Abstract
BRAF is a commonly mutated oncogene in various human malignancies and a target of a new class of anti-cancer agents, BRAF-inhibitors (BRAFi). The initial enthusiasm for these agents, based on the early successes in the management of metastatic melanoma, is now challenged by the mounting evidence of intrinsic BRAFi-insensitivity in many BRAF-mutated tumors, by the scarcity of complete responses, and by the inevitable emergence of drug resistance in initially responsive cases. These setbacks put an emphasis on discovering the means to increase the efficacy of BRAFi and to prevent or overcome BRAFi-resistance. We explored the role of p21-activated kinases (PAKs), in particular PAK1, in BRAFi response. BRAFi lowered the levels of active PAK1 in treated cells. An activated form of PAK1 conferred BRAFi-resistance on otherwise sensitive cells, while genetic or pharmacologic suppression of PAK1 had a sensitizing effect. While activation of AKT1 and RAC1 proto-oncogenes increased BRAFi-tolerance, the protective effect was negated in the presence of PAK inhibitors. Furthermore, combining otherwise ineffective doses of PAK- and BRAF-inhibitors synergistically affected intrinsically BRAFi-resistant cells. Considering the high incidence of PAK1 activation in cancers, our findings suggests PAK inhibition as a strategy to augment BRAFi therapy and overcome some of the well-known resistance mechanisms.
Keywords: vemurafenib, AZD6244, MAP Kinase Cascade, PF3758309, melanoma
BRAF, a serine-threonine kinase belonging to the RAF family, has been found to be mutated in a variety of cancers [1–4]. Commonly occurring mutations, such as the valine to glutamate change in position 600 (BRAF V600E) [4], produce an activated form of the protein. This leads to constitutive signaling through the MAP kinase cascade, which, in turn, contributes to enhanced growth, survival, and multiple other oncogenic properties of cancer cells. In cutaneous melanoma, numerous studies have reported activating mutations in BRAF at a high frequency, ranging approximately from 40% to 60% of all cases [4–8]. Development of clinically-usable inhibitors of activated BRAF, such as vemurafenib [9,10] and dabrafenib [11], revolutionized the care of melanoma, which is otherwise notoriously unresponsive to conventional therapy. The high selectivity of these agents against mutant BRAF may explain their relatively mild side effects. Although some tumor shrinkage could be observed in as many as 85% of the treated patients, only about half of all cases meet the definition of an “objective response” [9]. It is important to note that the category of responders in these studies predominantly includes partial responses and the duration of response was limited to several months, after which a drug-resistant disease emerges [12]. The situation is not better for other BRAF-mutant cancer types [13]. For example, Kepetz et al [14] reported only a 5% partial response rate and no complete responses in a Phase II colon cancer study of vemurafenib.
Limited initial success and the high eventual failure rate of anti-BRAF therapy stimulated an intense research effort into the molecular determinants of resistance and sensitivity to BRAF inhibition [12]. There is a growing understanding about the multiplicity and diversity of BRAFi-resistance pathways [15,16]. Early on, it became evident that activation of the PI3K–AKT signaling pathway, which is a long-known mechanism of stress tolerance in mammalian cells [17], may play a role in BRAFi- resistance as well [18–21]. The effect on AKT activity may also explain the protective effect of certain receptor tyrosine kinases [22–24]. Importantly, chemical inhibitors of this pathway sensitize BRAF-mutant cells to BRAFi [25–28], hinting at the possibility to increase the efficacy of therapy by using appropriate drug combinations.
Conceivable, an additional event that is functionally equivalent to BRAF activation would protect BRAF-mutant cells from BRAFi. In accordance with the theory that MAP kinase cascade activation is the key oncogenic function of BRAF, multiple alternative means of maintaining high ERK activity have been shown to circumvent BRAF inhibition in vitro and in vivo [23,29–31]. Generally, an evidence of functional equivalence could be mutual exclusivity with BRAF mutations despite frequent occurrence in the same cancer type. Activation of proto-oncogene NRAS, which is found in many melanomas, fits this profile well. Activated NRAS negates the effects of BRAFi when introduced into BRAF-mutant cells, and is found in BRAFi-resistant cases of BRAF-mutant malignancies [32]. Another event, which is seen in some melanomas with wild type BRAF and NRAS, is activation of a small GTPase, RAC1 [5]. Although the original research on RAC1 functions was focused on its role in cytoskeleton organization and cell motility, there are evidences connecting RAC1signaling to the MAP kinase cascade. Accordingly, it was reported that activated forms of RAC1 can protect BRAF-mutated cells from BRAFi [33].
Perhaps the best-known downstream effector of RAC1 is p21-activate kinase 1 (PAK1), a serine-threonine kinase, which is involved in a plethora of physiological and pathophysiological processes [34]. Among the reported functions of PAK1 is co-activation of the MAP kinase cascade through interactions with RAF proteins [35,36] and MEK1 [37]. PAK1 was also reported to interact directly with AKT [38], and is a critical co-factor in AKT-mediated oncogenic transformation [39]. Importantly, unlike RAS and RAC GTPases, PAK kinases are readily amenable to chemical inhibition, with multiple agents with various degrees of specificity currently used in pre-clinical and early-stage clinical studies [40]. We set forth to investigate the role of PAK1 in resistance and sensitivity to BRAFi in BRAF-mutated cells. We observed that hyperactive PAK1 provides substantial protection, while genetic and pharmacological inhibition of that protein had a sensitizing effect and reduced protection by active forms of RAC1 and AKT. These findings yield an insight into the avenues to augment the efficacy and extend the utility of an important class of anti-cancer agents.
Materials and Methods
Cell culture and reagents
All cell lines were cultured in humidified incubators at 37°C in the presence of 5% CO2. A375 (obtained from ATCC), HT29 (a gift from Dr. Yuri Ionov) and Colo205 (obtained from ATCC) were maintained in high-glucose Dulbecco’s Modified Eagle Medium (DMEM). SK-MEL-28 (obtained from ATCC) were maintained in Minimum Essential Media (MEM). B-CPAP (a gift from Dr. Katerina Gurova) were maintained in RPMI1640. All culture media were supplemented with penicillin (100 U/ml), streptomycin (100 µg/ml) and fetal bovine serum (10%).
The cells were authenticated by the Roswell Park Cancer Research Institute’s Genomics Shared Resource using Amp FLSTR Identifier Plus PCR Amplification Kit (A26182; ThermoFisher Scientific).
The cells were free of mycoplasma (as assessed by MycoAlert kit from Lonza, Inc.) and contaminating gamma-retroviruses [41].
PF3758309 was purchased from ChemieTek. Vemurafenib was obtained from LC Laboratories. IPA3 and AZD6244 were obtained from Selleck Chemicals.
Vectors and viral transduction
Lentiviral vector pLM-CMV-neo and its derivative pLM-CMV-mAKT-neo (expressing a myristoylated form of mouse Akt1) were gifts of Dr. Peter Chumakov. pBabe-PAK1-T423E was constructed by introducing PAK1 T423 coding fragment from pCMV6M-PAK1-T423E (a gift of Dr. Jonathan Chernoff, procured as Addgene plasmid #12208) into pBabeHygro [42]. pLX304 was a gift from Dr. David Root (procured as Addgene plasmid #25890). pLX304-RAC1-P29S was generated by PCR-based mutagenesis from pLX304-RAC1 (purchased from DNASU Plasmid Repository). The NRAS expression construct was a gift from Dr. Mikhail Nikiforov. RNA interference reagents were purchased from the Roswell Park Cancer Institute shRNA Core. Lentiviral transduction was performed as described elsewhere [43]. Gamma-retroviral vectors were used as before [44].
Antibodies and Immunoblotting
α-Tubulin (#SC-8035), p-ERK (#SC-7383), ERK1 (#SC-292838), PAK1 (#SC-881), and p-PAK1 Ser 199/204 (#SC-33531) were obtained from Santa Cruz. MEK2 (#9125S), p-MEK (#9121S), AKT (#9272S) and p-AKT (#4060S) were obtained from Cell Signaling. GAPDH (#AM4300) was purchased from Thermo-Fisher.
RIPA buffer (# 899900; Thermo Scientific) supplemented with protease/phosphatase inhibitor (#5872S; Cell Signaling) and EDTA solution (0.5mM [final]) was used to lyse cells. Membranes were probed overnight in 5% BSA at 4°C with gentle rocking with antibodies at manufacturers’ recommended dilutions.
Tissue microarray
An arrayed collection of metastatic melanoma samples [45] was probed using phospho-PAK Thr423 antibody (#NBP1-02914) from Novus Biologicals. The neoplastic cells for any given core were scored for intensity of staining (no staining - 0, weak staining −1, moderate staining - 2, strong staining −3) and the abundance of positive cells (none- 0, up to 10% - 1, between 10 and 50% - 2, above 50% - 3). The array, staining services and sample evaluation were provided by the Pathology Resource Network of the Roswell Park Cancer Institute.
Assays of cell numbers and proliferation
Drug response curves were carried out on 24-well plates in culture media supplemented with 5% FBS. Drug treatments began 24 hours after plating. After addition of drugs, the cells were cultured for 5 days and fixed with 100% methanol. Cells were stained with methylene blue followed by dye extraction with 10% SDS. Absorbance was measured at 595nm for quantification. IC50 values for each cell line were calculated using GraphPad Prism 6 (GraphPad Software, Inc.).
When indicated, cooperativity between the drugs was evaluated using Compusyn software (ComboSyn, Inc.), which relies on median-effect method of Chou–Talalay to define synergy [46]. The method involves the calculation of a “Combination Index”, CI, which compares the observed effect and a calculated additive effect [47]. CI values of 1, >1 and <1 indicate additivity, antagonism and synergism, respectively.
Cell proliferation was compared using an EdU incorporation assay. Cells were pulsed with EdU (10uM) for 1 hour and fixed with 3.7% paraformaldehyde for 15 min at room temperature and washed with PBS. Incubation in Triton X-100 (.5%) for 20 min was used to permeabilize cells. Subsequently, cells were stained with Click-iT™ Alexa Fluor 488 imaging kit (C10086; ThermoFisher Scientific) according to manufacturer’s protocol. 5 random fields of view were assessed under fluorescent microscopy to analyze EdU incorporation.
Results
The role of PAK1 in RAC1-induced resistance to BRAF inhibitors
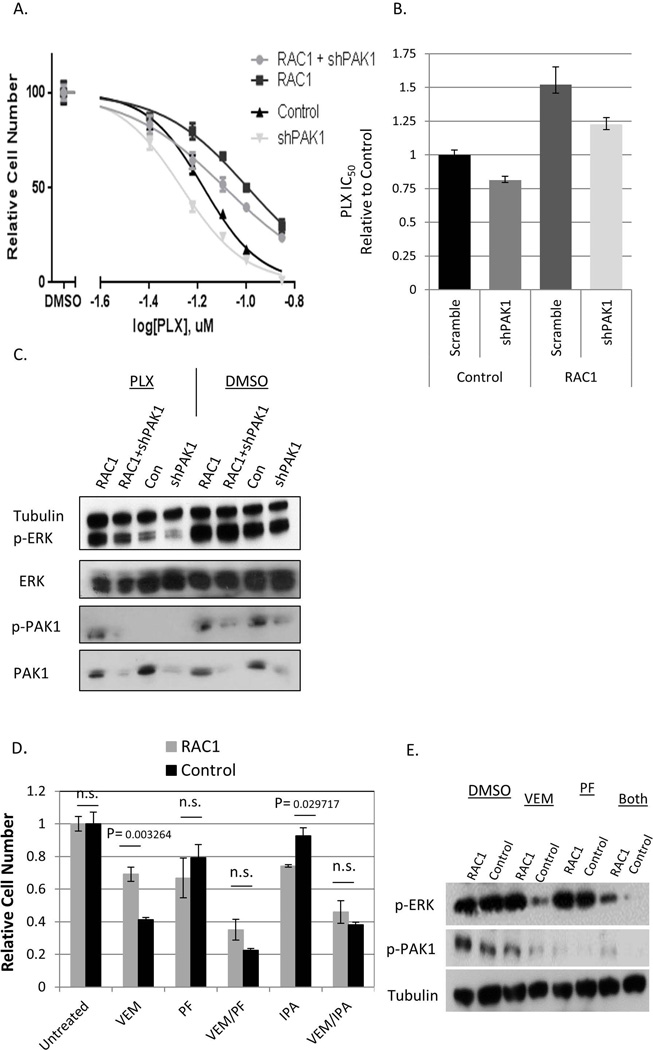
A375 is a cell line derived from metastatic melanoma that harbors an activating mutation in the BRAF gene (BRAF V600E). The cell line is known to be sensitive to BRAF inhibitors, including vemurafenib (aka PLX4032) and its analogue PLX4720 [48]. In agreement with earlier reports [33], ectopic expression of a cancer-derived RAC1 mutant (RAC1 P29S) increased resistance of these cells to PLX4720 and vemurafenib (Fig. 1A and Supplementary Fig. 1A). Resistance was associated with retention of higher activity of the MAP kinase cascade, as is indicated by higher levels of active (phosphorylated) forms of ERK1 and ERK2 (Fig. 1C, Fig. 1E).
Figure 1. The effects of inhibition of PAK on RAC1-mediated resistance to BRAFi.
(A) A375 cells were transduced with RAC1 P29S and shPAK1 (“RAC1+shPAK1”), pLX304 empty vector and shPAK1 (“shPAK1”), RAC1 P29S and a non-targeting “scrambled” shRNA (“RAC1”), or pLX304 and a “scrambled” shRNA (“Control”). The cells were treated for five days with the indicated doses of PLX4720 in quadruplicates, and the numbers of remaining cells were compared by methylene blue staining extraction method. The values with standard deviations are plotted as percentages of those in the corresponding cultures treated with the vehicle (DMSO) alone. (B) The IC50 concentrations were calculated for the data shown in (A) and are presented relatively to that of the “Control” culture. The error bars denote 95% confidence intervals. (C) The indicated cell lines were treated with 100nM PLX4720 or DMSO, and the corresponding lysates were probed by immunoblotting for the expression total and activated (phosphorylated) PAK1 and ERK1/2, as well as α-tubulin (loading control). (D) A375 cells transduced with RAC1-P29S (“RAC1”) or pLX304 (“Control”) were treated for 5 days with 50nM vemurafenib (“VEM”), 2.5uM IPA3 (“IPA”) or 10nM PF3758309 (“PF”), alone or in the indicated combinations. The numbers of remaining cells were compared using the methylene blue straining and extraction method and are shown as a fraction of the values in the corresponding “Untreated” (exposed to DMSO alone) cultures. (E) The lysates from the cells treated for 48h as in (D) were probed by immunoblotting for the levels of α-tubulin (loading control) and the activated (phosphorylated) forms of PAK1 and ERK1/2.
There are ample evidences demonstrating that multiple biological functions of RAC1 are mediated through activation of PAK kinases, and PAK1 in particular [34]. In order to explore whether BRAFi-resistance follows the same pattern, we reduced the levels of PAK1 using RNA interference. The results demonstrated that expression of anti-PAK1 shRNA caused considerable re-sensitization of the RAC1P29S -expressing cells (Fig. 1A, Fig. 1B), which was paralleled by lower ERK phosphorylation in drug-treated cells (Fig. 1C). Incomplete re-sensitization might have been caused by incomplete knockdown of PAK1, or by partial compensation of PAK1 functions by other kinases of the PAK family.
Importantly, a sensitizing effect of the shRNA was also seen in the absence of constitutively active RAC1 (Fig. 1A). This is not surprising, as the control cells have some endogenous PAK1 activity, which may be contributing to their drug tolerance.
SK-MEL-28 is another metastatic melanoma cell lines with an activating mutation in BRAF. In our experiments, SK-MEL-28 cells were somewhat more resistant to BRAFi than A375, but they too were sensitized by interference with PAK1 (Supplementary Fig. 4).
The data on genetic interference was corroborated by the results of chemical inhibition of PAK1. Two compounds were used: IPA3 prevents activation of group I PAKs by upstream G-proteins [49], while PF3758309 is a pan-PAK inhibitor, which competes with ATP binding [40]. Interestingly, both of these agents, when administered individually, were somewhat more toxic to cells with constitutively active RAC1 (Fig. 1D, Supplementary Fig. 1B, and 1C). Most importantly, constitutively active RAC1 offered no advantage in the presence of either vemurafenib-IPA3 or vemurafenib- PF3758309 combinations (Fig. 1D). Furthermore, the doses of vemurafenib and PF3758309, which alone were insufficient to affect ERK activity in cells with activated RAC1, achieved a pronounced affect when combined together (Fig. 1E).
Akin to the data on the effects of anti-PAK1 shRNA, addition of PF3758309 or IPA3 to vemurafenib also increased the efficacy against A375 cells, as was evident both by cell survival and ERK phosphorylation data (Fig. 1D and Fig. 1E). Overall, we concluded that inhibition of PAK, and PAK1 in particular, sensitizes BRAF-mutant melanoma cells to BRAFi and negated the protective benefit of RAC1 activation.
Interestingly, we consistently observed a decrease in the abundance of the active form of PAK1 in BRAFi-treated cells. This is consistent with the recently proposed BRAF-dependent, RAC1-independent mechanism of PAK1 activation [50]. The effect was diminished by concomitant expression of activated RAC1 (Fig. 1C and Fig. 1E), which also supports the hypothesis about BRAF and RAC1 affecting PAK1 status via parallel mechanisms. Considering the widespread involvement of PAK1 in cell growth, survival and metabolism [34], its eventual inhibition by BRAFi may be an important contributor to the anti-tumor effects of this class of compounds, while RAC-independent control of PAK1 by BRAF may explain some features of BRAF-mutant cells.
Activation of PAK1 conveys resistance to BRAF inhibitors
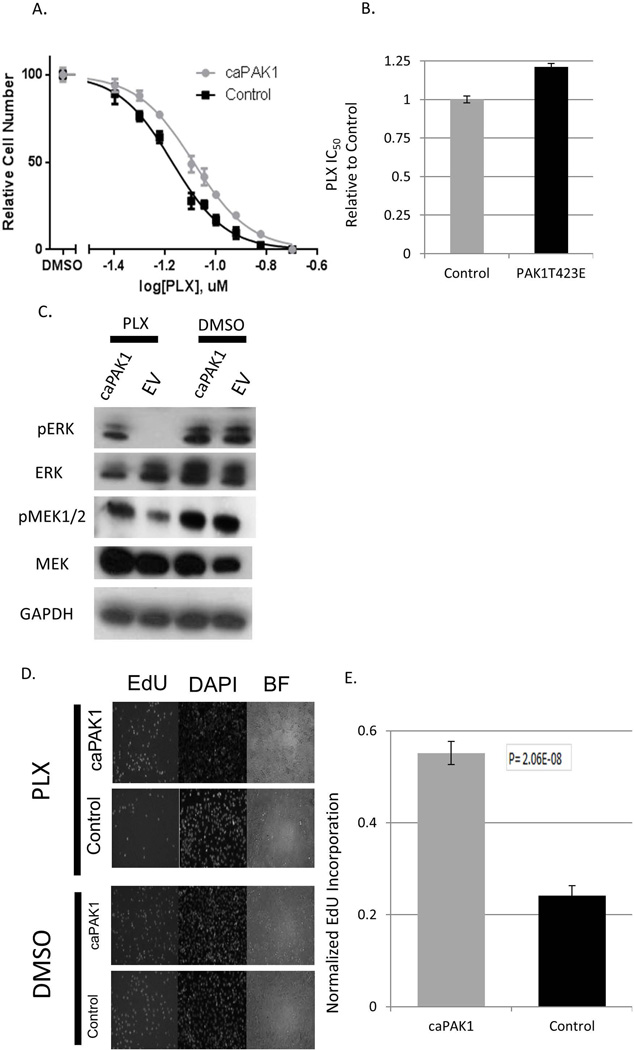
If PAK1 is an important downstream effector of RAC1 in the BRAFi- resistance phenomenon, then one may expect that PAK1 activation alone is able to recapitulate the protective effect. Indeed, we observed that ectopic expression of constitutively active PAK1 increased tolerance of A375 cells to PLX4720, resulting in higher IC50 values (Fig. 2A, Fig. 2B) and higher levels of active (phosphorylated) components of MAP kinase cascade (Fig. 2C). Accordingly, activated PAK1 allowed the cells to maintain a higher proliferative potential in the presence of modest doses of PLX4720, as attested by a higher proportion of cells undergoing DNA replication under these conditions (Fig. 2D, Fig. 2E). Importantly, BRAF-mutant colon carcinoma line Colo205, which is among the most sensitive colon cancer lines to RAF inhibition [51], was also protected by constitutively active PAK1 (Supplementary Fig. 5A, 5B). This indicates that the phenomenon is not limited to cells of melanoma origin.
Figure 2. Constitutive activation of PAK1 reduces the effects of BRAF inhibition.
(A) A375 cells transduced with PAK1 T423E (“caPAK1”) or the corresponding empty vector (“Control”) were treated in quadruplicates with the indicated doses of PLX4720. The numbers of remaining cells were compared using the methylene blue staining and extraction method. The values with standard deviations are plotted as percentages of those in the corresponding cultures treated with the vehicle (DMSO) alone. (B) The IC50 concentrations were calculated for the data shown in (A) and are presented relatively to that of the “Control” culture. The error bars denote 95% confidence intervals. (C) A375 cells transduced with PAK1 T423E (“caPAK1”) or the corresponding empty vector (“EV’) cells were treated for 48h with 60nM of PLX4720 or the corresponding vehicle (DMSO) alone. Cell lysates were probed by immunoblotting for GAPDH (loading control) and total and activated (phosphorylated) MEK1/2 and ERK1/2. (D) A375 transduced with caPAK1 or the corresponding empty vector (“Control”) were treated for 48h with PLX4720 or DMSO. EdU (10uM) was added and 1h later the cells were then stained as described in Materials and Methods. Representative images are shown. (E) For the cells treated as in (D), the fraction of EdU-labeled cells was scored on 5 randomly chosen fields of view. The bars show the values for PLX470-treated cultures as a fraction of the corresponding DMSO-treated controls. The error bars represent standard deviations.
Of note, activated PAK1 also increased resistance of A375 to a MEK inhibitor AZD6244 [52] (Supplementary Fig. 6). Thus, our observations argue that activation of PAK1 may serve as a protective mechanism against the anti-tumor activity of BRAFi, and, possibly, other anti-cancer agents that target the MAP kinase cascade.
Inhibition of PAK1 negates AKT-mediated resistance to BRAF inhibitors
Activation of the PI3K-AKT signaling pathway is a common occurrence in cancers, where it contributes to a plethora of biological and biochemical features of malignant cells [17]. As mentioned above, it is one of the better-known modes of resistance to BRAFi. It was reported that AKT directly interacts with PAK1 [53], and PAK1 function is important for the maintenance of transformed phenotypes in AKT-transformed cells [39]. We set forth to explore whether AKT-mediated protection is affected by PAK1 status.
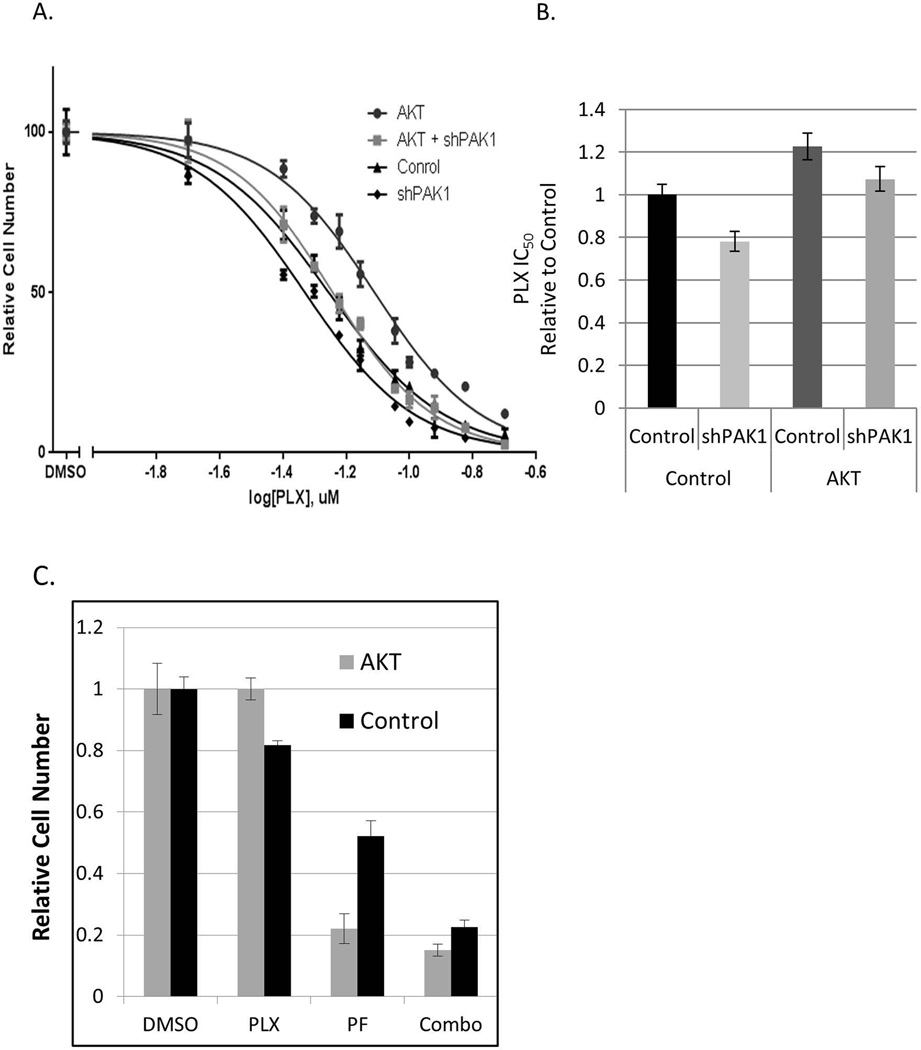
As expected, a constitutively active form of AKT (myristoylated, aka mAKT) increased resistance of A375 cells to PLX4720 (Fig. 3A, Fig. 3B). However, an shRNA against PAK1 was able to reduce the resistance of mAKT-bearing cells to the level of the parental cell line (Fig. 3B). Furthermore, cells harboring activated AKT were more sensitive to PF3758309, a pan-PAK chemical inhibitor (Fig. 3C, Supplementary Fig. 2B). Importantly, a combination of BRAF and PAK inhibitors (PLX4720 and PF3758309) was effective against mAKT-expressing and parental A375 cells (Fig. 3C). This observation suggests PAK inhibition as a strategy to negate the protective effects of AKT activation in BRAFi-treated cancers.
Figure 3. The effects of inhibition of PAK on AKT-mediated resistance to BRAFi.
(A) A375 cells were transduced with activated AKT and PAK1 shRNA (“AKT+shPAK1”), pLM-CMV-neo empty vector and PAK1 shRNA (“shPAK1”), activated AKT and a non-targeting “scrambled” shRNA (“AKT”), or pLM-CMV-neo and a “scrambled” shRNA (“Control”). The cells were treated in quadruplicates with the indicated doses of PLX4720. The numbers of remaining cells were compared using the methylene blue staining and extraction method. The values with standard deviations are plotted as percentages of those in the corresponding cultures treated with the vehicle (DMSO) alone. (B) The IC50 concentrations were calculated for the data shown in (A) and are presented relatively to that of the “Control” culture. The error bars denote 95% confidence intervals. (C) A375 cells transduced with activated AKT (“AKT”) or the corresponding vector control (“control”) were treated with 40nM PLX4720 or/and 8nM PF-3758309 over 5 days. The numbers of remaining cells were compared using the methylene blue staining and extraction method and are plotted after normalization to the respective drug-free populations (“Untreated”). The error bars denote standard deviations.
PAK inhibitor synergizes with BRAFi in intrinsically BRAFi-resistant cell lines
It is well known that some cancer cases, as well as some cancer-derived cell lines, are highly resistant to BRAFi, despite have activating mutations in the BRAF gene and no prior exposure to such drugs [12]. This intrinsic resistance limits the utility of BRAFi in some cancer types. While its mechanisms are not fully understood, the need to overcome it remains great. Considering the above-mentioned ability of PAK inhibitors to sensitize BRAFi-responsive cell lines and overcome the protective effects of ectopically expressed oncogenes, we explored whether targeting PAK could also sensitize BRAF-mutant cells that are intrinsically relatively resistant to BRAFi.
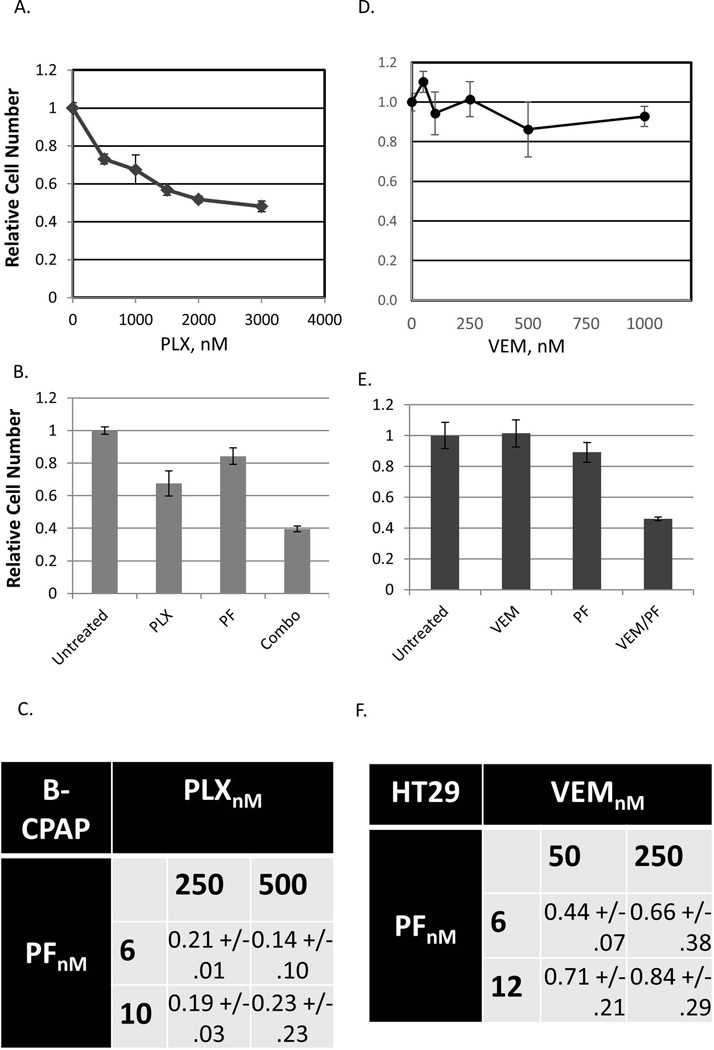
B-CPAP is a human papillary thyroid carcinoma-derived cell line, which harbors the BRAF V600E mutation. While this cell line is not completely insensitive to BRAF inhibitors, its PLX4720 IC50 is approximately 300-fold higher than that of A375 (Fig. 4A, compare to Fig. 1A). In these cells, PAK inhibitor PF3758309 greatly synergizes with PLX4720 at a range of doses (Fig. 4B and Fig. 4C).
Figure 4. Synergism between BRAF and PAK inhibitors in BRAFi-resistant cells.
(A) B-CPAP cells were treated for 5 days with the indicated doses of PLX4720. The numbers of remaining cells were compared by methylene blue staining and extraction method and are plotted relatively to those in untreated populations. (B) B-CPAP cells were treated for 5 days with PLX4720 (500nM; “PLX4720”) or PF3758309 (6nM; “PF”) or a combination thereof (“Combo”). The numbers of the remaining cells were compared by the methylene blue staining and extraction method and are shown relatively to those in untreated populations. (C) Combinational Index values were calculated using the Chou-Talalay method, as described in Materials and Methods, from a series of experiments conducted on B-CPAP cells essentially as in (B) and using the indicated doses of the drugs. Combinational Index values below 1 indicate synergy between the compounds. (D) HT29 cells were treated for 5 days with the indicated doses of vemurafenib. The numbers of remaining cells were compared by methylene blue staining and extraction method and are plotted relatively to those in untreated populations. (E) HT29 cells were treated for 5 days with vemurafenib (250nM; “VEM”) or PF3758309 (12nM; “PF”) or a combination thereof (“VEM/PF”). The numbers of the remaining cells were compared by the methylene blue staining and extraction method and are shown relatively to those in untreated populations. (F) Combinational Index values were calculated using the Chou-Talalay method, as described in Materials and Methods, from a series of experiments conducted on HT29 essentially as in (E) and using the indicated doses of the drugs. The values are shown as averages ± standard deviation from three independent experiments.
Similarly, a BRAF-mutant colorectal carcinoma cell line HT29 has demonstrated exceptional resistance to RAF inhibitor GDC-0879 [51]. It was also insensitive to vemurafenib in our experimental conditions (Fig. 4D), but was synergistically suppressed by a vemurafenib/ PF3758309 combination using the doses of the compounds that were ineffectual when applied individually (Fig. 4E and Fig. 4F).
Our observations suggest that, at least in some cases, a combination of PAK and BRAF inhibitors may improve the initial response to BRAFi therapy and might extend the utility of this therapy to a broader range of malignancies.
Discussion
Our findings strongly implicate PAK1 as a modulator of BRAFi response in cancer cells. The phenomenon is attested to by both genetic and pharmacological means, and consistent evidence come from melanoma, as well as from colon and thyroid cell lines. While a theoretical possibility of similar off-target effects cannot be ruled out for IPA3 and PF3758309, we find it unlikely, considering that the compounds are markedly different, both in terms of their chemical structure and the expected mechanisms of action [40].
In our experiments we relied on two BRAF inhibitors: vemurafenib and PLX4720. Although the compounds share considerable structural similarity and are expected to affect mutant BRAF in a similar manner, they are sufficiently distinct to differ in their interactions with some other biological molecules [54]. The fact that in our system qualitatively consistent results were obtained using both inhibitors (Supplementary Fig. 7 and data not shown) reduces the possibility that the observed phenomena are caused by off-target effects of these compounds.
Interestingly, PAK1 follows the mutual exclusivity pattern seen among other frequently mutated oncogenes: BRAF, NRAS and RAC1. While the latter oncogenes in melanomas are commonly afflicted by point mutations, PAK1 gene is amplified in 5% cases (Supplementary Fig. 8). PAK1 amplification is mutually exclusive with BRAF mutations (p=0.01), which is consistent with the hypothesis that the two events make equivalent contributions to oncogenesis. As discussed earlier [34], PAK1 is amplified as a part of a larger amplicon, which, typically, includes, among other genes, cyclin D1. PAK1 is also an essential oncogenic regulator of cyclin D1 [55], and, conceivably, co-amplification of the two provides a potent stimulus for tumor progression.
PAK kinases, and PAK1 in particular, have received considerable attention as therapeutic targets for various health conditions [34]. A large number of PAK inhibitors with various degrees of isoform specificity are currently used in preclinical and early clinical studies [40]. PAK1 can be activated by signals from oncogenic RAS [56] and represents a potential Achilles’ hill of RAS-transformed cells [55,57]. Our group has reported that in cancer types where the MAP kinase cascade could be alternatively activated by either RAS or BRAF mutations, BRAF-mutant cells are relatively resistant, while RAS-mutant cells are relatively sensitive to PAK inhibition [58]. This phenomenon was later confirmed by others [59], but the reason for differential sensitivity is still unclear. It is possible that RAS-mutant cells are “addicted” to PAK function because the latter is needed to alleviate some of the stress associated with RAS hyperactivity. Alternatively, the difference may be in the reliance of RAS cells on wild type CRAF, whose full activation is PAK1-dependent [60], while activated BRAF V600E mutant is fully functional without phosphorylation on the site targeted by PAKs [36]. Of note, increased reliance on CRAF has been previously reported in some BRAFi-resistant cells [25].
Importantly, the functions of PAK1 extend well beyond the regulation of MAP kinase cascade [34]. One might speculate that in RAS-mutant cells these functions are maintained via RAS- dependent activation of PAK1, while in BRAF cells this is fulfilled by alternative, PAK1-independent, mechanisms. However, a recent report suggests that BRAF has an activating effect on PAK1, and that small GTPases, traditional PAK1 activators, play no role in this phenomenon [50]. In this case, it is conceivable that the relative resistance of BRAF-mutant cells to PAK1 inhibition is simply the result of a more robust activation of PAK1. Of note, in our earlier study [58], we used IPA3, a PAK1 inhibitor, which acts through preventing PAK1 activation by upstream small GTPases [49] and would not be expected to affect the BRAF-mediated mechanism of activation. Interestingly, a small GTPase-independent mode of PAK activation has been previously proposed for AKT [38].
The data presented here is consistent with the existence of parallel BRAF- dependent and RAC1-dependent modes of PAK1 activation. Indeed, inhibition of BRAF in BRAF-mutant cells resulted in pronounced decrease in PAK1 activity, but this effect was abrogated by expression of constitutively active RAC1 (Fig. 1C, Fig. 1E). It is tempting to speculate that eventual reduction of PAK1 activity contributes to the efficacy of BRAF inhibitors, while an ability to maintain PAK1 would decrease the therapeutic response. Furthermore, since PAK1 activation alone is sufficient to protect cells from BRAFi, one may predict that many more direct or indirect PAK1 activators could enhance cell resistance to these drugs. In this regard, it is worth mentioning that ARGEF2 (Rho/Rac Guanine Nucleotide Exchange Factor 2) was found in an insertional-mutagenesis based screen for vemurafenib resistance genes [20]. ARHGEF2 is an activator of RAC1 [61]. It also interacts directly with PAK1 [62], although the biological significance of this interaction is still unclear. ARHGEF7 (aka β-PIX), another member of ARHGEF family and a classic RAC1 activator [63], exhibits a point mutation and a copy number gain in HT29 cells (data accessed via www.cbioportal.org). Bearing in mind that this cell line is relatively resistant to BRAFi and is sensitized by PAK inhibitors (Fig. 4D, Fig. 4E), it would be interesting to explore the role of ARHGEF7 changes in these phenomena. Also noteworthy is that the identity of the activating GEF determines the pattern of protein-protein interactions and, hence, the exact consequences of RAC1 activation [64]. Therefore, it is worthwhile exploring whether only a subset of GEFs can confer RAC1-mediated protection from vemurafenib.
In our collection of melanoma tissues samples (Supplementary Fig. 3), an antibody against activated (phosphorylated on threonine 423) PAK1 revealed immunoreactive material in 71 out of 92 cases (~77%), and the staining was considered moderate or high, at least in some of the cells, in 42 out of 92 cases (~46%). Considering the high incidence of PAK1 activation, the correlation between PAK1 activity and the clinical success of BRAF and, possibly, MEK inhibition is worth a closer look.
An important caveat to immunological detection of activated PAK1 is that the indicator threonine phosphorylation site is conserved among group I PAKs (PAK1, PAK2 and PAK3). Hence, the eventual signal is the cumulative contribution of multiple isoforms, rather than of PAK1 alone. The question of isoform-specificity of the observed phenomena remains open. None of the chemical PAK inhibitors tested in our studies are exclusively targeting PAK1: IPA3 is effective against all group I PAKs [49], while PF3758309 is considered a pan-PAK inhibitor [40]. Our genetic experiments with ectopic expression of activated PAK1 and with PAK1-specific RNA interference prove that PAK1 specifically can provide drug resistance and can be targeted for sensitization, at least in our model systems. However, evolutionary conservation in the PAK family makes it likely that the members share at least some of the targets, and a RAC1- dependent pro-proliferative function has been reported for PAK2 [65]. Thus, it is conceivable that activation of more than one distinct PAK isoform could lead to the drug-resistant phenotype. Indeed, involvement of other PAKs could be one of the explanations for why the sensitizing effect of chemical inhibitors noticeably exceeds that of PAK1 shRNA in RAC1-transduced cells. Furthermore, PAK3, which is functionally and structurally close to PAK1, was borderline-effective in a genetic screen of a collection of kinases for the ability to enhance vemurafenib resistance [29]. In the latter case, a wild-type PAK3 variant was expressed, leaving open a possibility that a stronger effect could have been achieved by an activated from or in the presence of an additional activating signal. The similarities and differences among the PAK proteins in regard to their roles in BRAFi response merits further investigation. However, it is important to note that all of the anti-PAK compounds currently reported in pre-clinical or early-clinical pipelines are active against, at least, all group I PAKs [40]. Thus, any functional redundancy among PAKs in cancer cells is unlikely to reduce the clinical prospects of these compounds.
The possibility that resistance to BRAFi may be coupled to a therapeutically exploitable vulnerability has been discussed in literature [66,67]. It is intriguing that activation of RAC1 or AKT in BRAF-mutant cells not only increases the resistance of these cells to BRAFi, but also sensitizes them to PAK inhibitors. While the molecular underpinning of this sensitivity is yet to be discovered, the phenomenon might have important practical implications: it offers a possibility to enhance BRAFi efficacy not by merely negating a protective mechanism, but by selectively eliminating potentially resistant cells.
Overall, our findings provide arguments in favor of a combined anti-BRAF/anti-PAK therapy for the malignancies with activating mutations in BRAF. Additional research into that issue is well warranted, as anti-PAK therapies are ready to enter the clinical arena.
Supplementary Material
Acknowledgments
We would like to thank Dr. Angela Omelian for expert processing of the melanoma tissue microarray. The work was supported by grants from the Roswell Park Alliance Foundation, Jennifer Linscott Tietgen Family Foundation and the National Institutes of Health (R21CA137708) to ESK. The use of Roswell Park Cancer Institute’s shared resources is supported through the National Cancer Institute (NCI) grant P30CA016056. The participation of HS and SJ was made possible through Science Research Program at Buffalo City Honors School.
Abbreviations
- BRAFi
BRAF inhibitor
- mAKT
myristoylated AKT
References
- 1.Abdel-Wahab O, Park CY. BRAF-mutant hematopoietic malignancies. Oncotarget. 2014;5(18):7980–7981. doi: 10.18632/oncotarget.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bollag G, Tsai J, Zhang J, et al. Vemurafenib: the first drug approved for BRAF-mutant cancer. Nature reviews Drug discovery. 2012;11(11):873–886. doi: 10.1038/nrd3847. [DOI] [PubMed] [Google Scholar]
- 3.Hertzman Johansson C, Egyhazi Brage S. BRAF inhibitors in cancer therapy. Pharmacology & therapeutics. 2014;142(2):176–182. doi: 10.1016/j.pharmthera.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 4.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 5.Hodis E, Watson IR, Kryukov GV, et al. A landscape of driver mutations in melanoma. Cell. 2012;150(2):251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alsina J, Gorsk DH, Germino FJ, et al. Detection of mutations in the mitogen-activated protein kinase pathway in human melanoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2003;9(17):6419–6425. [PubMed] [Google Scholar]
- 7.Goydos JS, Mann B, Kim HJ, et al. Detection of B-RAF and N-RAS mutations in human melanoma. Journal of the American College of Surgeons. 2005;200(3):362–370. doi: 10.1016/j.jamcollsurg.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 8.Libra M, Malaponte G, Navolanic PM, et al. Analysis of BRAF mutation in primary and metastatic melanoma. Cell cycle (Georgetown, Tex) 2005;4(10):1382–1384. doi: 10.4161/cc.4.10.2026. [DOI] [PubMed] [Google Scholar]
- 9.McArthur GA, Chapman PB, Robert C, et al. Safety and efficacy of vemurafenib in BRAF(V600E) and BRAF(V600K) mutation-positive melanoma (BRIM-3): extended follow-up of a phase 3, randomised, open-label study. The Lancet Oncology. 2014;15(3):323–332. doi: 10.1016/S1470-2045(14)70012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. The New England journal of medicine. 2011;364(26):2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hauschild A, Grob JJ, Demidov LV, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet (London, England) 2012;380(9839):358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 12.Spagnolo F, Ghiorzo P, Queirolo P. Overcoming resistance to BRAF inhibition in BRAF-mutated metastatic melanoma. Oncotarget. 2014;5(21):10206–10221. doi: 10.18632/oncotarget.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hyman DM, Puzanov I, Subbiah V, et al. Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. The New England journal of medicine. 2015;373(8):726–736. doi: 10.1056/NEJMoa1502309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kopetz S, Desai J, Chan E, et al. Phase II Pilot Study of Vemurafenib in Patients With Metastatic BRAF-Mutated Colorectal Cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2015;33(34):4032–4038. doi: 10.1200/JCO.2015.63.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roller DG, Capaldo B, Bekiranov S, et al. Combinatorial drug screening and molecular profiling reveal diverse mechanisms of intrinsic and adaptive resistance to BRAF inhibition in V600E BRAF mutant melanomas. Oncotarget. 2016;7(3):2734–2753. doi: 10.18632/oncotarget.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu D, Liu X, Xing M. Activities of multiple cancer-related pathways are associated with BRAF mutation and predict the resistance to BRAF/MEK inhibitors in melanoma cells. Cell cycle (Georgetown, Tex) 2014;13(2):208–219. doi: 10.4161/cc.26971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kandel ES, Hay N. The regulation and activities of the multifunctional serine/threonine kinase Akt/PKB. Experimental cell research. 1999;253(1):210–229. doi: 10.1006/excr.1999.4690. [DOI] [PubMed] [Google Scholar]
- 18.Cao J, Heijkants RC, Jochemsen AG, et al. Targeting of the MAPK and AKT pathways in conjunctival melanoma shows potential synergy. Oncotarget. 2016 doi: 10.18632/oncotarget.10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delmas A, Cherier J, Pohorecka M, et al. The c-Jun/RHOB/AKT pathway confers resistance of BRAF-mutant melanoma cells to MAPK inhibitors. Oncotarget. 2015;6(17):15250–15264. doi: 10.18632/oncotarget.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perna D, Karreth FA, Rust AG, et al. BRAF inhibitor resistance mediated by the AKT pathway in an oncogenic BRAF mouse melanoma model. Proc Natl Acad Sci U S A. 2015;112(6):E536–E545. doi: 10.1073/pnas.1418163112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shao Y, Aplin AE. Akt3-mediated resistance to apoptosis in B-RAF-targeted melanoma cells. Cancer research. 2010;70(16):6670–6681. doi: 10.1158/0008-5472.CAN-09-4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dugo M, Nicolini G, Tragni G, et al. A melanoma subtype with intrinsic resistance to BRAF inhibition identified by receptor tyrosine kinases gene-driven classification. Oncotarget. 2015;6(7):5118–5133. doi: 10.18632/oncotarget.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nazarian R, Shi H, Wang Q, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468(7326):973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Byeon HK, Na HJ, Yang YJ, et al. c-Met-mediated reactivation of PI3K/AKT signaling contributes to insensitivity of BRAF(V600E) mutant thyroid cancer to BRAF inhibition. Molecular carcinogenesis. 2016;55(11):1678–1687. doi: 10.1002/mc.22418. [DOI] [PubMed] [Google Scholar]
- 25.Su F, Bradley WD, Wang Q, et al. Resistance to selective BRAF inhibition can be mediated by modest upstream pathway activation. Cancer research. 2012;72(4):969–978. doi: 10.1158/0008-5472.CAN-11-1875. [DOI] [PubMed] [Google Scholar]
- 26.Penna I, Molla A, Grazia G, et al. Primary cross-resistance to BRAFV600E-, MEK1/2- and PI3K/mTOR-specific inhibitors in BRAF-mutant melanoma cells counteracted by dual pathway blockade. Oncotarget. 2016;7(4):3947–3965. doi: 10.18632/oncotarget.6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi H, Kong X, Ribas A, Lo RS. Combinatorial treatments that overcome PDGFRbeta-driven resistance of melanoma cells to V600EB-RAF inhibition. Cancer research. 2011;71(15):5067–5074. doi: 10.1158/0008-5472.CAN-11-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Villanueva J, Vultur A, Lee JT, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer cell. 2010;18(6):683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johannessen CM, Boehm JS, Kim SY, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468(7326):968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Allen EM, Wagle N, Sucker A, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014;4(1):94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Long GV, Fung C, Menzies AM, et al. Increased MAPK reactivation in early resistance to dabrafenib/trametinib combination therapy of BRAF-mutant metastatic melanoma. Nature communications. 2014;5:5694. doi: 10.1038/ncomms6694. [DOI] [PubMed] [Google Scholar]
- 32.Trunzer K, Pavlick AC, Schuchter L, et al. Pharmacodynamic effects and mechanisms of resistance to vemurafenib in patients with metastatic melanoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31(14):1767–1774. doi: 10.1200/JCO.2012.44.7888. [DOI] [PubMed] [Google Scholar]
- 33.Watson IR, Li L, Cabeceiras PK, et al. The RAC1 P29S hotspot mutation in melanoma confers resistance to pharmacological inhibition of RAF. Cancer research. 2014;74(17):4845–4852. doi: 10.1158/0008-5472.CAN-14-1232-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kichina JV, Goc A, Al-Husein B, Somanath PR, Kandel ES. PAK1 as a therapeutic target. Expert Opin Ther Targets. 2010;14(7):703–725. doi: 10.1517/14728222.2010.492779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beeser A, Jaffer ZM, Hofmann C, Chernoff J. Role of group A p21-activated kinases in activation of extracellular-regulated kinase by growth factors. The Journal of biological chemistry. 2005;280(44):36609–36615. doi: 10.1074/jbc.M502306200. [DOI] [PubMed] [Google Scholar]
- 36.Tran NH, Wu X, Frost JA. B-Raf and Raf-1 are regulated by distinct autoregulatory mechanisms. The Journal of biological chemistry. 2005;280(16):16244–16253. doi: 10.1074/jbc.M501185200. [DOI] [PubMed] [Google Scholar]
- 37.Frost JA, Xu S, Hutchison MR, Marcus S, Cobb MH. Actions of Rho family small G proteins and p21-activated protein kinases on mitogen-activated protein kinase family members. Molecular and cellular biology. 1996;16(7):3707–3713. doi: 10.1128/mcb.16.7.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang Y, Zhou H, Chen A, Pittman RN, Field J. The Akt proto-oncogene links Ras to Pak and cell survival signals. The Journal of biological chemistry. 2000;275(13):9106–9109. doi: 10.1074/jbc.275.13.9106. [DOI] [PubMed] [Google Scholar]
- 39.Somanath PR, Vijai J, Kichina JV, Byzova T, Kandel ES. The role of PAK-1 in activation of MAP kinase cascade and oncogenic transformation by Akt. Oncogene. 2009;28(25):2365–2369. doi: 10.1038/onc.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rudolph J, Crawford JJ, Hoeflich KP, Wang W. Inhibitors of p21-activated kinases (PAKs) Journal of medicinal chemistry. 2015;58(1):111–129. doi: 10.1021/jm501613q. [DOI] [PubMed] [Google Scholar]
- 41.Dong B, Silverman RH, Kandel ES. A natural human retrovirus efficiently complements vectors based on murine leukemia virus. PLoS One. 2008;3(9):e3144. doi: 10.1371/journal.pone.0003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morgenstern JP, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic acids research. 1990;18(12):3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singhal R, Deng X, Chenchik AA, Kandel ES. Long-distance effects of insertional mutagenesis. PLoS One. 2011;6(1):e15832. doi: 10.1371/journal.pone.0015832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singhal R, Bard JE, Nowak NJ, Buck MJ, Kandel ES. FOXO1 regulates expression of a microRNA cluster on X chromosome. Aging (Albany NY) 2013;5(5):347–356. doi: 10.18632/aging.100558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mannava S, Omilian AR, Wawrzyniak JA, et al. PP2A–B56alpha controls oncogene-induced senescence in normal and tumor human melanocytic cells. Oncogene. 2012;31(12):1484–1492. doi: 10.1038/onc.2011.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer research. 2010;70(2):440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 47.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Advances in enzyme regulation. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 48.Tsai J, Lee JT, Wang W, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci U S A. 2008;105(8):3041–3046. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deacon SW, Beeser A, Fukui JA, et al. An isoform-selective, small-molecule inhibitor targets the autoregulatory mechanism of p21-activated kinase. Chemistry & biology. 2008;15(4):322–331. doi: 10.1016/j.chembiol.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCarty SK, Saji M, Zhang X, et al. BRAF activates and physically interacts with PAK to regulate cell motility. Endocrine-related cancer. 2014;21(6):865–877. doi: 10.1530/ERC-14-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoeflich KP, Herter S, Tien J, et al. Antitumor efficacy of the novel RAF inhibitor GDC-0879 is predicted by BRAFV600E mutational status and sustained extracellular signal-regulated kinase/mitogen-activated protein kinase pathway suppression. Cancer research. 2009;69(7):3042–3051. doi: 10.1158/0008-5472.CAN-08-3563. [DOI] [PubMed] [Google Scholar]
- 52.Yeh TC, Marsh V, Bernat BA, et al. Biological characterization of ARRY-142886 (AZD6244), a potent, highly selective mitogen-activated protein kinase kinase 1/2 inhibitor. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13(5):1576–1583. doi: 10.1158/1078-0432.CCR-06-1150. [DOI] [PubMed] [Google Scholar]
- 53.Zhou GL, Zhuo Y, King CC, Fryer BH, Bokoch GM, Field J. Akt phosphorylation of serine 21 on Pak1 modulates Nck binding and cell migration. Molecular and cellular biology. 2003;23(22):8058–8069. doi: 10.1128/MCB.23.22.8058-8069.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Michaelis M, Rothweiler F, Nerreter T, et al. Differential effects of the oncogenic BRAF inhibitor PLX4032 (vemurafenib) and its progenitor PLX4720 on ABCB1 function. Journal of pharmacy & pharmaceutical sciences : a publication of the Canadian Society for Pharmaceutical Sciences, Societe canadienne des sciences pharmaceutiques. 2014;17(1):154–168. doi: 10.18433/j3tw24. [DOI] [PubMed] [Google Scholar]
- 55.Nheu T, He H, Hirokawa Y, Walker F, Wood J, Maruta H. PAK is essential for RAS-induced upregulation of cyclin D1 during the G1 to S transition. Cell cycle (Georgetown, Tex) 2004;3(1):71–74. [PubMed] [Google Scholar]
- 56.Tang Y, Yu J, Field J. Signals from the Ras, Rac, and Rho GTPases converge on the Pak protein kinase in Rat-1 fibroblasts. Molecular and cellular biology. 1999;19(3):1881–1891. doi: 10.1128/mcb.19.3.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chow HY, Jubb AM, Koch JN, et al. p21-Activated kinase 1 is required for efficient tumor formation and progression in a Ras-mediated skin cancer model. Cancer research. 2012;72(22):5966–5975. doi: 10.1158/0008-5472.CAN-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singhal R, Kandel ES. The response to PAK1 inhibitor IPA3 distinguishes between cancer cells with mutations in BRAF and Ras oncogenes. Oncotarget. 2012;3(7):700–708. doi: 10.18632/oncotarget.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ong CC, Jubb AM, Jakubiak D, et al. P21-activated kinase 1 (PAK1) as a therapeutic target in BRAF wild-type melanoma. Journal of the National Cancer Institute. 2013;105(9):606–607. doi: 10.1093/jnci/djt054. [DOI] [PubMed] [Google Scholar]
- 60.Chaudhary A, King WG, Mattaliano MD, et al. Phosphatidylinositol 3-kinase regulates Raf1 through Pak phosphorylation of serine 338. Current biology : CB. 2000;10(9):551–554. doi: 10.1016/s0960-9822(00)00475-9. [DOI] [PubMed] [Google Scholar]
- 61.Ren Y, Li R, Zheng Y, Busch H. Cloning and characterization of GEF-H1, a microtubule-associated guanine nucleotide exchange factor for Rac and Rho GTPases. The Journal of biological chemistry. 1998;273(52):34954–34960. doi: 10.1074/jbc.273.52.34954. [DOI] [PubMed] [Google Scholar]
- 62.Zenke FT, Krendel M, DerMardirossian C, King CC, Bohl BP, Bokoch GM. p21-activated kinase 1 phosphorylates and regulates 14-3-3 binding to GEF-H1, a microtubule-localized Rho exchange factor. The Journal of biological chemistry. 2004;279(18):18392–18400. doi: 10.1074/jbc.M400084200. [DOI] [PubMed] [Google Scholar]
- 63.Manser E, Loo TH, Koh CG, et al. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Molecular cell. 1998;1(2):183–192. doi: 10.1016/s1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- 64.Marei H, Carpy A, Macek B, Malliri A. Proteomic analysis of Rac1 signaling regulation by guanine nucleotide exchange factors. Cell cycle (Georgetown, Tex) 2016;15(15):1961–1974. doi: 10.1080/15384101.2016.1183852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.May M, Schelle I, Brakebusch C, Rottner K, Genth H. Rac1-dependent recruitment of PAK2 to G2 phase centrosomes and their roles in the regulation of mitotic entry. Cell cycle (Georgetown, Tex) 2014;13(14):2211–2221. doi: 10.4161/cc.29279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li YY, Wu C, Chen SM, et al. BRAF inhibitor resistance enhances vulnerability to arginine deprivation in melanoma. Oncotarget. 2016;7(14):17665–17680. doi: 10.18632/oncotarget.6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goulielmaki M, Koustas E, Moysidou E, et al. BRAF associated autophagy exploitation: BRAF and autophagy inhibitors synergise to efficiently overcome resistance of BRAF mutant colorectal cancer cells. Oncotarget. 2016;7(8):9188–9221. doi: 10.18632/oncotarget.6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.