Abstract
Background
Fc-gamma receptors (FCGRs) are expressed on immune cells, bind to antibodies, and trigger antibody-induced cell-mediated anti-tumor responses when tumor-reactive antibodies are present. The affinity of the FCGR/antibody interaction is variable and dependent upon FCGR polymorphisms. Prior studies of cancer patients treated with immunotherapy indicate that FCGR polymorphisms can influence antitumor response for certain immunotherapies that act via therapeutically administered mAbs or via endogenous tumor-reactive antibodies induced from tumor antigen vaccines. The previously published “SELECT” trial of high-dose aldesleukin (HD-IL2) for metastatic renal cell carcinoma (mRCC) resulted in an objective response rate (ORR) of 25%. We evaluated the patients in this SELECT trial to determine whether higher affinity FCGR polymorphisms are associated with outcome.
Methods
Single nucleotide polymorphisms (SNPs) in FCGR2A, FCGR3A, and FCGR2C were analyzed, individually and in combination, for associations between genotype and clinical outcome.
Results
When higher affinity genotypes for FCGR2A, FCGR3A and FCGR2C were considered together, they were associated with significantly increased tumor shrinkage and prolonged survival in response to HD-IL2.
Conclusions
While associations of higher affinity FCGR genotype with clinical outcome have been demonstrated with mAb therapy and with idiotype vaccines, to our knowledge, this is the first study to show associations of FCGR genotypes with outcome following HD-IL2 treatment. We hypothesize that endogenous anti-tumor antibodies may engage immune cells through their FCGRs, and HD-IL2 may enhance antibody-induced tumor destruction, or antibody-enhanced tumor antigen presentation, via augmented activation of innate or adaptive immune responses; this FCGR-mediated immune activity would be augmented through immunologically favorable FCGRs.
Keywords: FCGR, IL2, Immunotherapy, Renal Cell Carcinoma, Endogenous Antibody
INTRODUCTION
Patients with metastatic renal cell carcinoma (mRCC) show a 14%–22% response to the standard high-dose regimen of aldesleukin [interleukin-2 (IL2)]. Patients with mRCC were entered into the “SELECT” clinical trial of high-dose IL2 (HD-IL2) to prospectively determine if certain clinical and pathological criteria are associated with response to IL2. As noted in the clinical report, this study produced a response rate of 25% (1). In an effort to further identify genetic markers that might associate with efficacy of the HD-IL2 treatment for patients with mRCC, and potentially identify immunologic mechanisms involved in the response, we sought to identify genotypic factors that may influence the immune activity of HD-IL2 therapy. In this study we genotyped single nucleotide polymorphisms (SNPs) found in certain activating Fc-gamma Receptor (FCGR) genes (FCGR2A, FCGR3A and FCGR2C).
Variably expressed on immune cells, FCGRs bind the Fc fragment of IgG antibodies (2–4). Upon engagement and crosslinking, activating FCGRs transmit signaling within the immune cell and initiate immune activation (5–8). FCGR2A (expressed on dendritic cells, macrophages, monocytes, neutrophils, and eosinophils), FCGR3A (expressed on NK cells and macrophages), and FCGR2C (also expressed on NK cells) are all activating FCGRs (4, 9). The SNPs found in both FCGR2A and FCGR3A genes convey differential binding affinities for the Fc portion of antibody. The FCGR2A SNP encodes amino acids of either histidine (H) or arginine (R) at position 131 of the FCGR2A protein (FCGR2A-H131R, rs1801274), and the FCGR3A SNP encodes either valine (V) or phenylaline (F) at amino acid 158 of FCGR3A (FCGR3A-V158R, rs396991) (10–13). The FCGR2A-H and FCGR3A-V receptors each have higher binding affinities to human IgG than do the FCGR2A-R and FCGR3A-F receptors, respectively (2, 4, 12). This stronger binding affinity results in more potent in vitro antibody-dependent cell-mediated cytotoxicity (ADCC) and tumor cell death (14, 15). In some clinical trials involving various chimeric or humanized monoclonal antibodies (mAbs) specific for head and neck, colorectal, or B-cell malignancies, both FCGR2A-H and FCGR3A-V SNPs are associated with improved clinical response (14–17). Similarly, in a trial of an idiotypic vaccine for B-cell lymphoma, designed to induce endogenous anti-idiotypic antibody, better outcome was seen for patients with the higher affinity FCGR2A-H and FCGR3A-V SNPs (17). Alternatively, other studies have found no association of FCGR2A-H/R or FCGR3A-V/F SNP genotype with patient response to immunotherapy (18–20).
The FCGR2C gene has a SNP in exon 3 (c.169 C<T, rs759550223) that influences the expression of FCGR2C on NK cell surfaces (21–23). The presence of a “C” nucleotide in this SNP leads to an open reading frame, enabling the expression of the FCGR2C receptor. In contrast, a “T” nucleotide creates a stop codon, resulting in lack of expression, for that allele (21, 22, 24). A minority of individuals (20–40%) have the “C” allele (either FCGR2C-C/C or C/T genotype), and thus have FCGR2C expressed on their NK cells (24–27). When expressed, FCGR2C is capable of inducing ADCC after receptor crosslinking (24, 25, 27). While the SNP of FCGR2C genotype has been correlated with patient response to immunomodulatory therapy for autoimmune-based diseases (25, 28–32), little has been published regarding the role of FCGR2C expression in cancer immunotherapy.
In this study of patients with mRCC who received HD-IL2, we looked for associations of patient FCGR2A, FCGR3A and FCGR2C genotypes with clinical outcome. We found that higher affinity FCGR genotypes resulted in improved tumor shrinkage and overall survival (OS). These findings suggest a potential role for cells expressing these FCGRs in the clinical response of patients with mRCC to HD-IL2 therapy.
METHODS
DNA
A total of 106 patients from the SELECT trial had DNA available for genotyping, along with clinical data for correlative analyses. DNA was isolated from peripheral blood mononuclear cells (PBMCs) following the manufacturer’s protocol of the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA). DNA was kept at 4C during the time of analyses, and later was transferred to −80C for long term storage after completion of the analyses.
Genotyping
All SNP genotyping was performed on a StepOnePlus quantitative PCR machine (ABI/Life Technologies, Grand Island, NY). The FCGR2A SNP was determined using Taqman primer/probes available from ABI/Life Technologies and used per the manufacturers protocol. For both FCGR3A and FCGR2C, Rnase H primers and probes for each gene were developed in our lab to allow for specific amplification of each gene. For genotyping the FCGR3A SNP, Rnase H primers were developed to specifically amplify FCGR3A while not co-amplifying FCGR3B. These primers were paired with specific probes to determine the SNP (33). For genotyping the FCGR2C-C/T SNP, Rnase H primers were developed to specifically amplify this gene while not co-amplifying FCGR2B. Primers and probes for both FCGR3A and FCGR2C were designed through Integrated DNA Technologies (IDTDNA, Coralville, Iowa). Specific method details can be found in Erbe AK et al., 2016 (33). Genotyping was conducted in a blinded manner, where those individuals that determined the genotype of the patients did not have access to the clinical outcome data. Specific genotype results can be found in Supplementary Table 1.
Clinical data
The clinical results of the SELECT trial have been published (1). Clinical data for % tumor shrinkage and for OS were obtained from the clinical data set. Data were updated through October 31, 2013. The clinical characteristics of the patient subset we analyzed (106 patients of the 120 patients in the original trial) are similar to the clinical characteristics of those observed in the original study (Table 1). For our analysis of % tumor shrinkage, 2 patients did not have % tumor shrinkage clinical data available, and thus were excluded from the % tumor shrinkage analyses (104 for % tumor shrinkage).
Table 1.
Patient characteristics from original SELECT Trial and the subset of patients analyzed in this study.
| Total Patients Enrolled | FCGR Genotyped Pts | |
|---|---|---|
| Characteristics | n = 120 | N = 106 |
| Median age, y (range) | 56 (28–70) | 56 (28–70) |
| ECOG performance status (0/1), % | 72/24 | 71/25 |
| Prior nephrectomy, % | 99 | 99 |
| MSKCC risk factor, n (%) | ||
| 0 (favorable) | 23 (19) | 22 (21) |
| 1–2 (intermediate) | 84 (70) | 72 (68) |
| ≥3 (poor) | 13 (11) | 12 (11) |
| UCLA SANI Score, n (%) | ||
| Low | 10 (8) | 10 (9) |
| Intermediate | 102 (85) | 88 (83) |
| High | 8 (7) | 8 (8) |
Statistical methods
The clinical outcomes assessed included % tumor shrinkage and overall survival. The % tumor shrinkage was defined as the percent change in tumor size from baseline to maximum shrinkage. OS was defined as the time in months from the date of treatment initiation to the date of death, or was censored at the date of last contact with the patient. The association between % tumor shrinkage and genotyping predictors was evaluated using two-sample t-tests. The Kaplan-Meier method was used for estimation of the survival distribution for OS. For the survival plots, the tick marks along each line indicate patients censored; each drop of the line indicates a clinical event (i.e. patient death). Log-rank tests were used to assess the association between genotyping predictors and OS. The association between FCGR genotypes was assessed using Fisher’s exact test. Changes in tumor size were represented using box plots, which show the 25th percentile (Q1) (bottom of box), the 50th percentile (Q2) (bolded black line), the 75th percentile (Q3) (top of box), and the mean (red cross inside the box). The lower and upper short horizontal red lines represent the minimum and maximum values, excluding the outlying high and low values. Outlying values [i.e.: those that are a distance of more than 1.5x(Q3-Q1) from the box], are shown as circles outside the horizontal lines. No adjustments in reported p-values were made for multiplicity of testing.
RESULTS
Individual FCGR3A, FCGR2A, and FCGR2C genotypes show associations with clinical outcome
In these mRCC patients treated with HD-IL2, we found that individuals homozygous for the high-affinity FCGR3A-V/V allele had significantly prolonged OS compared to those having only 1 or no copy of this high affinity allele (FCGR3A-V/V: 73.4 months vs. FCGR3A-V/F or F/F: 40.6 months; p=0.03, Table 2). Additionally, although not significant, the % tumor shrinkage for FCGR3A-V/V patients was greater than that for FCGR3A-V/F or F/F patients, (FCGR3A-V/V: 32.8% vs. FCGR3A-V/F or F/F: 9.4%; p=0.21, Table 2). For FCGR2A, those with the higher-affinity-genotype compared to lower-affinity receptors (H/H vs. H/R or R/R, respectively) had increased tumor shrinkage, although these differences were not significant (FCGR2A-H/H: 25.8% vs. FCGR2A-H/R or R/R: 7.1%; p=0.18, Table 2). There was no difference in OS based on FCGR2A genotype. Patients that express the FCGR2C receptor [those with at least one copy of the C allele (C/C or C/T)] had prolonged OS as compared to those that did not express FCGR2C on their NK cell surface (those with an FCGR2C genotype of T/T), however these differences were not significant (FCGR2C-C/C or C/T: 73.3 months vs. FCGR2C-T/T: 40.6 months; p=0.12, Table 2). There were no differences in % tumor shrinkage based on FCGR2C genotype (Table 2).
Table 2.
Clinical Outcome by FCGR2A, FCGR3A and FCGR2C SNP. The amount of % tumor shrinkage and the duration of the OS for each FCGR were compared for individual FCGRs.
| Genotype Group | Tumor Shrinkage | Overall Survival | Disease Control Rate | |||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Mean (% )[Std Dev] | p-value | Median (months) [95% CI] | p-value | n | CR/PR/SD | p-value | ||
|
| ||||||||
| FCGR3A SNP | VV | 32.8 [45.5] (n=13) | 0.21 | 73.4 [32.5-NR]A (n=13; #Events=5) | 0.03 | 13 | 53.8% | 0.23 |
|
|
|
|||||||
| VF or FF | 9.4 [64.7] (n=91) | 40.6 [27.2–50.7] (n=93; #Events=65) | 91 | 35.2% | ||||
|
| ||||||||
| FCGR2A SNP | HH | 25.8 [39.8] (n=29) | 0.18 | 48.8 [26.7–61.9] (n=30; #Events=19) | 0.85 | 29 | 51.7% | 0.07 |
|
|
|
|||||||
| HR or RR | 7.1 [69.4] (n=75) | 40.6 [34.8–56.0] (n=76; #Events=51) | 75 | 32.0% | ||||
|
| ||||||||
| FCGR2C SNP | CC or CT | 12.0 [61.2] (n=31) | 0.97 | 73.3 [32.5-NR]A (n=31; #Events=17) | 0.12 | 31 | 35.5% | 1.00 |
|
|
|
|||||||
| TT | 12.4 [64.1] (n=73) | 40.6 [26.7–50.7] (n=75; #Events=53) | 73 | 38.4% | ||||
The value “NR” is reported where the Median Overall Survival is “Not Reached”.
Higher affinity FCGR2A and FCGR3A genotypes influence tumor shrinkage
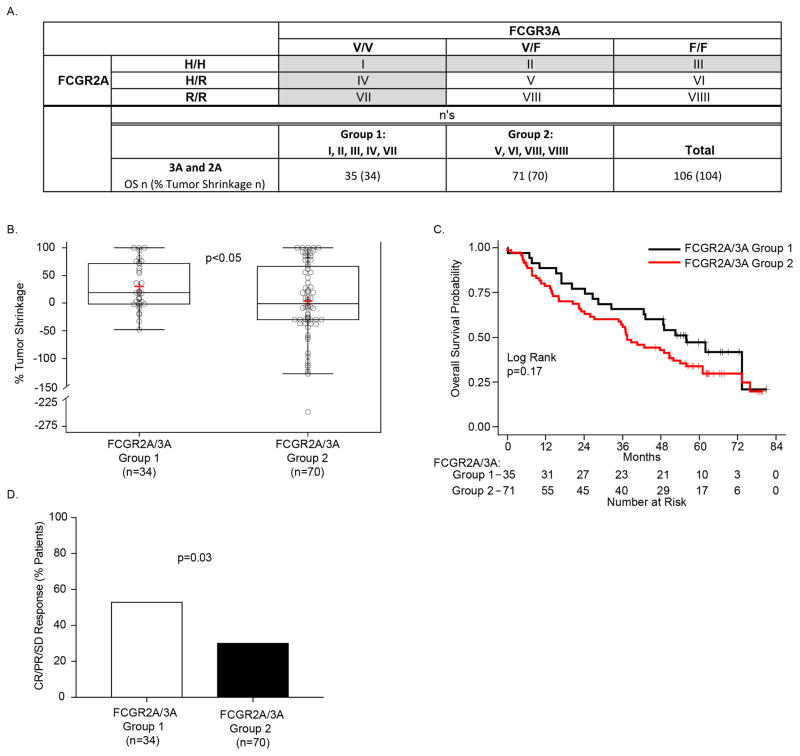
Upon antibody recognition and binding, there may be crosstalk between cells that express FCGR2A and cells that express FCGR3A that can influence NK cell response (34). Prior studies of patients treated with mAb have reported associations between FCGR SNPs and clinical outcome when both genotypes of FCGR2A and FCGR3A were combined for the analyses (35–37). We compared individuals that were homozygous for either the H allele of higher-affinity FCGR2A or for the V allele of higher-affinity FCGR3A (Group-1 in Fig. 1A) with individuals that were not homozygous for either the higher affinity allele of FCGR2A or FCGR3A (Group-2 in Fig. 1A). We found significantly improved tumor shrinkage in Group-1 vs. Group-2 (Fig. 1B, p<0.05). Additionally, Group-1 also showed prolonged OS vs. Group 2, but this was not significant (p=0.17, Fig. 1C).
Figure 1. FCGR2A and FCGR3A higher-affinity genotypes resulted in improved % tumor shrinkage.
A) The 3 separate genotypes for FCGR3A (V/V, V/F and F/F), when combined with the 3 separate genotypes for FCGR2A (H/H, H/R and R/R), yield 9 separate genotypes, each in a separate box designated: I–VIIII. Those patients with homozygous expression of either V/V or H/H (cells I, II III, IV, and VII) are included in Group-1 (n=35 patients for OS, n=34 for % Tumor Shrinkage), all remaining patients (neither V/V nor H/H) are included in Group-2 (n=71 patients for OS, n=70 for % Tumor Shrinkage). B) Patients in Group-1 (homozygous for either V/V or H/H) show a significant increase in the % tumor shrinkage as compared to those in Group-2 (not homozygous for either V/V or H/H). C) OS for Group-1 was prolonged vs. that for Group-2, although not significant.
Higher affinity FCGR3A and expression of FCGR2C genotypes influence OS
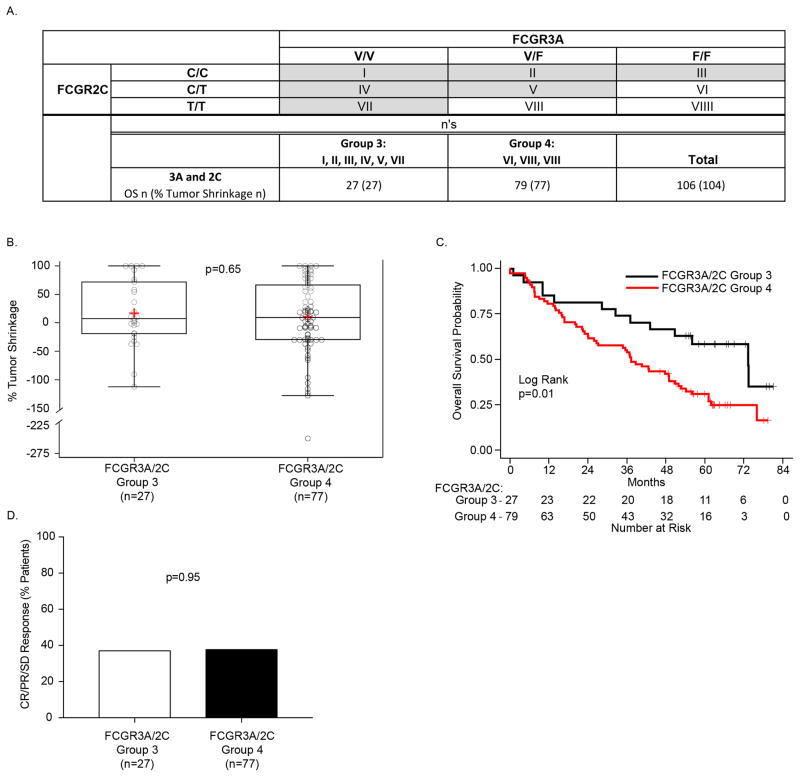
NK cell ADCC capabilities can be enhanced if FCGR2C is expressed on the cell surface (26). Since NK cells can express both FCGR3A and FCGR2C, we considered whether patient outcome was influenced by the combined genotypes for FCGR3A and FCGR2C. Patients that have 2 copies of the high affinity FCGR3A allele (V/V) or one copy of the high affinity FCGR3A allele (V/F) and at least one copy of FCGR2C (C/C or C/T), or 2 copies of FCGR2C (C/C) are identified as Group-3 (boxes I, II, III, IV, V and VII) in Fig. 2A. All other patients are identified as Group-4, and include those with genotypes that have only one copy of the high affinity FCGR3A allele (V/F) and have no copy of FCGR2C (T/T), and those that have no copy of the high affinity FCGR3A allele (F/F) and have only 1 or no copy of FCGR2C ( C/T or T/T), (boxes VI, VIII and VIIII in Fig. 2A). While there was no difference between Group-3 and Group-4 for % tumor shrinkage (Fig. 2B), Group-3 showed significantly prolonged OS compared to Group-4 (Fig. 2C, p=0.01).
Figure 2. FCGR3A and FCGR2C are associated with OS.
A) The 3 separate genotypes for FCGR3A (V/V, V/F and F/F), when combined with the 3 separate genotypes for FCGR2C (C/C, C/T and T/T), yield 9 separate genotypes, each in a separate box designated: I–VIIII. Since expression of FCGR2C (via C/C or C/T genotypes) can influence FCGR3A (2), we included those heterozygous for both FCGR2C and FCGR3A (box V). Thus, those patients with homozygous expression of either V/V or C/C (cells I, II III, IV, and VII) or with heterozygous expression for both (cell V) are included in Group-3 (n=27 patients for both OS and % Tumor Shrinkage), all remaining patients are included in Group-4 (n=79 patients for OS, n=77 for % Tumor Shrinkage). B) Patients in Group-3 show no difference in the % tumor shrinkage as compared to those in Group-4, however C) OS for Group-3 was significantly prolonged vs. that for Group-4.
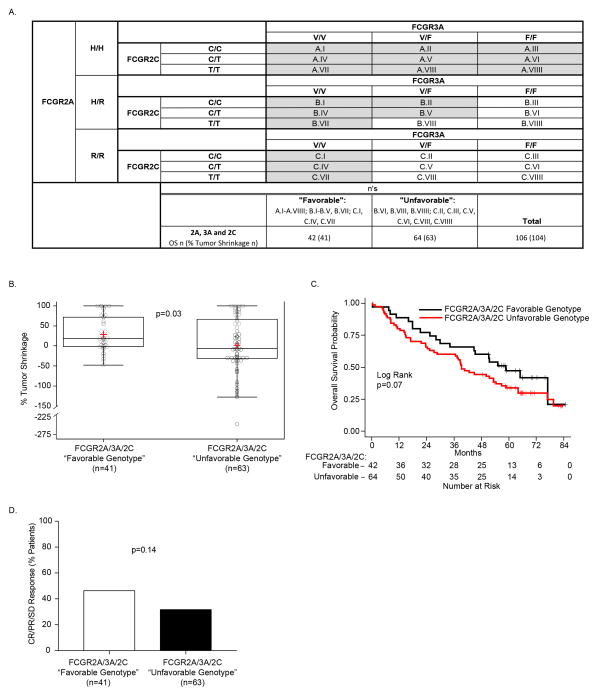
Favorable overall FCGR3A/2A/2C genotypes influence clinical outcome
Based on our findings that patients with FCGR2A and FCGR3A genotypes in homozygous form resulted in prolonged OS (although not statistically significant) and significantly improved % tumor shrinkage (Fig. 1), as well as our finding that high-affinity FCGR3A-V in combination with the expression of FCGR2C resulted in significant improvement in the length of OS (Fig. 2), we further assessed the combined influence of all three of these FCGR genotypes on patient response. In order to simultaneously consider the genotype combinations for all three FCGRs studied here, we categorized patients into “favorable” and “unfavorable” groups (Fig. 3A) based on the genotypic patterns presented in Figs. 1A and 2A. The favorable group (shaded in Fig. 3A) included all patients homozygous for FCGR3A V/V or FCGR2A H/H, as well as patients with at least 2 higher affinity alleles of FCGR3A or FCGR2A (at least one copy of FCGR3A-V and at least one copy of FCGR2A-H) with FCGR2C expression (C/C or C/T), namely V/F-H/R patients, if they also expressed FCGR2C (C/C or C/T). This corresponded to 42 favorable-genotype patients. The remaining 64 patients (unshaded in Fig. 3A) are designated as unfavorable genotype.
Figure 3. The combination of FCGR2A, FCGR3A and FCGR2C SNPs is associated with % tumor shrinkage and OS.
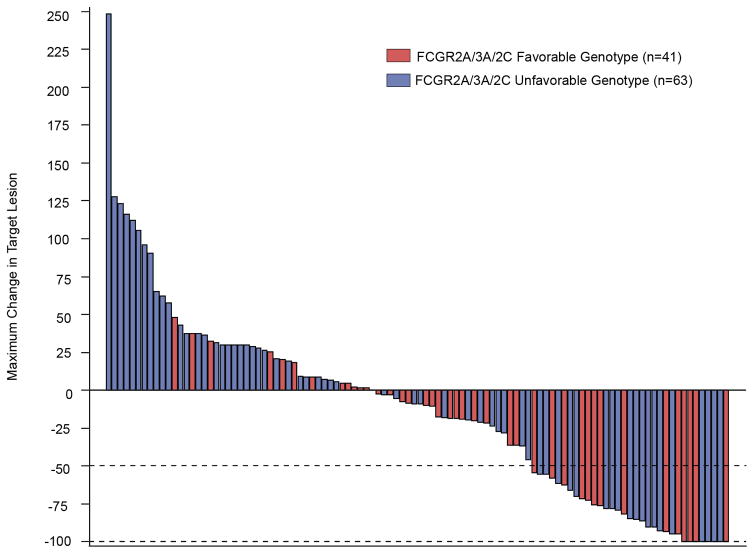
A) The 3 separate genotypes for FcgR3a (V/V, V/F and F/F), when combined with the 3 separate genotypes for FCGR2c (C/C, C/T and T/T) yield 9 separate genotypes. Here these are combined with the 3 separate genotypes for FcGR2A: H/H (upper panel); H/R (center panel); and R/R (lower panel). When genotypes for all 3 of these loci are combined (27 separate boxes), we divided them into favorable (shaded in Fig. 3A) vs. unfavorable (unshaded) genotypes. The favorable group includes all patients homozygous for H/H or V/V, as well as patients heterozygous for H/R if also expressing V/V or at least one copy of FCGR2C-”C”, and patients negative for H (ie: R/R patients) if they are V/V homozygotes, corresponding to 42 patients for OS, 41 patients for % Tumor Shrinkage. All others, namely those patients that do not have at least 2 copies of either high affinity allele (F/F-R/R, V/F-R/R or F/F-H/R), and those patients heterozygous for V/F and H/R but lacking any expression of FCGR2C are unshaded and labeled as unfavorable (n=64 patients for OS, n=63 patients for % Tumor Shrinkage). B) Patients with a favorable genotype show improved % tumor shrinkage as compared to those in the unfavorable group (p=0.03). C) This predominance of patients with favorable genotype (red) vs. unfavorable genotype (blue) amongst those with tumor shrinkage is also seen in the right side of the waterfall plot for % tumor shrinkage for all 105 patients evaluable for this analysis. D) A trend towards better OS was also seen for patients with favorable genotype (p=0.07)
Patients in the favorable FCGR genotype group had a significantly improved % tumor shrinkage as compared to those with “unfavorable” FCGR genotype (Fig. 3B, 28.5% vs. 1.7%; p=0.03). As depicted in the waterfall plot of % tumor shrinkage (Fig. 3C), those patients in the favorable group (red bars) are more prominent than those in the unfavorable group (blue bars) amongst those that showed tumor shrinkage rather than growth. Patients in the favorable group also showed a trend towards improved OS (Fig. 3D; 56.0 vs. 37.4 months for favorable vs. unfavorable groups; p=0.07).
DISCUSSION
While both the genotypes of the SNPs on FCGR2A and FCGR3A have been implicated in some analyses of the clinical anti-tumor response to tumor-reactive mAb immunotherapy, we believe this is the first study to show a potential association of favorable FCGR genotype with clinical outcome in the anti-tumor use of single-agent HD-IL2, without mAb administration. Moreover, FCGR2C expression based on SNP status has not yet been shown to influence clinical response to immunotherapeutics in cancer patients, in particular in patients not treated with mAb.
The data presented in Figs. 3B, C and D, show a significant association with % tumor shrinkage and a trend with OS when simultaneously considering genotypes for all 3 of these loci (FCGR2A, 3A and 2C). The finding that there are associations of FCGR genotype with the clinical outcome parameters of both tumor shrinkage and OS appears to involve all 3 of these FCGR genes. This is consistent with data in Table 2, showing a significant role for FCGR3A in OS, as well as trends in improved % tumor shrinkage for FCGR2A, and improved OS for FCGR2C (although not statistically significant).
Although we found significant associations of FCGR genotype with both of these clinical parameters (% tumor shrinkage and overall survival), we did not see significant associations of patient FCGR2A, 3A and 2C SNP genotype with patient overall response rate (data not shown). Since the overall response rate is based on data for % tumor shrinkage, but it is evaluated in binary form [responders (those with >50% tumor shrinkage) vs. non-responders (those with <50% tumor shrinkage)], it does not take into consideration the quantitative amount of tumor shrinkage. The waterfall analysis (Fig. 3C), which scores each patient based on their maximum amount of % tumor shrinkage, is based on quantitative measures and it is known to be more sensitive compared to overall response rate. This may account for the genotypic associations with % tumor shrinkage, but not with overall response status, found in this study.
Such associations of favorable FCGR genotypes and clinical outcome with HD-IL2 treatment do not prove a causal link. McDermott et al., 2015, reported that in the original cohort of patients treated with HD-IL2, in addition to the HD-IL2, 80 patients also received VEGF-targeted therapy. This additional VEGF-targeted therapy may have contributed to the OS length found in those individuals that were treated with it. Beyond the additional treatment measures (i.e. VEGF-targeted treatment) that may have influenced clinical response differences, genotypes that show an association with a clinical condition may do so because of their linkage disequilibrium to nearby loci that were not directly genotyped, yet influence the clinical associations seen. The FCGR genes are located in close proximity to each other on chromosome 1q23, with FCGR2A located upstream [with genomic coordinates (GRCh38): 1:161,505,41–161,524,048] of FCGR3A [with genomic coordinates (GRCh38): 1:161,541,759–161,550,623] followed by FCGR2C [with genomic coordinates: Genomic coordinates (GRCh38): 1:161,581,339–161,601,220] (38). Using the genotype data for this population (Supplementary Table 1), we found a trend towards linkage disequilibrium between FCGR3A and FCGR2A (p=0.08), a significant disequilibrium between FCGR3A and FCGR2C (p<0.01), and no significant disequilibrium between FCGR2A and FCGR2C (p=0.70) (data not shown). This linkage disequilibrioum involving these 3 genes could contribute to the favorable FCGR genotype grouping found in this study, as shown in Fig. 3. Furthermore, although unlikely, these favorable FCGR gene alleles that are associated with better outcome in this study could potentially be in linkage disequilibrium with a favorable allele for some separate (non-FCGR) gene that might actually be responsible for the improved outcome we observe with the favorable FCGR genotype. The fact that some of the associations that we have observed are significant while others are trends, suggests that the effect of the favorable FCGR genes is one of several factors involved in the anti-tumor activity of HD-IL2 in some patients (but not others) with mRCC.
This association of outcome with favorable FCGRs suggests that greater functionality of FCGRs, due to higher-affinity (for FCGR2A and FCGR3A) and expression of FCGR2C, may be playing a role in at least part of the anti-tumor activity of HD-IL2. Our current understanding of these FCGRs is that they function primarily through engaging antigen-bound IgG, transmitting an activating signal, and inducing cellular responses, such as the induction of ADCC (by NK cells, neutrophils and monocytes/macrophages) or antibody dependent cellular phagocytosis (ADCP), and the uptake of antigens by antigen presenting cells through immobilized, bound IgG molecules, resulting in antigen processing and presentation (3, 4, 6, 8, 26). In each of these settings, an antigen-reactive antibody (either endogenous or passively administered) is needed for antibody/FCGR-facilitated ADCC, ADCP or antigen processing. The data presented here, showing that HD-IL2 treated mRCC patients with more functional FCGR genotypes showed increased tumor shrinkage and prolonged OS compared to those with less-functional FCGR genotypes supports the hypothesis that some of these patients may have formed endogenous antibodies, reactive with their autochthonous mRCC that were capable of mediating ADCC, ADCP and/or antigen presentation. The in vivo antitumor activity of such endogenous anti-tumor antibodies would potentially be enhanced by the presence of more favorable FCGRs.
In 1955, Graham and Graham suggested that some gynecologic oncology patients formed endogenous antibodies recognizing autologous tumor antigens, but these endogenous antibodies did not recognize the tumor antigens derived from tumors of similar histology from other patients (39). Since that time, several endogenous antibodies that are reactive against well-described and conserved shared tumor antigens have been identified, including antigens on RCC (40–44). For example, Knutson et al. 2016 recently showed that for HER2+ breast cancer patients, a combination therapy that included chemotherapy together with trastuzumab (mAb against HER2) induced, in 69% of patients, endogenous IgG-antibodies directed against HER2 and a subset of endogenous shared tumor-associated antigens; this endogenous antibody response was associated with improved disease outcome (45). However, for most tumor types, methods to readily demonstrate and quantify the presence and functional activity of endogenous antibodies against the unique neo-antigens present on patients’ autochthonous tumors, for the full cohort of patients enrolled in a trial such as this one, remain elusive. Thus, in this retrospective study, with no access to patient sera or to patient tumor tissue, we have not attempted to evaluate the presence or functionality of endogenous antibody to autochthonous tumor.
The interplay of several immune cell types, through engagement of their FCGRs via antibody-bound-antigen recognition, creates the potential for a successful immunotherapeutic response following treatment with mAb (46). Based on the associations reported here, of more functional FCGRs being associated with improved outcome with HD-IL2 therapy, we hypothesize the following immunological pathways may be involved. First, the presence of pre-existing endogenous tumor-reactive IgG antibodies might enable IL2 to induce augmented ADCC and ADCP, which would be enhanced by the presence of more functional FCGR alleles through crosstalk of NK cells (expressing FCGR3A and potentially FCGR2C) and monocytes (expressing FCGR2A) (34). Alternatively, the pre-existing anti-tumor antibodies might facilitate tumor-antigen presentation and induction of an adaptive (dendritic cell, T-cell and potentially B-cell) response, which could be augmented by the IL2 treatment and more functional FCGR. Finally, in some patients, more than one of these mechanisms could be at work simultaneously. The FCGR genotype combinations identified here have the potential to serve as biomarkers to personalize immunotherapeutics for cancer treatment (47). Future studies validating this association of favorable FCGR genotype with outcome, as well as prospective efforts to evaluate sera from all treated patients for functional antibody reactive to tumor (particularly autochthonous tumor), will be needed to test these hypotheses, and determine whether they lead to actionable clinical modifications in this approach towards immunotherapy.
Supplementary Material
Figure 4.
Statement of Translational Relevance.
Associations with clinical outcome were found in this study in individuals that have a “favorable” FCGR genotype (higher-affinity alleles of FCGR2A and FCGR3A with expression of FCGR2C), suggesting that greater functionality of FCGRs plays a role in the anti-tumor activity of high-dose IL2 for patients with metastatic renal cell cancer (mRCC). The data presented in this report suggest that FCGRs may play a role in the in vivo antitumor effect seen in mRCC patients receiving high-dose IL2, raise important hypotheses for future research that may focus on the potential role of endogenous anti-tumor antibody, and indicate that future work should be pursued to test whether the combined analyses of FCGR3A/2A/2C genotypes may become a useful biomarker for prospective clinical planning and retrospective outcome analyses for other clinical trials of cancer immunotherapy that may involve NK cells or other FCGR-bearing immune cells.
Acknowledgments
Financial Support: This research was supported by The Institute for Clinical and Translational Research; Hyundai Hope on Wheels Grant; Midwest Athletes Against Childhood; Stand Up To Cancer – St. Baldrick’s Pediatric Dream Team Translational Cancer Research Grant, Grant Number SU2C-AACR-DT11-13 – Stand Up To Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research; University of Wisconsin-Madison Carbone Cancer Center; by NCI-Cytokine Working Group and supported in part by Public Health Service Grants CA014520, CA021115, CA023318, CA066636, CA180820, CA180794, CA021076, CA180799, CA14958, CA180816, CA166105, and CA197078, from the National Cancer Institute; the National Institutes of Health and the Department of Health and Human Services. Its content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute.
References
- 1.McDermott DF, Cheng SC, Signoretti S, Margolin KA, Clark JI, Sosman JA, et al. The high-dose aldesleukin “select” trial: a trial to prospectively validate predictive models of response to treatment in patients with metastatic renal cell carcinoma. Clin Cancer Res. 2015;21:561–8. doi: 10.1158/1078-0432.CCR-14-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruhns P, Iannascoli B, England P, Mancardi DA, Fernandez N, Jorieux S, et al. Specificity and affinity of human Fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood. 2009;113:3716–25. doi: 10.1182/blood-2008-09-179754. [DOI] [PubMed] [Google Scholar]
- 3.Nimmerjahn F, Ravetch JV. Analyzing antibody-Fc-receptor interactions. Methods Mol Biol. 2008;415:151–62. doi: 10.1007/978-1-59745-570-1_9. [DOI] [PubMed] [Google Scholar]
- 4.Nimmerjahn F, Ravetch JV. Fcgamma receptors as regulators of immune responses. Nat Rev Immunol. 2008;8:34–47. doi: 10.1038/nri2206. [DOI] [PubMed] [Google Scholar]
- 5.Amigorena S. Fc gamma receptors and cross-presentation in dendritic cells. J Exp Med. 2002;195:F1–3. doi: 10.1084/jem.20011925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bakema JE, van Egmond M. Fc receptor-dependent mechanisms of monoclonal antibody therapy of cancer. Curr Top Microbiol Immunol. 2014;382:373–92. doi: 10.1007/978-3-319-07911-0_17. [DOI] [PubMed] [Google Scholar]
- 7.Platzer B, Stout M, Fiebiger E. Antigen cross-presentation of immune complexes. Front Immunol. 2014;5:140. doi: 10.3389/fimmu.2014.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harbers SO, Crocker A, Catalano G, D’Agati V, Jung S, Desai DD, et al. Antibody-enhanced cross-presentation of self antigen breaks T cell tolerance. J Clin Invest. 2007;117:1361–9. doi: 10.1172/JCI29470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagelkerke SQ, Kuijpers TW. Immunomodulation by IVIg and the Role of Fc-Gamma Receptors: Classic Mechanisms of Action after all? Front Immunol. 2014;5:674. doi: 10.3389/fimmu.2014.00674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu J, Edberg JC, Redecha PB, Bansal V, Guyre PM, Coleman K, et al. A novel polymorphism of FcgammaRIIIa (CD16) alters receptor function and predisposes to autoimmune disease. J Clin Invest. 1997;100:1059–70. doi: 10.1172/JCI119616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark MR, Clarkson SB, Goldstein IM. Molecular basis for a polymorphism involving Fc receptor II on human monocytes. Trans Assoc Am Physicians. 1989;102:252–9. [PubMed] [Google Scholar]
- 12.Clark MR, Stuart SG, Kimberly RP, Ory PA, Goldstein IM. A single amino acid distinguishes the high-responder from the low-responder form of Fc receptor II on human monocytes. Eur J Immunol. 1991;21:1911–6. doi: 10.1002/eji.1830210820. [DOI] [PubMed] [Google Scholar]
- 13.Warmerdam PA, van de Winkel JG, Vlug A, Westerdaal NA, Capel PJ. A single amino acid in the second Ig-like domain of the human Fc gamma receptor II is critical for human IgG2 binding. J Immunol. 1991;147:1338–43. [PubMed] [Google Scholar]
- 14.Musolino A, Naldi N, Bortesi B, Pezzuolo D, Capelletti M, Missale G, et al. Immunoglobulin G fragment C receptor polymorphisms and clinical efficacy of trastuzumab-based therapy in patients with HER-2/neu-positive metastatic breast cancer. J Clin Oncol. 2008;26:1789–96. doi: 10.1200/JCO.2007.14.8957. [DOI] [PubMed] [Google Scholar]
- 15.Taylor RJ, Saloura V, Jain A, Goloubeva O, Wong S, Kronsberg S, et al. Ex vivo antibody-dependent cellular cytotoxicity inducibility predicts efficacy of cetuximab. Cancer Immunol Res. 2015;3:567–74. doi: 10.1158/2326-6066.CIR-14-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang W, Gordon M, Schultheis AM, Yang DY, Nagashima F, Azuma M, et al. FCGR2A and FCGR3A polymorphisms associated with clinical outcome of epidermal growth factor receptor expressing metastatic colorectal cancer patients treated with single-agent cetuximab. J Clin Oncol. 2007;25:3712–8. doi: 10.1200/JCO.2006.08.8021. [DOI] [PubMed] [Google Scholar]
- 17.Weng WK, Levy R. Two immunoglobulin G fragment C receptor polymorphisms independently predict response to rituximab in patients with follicular lymphoma. J Clin Oncol. 2003;21:3940–7. doi: 10.1200/JCO.2003.05.013. [DOI] [PubMed] [Google Scholar]
- 18.Kenkre VP, Hong F, Cerhan JR, Lewis M, Sullivan L, Williams ME, et al. Fc Gamma Receptor 3A and 2A Polymorphisms Do Not Predict Response to Rituximab in Follicular Lymphoma. Clin Cancer Res. 2016;22:821–6. doi: 10.1158/1078-0432.CCR-15-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Norton N, Olson RM, Pegram M, Tenner K, Ballman KV, Clynes R, et al. Association studies of Fcgamma receptor polymorphisms with outcome in HER2+ breast cancer patients treated with trastuzumab in NCCTG (Alliance) Trial N9831. Cancer Immunol Res. 2014;2:962–9. doi: 10.1158/2326-6066.CIR-14-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weng WK, Weng WK, Levy R. Immunoglobulin G Fc receptor polymorphisms do not correlate with response to chemotherapy or clinical course in patients with follicular lymphoma. Leuk Lymphoma. 2009;50:1494–500. doi: 10.1080/10428190903128660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Metes D, Ernst LK, Chambers WH, Sulica A, Herberman RB, Morel PA. Expression of functional CD32 molecules on human NK cells is determined by an allelic polymorphism of the FcgammaRIIC gene. Blood. 1998;91:2369–80. [PubMed] [Google Scholar]
- 22.Morel PA, Ernst LK, Metes D. Functional CD32 molecules on human NK cells. Leuk Lymphoma. 1999;35:47–56. doi: 10.3109/10428199909145704. [DOI] [PubMed] [Google Scholar]
- 23.Nagelkerke SQ, Tacke CE, Breunis WB, Geissler J, Sins JW, Appelhof B, et al. Nonallelic homologous recombination of the FCGR2/3 locus results in copy number variation and novel chimeric FCGR2 genes with aberrant functional expression. Genes Immun. 2015;16:422–9. doi: 10.1038/gene.2015.25. [DOI] [PubMed] [Google Scholar]
- 24.Ernst LK, Metes D, Herberman RB, Morel PA. Allelic polymorphisms in the FcgammaRIIC gene can influence its function on normal human natural killer cells. J Mol Med (Berl) 2002;80:248–57. doi: 10.1007/s00109-001-0294-2. [DOI] [PubMed] [Google Scholar]
- 25.Breunis WB, van Mirre E, Geissler J, Laddach N, Wolbink G, van der Schoot E, et al. Copy number variation at the FCGR locus includes FCGR3A, FCGR2C and FCGR3B but not FCGR2A and FCGR2B. Human mutation. 2009;30:E640–50. doi: 10.1002/humu.20997. [DOI] [PubMed] [Google Scholar]
- 26.Lejeune J, Piegu B, Gouilleux-Gruart V, Ohresser M, Watier H, Thibault G. FCGR2C genotyping by pyrosequencing reveals linkage disequilibrium with FCGR3A V158F and FCGR2A H131R polymorphisms in a Caucasian population. MAbs. 2012;4:784–7. doi: 10.4161/mabs.22287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Heijden J, Breunis WB, Geissler J, de Boer M, van den Berg TK, Kuijpers TW. Phenotypic variation in IgG receptors by nonclassical FCGR2C alleles. J Immunol. 2012;188:1318–24. doi: 10.4049/jimmunol.1003945. [DOI] [PubMed] [Google Scholar]
- 28.Dong C, Ptacek TS, Redden DT, Zhang K, Brown EE, Edberg JC, et al. Fcgamma receptor IIIa single-nucleotide polymorphisms and haplotypes affect human IgG binding and are associated with lupus nephritis in African Americans. Arthritis Rheumatol. 2014;66:1291–9. doi: 10.1002/art.38337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franke L, El Bannoudi H, Jansen DT, Kok K, Trynka G, Diogo D, et al. Association analysis of copy numbers of FC-gamma receptor genes for rheumatoid arthritis and other immune-mediated phenotypes. Eur J Hum Genet. 2016;24:263–70. doi: 10.1038/ejhg.2015.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mueller M, Barros P, Witherden AS, Roberts AL, Zhang Z, Schaschl H, et al. Genomic pathology of SLE-associated copy-number variation at the FCGR2C/FCGR3B/FCGR2B locus. Am J Hum Genet. 2013;92:28–40. doi: 10.1016/j.ajhg.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsang ASMW, Nagelkerke SQ, Bultink IE, Geissler J, Tanck MW, Tacke CE, et al. Fc-gamma receptor polymorphisms differentially influence susceptibility to systemic lupus erythematosus and lupus nephritis. Rheumatology (Oxford) 2016 doi: 10.1093/rheumatology/kev433. [DOI] [PubMed] [Google Scholar]
- 32.Wu J, Lin R, Huang J, Guan W, Oetting WS, Sriramarao P, et al. Functional Fcgamma receptor polymorphisms are associated with human allergy. PLoS One. 2014;9:e89196. doi: 10.1371/journal.pone.0089196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Erbe AK, Wang W, Gallenberger M, Hank JA, Sondel PM. Genotyping Single Nucleotide Polymorphisms and Copy Number Variability of the FCGRs Expressed on NK Cells. New York: Springer Science; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michel T, Hentges F, Zimmer J. Consequences of the crosstalk between monocytes/macrophages and natural killer cells. Front Immunol. 2012;3:403. doi: 10.3389/fimmu.2012.00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lode HN, Jensen C, Endres S, Pill L, Siebert N, Kietz S, et al. Immune activation and clinical responses following long-term infusion of anti-GD2 antibody ch14.18/CHO in combination with interleukin-2 in high-risk neuroblastoma patients. 2014 ASCO Annual Meeting: Journal of Clinical Oncology. [Google Scholar]
- 36.Lode HN, Valteau-Couanet D, Troschke-Meurer S, Manzitti C, Gray J, Castel V, et al. Phase II clinical trial with long-term infusion of anti-GD2 antibody ch14.18/CHO in combination with interleukin-2 (IL2) in patients with high risk neuroblastoma. 2016 ASCO Annual Meeting: Journal of Clinical Oncology. [Google Scholar]
- 37.Lode HN, Troschke-Meurer S, Valteau-Couanet D, Garaventa A, Gray J, Castel V, et al. Correlation of killer-cell Ig like receptor (KIR) haplotypes and Fcγ-receptor polymorphisms with survival of high-risk relapsed/refractory neuroblastoma patients treated by long-term infusion of anti-GD2 antibody ch14.18/CHO. 2016 ASCO Annual Meeting: Journal of Clinical Oncology. [Google Scholar]
- 38.Pruitt KD, Brown GR, Hiatt SM, Thibaud-Nissen F, Astashyn A, Ermolaeva O, et al. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014;42:D756–63. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graham JB, Graham RM. Antibodies elicited by cancer in patients. Cancer. 1955;8:409–16. doi: 10.1002/1097-0142(1955)8:2<409::aid-cncr2820080221>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 40.Chen W, Masterman KA, Basta S, Haeryfar SM, Dimopoulos N, Knowles B, et al. Cross-priming of CD8+ T cells by viral and tumor antigens is a robust phenomenon. Eur J Immunol. 2004;34:194–9. doi: 10.1002/eji.200324257. [DOI] [PubMed] [Google Scholar]
- 41.Finn OJ. Cancer immunology. N Engl J Med. 2008;358:2704–15. doi: 10.1056/NEJMra072739. [DOI] [PubMed] [Google Scholar]
- 42.Rescigno M, Avogadri F, Curigliano G. Challenges and prospects of immunotherapy as cancer treatment. Biochim Biophys Acta. 2007;1776:108–23. doi: 10.1016/j.bbcan.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 43.Smyth MJ, Godfrey DI, Trapani JA. A fresh look at tumor immunosurveillance and immunotherapy. Nat Immunol. 2001;2:293–9. doi: 10.1038/86297. [DOI] [PubMed] [Google Scholar]
- 44.Turk MJ, Wolchok JD, Guevara-Patino JA, Goldberg SM, Houghton AN. Multiple pathways to tumor immunity and concomitant autoimmunity. Immunol Rev. 2002;188:122–35. doi: 10.1034/j.1600-065x.2002.18811.x. [DOI] [PubMed] [Google Scholar]
- 45.Knutson KL, Clynes R, Shreeder B, Yeramian P, Kemp KP, Ballman K, et al. Improved Survival of HER2+ Breast Cancer Patients Treated with Trastuzumab and Chemotherapy Is Associated with Host Antibody Immunity against the HER2 Intracellular Domain. Cancer Res. 2016;76:3702–10. doi: 10.1158/0008-5472.CAN-15-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murillo O, Arina A, Tirapu I, Alfaro C, Mazzolini G, Palencia B, et al. Potentiation of therapeutic immune responses against malignancies with monoclonal antibodies. Clin Cancer Res. 2003;9:5454–64. [PubMed] [Google Scholar]
- 47.Whiteside TL, Demaria S, Rodriguez-Ruiz ME, Zarour HM, Melero I. Emerging Opportunities and Challenges in Cancer Immunotherapy. Clin Cancer Res. 2016;22:1845–55. doi: 10.1158/1078-0432.CCR-16-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.