Abstract
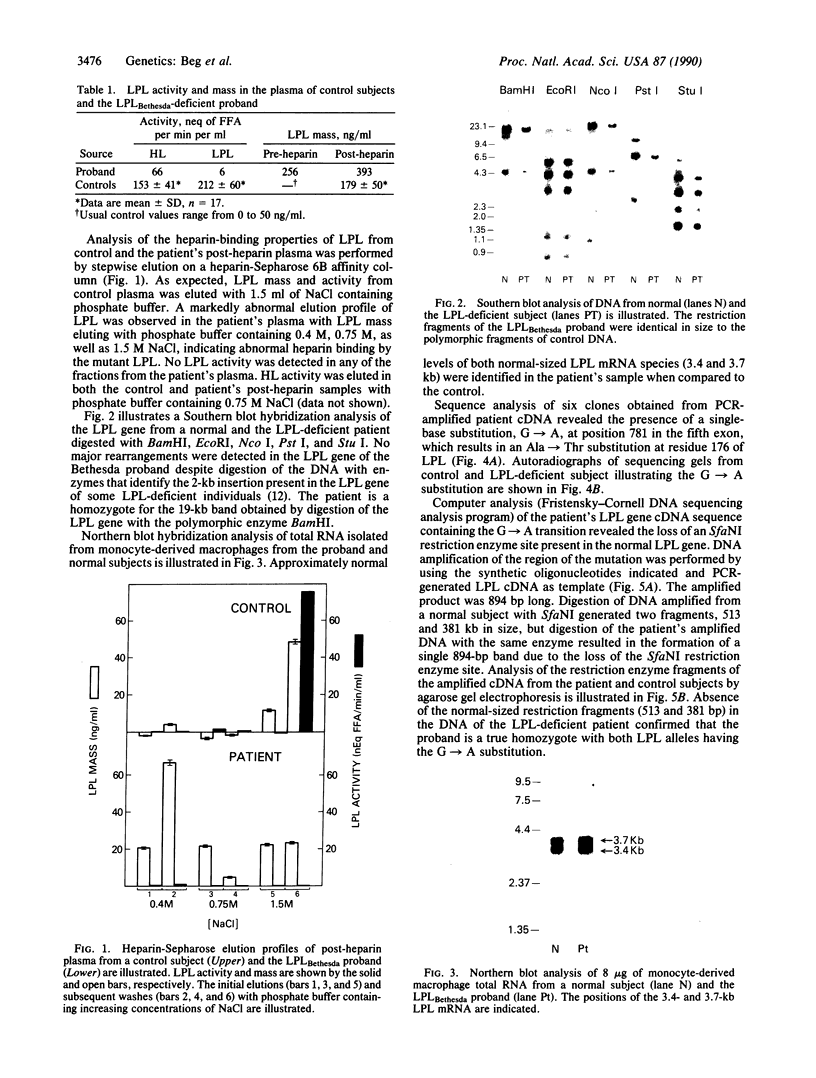
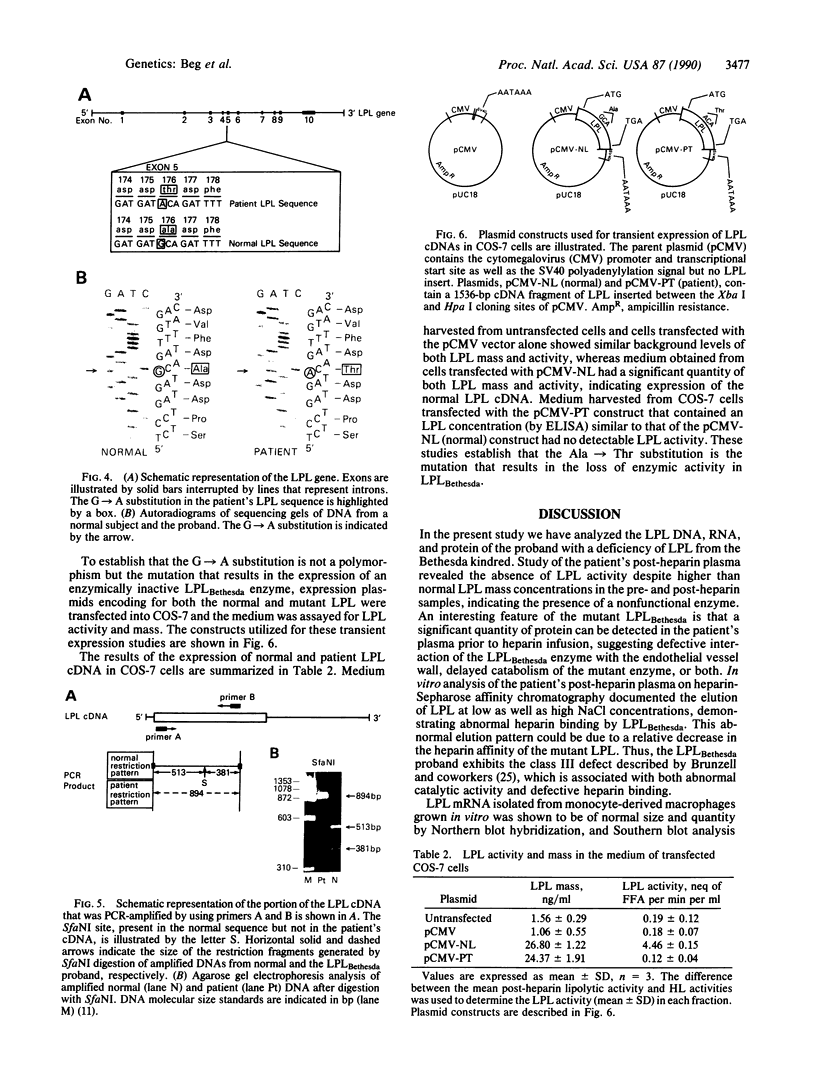
The molecular defect that leads to a deficiency of lipoprotein lipase (LPL) activity in the proband from a Bethesda kindred has been identified. The pre- and post-heparin plasma LPL mass in the proband was elevated when compared to controls; however, there was no detectable LPL activity, indicating the presence of a defective enzyme (termed LPLBethesda). Analysis of the patient's post-heparin plasma by heparin-Sepharose affinity chromatography demonstrated that the mutant LPL had an altered affinity for heparin. Southern blot hybridization of the gene for LPLBethesda revealed no major rearrangements. Northern blot analysis of LPLBethesda mRNA from patient monocyte-derived macrophages revealed normal-sized mRNAs (3.4 and 3.7 kilobases) as well as normal cellular mRNA levels when compared to control macrophages. Sequence analysis of polymerase chain reaction-amplified LPL cDNA revealed a G----A substitution at position 781 of the normal LPL gene that resulted in the substitution of an alanine for a threonine at residue 176 and the loss of an SfaNI site present in the normal LPL gene. Amplification of cDNA by the PCR followed by digestion with SfaNI established that the patient was a true homozygote for the mutation. Expression of LPL cDNA in COS-7 cells resulted in the synthesis of a nonfunctional LPL enzyme establishing that the Ala----Thr substitution was the mutation responsible for the inactive LPL. The identification of this mutation in the LPL gene defines a region of the LPL enzyme, at Ala-176, that is essential for normal heparin-binding and catalytic activity. We propose that an amino acid substitution in this critical region of LPLBethesda results in the synthesis of a nonfunctional enzyme that leads to the chylomicronemia syndrome expressed in this proband.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmad N., Venkatesan S. Nef protein of HIV-1 is a transcriptional repressor of HIV-1 LTR. Science. 1988 Sep 16;241(4872):1481–1485. doi: 10.1126/science.3262235. [DOI] [PubMed] [Google Scholar]
- Auwerx J. H., Babirak S. P., Fujimoto W. Y., Iverius P. H., Brunzell J. D. Defective enzyme protein in lipoprotein lipase deficiency. Eur J Clin Invest. 1989 Oct;19(5):433–437. doi: 10.1111/j.1365-2362.1989.tb00255.x. [DOI] [PubMed] [Google Scholar]
- Babirak S. P., Iverius P. H., Fujimoto W. Y., Brunzell J. D. Detection and characterization of the heterozygote state for lipoprotein lipase deficiency. Arteriosclerosis. 1989 May-Jun;9(3):326–334. doi: 10.1161/01.atv.9.3.326. [DOI] [PubMed] [Google Scholar]
- Ben-Avram C. M., Ben-Zeev O., Lee T. D., Haaga K., Shively J. E., Goers J., Pedersen M. E., Reeve J. R., Jr, Schotz M. C. Homology of lipoprotein lipase to pancreatic lipase. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4185–4189. doi: 10.1073/pnas.83.12.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson-Olivecrona G., Olivecrona T. Binding of active and inactive forms of lipoprotein lipase to heparin. Effects of pH. Biochem J. 1985 Mar 1;226(2):409–413. doi: 10.1042/bj2260409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Deeb S. S., Peng R. L. Structure of the human lipoprotein lipase gene. Biochemistry. 1989 May 16;28(10):4131–4135. doi: 10.1021/bi00436a001. [DOI] [PubMed] [Google Scholar]
- Devlin R. H., Deeb S., Brunzell J., Hayden M. R. Partial gene duplication involving exon-Alu interchange results in lipoprotein lipase deficiency. Am J Hum Genet. 1990 Jan;46(1):112–119. [PMC free article] [PubMed] [Google Scholar]
- Enerbäck S., Bjursell G. Genomic organization of the region encoding guinea pig lipoprotein lipase; evidence for exon fusion and unconventional splicing. Gene. 1989 Dec 14;84(2):391–397. doi: 10.1016/0378-1119(89)90513-1. [DOI] [PubMed] [Google Scholar]
- Enerbäck S., Semb H., Bengtsson-Olivecrona G., Carlsson P., Hermansson M. L., Olivecrona T., Bjursell G. Molecular cloning and sequence analysis of cDNA encoding lipoprotein lipase of guinea pig. Gene. 1987;58(1):1–12. doi: 10.1016/0378-1119(87)90023-0. [DOI] [PubMed] [Google Scholar]
- Fojo S. S., Law S. W., Sprecher D. L., Gregg R. E., Baggio G., Brewer H. B., Jr Analysis of the apoC-II gene in apoC-II deficient patients. Biochem Biophys Res Commun. 1984 Oct 15;124(1):308–313. doi: 10.1016/0006-291x(84)90953-7. [DOI] [PubMed] [Google Scholar]
- Havel R. J., Kane J. P., Kashyap M. L. Interchange of apolipoproteins between chylomicrons and high density lipoproteins during alimentary lipemia in man. J Clin Invest. 1973 Jan;52(1):32–38. doi: 10.1172/JCI107171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iverius P. H., Brunzell J. D. Human adipose tissue lipoprotein lipase: changes with feeding and relation to postheparin plasma enzyme. Am J Physiol. 1985 Jul;249(1 Pt 1):E107–E114. doi: 10.1152/ajpendo.1985.249.1.E107. [DOI] [PubMed] [Google Scholar]
- Iverius P. H., Ostlund-Lindqvist A. M. Lipoprotein lipase from bovine milk. Isolation procedure, chemical characterization, and molecular weight analysis. J Biol Chem. 1976 Dec 25;251(24):7791–7795. [PubMed] [Google Scholar]
- Kirchgessner T. G., Chuat J. C., Heinzmann C., Etienne J., Guilhot S., Svenson K., Ameis D., Pilon C., d'Auriol L., Andalibi A. Organization of the human lipoprotein lipase gene and evolution of the lipase gene family. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9647–9651. doi: 10.1073/pnas.86.24.9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchgessner T. G., Svenson K. L., Lusis A. J., Schotz M. C. The sequence of cDNA encoding lipoprotein lipase. A member of a lipase gene family. J Biol Chem. 1987 Jun 25;262(18):8463–8466. [PubMed] [Google Scholar]
- Langlois S., Deeb S., Brunzell J. D., Kastelein J. J., Hayden M. R. A major insertion accounts for a significant proportion of mutations underlying human lipoprotein lipase deficiency. Proc Natl Acad Sci U S A. 1989 Feb;86(3):948–952. doi: 10.1073/pnas.86.3.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinkoth J., Wahl G. Hybridization of nucleic acids immobilized on solid supports. Anal Biochem. 1984 May 1;138(2):267–284. doi: 10.1016/0003-2697(84)90808-x. [DOI] [PubMed] [Google Scholar]
- Osborne J. C., Jr, Bengtsson-Olivecrona G., Lee N. S., Olivecrona T. Studies on inactivation of lipoprotein lipase: role of the dimer to monomer dissociation. Biochemistry. 1985 Sep 24;24(20):5606–5611. doi: 10.1021/bi00341a048. [DOI] [PubMed] [Google Scholar]
- Ostlung-Lindqvist A. M., Boberg J. Purification of salt resistant lipase and lipoprotein lipase from human post-heparin plasma. FEBS Lett. 1977 Nov 15;83(2):231–236. doi: 10.1016/0014-5793(77)81011-9. [DOI] [PubMed] [Google Scholar]
- Rosenthal N. Identification of regulatory elements of cloned genes with functional assays. Methods Enzymol. 1987;152:704–720. doi: 10.1016/0076-6879(87)52075-4. [DOI] [PubMed] [Google Scholar]
- Ross R. S., Hoeg J. M., Higuchi K., Schumacher U. K., Fojo S., Gregg R. E., Brewer H. B., Jr Homozygous hypobetalipoproteinemia: transcriptional regulation and 5'-flanking sequence analysis in an apolipoprotein B deficiency state. Biochim Biophys Acta. 1989 Jul 17;1004(1):29–35. doi: 10.1016/0005-2760(89)90208-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Senda M., Oka K., Brown W. V., Qasba P. K., Furuichi Y. Molecular cloning and sequence of a cDNA coding for bovine lipoprotein lipase. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4369–4373. doi: 10.1073/pnas.84.13.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wion K. L., Kirchgessner T. G., Lusis A. J., Schotz M. C., Lawn R. M. Human lipoprotein lipase complementary DNA sequence. Science. 1987 Mar 27;235(4796):1638–1641. doi: 10.1126/science.3823907. [DOI] [PubMed] [Google Scholar]
- Wong C., Dowling C. E., Saiki R. K., Higuchi R. G., Erlich H. A., Kazazian H. H., Jr Characterization of beta-thalassaemia mutations using direct genomic sequencing of amplified single copy DNA. 1987 Nov 26-Dec 2Nature. 330(6146):384–386. doi: 10.1038/330384a0. [DOI] [PubMed] [Google Scholar]
- Yang C. Y., Gu Z. W., Yang H. X., Rohde M. F., Gotto A. M., Jr, Pownall H. J. Structure of bovine milk lipoprotein lipase. J Biol Chem. 1989 Oct 5;264(28):16822–16827. [PubMed] [Google Scholar]







