Abstract
Background
Techniques simultaneously assessing multiple levels of molecular processing are appealing because molecular signaling underlying complex neural phenomena occurs at complementary levels. The TRIzol method isolates RNA and DNA, but protein retrieval is difficult due to inefficient solubilization of precipitated protein pellets.
New Method
We optimized a protocol for the efficient solubilization of protein from TRIzol-precipitated brain tissue for Western blotting analysis, which was also more effective at directly homogenizing brain tissue than RIPA buffer.
Results
Protein yield during solubilization, in addition to protein yield via direct homogenization, is increased by optimizing concentrations of chemicals in a standard lysis buffer. Effective incubation parameters for both total protein yield and the analysis of post-translational modifications is remarkably flexible. Importantly, different neural cell types and protein classes are represented in solubilized protein samples. Moreover, we used dissociated mouse brain tissue to isolate microglia from other cell types and successfully resolved cell type-specific proteins from these small and difficult to attain samples.
Comparison with Existing Method(s)
Solubilization buffers to date have been comprised primarily of SDS or urea; the data herein demonstrate that components common to lysis buffers can also enhance protein solubilization both after direct homogenization and after precipitation.
Conclusions
This method is suitable for assessing gene and protein expression from a single brain sample, allowing for a more comprehensive evaluation of neural phenomena while minimizing the number of subjects.
Keywords: protein precipitation, protein solubilization, TRIzol, Western blotting, RIPA buffer
1. Introduction
Molecular processing occurs at different, complementary levels, which makes elucidating the molecular signaling underlying complex neural phenomena, such as cognition or the pathogenesis of neurological diseases and disorders, a substantial challenge. Thus, the advent of techniques to simultaneously assess multiple levels of molecular processing (e.g. DNA, RNA, protein) is an appealing prospect from both practical and theoretical perspectives in neuroscience research. From a practical perspective, small or difficult to attain samples can be maximally utilized without a prohibitive number of experimental subjects. Neuroscience research often involves complex behavioral or pharmacological manipulations in mammalian models, making the need to maximize data collection particularly important. Moreover, the inclusion (or justification of exclusion) of both male and female subjects, as mandated by the National Institutes of Health, can significantly enhance the translational capability of basic neuroscience research (Klein et al., 2015). From a theoretical perspective, neural communication and plasticity require highly integrated and dynamic molecular processing, and thus changes in any one protein or gene is difficult to interpret in isolation. For example, increased protein expression canonically results from nuclear signaling that initiates transcription of the corresponding gene, which is subsequently translated into protein. However, in the case of locally stored synaptic mRNAs, increased protein expression can also occur via rapid protein synthesis without the need for concurrent gene expression (Aakalu et al., 2001; Bramham and Wells, 2007). Conversely, increased gene expression does not always result in a subsequent increase in protein levels, which may indicate the critical involvement of post-transcriptional regulators such as miRNAs or RNA binding proteins (Fukao et al., 2015; Gebauer and Hentze, 2004; Ule and Darnell, 2006). Furthermore, several different signaling cascades can modulate the same gene and/or protein (for experimental example, see Kopec et al., 2015), in which case assessing the activity levels of upstream signaling brokers (e.g. phosphorylation state of receptors, kinases), which are best assessed with protein rather than gene expression analyses, are critical to understanding how the healthy brain functions, and how this goes awry in disease.
Kits are commercially available that can accomplish multi-level analyses, but their cost per sample can be prohibitive, and some reports suggest the representation of proteins may be skewed (Mathieson and Thomas, 2013). One solution is the guanidinium thiocyanate-phenol-chloroform extraction method (i.e. TRIzol method), which is widely used in molecular biology as a means to retrieve high quality RNA for gene expression analyses. It is additionally possible to precipitate DNA and protein from the same sample, but protein retrieval is difficult due to challenges in solubilizing the precipitated protein pellet. Several groups have reported solubilization strategies with varying degrees of protein yield including dialysis, buffers with varying concentrations of sodium dodecyl sulfate (SDS) and urea, different lengths of incubation at 50°C, and a combination of the aforementioned strategies plus sonication (Banerjee et al., 2003; Hummon et al., 2007; Likhite and Warawdekar, 2011; Man et al., 2006; Reddy et al., 2013; Simoes et al., 2013; Yamaguchi et al., 2013; Young and Truman, 2012).
Protein lysis and homogenization buffers, the most common of which is radioimmunoprecipitation (RIPA) buffer, contain several chemical components other than stringent denaturants, including NaCl, Tris, and ethylenediaminetetraacetic acid (EDTA), but the impact of these chemicals in solubilizing protein pellets after protein precipitation is unknown. Interestingly, RIPA buffer contains a relatively small concentration of SDS (0.1%), suggesting that solubilization of diverse cell components and proteins may be additionally facilitated by these chemicals. Previous reports have explored the effects of pH and detergents in the solubilization buffer (Banerjee et al., 2003; see Discussion), and thus we focused the present investigation on the role of SDS, NaCl, Tris, and EDTA concentrations in solubilization.
In addition, glia, including microglia, astrocytes, and oligodendrocytes, are now recognized as critical modulators of neural plasticity, cognition, and disease (Barres, 2008), and thus it is important that techniques for assessing molecular processing in the brain should include the potential contribution of these cell types in addition to neurons. However, it is unclear whether or not precipitated protein is representative of different classes of proteins (i.e. nuclear, cytoplasmic, membrane-bound), and in the case of brain tissue, if different neural cell types are represented in the recovered protein. Thus, our second goal was to determine if different cell types and protein classes are represented in precipitated protein samples. Moreover, we sought to expand upon this notion by isolating a specific glial cell type (microglia) from the rest of the neural population, in order to (i) support the claim that different cell types can be effectively analyzed from TRIzol-precipitated samples, and (ii) demonstrate the utility of this technique for very small (~400,000 microglial cells, ~10μm each in diameter) and difficult to attain samples.
Herein, we optimize the composition of a protein lysis buffer to establish a solubilization method for TRIzol-precipitated protein from brain tissue that is cost-effective, efficient, easily performed, and reproducible. We provide evidence that protein yield during solubilization can be increased by adjusting concentrations of the common lysis buffer chemicals, NaCl, EDTA, and SDS, while varying Tris concentration did not significantly impact protein yield. Using this optimized lysis buffer, we report that the protein yield and the analysis of post-translational modifications, specifically phosphorylation of the kinase ERK1/2, is stable across a variety of incubation lengths and temperatures. The optimized lysis buffer, to our surprise, was more effective than a very common homogenization buffer, RIPA buffer, in directly homogenized tissue samples. Moreover, protein loss due to TRIzol precipitation and solubilization in optimized lysis buffer (relative to direct homogenization in optimized lysis buffer) was equivalent to loss due to direct homogenization in RIPA buffer rather than optimized lysis buffer. Thus, if RIPA buffer is considered the ‘gold standard’ for tissue homogenization, solubilized samples with optimized lysis buffer constitute no loss in sample. Importantly, the solubilized protein obtained from this method represents a variety of cell types and protein classes. Finally, to test the utility of this technique for small, difficult to attain samples, we retrieved and analyzed protein from brain regions dissociated into a single cell suspension and sorted into CD11b positive (CD11b+; i.e. microglia) and CD11b negative (CD11b−; i.e. all other cell types) populations.
2. Materials and Methods
Tissue collection
For all buffer optimization experiments, male Sprague Dawley rats (Harlan; age Postnatal day 38–39 (P38–39)) were anesthetized with a ketamine-xylazine cocktail (80 mg/kg ketamine; 10mg/kg xylazine, i.p.) and transcardially perfused with cold 0.9% saline for ~5 mins. Brains were rapidly extracted and the hippocampus and prefrontal cortex was dissected (other hemisphere PFA-fixed for a different experiment), frozen in isopentane, and stored in 2mL microcentrifuge tubes at −80°C for 3–4 months until processing.
To determine protein yield between direct homogenization in RIPA or optimized lysis buffer, female rats (~P170–175 at time of sacrifice; purchased from Harlan within a certain age range) were euthanized with CO2 and transcardially perfused with cold 0.9% saline for ~8 mins. Bilateral frontal cortex and bilateral hippocampi were collected into pre-weighed 2mL microcentrifuge tubes, with one hemisphere used for one experimental condition (e.g. direct homogenization in RIPA buffer) and one hemisphere used as a within animal comparison to a separate condition (e.g. direct homogenization in optimized lysis buffer). Samples were stored on ice until tissue mass was measured, and then frozen until processing.
To test the utility of this method for small samples, single cell suspensions and cell type-specific sorting, male and female C57BL/6 mice (Charles River; P60) were euthanized with CO2 and transcardially perfused with cold 0.9% saline for ~3 mins. Bilateral hippocampi were dissected and stored on ice until further processing. All animals were treated compliant with Duke University’s and Harvard Medical School/Massachusetts General Hospital’s IACUC guidelines.
Single cell suspension and CD11b population enrichment
Bilateral mouse hippocampi were dissected and homogenized in an enzyme digestion mix containing collagenase A (1.5mg/mL; Roche) and DNase I (0.4mg/mL; Roche) in Hank’s Buffered Salt Solution (HBSS; ThermoFisher Scientific) for 45mins in a 37°C water bath. Every 15mins during the incubation, samples were removed from the water bath and passed through glass Pasteur pipettes multiple times to ensure complete dissociation. Samples were then filtered through 70μm nylon filters and centrifuged at 1200rpm for 10mins at 4°C. Cell pellets were resuspended in MACS buffer (1% bovine serum albumin (BSA), 1mM EDTA in 1x phosphate buffered saline (PBS; 137mM NaCl, 2.7mM KCl, 10mM Na2HPO4, 1.8mM KH2PO4, pH 7.4)) and incubated with cluster of differentiation molecule 11b (CD11b) antibody-conjugated magnetic beads (MACS Miltenyi Biotec) for 15mins at 4°C. After washing, cells were passed through magnetic bead columns (MACS Miltenyi Biotec) and CD11b+ and CD11b− populations were collected. Samples were washed with 1xPBS, centrifuged at 1200rpm for 10mins, resuspended in 800μL TRIzol, and stored at −80°C until further processing.
Protein extraction from TRIzol
Tissue was placed on dry ice until the addition of 500μL of TRIzol (Life Technologies) and homogenization with a Tissue Tearor (Biospec). The Tissue Tearor was placed on top of the tissue, and then moved up and down at top speed for ~2mins until the tissue was fully dissociated in TRIzol. Cell suspensions, already stored in TRIzol, were vortexed 2000rpm for 10mins (MixMate; Eppendorf), and then incubated at room temperature for 15mins. For both tissue and cell suspensions, RNA and DNA were precipitated as outlined in the TRIzol manufacturer’s manual. Briefly, chloroform (1:5 with TRIzol) was added to create a density gradient from which the aqueous solution containing RNA was removed and processed separately. DNA was precipitated from the remaining solution with 100% ethanol (3:10 with TRIzol), and the resulting phenol-ethanol phase containing protein was moved into new snap-cap locking tubes (FisherBrand). The hippocampal tissue solution was split into equal aliquotes as indicated in the text (either 16x 40μL aliquots from two combined hippocamus samples or 4x 80μL aliquots from one hippocampus sample), and all aliquots were processed in parallel. Solutions from both CD11b+ and CD11b− cell suspensions were processed in parallel.
To precipitate protein from all samples, an excess of isopropanol was added (at least 2x the phenol-ethanol solution) to the phenol-ethanol phase. Samples were thoroughly vortexed and incubated at room temperature for 10mins, after which protein was pelleted by centrifugation at 12,000xg for 10mins at 4°C. After discarding the supernatant into biohazard waste, pellets were washed with 500μL 95% ethanol and dislodged by inverting the tube or, if necessary, by gently prodding the pellet with a sterile pipette tip. Washed samples were centrifuged at 7600xg for 5mins at 4°C. The supernatant was discarded and an additional wash with 250μL 95% ethanol was performed as described. After decanting, tubes were inverted and slowly dabbed on a paper towel to remove excess ethanol, and then inverted on a Kimwipe to dry for 10mins.
Solubilization of the protein pellet
Tubes were checked for trace ethanol, and if needed, dried with a Kimwipe. Once dry, the protein pellet was dislodged in 40–100μL lysis buffer as indicated in the text (lysis buffer composition depend on sample size and experimental parameters, see Tables 1–3). To determine if the concentration of individual chemicals in the lysis buffer modulates protein solubilization and yield, only one chemical was varied while all other concentrations remained constant. Samples were then incubated in a heat block at either 100°C or 50°C for varying incubation times, dependent on the experiment. After incubation, samples were allowed to cool at room temperature for 2–3 mins.
Table 1.
Primary antibody details for Western blotting analysis
| Dilution | Company | Catalog # | Host | Clonality | |
|---|---|---|---|---|---|
| ERK1/2 | 1:5000 | Cell Signaling | 4696S | Mouse | Monoclonal |
| Phospho-ERK1/2 | 1:2000 | Cell Signaling | #4370S | Rabbit | Monoclonal |
| GFAP | 1:2000 | Millipore | MAB5628 | Mouse | Monoclonal |
| Iba1 | 1:2000a 1:500b |
Wako | 019-19741 | Rabbit | Polyclonal |
| NeuN | 1:2000a 1:500b |
Bioss | bs-1613R | Rabbit | Polyconal |
| PSD95 | 1:2000 | ThermoFischer Scientific | 51-6900 | Rabbit | Polyclonal |
| β-actin | 1:10,000 | Abcam | ab8227 | Rabbit | Polyclonal |
| IL-10 | 1:100 | Acris | AP52181PU-N | Rabbit | Polyclonal |
| Parvalbumin | 1:2000 | Abcam | ab11427 | Rabbit | Polyclonal |
| Olig2 | 1:500 | Abcam | ab109186 | Rabbit | Monoclonal |
| VGlut1 | 1:1000 | Millipore | Ab5905 | Guinea Pig | Polyclonal |
Concentration for tissue homogenate;
Concentration for dissociated cell sample
Table 3. Optimized lysis buffer formulation and finite concentration gradient to determine if protein yield can be further enhanced.
Protein pellet solubilization was performed in optimized lysis buffer and in buffers with small changes in EDTA, NaCl, and SDS concentrations. The concentration of one chemical component was varied, while other chemical concentrations remained at optimized lysis buffer levels. For example, while varying EDTA concentrations, NaCl and SDS remained at 140mM and 5%, respectively. Each lysis buffer tested included Tris pH 8.0 (100mM) and phosphatase (50mM NaF, 1mM activated NaOv) and protease (Roche tablet) inhibitors, the concentrations of which did not vary.
| Optimized lysis buffer | Concentrations examined | ||||
|---|---|---|---|---|---|
| EDTA (mM) | 20 | 10 | 20 | 30 | 40 |
| NaCl (mM) | 140 | 100 | 120 | 140 | 160 |
| SDS (%) | 5 | 3 | 4 | 5 | 6 |
Determination of protein concentration via SDS-PAGE and Coomassie stain
Sample buffer (5x; 4mL glycerol, 2.5mL β-mercaptoethanol, 2.5mL Tris pH 6.8, 1.15g SDS, 0.2% bromophenol blue) was added to samples (1X final concentration; exact volumes varied based on the total volume of lysis buffer used), which were then vortexed and incubated for 10mins at 100°C. Although not explored herein, practical substitutes for β-mercaptoethanol include DTT and/or TCEP. After cooling, samples were centrifuged at top speed at room temperature for 2mins, and then stored at −20°C until processing or immediately separated by molecular weight using SDS-PAGE. Because protein stains can vary based on incubation and wash times, homogenate with previously determined protein concentration (diluted to 0.33, 1, or 2μg/μL) was resolved on each SDS-PAGE gel, and a standard curve was used to estimate concentrations of experimental samples on the same gel. 5μL of standards and experimental samples were loaded onto 4–12% Bis-Tris 15-well gels (Invitrogen). SDS-PAGE was completed with MES buffer (Invitrogen) for 55–60mins at 180V. Gels were then incubated in Bio-Safe Coomassie Stain (Bio-Rad) for at least 2hrs, washed with deionized water for at least 2hrs, and imaged with a LiCor Odyssey Imaging system. These analyses could alternatively be performed with other protein stains, e.g. Sypro Ruby, if desired. Using LiCor analysis software, boxes were drawn to encompass all protein bands in a single well, and the integrated intensity of the standards were used to define a standard curve with which experimental sample yield was estimated.
Direct homogenization and determination of protein loss from TRIzol precipitation
To determine whether optimized lysis buffer or a standard homogenization buffer, RIPA buffer, was more effective for direct homogenization of brain tissue, frontal cortex was homogenized in 500μL buffer (one hemisphere in RIPA, one hemisphere in optimized lysis buffer) with a Tissue Tearor (Biospec). The Tissue Tearor was placed on top of the tissue, turned to max speed, and moved up and down for ~2mins to completely dissociate the sample. Both samples were then stored on ice for 15mins to complete cell lysis as directed in the RIPA buffer manual (Life Technologies), and then equivalent volumes were boiled with sample buffer for 10mins at 100°C and diluted 1:10 for Coomassie protein concentration determination. Protein yield estimated via Coomassie stain was normalized to starting milligrams of tissue.
In order to determine the magnitude of protein loss due to protein precipitation/solubilization relative to direct homogenization, one hippocampus was homogenized in optimized lysis buffer (1000μL) directly, and the other hippocampus was homogenization in TRIzol (as indicated above). One tenth of the TRIzol protein fraction was precipitated and solubilized in 100μL optimized lysis buffer for 14hrs at 50°C. The solubilized fraction and 100μL of the direct homogenate were boiled with sample buffer; protein yield was estimated via Coomassie stain and normalized to starting milligrams of tissue.
Western blotting
15μg protein was subjected to SDS-PAGE identical to protein concentration assessments described above. Separated proteins were then transferred to 0.22μm nitrocellulose membranes (LiCor) in cold Tris-Glycine buffer (Life Technologies) at 150mA for 90mins. Membranes were dried and labeled with pencil, then rinsed in PBS and blocked with either 5% bovine serum albumin (BSA) in 1xPBS or LiCor Blocking buffer (in the case of cell suspensions). Subsequently, they were incubated with primary antibodies in 5% BSA in PBS-T (PBS with 0.2% Tween-20) or LiCor Blocking buffer with 0.2% Tween-20 overnight at 4°C with agitation. The following day, membranes were washed three times (5mins each) in PBS-T and incubated with secondary antibodies in 5% BSA in PBS-T or LiCor Blocking buffer with 0.2% Tween-20. Membranes were washed four times (5mins each) in PBS-T, rinsed with PBS, and then stored in PBS until imaging on a LiCor Odyssey Imaging system. LiCor analysis software was used to determine the integrated intensity of individual bands.
Antibodies
Antibodies are described in Table 1. Each primary antibody incubation occurred overnight at 4°C, and secondary antibody (all 1:20,000; LiCor Biosciences) incubations occurred for 1hr at room temperature. Secondary antibodies matched the host species of the primary antibodies, comprising of IRDye 680LT goat anti-mouse IgG, IRDye 800CW goat anti-rabbit IgG, IRDye 800CW goat anti-mouse IgG, IRDy 800CW goat anti-rabbit IgG (all LiCor Biosciences), and Alexa Fluor 647 goat anti-guinea pig IgG (ThermoFisher Scientific A-21450).
Statistics
GraphPad Prism (version 7.01) was used for all statistical analyses. One-way ANOVAs were used to determine if there was a significant difference among groups, and positive results were further analyzed with planned post-hoc Fisher’s LSD tests. For within-animal comparisons, paired t-tests were performed. All histograms represent mean±standard deviation; asterisks indicate statistically significant results (p<0.05).
3. Results
3.1 EDTA, NaCl, and SDS concentrations modulate protein yield during solubilization of TRIzol-precipitated protein
Solubilization buffers used for TRIzol-precipitated protein are primarily composed of SDS or urea. However, protein lysis buffers for homogenization of brain tissue commonly contain EDTA, NaCl, and Tris in addition to SDS (individual concentrations vary). We first sought to determine if these different chemical components common lysis buffers could influence the efficiency of protein pellet solubilization after TRIzol precipitation. To do this, we combined protein-containing phenol-ethanol solutions from two different rat hippocampus samples, and then split the homogenized solution into 16 equal aliquots (40uL per aliquot). Protein from each aliquot was simultaneously precipitated and pelleted, and then incubated for 30mins at 100°C in 40μL lysis buffer. A standard lysis buffer formulation was selected due to the authors’ familiarity with it, which has been used for direct homogenization and Western blotting in work done previously by Kopec and colleagues (2015): 1mM EDTA, 140mM NaCl, 2% SDS, 10mM Tris pH 8.0, 50mM NaF, 1mM activated NaOv, protease inhibitor table (1 tablet per 50mL solution; Roche). Using this formulation as a starting point, we then varied the concentrations of either EDTA, NaCl, SDS, or Tris across individual experimental conditions (Table 2).
Table 2. Lysis buffer formulations used to determine if concentrations of chemicals commonly found in protein lysis buffers alter precipitated protein solubilization.
Protein pellet solubilization was performed in lysis buffers with varying concentrations of EDTA, NaCl, SDS, and Tris. The concentration of one chemical component was varied, while other chemical concentrations remained at standard lysis buffer levels. For example, while varying EDTA concentrations, NaCl, SDS, and Tris remained at 140mM, 2%, and 10mM, respectively. Each lysis buffer tested included phosphatase (50mM NaF, 1mM activated NaOv) and protease (Roche tablet) inhibitors, the concentrations of which remained unchanged.
| Standard lysis buffer | Concentrations examined | ||||
|---|---|---|---|---|---|
| EDTA (mM) | 1 | 1 | 5 | 20 | 100 |
| NaCl (mM) | 140 | 140 | 200 | 500 | 700 |
| SDS (%) | 2 | 0.2 | 2 | 5 | 7.5 |
| Tris (mM) | 10 | 10 | 50 | 100 | 500 |
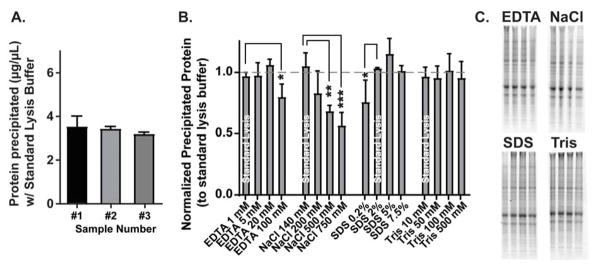
Four of the 16 experimental samples contained identical formulations to the standard lysis buffer recipe, and an analysis of protein from these samples alone indicated that precipitation in standard lysis buffer yields on average 3.35±0.30μg/μL from the hippocampal fraction as estimated by Coomassie stain (Fig. 1A). The standard lysis buffer samples were then averaged to serve as within sample controls to which all other buffer yields were normalized. This accounted for subtle differences in total protein between samples. One-way ANOVAs performed for each group (EDTA, NaCl, SDS, Tris) determined that concentrations of EDTA (F(3,8)=5.5, n=3, p<0.05), NaCl (F(3,8)=8.9, n=3, p<0.01, and SDS (F(3,8)=6.6, n=3, p<0.05) all significantly influenced protein solubilization, while different Tris concentrations did not significantly affect protein yield (Fig. 1B). Fisher’s LSD post-hoc analyses comparing experimental buffers with the standard buffer within each group determined that there were significantly increased protein yields in samples with 1mM EDTA vs. 100mM EDTA (Mean Difference=0.18, p<0.05), 140mM NaCl vs. 500mM NaCl (MD=0.38, p<0.01) and 750mM (MD=0.51, p<0.01), and 0.2% SDS vs. 2% SDS (MD=0.29, p<0.05). These data indicate that EDTA, NaCl, and SDS concentration can each individually modulate yield during protein pellet solubilization.
Figure 1. EDTA, NaCl, and SDS concentrations significantly alter protein yield during solubilization of TRIzol-precipitated protein.
(A) Solubilization with standard lysis buffer (1mM EDTA, 140mM NaCl, 2% SDS, 10mM Tris) after TRIzol precipitation from three separate samples (each containing two combined rat hippocampus samples) yields on average 3.4μg/μL protein. (B) Varying the concentration of EDTA, NaCl, and SDS individually, while all other chemical components were held at standard lysis buffer levels, significantly influenced the protein yield during solubilization. Tris concentration did not impact yield. (C) Representative Coomassie stained gels of the data quantified in (B).
To determine if simultaneously varying the concentrations of these chemicals could further enhance the solubilization process, we formulated an ‘optimized’ lysis buffer, defined as the combination of chemical concentrations that produced the nominally highest protein yields in Fig. 1B (20mM EDTA, 140mM NaCl, 5% SDS, 100mM Tris pH 8.0).
Because the initial concentration gradients that revealed the effect of concentration on solubilization (Fig. 1) covered a broad range, we next determined if smaller changes in concentration around the optimized level would further improve protein yield. The phenol-ethanol fractions from two hippocampus samples from different rats were pooled and split into 16 equal aliquots to replicate conditions in Fig. 1B, and then 12 of these aliquots were precipitated and incubated for 30mins at 100°C in 40μL lysis buffer (Table 3).
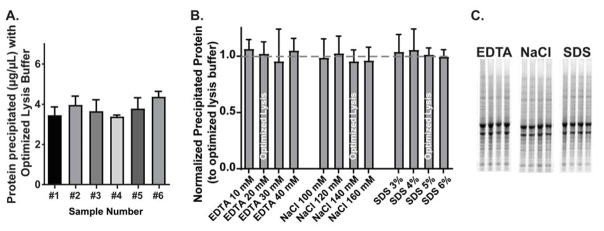
Similar to previous experiments, 3 aliquots from each sample were incubated in identical ‘optimized’ lysis buffers, and all other experimental conditions were normalized to these controls. The average protein yield using this optimized lysis buffer was on average 5.19±0.68μg/μL as measured by Coomassie stain (Fig. 2A), an increase in ~2μg/μL from the standard buffer. These data support the notion that optimizing each individual chemical component of the lysis buffer can in turn optimize the solubilization process as a whole. One-way ANOVAs were performed for each group (EDTA, NaCl, SDS) and revealed no significant differences between the concentrations tested (Fig. 2B; n=6), indicating that the experimentally determined optimized lysis buffer is likely to maximize protein yield from TRIzol-precipitated protein, at least within the parameters we tested. We thus continued to use this optimized lysis buffer for all subsequent experiments.
Figure 2. Simultaneous optimization of EDTA, NaCl, and SDS concentrations enhances protein yield during solubilization of TRIzol-precipitated protein.
(A) Solubilization with optimized lysis buffer (20mM EDTA, 140mM NaCl, 5% SDS, 100mM Tris) after TRIzol precipitation from six separate hippocampus samples yields on average 5.2μg/μL protein, compared to 3.4μg/μL yield with standard lysis buffer (Fig. 1A). (B) Varying the concentration of EDTA, NaCl, and SDS around the optimized values determined in Fig. 1B does not further enhance protein yield during solubilization. (C) Representative Coomassie stained gels of the data quantified in (B).
3.2 Optimized lysis buffer solubilizes TRIzol-precipitated protein more effectively than other protein lysis buffers in a variety of time and temperature incubations
We next wanted to confirm that the optimized lysis buffer was, indeed, a more efficient vehicle for protein pellet solubilization than other formulations of protein lysis buffers. Specifically, we compared the optimized lysis buffer to the standard lysis buffer formulation used in Fig. 1 and the widely used homogenization buffer, RIPA buffer. Additionally, we added a group to test the efficacy of these protein lysis buffers against the primary recommended solubilization buffer by the TRIzol manufacturer, 1% SDS (see Table 4 for a comparison of these buffers). The other common class of solubilization buffers for TRIzol-precipitated protein reported in the literature are urea-containing buffers (Man et al., 2006; Reddy et al., 2013; Simoes et al., 2013; Yamaguchi et al., 2013; Young and Truman, 2012); however, we chose not to test urea buffers here due to the ability of urea to modify proteins upon heating (Kollipara and Zahedi, 2013; see Discussion).
Table 4.
Comparison of chemical composition of buffers commonly used for protein analyses
| Optimized | RIPA | 1% SDS | Standard | |
|---|---|---|---|---|
| EDTA (mM) | 20 | 1 | ||
| NaCl (mM) | 140 | 150 | 140 | |
| SDS (%) | 5 | 0.1 | 1 | 2 |
| Tris (mM) | 100 | 50 | 10 | |
| Triton-X100 (%) | 1 | |||
| Sodium Deoxycholate (%) | 0.5 |
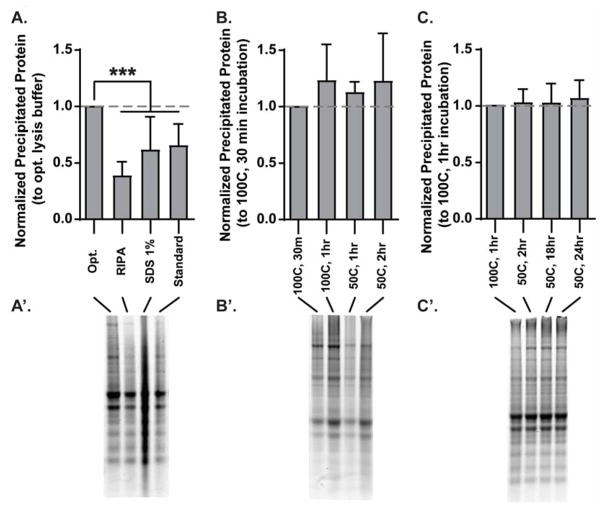
The phenol-ethanol solution from a single rat hippocampus was split into 4 equal aliquots (80μL), precipitated in parallel, and then incubated for 30mins at 100°C in 100μL buffer. The standard and optimized lysis buffer incubation resulted in an average yield of 3.57±1.35μg/μL and 5.93±2.59μg/μL, respectively (data not shown), which replicates (albeit with more inter-sample variability) the average yield produced by earlier experiments using these buffers (Figs. 1A and 2A). To account for variations in the total protein in different hippocampus samples, protein yield from all experimental conditions was normalized to that of the optimized lysis buffer within the same sample. A one-way ANOVA revealed a significant difference between groups (Fig. 3A; F(3,20)=12.03, n=6, p<0.01), and post-hoc analyses determined that the optimized lysis buffer yielded significantly more protein than all other buffers (Opt. vs RIPA: MD=0.66; Opt. vs SDS: MD=0.41; Opt. vs. Standard: MD=0.37, n=6, all p<0.01). Interestingly, the samples solubilized in 1% SDS often resulted in a smear during SDS-PAGE (e.g. representative image in Fig. 3A), suggesting that the protein in the sample may not be fully denatured despite receiving the same denaturing treatment that was effective for samples solubilized in other buffers (10min incubation at 100°C in 1x sample buffer). These data collectively support our previous results indicating that the composition of the lysis buffer can significantly impact the efficiency of pellet solubilization and highlight the efficiency of the experimentally determined optimized lysis buffer.
Figure 3. Optimized lysis buffer solubilizes TRIzol-precipitated protein more effectively than commonly used buffers in a variety of time and temperature incubations.
(A) Optimized lysis buffer significantly increased protein yield during solubilization (30mins at 100°C) relative to other lysis buffers (RIPA, 1% SDS, and standard lysis buffer [see Fig. 1]). Protein yield via solubilization in optimized lysis buffer does not significantly differ when performed at a variety of temperatures (B) or for varying lengths of time (C). Representative Coomassie stained gels of the data quantified are depicted in A′–C′.
Upon confirmation of the utility of optimized lysis buffer, we sought to determine the optimal time and temperature incubations necessary to provide the highest yield of precipitated protein. A 30min incubation at 100°C was initially chosen based on various reports indicating that modest heat (50°C) aided in, but did not completely solubilize precipitated protein pellets. However, the combination of very high temperatures and pressure in the microcentrifuge incubation tubes can cause the tops to burst open, resulting in a loss of volume. To determine if protein solubilization could be effective at lower temperature incubations, we split a single TRIzol-homgenated hippocampus into 4 equal aliquots, precipitated the protein, and incubated the pellets in 100μL optimized lysis buffer for (i) 30mins at 100°C, (ii) 60mins at 100°C, (iii) 60mins at 50°C, and (iv) 120mins at 50°C. Protein yield was normalized to that of the 30min, 100°C incubation within each sample. A one-way ANOVA revealed no significant differences between the different incubation parameters (Fig. 3B; n=6). These unexpected data suggest that the optimized lysis buffer can yield equivalent protein concentration over a 50°C temperature range (i.e. 50–100°C). To determine if there is similar flexibility in incubation time, we compared protein yield from (a) 1hr 100°C, (b) 2hr 50°C, (c) 18hr 50°C, and (d) 24hr 50°C incubations. A one-way ANOVA revealed no significant differences between groups (Fig. 3C, n=6), indicating that protein yield is not affected by the length of incubation, at least up to 24 hrs.
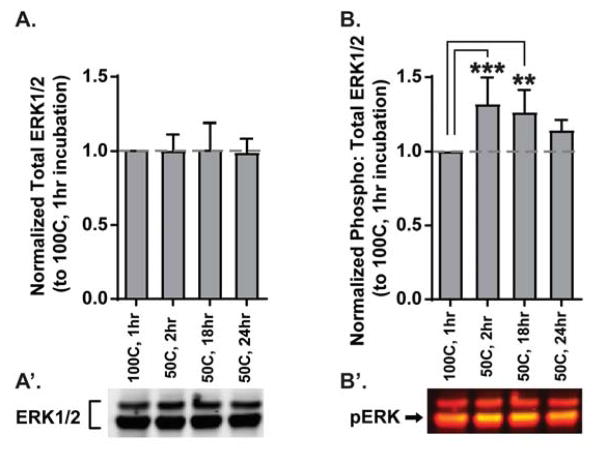
Long incubations increase the risk of losing the resolution of post-translational modifications, like phosphorylation status. Thus, we subjected 15μg of the protein depicted in Fig. 3C to Western blotting to analyse the phosphorylation state of ERK1/2, a kinase that becomes activated by several upstream signals to mediate a wide variety of molecular events (Adams and Sweatt, 2002), making it a ubiquitous component of most neural phenomena. Phospho-specific ERK1/2 signal was normalized to total ERK1/2 within the same sample to calculate an activity ratio. The activity ratios across experimental conditions were then normalized to that of 1hr 100°C. While there was no difference in total ERK1/2 across the groups (Fig. 4A, n=6), a one-way ANOVA indicated there was a significant difference between groups in the level of ERK1/2 phosphorylation (Fig. 4B; F(3,20)=7.9, n=6, p<0.01), and post-hoc analyses revealed that the 50°C 2hr and 18hr groups had significantly higher phospho-ERK1/2 than the 1hr 100°C group (100°C 1hr vs 50°C 2hr: MD=−0.34; 100°C vs 50°C 18hr: MD=−0.28, n=6, all p<0.01), while 50°C 24hr was not significantly different.
Figure 4. Solubilization with optimized lysis buffer preserves phospho-specific signals for incubations up to 18 hrs.
(A) Total ERK1/2 protein does not change despite extended incubations, consistent with no changes in total protein during these same incubations (Fig. 3C). (B) Phospho-specific ERK1/2 signal is significantly enhanced by incubations at lower temperatures for up to 18 hrs. Representative Western blots of the data quantified are depicted in A′–B′. In B′, total ERK1/2 appears red and phospho-ERK1/2 (pERK) appears yellow (green overlaid on red) and is most strongly recognized in the 42kDa band (arrow).
Taken together, these data indicate that protein pellet solubilization in optimized lysis buffer is best achieved with a 50°C incubation for 2–18 hrs to preserve phospho-specific signals. This affords a remarkable amount of flexibility for experimental design and scheduling.
3.3 Effect of optimized lysis buffer on protein yield during direct homogenization versus TRIzol-precipitation and solubilization
Having identified an optimized lysis buffer formulation and solubilization protocol for TRIzol-precipitated protein, we next determined whether protein precipitation and subsequent solubilization would result in protein loss compared to a directly homogenized tissue sample. The most direct comparison of protein yield after solubilization in optimized lysis buffer is tissue directly homogenized in optimized lysis buffer. However, it was unclear whether or not the formulation of optimized lysis buffer would be effective for direct homogenization. Thus, we first wanted to determine if homogenization in RIPA buffer, considered the standard for direct homogenization, produced better protein yield than our optimized lysis buffer. Thus, solubilized protein yield could then be compared to the most effective direct homogenization alternative.
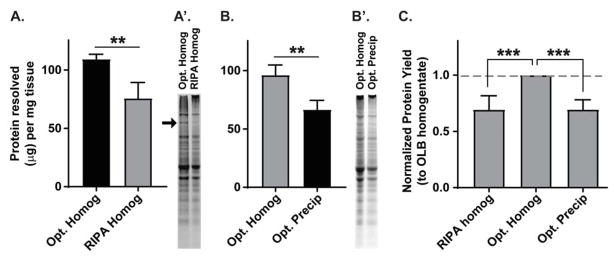
Frontal cortex from a single animal was collected and separated into two samples (one from each hemisphere) and its mass was recorded for within-animal comparisons. Samples were homogenized either in 500μL of optimized lysis buffer or RIPA buffer and diluted 1:10 in the corresponding buffer for concentration analyses. To our surprise, optimized lysis buffer resolved on average 109.3±4.22μg protein per milligram of tissue, which was significantly higher than protein resolved with RIPA buffer homogenation (75.72±13.46μg protein resolved per milligram tissue; paired t-test t(5)=5.98, n=6, p<0.01; Fig. 5A). Furthermore, in every set of samples, there was a high molecular weight band (~200kDa) present in optimized lysis buffer samples but not in RIPA buffer samples (representation in Fig. 5A′, arrow), suggesting an entire class of proteins may be not be effectively solubilized by RIPA buffer.
Figure 5. Comparison of optimized lysis buffer protein yield in directly homogenized and precipitated samples.
(A) Direct homogenization of frontal cortex with optimized lysis buffer (Opt. Homog) resolves significantly more protein per milligram of tissue than RIPA buffer (RIPA Homog). (A′) Representative Coomassie stained gels normalized for starting tissue mass. An arrow depicts a high molecular weight protein band that is only present if tissue is homogenized in optimized lysis buffer, and not RIPA buffer. (B) Optimized lysis buffer solubilization of TRIzol-precipitated protein (Opt. Precip) yields ~70% of the protein from hippocampal samples than if optimized lysis buffer is used for direct homogenization (Opt. Homog). (B′) Representative Coomassie stained gels normalized for starting tissue mass. Importantly, both the homogenized and precipitated protein samples resolve the protein band absent from RIPA buffer-homogenized protein. (C) Protein yields from (A) and (B) were normalized to optimized lysis buffer homogenate (Opt. Homog) within the same animal. Protein yield due to solubilization (Opt. Precip) is equivalent to protein yield in RIPA buffer homogenate (RIPA homog).
We next determined the level of protein loss from TRIzol precipitation and pellet solubilization in optimized lysis buffer relative to direct homogenization in optimized lysis buffer. Hippocampal samples were collected from a single animal and separated into two samples (one from each hemisphere) and its mass was recorded for within-animal comparisons. One sample was directly homogenized in 1mL optimized lysis buffer as described, and one sample was homogenized in 500μL TRIzol, and 1/10 the protein-containing solution was precipitated and solubilized (~14hrs at 50°C) in 100μL optimized lysis buffer so the fraction solubilized would represent an equivalent dilution to the directly homogenized sample. Direct homogenization with optimized lysis buffer resolved on average 96.18±8.60μg protein per milligram of tissue, while TRIzol precipitation and solubilization resolved on average 66.55±7.95μg protein per milligram of tissue (paired t-test, t(5)=6.87, n=6, p<0.01; Fig. 5B). The average protein loss due to either RIPA buffer homogenization or protein precipitation and solubilization in optimized lysis buffer (each relative to optimized lysis buffer homogenization) appeared roughly equivalent. In order to compare the protein yield from RIPA buffer homogenization with optimized lysis buffer solubilization directly, we scaled the common factor in each experiment, optimized lysis buffer homogenate, to “1,” and adjusted the matched within-animal comparison (either RIPA homogenate or precipitated protein) equivalently. A one-way ANOVA (F(2,21)=52.08, p<0.01) and subsequent comparison of all groups confirmed a significant difference between samples directly homogenized with optimized lysis buffer with (i) those homogenized with RIPA buffer (MD=−0.31, p<0.01) and (ii) those precipitated and solubilized in optimized lysis buffer (MD=0.31, p<0.01). However, there was no difference between samples homogenized in RIPA buffer and samples precipitated and solubilized in optimized lysis buffer (MD=0.002, p=0.97, all n=6; Fig. 5C). These data have important implications for (i) the establishment of a baseline, or ‘best’ practice for protein analyses, and (ii) defining the expected protein yield used to compare other methods of extraction/solubilization (see Discussion).
3.4 Solubilized protein is representative of different neural cell types and protein classes
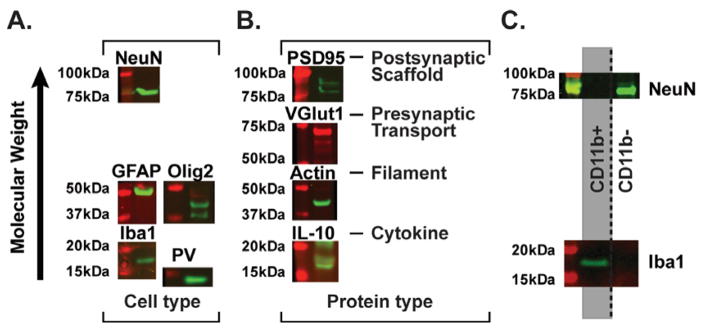
We next determined the cell type-specific and protein class representation of precipitated protein via Western blot. Using a Coomassie gel to determine total protein concentration as described above, 15μg protein from rat prefrontal cortex or hippocampus was analyzed via SDS-PAGE, transferred to nitrocellulose membranes, and subsequently incubated with primary and secondary antibodies. Specifically, to determine if multiple cell types were represented in precipitated protein, we probed the blots for (i) NeuN, a nuclear and neuron-specific marker, (ii) glial fibrillary acidic protein (GFAP), a filament protein in astrocytes, (iii) oligodendrocyte transcription factor 2 (Olig2), (iv) ionized calcium-binding adaptor molecule 1 (Iba1), a ubiquitious protein in microglia, and (v) parvalbumin (PV), a calcium-binding albumin protein characteristic of some classes of inhibitory interneurons. All cell types represented by these proteins were present in TRIzol-precipitated hippocampal protein (Fig. 6A). Next, because neural communication requires coordinated pre- and post-synaptic signaling as well as secreted proteins from both neurons as well as other cell types, we probed for (1) post-synaptic density protein 95 (PSD95), a post-synaptic scaffolding molecule at excitatory synapses, (2) vesicular glutamate transporter 1 (VGlut1), a pre-synaptic transporter at excitatory synapses, (3) actin, a cell-wide filament, and (4) interleukin-10 (IL-10), a secreted anti-inflammatory cytokine primarily released by microglia. In addition to the expression of the nuclear-specific NeuN (Fig. 6A) and total and phospho-ERK1/2 (Fig. 4), which can be found in all subcellular compartments depending on the context, all classes of proteins tested, including secreted proteins normally very difficult to quantify in brain (e.g. IL-10), were represented in precipitated protein samples (Fig. 6B). Additional information on each antibody (e.g. host species, clonality, concentration) can be found in the Methods (Table 1), and a comparison of the predicted kDa molecular weight based on the amino acid sequence versus observed molecular weight for each antibody is presented in Table 5. Importantly, changes in molecular weight as measured via Western blot are common due to a variety of post-translational modifications to which many proteins are subjected.
Figure 6. TRIzol-precipitated protein from brain tissue represents multiple cell types and protein classes.
(A) Different cell types inherent in brain tissue are detected in Western blot analysis of precipitated protein. NeuN=neurons, GFAP=astrocytes, Olig2=oligodendrocytes, Iba1=microglia, PV=inhibitory interneurons. (B) Different protein classes are represented in precipitated protein, including synaptic (PSD95, VGlut1), membrane bound (Iba1), nuclear (NeuN), cytoplasmic (actin and ERK1/2 in Fig. 4), and secreted (IL-10) proteins. (C) Protein from specific populations of a single cell suspension (i.e. microglia in the CD11b+ population and other neural cell types in the CD11b− population) is successfully analyzed after solubilization in optimized lysis buffer. The CD11b+ population contains Iba1, but not NeuN protein, while the CD11b− population contains NeuN, but not Iba1 protein.
Table 5.
Predicted and observed molecular weights of proteins detected via Western blotting
| Expected Molecular Weight (kDa) | Observed Molecular Weight (kDa) | |
|---|---|---|
| ERK1/2 | 42, 44 | 42, 44 |
| Phospho-ERK1/2 | 42, 44 | 42, 44 |
| GFAP | 50 | 50 |
| Iba1 | 17 | 17 |
| NeuN | 48 | 75 |
| PSD95 | 95 | 95,85 |
| β-actin | 42 | 42 |
| IL-10 | 18 | 17 |
| Parvalbumin | 12 | 12* |
| Olig2 | 32 | 42, 37 |
| VGlut1 | 62 | 70, 62 |
Additional diffuse signal
In a final set of experiments, we sought to determine the practical application of TRIzol precipitation and solubilization in optimized lysis buffer for small and difficult to attain brain samples. We used enzymatic and mechanical dissociation to create a single cell suspension from bilateral mouse hippocampus tissue followed by cell type-specific isolation. Specifically, we sorted the suspension into CD11b+ and CD11b− populations using CD11b-conjugated microbeads (MACS Miltenyi Biotec). CD11b is a component of the complement receptor 3 heteromer that is expressed on a variety of immune cells, including microglia. Saline perfusion of tissue prior to dissection decreases the contribution of immune cells in blood vessels, and thus a CD11b+ population from brain tissue should be enriched for microglia, while the CD11b− population should contain all other cell types, which has been confirmed via fluorescence activated cell sorting (FACS) and gene expression analyses (Schwarz et al., 2013). Protein from CD11b+ (estimated at ~400,000 cells) and CD11b− (estimated at ~1,000,000 cells) populations was precipitated and solubilized in 40μL optimized lysis buffer at 50°C for 2hrs, and protein yield was measured via Coomassie stain. 15μg protein was analyzed via Western blot and probed with microglia-specific Iba1 and neuron-specific NeuN antibodies. As predicted, the CD11b+ cell population, i.e. microglia, had Iba1, but not NeuN immunoreactivity. Moreover, the CD11b− population, consisting of other neural cell types, had NeuN, but not Iba1 immunoreactivity (Fig. 6C). These data broaden the applicability of solubilization in optimized lysis buffer to the analysis of protein in enriched cell type-specific populations from dissociated brain regions, thus maximizing the data available from difficult techniques or small samples. This is particularly advantageous as it permits the analysis of cell type-specific alterations in a biologically and behaviorally relevant context, while immortalized cell culture lines cannot completely recapitulate the dynamics of in vivo neural networks.
4. Discussion
We have experimentally defined an optimized buffer and incubation protocol for analysing brain tissue via Western blot by solubilizing protein pellets precipitated from TRIzol. This technique permits a comprehensive examination of the molecular signalling underlying neural phenomena via the simultaneous analysis of gene and protein expression levels. Furthermore, the retrieval of protein adds a level of analysis that is difficult to infer using gene expression analyses alone: the activation state of signalling brokers and other post-translational modifications. In essence, this technique doubles the available data without increasing the number of subjects. Importantly, solubilization in optimized lysis buffer yields samples reflective of different neural cell types and protein classes, and can successfully resolve protein from small and difficult to attain brain samples in both mice and rats.
4.1 Outline of the optimized protein solubilization protocol
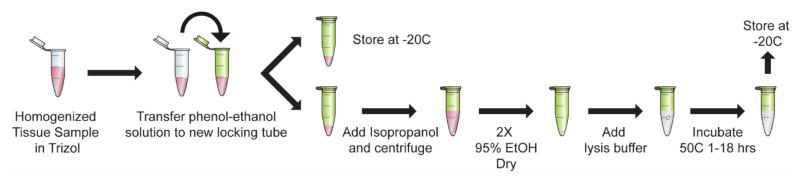
Our protocol is graphically depicted in Fig. 7. Tissue is homogenized in 500μL TRIzol with a Tissue Tearor (BioSpec). The Tissue Tearor is placed on the tissue (off), and then turned on to the max speed and slowly moved up and down for ~2mins to fully homogenize the tissue. RNA and DNA are then precipitated according to the manufacturer’s protocol (Life Technologies), and the remaining phenol-ethanol solution is transferred to new snap-cap locking tubes. We recommend experimentally determining the optimal fraction of the phenol-ethanol solution per brain region to use for precipitation, as very large protein pellets are prohibitive to full solubilization. The remaining TRIzol-protein fraction can be frozen and precipitated for future experiments if needed. At least 2x volume of isopropanol is added to the solution to precipitate protein, which is subsequently pelleted by centrifugation (12,000xg for 15mins at 4°C). The pellet is washed twice in 95% ethanol and dried. During washes, it is important to physically dislodge the pellet for complete washing. This can be achieved by using a pipette tip to nudge it, and then inverting the tube. We have noticed that the extent to which trace ethanol is removed from the tube influences the efficiency of solubilization, with a larger volume of trace ethanol interfering with this process. We encourage experimenters to dry tubes via vacuum or swabs if they notice excessive ethanol remaining after drying. Pellets are then dislodged in optimized lysis buffer (they often float on their own, but can be lightly nudged with a sterile pipette tip to facilitate this process). The volume of lysis buffer should additionally be experimentally determined. As presented here, up to 25% of a unilateral rat hippocampus efficiently solubilizes in 100μL lysis buffer and a single cell suspension enriched for microglia from bilateral hippocampi efficiently solubilizes in 40μL. Samples should be incubated in lysis buffer at 50°C for 1–18hrs. As long as the experimenter is consistent with the incubation length between experimental groups, our data indicate these incubation lengths are equally effective. After incubation, protein can be stored at −20°C until further processing.
Figure 7. Graphical depiction of protein precipitation and solubilization procedure.
Tissue is homogenized in TRIzol, and a fraction of the phenol-ethanol solution (following RNA isolation) is used for protein precipitation, while the remaining fraction can be stored at −20 to −80°C indefinitely. Isopropanol precipitates protein, after which it is pelleted and washed with 95% ethanol. The pellets are dried, suspended in optimized lysis buffer (20mM EDTA, 140mM NaCl, 5% SDS, 100mM Tris pH 8.0, 50mM NaF, 1mM activated NaOv, Roche protease inhibitors), and incubated at 50°C for 1–18hrs. The temperature and incubation time is flexible and can be defined by the experimenter as long as all samples are consistently treated.
4.2 Deviations from established solubilization protocols
Our protocol has a few differences from other protocols that are worth discussing. First, pellet washes are completed in 95% ethanol, while many published protocols add guanidine hydrochloride to the wash solution. Guanidine hydrochloride reacts with SDS to form a precipitant (Arnold and Ulbrich-Hofmann, 1999), and despite subsequent washes in ethanol alone to limit this reaction, we found that a precipitant formed in optimized lysis buffer. A pilot experiment determined that there was no significant difference between washes in 95% ethanol or 95% ethanol with guanidine hydrochloride (data not shown), and thus we eliminated guanidine hydrochloride. An additional difference is the recommendation to precipitate only a portion of the phenol-ethanol solution. While this may require multiple precipitation procedures depending on the needs of the experiment, the protocol detailed here reliably yields enough protein for several 15–30μg Western blots. If the targets to be probed with primary antibodies are relatively high abundance, this number increases as fewer μg of protein per sample are needed for accurate Western blotting assessment. Moreover, Western blots can withstand antibody stripping 1–2 times, and thus experiments can be strategically planned to measure lowest abundance targets first, and higher abundance targets after antibody removal.
The primary deviation differentiating this protocol from those in the literature is the chemical composition of the solubilization buffer. The most common solubilization buffers are either varying percentages of SDS alone or a urea-containing solution. In fact, the TRIzol protocol (Life Technologies) recommends solubilization in 1% SDS, and indicates that dialysis and subsequent reconstitution in an SDS-urea solution may improve yield (Life Technologies; November 9, 2016 edition). Solutions containing SDS alone are not very effective, even with 50°C incubations and sonication (Banerjee et al., 2003; Likhite and Warawdekar, 2011). Urea-containing buffers do not result in complete solubilization, but are more effective than SDS solutions (Man et al., 2006; Reddy et al., 2013; Simoes et al., 2013; Yamaguchi et al., 2013; Young and Truman, 2012). However, urea should not be heated over 37°C due to its hydrolysis into isocyanate, which is known to interfere with proteins via carbamylation (Kollipara and Zahedi, 2013). Carbamylation, like other post-translational modifications, can occlude amino acid residues and change protein size, but is additionally a natural molecular process exacerbated in the brain during aging (Gorisse et al., 2016). This makes introducing excess carbamylation problematic for protein analysis in neural phenomena. Carbamylation has been demonstrated to be effectively inhibited by ammonium containing buffers like ammonium bicarbonate (Sun et al., 2014), but ammonium bicarbonate is similarly thermally unstable. Diethylamine (DEA), a reagent often used to solubilize amyloid beta particles, has also been reported to be effective in protein pellet solubilization (Nolan and Teller, 2006), though the need to neutralize DEA by a volume increase prior to SDS-PAGE analysis could potentially make it more difficult to use with samples requiring a small volume to concentrate proteins prior to detection. Lastly, one group, to which the TRIzol manual is referring, used dialysis, protein concentration via centrifugation, and then reconstitution with urea and/or SDS buffers to obtain very efficient solubilization (Hummon et al., 2007). However, with dialysis solution changes after 16, 20, and 22hrs in addition to protein concentration equipment, this technique is difficult in practice.
The advantage of the solubilization buffer experimentally determined here is that it quickly and efficiently solubilizes protein while being readily amenable to heat-mediated denaturing important for Western blotting. What remains unclear is the exact chemistry underlying the increased efficiency of protein pellet solubilization. Our data indicate that EDTA, SDS, and NaCl concentrations significantly impact protein yield (Fig. 1). While SDS has a well described role in protein denaturation (Anderson et al., 2009; Otzen, 2011; though it is less effective for proteins enriched with β-sheet structure; Nielson et al., 2007), it is less clear how changes in the concentration of EDTA and NaCl affect this process. EDTA binds divalent metal cations like magnesium and calcium. Interestingly, divalent cations have also been demonstrated to be critical for protein crystallization (Trakhanov and Quiocho, 1995), disulfide bond formation (which was reversed by EDTA; Price et al., 1969), and metalloprotein stabilization (also reversed by EDTA; Janecki and Reilly, 2005). Thus the ability of proteins to maintain states that make them insoluble (e.g. quaternary or tertiary structure or protein-protein aggregate interactions) may be reduced by virtue of cation chelation by EDTA. A phenomenon known as “salting out” occurs when the salt concentration of a solvent is increased in a protein-containing solution, which alters the charge and/or conformation of proteins, in effect forcing protein out of solution (Arakawa and Timasheff, 1984; Ghosh et al., 2005; Kramer et al., 2012). Conversely, lowering the salt concentration can enhance protein solubility (i.e. “salting in”), which may account for significantly increased protein yield at lower NaCl concentrations (Fig. 1B). We thus propose that a combination of these processes may in part account for the superior ability of optimized lysis buffer to solubilize protein pellets relative to other buffers.
4.3 Limitations to and advantages of the current method
Like all techniques, there are limitations and advantages inherent in the process. The main limitation of the technique presented here is that it is primarily compatible with Western blotting. We suspect that the solubilized protein is not entirely in its native form, due to high levels of SDS and EDTA, which can modify protein structure as discussed above. Although the solution may not consist of completely denatured proteins, this is likely to be incompatible with other protein analyses that require native protein, like ELISA or mass spectrometry. Reports have demonstrated that dilution of high-SDS solutions to less than 0.2% SDS results in comparable ELISA signal to solutions with much lower SDS concentrations (Lechtzier et al., 2002; Winkler et al., 1987). Furthermore, SDS can be removed via filter-aided sample preparation (FASP) to make solutions compatible with mass spectrometry analysis (Wisniewski et al., 2009). This same notion pertains to the method of determining protein concentration. The optimized lysis buffer contains significantly higher SDS concentrations (5% vs. 0.125%) than is recommended for Bradford assays, and slightly higher EDTA concentrations (20mM vs 10mM) than is recommended for BCA assays. NanoDrop is a fast and easy technique for measuring purified proteins that preserves sample volume (1–2μL per measurement), but is not often used for protein measurements because (i) the sample is often not pure, and (ii) many common buffers, such as RIPA buffer, contain detergents that have significant absorption profiles at 280nm, the wavelength at which protein concentration is primarily determined. The solubilized solutions using the presented method are, in theory, pure protein samples, and optimized lysis buffer does not have significant 280nm absorption (data not shown). Pierce 660nm assays have an additional reagent that can be added to incubation buffers to help account for high EDTA and SDS content. However, a preliminary screen comparing RIPA buffer-homogenized and optimized lysis buffer-homogenized tissue (samples depicted in Fig. 5A) indicate that RIPA buffer yield was much higher than optimized lysis buffer yield (data not shown), which was clearly not the case when assessed by Coomassie-stained gels (Fig. 5A′). These data suggest that a substantial amount of protein (~2/3) is at least partially denatured, and thus not reliably measured by any assessment of native protein. Additional analyses will need to be completed to confirm this notion, as well as to determine if denatured proteins can be returned to their native confirmation by removal of denaturing reagents in order to be compatible with other protein analysis techniques.
There are also several advantages to the proposed technique. First, the ability to analyze gene and protein expression levels from the same sample is very beneficial, as discussed above. The solubilized protein solution, while not being quite as effective as direct homogenization in optimized lysis buffer, was as effective as direct homogenization in RIPA buffer (Fig. 5). Moreover, a ~200kDa band, the proteins of which it is comprised are as yet undetermined, was present in optimized lysis buffer-homogenized samples and entirely absent from RIPA-homogenized samples (Fig. 5 A′ versus B′). Our data indicate that several different neural cell types and protein classes are represented in solubilized protein samples. Thus, as long as both control and experimental samples are treated equivalently, direct comparisons of both gene expression, protein levels, and protein activation status for several different cell types should be possible for each sample, including small cell type-specific suspensions.
Importantly, different brain regions or tissue from different organs may require slightly different preparation/optimization. Our lab has performed protein precipitation using the optimized lysis buffer in hippocampus, nucleus accumbens, prefrontal cortex, spleen, liver, and dissociated cell suspensions from hippocampal tissue, all with similar success. It is important to note that while our cell type-specific isolations are proof of principle that small samples can be analyzed via precipitation and solubilization, we did not determine the minimum number of cells required to reliably obtain data. By extrapolation (a sample of the cell suspension was stained and counted with a hemocytometer), our samples (Fig. 6C) represent ~400,000 microglial cells in the CD11b+ population and 1,000,000 other neural cells in the CD11b− population. Neural cell types vary in size considerably, even within a single cell type (i.e. all neurons): neurons can range in cell body diameter from 5–100μm, while microglia, the immune cells of the brain, can range in cell body diameter from 5–20μm. Thus, the minimum number of cells required may depend on the cell type of interest (i.e. larger cells will in theory yield more protein), and should be experimentally determined. Moreover, some brain regions have distinct protein expression in rostral-caudal, dorsal-ventral, or medial-lateral gradients, which will not be represented in protein homogenate via either standard techniques (i.e. direct homogenization) or this solubilization technique. If experimenters wish to evaluate these parameters, care should be taken to dissect the regions of interest into separate samples, or tissue punches should be acquired from the region of interest.
Second, there is a great deal of experimental flexibility inherent in this protocol. The volume for solubilization can be determined by the needs of the experimenter. We recommend making a more concentrated solution initially, and then diluting to concentrations appropriate for the amount of total protein to be analyzed. This is particularly useful for small or precious samples, when over-diluting can be disadvantageous. Moreover, the temperature and length of incubation can be dictated by the needs and schedule of the experimenter. We recommend a moderately long incubation at 50°C to maximize protein yield without compromising phospho-specific signal. However, if the experimenter is working with very abundant tissue (e.g. hippocampus), using 10–25% of the total phenol-ethanol fraction with a short incubation at 100°C may be the best use of time. Alternatively, an overnight incubation may be advantageous. Importantly, we report that the phosphorylation status of an abundant kinase, ERK1/2, is relatively stable during long incubations, but it is possible that other kinases may exhibit different temporal profiles of de-phosphorylation in this lysis buffer. This should be experimentally determined. If the loss of phospho-specific signal is a concern, we recommend increasing the concentration of the phosphatase inhibitors in the buffer (NaF and NaOv). Moreover, protein yield may be enhanced during short incubations if a Thermomixer is utilized to continually agitate the pellet at a permissive incubation temperature.
Lastly, high protein yield from fractions of brain tissues or from small samples such as isolated cell populations, coupled with dual-channel analysis in LiCor systems and blot stripping for subsequent re-probing, will facilitate the analysis of many proteins of interest, while the remaining fraction can be stored for future precipitation and analysis. Moreover, this procedure can be used in several different contexts. Tissue can be homogenized in TRIzol directly, or if ELISAs or mass spectrometry is a desired endpoint, the tissue could be homogenized in solutions compatible with those analyses (e.g. RIPA buffer), and part of the homogenate could then be mixed with an excess of TRIzol for subsequent RNA and protein isolation. Furthermore, there are several procedures published for analyzing synaptosomes to determine if there is important molecular signaling occurring exclusively at the synapse (Villasana et al., 2006). These samples could then be mixed with TRIzol, and gene expression analyses could determine if there are changes in the content or amount of mRNAs stored locally at synapses (e.g. in association with synaptic RNA binding proteins like the fragile X mental retardation protein, FMRP). This could be a powerful addition to quantifying molecular signaling at the synapse.
4.4 Choosing a simultaneous extraction method
There are several different options available for completing simultaneous RNA and protein extraction, both in the form of commercially available kits as well as permutations of TRIzol precipitation and solubilization, and each option has both strengths and weaknesses. The predominant weaknesses for commercially available kits are cost per sample and some reports indicating that extraction kits can introduce bias or inconsistencies in RNA, DNA, and/or protein quality or retrieval (Mathieson and Thomas, 2013). Alternatively, column-based kits are often quite fast (an hour or less). TRIzol-based extractions of RNA and DNA are robust, reliable, and produce very good quality samples for a fraction of the cost of column-based kits (see Table 6), but can take much longer from start to finish. For some labs, the prohibitive cost of commercially available kits is reason enough to rely on TRIzol-based techniques.
Table 6.
Comparison of methods for isolation of DNA, RNA, and protein from a single biological sample
| Product | Company | Method | Price (USD) | Size | Cost per sample (USD) |
|---|---|---|---|---|---|
| TRIzol | Life Technologies | Phase separation | $326 | 200mL | $0.82* |
| TriplePrep | GE Healthcare | Spin columns | $503 | 50 reactions | $10.06 |
| AllPrep | Qiagen | Spin columns | $610 | 50 reactions | $12.20 |
| NucleoSpin TriPrep | Takara Bio | Spin columns | $405 | 50 reactions | $8.10 |
Prices obtained from company websites in January 2017; prices may vary based on provider.
TRIzol cost based one 500μL/reaction
As previously discussed, SDS and urea solubilization buffers result in varying success, and have some important caveats (e.g. possible induction of posttranslational modifications in the case of heated urea buffers). There are, however, alternative methods for either the precipitation procedure or the solubilization buffer composition that have been shown to enhance protein yield. The most effective reported SDS/urea solubilization protocol comes from Hummon and colleagues (2007), and involves dialysis and subsequent protein concentration via iCON protein concentrator columns. In their study, this procedure results in 98% recovery of protein compared to direct homogenization in their formulation of TNE (Tris-NaCl-EDTA) lysis buffer. Banerjee and colleagues (2003) have reported that adding 20μg glycogen, a ‘carrier’ used to increase RNA yield during precipitation, to the protein-containing TRIzol solution prior to precipitation with isopropanol can increase protein yield. They go on to demonstrate that higher pH (50mM Tris pH 8.0–8.8) and the addition of the detergent sarkosyl (62.5mM) to a 1% SDS solution can significantly increase protein yield. Additionally, Chey and colleagues (2011) used an ethanol-bromochloropropane-water solution to precipitate protein from TRIzol rather than isopropanol, after which ~95% protein (compared to RIPA-homogenized samples) could be quickly solubilized in 4% SDS. Our data add to this literature by demonstrating that a protein lysis buffer containing certain NaCl, EDTA, and SDS concentrations (and perhaps their additive effect on proteins), can also enhance protein yield via solubilization. Thus there are certainly more ways in which both our technique as well as other techniques for effective protein solubilization can be enhanced.
One important caveat remains: what is the correct comparison for determining protein loss and thus objectively quantifying the advantages of one method over another? RIPA buffer is very often used as a representation of the accurate composition of a tissue samples, but our data clearly demonstrate that direct homogenization with RIPA buffer can be improved upon (Fig. 5A). Moreover, if the experimental protein of interest happens to be in the ~200kDa band that was absent from RIPA homogenate (Fig. 5A′), then Western blotting may have not ever been a viable option with the existing buffers despite high protein yields from either direct homogenization or solubilization. RIPA buffer, however, is not the only lysis buffer utilized for direct homogenization. The dialysis data described above used a variation of TNE lysis buffer, the optimization described herein began with a lysis buffer reported by Kopec and colleagues (2015) and there are additional cell lysis buffers available for purchase from different companies. Thus, we suggest that protein yield and protein loss are not absolute quantities, but rather are determined by the choice of comparison buffers and methods.
In summary, we have made use of a protein lysis buffer formulation that optimizes both the solubilization step of protein precipitated from TRIzol-homogenized brain tissue, as well as the resolution of protein via direct homogenization (relative to RIPA buffer). The technique outlined here results in high protein yield over a variety of different time and temperature incubations, and is representative of different cell types and classes inherent in brain tissue. We propose that this technique can help to maximize the data collected from any one sample, making it both cost effective as well as advantageous to acquiring a multi-level molecular assessment of research questions. This will, in turn, benefit our understanding of the molecular contexts in which neural phenomena occur.
Acknowledgments
The authors declare no competing financial interest. This work was supported by the National Institutes of Health [R01-DA034185 and R01-MH101183].
6. Bibliography
- Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. doi: 10.1016/s0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Adams JP, Sweatt JD. Molecular psychology: roles for the ERK MAP kinase cascade in memory. Annu Rev Pharmacol Toxicol. 2002;42:135–63. doi: 10.1146/annurev.pharmtox.42.082701.145401. [DOI] [PubMed] [Google Scholar]
- Andersen KK, Oliveira CL, Larsen KL, Poulsen FM, Callisen TH, Westh P, Pedersen JS, Otzen D. The role of decorated SDS micelles in sub-CMC protein denaturation and association. J Mol Biol. 2009;391:207–26. doi: 10.1016/j.jmb.2009.06.019. [DOI] [PubMed] [Google Scholar]
- Arakawa T, Timasheff SN. Mechanism of protein salting in and salting out by divalent cation salts: balance between hydration and salt binding. Biochemistry. 1984;23:5912–23. doi: 10.1021/bi00320a004. [DOI] [PubMed] [Google Scholar]
- Arnold U, Ulbrich-Hofmann R. Quantitative protein precipitation from guanidine hydrochloride-containing solutions by sodium deoxycholate/trichloroacetic acid. Anal Biochem. 1999;271:197–9. doi: 10.1006/abio.1999.4149. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Smallwood A, Chambers AE, Nicolaides K. Quantitative recovery of immunoreactive proteins from clinical samples following RNA and DNA isolation. Biotechniques. 2003;35:450–2. 4, 6. doi: 10.2144/03353bm02. [DOI] [PubMed] [Google Scholar]
- Barres BA. The mystery and magic of glia: a perspective on their roles in health and disease. Neuron. 2008;60:430–40. doi: 10.1016/j.neuron.2008.10.013. [DOI] [PubMed] [Google Scholar]
- Bramham CR, Wells DG. Dendritic mRNA: transport, translation and function. Nat Rev Neurosci. 2007;8:776–89. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- Chey S, Claus C, Liebert UG. Improved method for simultaneous isolation of proteins and nucleic acids. Anal Biochem. 2011;411:164–6. doi: 10.1016/j.ab.2010.11.020. [DOI] [PubMed] [Google Scholar]
- Fukao A, Aoyama T, Fujiwara T. The molecular mechanism of translational control via the communication between the microRNA pathway and RNA-binding proteins. RNA Biol. 2015;12:922–6. doi: 10.1080/15476286.2015.1073436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–35. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh T, Kalra A, Garde S. On the salt-induced stabilization of pair and many-body hydrophobic interactions. J Phys Chem B. 2005;109:642–51. doi: 10.1021/jp0475638. [DOI] [PubMed] [Google Scholar]
- Gorisse L, Pietrement C, Vuiblet V, Schmelzer CE, Kohler M, Duca L, Debelle L, Fornes P, Jaisson S, Gillery P. Protein carbamylation is a hallmark of aging. Proc Natl Acad Sci U S A. 2016;113:1191–6. doi: 10.1073/pnas.1517096113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummon AB, Lim SR, Difilippantonio MJ, Ried T. Isolation and solubilization of proteins after TRIzol extraction of RNA and DNA from patient material following prolonged storage. Biotechniques. 2007;42:467–70. 72. doi: 10.2144/000112401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janecki DJ, Reilly JP. Denaturation of metalloproteins with EDTA to facilitate enzymatic digestion and mass fingerprinting. Rapid Commun Mass Spectrom. 2005;19:1268–72. doi: 10.1002/rcm.1924. [DOI] [PubMed] [Google Scholar]
- Klein SL, Schiebinger L, Stefanick ML, Cahill L, Danska J, de Vries GJ, Kibbe MR, McCarthy MM, Mogil JS, Woodruff TK, Zucker I. Opinion: Sex inclusion in basic research drives discovery. Proc Natl Acad Sci U S A. 2015;112:5257–8. doi: 10.1073/pnas.1502843112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollipara L, Zahedi RP. Protein carbamylation: in vivo modification or in vitro artefact? Proteomics. 2013;13:941–4. doi: 10.1002/pmic.201200452. [DOI] [PubMed] [Google Scholar]
- Kopec AM, Philips GT, Carew TJ. Distinct Growth Factor Families Are Recruited in Unique Spatiotemporal Domains during Long-Term Memory Formation in Aplysia californica. Neuron. 2015;86:1228–39. doi: 10.1016/j.neuron.2015.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer RM, Shende VR, Motl N, Pace CN, Scholtz JM. Toward a molecular understanding of protein solubility: increased negative surface charge correlates with increased solubility. Biophys J. 2012;102:1907–15. doi: 10.1016/j.bpj.2012.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtzier V, Hutoran M, Levy T, Kotler M, Brenner T, Steinitz M. Sodium dodecyl sulphate-treated proteins as ligands in ELISA. J Immunol Methods. 2002;270:19–26. doi: 10.1016/s0022-1759(02)00214-4. [DOI] [PubMed] [Google Scholar]
- Likhite N, Warawdekar UM. A unique method for isolation and solubilization of proteins after extraction of RNA from tumor tissue using trizol. J Biomol Tech. 2011;22:37–44. [PMC free article] [PubMed] [Google Scholar]
- Man TK, Li Y, Dang TA, Shen J, Perlaky L, Lau CC. Optimising the use of TRIzol-extracted proteins in surface enhanced laser desorption/ ionization (SELDI) analysis. Proteome Sci. 2006;4:3. doi: 10.1186/1477-5956-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson W, Thomas GA. Simultaneously extracting DNA, RNA, and protein using kits: is sample quantity or quality prejudiced? Anal Biochem. 2013;433:10–8. doi: 10.1016/j.ab.2012.10.006. [DOI] [PubMed] [Google Scholar]
- Nielsen MM, Andersen KK, Westh P, Otzen DE. Unfolding of beta-sheet proteins in SDS. Biophys J. 2007;92:3674–85. doi: 10.1529/biophysj.106.101238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan RL, Teller JK. Diethylamine extraction of proteins and peptides isolated with a mono-phasic solution of phenol and guanidine isothiocyanate. J Biochem Biophys Methods. 2006;68:127–31. doi: 10.1016/j.jbbm.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Otzen D. Protein-surfactant interactions: a tale of many states. Biochim Biophys Acta. 2011;1814:562–91. doi: 10.1016/j.bbapap.2011.03.003. [DOI] [PubMed] [Google Scholar]
- Price PA, Stein WH, Moore S. Effect of divalent cations on the reduction and re-formation of the disulfide bonds of deoxyribonuclease. J Biol Chem. 1969;244:929–32. [PubMed] [Google Scholar]
- Reddy PJ, Rao AA, Malhotra D, Sharma S, Kumar R, Jain R, Gollapalli K, Pendharkar N, Rapole S, Srivastava S. A simple protein extraction method for proteomic analysis of diverse biological specimens. Current Proteomics. 2013;10:298–311. [Google Scholar]
- Schwarz JM, Smith SH, Bilbo SD. FACS analysis of neuronal-glial interactions in the nucleus accumbens following morphine administration. Psychopharmacology (Berl) 2013;230:525–35. doi: 10.1007/s00213-013-3180-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes AE, Pereira DM, Amaral JD, Nunes AF, Gomes SE, Rodrigues PM, Lo AC, D’Hooge R, Steer CJ, Thibodeau SN, Borralho PM, Rodrigues CM. Efficient recovery of proteins from multiple source samples after TRIzol((R)) or TRIzol((R))LS RNA extraction and long-term storage. BMC Genomics. 2013;14:181. doi: 10.1186/1471-2164-14-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Zhou JY, Yang W, Zhang H. Inhibition of protein carbamylation in urea solution using ammonium-containing buffers. Anal Biochem. 2014;446:76–81. doi: 10.1016/j.ab.2013.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trakhanov S, Quiocho FA. Influence of divalent cations in protein crystallization. Protein Sci. 1995;4:1914–9. doi: 10.1002/pro.5560040925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Darnell RB. RNA binding proteins and the regulation of neuronal synaptic plasticity. Curr Opin Neurobiol. 2006;16:102–10. doi: 10.1016/j.conb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Villasana LE, Klann E, Tejada-Simon MV. Rapid isolation of synaptoneurosomes and postsynaptic densities from adult mouse hippocampus. J Neurosci Methods. 2006;158:30–6. doi: 10.1016/j.jneumeth.2006.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler GC, Brown GM, Lutz H. Detection of antibodies to Anaplasma marginale by an improved enzyme-linked immunosorbent assay with sodium dodecyl sulfate-disrupted antigen. J Clin Microbiol. 1987;25:633–6. doi: 10.1128/jcm.25.4.633-636.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–62. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Hasegawa K, Esumi M. Protein from the fraction remaining after RNA extraction is useful for proteomics but care must be exercised in its application. Exp Mol Pathol. 2013;95:46–50. doi: 10.1016/j.yexmp.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Young C, Truman P. Proteins isolated with TRIzol are compatible with two-dimensional electrophoresis and mass spectrometry analyses. Anal Biochem. 2012;421:330–2. doi: 10.1016/j.ab.2011.10.045. [DOI] [PubMed] [Google Scholar]