Abstract
Genes crucial for cancer development can be mutated via various mechanisms, which may reflect the nature of the mutagen. In thyroid papillary carcinomas, mutations of genes coding for effectors along the MAPK pathway are central for transformation. BRAF point mutation is most common in sporadic tumors. By contrast, radiation-induced tumors are associated with paracentric inversions activating the receptor tyrosine kinases RET and NTRK1. We report here a rearrangement of BRAF via paracentric inversion of chromosome 7q resulting in an in-frame fusion between exons 1–8 of the AKAP9 gene and exons 9–18 of BRAF. The fusion protein contains the protein kinase domain and lacks the autoinhibitory N-terminal portion of BRAF. It has elevated kinase activity and transforms NIH3T3 cells, which provides evidence, for the first time to our knowledge, of in vivo activation of an intracellular effector along the MAPK pathway by recombination. The AKAP9-BRAF fusion was preferentially found in radiation-induced papillary carcinomas developing after a short latency, whereas BRAF point mutations were absent in this group. These data indicate that in thyroid cancer, radiation activates components of the MAPK pathway primarily through chromosomal paracentric inversions, whereas in sporadic forms of the disease, effectors along the same pathway are activated predominantly by point mutations.
Introduction
Thyroid papillary carcinoma is the most common type of endocrine malignancy. In this tumor, mutations of genes coding for effectors along the MAPK pathway are central for transformation. Indeed, activating mutations of BRAF, RET, or RAS are found in 70% of all cases and rarely overlap in the same tumor (1–3).
The RET gene codes for a cell membrane receptor tyrosine kinase (4, 5). In papillary carcinomas, it is activated via chromosomal rearrangement, which results in fusion of the 3′ portion of the gene coding for the tyrosine kinase domain to the 5′ portion of various genes, creating chimeric oncogenes named RET/PTC (6, 7). The 2 most common rearrangement types, RET/PTC1 and RET/PTC3, are paracentric inversions, since both RET and its respective fusion partner, H4 or NCOA4 (ELE1), reside on the long arm of chromosome 10 (8, 9). The fusion leads to ligand-independent activation of the truncated RET receptor. Binding of activated RET to SHC via Y1062 has been shown to be critical to RET-mediated transformation and dedifferentiation, primarily through the RAS-RAF-MAPK pathway (10, 11). RAS, in turn, can be independently activated by point mutations, primarily in the follicular variant of papillary thyroid cancer (12).
Most recently, point mutations activating the BRAF serine/threonine kinase have surfaced as the most common genetic event in sporadic thyroid papillary carcinomas (1, 13). RAF proteins, including ARAF, BRAF, and CRAF or RAF-1, phosphorylate MAPK/ERK kinase (MEK) within the MAPK cascade upon stimulation by RAS. BRAF somatic mutations were first discovered in malignant melanomas and in a smaller subset of colorectal and ovarian cancers (14). The majority of mutations in those tumors and virtually all mutations in thyroid carcinomas result in a valine-to-glutamate substitution at residue 600 (V600E), formerly designated as V599E. This mutation is believed to produce a constitutively active kinase by disrupting hydrophobic interactions between residues in the activation loop and residues in the ATP binding site that maintain the inactive conformation, allowing development of new interactions that fold the kinase into a catalytically competent structure (15). Correspondingly, BRAFV600E exhibits elevated basal kinase activity and transforms NIH3T3 cells with high efficiency (14).
Conceivably, other alterations that would release the inhibitory constrains of the catalytic domain of BRAF could also result in kinase activation. Indeed, loss of the N-terminal regulatory domains as a result of fusion to different genes during in vitro transfection has been shown to activate BRAF kinase and transform NIH3T3 cells (16, 17). Despite those reports, which appeared more than a decade ago, this mechanism of BRAF activation has not been identified in human tumors. Here, we report the activation of BRAF by chromosomal rearrangement that results in its N-terminal truncation in several papillary thyroid carcinomas. We show that the fusion gene exhibits elevated basal kinase activity, stimulates ERK phosphorylation, and induces transformation of NIH3T3 cells, which is consistent with its functioning as an oncogene. This observation also provides insights into distinct mechanisms of activation of components of the MAPK pathway in radiation-induced cancers, which involve chromosomal inversions, in contrast to sporadic cancers, where it happens predominantly by point mutations.
Results
Identification of BRAF rearrangement.
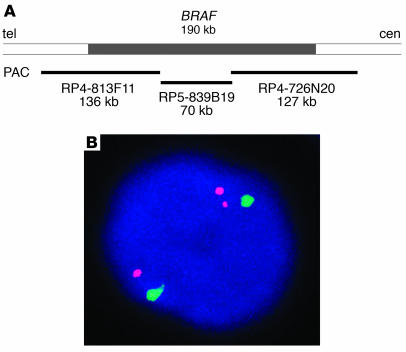
The possibility of BRAF involvement in chromosomal rearrangement was studied by fluorescence in situ hybridization (FISH). A contig of 3 P1 artificial chromosome (PAC) clones of approximately 330 kb, spanning the entire BRAF gene, was assembled to use as a probe (Figure 1A). Benign thyroid cells showed, as expected, 2 signals. Analysis of 32 papillary thyroid carcinomas revealed 1 tumor containing 3 signals, whereas subsequent hybridization with a chromosome 7 centromeric probe demonstrated the presence of 2 copies of the chromosome (Figure 1B). Since the split of one BRAF signal resulted in a pair of signals located at a relatively constant distance from each other, the likely mechanism for this event was intrachromosomal inversion.
Figure 1.
Identification of the BRAF gene rearrangement. (A) Genomic region on 7q containing the BRAF gene and position of PAC clones used as a probe for FISH. (B) Interphase nucleus from the index tumor showing split of 1 BRAF signal (red) and preservation of 2 chromosome 7 centromeric signals (green), which indicates the rearrangement of the BRAF gene.
Identification and characterization of the fusion partner.
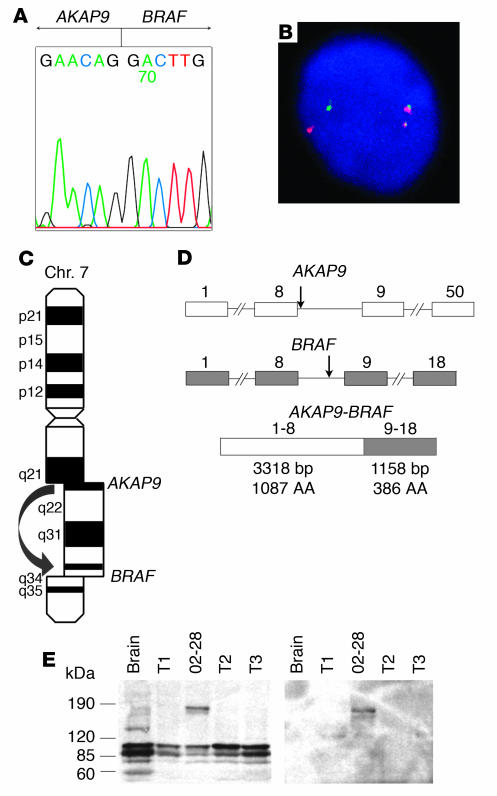
The identification of the fusion partner was achieved by 5′ rapid amplification of cDNA ends (RACE) using tumor poly(A)+ RNA and a pool of primers designed along the 3′ end of the BRAF gene. RACE products were sequenced, and several of them revealed a fusion between exon 9 of BRAF and exon 8 of the A-kinase anchor protein 9 (AKAP9) gene (Figure 2A). Confirmation of the fusion was obtained by RT-PCR with primers located in exon 8 of AKAP9 and exon 10 of BRAF, which yielded the expected product of 181 bp. Genomic DNA was used for PCR to identify the fusion at the genomic level, which was found to reside between nucleotide 364 of AKAP9 intron 8 and nucleotide 4,930 of BRAF intron 8. The reciprocal fusion gene product was subsequently identified by RT-PCR using tumor cDNA, indicating the reciprocal nature of the rearrangement. Further confirmation was obtained by double-color FISH with probes corresponding to the BRAF and AKAP9 genes (Figure 2B). Since the BRAF gene is located on chromosome 7q34 (18) and AKAP9 on 7q21–q22 (19–21), the AKAP9-BRAF fusion results from inv(7)(q21–22q34) (Figure 2C). Using primers located within the 5′ and 3′ untranslated region of AKAP9 and BRAF, respectively, the AKAP9-BRAF coding cDNA sequence was amplified from the tumor, cloned into the pCR-XL-TOPO vector, and sequenced (GenBank accession number AY803272). Analysis of the sequence (4,567 bp total size) revealed a 4,476-bp open reading frame (ORF) containing exons 1–8 of AKAP9 fused in-frame with exons 9–18 of BRAF (Figure 2D). Based on the known sequences of the untranslated regions of both genes, the predicted length of the complete AKAP9-BRAF cDNA is 4,849 bp. Four transcript variants of human AKAP9 have been previously identified based on several coding region differences. The AKAP9 sequence of the chimeric cDNA lacked 36 nucleotides at position 269–304 of transcript version 1 (GenBank accession number NM_147171), which indicates that it corresponded to either transcript version 2 or 3 of the gene. Multiple RT-PCR reactions revealed no splice variants of the fusion transcript in the tumor cDNA. Western blot analysis of the tumor using an antibody to the C-terminus of BRAF showed a band of approximately 170 kDa, corresponding to the predicted molecular weight of 172 kDa for the fusion protein (Figure 2E). The same band was detected with an antibody against the N-terminus of AKAP9.
Figure 2.
BRAF is recombined with the AKAP9 gene and results in expression of a fusion protein. (A) Sequence of the 5′ RACE product showing a fusion point between AKAP9 and BRAF. (B) Confirmation of the reciprocal fusion by FISH with probes corresponding to the BRAF (red) and AKAP9 (green) genes. (C) The fusion is a result of paracentric chromosomal inversion inv(7)(q21–22q34). Chr., chromosome. (D) Genomic structure of the BRAF and AKAP9 genes showing the location of breakpoints (arrows) and the organization of the chimeric cDNA. Exons are represented by boxes and introns by lines. Numbers above indicate exon numbers. (E) Western blot analysis using BRAF antibody (left) and AKAP9 antibody (right), showing an approximately 170-kDa protein, corresponding to the predicted molecular weight of the fusion protein in the index case (number 02-28). Three other papillary carcinomas (T1–T3) are shown for comparison. Wild-type AKAP9 is 453 kDa in size and was not detected in this Western blot.
The wild-type AKAP9 gene encodes a 453-kDa protein capable of binding the type II regulatory subunit (RII) of protein kinase A (PKA) as well as other signaling proteins, anchoring them to the centrosome and Golgi apparatus (19–21). Several isoforms arising through alternative splicing are selectively expressed in most human tissues at low abundance (19, 20). The AKAP9 protein localizes by immunofluorescence as a single perinuclear dot and some surrounding dispersed fluorescence, corresponding to the centrosome and Golgi compartments, respectively, in interphase HeLa and SaOS2 osteosarcoma cells (19, 20). Based on the sequence of the chimeric cDNA, the fusion protein is predicted to lack 2 putative RII binding regions and the centrosome localization domain of AKAP9 (20, 22, 23).
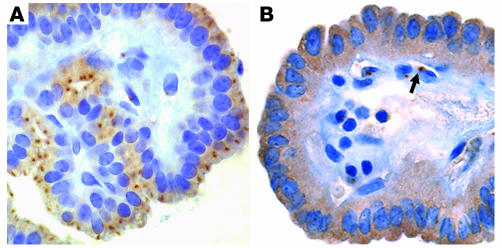
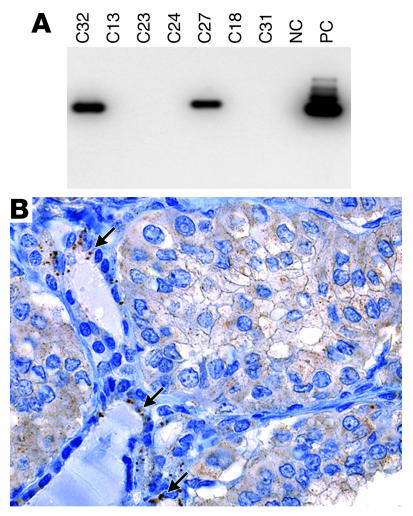
In papillary thyroid carcinomas negative for the fusion and in non-neoplastic thyroid tissue, AKAP9 expression was readily detectable by RT-PCR (data not shown), and the protein was seen as a single perinuclear dot with faint diffuse cytoplasmic staining (Figure 3A). In the tumor expressing the AKAP9-BRAF protein, the perinuclear dot was no longer identifiable, whereas diffuse staining was observed in the cytoplasm (Figure 3B). However, the presence of intact centrosomes in the tumor cells was confirmed by immunohistochemistry with an antibody against the centrosomal protein γ–tubulin (data not shown). This is consistent with the absence of the centrosomal localization domain of AKAP9 in the fusion protein, which prevents its preferential targeting to the centrosomal compartment.
Figure 3.
Subcellular localization of AKAP9 and AKAP9-BRAF detected by immunohistochemistry with an AKAP9 antibody. (A) In a papillary thyroid carcinoma negative for the fusion, the protein is predominantly targeted to centrosomes, seen as a single perinuclear dot. There is also weak diffuse cytoplasmic staining. (B) The tumor expressing AKAP9-BRAF reveals diffuse cytoplasmic distribution of the chimeric protein and the perinuclear dot is no longer visible. The shift in localization involves only tumor cells but not adjacent vascular endothelial cells, which preserve the dotted pattern of staining (arrow). Magnification, ×200.
Functional characterization of the AKAP9-BRAF fusion.
The wild-type human BRAF gene codes for an 84-kDa serine/threonine kinase in the MAPK pathway (16, 24). Structurally, BRAF, similar to other RAF kinases, contains 2 highly conserved N-terminal regulatory domains, CR1 and CR2, that mediate autoinhibition of BRAF and a C-terminal protein kinase domain (CR3). CR1 includes the RAS-GTP binding domain (RBD), which mediates RAS binding, triggering the recruitment of the protein to the cell membrane and kinase activation (25). Based on the cDNA sequence, the AKAP9-BRAF fusion protein is predicted to lack the BRAF N-terminal regulatory domains, including RBD, and to retain an intact kinase domain. Loss of the regulatory domains would be expected to lead to constitutive activation of BRAF kinase in a RAS-independent manner.
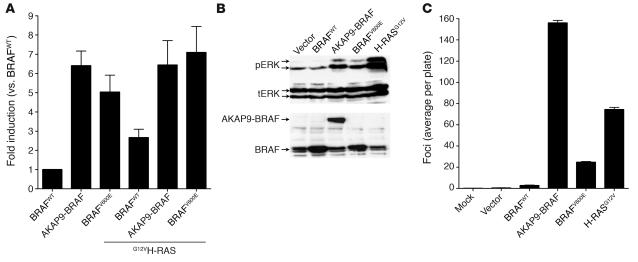
We characterized the kinase activity of the AKAP9-BRAF fusion protein by transient transfection of myc epitope–tagged versions of cDNAs for AKAP9-BRAF, BRAFV600E, or wild-type BRAF into COS7 cells, using bacterially purified MEK1 as a substrate for phosphorylation (Figure 4A). Basal BRAF kinase activity of the fusion protein was 6-fold greater than BRAFWT activity and not further increased by coexpression of activated RAS. Basal activity of BRAFV600E was also increased, but to a lesser extent, and was modestly increased by H-RASG12V. By contrast, H-RASG12V increased BRAFWT kinase activity by about 2.5-fold. The fusion protein stimulated endogenous ERK1/2 phosphorylation to a similar extent as BRAFV600E in pooled stably transfected NIH3T3 cells, whereas overexpression of BRAFWT had no discernible effect (Figure 4B).
Figure 4.
The AKAP9-BRAF chimeric protein has increased BRAF kinase activity and results in NIH3T3 cell transformation. (A) In vitro kinase activity of myc-tagged BRAFWT, AKAP9-BRAF, and BRAFV600E in the absence or presence of H-RASG12V in COS7 cells. BRAF kinase activity was measured in myc-IgG immunoprecipitates using MEK1 substrate phosphorylation as a read-out. Each sample was assayed in triplicate, and bars represent the standard deviations from the mean. Similar results were obtained in 2 independent transfections. (B) Western blots of lysate from cells stably transfected with pEFP, pEFP-BRAFWT, pEFP–AKAP9-BRAF, pEFP-BRAFV600E, or pBabe puro–H-RASG12V probed with antibodies to phospho-ERK1/2 (pERK), total ERK (tERK), or C-terminus of BRAF. Similar results were obtained in 3 additional independent experiments. (C) Foci formation of NIH3T3 cells transfected with pEFP-BRAFWT, pEFP–AKAP9-BRAF, pEFP-BRAFV600E, or pBabe puro–H-RASG12V. Bars represent the average number of foci per plate from 6 plates. Similar results were obtained in 2 independent experiments.
The ability of AKAP9-BRAF fusion protein to induce transformation was studied in an NIH3T3 cell focus assay (Figure 4C). Vector and BRAFWT transformed cells at very low efficiency. AKAP9-BRAF showed the highest transformation activity in this assay, followed by H-RASG12V and BRAFV600E. In addition, NIH3T3 cells stably transfected with empty vector, BRAFWT, AKAP9-BRAF, BRAFV600E, and H-RASG12V were assayed for tumorigenicity in nude mice. Two weeks after injection of 106 cells, tumors of at least 5 mm in diameter were formed with complete penetrance (6 out of 6 animals, repeated in 2 independent experiments) for AKAP9-BRAF, BRAFV600E, and H-RASG12V constructs, whereas vector and BRAFWT did not lead to the development of tumors (0 our of 6 animals). These results demonstrate that AKAP9-BRAF fusion leads to activation of BRAF kinase and transformation of NIH3T3 cells.
Prevalence of AKAP9-BRAF fusion and BRAF point mutation in thyroid cancer.
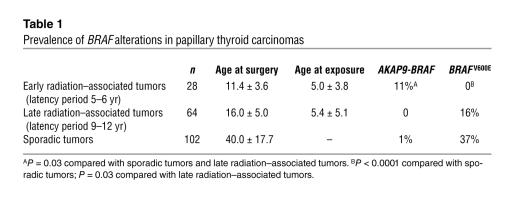
To determine the prevalence of AKAP9-BRAF fusion in various populations, we used RT-PCR with primers flanking the fusion point to study a large series of papillary thyroid carcinomas (Table 1). In a group of 102 sporadic papillary tumors, no additional case of the fusion was found. We also examined papillary carcinomas from 2 cohorts of Belarussian children and adolescents exposed to radiation after the Chernobyl nuclear accident in 1986. Among 28 tumors that developed 5–6 years after exposure, 3 (11%) were positive for the fusion, whereas 64 tumors developing 9–12 years after the accident were all negative (Figure 5A). Paraffin blocks were available for immunohistochemical analysis with AKAP9 antibody in 2 out of 3 positive cases. Both cases showed a shift in subcellular localization of AKAP9 from the centrosomal pattern seen in adjacent non-neoplastic thyroid tissue to diffuse cytoplasmic distribution in tumor cells (Figure 5B).
Table 1.
Prevalence of BRAF alterations in papillary thyroid carcinomas
Figure 5.
Analysis of post-Chernobyl tumors that developed 5–6 years after exposure. (A) Southern blot of RT-PCR products amplified with primers Ex8A and Ex10B and hybridized with probe PR showing 2 positive cases (C32 and C27). Other tumor samples are negative (C13, C23, C24, C18, and C31). NC, negative control; PC, positive control (index case). (B) Immunohistochemical analysis of tumor C32 with AKAP9 antibody showing a shift in subcellular localization of the protein from a single dot/centrosomal pattern observed in the entrapped non-neoplastic follicles (arrows) to diffuse cytoplasmic staining in tumor cells. Magnification, ×200.
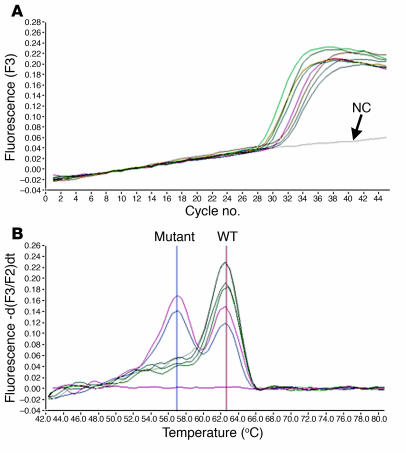
We also tested these tumors for BRAF point mutation leading to the V600E substitution. The mutation was identified in 38 (37%) sporadic tumors, in 10 (16%) tumors developing 9–12 years after exposure, but in none of the tumors that developed 5–6 years after exposure (Figure 6). The differences in prevalence of the 2 types of BRAF activation were statistically significant between the early radiation group and both late radiation and sporadic groups (Table 1).
Figure 6.
Detection of V600E BRAF mutation cDNA from post-Chernobyl tumors that developed 9–12 years after exposure using LightCycler real-time PCR followed by fluorescence melting curve analysis. (A) Real-time amplification of cDNA samples. (B) Detection of point mutations by the postamplification fluorescence melting curve analysis based on a distinct melting temperature of duplexes formed between the WT probe and either WT (62.8°C) or mutant (57.0°C) sequences. The mutations in both samples are heterozygous, since both mutant and WT peaks are detectable.
Discussion
The MAPK signaling pathway is a ubiquitously present regulator of cell growth, proliferation, apoptosis, and differentiation. Recently, BRAF, an essential component of the kinase cascade, has received renewed attention since mutations in the gene were identified in several types of human cancer. Indeed, activating point mutations of BRAF have been found in more than 60% of malignant melanomas, 10–14% of colorectal and ovarian cancer, and in very few cases of lung cancer, soft tissue sarcomas, and other cancer cell lines (14, 26, 27). Thyroid cancer was promptly added to the list with 36–69% prevalence of BRAF mutation (1, 2, 13, 28). However, neither BRAF nor other intracellular effectors along the MAPK pathway have been previously implicated in human carcinogenesis through mechanisms other than point mutation. In the present study, we demonstrate BRAF activation by chromosomal rearrangement that results in BRAF fusion to the AKAP9 gene in thyroid papillary carcinomas. The fusion protein lacks the regulatory domains of BRAF, which results in constitutive kinase activation in a RAS-independent manner, and exhibits transforming activity similar to that of the most common BRAFV600E.
The fusion partner of BRAF, the AKAP9 gene, belongs to the group of A-kinase anchor proteins that have the common function of binding to the regulatory subunit of PKA and targeting it to discrete locations within the cell (29). AKAP9, in particular, has predominantly centrosomal and Golgi compartmentalization (19–21). The fusion protein lacks the C-terminal centrosomal domain and, as expected, loses its centrosomal localization in cancer cells. Moreover, immunohistochemical analysis with AKAP9 antibody revealed no evidence of wild-type AKAP9 within centrosomes of cancer cells in all cases positive for the fusion by RT-PCR. This may suggest the competitive inhibition of wild-type AKAP9 by AKAP9-BRAF, although further studies are required to confirm this. AKAP9 is an important regulator of c-AMP–dependent PKA and interacts with many other signaling proteins, including serine/threonine kinase protein kinase N, protein phosphatase 1, protein phosphatase 2A, and PKC-ε (20, 30). Thus, deregulation of its function in cells harboring AKAP9-BRAF fusion may affect a variety of physiological functions. On the other hand, neither AKAP9 nor other AKAP genes have been implicated in carcinogenesis so far. Therefore, it remains unclear whether or not the AKAP9 portion of the fusion gene contributes independently to its transforming potential, besides providing an active promoter driving the expression of truncated BRAF.
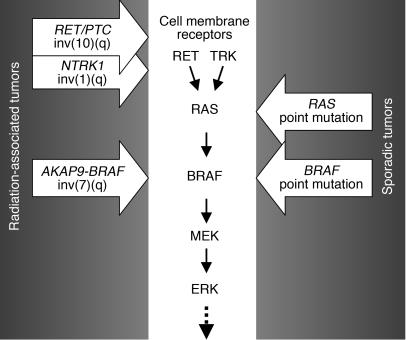
Over the last decade, it has become apparent that activation of components of the MAPK pathway in thyroid cancer can be achieved through several distinct genetic events, i.e., RET/PTC (or, less frequently, TRK) rearrangements, RAS point mutations, or BRAF point mutations. However, the relative frequency of each event varies significantly in different populations. BRAF point mutation is the most common change in sporadic adult papillary carcinomas. In contrast, tumors associated with accidental or therapeutic irradiation have exceedingly high frequency of RET/PTC rearrangements and absence or low prevalence of BRAF point mutations (31–38). The 2 most common RET/PTC types found in up to 80% of radiation-associated tumors, RET/PTC1 and RET/PTC3, are both paracentric inversions of chromosome 10q (39, 40). Another rearrangement, which contributes to about 7% of radiation-induced papillary carcinomas, involves the nerve growth factor gene NTRK1 and results from paracentric inversion of chromosome 1q (41, 42). Here, we report the identification of another paracentric inversion, involving 7q, which is also more common in radiation-associated tumors, especially those appearing with a short latency. These tumors, however, revealed no BRAF point mutations. The sharp contrast in mechanisms of activation of the same gene in radiation-induced and sporadic tumors points to the association between environmental factors and distinct genetic mechanisms of activation of MAPK pathway components: through chromosomal rearrangement in patients exposed to radiation or through point mutations in patients with no radiation history (Figure 7).
Figure 7.
Activation of MAPK pathway in papillary thyroid carcinogenesis involves predominantly point mutations in sporadic tumors or chromosomal rearrangements in radiation-associated tumors.
The vast majority of RET/PTC and all TRK and AKAP9-BRAF chromosomal rearrangements in radiation-induced tumors are paracentric inversions. In epithelial cancers as a whole, point mutations are more prevalent than chromosomal rearrangements, and the latter are usually translocations or interchromosomal exchanges. The reason for the striking predisposition of radiation-induced thyroid cancers to paracentric inversions is not entirely clear, although some evidence suggests that this may be related to the organization of chromosomal domains and arm domains and spatial interaction between pairs of various chromosomal regions. Indeed, there is a general consensus that free DNA ends produced in the nuclear volume after passage of radiation tracks have to be within a certain distance of each other in order to recombine (43, 44). Residing within the same chromosomal arm domain would limit their maximum separation and help to fulfill this requirement. Indeed, the yields of intra-arm chromosomal exchanges after both low– and high–linear energy transfer radiation has been shown to exceed the expected levels, calculated based on spatial randomness, by up to 10-fold (45). The conditions would be even more favorable if potentially recombinogenic foci within the arm domain were frequently juxtaposed to each other, as was demonstrated in human thyroid cells for RET and H4, the RET/PTC1 partners (46).
In this study, we observed significant difference in mechanisms of BRAF activation not only between early radiation–associated tumors and sporadic cancers, but also between early and late post-radiation tumors. Such a difference is not without precedent. Similar variation has been previously observed with respect to RET/PTC rearrangements between post-Chernobyl tumors developing 10 years or less after exposure and those manifesting after a longer latency (34, 35). Indeed, the overall prevalence of RET/PTC was significantly higher in the former group, and RET/PTC3 was the most common type. By contrast, tumors arising after a longer latency had lower prevalence of RET/PTC, and the predominant type was RET/PTC1, both of which are features more typical of sporadic tumors. The present study demonstrates that late post-radiation tumors also occupy an intermediate position with respect to the predominant mechanism of BRAF activation. This suggests that tumors arising early after exposure are likely to be initiated through direct radiation damage–related genetic events, such as chromosomal rearrangements occurring after double-stranded DNA breaks. As for the tumors developing after a longer latency, their pathogenesis may involve a combination of radiation-related and -unrelated mechanisms and rely on molecular alterations which develop after repair of radiation-induced DNA damage that are stimulated by genomic instability that persists in the progeny of the exposed cells (47, 48).
In summary, we report here a novel mechanism of in vivo BRAF activation via inversion of chromosome 7q, resulting in BRAF fusion to the AKAP9 gene. AKAP9-BRAF functions as an oncogene and is more prevalent in thyroid carcinomas with recent history of radiation exposure. The significance of this study is 2-fold. It demonstrates a novel mechanism of BRAF activation in human tumors, showing for the first time that intracellular effectors along the MAPK pathway can be activated by recombination. In addition, it shows correlation between the nature of the mutagen and the mechanism of mutation and suggests that paracentric chromosomal inversions represent a common genetic mechanism of radiation-associated thyroid carcinogenesis.
Methods
Tumor samples.
Frozen tissues from sporadic thyroid carcinomas were obtained from the Department of Pathology, University of Cincinnati, following an institutionally approved tissue procurement protocol and through the Cooperative Human Tissue Network. Post-Chernobyl tumors from Belarussian patients who underwent surgery in 1991–92 were available as archival paraffin-embedded tissue. Samples from Belarussian patients who underwent surgery in 1995–1998 were from the Experimental Division of the Institute of Pathology, University of Munich, and available as RNA extracted from frozen tissue; they were negative for RET/PTC and TRK rearrangements (35). The study was approved by the University of Cincinnati Institutional Review Board.
FISH analysis.
Tumor touch preparations were made from snap-frozen tissues. PAC clones for BRAF (RP4-726N20, RP5-839B19, RP4-813F11) were obtained from BAC/PAC Resources, Children’s Hospital Oakland, and bacterial artificial chromosome (BAC) clones for AKAP9 (CTB-104F4, CTB-161K23) from Open Biosystems. The probes were labeled by nick translation using SpectrumOrange-dUTP and SpectrumGreen-dUTP (Vysis Inc.), respectively. An α-satellite DNA centromeric probe for chromosome 7 was obtained from Vysis Inc. Nuclei were counterstained with DAPI. Microscopy was performed with a Leica Microsystems TCS 4D confocal laser scanning fluorescence microscope with digital image capture. One hundred nuclei were scored for each preparation.
5 ′ RACE.
Fifty micrograms of poly(A)+ RNA from the index case were used for 5′ RACE using the 5′ RACE System for Rapid Amplification of cDNA Ends (Invitrogen Corp.). The BRAF gene–specific primers were designed for cDNA synthesis (5′-GATGACTTCTGGTGCCATCC-3′, 5′-AACTGCTGAGGTGTAGGTGCTGTC-3′, and 5′-GTCTCGTTGCCCAAATTGAT-3′) and for subsequent PCR reactions (5′-TTTCACTGCCACATCACCAT-3′ for first PCR and 5′-TCACTCGAGTCCCGTCTACC-3′ for nested PCR). The 5′ RACE products were excised from the gel, purified, and sequenced using an automated ABI 377 sequencer (PerkinElmer).
Cloning and sequencing analysis.
Total RNA was used to amplify the ORF of the chimeric cDNA using primers located in the 5′-untranslated region of AKAP9 (5′-ACCACCCCTCAACCCCTGTTTT-3′) and 3′-untranslated region of BRAF (5′-TTCCTTTTGTTGCTACTCTCCTGA-3′). The PCR product was electrophoresed in an agarose gel, purified from the gel, and cloned into the pCR-XL-TOPO vector (Invitrogen Corp.). Several clones were fully sequenced. The genomic AKAP9-BRAF fusion was amplified with primers 5′-GCAACTCAACCAAGTGAAAATG-3′ and 5′-GGTTGATCCTCCATCACCAC-3′, yielding a product of approximately 2.4 kb, and the reciprocal fusion product cDNA was amplified with primers 5′-ACCCGCCTCGGACTCTATT-3′ and 5′-GTTGGGCTTCCATCTGTAGC-3′, yielding a product of 242 bp.
Western blot and immunohistochemistry.
Frozen tissues from papillary carcinomas were used for Western blotting, which was performed as previously described (49). We loaded 50 μg of protein on a 5% polyacrylamide gel. After transfer, the membranes were incubated with anti–C-terminus BRAF polyclonal (Raf-B, C-19; Santa Cruz Biotechnology Inc.) or anti–N-terminus AKAP9 monoclonal (AKAP450; BD Biosciences) antibodies. The same AKAP9 antibody in 1:50 dilution was used for immunohistochemical analysis, which was performed on paraffin sections after overnight pretreatment in borate buffer using avidin-streptavidin immunoperoxidase detection. Rabbit anti–γ-tubulin polyclonal antibody was provided by K. Fukasawa (University of Cincinnati College of Medicine, Cincinnati, Ohio, USA) (50) and was used in 1:800 dilution on paraffin sections after pretreatment in borate buffer.
In vitro kinase assay.
COS7 cells were transiently cotransfected with either pEFP vector or pEFP-containing myc-tagged BRAFWT, AKAP9-BRAF, or BRAFV600E and either pBabe puro or pBabe puro–HRASG12V using Lipofectamine 2000 (Invitrogen Corp.). After 48 hours, cell extracts were prepared, immunoprecipitated with mouse anti–myc-IgG (Upstate Cell Signaling Solutions), and kinase activity determined as directed by the manufacturer using bacterially purified MEK1 as substrate, and Western blotting with pMEK1 antibody as a read-out (B-RAF Kinase Assay Kit; Upstate Cell Signaling Solutions). To determine the effects of the BRAF mutants on endogenous ERK1/2 phosphorylation, we incubated pools of stably transfected cells for 12 hours without serum prior to lysate preparation. Lysates were centrifuged, the supernatant collected, and 100 μg of extract subjected to SDS-PAGE. Proteins were transferred to a nitrocellulose membrane and subject to Western blot analysis with anti–phospho-ERK1/2, anti–total ERK1/2, or anti-BRAF (Santa Cruz Biotechnology Inc.) antibodies.
Transformation assays.
For focus assays, NIH3T3 cells were transfected with empty vector or the indicated BRAF and H-RAS expression plasmids using lipofectamine. After 2 days, cells were split into six 100-mm3 dishes and incubated with DMEM containing 5% calf serum for 21 days. Cells were stained with 0.4% crystal violet and foci counted. For the mouse xenograft experiments, NIH3T3 cells were cotransfected with pBabe puro and pEFP vector, pEFP–myc BRAFWT, pEFP–AKAP9-BRAF, pEFP–myc BRAFV600E, or pBabe puro–H-RASG12V and mass selected by incubation with DMEM containing 5% calf serum and 1 μg/ml puromycin for 3 weeks. A total of 106 pooled cells was injected into nude mice. Animals were killed after 2 weeks, and nodules larger than 5 mm in diameter were scored as positive.
Mutation analysis.
For the detection of AKAP9-BRAF fusion, cDNA amplification was achieved by PCR using primers located in exon 8 of AKAP9 (Ex8A, 5′-AGCAAGAACAGTTGATTTTGGA-3′) and exon 10 of BRAF (Ex10B, 5′-GCAGACAAACCTGTGGTTGA-3′), with the expected product of 181 bp. The PCR products were electrophoresed on agarose gels and bands visualized by staining with ethidium bromide (frozen tissues) or by transfer to a nylon filter and hybridization with an internal oligonucleotide probe (PR, 5′-AAACTTCAGAAAGAACTCAATGTACTT-3′) labeled with 50 μCi of [γ-32P]-ATP (paraffin-embedded tissue). Detection of V600E BRAF mutation was performed from DNA using real-time PCR and fluorescence melting curve analysis, as previously reported (51), or from cDNA using real-time RT-PCR with primers 5′-CGACAGACTGCACAGG-3′ and 5′-TGACTTCTGGTGCCAT-3′ and the same probes.
Statistical analysis.
The comparison between groups was performed using a 2-tailed Fisher exact test. Differences were considered significant when P was less than 0.05.
Acknowledgments
This study was supported by NIH grant R01 CA88041 and American Cancer Society grant RSG-03-027-01-CCE (to Y.E. Nikiforov), NIH grant R01 CA50706 (to J.A. Fagin), a grant from the American Cancer Society Ohio Chapter #06001011 (to J.A. Knauf), and a grant from Deutsche Krebshilfe, Bonn, Germany (to H.M. Rabes). Collection of tissue samples was supported in part by a grant from the NIH (PHS M01 RR08084) and through the Cooperative Human Tissue Network funded by the National Cancer Institute. We thank Kenji Fukasawa for γ-tubulin antibody, Mickey Croyle for technical assistance, and Richard Marais for the BRAFWT and BRAFV600E expression vectors.
Footnotes
See the related Commentary beginning on page 20.
Nonstandard abbreviations used: BAC, bacterial artificial chromosome; FISH, fluorescence in situ hybridization; MEK, MAPK/ERK kinase; ORF, open reading frame; PAC, P1 artificial chromosome; PKA, protein kinase A; RACE, rapid amplification of cDNA ends; RBD, RAS-GTP binding domain; RII, type II regulatory subunit.Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Kimura ET, et al. High prevalence of BRAF mutations in thyroid cancer: genetic evidence for constitutive activation of the RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma. Cancer Res. 2003;63:1454–1457. [PubMed] [Google Scholar]
- 2.Soares P, et al. BRAF mutations and RET/PTC rearrangements are alternative events in the etiopathogenesis of PTC. Oncogene. 2003;22:4578–4580. doi: 10.1038/sj.onc.1206706. [DOI] [PubMed] [Google Scholar]
- 3.Frattini M, et al. Alternative mutations of BRAF, RET and NTRK1 are associated with similar but distinct gene expression patterns in papillary thyroid cancer. Oncogene. 2004;23:7436–7440. doi: 10.1038/sj.onc.1207980. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi M, Ritz J, Cooper GM. Activation of a novel human transforming gene, ret, by DNA rearrangement. Cell. 1985;42:581–588. doi: 10.1016/0092-8674(85)90115-1. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi M. Structure and expression of the ret transforming gene. IARC Sci. Publ. 1988;1988:189–197. [PubMed] [Google Scholar]
- 6.Fusco A, et al. A new oncogene in human thyroid papillary carcinomas and their lymph-nodal metastases. Nature. 1987;328:170–172. doi: 10.1038/328170a0. [DOI] [PubMed] [Google Scholar]
- 7.Grieco M, et al. PTC is a novel rearranged form of the ret proto-oncogene and is frequently detected in vivo in human thyroid papillary carcinomas. Cell. 1990;60:557–563. doi: 10.1016/0092-8674(90)90659-3. [DOI] [PubMed] [Google Scholar]
- 8.Pierotti MA, et al. Characterization of an inversion on the long arm of chromosome 10 juxtaposing D10S170 and RET and creating the oncogenic sequence RET/PTC. Proc. Natl. Acad. Sci. U. S. A. 1992;89:1616–1620. doi: 10.1073/pnas.89.5.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minoletti F, et al. The two genes generating RET/PTC3 are localized in chromosomal band 10q11.2. Genes Chromosomes Cancer. 1994;11:51–57. doi: 10.1002/gcc.2870110108. [DOI] [PubMed] [Google Scholar]
- 10.Asai N, Murakami H, Iwashita T, Takahashi M. A mutation at tyrosine 1062 in MEN2A-Ret and MEN2B-Ret impairs their transforming activity and association with shc adaptor proteins. J. Biol. Chem. 1996;271:17644–17649. doi: 10.1074/jbc.271.30.17644. [DOI] [PubMed] [Google Scholar]
- 11.Knauf JA, Kuroda H, Basu S, Fagin JA. RET/PTC-induced dedifferentiation of thyroid cells is mediated through Y1062 signaling through SHC-RAS-MAP kinase. Oncogene. 2003;22:4406–4412. doi: 10.1038/sj.onc.1206602. [DOI] [PubMed] [Google Scholar]
- 12.Zhu Z, Gandhi M, Nikiforova MN, Fischer AH, Nikiforov YE. Molecular profile and clinical-pathologic features of the follicular variant of papillary thyroid carcinoma. An unusually high prevalence of ras mutations. Am. J. Clin. Pathol. 2003;120:71–77. doi: 10.1309/ND8D-9LAJ-TRCT-G6QD. [DOI] [PubMed] [Google Scholar]
- 13.Cohen Y, et al. BRAF mutation in papillary thyroid carcinoma. J. Natl. Cancer Inst. 2003;95:625–627. doi: 10.1093/jnci/95.8.625. [DOI] [PubMed] [Google Scholar]
- 14.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 15.Wan PT, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 16.Ikawa S, et al. B-raf, a new member of the raf family, is activated by DNA rearrangement. Mol. Cell. Biol. 1988;8:2651–2654. doi: 10.1128/mcb.8.6.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miki T, et al. Development of a highly efficient expression cDNA cloning system: application to oncogene isolation. Proc. Natl. Acad. Sci. U. S. A. 1991;88:5167–5171. doi: 10.1073/pnas.88.12.5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eychene A, Barnier JV, Apiou F, Dutrillaux B, Calothy G. Chromosomal assignment of two human B-raf(Rmil) proto-oncogene loci: B-raf-1 encoding the p94Braf/Rmil and B-raf-2, a processed pseudogene. Oncogene. 1992;7:1657–1660. [PubMed] [Google Scholar]
- 19.Witczak O, et al. Cloning and characterization of a cDNA encoding an A-kinase anchoring protein located in the centrosome, AKAP450. EMBO J. 1999;18:1858–1868. doi: 10.1093/emboj/18.7.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahashi M, et al. Characterization of a novel giant scaffolding protein, CG-NAP, that anchors multiple signaling enzymes to centrosome and the golgi apparatus. J. Biol. Chem. 1999;274:17267–17274. doi: 10.1074/jbc.274.24.17267. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt PH, et al. AKAP350, a multiply spliced protein kinase A-anchoring protein associated with centrosomes. J. Biol. Chem. 1999;274:3055–3066. doi: 10.1074/jbc.274.5.3055. [DOI] [PubMed] [Google Scholar]
- 22.Gillingham AK, Munro S. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 2000;1:524–529. doi: 10.1093/embo-reports/kvd105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takahashi M, Yamagiwa A, Nishimura T, Mukai H, Ono Y. Centrosomal proteins CG-NAP and kendrin provide microtubule nucleation sites by anchoring gamma-tubulin ring complex. Mol. Biol. Cell. 2002;13:3235–3245. doi: 10.1091/mbc.E02-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sithanandam G, Kolch W, Duh FM, Rapp UR. Complete coding sequence of a human B-raf cDNA and detection of B-raf protein kinase with isozyme specific antibodies. Oncogene. 1990;5:1775–1780. [PubMed] [Google Scholar]
- 25.Avruch J, et al. Ras activation of the Raf kinase: tyrosine kinase recruitment of the MAP kinase cascade. Recent Prog. Horm. Res. 2001;56:127–155. doi: 10.1210/rp.56.1.127. [DOI] [PubMed] [Google Scholar]
- 26.Brose MS, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62:6997–7000. [PubMed] [Google Scholar]
- 27.Rajagopalan H, et al. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 28.Xu X, Quiros RM, Gattuso P, Ain KB, Prinz RA. High prevalence of BRAF gene mutation in papillary thyroid carcinomas and thyroid tumor cell lines. Cancer Res. 2003;63:4561–4567. [PubMed] [Google Scholar]
- 29.Edwards AS, Scott JD. A-kinase anchoring proteins: protein kinase A and beyond. Curr. Opin. Cell Biol. 2000;12:217–221. doi: 10.1016/s0955-0674(99)00085-x. [DOI] [PubMed] [Google Scholar]
- 30.Takahashi M, Mukai H, Oishi K, Isagawa T, Ono Y. Association of immature hypophosphorylated protein kinase cepsilon with an anchoring protein CG-NAP. J. Biol. Chem. 2000;275:34592–34596. doi: 10.1074/jbc.M005285200. [DOI] [PubMed] [Google Scholar]
- 31.Fugazzola L, et al. Oncogenic rearrangements of the RET proto-oncogene in papillary thyroid carcinomas from children exposed to the Chernobyl nuclear accident. Cancer Res. 1995;55:5617–5620. [PubMed] [Google Scholar]
- 32.Klugbauer S, Lengfelder E, Demidchik EP, Rabes HM. High prevalence of RET rearrangement in thyroid tumors of children from Belarus after the Chernobyl reactor accident. Oncogene. 1995;11:2459–2467. [PubMed] [Google Scholar]
- 33.Nikiforov YE, Rowland JM, Bove KE, Monforte-Munoz H, Fagin JA. Distinct pattern of ret oncogene rearrangements in morphological variants of radiation-induced and sporadic thyroid papillary carcinomas in children. Cancer Res. 1997;57:1690–1694. [PubMed] [Google Scholar]
- 34.Smida J, et al. Distinct frequency of ret rearrangements in papillary thyroid carcinomas of children and adults from Belarus. Int. J. Cancer. 1999;80:32–38. doi: 10.1002/(sici)1097-0215(19990105)80:1<32::aid-ijc7>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 35.Rabes HM, et al. Pattern of radiation-induced RET and NTRK1 rearrangements in 191 post-Chernobyl papillary thyroid carcinomas: biological, phenotypic, and clinical implications. Clin. Cancer Res. 2000;6:1093–1103. [PubMed] [Google Scholar]
- 36.Bounacer A, et al. High prevalence of activating ret proto-oncogene rearrangements, in thyroid tumors from patients who had received external radiation. Oncogene. 1997;15:1263–1273. doi: 10.1038/sj.onc.1200206. [DOI] [PubMed] [Google Scholar]
- 37.Collins BJ, et al. RET expression in papillary thyroid cancer from patients irradiated in childhood for benign conditions. J. Clin. Endocrinol. Metab. 2002;87:3941–3946. doi: 10.1210/jcem.87.8.8748. [DOI] [PubMed] [Google Scholar]
- 38.Nikiforova MN, et al. Low prevalence of BRAF mutations in radiation-induced thyroid tumors in contrast to sporadic papillary carcinomas. Cancer Lett. 2004;209:1–6. doi: 10.1016/j.canlet.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 39.Pierotti MA, et al. Characterization of an inversion on the long arm of chromosome 10 juxtaposing D10S170 and RET and creating the oncogenic sequence RET/PTC. Proc. Natl. Acad. Sci. U. S. A. 1992;89:1616–1620. doi: 10.1073/pnas.89.5.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minoletti F, et al. The two genes generating RET/PTC3 are localized in chromosomal band 10q11.2. Genes Chromosomes Cancer. 1994;11:51–57. doi: 10.1002/gcc.2870110108. [DOI] [PubMed] [Google Scholar]
- 41.Greco A, et al. Chromosome 1 rearrangements involving the genes TPR and NTRK1 produce structurally different thyroid-specific TRK oncogenes. Genes Chromosomes Cancer. 1997;19:112–123. [PubMed] [Google Scholar]
- 42.Beimfohr C, Klugbauer S, Demidchik EP, Lengfelder E, Rabes HM. NTRK1 re-arrangement in papillary thyroid carcinomas of children after the Chernobyl reactor accident. Int. J. Cancer. 1999;80:842–847. doi: 10.1002/(sici)1097-0215(19990315)80:6<842::aid-ijc7>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 43.Sachs RK, Chen AM, Brenner DJ. Review: proximity effects in the production of chromosome aberrations by ionizing radiation. Int. J. Radiat. Biol. 1997;71:1–19. doi: 10.1080/095530097144364. [DOI] [PubMed] [Google Scholar]
- 44.Savage JR. Cancer. Proximity matters. Science. 2000;290:62–63. doi: 10.1126/science.290.5489.62. [DOI] [PubMed] [Google Scholar]
- 45.Sachs RK, Brenner DJ, Chen AM, Hahnfeldt P, Hlatky LR. Intra-arm and interarm chromosome intrachanges: tools for probing the geometry and dynamics of chromatin. Radiat. Res. 1997;148:330–340. [PubMed] [Google Scholar]
- 46.Nikiforova MN, et al. Proximity of chromosomal loci that participate in radiation-induced rearrangements in human cells. Science. 2000;290:138–141. doi: 10.1126/science.290.5489.138. [DOI] [PubMed] [Google Scholar]
- 47.Smith LE, Nagar S, Kim GJ, Morgan WF. Radiation-induced genomic instability: radiation quality and dose response. Health Phys. 2003;85:23–29. doi: 10.1097/00004032-200307000-00006. [DOI] [PubMed] [Google Scholar]
- 48.Little JB. Genomic instability and radiation. J. Radiol. Prot. 2003;23:173–181. doi: 10.1088/0952-4746/23/2/304. [DOI] [PubMed] [Google Scholar]
- 49.Kunzli BM, et al. Influences of the lysosomal associated membrane proteins (Lamp-1, Lamp-2) and Mac-2 binding protein (Mac-2-BP) on the prognosis of pancreatic carcinoma. Cancer. 2002;94:228–239. doi: 10.1002/cncr.10162. [DOI] [PubMed] [Google Scholar]
- 50.Kawamura K, et al. Induction of centrosome amplification and chromosome instability in human bladder cancer cells by p53 mutation and cyclin E overexpression. Cancer Res. 2004;64:4800–4809. doi: 10.1158/0008-5472.CAN-03-3908. [DOI] [PubMed] [Google Scholar]
- 51.Nikiforova MN, et al. BRAF mutations in thyroid tumors are restricted to papillary carcinomas and anaplastic or poorly differentiated carcinomas arising from papillary carcinomas. J. Clin. Endocrinol. Metab. 2003;88:5399–5404. doi: 10.1210/jc.2003-030838. [DOI] [PubMed] [Google Scholar]