Abstract
Background
HPV-positive oropharyngeal cancer is generally associated with excellent response to therapy, but some HPV-positive tumors progress despite aggressive therapy. This study evaluates viral oncogene expression and viral integration sites in HPV16 and HPV18-positive squamous carcinoma cell lines.
Methods
E6-E7 alternate transcripts were assessed by RT-PCR. Detection of integrated papillomavirus sequences (DIPS-PCR) and sequencing identified viral insertion sites and affected host genes. Cellular gene expression was assessed across viral integration sites.
Results
All HPV-positive cell lines expressed alternate HPVE6/E7 splicing indicative of active viral oncogenesis. HPV integration occurred within cancer-related genes TP63, DCC, JAK1, TERT, ATR, ETV6, PGR, PTPRN2, and TMEM237 in 8 HNSCC lines but UM-SCC-105 and UM-GCC-1 had only intergenic integration.
Conclusions
HPV integration into cancer-related genes occurred in 7/9 HPV-positive cell lines and of these six were from tumors that progressed. HPV integration into cancer-related genes may be a secondary carcinogenic driver in HPV-driven tumors.
Keywords: HPV, Integration, HNSCC, Cancer, Oropharynx
INTRODUCTION
High-risk human papillomaviruses (hrHPV) are frequently identified as etiologic factors in the increasing incidence of head and neck cancer, particularly hrHPV-positive oropharynx cancers1,2. In contrast, rates of HPV-negative oropharyngeal tumors, which are more often smoking and alcohol related, are declining in frequency. Among patients studied at the University of Michigan, over 80% of oropharyngeal cancers, 33% of nasopharynx cancers, 14% of larynx cancers and 10% of oral cavity cancers are positive for hrHPV3. In the oropharynx, hrHPV is generally considered to be associated with better prognosis4,5.
Clinical trial data from the University of Michigan shows that selected patients with stage 3 and 4 oropharynx cancer who could be treated with radiation fields that spare a parotid gland and the swallowing musculature had 88% three-year progression-free survival after treatment with concurrent platinum-taxol based chemotherapy and intensity modulated radiation therapy (chemo-RT)6. Similarly, a retrospective analysis of an RTOG (Radiation Therapy Oncology Group) randomized trial of tumor HPV status and survival in patients with stage III and IV oropharyngeal cancer found patients to have three year overall survival of 82.4%7. These high rates of response and outcome data have stimulated a national dialogue on de-escalating treatment intensity to reduce treatment-related morbidity in patients with HPV-positive oropharyngeal squamous cell carcinomas8,9. However, there are no indicators that distinguish those HPV-positive tumors most likely to respond from those that progress even after intensive therapeutic regimens.
There are relatively few cell lines established from HPV-positive head and neck tumors. To better understand the characteristics of HPV-positive tumors, we collected and studied HPV oncogene transcription, physical status, integration sites, and identification of the cellular genes affected by integration, in seven HPV16-positive cell lines, one HPV18-positive HNSCC cell line and one HPV16-positive cervical carcinoma cell line (UD-SCC-2, UM-SCC-47, UM-SCC-104, UM-SCC-105, UPCI:SCC090, UPCI:SCC152, UPCI:SCC154, VU-SCC-147 and UM-GCC-1). It is well established that the viral oncogenes, E6 and E7, are potent drivers of malignant behavior10,11, chromosomal instability, and that viral integration into the host genome is associated with malignant transformation, progression to high grade CIN, and invasion in cervical lesions12,13, as well as high level expression of viral oncogene transcripts14. Surveys of anogenital HPV-related tumor specimens commonly exhibit integration into either intragenic regions of the cellular genome or integration into fragile sites15–17. Viral integration is reported to occur in 65–75% of HPV16-positive head and neck tumors18,19, but until recently identification of cellular sites of integration and effects on cellular gene expression has been lacking. Ragin et al.15 examined the HPV16-positive oropharyngeal cell line UPCI:SCC090 and observed multiple viral integration sites in chromosomes 3, 6, 9q, 13q and a t(1;8)(q;?) and suggested that these occurred at common fragile sites. Wald et al.20 examined the HPV-positive oropharyngeal cancer lines UD-SCC-2, UM-SCC-47, UPCI:SCC090, and 93-VU-147T (also known as VU-SCC-147) and found that characteristic microarray profiles were observed in the HPV-positive lines that distinguished them from HPV-negative lines from the oropharynx. Akagi et al.21 analyzed genome wide analysis of HPV integration in several cervical and oropharyngeal cell lines and found significant clastogenic effects of HPV integration, including extensive host genomic amplifications, rearrangements, deletions, inversions, and chromosomal translocations which they linked to a looping HPV integrant-mediated DNA replication and recombination which were associated with gene disruption and amplification of viral oncogenes. We previously investigated UD-SCC-2, UM-SCC-47, UM-SCC-104, UPCI:SCC090, UPCI:SCC152, and UPCI:SCC154 for HPV copy number and virus-host fusion-transcripts using primarily Amplification of Papillomavirus Oncogene Transcripts (APOT)22. In that study, fusion transcripts were found in 4 of the cell lines22, but viral loads were not associated with integration status . UD-SCC-2 expressed a fusion transcript into exon 20 of DIAPH2, UM-SCC-47 expressed a fusion transcript into exon 7 of TP63, and UPCI:SCC090 and UPCI:SCC152 both expressed a fusion transcript into NAP1. In the present study we assessed HPV viral oncogene alternate transcript expression, the cellular genomic site of viral integration in eight head and neck and one cervical cancer HPV-induced human tumor cell lines, and determined the effect of viral integration on effected host genes.
MATERIALS AND METHODS
Cell lines
Seven HPV16-positive and one HPV18-positive HNSCC tumor cell lines and one cervical carcinoma cell lines were studied. Four were developed in our lab: UM-SCC-4723,24, UM-SCC-10423 and UM-SCC-105 all HNSCC and UM-GCC-1 a cervical glassy cell carcinoma variant of SCC. UD-SCC-225, was obtained from H. Bier and T. Hoffmann, University of Düsseldorf25, VU-SCC-147 (previously called 93-VU-147T)26, from R. Brakenhoff, Vrije Universiteit, Amsterdam; and UPCI:SCC090, UPCI:SCC152, and UPCI:SCC154, from S. Gollin, University of Pittsburgh Cancer Institute27–29. UM-SCC-47 was established in 1985 from a surgical resection of a previously untreated T3N1M0 carcinoma of the lateral tongue in a 53-year-old male smoker. The patient was referred for radiation closer to his home but died within a year of diagnosis. UM-SCC-104 was from a 56-year-old male heavy smoker with recurrent T4N2bM0 SCC of the floor of mouth after prior treatment at an outside hospital. He was treated at U of M with surgical resection in 2009 and had post-operative radiation, but the tumor persisted and he succumbed in 2010. UM-SCC-105 was derived from a biopsy of a T4N0M0 glottic mass in a 51 year never smoker male patient who had been treated symptomatically for hoarseness for 18 months before referral to an otolaryngologist who discovered the large laryngeal tumor. The patient was treated with radiation therapy and remains free of disease 5 years later. UM-GCC-1, an HPV16-positive cell line was derived from a glassy cell cervical carcinoma from a 27 year old female patient, who was treated with surgical resection and remains healthy and free of disease 30 years later. UD-SCC-2 was derived from surgical resection from a 58-year-old male smoker with T1N3 carcinoma of the pyriform sinus who died from pulmonary metastases 1 year after diagnosis30. UPCI:SCC090 and UPCI:SCC152 were established from a 46-year-old male smoker with an oropharyngeal T2N1M0 SCCHN arising in the base of tongue. The histology was moderately to poorly differentiated invasive squamous cell carcinoma with basaloid features. UPCI:SCC090 was from the surgical resection of the primary; UPCI:SCC152 was established from a recurrent tumor in the same patient I year later. The patient died of his disease 4 years after diagnosis. VU-SCC-147 was derived from a 57-year-old male smoker with a T4N2 carcinoma of the floor of mouth/lower alveolar ridge . He was treated with surgery and postoperative radiotherapy, but developed an untreatable second primary tumor after 6.5 years and was lost to follow-up after 7 years. UPCI:SCC154 was derived from a 52-year-old male smoker with T4N2 previously untreated squamous cell carcinoma of the tongue The patient was alive at most recent follow up, 10 years and 2 months after surgery
All cell lines established at the University of Michigan were from donors who gave written informed consent to use the resected tissue from their tumors for laboratory study, including cell line development. The IRBMED institutional review board approved the studies. The cell lines from other institutions were obtained directly from the originators in 2010. Primary tumor tissue from which the cell lines were derived is unavailable, with the exception of UM-SCC-104, UM-SCC-105 and UM-GCC-1. Like the cell lines, these patients’ tumor tissue was positive for HPV16, HPV18 and HPV16 respectively. All lines were genotyped in the University of Michigan Genomics Core using ProfilerPlus, which interrogates 10 tetranucleotide short tandem repeats (STR), and were confirmed to have unique genotypes. UPCI:SCC090 and UPCI:SCC152 share the same genotype, as they are derived from separate tumors in the same patient. All lines were tested upon receipt from the originators and repeat confirmatory tests were performed immediately prior to the integration experiments carried out between 2012 and 2013. Since there was only one of the HPV-positive HNSCC cell line was obtained from a known survivor, UM-GCC-1, an HPV16-positive cervical carcinoma cell line, and UM-SCC-105 an HPV18 positive laryngeal carcinoma were also tested for viral integration site. Genomic DNA was extracted from cells using the DNeasy Spin Column kit (Qiagen). RNA was isolated from cells using the RNeasy Mini Kit (Qiagen), followed by on-column DNase treatment.
Human Papillomavirus detection
All cell lines were grown on glass slides and examined for expression of p16INK4A (inhibitor of cyclin-dependent kinase 4) using the CINtec (Roche/Ventana, Tucson, Arizona,) assay per supplier protocol. HPV in situ hybridization (ISH) was performed using the INFORM HPVIII assay (Ventana, Tucson, Arizona,) (detects 12 hrHPV types: HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58 and 66) per supplier protocol. All cell lines were tested for the presence and type of HPV using the HPV PCR-MassArray assay3,23,31–33. As part of an earlier study to characterize UM-SCC-47, single color fluorescence in situ hybridization (FISH) was performed on UM-SCC-47 using a fluorescein labeled HPV16 bacterial artificial chromosome (BAC). Metaphase spreads were harvested from UM-SCC-47 cells in their 34th to 36th passages and treated with 0.075M potassium chloride hypotonic solution. Spectral Karyotyping of UM-SCC-47 was performed at the Van Andel Institute (Grand Rapids, MI).
Human Papillomavirus E6 and E7 transcript analysis
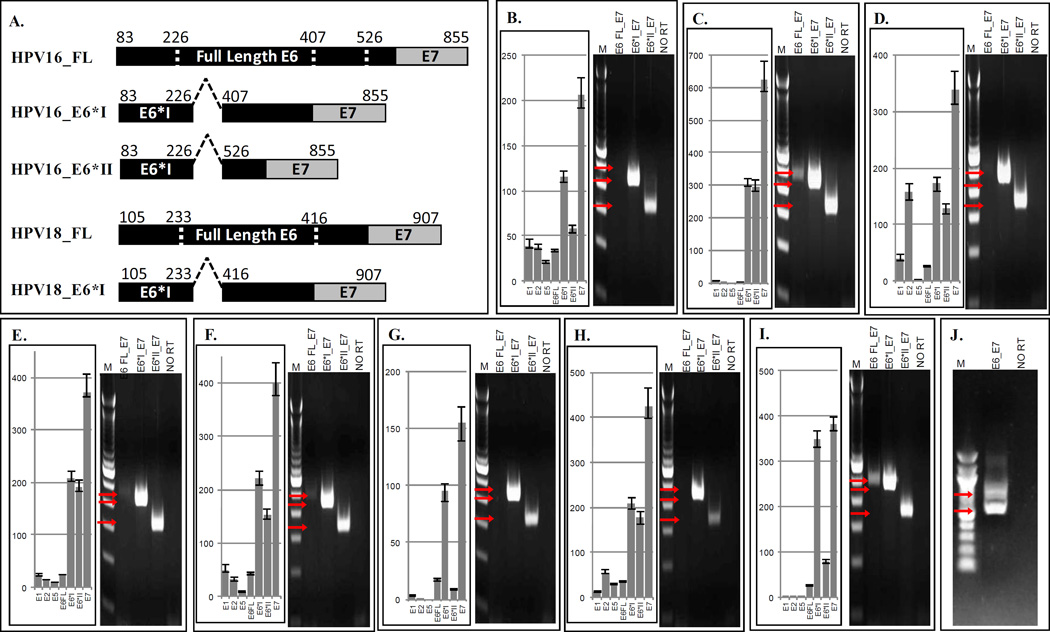
Two complementary methods were used to evaluate and quantify the relative expression of the viral transcripts. The E6*I and E6*II alternate transcripts result from a single donor site at nucleotide (nt) 226 of the viral genome and two acceptor sites at nt 407 (E6*I) and at nt 526 (E6*II). To examine the expression of HPV E6 and E7 transcripts, primer sets were designed that specifically and discretely amplify the intact, non-spliced, full-length E6-E7 transcript, the spliced E6*I-E7 transcript, and the spliced E6*II-E7 transcript, as illustrated in Figure 2A. The full-length E6-E7 transcript was generated using a forward primer located within the region that is eliminated by splicing, while the transcripts for the alternate splice forms were generated using unique forward primers that span the respective splice junctions. (Primer sets are listed in Supplemental Table 1). Primers for GAPDH were used as a negative control in the no reverse transcriptase lane to confirm the absence of contaminating genomic DNA.
Figure 2.
HPV Oncogene Transcript- Specific Quantitative RT-PCR and E6–E7 RT-PCR in HPV-Positive HNSCC Cell Lines. Bar graphs represent TaqMan quantitative PCR relative expression, and electrophoretic gel images represent E6-E7 RT-PCR. Panel A. Depiction of HPV16 and HPV18 splicing. Primers within the splice region or across splice junctions allow for exclusive amplification of full length E6 or alternate E6 transcripts. Panel B. UD-SCC-2, Panel C. UM-SCC-47, Panel D. UM-SCC-104, Panel E. UPCI:SCC90, Panel F. UPCI:SCC152, Panel G. UPCI:SCC154, Panel H. VU-SCC-147, Panel I. UM-GCC-1, Panel J. UM-SCC-105. Arrows indicate sizes of expected amplicon bands: HPV16 E6 FullLength_E7=499bp, HPV16 E6*I_E7= 454bp, and HPV16 E6*II_E7= 338bp, HPV18 E6_E7 FullLength=723bp, HPV18 E6*I_E7= 540bp, NO RT=no reverse transcriptase negative control.
Quantitative RT-PCR was similarly performed using TaqMan assays designed to exclusively amplify each HPV early gene transcript: E1, E2, E5, non-spliced full length E6, spliced E6*I, spliced E6*II and E7 (Primers listed in Supplemental Table 2). A prepared GAPDH endogenous control primer/probe assay was used to quantify relative viral gene expression.
Detection of Integrated Papillomavirus Sequences-Polymerase Chain Reaction
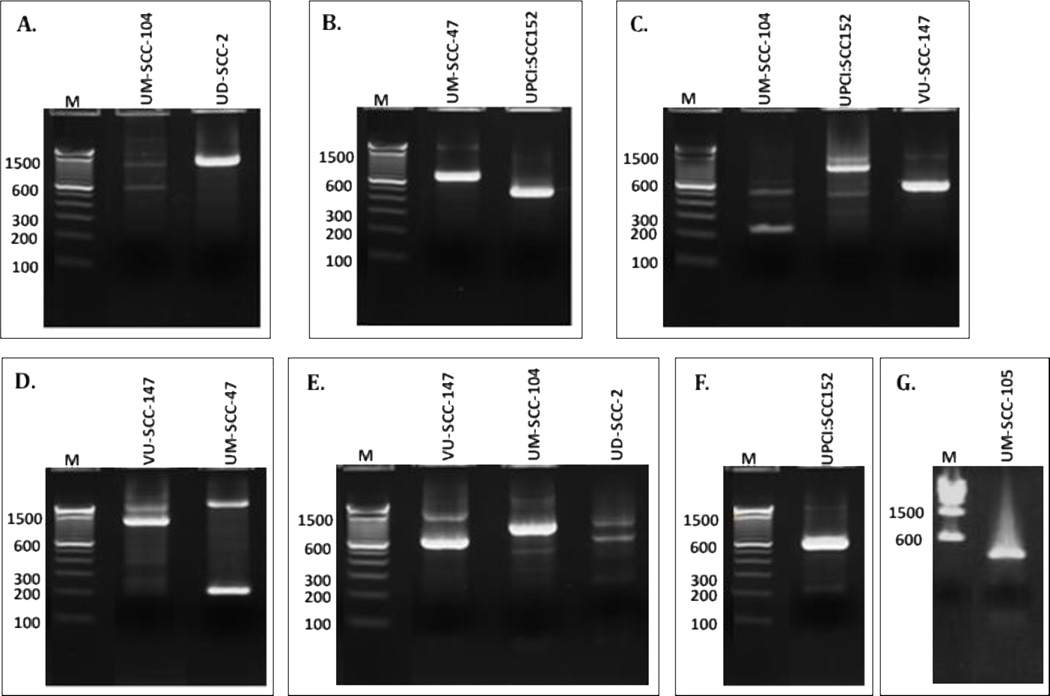
Integration analysis was performed using a modified method (DIPS-PCR) based on previously published work34,35. Briefly, genomic DNA was isolated from each cell line, and digested with the restriction enzyme, Taqα1, which cuts the primary HPV16 viral genome only once at position 505 within E6 (additional Taqα1 restriction sites have been described in HPV16 variants at positions 311 and 2608) and cuts the cellular genomic DNA at approximately 1.5 million sites. After ligating a double-strand adapter oligo (5’-CGCAACGTGTAAGTCTG-NH2-3’ annealed to 5’-GGGCCATCAGTCAGCAGTCGTAGCCGGATCCAGACTTACACGTTG-3’) to the overhanging ends of each fragment, linear PCR amplification with 11 viral-specific primers was followed by a second logarithmic PCR using 11 nested viral primers and a reverse adapter-specific primer (Supplemental Table 3). Thermocycling conditions used for both rounds of PCR included 3 minute extension cycles that limited amplification of large (>3kb), episome-only fragments. PCR products were separated by agarose gel electrophoresis. To search for a previously reported HPV insertion into 9q3115 that was not detected by DIPS-PCR in UPCI:SCC090, we used primers from multiple regions of HPV16 and within 9q31 for direct PCR using DNA from UPCI:SCC090 and the second cell line from the same patient, UPCI:SCC152 (Primers listed in Supplemental Table 4). PCR products were separated by gel electrophoresis; bands were purified and sequenced with the appropriate primer sets.
Sequence analysis of cellular genes with integrated virus
Fragments generated exclusively from non-integrated virus were excluded based on amplicon sizes predicted for episome-only bands, which were based on viral-specific primer locations in relation to the Taqα1 restriction site in the viral genome. Viral-cellular amplicons were identified, excised from the gels, purified, and sequenced. Viral integrations into known genes were verified by direct PCR and sequencing of the otherwise unmodified cell line genomic DNA, using primers specific to each viral and cellular region.
Integration site transcript analysis
Cell line RNA was evaluated for viral-cellular fusion transcripts and cellular gene transcripts affected by confirmed viral integrations. RT-PCR assays were used that amplified virus-cellular fusion transcripts from HPV open reading frames into cellular gene exons, cellular gene exon-exon transcripts across the integration site, and distant cellular gene transcripts. All amplified transcripts were separated by agarose gel electrophoresis sequenced for confirmation.
Protein isolation and western blot analysis
The cells were lysed with 1% Nonidet-P40 lysis buffer containing protease and phosphotase inhibitors (Peirce, Rockford, IL). The supernatant was collected and the protein content was measured using the BCA Protein Assay Kit (Pierce, Rockford, IL). Equal amounts of protein were electrophoresed on NuPAGE Bis-Tris gels and transferred to Immoblin-P (Millipore Corporation, Billerica, MA). Membranes were blocked in 5% milk in TBST (Tris Buffered saline with 0.1% Tween). The membranes were incubated overnight with primary TP63 C-Terminus (Boster Biological Technology, Pleasanton, CA) and p63 delta N (Biolegend, San Diego, CA) rabbit antibodies at 4°C for two hours at room temperature, followed by incubation with secondary anti-antibody horseradish peroxidase conjugate (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) and detected by chemiluminescence (Amersham, Little Chalfont, UK).
RESULTS
All nine cell lines were verified to contain HPV16 or HPV18 by PCR-MassArray and all cell lines strongly expressed viral oncogene transcripts.
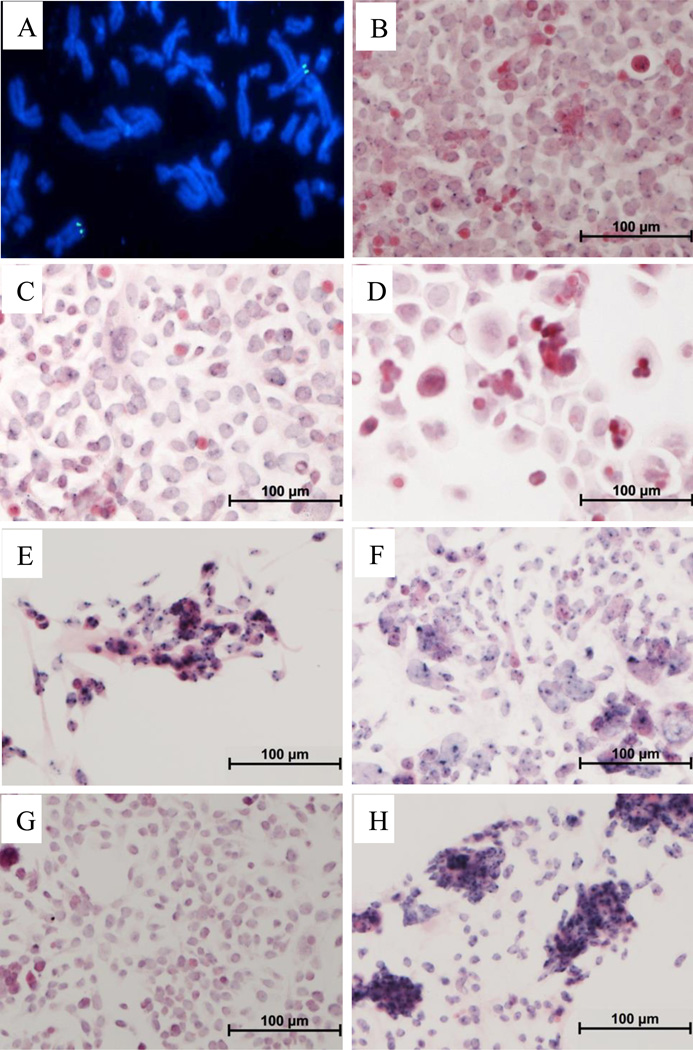
Metaphase chromosome spreads of UM-SCC-47 were examined by HPV16 FISH, which revealed a strong signal, likely representing multiple copies of the viral genome, integrated into the distal long arm of a single autosomal chromosome (Figure 1A). Spectral karyotype analysis of UM-SCC-47 shows rearranged chromosome 3 (Figure S1). All of the cell lines were examined for nuclear viral DNA by ISH (Figure 1B-1H) with deep blue hybridization signals indicating the presence of HPV DNA. UM-SCC-104 (Figure 1D) and UPCI:SCC154 (Figure 1G) have very faint hybridization signals, consistent with the low viral copy number in these cell lines reported previously22. The Ventana ISH assay was no longer available for testing UM-SCC-105 or UM-GCC-1.
Figure 1.
HPV In Situ Hybridization in HNSCC cell lines. Panel A: UM-SCC-47 fluorescence in situ hybridization for HPV16, Panels B-H show hrHPV In Situ Hybridization indicated by dark blue signals. Panel B. UD-SCC-2 (43 HPV copies/cell), Panel C. UM-SCC-47 (5 HPV copies/cell), Panel D. UM-SCC-104 (1.6 HPV copies/cell), Panel E. UPCI:SCC90 (284 HPV copies/cell), Panel F. UPCI:SCC152 (150 HPV copies/cell), Panel G. UPCI:SCC154 (1.3 HPV copies/cell), Panel H. VU-SCC-147 (218 HPV copies/cell).
Viral oncogene expression
All HPV16-positive[c1] HNSCC cell lines strongly express viral oncogene transcripts, particularly the alternate E6-E7 transcripts expressed in hrHPV transformed tumor cells (Figure 2). The HPV16 E6 gene contains two introns that can be spliced out, generating alternate E6*I-E7 and E6*II-E7 transcripts that have been linked to increased expression of E7 at the expense of full length E636. The E6*I and E6*II alternate transcripts result from a single donor site at nucleotide (nt) 226 of the viral genome and two acceptor sites at nt 407 (E6*I) and at nt 526 (E6*II). The HPV18 E6 alternate transcript results from a donor site at nucleotide (nt) 233 and acceptor site at nt 416 (E6*I) of the viral genome. As shown in Figure 2B–I, all of the HPV16-positive cell lines strongly express the viral oncogene transcripts and all express the alternate E6-E7 transcripts, primarily E6*I, and to a lesser extent E6*II, at the expense of full length E6 (qRT-PCR) or full length E6-E7 (RT-PCR). The HPV 18-positive cell line UM-SCC-105 also shows the alternate E6*I transcript as a smaller band when E6 and E7 is amplified end to end (Figure 2J). These findings are consistent with the viral oncogenes particularly the alternate E6* transcripts and E7 as drivers of tumor development. In all of the cell lines, the expression of E1 and E2 is severely reduced compared to the E6-E7 transcripts, as measured by TaqMan qRT-PCR relative to GAPDH. Only UM-SCC-104 showed moderate levels of E2, but still expressed very low E1, E5 and full length E6 expression. These results support an important role for alternate E6 transcripts in viral oncogenesis together with disruption of the viral E1-E2 region and loss of full length E6 expression.
Identification of viral integration sites
Separated amplicon DIPS-PCR bands are shown in the representative gels in Figure 3. A total of 104 hybrid viral-cellular amplicons were isolated and sequenced, ranging from 5 to 16 amplicons for each cell line. Viral-host DNA fusions were identified by sequence and BLAST analysis. All sequence reads were studied. Reads mapped to viral-only sequence, viral-cellular hybrids as described below, or were unmapped due to poor sequence resolution. Diagrammatic representations of the viral insertion sites determined by this method are shown in Figure 4, and Table 1 summarizes the integration results for all nine cell lines, indicating the HPV integration site, chromosome locus, cellular gene, and the cellular gene region of integration.
Figure 3.
Representative Cell Line DIPS PCR gels. A. HPV16-E1a/Adapter primers, B. HPV16-E1c/Adapter primers, C. HPV16-E5a/Adapter primers, D. HPV16-L2a/Adapter primers, E. HPV16-E2b/Adapter primers, F. HPV16-E6a/Adapter primers, G. HPV18-L2f/Adapter primers. M=100 bp marker ladder.
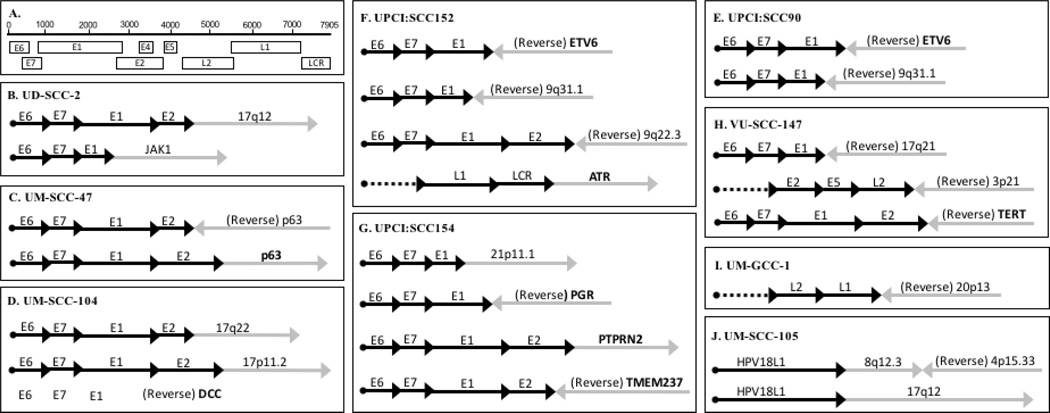
Figure 4.
Schematic representation of integration events in HPV-positive HNSCC cell lines. Panel A. Linear organization of the HPV genome, Panel B. UD-SCC-2, Panel C. UM-SCC-47, Panel D. UM-SCC-104, Panel E. UPCI:SCC90, Panel F. UPCI:SCC152, Panel G. UPCI:SCC154, Panel H. VU-SCC-147, Panel I. UM-GCC-1, Panel J. UM-SCC-105.
Table 1.
Summary of integration events in HPV-positive cell lines.
| Cell Line | Patient Information | HPV Site |
Cellular Integration Site | ||
|---|---|---|---|---|---|
| Locus | Gene | Region | |||
| UD-SCC-2 | 58-year-old male smoker with T1N3 SCC of the pyriform sinus, died by pulmonary metastases 1 year after diagnosis |
E2 | 17q12 | Intergenic | |
| E1 | 1p32.3 | JAK1 | Intron 14 | ||
| UM-SCC-47 | 53-year-old male smoker with T3N1M0 lateral tongue SCC, died from disease within a year of diagnosis |
E2 | 3q28 | TP63 | Intron 10 |
| E2 | 3q28 | TP63 | Exon 14 | ||
| UM-SCC-104 | 56-year-old male smoker with recurrent floor of mouth T4N2bM0 SCC, died from disease within a year after treatment |
E2 | 17q22 | Intergenic | |
| E2 | 17p11.2 | Intergenic | |||
| E1 | 18q21.3 | DCC | Intron 1 | ||
| UPCI:SCC90 | 46-year-old male smoker with recurrent base of tongue T2N1M0 SCC, died from disease 4 years after diagnosis* |
E1 | 9q31.1 | Intergenic | |
| E1 | 12p13 | ETV6 | Intron 1 | ||
| UPCI:SCC152 | 47-year-old male smoker (donor of UPCI:SCC90) with recurrent hypopharynx SCC, died from disease 4 years after diagnosis* |
E2 | 9q22.33 | Intergenic | |
| E1 | 9q31.1 | Intergenic | |||
| LCR | 3q23 | ATR | Intron 36 | ||
| E1 | 12p13 | ETV6 | Intron 1 | ||
| UPCI:SCC154 | 52-year-old male smoker with base of tongue T4N2 SCC, alive 10 years after surgery |
E1 | 21p11.1 | Intergenic | |
| E2 | 2q33.2 | TMEM237 | Exon 14 | ||
| E2 | 7q36 | PTPRN2 | Intron 3 | ||
| E1 | 11q22–23 | PGR | Intron 3 | ||
| VU-SCC-147 | 57-year-old male smoker with floor of mouth T4N2 SCC, developed untreatable second primary after 6.5 years |
E1 | 17q21 | Intergenic | |
| L2 | 3p21 | Intergenic | |||
| E2 | 5p15.33 | TERT | Promoter | ||
| UM-GCC-1 | 26-year old female, stage IB cervical carcinoma, alive without disease 30 years after diagnosis |
L1 | 20p13 | Intergenic | |
| UM-SCC-105 | 51-year old male never smoker with T4N0M0 larynx carcinoma, alive without disease 5 years after treatment |
L1 | 8q12.3/4p15.33 | Intergenic | |
| L1 | 17q12 | Intergenic | |||
UPCI:SCC90 and UPCI:SCC152 are from the same patient
A single integration event was identified in UM-GCC-1. The L1 region of HPV16 was joined to an intergenic region of chromosome 20p13 (Fig 4I). Two integration events were found in UM-SCC-105; both involved the HPV18 L1 region. One involved a complex rearrangement with the viral read going into 8q12.3/4p15.33. Both host regions were in non-coding regions of the cellular genome. The other integration event HPV18 L1 read into 17q12, also a non-coding region of chromosome 17 (Table 1, Figure 4J). Two fusion events were detected in UD-SCC-2. The first was integration of HPV from E2 into an intergenic region of chromosome 17q12, and a second fusing HPV E1 to intron 14 of JAK1. JAK1 is a large membrane protein tyrosine kinase involved in the proper function of the interferon receptor complexes and signaling through the Signal Transducers and Activators of Transcription (STAT1–4) pathway. How integration of HPV into JAK1 might be advantageous to an HPV-positive tumor is uncertain, but could relate to loss of interferon signaling within a virally infected/transformed cell. This hypothesis is supported by the association of STAT1 with impaired induction of INFβ37. UM-SCC-47 exhibited two HPV integration events with breakpoints within E2 each extending into TP63, one into TP63 reverse intron 10 and the second into TP63 exon 14. As TP63 is located at chromosome 3q28, this is finding is consistent with the FISH result (Figure 1A) showing a strong signal on the distal arm of an aberrant chromosome that is likely a t(3;7) chromosome rearrangement identified by SKY (Supplemental Figure 1). Integration into TP63 has been observed in cervical cancers and Schmitz et al38 reported a region of homology between the HPV16 E1 region and a segment of chromosome 3q28 within TP63 that may facilitate this integration. We detected a fusion transcript between HPV16 E2 and TP63 (Fig 5B). To investigate this further we investigated the expression of TP63 protein using western blotting. A subset of HPV positive cell lines and UM-SCC-38, an HPV-negative oropharynx cancer cell line, were tested (Figure S2). UD-SCC-2, UM-SCC-104, UPCI:SCC-154, VU-SCC-147 and UM-SCC-38 express TP63-alpha as detected by the c-terminus antibody, but UM-SCC-47 expresses only a truncated version of this TP63α isoform. Further, UD-SCC-2, UM-SCC-104, and UM-SCC-38 strongly express the alpha isoform of ΔNp63, but UM-SCC-47, as well as VU-SCC-147, and UPCI:SCC-154 fail to express ΔNp63. This suggests that the HPV integration in UM-SCC-47 affects the size of the tumor suppressor form of TP63. It is accompanied by loss of the oncogenic ΔNp63 isoform which is also observed in two other HPV-positive tumor cell lines that did not exhibit viral integration into TP63. Clearly this is an area for further investigation. TP63 is a homolog to TP53 and TP73, and is a tumor suppressor gene, functioning as both a sequence-specific DNA binding transcriptional repressor and activator. The p63 protein product of TP63 is involved in differentiation and cell-cycle regulation, as well as TGFβ and WNT signaling39. In contrast, ΔNp63 lacks the transactivation domain and acts as a dominant negative inhibitor of the transactivating p63 isoforms40. Thus, HPV integration may cause reduced p63 signaling and increased WNT function leading to increased proliferative signaling. UM-SCC-104 exhibited multiple integration events including two HPV E2 integration events into intergenic regions of 17q22 and 17p11.2 (Fig 4D). Additionally, in UM-SCC-104, HPV E1 integrated into reverse DCC intron 1. DCC is a receptor for netrin-1, and when not bound, functions as a tumor suppressor in the caspase-9 dependent apoptosis pathway. DCC is located in a region of chromosome 18q that is frequently lost in squamous cell carcinomas41,42. In UM-SCC-104 there was no DCC transcript detectable (data not shown), suggesting that one copy may have been disrupted by HPV integration and the other lost or silenced by methylation42. UPCI:SCC090 and UPCI:SCC152 (tumors from the same patient) are interesting in that both share the identical integration from HPV E1 into intron 1 of ETV6, which is consistent with this being an early event before the primary tumor and recurrent populations diverged. ETV6 is a transcription factor involved primarily in development and hematopoiesis. Gene fusions involving ETV6 have been discovered in multiple hematological malignancies, including ETV6-PDGFRB, ETV6-NTRK3, ETV6-ABL1, ETV6-ABL2, ETV6-JAK2, and ETV6-EVI1 fusions43. There is also evidence suggesting mutational inactivation of ETV6 in prostate carcinoma44. Interestingly, an ETV6 fusion oncogene was recently identified in a subset of salivary gland tumors45. These tumors, designated mammary analogue secretory carcinomas, have been shown to contain a clinically significant ETV6-NTRK3 gene fusion that is also present in secretory carcinoma of the breast45,46. It is possible that in UPCI:SCC090 and UPCI:SCC152 cell lines, viral integration into ETV6 causes alteration of gene expression. Previous studies of UPCI:SCC090 had reported a complex rearrangement of HPV into a rearranged chromosome 9 with fusions between HPV16 and 9q31.1 and 9p2415,21,22. Because we did not find this by integration by DIPS-PCR using Taqα1, we confirmed its presence by targeted PCR. Sequence analysis revealed HPV E1 integrated into the same sequence as reported by Ragin et al.15, which was confirmed by NCBI BLAST analysis to map to 9q31.1. DIPS-PCR in UPCI:SCC152, in addition to the HPV E1- ETV6 integration, identified a viral rearrangement resulting in fusion of HPV E2 into an intergenic region of reverse chromosome 9q22.33 which is similar to the integration reported by Olthof et al.22 and Akagi et al.21. In addition, DIPS-PCR revealed integration of HPV LCR into ATR intron 36 on chromosome 3q23. ATR codes for a cell-cycle checkpoint protein kinase required for arrest and repair in response to DNA damage. Disruption of this gene by integration of HPV at the viral non-coding LCR region could result in uncontrolled cell-cycle progression and uninhibited tumor cell replication and growth. UPCI:SCC152 was also evaluated for the 9q31.1 integration that was previously reported, and was detected by direct PCR exactly as in UPCI:SCC090. The multiple viral integrations into chromosome 9 in UPCI:SCC090 and UPCI:SCC152 appear to be complex, involving both the 9p and 9q arms. Thus far, our indications are that these chromosome 9 integrations involve exclusively intergenic regions of the chromosome. UPCI:SCC154 exhibited four integration events detected by DIPS-PCR, including one from HPV E1 into an intergenic region of chromosome 21p11.1, HPV E1 into reverse PGR, and two involving HPV E2; one into PTPRN2 intron 3 and the second into reverse TMEM237 exon 14. PGR is a steroid receptor for progesterone, and participates in estrogen and glucocorticoid receptor pathways as well as signaling by binding to transcription factors such as NF-κB, AP-1 or STAT. Overexpression of PGR has been associated with disease-related mortality and recurrence in breast and gastric cancers47,48. PTPRN2 (protein tyrosine phosphatase, receptor type, N2) belongs to the transmembrane protein tyrosine phosphatase family, and is reported to be a tumor suppressor involved in the regulation of the cell cycle, as well as growth, differentiation, and oncogenic transformation. It has been demonstrated that PTPRN2 is hypermethylated and subsequently inactivated in squamous cell lung cancer49. This suggests that a similar inactivating event such as gene disruption by integrated HPV, alone or in combination with methylation, may also result in functional loss of PTPRN2 tumor suppressor activity and contribute to tumor cell malignancy and resistance to therapy. TMEM237 is a tetraspanin membrane protein that is thought to participate in the WNT signaling pathway. While the specific interactions of this protein are not yet entirely understood, it is feasible that disruption of this gene by HPV integration would affect the WNT signaling pathway, possibly resulting in dysregulated differentiation and proliferation of the tumor cells. Three integration sites were identified in VU-SCC-147, one from HPV E1 into reverse chromosome 17q21, a second from HPV L2 into reverse chromosome 3p21, and a third from HPV E2 into reverse TERT (telomerase reverse transcriptase) in the promoter region. Disruption in the promoter region of TERT leading to increased telomerase reverse transcriptase expression could provide a growth advantage to tumor cells. In a study that evaluated the frequency of TERT promoter mutations in 60 tumor types, squamous cells carcinomas of the head and neck were among the highest, with 17% of tumors having mutations in the promoter region of the gene50. These results show that in every cell line, viral integration into one or more cancer related genes was identified. Table 1 summarizes the integration results for all seven cell lines, indicating the chromosome locus, known genes, and the regions of integration into the cellular gene.
Figure 5.
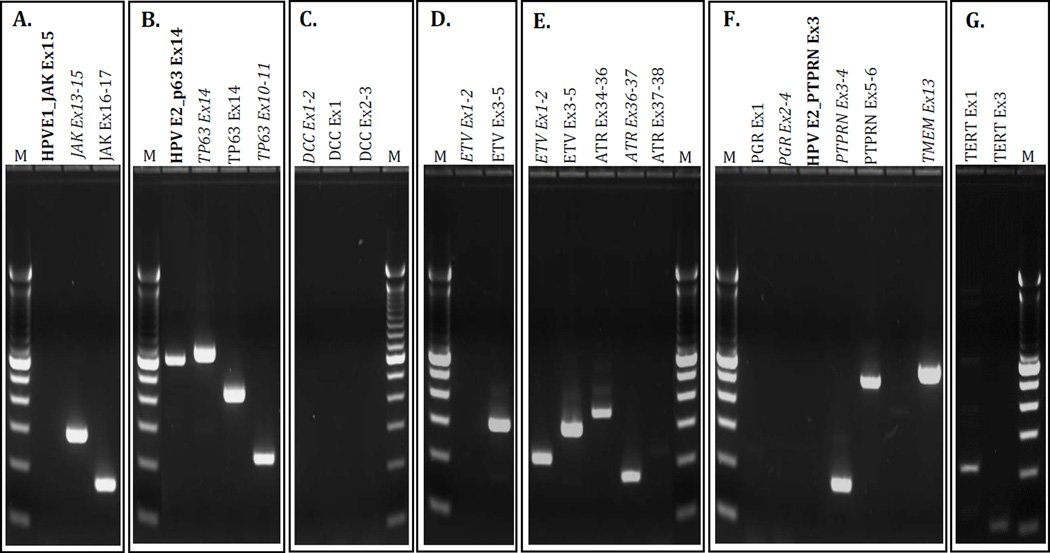
Gel Electrophoresis of Cell Line Integration Transcript Analysis. Bold text= Viral/cellular fusion transcript, Italicized text=Transcript spans integration site. Panel A. UD-SCC-2, Panel B. UM-SCC-47, Panel C. UM-SCC-104, Panel D. UPCI:SCC090, Panel E. UPCI:SCC152, Panel F. UPCI:SCC154, Panel G. VU-SCC-147. M= 100bp ladder.
Integration site transcript analysis
Based on the DIPS-PCR integration results, RT-PCR assays were designed to assess virus-cellular fusion transcripts from HPV ORFs into cellular gene exons, cellular gene exon-exon transcripts across the integration site, and distant cellular gene transcripts. HPV fusion transcript and cellular gene transcript RT-PCR amplicon products are shown in Figure 5.
Transcript analysis in the UD-SCC-2 cell line revealed that there was no HPV-E1/JAK exon 15 fusion transcript generated, but there were transcripts generated across the splice insertion site, including JAK exons13–15, as well as downstream from the integration site for JAK exons 16–17. In the UM-SCC-47 cell line, transcripts across the integration sites in TP63 exon 14 and intron 10 were generated, as well as a transcript for TP63 exon 14 downstream of the integration site. There was also an HPV-E2/TP63 exon 14 fusion transcript generated for which the sequence was correct but the splicing was out of frame. In the UM-SCC-104 cell line, DCC transcripts within exon 1, across exons 1 and 2 that spanned the HPV integration site in intron 1, and across exons 2 and 3 were interrogated, but no DCC transcripts were detected. This suggests that one copy may have been disrupted by HPV integration and the other lost or silenced by methylation42.
In both UPCI:SCC090 and UPCI:SCC152/3/2017 3:50PM2 cell lines, ETV6 transcripts were found outside of the intron 1 integration site, across exons 3 and 5. Interestingly, the ETV6 transcript across exons 1 and 2, spanning the integration site in intron 1 was produced in UPCI:SCC152, but not in UPCI:SCC090. The transcripts that were generated in UPCI:SCC090 and UPCI:SCC152 were all correct and in-frame. Evaluation of the second integration event in UPCI:SCC152 revealed the correct, in-frame, ATR transcript present upstream of the integration site in intron 36 (across exons 34 to 36). However, the transcript across exons 36 and 37, spanning the integration site, was generated but was not spliced in-frame. Furthermore, the ATR transcript across exons 37 and 38, downstream from the intron 36 integration site, was not generated at all. In the UPCI:SCC154 cell line, neither the PGR transcript across exons 2 to 4, spanning the integration site in intron 3, nor the exon 1 transcript (outside of the integration region) was generated. There was no HPV/PTPRN2 fusion transcript produced, but the PTPRN2 transcript across exons 3 and 4, spanning the integration junction, was produced, as was the PTPRN2 transcript across exons 5 and 6, located downstream of the viral integration site. Both PTPRN2 exon-exon transcripts were in frame. Similarly, there was no HPV/TMEM237 fusion transcript, but the TMEM237 transcript generated within exon 13 that spanned the integration site was the correct, in-frame sequence. In VU-SCC-147, the correct, in-frame, TERT exon 1 and exon 3 transcripts were produced.
Discussion
The incidence of HPV-positive oropharyngeal cancer is increasing2,33,51. Unlike cervical cancers that are detected early by Pap smear screening programs and often cured by colposcopy, there is no method for early detection of HPV-related head and neck cancer, and most such tumors present in an advanced state. The incidence of invasive cervical cancer is declining in western countries secondary to early detection and intervention. In contrast, largely due to high-risk HPV, the incidence of oral, oropharyngeal, and laryngeal cancers is increasing and the incidence of oropharynx cancers exceeded that of cervix cancer in 201352.
HPV-related oropharyngeal cancers are significantly more responsive to current therapeutic regimens than are HPV-negative cancers arising at the same anatomic sites5,53–56, prompting interest in reducing the intensity of treatment for this disease9. However, even with rigorous therapeutic approaches combining concurrent chemotherapy with radiation, 20–30 percent of HPV-positive cancers progress and become unresponsive to further treatment efforts33,54,57. Thus, it is important to understand why some tumors respond and others progress.
Only a small number of HPV-positive head and neck cancer cell lines have been developed. Most of the HPV-positive HNSCC cell lines studied thus far are derived from tumors that failed to respond to therapy, and may represent a more aggressive subset of tumors with features consistent with tumor progression. All of the HPV16-positive head and neck cancer cell lines express p16INK4a, and exhibit HPV E6-E7 viral oncogene expression, with dominant expression of the E6-E7 alternate transcripts. UM-SCC-105 which contains HPV18, also strongly expresses p16ink4a and expresses HPV18 E6 and E7 transcripts. In addition, all exhibit viral integration into the host cellular genome. As shown in this study, the integration is often complex, with rearrangements and multiple cellular sites of integration involving different segments of the viral genome. In UM-SCC-105 we found HPV18 integration of L1 into a complex t(8;4) translocation. Similarly, HPV16 integration into TP63 on chromosome 3 was also located near the site of a t(3;7) translocation. Similar clastogenic events were also reported for complex rearrangements at sites of HPV integration in head and neck tumors as reported by Seiwert et al58. A somewhat surprising finding in our study was that in 8 of the cell lines, the virus had integrated into cellular genes involved in cancer-related pathways. These findings suggest that assessment of cellular sites affected by viral integration in HNSCC may provide a second mechanism of oncogenesis through cellular gene disruption. Such a mechanism has been reported for oncogenesis by low-risk HPV types59 which lack the transforming ability of the high-risk E6 and E7 genes60. It should be noted that of the HNSCC cell lines all but UM-SCC-105 were obtained from patients with a history of heavy smoking; we found no evidence of smoking by the donor of UM-GCC-1, but it is unknown if a smoking history was elicited at the time of diagnosis. Contribution of additional genomic alterations resulting from the added carcinogenic effects of tobacco use is not unlikely33.
High-risk HPV integration has been widely examined in uterine cervix samples, and is strongly associated with high grade cervical intraepithelial neoplasia (CIN) and cancer development61. HPV E2, a transcriptional repressor of E6 and E7, is frequently reported to be disrupted upon integration, resulting in prolific expression of E6 and E733, 34. In cervical cancer studies, as well as a small number of studies on HNSCC, viral integration has been found primarily in intragenic sites (∼90% of the genome is intragenic), and in chromosome fragile sites15,16, although integration into cellular genes has also been reported in a minority of cases35,38,62.
In this study, we detected integration sites that differed from other investigators studying the same cell lines15,21,22. Studies using DIPS-PCR may detect different sites of integration depending on the restriction enzymes used for DNA digestion, the amplification primers used in the PCR steps, the thermocycling conditions, and amplicon bands selected for sequence analysis. The DNA digest is typically performed with Taqα1, which has a single restriction site within the HPV genome, or Sau3AI, with 10 restriction sites in the HPV genome. Both enzymes cut at numerous sites in the host cellular genome, but since the sites occur at different locations in the genome, the enzyme used will determine the cellular regions amplified in the assay. In fact, Olthof et al.22 used Sau3AI whereas in the current study we used Taqα1 and found different integration sites in the same cell lines. Subsequent PCR steps include viral-specific primers intended to amplify from the virus into the adjacent cellular sequence. The number and location of these primers direct the generation of viral-cellular amplicon products; when few primers are used, or the primers are exclusive to the E2 region, integration events will be missed, particularly if the viral disruption occurs outside of the E2 region, or the viral-specific primers are too far from the viral-cellular junction for efficient amplification and sequencing, or viral rearrangements preclude primer annealing. Furthermore, failure to detect integration events that involve multiple concatenated viral genomes may occur if amplicon separation by gel electrophoresis and sequencing are not adequate to discriminate within-viral from viral-cellular amplicon products. In this study we selected and sequenced all bands less than 2kb to minimize analysis of virus-only amplicons. The DIPS-PCR approach used in this study found integration sites previously unreported using Taqα1, but did not find the chromosome 9 intergenic insertion in UPCI:SCC090 that was found by a similar approach using Sau3AI22, a focal sequencing approach15 and WGS21. With directed PCR we confirmed the presence of this insertion in the UPCI:SCC090 cells we studied. Another common method used to detect HPV integration, APOT (Amplification of Papillomavirus Oncogene Transcripts)63, which detects fusion transcripts from integrated HPV, has similar challenges in that this method will detect some but not all events due to limitations of viral primer location, possible gene rearrangement, absence of fusion transcripts, or insufficient assay sensitivity. Even a WGS approach failed to find some of the cell line integration sites detected by DIPS-PCR. Clearly viral integration can be complex and affect multiple cellular sites64. Evaluation is further complicated by the possibility of multiple viral copies existing in episomal or integrated forms as complete or partial genomes.
Disruption of a cellular gene due to viral integration may or may not determine knockout of the gene, depending on whether the second copy (or multiple copies, in the case of aneuploid tumor cells) is affected. The affected cellular gene may be upregulated, disrupted, or unchanged, contingent on strand orientation, as well as the precise viral-cellular junction relative to sequence elements such as promoters and splice sites. Genomic amplification at HPV integration sites is not uncommon21,64.
Our assessment of cellular transcripts affected by viral integration provides important but limited information on the consequence of HPV integration on cellular gene expression. In the most straightforward cases, viral integration into DCC in UM-SCC-104 and PGR in UPCI:SCC154, our analysis indicates that there are no transcripts generated for either of these cellular genes. DCC can function as a tumor suppressor, so it is feasible that disruption of this gene through HPV integration could provide a growth advantage for tumor cells, but the clinical relevance of PGR deficiency in these tumors is yet uncertain.
The HPV integration into ATR is of special interest. In this case the integration into intron 36 did not abrogate transcription across exons 34 and 36, but was associated with out of frame splicing in exons 36–37 and absence of transcription across exons 37 and 38. It will be necessary to expand the evaluation of each integration event to fully examine the effects on the complete cellular gene transcript.
In the remaining cases, further investigation is needed to fully understand the effect HPV integration has on cellular gene expression. No in-frame HPV-cellular fusion transcripts were identified, and in nearly all cases, in-frame sequence of cellular transcripts across viral-cellular integration junctions suggests the existence of at least one intact copy of the genes evaluated (UD-SCC-2 JAK1; UM-SCC-47 p63; UPCI:SCC090 ETV6; UPCI:SCC152 ETV6; and ATR; and UPCI:SCC154 PTPRN2). In the majority of these cases, the viral integration occurs in an intron, and we speculate that perhaps the virus is contained within the intron and is spliced out upon cellular RNA processing. A probable explanation for retained exon-exon transcription of genes with integrated HPV is the presence of additional unaffected gene copies that can generate the intact transcripts. Another possibility in cases with viral integration into either cellular introns or exons may be unanticipated splicing from upstream viral regions into cellular exons, such that the transcripts generated do not contain viral regions proximal to the DNA integration sites. In addition to further analysis of the cellular transcripts and protein expression, it may be useful to examine the HPV genome distal to the integration site in order to determine whether the virus has integrated into 2 different sites in possibly rearranged chromosomes. It is of interest that of the studies examining HPV16 integration in TP63 in the UM-SCC-47 cell line, several different loci within the gene were affected. Olthof et al.22 reported integration of HPV E1 into exon 7 of TP63, Akagi et al.21 reported HPV E2 integration into TP63 intron 13 reading into a rearranged repeated segment of exon 9a and fusion transcripts of E2-TP63, as well as E2 fusion transcripts reading through exon 9, 10, 11 and 12, while in the current study we found integration of HPV E2 into TP63 exon 14 and intron 10. Akagi et al.21 presented a rolling loop model of integration to explain their observations. We speculate that the viral integrations are unstable or that there may be multiple clones with varying integration sites in TP63. We assessed expression of the different p63 isoforms in the UM-SCC-47 cell line, using some of the other cell lines as a reference. While the oncogenic ΔNp63 is the most prominent isoform expressed in HNSCC 65,66, other studies have demonstrated that the TAp63 tumor suppressor isoform is expressed at a higher level than the ΔNp63 oncogenic isoform in HNSCC cell lines25. ΔNp63 has also been implicated in blockade of keratinocyte differentiation as well as acting as a positive and negative transcriptional regulator67. Our analysis of protein expression indicates that the ΔNp63 isoform is absent from the UM-SCC-47 cell line and that the TAp63 isoform is present as a truncated protein. Further research is underway to determine the nature of this truncated form and its possible oncogenic function. Interestingly, previous work in this cell line has shown HPV integration-mediated gene amplification resulting in aberrant expression of a novel truncated p63 protein that functions as a dominant-negative regulator of the TAp63 tumor suppressor isoform.21
The integration of hrHPV into cancer-related genes in seven of the HNSCC cell lines, but only intergenic integration into UM-SCC-105 and UM-GCC-1, suggests a basis for further investigation of this as one possible factor in tumor progression and response to therapy. However, ascertaining the true impact of viral integration on the expression or activity of cellular genes is complicated by both irregular patterns of viral integration (multiple concatenated copies, alternating forward/reverse copies, and rearrangements within integrated viral copies) and atypical, disordered, and likely aneuploid, cellular genomes.
We postulate that integration into gene poor or chromosome fragile sites probably occurs in the majority of HPV-driven cancers, but that secondary integration events into cellular genes, such as tumor suppressor genes or genes involved in cancer pathways may be linked to more aggressive malignant behavior. Studies of the tumors submitted to TCGA had a bias for large primary cancers although many HPV induced primary tumors are small and may be detected by the appearance of a nodule in the neck from early lymphatic spread. Responses of T1-T3 HPV positive tumors even with positive N status are generally better than those with high T-class9. Thus, those larger primary tumors may be enriched for complex HPV integration64. Design of a model to distinguish responsive from non-responsive HPV-positive head and neck tumors assumes viral integration as a primary carcinogenic event, associated with disruption of the E1/E2 region, and alternate E6*I, E6*II transcription, which leads to increased E7 viral oncogene expression68–70. In such a model, tumors with HPV integration into intergenic chromosome sites or fragile sites are maintained as primarily HPV-driven tumors and are likely to respond to current or reduced-intensity treatment, but tumors with HPV integration into cancer-related genes may acquire secondary alterations in cellular gene expression or dysfunction, resulting in a more aggressive malignant phenotype resistant to current therapies.
Comprehensive investigation to understand the specific cellular alterations caused by HPV integration may provide insight for development of alternate therapies for non-responsive tumors. Implementation of viral integration analysis to differentiate responsive from non-responsive HPV-positive head and neck tumors may provide further insight into the factors that distinguish responsive and non-responsive oropharyngeal cancers. Improved knowledge of genomic factors may be valuable in patient selection to avoid under-treatment of patients selected to receive reduced-intensity therapy and to improve treatment of those with more aggressive tumors who fail to respond to intensive treatment.
Supplementary Material
Supplemental Figure I. Spectral Karyotyping (SKY) Chromosome Analysis of UM-SCC-47, Showing the t(3;7) Rearrangement. UM-SCC-47 is a pseudotetraploid cell line from a male donor with several characteristic chromosomal rearrangements. 72,XX,+1,+del(1q),+2,+der(2)t(2;7),+der(2)t(2;7),+der(3)t(3;7),+i(5p),+der(5)t(5;7),+7,+8,+8,9,+10,+11,+der(12)t(8;12),+i(13q),der(13)t(13;16;22),der(13)t(13;21),+14,+14,+14,+15,i(17p),i(17q),+19,+19,+20,der(21) t(21;22),+22. (Y chromosome is lost in most cells including the one shown).
Supplemental Figure 2. Western blot analysis of TP63 isoforms.
Supplemental Table 1: RT-PCR assay primer sequences and corresponding amplicon sizes.
Supplemental Table 2: HPV16 TaqMan quantitative RT-PCR assay primer and probe sequences.
Supplemental Table 3: HPV16 and HPV18 DIPS primer sequences and predicted episome-only amplicon sizes.
Supplemental Table 4: HPV16 and chromosome 9q31.1 PCR primer sequences used to interrogate UPCI:SCC90 and UPCI:SCC152 cell lines.
Acknowledgments
The authors acknowledge the assistance of the University of Michigan DNA core, especially Dr. Robert Lyons and Ellen Pedersen, and the contribution of Jason Yuhase who helped with the Spectral Karyotyping of UM-SCC-47.
Financial Support for each author: Supported by NIH-NCI Head and Neck SPORE P50 CA097248, NIH-NIDCR R01 DE019126, NIH-NCI P30 Cancer Center Support Grant P30 CA046592, and NIH-NIDCD P30 DC05188. HMW supported by the Cancer Biology Training Grant NIH-NCI T32 CA09676, and the Eleanor Lewis Scholarship through the University of Michigan Rackham Graduate School.
Footnotes
The authors disclose no potential conflicts of interest.
References
- 1.Gillison ML. Human papillomavirus-associated head and neck cancer is a distinct epidemiologic, clinical, and molecular entity. Semin Oncol. 2004;31:744–754. doi: 10.1053/j.seminoncol.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Gillison ML, Koch WM, Capone RB, et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92:709–720. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 3.Walline HM, Komarck CM, McHugh JB, et al. High-risk human papillomavirus detection in oropharyngeal, nasopharyngeal, and, oral cavity cancers: Comparison of multiple methods. JAMA Otolaryngology. 2013 doi: 10.1001/jamaoto.2013.5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar B, Cordell KG, Lee JS, et al. Response to therapy and outcomes in oropharyngeal cancer are associated with biomarkers including human papillomavirus, epidermal growth factor receptor, gender, and smoking. Int J Radiat Oncol Biol Phys. 2007;69:S109–S111. doi: 10.1016/j.ijrobp.2007.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gillison ML, D’Souza G, Westra W, et al. Distinct risk factor profiles for human papillomavirus type 16-positive and human papillomavirus type 16-negative head and neck cancers. J Natl Cancer Inst. 2008;100:407–420. doi: 10.1093/jnci/djn025. [DOI] [PubMed] [Google Scholar]
- 6.Feng FY, Kim HM, Lyden TH, et al. Intensity-modulated chemoradiotherapy aiming to reduce dysphagia in patients with oropharyngeal cancer: clinical and functional results. J Clin Oncol. 2010;28:2732–2738. doi: 10.1200/JCO.2009.24.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adelstein DJ, Ridge JA, Gillison ML, et al. Head and neck squamous cell cancer and the human papillomavirus: Summary of a National Cancer Institute State of the Science Meeting, November 9–10, 2008. Vol. 31. Washington, D.C: Head Neck; 2009. pp. 1393–1422. [DOI] [PubMed] [Google Scholar]
- 9.Adelstein DJ, Ridge JA, Brizel DM, et al. Transoral resection of pharyngeal cancer: summary of a National Cancer Institute Head and Neck Cancer Steering Committee Clinical Trials Planning Meeting, November 6–7, 2011. Vol. 34. Arlington, Virginia: Head Neck; 2012. pp. 1681–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howley PM, Munger K, Werness BA, Phelps WC, Schlegel R. Molecular mechanisms of transformation by the human papillomaviruses. Princess Takamatsu Symp. 1989;20:199–206. [PubMed] [Google Scholar]
- 11.Munger K, Phelps WC, Bubb V, Howley PM, Schlegel R. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J Virol. 1989;63:4417–4421. doi: 10.1128/jvi.63.10.4417-4421.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Marco L, Gillio-Tos A, Bonello L, Ghisetti V, Ronco G, Merletti F. Detection of human papillomavirus type 16 integration in pre-neoplastic cervical lesions and confirmation by DIPS-PCR and sequencing. J Clin Virol. 2007;38:7–13. doi: 10.1016/j.jcv.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Nambaru L, Meenakumari B, Swaminathan R, Rajkumar T. Prognostic significance of HPV physical status and integration sites in cervical cancer. Asian Pac J Cancer Prev. 2009;10:355–360. [PubMed] [Google Scholar]
- 14.Ho CM, Lee BH, Chang SF, et al. Integration of human papillomavirus correlates with high levels of viral oncogene transcripts in cervical carcinogenesis. Virus Res. 2011;161:124–130. doi: 10.1016/j.virusres.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 15.Ragin CC, Reshmi SC, Gollin SM. Mapping and analysis of HPV16 integration sites in a head and neck cancer cell line. Int J Cancer. 2004;110:701–709. doi: 10.1002/ijc.20193. [DOI] [PubMed] [Google Scholar]
- 16.Wentzensen N, Vinokurova S, von Knebel Doeberitz M. Systematic review of genomic integration sites of human papillomavirus genomes in epithelial dysplasia and invasive cancer of the female lower genital tract. Cancer Res. 2004;64:3878–3884. doi: 10.1158/0008-5472.CAN-04-0009. [DOI] [PubMed] [Google Scholar]
- 17.Ziegert C, Wentzensen N, Vinokurova S, et al. A comprehensive analysis of HPV integration loci in anogenital lesions combining transcript and genome-based amplification techniques. Oncogene. 2003;22:3977–3984. doi: 10.1038/sj.onc.1206629. [DOI] [PubMed] [Google Scholar]
- 18.Koskinen WJ, Chen RW, Leivo I, et al. Prevalence and physical status of human papillomavirus in squamous cell carcinomas of the head and neck. Int J Cancer. 2003;107:401–406. doi: 10.1002/ijc.11381. [DOI] [PubMed] [Google Scholar]
- 19.Deng Z, Hasegawa M, Kiyuna A, et al. Viral load, physical status, and E6/E7 mRNA expression of human papillomavirus in head and neck squamous cell carcinoma. Head Neck. 2013;35:800–808. doi: 10.1002/hed.23034. [DOI] [PubMed] [Google Scholar]
- 20.Wald AI, Hoskins EE, Wells SI, Ferris RL, Khan SA. Alteration of microRNA profiles in squamous cell carcinoma of the head and neck cell lines by human papillomavirus. Head Neck. 2010 doi: 10.1002/hed.21475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akagi K, Li J, Broutian TR, et al. Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Res. 2013 doi: 10.1101/gr.164806.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olthof NC, Huebbers CU, Kolligs J, et al. Viral load, gene expression and mapping of viral integration sites in HPV16-associated HNSCC cell lines. Int J Cancer. 2015;136:E207–E218. doi: 10.1002/ijc.29112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang AL, Hauff SJ, Owen JH, et al. UM-SCC-104: a new human papillomavirus-16-positive cancer stem cell-containing head and neck squamous cell carcinoma cell line. Head Neck. 2012;34:1480–1491. doi: 10.1002/hed.21962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brenner JC, Graham MP, Kumar B, et al. Genotyping of 73 UM-SCC head and neck squamous cell carcinoma cell lines. Head Neck. 2010;32:417–426. doi: 10.1002/hed.21198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gwosdz C, Balz V, Scheckenbach K, Bier H. p53, p63 and p73 expression in squamous cell carcinomas of the head and neck and their response to cisplatin exposure. Adv Otorhinolaryngol. 2005;62:58–71. doi: 10.1159/000082473. [DOI] [PubMed] [Google Scholar]
- 26.Steenbergen RD, Hermsen MA, Walboomers JM, et al. Integrated human papillomavirus type 16 and loss of heterozygosity at 11q22 and 18q21 in an oral carcinoma and its derivative cell line. Cancer Res. 1995;55:5465–5471. [PubMed] [Google Scholar]
- 27.Ferris RL, Martinez I, Sirianni N, et al. Human papillomavirus-16 associated squamous cell carcinoma of the head and neck (SCCHN): a natural disease model provides insights into viral carcinogenesis. Eur J Cancer. 2005;41:807–815. doi: 10.1016/j.ejca.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 28.White JS, Weissfeld JL, Ragin CC, et al. The influence of clinical and demographic risk factors on the establishment of head and neck squamous cell carcinoma cell lines. Oral Oncol. 2007;43:701–712. doi: 10.1016/j.oraloncology.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin CL, Reshmi SC, Ried T, et al. Chromosomal imbalances in oral squamous cell carcinoma: examination of 31 cell lines and review of the literature. Oral Oncol. 2008;44:369–382. doi: 10.1016/j.oraloncology.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ballo H, Koldovsky P, Hoffmann T, et al. Establishment and characterization of four cell lines derived from human head and neck squamous cell carcinomas for an autologous tumor-fibroblast in vitro model. Anticancer Res. 1999;19:3827–3836. [PubMed] [Google Scholar]
- 31.Yang H, Yang K, Khafagi A, et al. Sensitive detection of human papillomavirus in cervical, head/neck, and schistosomiasis-associated bladder malignancies. Proc Natl Acad Sci U S A. 2005;102:7683–7688. doi: 10.1073/pnas.0406904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maxwell JH, Kumar B, Feng FY, et al. HPV-positive/p16-positive/EBV-negative nasopharyngeal carcinoma in white North Americans. Head Neck. 2010;32:562–567. doi: 10.1002/hed.21216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maxwell JH, Kumar B, Feng FY, et al. Tobacco use in human papillomavirus-positive advanced oropharynx cancer patients related to increased risk of distant metastases and tumor recurrence. Clin Cancer Res. 2010;16:1226–1235. doi: 10.1158/1078-0432.CCR-09-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luft F, Klaes R, Nees M, et al. Detection of integrated papillomavirus sequences by ligation-mediated PCR (DIPS-PCR) and molecular characterization in cervical cancer cells. Int J Cancer. 2001;92:9–17. [PubMed] [Google Scholar]
- 35.Matovina M, Sabol I, Grubisic G, Gasperov NM, Grce M. Identification of human papillomavirus type 16 integration sites in high-grade precancerous cervical lesions. Gynecol Oncol. 2009;113:120–127. doi: 10.1016/j.ygyno.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 36.Tang S, Tao M, McCoy JP, Jr, Zheng ZM. The E7 oncoprotein is translated from spliced E6*I transcripts in high-risk human papillomavirus type 16- or type 18-positive cervical cancer cell lines via translation reinitiation. J Virol. 2006;80:4249–4263. doi: 10.1128/JVI.80.9.4249-4263.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reiser J, Hurst J, Voges M, et al. High-risk human papillomaviruses repress constitutive kappa interferon transcription via E6 to prevent pathogen recognition receptor and antiviral-gene expression. J Virol. 2011;85:11372–11380. doi: 10.1128/JVI.05279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitz M, Driesch C, Jansen L, Runnebaum IB, Durst M. Non-random integration of the HPV genome in cervical cancer. PLoS One. 2012;7:e39632. doi: 10.1371/journal.pone.0039632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pozzi S, Zambelli F, Merico D, et al. Transcriptional network of p63 in human keratinocytes. PLoS One. 2009;4:e5008. doi: 10.1371/journal.pone.0005008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Serber Z, Lai HC, Yang A, et al. A C-terminal inhibitory domain controls the activity of p63 by an intramolecular mechanism. Mol Cell Biol. 2002;22:8601–8611. doi: 10.1128/MCB.22.24.8601-8611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frank CJ, McClatchey KD, Devaney KD, Carey TE. Evidence that loss of chromosome 18q is associated with tumor progression. Cancer Res. 1997;57:824–827. [PubMed] [Google Scholar]
- 42.Carvalho AL, Chuang A, Jiang WW, et al. Deleted in colorectal cancer is a putative conditional tumor-suppressor gene inactivated by promoter hypermethylation in head and neck squamous cell carcinoma. Cancer Res. 2006;66:9401–9407. doi: 10.1158/0008-5472.CAN-06-1073. [DOI] [PubMed] [Google Scholar]
- 43.De Braekeleer E, Douet-Guilbert N, Morel F, Le Bris MJ, Basinko A, De Braekeleer M. ETV6 fusion genes in hematological malignancies: a review. Leuk Res. 2012;36:945–961. doi: 10.1016/j.leukres.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 44.Kibel AS, Faith DA, Bova GS, Isaacs WB. Mutational analysis of ETV6 in prostate carcinoma. Prostate. 2002;52:305–310. doi: 10.1002/pros.10112. [DOI] [PubMed] [Google Scholar]
- 45.Stenman G. Fusion Oncogenes in Salivary Gland Tumors: Molecular and Clinical Consequences. Head Neck Pathol. 2013 doi: 10.1007/s12105-013-0462-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Z, Tognon CE, Godinho FJ, et al. ETV6-NTRK3 fusion oncogene initiates breast cancer from committed mammary progenitors via activation of AP1 complex. Cancer Cell. 2007;12:542–558. doi: 10.1016/j.ccr.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andres SA, Brock GN, Wittliff JL. Interrogating differences in expression of targeted gene sets to predict breast cancer outcome. BMC Cancer. 2013;13:326. doi: 10.1186/1471-2407-13-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kominea A, Konstantinopoulos PA, Kapranos N, et al. Androgen receptor (AR) expression is an independent unfavorable prognostic factor in gastric cancer. J Cancer Res Clin Oncol. 2004;130:253–258. doi: 10.1007/s00432-003-0531-x. [DOI] [PubMed] [Google Scholar]
- 49.Anglim PP, Galler JS, Koss MN, et al. Identification of a panel of sensitive and specific DNA methylation markers for squamous cell lung cancer. Mol Cancer. 2008;7:62. doi: 10.1186/1476-4598-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Killela PJ, Reitman ZJ, Jiao Y, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110:6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chaturvedi AK, Engels EA, Anderson WF, Gillison ML. Incidence trends for human papillomavirus-related and -unrelated oral squamous cell carcinomas in the United States. J Clin Oncol. 2008;26:612–629. doi: 10.1200/JCO.2007.14.1713. [DOI] [PubMed] [Google Scholar]
- 52.Gillison ML, Castellsague X, Chaturvedi A, et al. Comparative epidemiology of HPV infection and associated cancers of the head and neck and cervix. Int J Cancer. 2013 doi: 10.1002/ijc.28201. [DOI] [PubMed] [Google Scholar]
- 53.Hafkamp HC, Manni JJ, Haesevoets A, et al. Marked differences in survival rate between smokers and nonsmokers with HPV 16-associated tonsillar carcinomas. Int J Cancer. 2008;122:2656–2664. doi: 10.1002/ijc.23458. [DOI] [PubMed] [Google Scholar]
- 54.Licitra L, Bossi P, Locati LD. A multidisciplinary approach to squamous cell carcinomas of the head and neck: what is new? Curr Opin Oncol. 2006;18:253–257. doi: 10.1097/01.cco.0000219254.53091.35. [DOI] [PubMed] [Google Scholar]
- 55.Fakhry C, Gillison ML. Clinical implications of human papillomavirus in head and neck cancers. J Clin Oncol. 2006;24:2606–2611. doi: 10.1200/JCO.2006.06.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fakhry C, Westra WH, Li S, et al. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100:261–269. doi: 10.1093/jnci/djn011. [DOI] [PubMed] [Google Scholar]
- 57.Bonner JA, Harari PM, Giralt J, et al. Radiotherapy plus cetuximab for locoregionally advanced head and neck cancer: 5-year survival data from a phase 3 randomised trial, and relation between cetuximab-induced rash and survival. Lancet Oncol. 2010;11:21–28. doi: 10.1016/S1470-2045(09)70311-0. [DOI] [PubMed] [Google Scholar]
- 58.Seiwert TY, Zuo ZX, Keck MK, et al. Integrative and Comparative Genomic Analysis of HPV-Positive and HPV-Negative Head and Neck Squamous Cell Carcinomas. Clinical Cancer Research. 2015;21:632–641. doi: 10.1158/1078-0432.CCR-13-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huebbers CU, Preuss SF, Kolligs J, et al. Integration of HPV6 and downregulation of AKR1C3 expression mark malignant transformation in a patient with juvenile-onset laryngeal papillomatosis. PLoS One. 2013;8:e57207. doi: 10.1371/journal.pone.0057207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pim D, Banks L. Interaction of viral oncoproteins with cellular target molecules: infection with high-risk vs low-risk human papillomaviruses. APMIS : acta pathologica, microbiologica, et immunologica Scandinavica. 2010;118:471–493. doi: 10.1111/j.1600-0463.2010.02618.x. [DOI] [PubMed] [Google Scholar]
- 61.Pett M, Coleman N. Integration of high-risk human papillomavirus: a key event in cervical carcinogenesis? J Pathol. 2007;212:356–367. doi: 10.1002/path.2192. [DOI] [PubMed] [Google Scholar]
- 62.Lace MJ, Anson JR, Klussmann JP, et al. Human papillomavirus type 16 (HPV-16) genomes integrated in head and neck cancers and in HPV-16-immortalized human keratinocyte clones express chimeric virus-cell mRNAs similar to those found in cervical cancers. J Virol. 2011;85:1645–1654. doi: 10.1128/JVI.02093-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klaes R, Woerner SM, Ridder R, et al. Detection of high-risk cervical intraepithelial neoplasia and cervical cancer by amplification of transcripts derived from integrated papillomavirus oncogenes. Cancer Res. 1999;59:6132–6136. [PubMed] [Google Scholar]
- 64.Parfenov M, Pedamallu CS, Gehlenborg N, et al. Characterization of HPV and host genome interactions in primary head and neck cancers. P Natl Acad Sci USA. 2014;111:15544–15549. doi: 10.1073/pnas.1416074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lu H, Yang XP, Duggal P, et al. TNF-alpha Promotes c-REL/Delta Np63 alpha Interaction and TAp73 Dissociation from Key Genes That Mediate Growth Arrest and Apoptosis in Head and Neck Cancer. Cancer Research. 2011;71:6867–6877. doi: 10.1158/0008-5472.CAN-11-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hoadley KA, Yau C, Wolf DM, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158:929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.King KE, Ponnamperuma RM, Yamashita T, et al. deltaNp63alpha functions as both a positive and a negative transcriptional regulator and blocks in vitro differentiation of murine keratinocytes. Oncogene. 2003;22:3635–3644. doi: 10.1038/sj.onc.1206536. [DOI] [PubMed] [Google Scholar]
- 68.Cricca M, Venturoli S, Leo E, Costa S, Musiani M, Zerbini M. Molecular analysis of HPV 16 E6I/E6II spliced mRNAs and correlation with the viral physical state and the grade of the cervical lesion. J Med Virol. 2009;81:1276–1282. doi: 10.1002/jmv.21496. [DOI] [PubMed] [Google Scholar]
- 69.Zheng ZM, Baker CC. Papillomavirus genome structure, expression, and post-transcriptional regulation. Front Biosci. 2006;11:2286–2302. doi: 10.2741/1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jeon S, Lambert PF. Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: implications for cervical carcinogenesis. Proc Natl Acad Sci U S A. 1995;92:1654–1658. doi: 10.1073/pnas.92.5.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure I. Spectral Karyotyping (SKY) Chromosome Analysis of UM-SCC-47, Showing the t(3;7) Rearrangement. UM-SCC-47 is a pseudotetraploid cell line from a male donor with several characteristic chromosomal rearrangements. 72,XX,+1,+del(1q),+2,+der(2)t(2;7),+der(2)t(2;7),+der(3)t(3;7),+i(5p),+der(5)t(5;7),+7,+8,+8,9,+10,+11,+der(12)t(8;12),+i(13q),der(13)t(13;16;22),der(13)t(13;21),+14,+14,+14,+15,i(17p),i(17q),+19,+19,+20,der(21) t(21;22),+22. (Y chromosome is lost in most cells including the one shown).
Supplemental Figure 2. Western blot analysis of TP63 isoforms.
Supplemental Table 1: RT-PCR assay primer sequences and corresponding amplicon sizes.
Supplemental Table 2: HPV16 TaqMan quantitative RT-PCR assay primer and probe sequences.
Supplemental Table 3: HPV16 and HPV18 DIPS primer sequences and predicted episome-only amplicon sizes.
Supplemental Table 4: HPV16 and chromosome 9q31.1 PCR primer sequences used to interrogate UPCI:SCC90 and UPCI:SCC152 cell lines.