or more synoptic reports, each correspond to 1 lesion in the surgical specimen. The College for American Pathologists had recommendations for clear and consistent reporting of tumorous lesions in the synoptic reports; therefore, the set of data elements was well defined both medically and semantically to be extracted into a Warehouse, with limited free texts. One caveat is that these synoptic reports are consistently updated, at times with differences in staging between different versions. In many cases, pathology report included result of molecular studies, which, because it is a young and growing field, the reports are generally more standardized, allowing for straightforward extraction. The CoPath implementation (Cerner Corporation, Kansas City, MO) used in Robert Wood Johnson University Hospital closely adhered to the guidelines so that our system can be easily generalized to other hospitals and institutions.
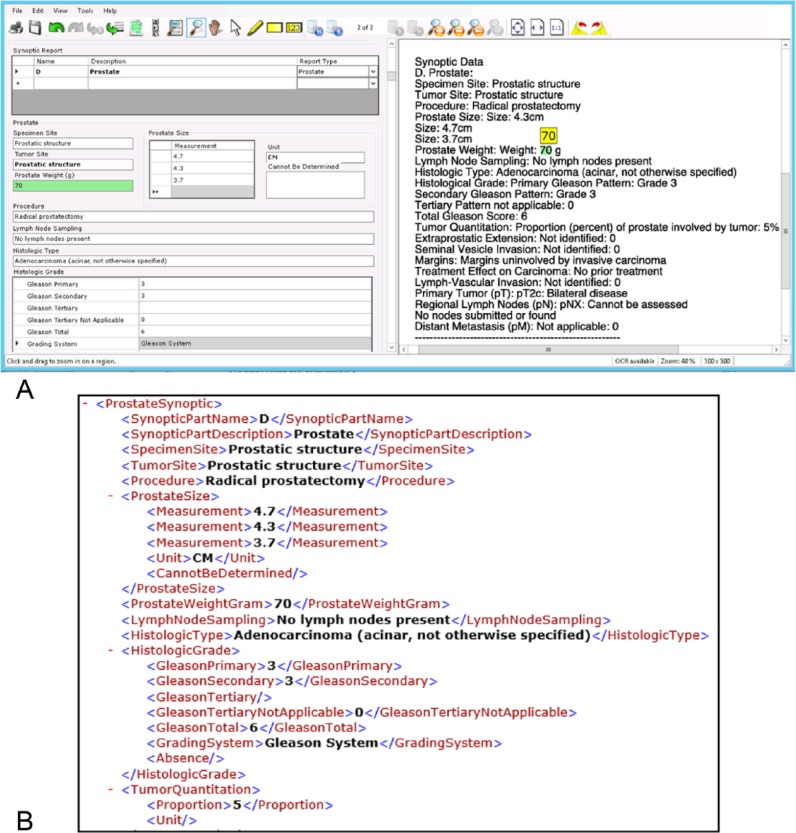
In an assessment of Extract Systems implementation, a balanced set of surgical pathology reports, including approximately 20 synoptic reports of each recognized type, a small set of reports of other synoptic forms, as well as faxed and/or scanned Foundation Medicine sequencing reports, was submitted for automatic data extraction based on rules established at the Extract Systems software. Trained personnel used the provided quality-control interface to examine each extracted data element. The quality-control interface was specifically designed for showing all extracted data elements in a user-friendly electronic form, with the original report displayed side by side for review as shown in Figure 3. As the quality-control personnel hit the Tab key to traverse through each data element in the form, the corresponding text from the original report, where extraction of the very element originated, was highlighted. If the extraction is not accurate, user can overwrite the element, or, more easily, use the software tool to swipe the correct area of the original report to re-extract the data. Text highlighting is most often in green color, signaling high confidence of the detection. Occasionally, orange highlights appear when further attention is required from the human operator in case when the recognition and extraction may be ambiguous to the algorithm. We found this display of confidence interval highly useful to achieve efficiency and maintain operator’s attention during the quality-control process.
Figure 3.
An example of pathology report data extraction using Extract Systems software. (A) After the software performs automatic data detection and extraction, the verification software interface displays the report-in-process on right-hand side of screen, to be compared with the extracted information on dynamically generated data form on the left side. Screen capture shows an example section containing part of a synoptic pathology report. Please note one selected data element (in green) with its highlighted counterpart on the original report for easy verification. (B) Corresponding section of the resulting XML output file was subsequently loaded into the Warehouse.
Through Extract Systems platforms’ built-in quality monitoring functionality, we tracked the generation and quality-control process of a total of 18 530 data elements, which originated from 26 genetic analysis reports and 212 pathology reports, including 164 synoptic tumor forms. For performance evaluation of the Extract Systems algorithms, we considered the quality-controlled data to be accurate, and any data field that was touched up, corrected, or added/deleted by human operators in the quality-control process was captured error. As a result, the overall accuracy of automatic data element extraction by Extract Systems platform was determined to be 96.65%. Of course, the actual data extraction work flow included a quality-control team that corrects any errors that may have been introduced during the recognition process and ensures quality of the resulting data set.
Discussion and Conclusion
During the course of constructing, developing, and exercising the oncology data warehouse, 1 key challenge that was regularly confronted was how to maintain a balance between keeping the database concise and focused, versus providing views of the data that offer a comprehensive summary of the salient clinical information. Although the most common usage of the Warehouse pertains to querying to identify specific patient cohorts with an emphasis on positive diagnostic tests (eg, pathology synoptic) and summarized data (such as cancer-specific diagnoses, annotated genetic variations, and therapeutic outcomes), our team also contended with demands from our researchers to support big data mining research, which often required thorough and temporal assembly of patient visits, lab tests, low-level sequencing reads in the form of Binary Alignment/Map (BAM) files, or variation calling files.
Reflecting on our strategy of implementing the “backward-in” strategy to develop the Warehouse, we recognize that it offered many merits, including drawing quick successes to gain wider support from management and clinical users, prioritizing the ETL on the most essential components of clinical data, as well as allowing us to lay out and exercise the Warehouse in parallel to the development of use-case–based applications. This strategy of building out the Warehouse framework based on key use cases, if not carefully implemented, could potentially lead to a limited ETL that will not support the addition of new use cases, and hence reduce usability of the Warehouse going forward. The success of this project leveraged the expert knowledge and guidance provided by our Data Governance Council through continuous communication, regular brainstorming sessions, and iterative prototyping. Another unique effort to ensure long-term success of the Warehouse was the parallel development of the feeder databases to prototype and evaluate data types and modalities as candidate expansions to the core Warehouse schema. Other possible technical extensions of the Warehouse include expansion of the data elements collected and implementation of natural language processing modules to allow automated processing of clinical free text to incorporate searchable information, integration of image analysis modules to support generation and indexing of image-based features to support content-based image retrieval and computer-aided detection, as well as online processing and visualization tools for genetic data.
A clinical data warehouse is composed of many components; most of them run on commercial software systems. When selecting private vendors for each component, the team at Rutgers Cancer Institute of New Jersey emphasized not only on their experience in related fields but also on the flexibility of their products in accommodating data and applications suitable for the Warehouse project. The vendors’ cooperation provided a substantial boost to the continued progress of the project.
A wide range of clinical data warehouse solutions are being developed and evaluated at leading institutions across the country to support the steady rise in the number of Precision Medicine and Translational programs that are being established. The success of establishing a comprehensive clinical data warehouse at our Institution in which we have included pathology, genetics, and imaging data in a relatively short time in Rutgers Cancer Institute of New Jersey is attributed to careful planning and governance from institute executives, close collaboration with key clinical and operational departments, careful selection of software partners, as well as excellent communication and orchestration from the development and research team. The colocalization of such a broad number of correlated data elements representing the full spectrum of clinical information, imaging studies, and genomic information coupled with our experience and expertise in advanced pattern recognition, high-performance computing, and data mining has positioned our team with unique opportunities to optimize personalizing treatment, refining best practices, and providing objective, reproducible insight as to the underlying mechanisms of disease onset and progression.
Acknowledgments
We thank the patients and appreciate services provided by the Biospecimen Repository Service and the Office for Human Research Service at Rutgers Cancer Institute of New Jersey.
Footnotes
Peer Review:Six peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1308 words, excluding any confidential comments to the academic editor.
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by Hugs for Brady, The Val Skinner Foundation, and the National Institutes of Health through contracts P30CA072720, 5R01CA156386-10, and 7R01CA161375-06 from the National Cancer Institute; and contract 4R01LM009239-08 from the National Library of Medicine. Additional support was provided by a generous gift to the Genetics Diagnostics to Cancer Treatment Program of the Rutgers Cancer Institute of New Jersey and Rutgers University Cell and DNA Repository Infinite Biologics.
Declaration Of Conflicting Interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: DJF contributed in overall design, concept, and oversight. WC and DJF wrote and edited the manuscript. HC and ES contributed to the writing of the manuscript. DL and HC designed and implemented core warehouse, WC designed feeder databases and developed ETL for pathology reports and sequencing reports, WC and HC incorporated imaging components. LR, DJF, WC, and DL established and exercised the honest broker system. RSD and LR managed administrative and clinical adoption. SG, KH, LAG, ES, and GR contributed in data elements selection and clinical evaluation.
Disclosures and Ethics: The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. The authors have no affiliations with, nor has any financial or nonfinancial interest with, the private vendors (Informatica, BioFortis, and Extract Systems) mentioned in this manuscript.
References
- 1. Winkler SJ, Witte E, Bierer BE. The Harvard catalyst common reciprocal IRB reliance agreement: an innovative approach to multisite IRB review and oversight. Clin Transl Sci. 2015;8:57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Weber GM, Murphy SN, McMurry AJ, et al. The Shared Health Research Information Network (SHRINE): a prototype federated query tool for clinical data repositories. J Am Med Inform Assoc. 2009;16:624–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chute CG, Beck SA, Fisk TB, Mohr DN. The enterprise data trust at Mayo Clinic: a semantically integrated warehouse of biomedical data. J Am Med Inform Assoc. 2010;17:131–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Evans RS, Lloyd JF, Pierce LA. Clinical use of an enterprise data warehouse. AMIA Annu Symp Proc. 2012;2012:189–198. [PMC free article] [PubMed] [Google Scholar]
- 5. Hollis J. Deploying an HMO’s data warehouse. Health Manag Technol. 1998;19:46–48. [PubMed] [Google Scholar]
- 6. Zhang Q, Matsumura Y, Teratani T, et al. The application of an institutional clinical data warehouse to the assessment of adverse drug reactions (ADRs). Evaluation of aminoglycoside and cephalosporin associated nephrotoxicity. Methods Inf Med. 2007;46:516–522. [PubMed] [Google Scholar]
- 7. Cho IS, Haug PJ. The contribution of nursing data to the development of a predictive model for the detection of acute pancreatitis. Stud Health Technol Inform. 2006;122:139–142. [PubMed] [Google Scholar]
- 8. Evans RS, Lloyd JF, Pierce LA. Clinical use of an enterprise data warehouse. Paper presented at: AMIA Annual Symposium Proceedings November 3–7, 2012; Chicago, IL. [PMC free article] [PubMed] [Google Scholar]
- 9. MacKenzie SL, Wyatt MC, Schuff R, Tenenbaum JD, Anderson N. Practices and perspectives on building integrated data repositories: results from a 2010 CTSA survey. J Am Med Inform Assoc. 2012;19:e119–e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blobel BG, Engel K, Pharow P. Semantic interoperability—HL7 Version 3 compared to advanced architecture standards. Methods Inf Med. 2006;45:343–353. [PubMed] [Google Scholar]
- 11. Dolin RH, Alschuler L. Approaching semantic interoperability in Health Level Seven. J Am Med Inform Assoc. 2011;18:99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Srigley JR, McGowan T, Maclean A, et al. Standardized synoptic cancer pathology reporting: a population-based approach. J Surg Oncol. 2009;99:517–524. [DOI] [PubMed] [Google Scholar]
- 13. O’Leary DE. Embedding AI and crowdsourcing in the big data lake. IEEE Intell Syst. 2014;29:70–73. [Google Scholar]
- 14. Boyd AD, Hosner C, Hunscher DA, Athey BD, Clauw DJ, Green LA. An “Honest Broker” mechanism to maintain privacy for patient care and academic medical research. Int J Med Inform. 2007;76:407–411. [DOI] [PubMed] [Google Scholar]
- 15. Markel SF, Hirsch SD. Synoptic surgical pathology reporting. Hum Pathol. 1991;22:807–810. [DOI] [PubMed] [Google Scholar]
- 16. Gill AJ, Johns AL, Eckstein R, et al. Synoptic reporting improves histopathological assessment of pancreatic resection specimens. Pathology. 2009;41: 161–167. [DOI] [PubMed] [Google Scholar]
- 17. Edge S, Byrd D, Compton C, Green F, Trotti A. AJCC Cancer Staging Manual. 7th ed. New York, NY: Springer; 2010. [Google Scholar]
- 18. Dolin RH, Alschuler L, Beebe C, et al. The HL7 clinical document architecture. J Am Med Inform Assoc. 2001;8:552–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dolin RH, Alschuler L, Boyer S, et al. HL7 clinical document architecture, release 2. J Am Med Inform Assoc. 2006;13:30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Beeler GW. HL7 version 3—an object-oriented methodology for collaborative standards development. Int J Med Inform. 1998;48:151–161. [DOI] [PubMed] [Google Scholar]
- 21. Anderson NR, Lee ES, Brockenbrough JS, et al. Issues in biomedical research data management and analysis: needs and barriers. J Am Med Inform Assoc. 2007;14:478–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bhagavathi S, Goodell L, Chan C, et al. Validation of a droplet PCR-based assay for the detection of somatic variants in solid tumors. Paper presented at: Association for Molecular Pathology (AMP) 2015 Annual Meeting; 2015 November 5–7; Austin, TX. [Google Scholar]
- 23. Hirshfield KM, Tolkunov D, Zhong H, et al. Clinical actionability of comprehensive genomic profiling for management of rare or refractory cancers [published online ahead of print August 26, 2016]. Oncologist. doi: 10.1634/theoncologist.2016-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rodriguez-Rodriguez L, Hirshfield KM, Rojas V, et al. Use of comprehensive genomic profiling to direct point-of-care management of patients with gynecologic cancers. Gynecol Oncol. 2016;141:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Overhage JM, Ryan PB, Reich CG, Hartzema AG, Stang PE. Validation of a common data model for active safety surveillance research. J Am Med Inform Assoc. 2012;19:54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kallioniemi O-P, Wagner U, Kononen J, Sauter G. Tissue microarray technology for high-throughput molecular profiling of cancer. Hum Mol Genet. 2001;10:657–662. [DOI] [PubMed] [Google Scholar]
- 27. Kononen J, Bubendorf L, Kallioniemi A, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–847. [DOI] [PubMed] [Google Scholar]