SUMMARY
The BabA adhesin mediates high-affinity binding of Helicobacter pylori to the ABO blood-group antigen-glycosylated gastric mucosa. Here we show that BabA is acid responsive but binding is restored by pH neutralization. Acid responsiveness differs among strains; often correlates with different intragastric regions; changes during chronic infection and disease progression; and depends on pH sensor sequences in BabA and on pH reversible formation of high-affinity binding BabA multimers. We propose that BabA’s extraordinary reversible acid-responsiveness enables tight mucosal bacterial adherence while at the same time allowing an effective escape from epithelial cells and mucus that are shed into the acidic bactericidal lumen and that bio-selection and changes in BabA binding properties through mutations and recombination with babA-related genes are selected by differences among individuals and by changes in gastric acidity over time. These processes generate diverse H. pylori subpopulations, and BabA’s adaptive evolution contributes importantly to H. pylori persistence and to overt gastric disease.
Keywords: Helicobacter pylori, blood group antigen-binding adhesin (BabA), gastric acidity, acid-responsiveness, adaptation, diversity, polymorphism, subpopulations, multimerization, gastric cancer
In Brief
Helicobacter pylori bind with high strength to ABO/Leb blood group antigens to persist in the stomach mucosa. Bugaytsova et al. show that its adherence is acid responsive but fully reversible, activated by BabA pH sensors and adapts during chronic infection and disease to allow for functional recycling of infection.
INTRODUCTION
A great challenge for any microbe is how to best exploit hospitable niches while avoiding nearby deleterious ones. This is especially the case for Helicobacter pylori, which chronically infects billions of people worldwide and is implicated in peptic ulcer disease and gastric cancer (Peek and Blaser, 2002). H. pylori thrives in the gastric epithelium and in the ~300 micron thick overlying mucus, a special niche that is hostile to nearly all other microbes (Figure S1I). The mucus layer normally contains a pH gradient from pH ~6 to pH 2 between the epithelial and luminal boundaries and is intrinsically unstable with mucus and epithelial cells continuously being shed into the lumen (Schreiber et al., 2004; Lee, 1985). H. pylori is partially protected from luminal acidity by epithelial secretions of buffering bicarbonate and urea and the ammonia generated by its urease (Sachs et al., 2003), by chemotaxis away from acidity (reviewed in Keilberg and Ottemann, 2016), and by tight adherence to mucosal glycan receptors (Ilver et al., 1998; Aspholm-Hurtig et al., 2004) mediated by attachment proteins (adhesins) that are extraordinary in being acid responsive, as detailed here. The best-studied H. pylori adhesin is BabA, which mediates high-affinity bacterial binding to ABO/Leb blood-group antigens (Leb) that are abundant on gastric epithelial cells and mucins (Aspholm-Hurtig et al., 2004; Lindén et al., 2002). BabA binding affinities (Ka ~107–1012 M−1) are orders of magnitude greater than most carbohydrate-binding proteins (Aspholm-Hurtig et al., 2004; Imberty et al., 2005) and aid H. pylori’s delivery of effector molecules that subvert host defenses (Ishijima et al., 2011) and are necessary for H. pylori replication (Tan et al., 1995) and nutrient acquisition (Kirschner and Blaser, 1995). However, if such binding were unalterable, this would prevent the bacteria from escaping from the mucosal debris that is shed into the acidic and bactericidal gastric lumen.
Here we show that BabA-mediated binding is acid sensitive and responsive, but also fully reversible and restored by pH neutralization. The acid sensitivity profiles change by mutations and recombination with babA-related sequences driven by adaptation to differences in acidity in gastric regions and also in different individuals.
RESULTS
Acid Response and Reversal of Leb Binding
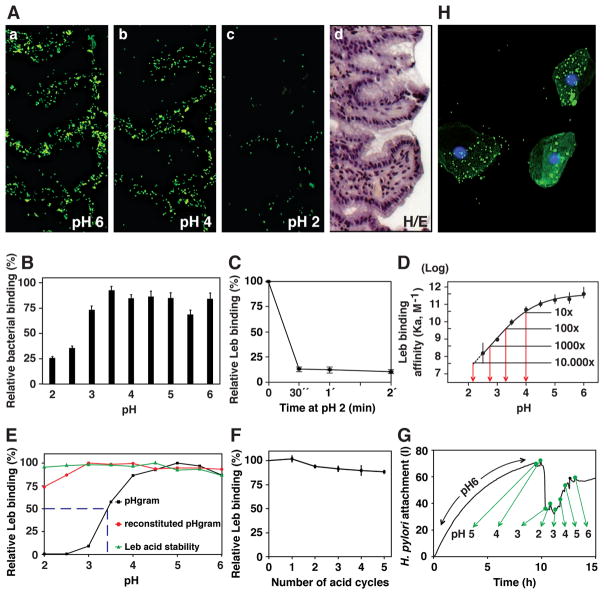
We found that adherence of H. pylori strain 17875/Leb to human gastric mucosal epithelium was acid sensitive, with 2- and 20-fold less binding at pH 4 and 2, respectively, compared to pH 6 (Figure 1A). Equivalent acid-sensitive binding was seen with human MUC5AC gastric mucin and a clinical H. pylori isolate (Figure 1B). A quantitative dissociation assay showed detachment of >85% of previously bound Leb within 30 seconds of a pH 6 to pH 2 shift (Figure 1C). Leb binding at equilibrium showed 10-fold lower affinity at pH 4 than at pH 6 and a further 2000-fold reduction at pH 2, i.e. >20,000-fold decrease in total affinity (Figure 1D). Four sets of experiments showed that acid inactivation of H. pylori binding is reversible. First, H. pylori was preincubated at several low pHs and then restored to pH 5 (which is optimal for its Leb binding). More than 95% of the binding activity was recovered after initial incubation at ≥pH 3, and 87% and 74% after incubation at pH 2.5 and 2.0, respectively (in red, Figure 1E). Second, prior acidification did not affect Leb binding kinetics (Figure S1B). Third, 87% of Leb binding was retained after five full cycles of pH 2.5 acidification and pH 5 restoration (this is referred to as a pH-cycle) (Figure 1F). Fourth, H. pylori were applied to a LigandTracer dish (Figures S1C–S1H) coated with Leb-expressing buccal epithelial cells (BECs) from a Leb-expressing volunteer that represent an authentic epithelial glycosylation pattern typical of human primary gastro-intestinal epithelium collected (Figure 1H) or with Leb-expressing recombinant CHO-cells (Figure S1G), and adherence was LigandTraced to monitor bacterial binding as a function of pH in real time. Adherence to both cell types was again lost at pH 2 and regained at pH 6 (Figure S1H; Figure 1G), though the response to acidification was more pronounced with Leb-CHO cells that overexpress the Leb antigen compared to the authentic and more complex BECs.
Figure 1. H. pylori Binding to Leb in Human Gastric Mucosa is Acid Sensitive, Responsive, Reversible, and Robust.
(A) In vitro binding of fluorescent H. pylori 17875/Leb (binds only to Leb) at (a) pH 6, (b) pH 4, and (c) pH 2 (quantified in Figure S1A); (d) H/E-stained adjacent section.
(B) H. pylori bacterial cells demonstrate pH dependence in binding to mucins (Lindén et al., 2004). Purified human gastric MUC5AC mucin was probed with gastric cancer (GC)-patient isolate (Y0 in Figure 6D) (means + SEM, n = 13 replicates).
(C) 17875/Leb bacteria were mixed with 125I-Leb-conjugate (125I-Leb) at pH 6 then acidified to pH 2. The proportion of Leb that remained bound was scored (means ± SD for n = 2).
(D) The 4 × 1011 M−1 affinity (Ka) at pH 6 showed log-fold reductions at pH 4, 3.3, 2.8, and 2.1 (arrows), and these are shown with 95% confidence intervals (CI).
(E) The acid sensitivity profile, here denoted pHgram (in black), was determined by a 1 h incubation with 125I-Leb at pH from 6 to 2. The dashed line shows when half of the Leb binding remained, i.e. 17875/Leb pH50 = 3.4. Reconditioning of bacterial cells to pH 5 after a 1 h exposure to acidic conditions and testing reactivation of Leb binding (in red) showed that the Leb glycan conjugate exhibited full acid stability at pH 2 (in green), which suggests that acid sensitivity involves the BabA protein itself.
(F) 17875/Leb bacteria were tested for Leb binding after each pH-cycle (means ± SD, n = 2 replicates).
(G and H) 17875/Leb bacterial binding in real time to recLeb-CHO cells (G and Figure S1G) or human BECs (H and Figure S1H) as measured by LigandTracer. The integrity of BECs (H) and recLeb-CHO cells (Figure S1G) was assessed at the end of the pH-cycle. Alexa488 was acid stabile (Figure S1E), and the restoration of Leb binding after neutralization resulted from pre-existing BabA protein, not de novo synthesis, because these experiments used previously frozen (dead) H. pylori.
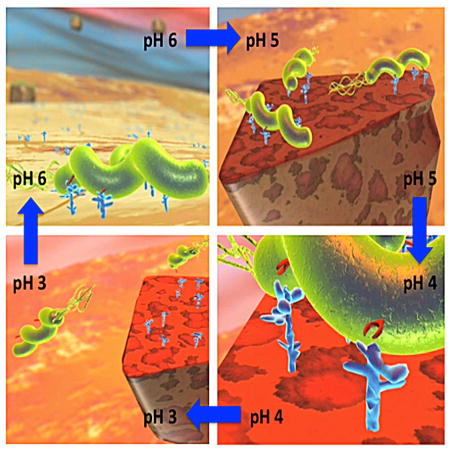
These results inspired our dynamic model in which H. pylori escape from shed mucosal debris as it moves towards the acidic and bactericidal lumen and then return to the epithelial surface and proximal mucus through chemotaxis where the bacteria with best adapted ability in adherence reattach to their Leb binding sites (Movie S1).
BabA’s Polymorphic and Reversible Acid Sensitivity in Binding
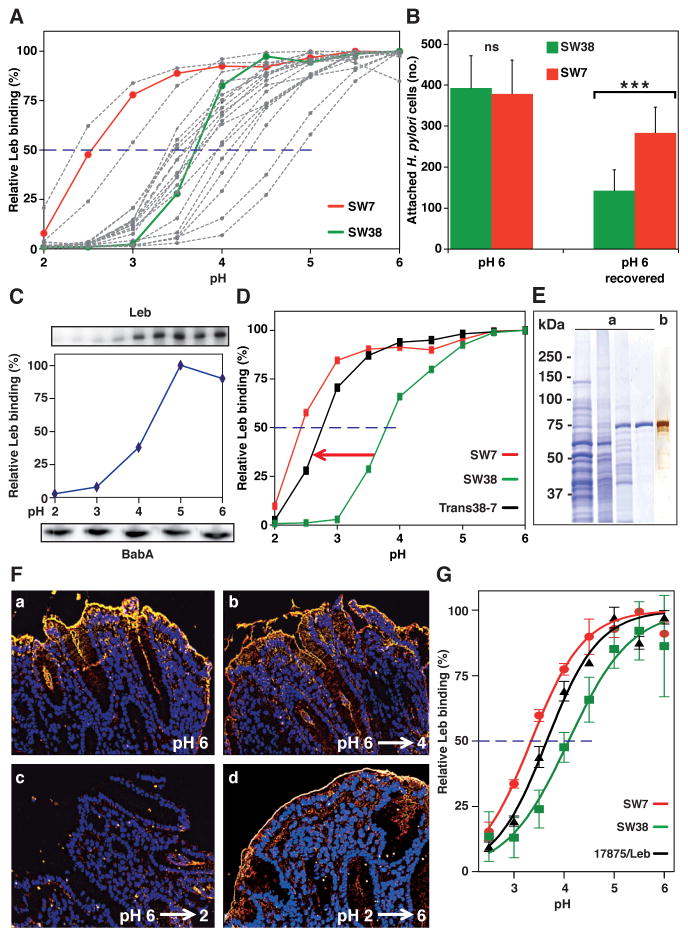
Acid sensitivity of 21 Swedish clinical isolates (SW) was scored as the pH at which half the maximal Leb binding was lost (pH50). The pH50 values ranged from 2.3 (the most acid resistant) to 4.9 (the most acid sensitive) (Figure 2A) (median = 3.7, Figure 6F). Thus, acid-sensitive Leb binding is a general trait but with differences among strains, which suggests that H. pylori can adapt to individual acid secretion patterns. To test in real time for adaptation by selection and gain of function, bacterial cells of the acid-resistant strain SW7 and the more acid-sensitive strain SW38 (Figure 2A) were mixed and exposed to a pH-cycle and LigandTraced. The two strains adhered most similarly to the Leb-coated dish at neutral pH, but after step-wise acidification to pH 3, followed by reconditioning to pH 6, the SW7 strain had a 2-fold increase in binding compared to SW38 (Figure 2B; Figure S2C).
Figure 2. Diversity in Acid Sensitivity in Binding to Leb Among Clinical Isolates is Encoded by the Protein Sequence of BabA.
(A) Leb binding is displayed by pHgram in relative terms to facilitate inter-strain comparison in acid sensitivity in binding (absolute Leb binding is shown in Figure S2A). Each individual pHgram and pH50 value is highly reproducible (Figure S2B).
(B) Attachment at pH 6 and re-attachment after a full pH-cycle (Figure S2C, similar to Figure 1G) by strains SW7 (Alexa555, red) and SW38 (Alexa488, green) was significantly different as measured by Bonferroni post-hoc tests; means ± SD, n = 7 and n = 10 field views for SW7 and SW38, respectively.
(C) 17875/Leb whole-cell proteins were separated by SDS-PAGE, transferred to a membrane, and strips were probed with Leb (Ilver et al., 1998) at pH 2–6 (Leb-binding is shown at the top and quantified in the middle, and the confirmation of full BabA membrane retention is shown at the bottom).
(D) pHgrams of babA donor strain SW7, strain SW38, and the Trans38-7 transformant. SW7 and SW38 were chosen because of their divergence in pH50 (A) and sequence difference (44 amino acids (6%)) (Table S5; Figure S2F) but similar binding affinities (Aspholm-Hurtig at al., 2004). The Trans38-7’s ~25% reduction in gained acid resistance might come from its lower BabA expression, which was more similar to SW38 (Figure S2E).
(E) Protein purification of BabA from H. pylori bacterial cells. SDS-PAGE of (a) whole bacterial cell protein extract (lane 1), ZW-12 detergent bacterial extract (lane 2), proteins eluted from the CEX column (lane 3), and BabA eluted from the Leb column (lane 4) (Figure S2G). (b) Silver staining and MALDI–MS (Figure S2H) shows BabA purified to homogeneity with high yield (Table S1).
(F) Purified BabA was applied to gastric mucosa at pH 6 and then exposed to (a) pH 6, (b) pH 4, or (c) pH 2 followed by immunostaining (yellow) (quantified in Figure S2I); (d) BabA was acidified at pH 2, reconditioned at pH 6, then applied to gastric mucosa and immunostained.
(G) pHgrams of Leb-ELISA of BabA protein purified from H. pylori strains SW7, 17875/Leb and SW38 with pH50 values of 3.3, 3.7, and 4.1, respectively (means ± SD for n = 2).
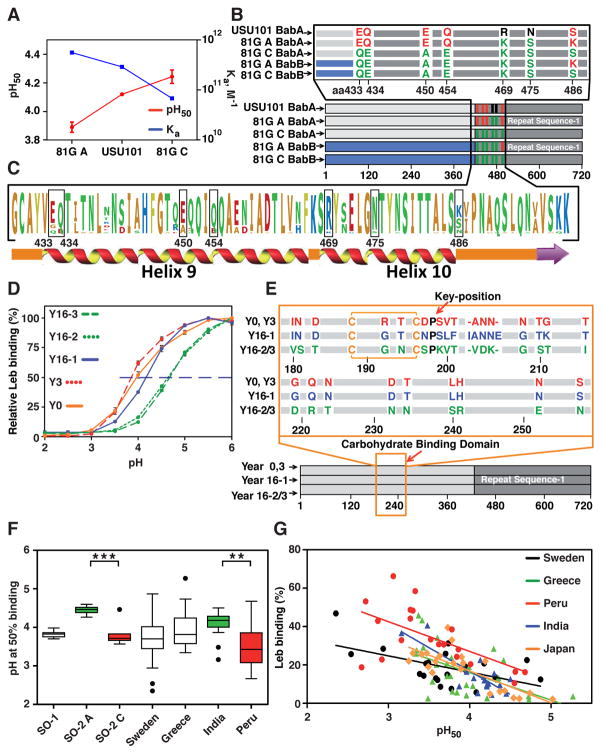
Figure 6. BabA Adaptation in Acid Sensitivity During Severe Disease Progression.
(A) The 81G antrum and 81G corpus clones had pH50 values of 3.9 and 4.25 and Ka values of 6.8 × 1010 M−1 and 5.6 × 1011 M−1, respectively, compared to 4.15 and 2.9 × 1011 M−1 for the USU101 parent strain.
(B) Amino acid replacements in BabA due to babA/babB recombinations. The 81G corpus and antrum BabAs had six and three amino acid substitutions, respectively, compared to USU101 (Figure S6G).
(C) UniProt alignment of 98 BabA Velcro Domain sequences with the seven 81G substitutions indicated.
(D) pHgrams of H. pylori isolated from the GC-patient at Year (Y)0, Y3, Y16-1, Y16-2, and Y16-3 (means ± SD, n = 2).
(E) BabA alignment of the Y0/Y3 (identical) isolates, the Y16-1 isolate, and the more acid-sensitive Y16-2/3 (identical) isolates, where all but one substitution (aa343) are located in the CBD (in red, blue, and green, respectively, see panel D).
(F) Box plots of pH50 for 26 SO-1 (non-reflux dyspepsia) isolates, 20 SO-2 (gastric reflux) antrum (A) and 10 SO-2 corpus (C) isolates (Figure 3A), and 21 Swedish (Figure 2A), 30 Greek (Figure S4B), 17 Indian, and 20 Peruvian (Figure 5A) strains with values outside the box shown as individual points. Mann–Whitney U-tests show significant differences (** p < 0.01; *** p < 0.001).
(G) Tests of strains from Sweden (n = 21,) Peru (n = 20), India (n = 17), Greece (n = 30), and Japan (n = 22) showed that acid sensitivity (pH50) correlates with Leb-binding capacity for the combined set (rs = −0.68, n = 110, p < 0.0001), see also Figure S7F.
Seven experiments tested if reversible acid sensitivity and pH50 differences among strains stem from differences in BabA protein sequence. First, a Far-Western test showed that Leb binding to size-separated BabA protein was maximal at pH 5–6 and negligible at <pH 3 (Figure 2C). Second, immunoblotting showed that the BabA protein was stable at pH 2 for 24 h (Figure S2D). Third, a babA deletion mutant of strain SW38 (pH50 = 3.8) was transformed with babA from acid-resistant SW7 (pH50 = 2.4). The transformant with SW7’s complete babA gene, “Trans38-7”, had a pH50 of 2.8 and thus had gained ~75% of SW7’s acid resistance (Figure 2D). Fourth, native 17875/Leb BabA protein purified to homogeneity (Figure 2E; Figures S2G and S2H) bound specifically to human gastric foveolar epithelium at pH 6 (Figure 2Fa). Acidification to pH 4 and pH 2 caused 15% and >99% detachment, respectively (Figure 2Fb–c; Figure S2I), with binding at pH 6 being unaffected by prior incubation at pH 2 (Figure 2Fd). Fifth, BabA purified from strains SW7, 17875/Leb, and SW38 showed pH50 values with the same rank order as those of their source bacteria, although with 0.9, 0.2, and 0.3 pH units higher acid sensitivities, respectively (Figure 2G). Sixth, circular dichroism (CD) spectra showed that acidification from pH 7 to pH 4 and pH 2.5 caused only minor local shifts in native BabA protein secondary and tertiary structure (Figure S2J). Seventh, for comparison, the common blood group O antigen binding lectins UEA and AAA were tested, and unlike BabA, acid inactivation was found to be irreversible for both lectins (Figure S2K). Thus, the BabA protein sequence determines acid sensitivity in Leb binding, its remarkable reversibility, and the diversity of sensitivity among strains.
Gastric Antrum–Corpus Differences in Acid-Sensitivity in Binding
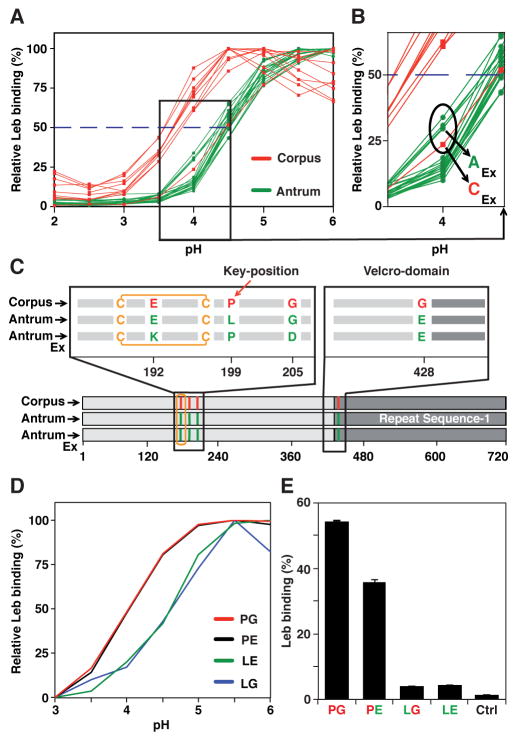
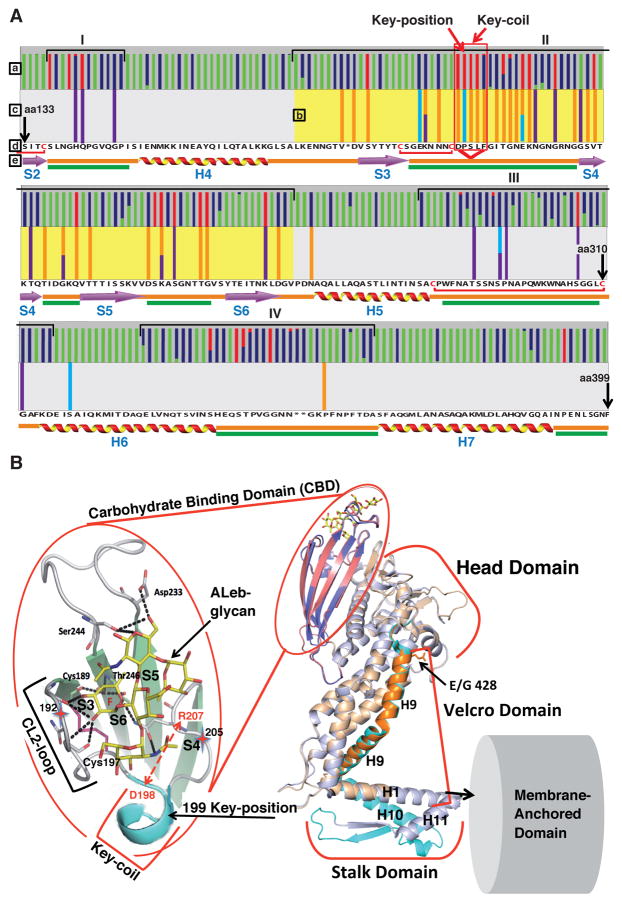
The gastric antrum is less acidic and has a thicker protective mucus layer than the gastric corpus, which contains the acid-producing parietal cells (Figure S3A). To assess if regional gastric physiology might select for pH50 differences in resident H. pylori strains, we studied 20 antrum and 10 corpus isolates from Swedish patient SO-2 with reflux dyspepsia. These isolates had a median difference of 0.7 pH units (pH50 4.4 in the antrum and 3.7 in the corpus, Mann–Whitney U-test, p < 0.0001) (Figure 3A). All SO-2 isolates had identical glr and cysS housekeeping gene sequences (Table S5), indicating recent descent from the same ancestral strain. In contrast, no significant pH50 differences were found among isolates from another patient, SO-1, with non-reflux dyspepsia (Figure 6F). We propose that the process of BabA adaptation to antrum vs. corpus acidity would be more discriminating in patients such as SO-2 with hyper-secretion of acid (pH50 of patient SO-2 vs. SO-1 (Figure 6F)). Several babA alleles were found among the SO-2 isolates, and these differed mainly at only two amino acid positions – Leu(L)199 and Glu(E)428 in the antrum isolates vs. Pro(P)199 and Gly(G)428 in the corpus isolates (Table S5; Figure 3C; Figure S3B; Figure S3F). Two tests showed that the residue at position 199 is functionally important. First, surface plasmon resonance (SPR) of recombinant BabA (recBabA) proteins showed that residue P199 (corpus) conferred a 0.5 pH unit higher acid resistance than L199 (antrum), but without detectable contribution from E428 vs. G428 (Figure 3D; Figure S3C). Second, full-length P199 BabA expressed on the surface of E. coli had >20-fold higher Leb binding than L199 BabA (Figure 3E) due to P199 BabA’s 10-fold higher affinity and ~40-fold higher binding capacity (equilibrium data not shown). Furthermore, chimeric BabA with P199 (corpus) and E428 (antrum) had 30% lower binding than corpus BabA (Figure 3E) due to 1.5-fold lower binding capacity and 1.15-fold lower affinity (equilibrium data not shown), and this contribution by E428 was also seen by SPR (Figure S3C). Thus, position 199 is an important determinant of both acid sensitivity and binding affinity and will henceforth be denoted as “Key-position”. This is the first residue in a short α-helix (amino acids 199 to 202) called the “Key-coil” because it affects the acid sensitivity in Leb binding (Figures 4A and 4B; Figure S4A). Replacement of P199 with L can turn Key-coil into a random coil (Krieger et al., 2005). Similar L to P substitutions with subsequent structural alterations are recognized human cancer markers (Kundu et al., 2013). We also found that 5 of the 30 SO-2 isolates were “Exceptional” in having an antrum-like median pH50 of 4.4 (Figures 3A and 3B) but corpus-like affinities 20-fold and 50-fold higher than other antrum isolates at pH 6 and pH 4, respectively (Figure S3E). This unusual pH50 and affinity combination revealed a corpus-like P199 paired with E192K and G205D substitutions. Positions 192 and 205 are both located next to the Key-position in BabA’s carbohydrate binding domain (CBD) (Figure 4B). The second set of substitutions that influence the acid sensitivity of SO-2 BabA is G/E428 in Helix-9 (H9), which is located ~40 Å away from the CBD. H9, together with H1 and H10, form a coiled-coil Stalk Domain connecting BabA’s surface-exposed domains and membrane-anchored domains (Figure 4B) (Hage et al., 2015). We denote this trio of helices as BabA’s “Velcro Domain” (Figure 4B). We suggest that the Velcro Domain contributes to BabA multimerization and thus to its binding avidity (Figure 6; Figure 7).
Figure 3. Gastric Antrum vs. Corpus Adaptation in Acid Sensitivity and Affinity in Binding is Encoded in BabA.
(A) pHgrams of 30 isolates from patient SO-2 from Orebro University Hospital, Sweden. The most and least acid sensitive isolates displayed a full pH unit difference.
(B) The Exceptional Antrum (AEx) and Corpus (CEx) isolates are circled.
(C) Location of the SO-2 BabA substitutions: P199L in Key-position and E192K/G205D in the Exceptional (Ex) isolates are all in the proximity of the fucose-binding CL2 disulfide-clasped loop (yellow) that constitutes the basis for the CBD. The G428E substitution is located diametrically opposite the CBD where it is part of Velcro Domain and is situated at the very junction of the Repeat-Sequence-1, i.e. the C-terminal 200 amino acid segment including the membrane anchored domain (Figure 4B) that is conserved between BabA, BabB and BabC (Alm et al., 2000).
(D) SPR-derived pHgram analyses of Leb-binding by E. coli recBabA526 protein from SO-2 corpus (P199/G428 (PG)), antrum (L199/E428 (LE)) and chimeric variants L199/G428 (LG) and P199/E428 (PE) (sensograms in Figure S3C).
(E) Leb binding by E. coli bacterial cells that express full-length recBabA720 from SO-2 corpus (PG), antrum (LE), and chimeric variants LG and PE was assessed by RIA. E. coli with no expression of recBabA was used as the control (Ctrl) (means + SD for n = 2).
Figure 4. Amino Acid Replacements for Adaptation in Acid Sensitivity in Leb-Binding.
(A) The BabA CBD is under both positive and adaptive selection. Here, the BabA aa133–399 segment is shown with the Key-position and Key-coil (red vertical box) indicated in close proximity to the fucose-binding CL2 disulfide-clasped loop (red bracket). (a) The colored bars represent amino acid residues under positive selection, i.e. single nucleotide mutations that result in high level (red) or moderate level (blue) of amino acid substitutions or conservation (green) of amino acids (Aspholm-Hurtig et al., 2004). The high-level positive selection clusters are indicated as I–IV. (b) The pH50-dependent substitutions co-localize the strongest with Cluster II (CBD, aa175–255 (yellow)), but not with Cluster IV. (c) The series of pH50 substitutions in Greek strains (purple bars (Figure S4D)), SO-2 isolates (light-blue bars (Figure S3B)), and the GC-patient isolates (orange bars (Figure 6E)) are located in Clusters I–III. (d) The reference sequence is P448 BabA, and stars indicate positions missing in this strain’s BabA sequence (Aspholm-Hurtig et al., 2004). (e) Intrinsically unstructured regions (IUR) (green bars) were predicted by DisEMBL and GlobPlot. In BabA, the IUR segments join β-strands (lilac arrows) and α-helices (ribbons) and exhibit higher prevalence of polar amino acids and prolines for rapid and precise responses to changes in the local environment (Dyson and Wright, 2005; Rauscher et al., 2006).
(B) Structural organization of the CBD and Velcro Domain acid-sensitivity determinants. The CBD is carried by the Head Domain. The coiled-coil Stalk Domain connects the Head Domain with the predicted Membrane-Anchored Domain (Hage et al., 2015; Moonens et al., 2016). The four β-strands S3–S6 with IUR loops constitute the CBD along with the disulfide (C189/C197)-clasped (in magenta) fucose-binding loop CL2, which is immediately followed by the Key-coil (aa198–202) and Key-position (aa199) in most BabAs. D198 in the Key-coil forms a salt bridge with R207 that acts as a structural clamp (dashed red arrow). The salt bridge acts as a pH sensor and breaks open due to protonation of the aspartic acid at pH <4. H9 in the Head Domain and H1/10 in the Stalk Domain together comprise the Velcro Domain. The K192/D205 Exceptional (Ex) motif (red stars) seems to constitute an additional mechanism for increased acid sensitivity in binding.
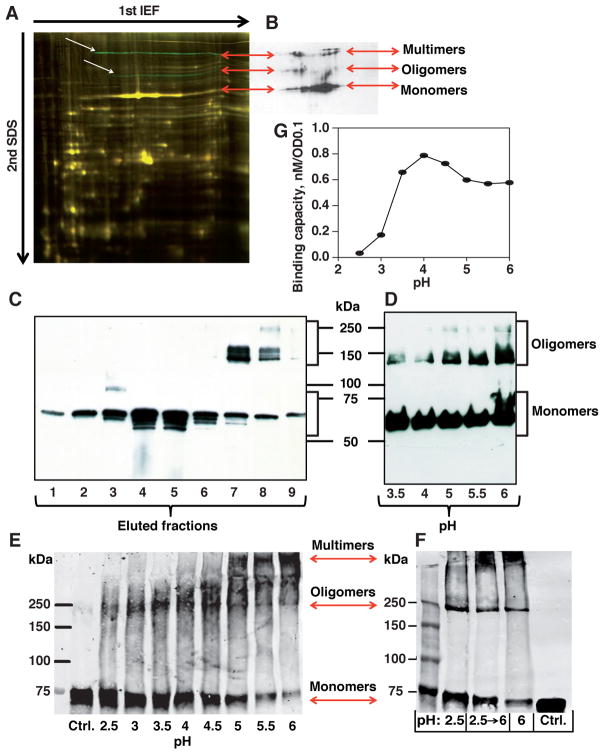
Figure 7. BabA Multimerization and Reversible Acid Dissociation.
(A) H. pylori J166 whole-protein extracts analyzed by 2D-DIGE. The two green bands (white arrows) that were missing or significantly reduced after 8 weeks of infection in a rhesus macaque (presumable BabA oligomers and multimers) were, together with the dominant yellow band that corresponds to the BabA monomer, positive for BabA by immunoblot (B) (see also differential fluorescence in Figure S7A).
(B) BabA immunoblot of J166 wild-type whole-protein extract separated by 2D-DIGE revealed three groups of bands – BabA monomer, oligomers, and HMM (double-band) multimers (arrows).
(C) Non-reducing SDS-PAGE of CEX fractions from BabA purification (Figure S2Ga). BabA eluted as a monomer followed by HMM oligomers and possible multimers.
(D) Non-reducing SDS-PAGE of ZW-12 detergent extracts of 17875/Leb bacterial cells in a pH range from 3.5 to 6. Acidification of the bacterial extract caused dissociation of BabA oligomers and multimers into monomers.
(E) Glutaraldehyde crosslinking of 17875/Leb bacterial cells after exposure to pH 2.5–6. Digital integration of the BabA fluoro-Infra-Red (IR)-signal is shown in Figure S7D.
(F) Glutaraldehyde crosslinking of 17875/Leb bacterial cells after exposure to pH 2.5 with BabA dissociation into monomers (pH 2.5); reconstitution of BabA into multimers by pH 6 reconditioning after prior pH 2.5 acidification (pH 2.5→6); and pH 6 exposure for visualization of the predominant multimers (pH 6). Digital integration of the BabA IR-signal is shown in Figure S7E.
(G) Correlation between acid dependence (pH50) and maximal Leb-binding capacity (as an approximation of active and available BabA adhesin protein) for 17875/Leb bacterial cells was estimated by equilibrium-in-binding affinity analysis.
BabA differences that reflect adaptations to local gastric sites were also seen in the analysis of 30 Greek isolates that, similar to the Swedish isolates, exhibited a range of pH50 values from 3.3 to 5.3 (Figure S4Ba), with a median pH50 of 3.8 (Figure 6F). Of the 13 strains that demonstrated Leb binding in both antrum and corpus isolates, four strains exhibited pH50 antrum-corpus differences, from + 0.9 to −0.6 pH units. The antrum isolates from two patients (G0017 and G1055) were more acid sensitive, whereas the corpus isolates from two other patients (G1034 and G2030) were more acid sensitive (Figure S4Bb–f). For comparison, an equivalent difference in acid-sensitive Leb-binding of the G1007 antrum vs. corpus isolates was seen with human MUC5AC gastric mucin (Figure S4C). The antrum and corpus isolate pairs were 3 to 79 amino acids different (Figure S4D), and each isolate pair had identical glr and cysS housekeeping gene sequences (Table S5). These substitutions tightly overlap the BabA Clusters I–III, i.e. domains that are high in positive-selection activity (with amino acid substitution as a consequence of nucleotide mutation), including the CBD (Figure 4A). Also present are three amino acid substitutions in the Velcro Domain of strain G1007, including E428G (Figure S4D), which is identical to the Velcro Domain substitution in patient SO-2 (Figure 3C). The positions of these substitutions suggest that they have direct acid-responsive effects on both monomeric BabA-Leb interactions (through the CBD) and monomer-oligomer transitions (through the Velcro Domain).
BabA Acid Sensitivity and Geographic Disease Patterns
The babA genes are far more diverse than other H. pylori genes, as illustrated by the unique sequences of ~100 full-length BabAs (Figure S5A), and this diversity is shaped by multiple selective forces, including ABO blood group phenotypes and differences in the gastric physiology among human populations. Because H. pylori infection is associated with pangastritis, corpus atrophy, hypochlorhydria (higher gastric pH), and elevated gastric cancer risk in Peru (Correa, 2013) vs. antrum-predominant gastritis (Misra et al., 2000), hyperchlorhydria (lower gastric pH), increased duodenal ulcer risk, and reduced gastric cancer risk in India (Lam, 2000), we hypothesized that Leb binding by Indian strains would be more acid resistant. However, we found that 14 of 16 Indian strains were more acid sensitive compared to Peruvian strains (Figure 5A), with a median difference of 0.8 pH units (pH50, 4.2 vs. 3.4) (Mann–Whitney U-test, p = 0.003).
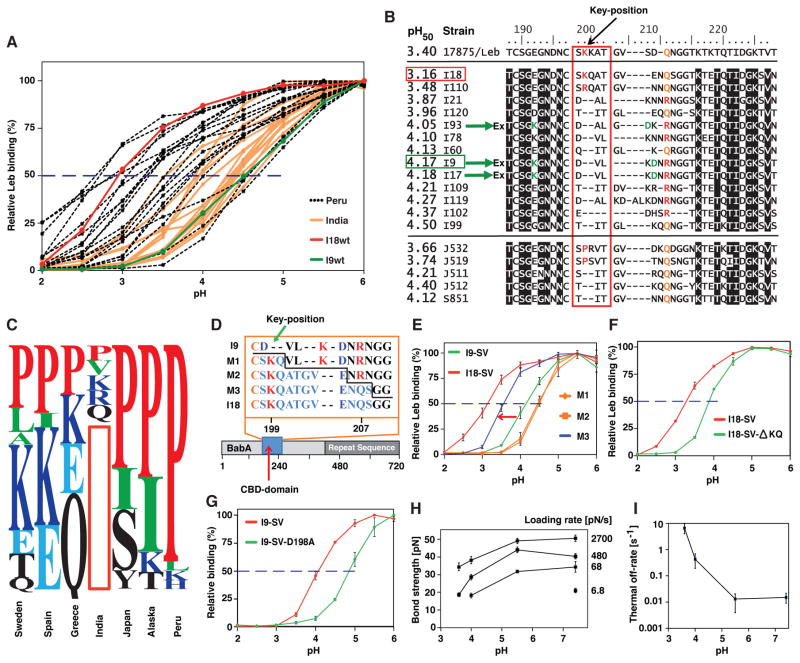
Figure 5. Acid Inactivation of Leb Binding Depends on the pH Response-Sensor in the Key-Coil.
(A) pHgrams of 17 Asian Indian and 20 Peruvian strains with acid-resistant Indian I18 shown in red and more acid-sensitive Indian I9 shown in green.
(B) BabA sequences from 17875/Leb, the Asian-Indian strains (I), and strains from Japan (J) and Spain (S) with/without Key-coil positions 199 and 200 ranked according to their pH50. A red vertical box indicates the Key-coil. The acid-resistant strain I18 (horizontal red box) and acid-sensitive strain I9 (horizontal green box) are indicated. The three green arrows show strains that carry the K192/D205 Exceptional (Ex) motif for increased acid sensitivity in binding (similar to Ex motif in Figure 3C).
(C) The prevalence of P in the Key-position is ~25% in Europe, ~50% among North American Amerindians (Alaska), and ~90% among South American Amerindians (Peru) (Table S2). The red vertical box illustrates the missing Key-position in most Indian strains.
(D) The three I9-SV BabA mutants (M) with 3, 8, or 11 amino acids from the Key-coil region of strain I18.
(E) pHgrams show that BabA I9 with the S198–S208 (11 aa) segment, expressed in H. pylori strain P1ΔbabA, gave strain M3 a 60% increase in acid resistance (arrow) compared to the original I9 strain (pH50 4.17). M3 pH50 = 3.56 (blue), whereas the M1 and M2 mutants (orange) showed contrasting increased acid sensitivity (pH50 = 4.46) (means ± SD for n = 5 (M1), n = 6 (M2 and M3), or n = 3 (I9 and I18) individual clones).
(F) Recombinant expression of BabA I18-SV and I18-SV- KQ in H. pylori strain P1ΔbabA, demonstrated increased acid sensitivity from pH50 3.3 to 3.9 (means ± SD for n = 4) due to deletion of the two residues 199K and 200Q.
(G) Recombinant expression of BabA I9-SV and I9-SV-D198A in H. pylori strain P1ΔbabA demonstrated increased acid sensitivity from pH50 = 4.1 to 4.9 (means ± SD for n = 4) due to inactivation of the salt bridge’s D-node by the D198A substitution.
(H) Bond strengths for twelve combinations of acidity and loading rate. These are the effective loading rates for the binding when the influence of the elasticity of the bacterium body has been compensated for (Bell, 1978).
(I) The off-rate was assessed by a linear fit of the measured bond strength versus the logarithm of the loading rate (from H), where the off-rate is calculated from the slope of this line (Björnham et al., 2009). The measured increase in off-rate (dissociation) is comparable to the decrease in bacterial binding affinity at pH 3.6 (Figure 1D). The results are based on >4000 single measurements.
The Key-position is P in 20 of 23 Peruvian strains (Aspholm-Hurtig et al., 2004), but in only 1 of 14 Indian strains with acid-sensitive binding (Table S5). In addition, 11 of these 14 Indian BabAs lack residues 199 and 200, and thus have a truncated Key-coil compared to most other strains (Figure 5B). BabAs of acid-sensitive Indian strains I93, I9, and I17 also contain the K192D205-motif (Figure 5B), which similarly makes the Exceptional SO-2 isolates more acid sensitive (Figures 3B and 3C). In contrast, BabAs of the two acid-resistant Indian strains (I18 and I110) with pH50 = 3.16 and 3.48 (Figure 5B) contain full Key-coils, but with K or R in the Key-position (red in Figure 5B), which is reminiscent of charged (K, E) or polar (Q) residues that are present in the majority of BabAs from European H. pylori isolates and which is distinct from the almost universal P in BabAs from East Asian and Amerindian isolates (Figure 5C).
Six experiments were performed to determine the contribution of the Key-coil to acid sensitivity. First, BabAs from Indian acid-sensitive (I9) and acid-resistant (I18) strains (Figures 5A and 5B) were expressed from a plasmid shuttle vector (SV) in H. pylori strain P1 babA. The resulting pH50 profiles closely matched those of the source strains (Figure S5B), further showing that the BabA sequence determines its acid sensitivity. Second, 3 (M1), 8 (M2), or 11 (M3) amino acid residues in and near the Key-coil of acid-resistant strain I18 were used to replace the corresponding BabA residues in acid-sensitive strain I9 (pH50 = 4.17). The H. pylori I9 BabA-M3-mutant, containing I18’s S198–S208 segment, became 0.6 pH units more acid resistant than the original I9 isolate (Figures 5D and 5E). Third, two strains from Japan (J511, J512) and one from Spain (S851) (Aspholm-Hurtig et al., 2004), which similarly lack Key-coil positions 199 and 200, were found to be more acid sensitive (pH50 4.12–4.40) than phylogenetically closely related Japanese BabAs with intact Key-coils (J532, J519) (pH50 = 3.66 and 3.74) (Figure 5B). Fourth, the importance of Key-coil positions 199 and 200 was further demonstrated by deletion of K199 and Q200 in strain I18, which increased the pH50 by 0.6 units (Figure 5F), i.e. a gain in acid sensitivity that is very similar to the three strains from Japan and Spain that naturally lack these two residues (Figure 5B). Fifth, the increase in acid resistance caused by replacement of D198–R207 with S198–Q207 (R207 vs. Q207 indicated in Figure 5B) in the M3-mutant (in the second experiment) is noteworthy because charged residues D198 and R207 are predicted to form a salt bridge (Figure 4B) (Moonens et al., 2016). In contrast to M3, the M1 and M2 mutants that lost the salt bridge D-pole became much more acid sensitive (Figure 5E). The contribution of the putative salt bridge in strain I9 was analyzed by a D198A substitution, which increased acid sensitivity by 0.8 pH units (Figure 5G). Sixth, we used Force Measuring Optical Tweezers (FMOT) to understand how acid-induced Key-coil relaxation relates to binding kinetics. Important in this context is that BabA is largely unaffected by BabA multivalency because the bacterial cell and Leb make distinct contacts due to the use of spherical beads as handles during the measurements (Björnham et al., 2005), which is very different from SPR measurements where the interpretation of bacterial cell binding is complicated by extremely slow dissociation at pH optimal for Leb binding i.e. the nearly zero off-rate in binding (Figure S3D). However, with FMOT we subjected BabA-Leb bonds to increasing external force and exponentially increased loading rates from pH 7.4 to 3.6 (Figure 5H), and we found that the off-rate increased >500-fold at pH 3.6 (Figure 5I). We conclude that BabA’s rapid acid-induced decrease in Leb binding and affinity stems mainly from an increased off-rate. Thus the D198/R207 salt bridge clamps the Key-coil at higher pH, but it acts as a pH sensor where the salt bridge is disrupted at pH <4 (~pKa of D). This mechanism brings about immediate Key-coil relaxation, distortion of the proximal fucose-binding CL2 loop, and a hugely increased off-rate in binding because CBD loses its ability to hold Leb in place upon acidification.
Adaptation of BabA Acid Sensitivity During Disease Progression Into Gastric Cancer
We next tested for BabA evolution driven by the gastric changes that H. pylori itself elicits. Rhesus macaques resemble humans in gastric physiology and mucosal ABO/Leb-glycosylation (Lindén et al., 2008) and are naturally infected by H. pylori, although babA and hence Leb binding tend be lost during infection (Solnick et al., 2004). Two rhesus macaques (54H and 81G) had been infected by H. pylori USU101, a strain originating from a patient with gastric cancer, initially to test if the alimentary carcinogen ethyl-nitro-nitrosoguanidine (ENNG) exacerbated H. pylori-induced gastritis development into gastric cancer (Liu et al., 2009). After 6 years of USU101 infection, the 81G animal that had also received the ENNG carcinogen developed gastric atrophy, dysplasia, and gastric cancer (Figure S6A; Table S3A). In contrast, the 54H animal with no ENNG supplementation was only diagnosed with gastritis (Table S3B). As expected, most H. pylori recovered from each animal had lost Leb binding activity. However, distinct subpopulations of Leb-binding bacterial cells were detected in both animals (Figure S6B) and were isolated from both the antrum and corpus (Figure S6C). In the ENNG-treated 81G animal, coexisting subpopulations of Leb binders developed adapted binding properties during the progression into gastric cancer. Here, the corpus isolates (81G-C) were more acid sensitive (mean pH50 = 4.25) and the antrum isolates (81G-A) were less acid sensitive (mean pH50 = 3.9) compared to the US101 parent strain (pH50 = 4.1) (Figure 6A; Figure S6D). We suggest that the higher acid sensitivity of the corpus clones reflects gastric hypochlorhydria caused by corpus atrophy, depletion of acid-producing parietal cells, and BabA adaptation to the elevated corpus pH. Such increased acid sensitivity was also seen with corpus isolates from two Greek patients (G1034 and G2030) (Figure S4Bb). In contrast, the pH50 difference in isolates from the 54H animal (not ENNG treated) was only 0.2 units (pH50 = 4.2 for corpus and 4.4 for antrum isolates). Such a more acid sensitive and hence health-associated antrum clone, suggests adaptation to a healthier gastric environment such as the 54H animal (Figure S6E). This was similar to the SO-1 patient with non-reflux dyspepsia with a ~0.2 difference in pH50 between corpus clones and more acid sensitive antrum clones (Figure 6F). SPR binding tests of the corresponding recBabA proteins confirmed the differences in acid sensitivity (Figure S6F). Equilibrium analysis revealed ~10-fold lower Leb affinity (Figure 6A) and 30% reduced binding capacity (fewer BabA/bacterial cell) for 81G-C compared to 81G-A isolates and to the more intermediate US101 parent strain. The CD spectra showed that the recBabA 81G-A and C proteins require acidification to pH 2.5 and 3.5, respectively, to reach similar helical features in acid-induced secondary structure, which suggests that the antrum and corpus proteins have differences in structural stability (Figure S7B). Seven amino acid differences between the antrum and corpus isolates and/or the USU101 ancestor were found (Table S5; Figure 6B), all of which clustered in the highly polymorphic H9/H10 Velcro Domain (Figure 6C). The seven substitutions in BabA were aa469 and aa475 in Helix-10 in both 81G-A and 81G-C isolates (Figures 6B and 6C; Figure S6G, yellow box), four substitutions in the aa433–454 segment in H9 uniquely in 81G-C (Figures 6B and 6C; Figure S6G, green box), and a single aa486 substitution unique to 81G-A in H10 (Figures 6B and 6C; Figure S6G, red box). Sequencing of the related but divergent babB gene (BabB does not bind to Leb) (Ilver et al., 1998) argues that the 81G-A and 81G-C BabA proteins adapted due to recombinational import-based gene conversion of babB DNA sequences into babA (Table S5; Figure 6B; Figure S6G), a phenomenon also seen with SO-2 isolates (Table S5; Figure S3F) and similar to the E428G substitution in the Greek G1007 isolates (Figure S4D). Reciprocally, 81G-C babB contained a ≥50 bp segment with six nucleotide differences derived from the original babA gene (dashed arrows in the red box in Figure S6G). Importantly, because the 81G-A and 81G-C BabA CBDs are identical, the divergence between these BabAs in pH50 acid sensitivity and affinity can be ascribed to their differences in Velcro Domain strength. Thus multiple recombinations between babA, babB, and possibly babC gene sequences present in the same genome can create changes in Velcro Domain sequence and influence BabA binding properties.
We also studied BabA adaptation during chronic infection in a patient who developed gastric cancer (here denoted the GC-patient) using isolates from the time of the first intestinal metaplasia at year Y0 and Y3 and at Y16 when the metaplasia had progressed to dysplasia (Kennemann et al., 2011). The single available Y0 and Y3 isolates and one of three Y16 isolates were each more acid resistant in Leb binding (pH50 = 4.0, 3.9, and 4.2, respectively) than two other Y16 isolates (pH50 = 4.65) (Figure 6D), although all isolates were similar in binding capacity and affinity (Figure S6H). Again, the identical glr and cysS gene sequences in the isolates verify that they derived from the same ancestral strain (Table S5). The BabA sequences from Y0 and Y3 were identical and differed from Y16-1 by 9 substitutions and from Y16-2 and Y16-3 by 25 substitutions (Figure 6E; Table S5). All but one of these acid sensitivity-determining amino acid substitutions stem from single nucleotide differences, cluster in the CBD (yellow segment in Figure 4A), and are specifically located in intrinsically unstructured regions (green bars in Figure 4A) where they can modulate the reactivity of these loops. The dual clustering of apparently adaptive BabA substitutions in the CBD and Velcro Domain during chronic infection and progression to gastric cancer illustrates the importance of these two domain in determining BabA’s acid sensitivity and Leb affinity.
Acid Sensitive and Reversible BabA Multimerization
We investigated whether BabA’s reciprocity in acid sensitivity vs. high affinity in Leb binding depends on the CBD’s intrinsic affinity seen in BabA monomers combined with multimeric avidity in binding through Velcro Domain interactions. First, 2D-electrophoresis showed that BabA proteins migrate as oligomeric and high molecular mass (HMM) multimeric complexes (Figures 7A and 7B; Figure S7A). Second, BabA eluted as oligomeric complexes during cation exchange (CEX) chromatography (Figure 7C). Third, the bacterial whole-cell protein level of BabA (Table S1) compared to bacterial cell Leb-binding capacity (Figure 1D) gave a ratio of 2.8 BabA proteins per binding site, which suggests that the high-affinity form of BabA is a trimer. Fourth, in contrast to the acid stability of the native BabA protein, CD spectra of the 81G antrum and corpus recBabA526 fragments show structural transitions at pH 4.5, which suggests that the C-terminal β-barrel membrane domain and proximal Velcro and Stalk domains indeed contribute to structural support (Figures S2J vs. S7B). Fifth, small-angle X-ray scattering (SAXS) showed recBabA in pure monomer form below pH 4, whereas BabA forms discrete higher molecular weight species above pH 6 (Figure S7C). Sixth, non-reducing (less denaturing) SDS-PAGE showed that BabA released from H. pylori membranes by the mild detergent ZW-12 is oligomeric but is monomerized by acidification to <pH 4 (Figure 7D). Seventh, crosslinker treatment of H. pylori bacterial cells at different pHs identified BabA in HMM multimers at pH 6–5 that progressively dissociated into ~250 kDa oligomers at pH <4 and ultimately into 75 kDa monomers at lower pHs (Figure 7E; Figure S7D). Eight, to determine how acid-induced dissociation of BabA multimers relates to its highly reversible Leb-binding mode, we again used crosslinker treatment to show that reconditioning of the cells to pH 6 from prior pH 2.5 exposure resulted in ~70% recovery of the BabA HMM multimers and a reciprocal reduction in BabA monomers (Figure 7F; Figure S7E). Ninth, we found a ~35% increase in H. pylori binding capacity during the initial acidification to pH 4 (Figure 7G), which we interpret as an increase in the number of independent Leb-binding sites due to acid-induced dissociation of BabA multimers to monomers. These results show that BabA contains two distinct domains that together determine acid sensitivity, affinity, and avidity of Leb binding – the CBD for direct glycan receptor binding and the Velcro Domain that controls BabA multimerization, and thus Leb-binding multivalency, with increased avidity that is interpreted in terms of increased binding strength and acid resistance.
DISCUSSION
Here we show that BabA-mediated H. pylori adherence is acid sensitive, which allows the infection to adapt to changes in gastric acidity due to BabA’s responsive and fully reversible binding by pH neutralization. This adaptation is reflected in the spectra of acid sensitivity profiles among clinical isolates. BabA acid sensitivity allows the H. pylori infection to adapt to the gradual shifts in gastric acidity that result from life-long chronic inflammation, and this adaptation occurs through mutations and recombination events with divergent babA-related genes that are duplicated in the same genome and/or genes from other strains. Gain of function is achieved by natural selection for the optimum combination of acid sensitivity in binding combined with chemotaxis-driven recycling of infection. Such adaptation and selection allows H. pylori to occupy its niche in the thicker and more buffered antrum mucosa, even during elevated acid secretion, as well as in the thinner and more acidic corpus mucosa. Given the heterogeneity of the inflamed, and sometimes dysplastic, mucosa, H. pylori also develops heterogenic subpopulations of bacterial cells that are optimized for different gastric habitats. Such selection pressures adapt BabA for increased acid secretion (lower pH) or decreased acid secretion (higher pH), conditions that are pathognomonic in the development of peptic ulcers and gastric cancer, respectively.
BabA Adaptation to Global Disease Patterns
In principle, evolution should favor H. pylori infections that fully occupy the gastric mucosa (as they do in isolates from Peru), so why is it that the Indian strains have developed antrum-restricted tropism combined with high acid sensitivity in binding? We suggest that due to the Indian strains’ preference for high acid sensitivity, the infection relocates closer to the more buffered epithelium and away from the acidic mucus layer and the brunt of the gastric lumen acidic juice. However, gastrin production increases during antrum inflammation with resulting hyper secretion of acid in the corpus through a paracrine loop. It follows that the Indian strains prefer to avoid this acid rain, which is reflected in their acquired acid sensitivity and predominantly antrum location. However, exacerbation of antral inflammation results in increased gastrin secretion, hyper secretion of acid and progression into peptic ulcer disease. Indeed, peptic ulcer disease is common in Asian Indians (Misra et al., 2000; Lam, 2000). Thus, the sheltered antrum location is augmented by the hyperchlorhydria and the resultant low-pH gastric juice. We suggest that such a hallmark might have been evolutionarily selected for by the partial protection that high gastric acidity confers against lethal gastrointestinal infections such as Vibrio cholera, which has likely been endemic in the Bay of Bengal region for centuries or millennia. This is also in accord with the unusually high prevalence of blood group B among Indians, which is similarly adaptive in decreasing cholera susceptibility (Glass et al., 1985). In contrast, the ~1 pH unit increase in acid resistance in binding in Peruvian strains (Figure 6F) allows for pangastric dissemination with resultant corpus atrophy and increased gastric cancer risk (Correa, 2013). On the geographic level, our results suggest that changes in acid sensitivity in binding are driven by external selection pressures such as pandemic infectious diseases. This predisposes for differences in H. pylori regional antrum vs. corpus tropism and shapes the types of disease that will develop within a population. Future work will elucidate the molecular mechanisms that prevent H. pylori in India from pangastric spread, i.e. whether antrum tropism is due to local host factors or H. pylori colonization properties.
BabA Adaptation to Disease Progression
Over time, pangastritis and corpus infection predispose for the loss of acid-secreting parietal cells (atrophy) with subsequent reduction in acid secretion and elevated pH. We propose that such degenerative processes select for increased acid sensitivity in binding as illustrated by BabA from Greek patients (Figure S4Bbef), the dysplastic GC-patient (Figure 6D), and the rhesus macaque with gastric cancer (Figure 6A). However, gastric regional heterogeneity in terms of inflammation, metaplasia, atrophy, and sometimes also neoplastic dysplasia provides ample opportunities for expansion of H. pylori subpopulations. Such subpopulations as identified in the rhesus macaques (Figure S6B), the GC-patient (Figure 6D), and the Exceptional-SO2 isolates (Figure 3B) have evolved binding characteristics for optimal conformity to additional mucosal micro-niches in the gastric landscape during long-term infection. The extraordinarily high diversity in pH50 (ranging from pH 2.3 to pH 5.3) among clinical strains shows that BabA adapts over time to changes in the secretion of acid, i.e. to local pH conditions. Such adaptation is illustrated by the elevated pH50 in both the rhesus macaque isolates and the GC-patient isolates during progression of gastric disease over many years into cancer. But how does H. pylori maintain its Leb binding at higher pH, sometimes even at neutral pH that occurs as a consequence of atrophy progression and gastric cancer? Tests of 110 strains from populations around the world showed that acid sensitivity in binding correlates with binding capacity (Figure 6G; Figure S7Fab), but not with binding affinity (Figure S7Fc), i.e. strains with high binding capacity, and hence high level of BabA expression, are often more acid resistant in binding. In contrast, acid-sensitive strains demonstrate low binding capacity and low numbers of BabA adhesins. This suggests that the structural plasticity of BabA exhibits an upper pH50 limit of ~5 for adaptation in acid sensitivity in binding to disease-associated elevations in gastric pH. This conclusion is further supported by the D198A mutant where inactivation of its pH sensor mechanism resulted in an exceedingly acid-sensitive phenotype, approaching pH50 = 5. Our results argue for a critical role of the spring-activated (but clamped) Key-coil and its acid responsiveness through structural relaxation, synergistic mechanisms that together form the BabA pH-sensor. Thus, further adaptation in acid responsiveness necessitates a reduction in overall bacterial binding strength through lowered avidity by reduced expression of BabA and hence lowered binding capacity. Our new understanding of H. pylori adaptation to changes over time in gastric pH suggests that BabA-mediated gastric colonization patterns can also adapt its adherence properties to long-term acid-secretion inhibition therapy. Numerous conditions, including esophageal reflux disease and dyspepsia, are symptomatically treated with proton pump inhibitors (PPIs) to reduce gastric acidity, often for the patient’s lifetime. When these patients are also infected with H. pylori, there is often a redistribution of H. pylori infection and gastritis from predominantly in the antrum to stomach wide, and this increases the risk of pan-gastritis, corpus atrophy, and gastric cancer (Kuipers et al., 1996). Similar disease developments by PPI treatment have been confirmed in animal models (Hagiwara et al., 2011). Much of the H. pylori movement into the corpus in such cases might reflect the search for more acidic mucus pH gradients that would better support the function of its acid-responsive BabA-mediated adherence, and by doing so H. pylori might circumvent PPI therapy. The often-long duration and consequences of acid-suppression therapies necessitates the search for alternative therapies and new diagnostics. For example, acid sensitivity-linked changes in BabA might be used as a real-time marker for changes in gastric pH over time and for the risk of severe disease.
EXPERIMENTAL PROCEDURES
In vitro H. pylori Adherence to Gastric Mucosa
Fluorescence labeled H. pylori were pre-incubated in citrate-phosphate buffers at pH 2, 4, and 6 and then applied to gastric mucosal histo-tissue sections. Bacterial adherence was digitalized with a Zeiss AXIOcam MRm (Carl Zeiss AB, Stockholm, Sweden) with optical magnification of 200×. Zeiss AxioVision software v.4.5 was used to quantify bacterial adherence. ApoTome (Carl Zeiss AB) was used in Figure 1A to increase sharpness.
Acid Sensitivity in Leb-binding (pH50) i.e. pHgram
H. pylori Leb binding properties were analyzed by radioimmunoassay (RIA). Leb-HSA conjugate (IsoSep AB, Tullinge, Sweden) was 125I-labeled by the chloramine T method (Ilver et al., 1998). 125I-Leb-conjugate was incubated with bacteria, and then pelleted by centrifugation and the 125I in the pellet vs. the supernatant was measured using the 2470 Wizard2 Automatic Gamma counter (PerkinElmer, Waltham, MA, USA). pH50 was assessed in citrate-phosphate buffers pH 2 – 6. 125I-Leb bound to pelleted cells was determined and presented as “pHgrams”. The sensitivity of different strains to pH was expressed as their midpoints, pH50 (interpolated pH value at a 50% decrease in binding relative to maximal binding).
Real-Time Attachment of H. pylori was performed with LigandTracer Green (Ridgeview, Uppsala Sweden). Leb-conjugate, recLeb-CHO cells, or BECs were immobilized on a dish to which fluoro-labeled H. pylori was applied and then exposed to a pH gradient. Attached bacteria were quantified in real time by the fluorescence detector as described in the SEP:
Real-time adhesion to Leb conjugates. The Petri dish was coated with Leb-conjugate. Alexa488 (acid stabile)-H. pylori cells were applied and placed in LigandTracer Green with slow rotation of the dish. After 10 h, pH was decreased to pH 3, and then reconditioned by NaOH. The fluorescent signal represents number of H. pylori bacterial cells bound to the Leb conjugate;
Leb-CHO cells were applied to a Nunclone™ Surface Petri dish (Nunc A/S, Roskilde, Denmark) and fixed with paraformaldehyde. Alexa488-labeled H. pylori was applied to the dish, and placed in LigandTracer Green system for 9 h at RT. The pH was acidified and then reconstituted. Bacterial cells attached were registered as described above.
Real-time adhesion to buccal epithelial cells (BECs) The buccal mucosal surface of an (ABO/Leb)-secretor positive individual was scraped with a Cytobrush Plus Cell collector (CooperSurgical. Inc. Berlin, Germany). For immobilization of BECs to the Petri dish, a new method was developed, where BEC cells were re-suspended in N-hydroxy-succinimide ester (biotin) (Sigma, Steinheim, Germany). A Petri dish was coated with MegaCellR-Streptavidin (Cortex Biochem, San Leonardo, CA, USA). BECs were added, fixed with paraformaldehyde and LigandTraced with Alexa488-labeled H. pylori for 9h with slow rotation in a pH 2–6 gradient. Bacterial cells attached to the BECs were measured as above.
Purification of Native BabA Adhesin Protein from H. pylori
Bacterial cells were detergent solubilized in ZW 3–12 (Sigma Aldrich, St. Louis, MO, USA), and the protein extract was applied to a Source 30S (GE Healthcare, Uppsala, Sweden) cation exchange column. BabA protein was eluted with a linear gradient of 0–200 mM NaCl. BabA fractions were identified by immunoblotting, pooled diluted with octyl-glucoside, and Leb-affinity purified. BabA protein was eluted by an acid gradient and quickly neutralized.
pH-Dependent In Vivo BabA Oligomerization
H. pylori was incubated in buffers pH 2.5 to 6 at RT for 30 min followed by glutaraldehyde crosslinking of BabA oligomers at pH 7 for 2 min. BabA oligomers were separated by SDS-PAGE, immunoblot detected, visualized with IRDye 800CW 2nd antibody, scanned by Odyssey Sa, and quantified by Image StudioTM Lite Ver 4 (LI-Cor Bioscience) as described in the SEP.
Statistical Analysis was performed with GraphPad Prism 6 as described in the SEP.
Supplementary Material
HIGHLIGHTS.
H. pylori adherence is acid responsive, with inactivation fully reversed by increased pH
Adhesin diversity in acid sensitivity is driven by inflammation and disease progression
pH sensor sequences in BabA’s binding domains determine its pH responsiveness
BabA adaptation to mucosal atrophy and resulting elevated pH can promote gastric cancer
Acknowledgments
This paper is dedicated to the memory of our friend and collaborator and co-author Dr. Andre Dubois, a great scientist who contributed importantly both intellectually and materially to this project.
We thank S. Michopoulos, G. Mantzaris for H. pylori clinical isolates; Ö. Furberg (NoPolo.se), N. Ulander, S. Lindström, and M. Borén for the digital movie, tech, art, and figure work, respectively. This work was supported by grants from Vetenskapsrådet (VR/M) to TB and SKL, Cancerfonden to TB and AA, VR/NT to AA and SSc, Formas to SKL, the J.C. Kempe and Seth M. Kempe Memorial Foundation, the Knut and Alice Wallenberg Foundation (2012.0090) to TB and ML, and European Union Seventh Framework Program GastricGlycoExplorer ITN grant number 316929 to TB and YAC, Magn. Bergvall’s Foundation to SSc, DFG (SFB 900/A1) to SSu, DFG (HA2697/16-1) to RH, FP6 ANR-06-PATHO-00701 ERA-NET and Actions Concertées Inter-Pasteuriennes (ACIP) (2006) to DS, NIH R01DK063041 to DEB, NIH CA082312 to DSM, and was in part performed within the Adhesion Centre Umeå, Umeå Centre for Microbial Research (UCMR), and the Biochemical Imaging Centre Umeå (BICU) within the National Microscopy Infrastructure (NMI). Animal experiments were performed in accordance with NIH guidelines, the Animal Welfare Act, and US federal law. The experimental challenge infection with strain J166babB was carried out at the University of California, Davis under protocol #15597 approved by the UC Davis Institutional Animal Care and Use Committee (IACUC), which has been accredited by the Association of Assessment and Accreditation of Laboratory Animal Care (AAALAC). All animals were housed under these guidelines in an accredited research animal facility fully staffed with trained personnel. The USU101/81G series of experiments were carried out at the Uniformed Services University of the Health Sciences, Bethesda, and conducted according to the principles in the “Guide for the Care and Use of Laboratory Animals”, Institute of Laboratory Animal Resources, NRC, HHS/NIH Pub. No. 85-23. All procedures involving animals were reviewed and approved by the AFRRI animal care and use committee.
Footnotes
ACCESSION NUMBERS
The babA, babB, cysS and glr sequences have been deposited in GenBank under the accession numbers Y613381-KY613496 (Table S5).
Supplemental Information includes Supplemental Experimental Procedures, seven figures, five tables, and one movie, which can be found with the online article at http://xxxxxxxxxxx.
AUTHOR CONTRIBUTIONS
All authors commented on the different stages of the ms, and read and approved the final version of the manuscript. DI, RS, JB and TB conceptualized the study. In vitro binding of bacterial cells and native BabA to tissue sections by SH. In vitro binding to mucins by MQPH, SKL. RIA, pHgrams and affinity analyses by RS, ASh. LigandTracer by JB, ASh. pH stability of fluorochromes by ASh, AEs, NS. Far-Western blot by JM, JÖ. H. pylori transformation by GB. Native BabA purification by JB, SV. Native BabA pH50 by JB. Native BabA MALDI-MS by AH. Acid stability of H. pylori BabA content by AÅ. Circular dichroism of BabA by CA, JB, YAC, JÅ, GG. BabA oligomerization analyzed with SAX by KM, HR. Clinical isolate sequences of babA, babB and housekeeping genes by YAC, MM, GB, JB. H. pylori and BabA protein binding with SPR by KB, AO, JSY, CGK. E. coli recBabA with pOPE-vector by SH, ASc, LR, YAC. Antibody “Vite” by ASc. H. pylori recBabA with shuttle vector by PG, YAC, VK, RH. Acid-dependent amino acid substitutions aligned with positive selection clusters and secondary structure predictions by JB. Worldwide prevalence of P199 and BabA alignments by JB. FMOT measurements by OB, SSc. Isolation of H. pylori subpopulations by MM, YAC. Patho-morphological analysis of rhesus macaque gastric tissues by RM, ML, AD, DSM. Subpopulation detection with Alexa555-Leb by JB. 2D-DIGE by BAC, AEl. Acid-dependent oligomerization by YAC, JB. H. pylori isolates provided by RHG, AC, AKM, BGN, KSP, BMG, DNS, LE, MU, DD, SSu. H. pylori obtained from infected rhesus macaque by AD, DSM, JMW, HL, MG, LMH, JVS. GenBank depositions by LMH, JVS. Synthesis of Leb-conjugate by SO. Recombinant CHO cells by JH. Statistical analysis by ASh. TB and JB supervised YAC. TB supervised MM, SH, JÖ, DI. AA, JS, RH, and TB supervised PG. RH supervised VK. SSc supervised OB. LAMR supervised RM. JB, YAC, PG, DI, HR, AA, SSc, DEB, and TB wrote the manuscript. JB, DEB, and TB reviewed and edited the final version of the paper. TB is one of the founders of Helicure and is a member of its scientific advisory board.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alm RA, Bina J, Andrews BM, Doig P, Hancock REW, Trust TJ. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect Immun. 2000;68:4155–4168. doi: 10.1128/iai.68.7.4155-4168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspholm-Hurtig M, Dailide G, Lahmann M, Kalia A, Ilver D, Roche N, Vikström S, Sjöström R, Lindén S, Bäckström A, et al. Functional adaptation of BabA, the H. pylori ABO blood group antigen binding adhesin. Science. 2004;305:519–522. doi: 10.1126/science.1098801. [DOI] [PubMed] [Google Scholar]
- Bell MG. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- Björnham O, Fällman E, Axner O, Ohlsson J, Nilsson UJ, Borén T, Schedin S. Measurements of the binding force between the Helicobacter pylori adhesin BabA and the Lewis b blood group antigen using optical tweezers. J Biomed Opt. 2005;10:44024-1–9. doi: 10.1117/1.1989227. [DOI] [PubMed] [Google Scholar]
- Björnham O, Bugaytsova J, Borén T, Schedin S. Dynamic force spectroscopy of the Helicobacter pylori BabA-Lewis b binding. Biophys Chem. 2009;143:102–105. doi: 10.1016/j.bpc.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Correa P. Gastric cancer. Gastroenterol Clin North Am. 2013;42:211–217. doi: 10.1016/j.gtc.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their function. Nat Rev Mol Cell Biol. 2005;3:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Glass RI, Holmgren J, Haley CE, Khan MR, Svennerholm AM, Stoll BJ, Belayet Hossain KM, Black RE, Yunus M, Barua D. Predisposition for cholera of individuals with O blood group. Possible evolutionary significance. Am J Epidemiol. 1985;121:791–796. doi: 10.1093/oxfordjournals.aje.a114050. [DOI] [PubMed] [Google Scholar]
- Hage N, Howard T, Phillips C, Brassington C, Overman R, Debreczeni J, Gellert P, Stolnik S, Winkler GS, Falcone FH. Structural basis of Lewis(b) antigen binding by the Helicobacter pylori adhesin. BabA Adv. 2015;1:e1500315. doi: 10.1126/sciadv.1500315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara T, Mukaisho K, Nakayama T, Sugihara H, Hattori T. Long-term proton pump inhibitor administration worsens atrophic corpus gastritis and promotes adenocarcinoma development in Mongolian gerbils infected with Helicobacter pylori. Gut. 2011;60:624–630. doi: 10.1136/gut.2010.207662. [DOI] [PubMed] [Google Scholar]
- Ilver D, Arnqvist A, Ögren J, Frick IM, Kersulyte D, Incecik ET, Berg DE, Covacci A, Engstrand L, Boren T. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279:373–377. doi: 10.1126/science.279.5349.373. [DOI] [PubMed] [Google Scholar]
- Imberty A, Mitchell EP, Wimmerova M. Structural basis of high-affinity glycan recognition by bacterial and fungal lectins. Curr Opin Struct Biol. 2005;15:525–534. doi: 10.1016/j.sbi.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Ishijima N, Suzuki M, Ashida H, Ichikawa Y, Kanegae Y, Saito I, et al. BabA-mediated Adherence Is a Potentiator of the Helicobacter pylori Type IV Secretion System Activity. J Biol Chem. 2011;286:25256–25264. doi: 10.1074/jbc.M111.233601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilberg D, Ottemann KM. How Helicobacter pylori senses, targets and interacts with the gastric epithelium. Environ Microbiol. 2016;18:791–806. doi: 10.1111/1462-2920.13222. [DOI] [PubMed] [Google Scholar]
- Kennemann L, Didelot X, Aebischer T, Kuhn S, Drescher B, Droege M, Reinhardt R, Correa P, Meyer TF, Josenhans C, et al. Helicobacter pylori genome evolution during human infection. Proc Natl Acad Sci USA. 2011;108:5033–5038. doi: 10.1073/pnas.1018444108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner DE, Blaser MJ. The dynamics of Helicobacter pylori infection of the human stomach. J Theor Biol. 1995;176:281–290. doi: 10.1006/jtbi.1995.0198. [DOI] [PubMed] [Google Scholar]
- Krieger F, Möglich A, Kiefhaber T. Effect of proline and glycine residues on dynamics and barriers of loop formation in polypeptide chains. J Am Chem Soc. 2005;127:3346–3352. doi: 10.1021/ja042798i. [DOI] [PubMed] [Google Scholar]
- Kuipers J, Lundell L, Klinkenberg-Knol EC, Havu N, Festen HP, Liedman B, Lamers CB, Jansen JB, Dalenback J, Snel P, et al. Atrophic gastritis and Helicobacter pylori infection in patients with reflux esophagitis treated with omeprazole or fundoplication. N Eng J Med. 1996;334:1018–1022. doi: 10.1056/NEJM199604183341603. [DOI] [PubMed] [Google Scholar]
- Kundu A, Bag S, Ramaiah S, Anbarasu A. Leucine to proline substitution by SNP at position 197 in Caspase-9 gene expression leads to neuroblastoma: a bioinformatics analysis. 3. Biotech. 2013;3:225–234. doi: 10.1007/s13205-012-0088-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam SK. Differences in peptic ulcer between East and West. Baillieres Best Pract Res Clin Gastroenterol. 2000;14:41–52. doi: 10.1053/bega.1999.0058. [DOI] [PubMed] [Google Scholar]
- Lee ER. Dynamic histology of the antral epithelium in the mouse stomach: III. Ultrastructure and renewal of pit cells. Am J Anat. 1985;172:225–240. doi: 10.1002/aja.1001720305. [DOI] [PubMed] [Google Scholar]
- Lindén S, Nordman H, Hedenbro J, Hurtig M, Borén T, Carlstedt I. Strain- and blood group-dependent binding of Helicobacter pylori to human gastric MUC5AC glycoforms. Gastroenterology. 2002;123:1923–1930. doi: 10.1053/gast.2002.37076. [DOI] [PubMed] [Google Scholar]
- Lindén S, Mahdavi J, Hedenbro J, Borén T, Carlstedt I. Effect of pH on Helicobacter pylori binding to human gastric mucins; identification of binding to non-MUC5AC mucins. Biochem J. 2004;384(part 2):263–270. doi: 10.1042/BJ20040402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindén S, Mahdavi J, Semino-Mora C, Olsen C, Carlstedt I, Borén T, Dubois A. Role of ABO secretor status in mucosal innate immunity and H. pylori infection. PLoS Pathog. 2008;4:e2. doi: 10.1371/journal.ppat.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Merrell DS, Semino-Mora C, Goldman M, Rahman A, Mog S, Dubois A. Diet synergistically affects Helicobacter pylori-induced gastric carcinogenesis in nonhuman primates. Gastroenterology. 2009;137:1367–1379. doi: 10.1053/j.gastro.2009.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra V, Misra S, Dwivedi M, Singh UP, Bhargava V, Gupta SC. A topographic study of Helicobacter pylori density, distribution and associated gastritis. J Gastroenterol Hepatol. 2000;15:737–743. doi: 10.1046/j.1440-1746.2000.02240.x. [DOI] [PubMed] [Google Scholar]
- Moonens K, Gideonsson P, Subedi S, Bugaytsova J, Romaõ E, Mendez M, Nordén J, Fallah M, Rakhimova L, Shevtsova A, et al. Structural insights into Polymorphic ABO Glycan Binding by Helicobacter pylori. Cell Host & Microbe. 2016;19:55–66. doi: 10.1016/j.chom.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peek RM, Jr, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2:28–37. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- Rauscher S, Baud S, Miao M, Keeley FW, Pomés R. Proline and glycine control protein self-organization into elastomeric or amyloid fibrils. Structure. 2006;14:1667–1676. doi: 10.1016/j.str.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Sachs G, Weeks DL, Melchers K, Scott DR. The gastric biology of Helicobacter pylori. Annu Rev Physiol. 2003;65:349–369. doi: 10.1146/annurev.physiol.65.092101.142156. [DOI] [PubMed] [Google Scholar]
- Schreiber S, Konradt M, Groll C, Scheid P, Hanauer G, Werling HO, Josenhans C, Suerbaum S. The spatial orientation of Helicobacter pylori in the gastric mucus. Proc Natl Acad Sci USA. 2004;101:5024–5029. doi: 10.1073/pnas.0308386101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solnick JV, Hansen LM, Salama NR, Boonjakuakul JK, Syvanen M. Modification of Helicobacter pylori outer membrane protein expression during experimental infection of rhesus macaques. Proc Natl Acad Sci USA. 2004;101:2106–2111. doi: 10.1073/pnas.0308573100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S, Tompkins LS, Amieva MR. Helicobacter pylori usurps cell polarity to turn the cell surface into a replicative niche. PLOS Pathog. 2009;5:e1000407. doi: 10.1371/journal.ppat.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.