Abstract
The TS ribozyme (originally called twister-sister) is a nucleolytic catalytic RNA. We present a crystal structure of the ribozyme in a pre-reactive conformation. Two co-axial helical stacks are organized by a three-way junction and two tertiary contacts. Five divalent metal ions are directly coordinated to RNA ligands, making important contributions to the RNA architecture. The scissile phosphate lies in a quasi-helical loop region that is organized by a network of hydrogen bonding. A divalent metal ion is directly bound to the nucleobase 5' to the scissile phosphate, with an inner-sphere water molecule positioned to interact with the O2' nucleophile. The rate of ribozyme cleavage correlated in a log-linear manner with divalent metal ion pKa, consistent with proton transfer in the transition state, and we propose the bound metal ion is a likely general base for the cleavage reaction. Our data indicate that the TS ribozyme functions predominantly as a metalloenzyme.
Introduction
The nucleolytic ribozymes are a set of diverse catalytic RNA species that cleave a specific phosphodiester linkage within the RNA, in order to process replication intermediates, transcripts or to control gene expression 1,2. Despite having no structural similarity, these ribozymes all achieve a site-specific cleavage reaction by the nucleophilic attack of the O2' on the adjacent phosphate in an SN2 reaction with departure of the O5' oxyanion leaving group, generating cyclic 2',3'-phosphate and 5'-OH products (Supplementary Results, Supplementary Figure 1). In some ribozymes (where the product of cleavage is held in place) the reverse, ligation reaction is also catalyzed 3–6.
In principle these transesterification reactions can be catalyzed by facilitation of in-line nucleophilic attack, by stabilization of the phosphorane transition state, and by using a general base and acid to deprotonate the nucleophile and protonate the oxyanion leaving-group respectively. In RNA (in contrast to protein enzymes) the variety of groups that might participate in the reactions is quite limited, restricted to the nucleobases, hydrated metal ions and the 2'-hydroxyl group. Nucleobase-mediated general acid-base catalysis is very common in the nucleolytic ribozymes, but not universal as exemplified by the roles of a metal ion in the HDV ribozyme 7,8, a 2'-hydroxyl group in the hammerhead ribozyme 9,10, and bound glucosamine-6-phosphate in GlmS 11–13. Interestingly, almost all the known ribozymes exhibit mechanistic differences (perhaps with the exception of the similarity between the hairpin and VS ribozyme mechanisms 14), and each new ribozyme provides a fresh challenge to understand its mechanism.
After the initial discovery of a number of self-cleaving ribozymes in the 1980s and 90s, the first decade of the current millennium saw only one new ribozyme identified, GlmS 15, although it was found that well-studied ribozymes like the hammerhead and HDV were much more widespread than previously supposed 16–19. However, recently bioinformatic methods have been used to search for new classes of structured RNA in the vicinity of known ribozymes and protein-coding sequences in bacterial genomes. This has led to the identification of four new nucleolytic ribozymes 20,21.
The first of these to be discovered was the twister ribozyme 20, another nucleolytic ribozyme widespread in bacterial and eukaryotic genomes. We 22 and others 23,24 have recently solved the crystal structure of the twister ribozyme. A more recently-discovered ribozyme 21 was proposed to resemble twister in terms of the connectivity of helices and the sequence of a conserved loop, and thus called twister-sister. However this name is misleading both from structural and mechanistic points of view, and we shall subsequently refer to this as the TS ribozyme. Like twister, the TS ribozyme is found in a variety of forms with or without some of the helices (Figure 1A, Supplementary Figure 2). Eleven very highly conserved nucleotides (> 97%) clustered within loops L1 and L4 of the TS ribozyme sequence were identified 21, and a further 3 nucleotides that are > 90% conserved (Figure 1A). They also showed that in the TS-1 ribozyme site-specific cleavage generated the standard cyclic 2',3' phosphate and 5'-OH products, and that the reaction rate increased with pH, with a very steep dependence on Mg2+ ion concentration. Mg2+ ions could be substituted by Ca2+ (but activity in Sr2+ was low) and some divalent transition metal ions such as Co2+ and Ni2+. However, no reaction occurred in 1M monovalent ions except for weak activity in Li+, and there was essentially no activity in Co(NH3)6 (III) ions to 10 mM. These properties suggest a possible direct role for a hydrated metal ion in the cleavage reaction.
Figure 1. The overall structure of the TS ribozyme.
A. The sequence and secondary structure of the RNA. Nucleotides conserved >97% (ref. 21) are colored red. The helical (P) and loop (L) sections are labeled, and the two tertiary interactions are indicated by the lines labeled T1 and T2. The helices are colored, and this scheme is used throughout.
B. A schematic model of structure observed in the crystal. The overall structure is formed from two coaxially aligned assemblies of helices, comprising P1-L1-P2-T2 and P4-P5 connected by the three-way helical junction via A15 and C47. The terminal loop of L4 makes the T1 tertiary interaction involving A8. Loop L1 is quasi-helical and includes the scissile phosphate indicated by the red circle.
C. A parallel-eye stereoscopic view of complete ribozyme structure. The nucleotides are shown in cartoon form, with a ribbon tracing the pathway of the phosphodiester backbone through the structure. Metal ions are shown as green spheres with inner-sphere water molecules as red spheres. The scissile phosphate (P) is labeled in red, and the flanking nucleotides (C54 and A55) colored cyan.
The maximum cleavage rate of TS-1 was relatively slow, with the rate achieving a rather abrupt maximum as a function of Mg2+ ion concentration. This suggests that the rate might be limited by conformation. However, some variation was noted amongst the different TS ribozyme isolates. TS-3 (Supplementary Figure 2), like TS-1, contains a four-way junction including helices P3 and P5, but exhibited higher rates of cleavage, that increased with pH over the whole range up to 8.0. TS-4 was found to be still faster, achieving near-complete cleavage in less than a minute. We have therefore chosen to study the TS-4 ribozyme sequence, and have crystallized and solved the molecular structure of this ribozyme in a likely pre-catalytic conformation. Furthermore, mechanistic studies are consistent with the direct participation in the cleavage reaction of a hydrated Mg2+ ion observed in the structure.
Results
Crystallization and overall structure of the TS ribozyme
TS ribozyme used for crystallization was a two-piece RNA construct that is a shortened version of TS-4 (Supplementary Fig. 3)21. It contained 2'H substitution at C54 to prevent cleavage and two bromocytosine substitutions in P5 to provide phase information. The RNA crystallised in tetragonal space group P41212, and data were collected to a resolution of 2.0 Å (Supplementary Table 1). The bromine atoms were located using single-wavelength anomalous diffraction, from which models of the complete structure were then built and refined. The quality of the electron density map is very high, with all sections of the RNA and bound hydrated metal ions well defined except for U56 (Supplementary Figures 4-8).
The overall fold of the TS ribozyme is based upon two coaxially-stacked helical sections, connected by a three-way helical junction and the formation of two tertiary contacts (Figure 1). Loop L1 containing the scissile phosphate in the cleavage reaction adopts a quasi-helical character (see below), and is located in a continuous coaxial stack in between P1 and P2. The P1-L1-P2 stack is capped off by the single Watson-Crick basepair C14-G27 (part of the T2 tertiary interaction). P4 and P5 also form a coaxially-stacked pair of helices, and are connected to the P1-L1-P2-T2 stack by the three-way junction, with A15 (stacked onto the T2 basepair) and C47 forming the connecting sections of the junction. The folding of the ribozyme brings L4 into proximity with L1 resulting in the formation of the A8-G23 base pair (the T1 tertiary interaction).
Seven hydrated metal ions are present in the structure (Supplementary Table 2; Supplementary Figure 8). These are assigned as Mg2+ ions, with octahedral coordination and metal-O distances of 2.2 Å. Two are outer sphere complexes, while the remaining ions have exchanged one (three ions) or two (two ions) inner-sphere water molecules with RNA ligands. All but one are non-bridging phosphate O atoms. In general one or more water ligands are H-bonded to RNA. The environment of specific ions will be discussed in context below.
The L4 terminal loop forms key tertiary contacts
Nucleotides of the L4 terminal loop are in involved in both of the tertiary contacts stabilizing the fold of the ribozyme. Four of the nine nucleotides assigned to L4 (ref. 21) form the two terminal base pairs of P4; the invariant nucleotides G21 and C29 form a Watson-Crick base pair and the highly conserved (>95 %) nucleotides U22 and A26 form a trans Watson-Crick – Hoogsteen base pair (Figure 2A). The nucleotides G23, C24 and A25 participate in the T1 tertiary interaction. These nucleotides form a receptor for A8 that is extruded from L1, the four nucleotides adopting a conformation similar to a GNRA tetraloop. A8 is stacked between A25 and A26, and its N3 accepts a H-bond from G23 N2. This interaction is supported by additional H-bonds from G23 O2' to A25 N7 and A8 O2' to A25 N3 (Figure 2A, Supplementary Figure 5).
Figure 2. Close view of special features of the TS ribozyme structure.
Parallel-eye stereoscopic views are shown in cartoon form for each. Key nucleotides are shown in stick form. H-bonds are indicated by broken lines.
A. The terminal loop of L4 and its T1 tertiary interaction with A8. Note the H-bonding network in L4, and the interaction between A8 and G23.
B. The T2 tertiary interaction. G27 is looped out from P4, and H-bonds with C14. Hydrated metal ions are not shown in this view for clarity.
C. The three-way helical junction. Metal ions are shown as green spheres, with inner-sphere water molecules as red spheres. Note that the view shown here is of the side opposite to that shown in part B.
Bioinformatic data 21 and atomic mutagenesis (Supplementary Table 3) support the maintenance and importance of this structure in the active ribozyme. C24 is in an open position at the lower end of the loop, making no intramolecular interactions; substitution by zebularine (C24Z), which lacks the exocyclic N4 amine of cytosine, led to no loss of activity and all four nucleotides occupy position 24 in the alignment of TS sequences. A25 accepts hydrogen bonds at N7 and N3. While substitution by either purine (A25P) or inosine (A25I) resulted in rather small reductions of activity, substitution of N7 by CH (A25N7C) led to a 20-fold loss of activity (Supplementary Table 3). G23 is invariant, yet a 2-aminopurine substitution that retains the ability to donate a H-bond to A8 had a minimal effect on activity. Adenine is the predominant nucleotide at position 8 in the alignment of TS sequences; C and U are also found, but G is never observed at that position. The O2 of the pyrimidines could accept a H-bond from G23 whereas substitution of A8 with a G would lead to a steric clash with G23.
Between A26 and C29 a loop containing G27 and C28 extrudes from P4 (although formally part of L4), and G27 pairs with C14 to make the T2 tertiary interaction (Figure 2B, Supplementary Figure 6). This base pair is fully conserved amongst those TS ribozymes that have a three-way junction. In order to allow G27 to make an antiparallel Watson-Crick basepair the looped strand must make a 360° turn before re-entering the P4 helix, and this is stabilized by several structural features. C28 is directed into the center of the ribozyme, where its N3 and N4 are H-bonded to two inner-sphere water molecules of Mg2+ ion M3. This ion is directly coordinated to the proR non-bridging O atoms of A9 and G10, contributing to the interaction between P2 and L4. C28 is also fully conserved in the three-way junction form of the ribozyme. Mg2+ ions M2 and M5 bind in the major groove of P4 and are directly coordinated to the proR non-bridging O atom of G27 and the proS non-bridging O atoms of C28, respectively. M2 bridges the major groove, with inner-sphere water molecules H-bonding with C20 and the invariant G21. It also helps stabilize the reversal of the backbone with another inner-sphere water donating an H-bond to the proR non-bridging O atom of C28. The proR O of C28 also accepts a H-bond from N4 of the invariant C29. The major groove interactions of the G21-C29 base pair presumably account for the conservation of sequence. M5 also interacts with the backbone by inner-sphere water molecules bonding to the Hoogsteen edge of G30 and G31, and contributes to the interaction between P2 and P4 with another inner-sphere water molecule donating a H-bond to the proR non-bridging O atom of G48.
The three-way helical junction
The 2HS1HS2 (ref. 25) three-way helical junction connects the T2 end of the P1-L1-P2-T2 stack with the P4-P5 stack, with the connecting strands entering and leaving in between the P4 and P5 helices (Figure 2C). The strand running from the P2-T2 helix passes into the P4 helix via A15, which is stacked onto the T2 basepair. That running from P5 into T2 makes a tight turn at C47, which is stacked onto the ribose of G27. Both A15 and C47 are very highly conserved in TS ribozymes with a three-way junction 21. Two metal ions are bound close to the turn at C47 (Supplementary Figure 8). The first (M5) is described above. The second (M6) is located in the major groove of P4, with a complete octahedral inner coordination sphere of water, two of which are H-bonded to the proS phosphate oxygen of G48. The axes of the P1-L1-P2-T2 and P4-P5 stacks are inclined at an angle of 40° to each other.
Loop L1 and the environment around the scissile phosphate
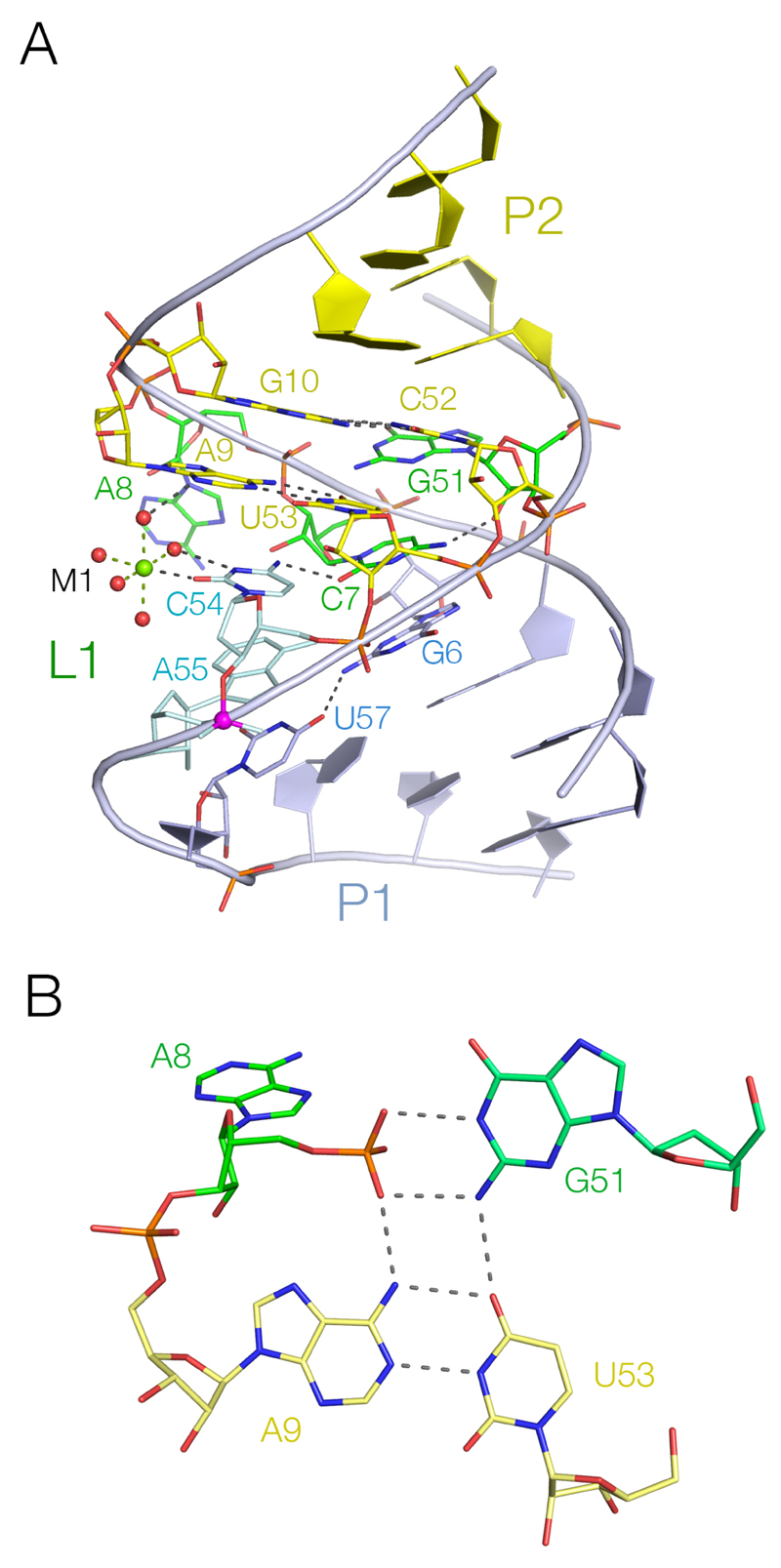
The L1 loop adopts a quasi-helical conformation (Figure 3A), presenting a very open minor groove facing the turn of the L4 loop. This is widened by a compression of the adjacent major groove (P5-P52 = 5.1 Å), stabilized by metal ion M4 (Supplementary Figure 8) that is directly bonded to non-bridging phosphate O atoms of C52 and G5 thus bridging the opposing strands, together with additional interactions with both strands through inner-sphere water molecules. Parts of L1 are basepaired and assigned to the flanking helices (Figure 3A, Supplementary Figure 7). At the lower end G6 and U57 are coplanar and G6 N2 donates a hydrogen bond to U57 O4; it is assigned to P1. No density for U56 was observed, and this base is probably extrahelical and mobile. Deletion of this nucleotide (U56∆) resulted in only a minor loss of activity (Supplementary Table 3), consistent with the observation that most TS ribozymes only have two nucleotides between the scissile phosphate and P1 whereas TS-4 has three. At the upper end of the loop A9 and U53 are basepaired, as are G10 and C52, and therefore assigned to P2. The A9-U53 basepair forms a triple base interaction with G51, with a hydrogen bond donated from G51 N2 to U53 O4. Both non-bridging O atoms of the phosphate connecting C7 and A8 are H-bonded to G51 and the proR O atom is H-bonded to A9 N6 generating a square array of H-bonds (Figure 3B) in an apparently very stable conformation. This could be regarded as the scaffold that supports the structure of the L1 loop and the active center. All three nucleobases participating in this interaction are fully conserved.
Figure 3. Loop L1 is quasi-helical, and coaxial with P1 and P2.
A. A view of L1, with key nucleotides shown in stick form. The scissile phosphate is colored magenta. Note the extensive H-bonding network involving C54, C7, G51 and A9. Metal ion M1 is directly coordinated to C54 O2, and inner-sphere water molecules are H-bonded to C54 and A9.
B. The network of H-bonding interactions between A9-U53-G51-P8.
The two nucleotides 5' of A9 are also important; A8 is extruded from the helix to make the T1 tertiary contact with L4 discussed above, while C7, which is also invariant, is stacked on G6 and makes H-bonds to G51 O2' (from N4), C54 N4 (from O2) and A55 N6 (from O2). Ribozyme activity is particularly sensitive to atomic substitution at the nucleobase of C7 (Supplementary Table 3). No cleavage activity was detectable with uracil (C7U) or zebularine (C7Z) substitutions, and 5-fluoroC (C7 5F) also exhibited markedly reduced activity. These substitutions result in the loss or weakening of the H-bond to G51 O2', and a deoxyribose substitution at G51 was also highly deleterious. C7 and C54 form a trans basepair, with C54 stacked under the A9-U53 pair. However uridine is often found at position 54, and C54 can be substituted by uridine (C54U) without loss of activity (Supplementary Table 3), which suggests that the bond between C7 and C54 may not be important.
A Mg2+ ion (M1) is directly coordinated to C54 O2 in the minor groove (Figure 3A, Supplementary Figure 8) and an inner-sphere water of hydration is H-bonded to N3 of C54. Uridine at position 54 could make equivalent interactions. A second inner-sphere water molecule donates an H-bond to A9 N3. To assess the importance of this bond, we made an A9 3-deaza substitution (A9N3C), resulting in a cleavage rate lowered by three orders of magnitude (Supplementary Table 3). This substitution required the use of a simultaneous O2'H modification, but the corresponding single O2'H substitution led to only a minor loss of activity.
The scissile phosphate lies between C54 and A55. The two nucleobases are stacked, and both directed towards the axis. Thus the local geometry is far from the in-line conformation that would be required for nucleophilic attack of the C54 O2' atom on the phosphate, and plainly some conformational change would be required to generate the active conformation. While C54 is held in place by its interaction with M1 and C7, there are no H-bonds holding the nucleobase of A55 in place. It is therefore probable that A55 rotates to generate the required in-line trajectory. Moreover A55 exhibits an elevated flexibility in the crystal, suggesting a low barrier to local rearrangement. Whereas the overall B-factor of the ribozyme and that for C54 are both ~30, that of A55 is > 60. Substitution of A55 by 2'-deoxyA (AO2'H), or N3-deazaA (A55N3C) led to substantial reductions in activity (Supplementary Table 3); these effects are not explicable in terms of the structure observed in the crystal, consistent with a rearranged structure in the active conformation.
The role of hydrated metal ions in the cleavage reaction
The TS ribozyme was found to exhibit no cleavage activity in monovalent ions, aside from a low level in 1M Li+ ions, and very weak activity in Co(NH3)63+ ions 26. We have explored the activity of our construct as a function of the ionic composition of the medium (Figure 4A,B, Supplementary Table 3). While the ribozyme was active in divalent metal ions such as Mg2+ and Mn2+, activity in 1 M Li+ or 1 mM Co(NH3)63+ ions was extremely low (kobs = 5.7x10-5 and <1x10-5 s-1 respectively).
Figure 4. The effect of metal ions on cleavage rates by the TS ribozyme.
A. Cleavage reaction progress in the presence of Mg2+ (filled circles) and Mn2+ (open squares) ions. The data have been fitted to exponential functions. Supplementary Figure 12 shows an example of the separation of substrate and product by gel electrophoresis at various times in the presence of Mg2+ ions.
B. A plot of the logarithm of the cleavage rate (taken from Supplementary Table 3) as a function of the pKa values for the divalent metal ions present in the reaction 57. These data have been fitted to a straight line (gradient = -1.26, R = 0.965). Three (Mn2+and Ca2+) or four (Mg2+ and Sr2+) independent measurements were made, and the error bars show twice the standard deviation.
This suggests a direct role for a water molecule bound to a divalent metal ion in the cleavage reaction. In our crystal structure an octahedral metal ion (M1) is directly bound to C54 O2 (Figure 3A, Supplementary Figure 8). One of the inner-sphere water molecules is relatively close to where the C54 O2' would be if it were present (i.e. the nucleophile of the transesterification reaction) (Figure 5), so this could conceivably act as a general base in the cleavage reaction. We have measured the rate of cleavage in the presence of a series of different divalent metal ions, and find a linear correlation between cleavage rate and the pKa of the hydrated metal ion (Figure 4C). This provides evidence for proton transfer involving metal ion-bound water in the reaction, consistent with a role in general acid-base catalysis.
Figure 5. The hydrated metal ion M1 is bound to C54 in proximity to the nucleophile of the cleavage reaction.
A view of the interactions in loop 1, with an oxygen atom modeled on to C54 (red sphere) at the 2' position, i.e. the nucleophile in the cleavage reaction that was absent in the construct used for crystallography. This O2' lies 3.8 Å from one of the inner sphere hydration water molecules of M1 (denoted by broken red line). H-bonds linking A9-M1-C54-C7-G51 are indicated. The nucleobases of C52, U53 and G10 are not displayed to provide an unobstructed view of the key interactions in this region.
Discussion
The TS ribozyme was named twister-sister 21 because it was thought to resemble the twister ribozyme. The connectivity of the helices is strikingly similar, shown schematically in Supplementary Figure 9 where corresponding helices are colored the same. The structures are compared in Supplementary Figure 10. The structure of L4 is also very similar (Supplementary Figure 11). In both ribozymes the nucleotides of L4 form the terminal two base pairs of P4; a trans Watson Crick-Hoogsteen pair (U22-A26 in TS) and a cis Watson Crick pair (G21-C29). This latter pair also makes the same major groove interactions in both ribozymes. Two nucleotides are extruded between these two base pairs to form the T2 interaction (G27 and C28) and the nucleotides (G23-A25) between those that form the closing base pair comprise the terminal loop and participate in the T1 interaction. Yet these ribozymes exhibit substantial differences so that the name “twister-sister” is misleading. The scissile phosphate is located near the 5' end in twister, but near the 3' end of the TS ribozyme, and the mechanisms of catalysis are clearly different (see below).
Furthermore, there is a more intimate association between L1 and L4 in the twister ribozyme, where the four base pairs comprising T1 form a helix that is located between and coaxial with P1 and P2 (ref. 22–24). In the TS ribozyme, L1 is quasi-helical and stacked between P1 and P2. Thus the structure in the crystal may represent a pre-reaction conformation that must undergo substantial remodeling to form the active structure. Some reorganization must occur because the conformation around the scissile P is far from in-line and not poised for nucleophilic attack. Given the flexibility of A55 in the crystal and the sensitivities of the cleavage reaction to substitutions at A55 that should not affect the structure observed in the crystal, this nucleotide may undergo substantial displacement to form the active structure. Other nucleotides in L1 may also relocate to some extent. The substantial compression of the major groove stabilized by metal ion M4 (Supplementary Figure 8) may arise in part from crystal packing, and its relaxation may reposition nucleotides in the lower half of L1. Yet the remaining effects of atomic mutagenesis are consistent with the crystal structure, suggesting that remodeling of the structure is unlikely beyond a conformational change in L1. A small reorientation of the two helical stacks may occur, bringing L4 closer to L1. This may result in the Watson-Crick edge of the highly conserved A25, which is directed towards L1, making an interaction not observed in the crystal.
Considerable evidence points to the involvement of divalent metal ions in the function of the TS ribozyme. TS activity exhibits a strong dependence on Mg2+ ion concentration below 1 mM, with the plot of rate versus log[MgCl2] having a slope of ~3 (ref. 21). This indicates highly cooperative Mg2+ ion-dependent folding of the ribozyme. Although this does not necessarily require specific Mg2+ ion binding, it seems likely in this case. In particular, we hypothesize that the Mg2+ ions that stabilize the T2 interaction (M2, M3 and M5) could bind cooperatively.
A metal ion is a probable participant in the catalytic process. The TS ribozyme exhibits very weak activity in the absence of divalent metal ions. This is similar to the HDV ribozyme 7,27, which utilizes a bound Mg2+ ion to activate the nucleophile7,8,28–30. Linear correlation between ribozyme cleavage rate and the pKa of the metal ion (Figure 4C) is good evidence for proton transfer involving metal ion-bound water occurring in the reaction. A similar relationship has been observed for the hammerhead ribozyme 31; in this case the metal ion is thought to activate a 2'-hydroxyl group 9,10,32, lowering its pKa so that it might better function as a proton donor to the 5'-oxyanion leaving group. However, the hammerhead ribozyme is more active in monovalent cations than the HDV ribozyme because the 2'-hydroxyl group is still able to function as a general acid, albeit more weakly. Thus the properties of the TS ribozyme are more consistent with the direct participation of a hydrated metal ion in the cleavage reaction as a general base rather than metal ion activation of a 2'-hydroxyl as a general acid. Furthermore, there are no 2'-hydroxyl groups close to the scissile phosphate that could function as a general acid, the closest being that of C7 at a distance of 11.7 Å.
While the kinetic data support the direct participation of a hydrated divalent cation in the catalysis they cannot distinguish between activation of the nucleophile as a general base or protonation of the leaving group. Mg2+ ion M1 is bound at the active site in the crystal structure with one of its inner-sphere water molecules only 3.8 Å from the modeled position of the O2' nucleophile (Figure 5). Disruption of the bonding of M1 to A9 N3 led to a 1000-fold loss of activity, supporting a critical role for this cation. The simplest explanation consistent with structural and kinetic data is nucleophile activation by the ion acting as a general base.
The high rate of TS-catalyzed cleavage strongly indicates that a general acid is contributing to catalysis, but its identity is presently unknown. If a nucleobase fulfills this role it would likely be highly conserved, and only C7 and A25 are close to the active site and have ring nitrogen atoms free to undergo proton transfer. The ribozyme is very sensitive to changes at C7, yet for this nucleobase to participate it would have to rotate towards the scissile phosphate, which would break the H-bond from C7 N4 to G51 O2'. Substitutions at C7 and G51 are consistent with this bond remaining intact in the active ribozyme. However, it is possible that the loss of activity arising from a deoxyribose substitution at G51 arises solely from the loss of the H-bond donated by G51 O2' to the proR O of G6. Thus our data do not eliminate C7 as a general acid. Substitution of purine or inosine at position 25 had little effect on activity. Compared to adenine, these nucleobases have a substantially lower and higher pKa, respectively, and thus a greater effect would be expected if A25 were the general acid. A substantial displacement of L4 towards L1 would also be required. A55, immediately 3' to the scissile phosphate, could act as a general acid through its N3 in a manner analogous to A1 in the twister ribozyme 33. A55 is almost as highly conserved as C7 and A25, and atomic mutagenesis of N3 resulted in a substantial reduction in activity. However the effect is much smaller than the 105-fold reduction observed for twister and most probably arises from structural destabilization. Apart from the nucleobases, the other possible general acids are a 2'-hydroxyl group, which seems unlikely (see above), and a hydrated metal ion. This could be the same metal ion that acts as a general base or a second ion not present in the crystal structure.
In general, ribozymes employ two catalytic strategies to catalyze phosphoryl transfer reactions 34. The self-splicing introns 35–38 and RNaseP 39,40 bind metal ions that directly participate in the reaction chemistry. By contrast, the nucleolytic ribozymes all use general acid-base catalysis. In most cases the catalytic participants are nucleobases, with guanine being the most common general base 41–49. The main exception is the HDV ribozyme, where a hydrated metal ion is the probable general base 7,8,28–30. We have now shown that the TS ribozyme makes extensive use of metal ions in both structural and catalytic roles. We observe a divalent metal ion bound in the active site, with a coordinated water molecule well positioned to act as the general base in cleavage. This is consistent with our mechanistic data. This contrasts with the twister ribozyme, where the nucleobases of adenine and guanine function as general acid and base, and there is no direct role for a metal ion in catalysis 22,33. The identity of a general acid in the TS ribozyme is unclear at present, but it remains possible this is also the same or a different hydrated metal ion. Each new nucleolytic ribozyme that is discovered provides a fresh perspective on RNA catalysis, and broadens their range of catalytic strategies.
On-Line Methods
RNA synthesis
Oligonucleotides were synthesized using t-BDMS phosphoramidite chemistry 50 as described in Wilson et al. 51, implemented on an Applied Biosystems 394DNA/RNA synthesizer. RNA was synthesized using UltraMILD ribonucleotide phosphoramidites with 2'O-tert-butyldimethyl-silyl (t-BDMS) protection 52,53 (Link Technologies). Oligoribonucleotides containing 5-bromocytidine (ChemGenes corp.) were deprotected using anhydrous 2 M ammonia in methanol (Sigma-Aldrich) for 36 h. Unmodified sequences and oligonucleotides incorporating other modifications were deprotected in either 25% ethanol/ammonia solution at room temperature for 3 h. or in 1:1 ammonia/methylamine at 60°C for 20 min. according to the manufacturer’s recommendations, and evaporated to dryness. All oligoribonucleotides were redissolved in 115 μL of anhydrous DMSO and 125 μL triethylamine trihyrofluoride (Aldrich) to remove t-BDMS groups, and agitated at 65°C in the dark for 2.5 h prior to butanol precipitation. Full-length RNA was then purified as described previously 33.
Crystallization
The construct used for crystallization comprised two strands of sequence (all sequences written 5' to 3'): GCAGGGCAAGGCCCAGUCCCGUGCAAGCCGGGACCGBrCCBrCC and GGGGCGCGGCGCUdCAUUCCUGC (Supplementary Figure 3A). 2'H substitution at C54 was included to prevent ribozyme cleavage, and 5-bromocytosine substitution at C37 and C39 to provide phase information. Purified RNA was annealed by slow cooling from 60°C, and its concentration was adjusted to 0.1 - 0.2 mM before mixing with an equal volume of 40 mM Mg acetate, 50 mM Na cacodylate (pH 6.0) and 30% MPD. This was sealed with 0.5 ml mother liquid at 16°C in hanging drop mode and crystals of 20 x 20 x 50 μm of space group P41212 grew in two weeks. Crystals were flash frozen and stored in liquid nitrogen.
Data collection and structure determination
A 2.0 Å data set at the Br absorption K edge was acquired at the European Synchrotron Radiation Facility (Grenoble) beamline ID23-1. Data sets were processed using the CCP4 suite 54. The asymmetric unit contains a single ribozyme molecule. Initial phases were acquired from the SAD data by locating the two bromine atoms with Autosol in the PHENIX suite 55. The initial model was generated automatically by PHENIX autobuild wizard, and refined in multiple rounds of manual model building in Coot 56 and refinement in Phenix.refine. The quality of the electron density map was very high and all nucleotides except U56 could be placed with confidence, together with a number of Mg2+ ions. Crystallographic statistics are shown in Supplementary Table 1. Debye-Waller B factors were calculated as B = 8π2 <u2>, where u is the mean atomic displacement. Coordinates have been deposited in the PDB with accession number 5T5A.
Analysis of ribozyme kinetics
The TS-4 ribozyme 21 used for kinetic analysis is assembled from two oligonucleotides (Suppelementary Figure 3), termed ribozyme and substrate: ribozyme (47 nt) : 5' UUUCGCAGGGCAAGGCCCAGUCCCGUGCAAGCCGGGACCGCCCCUUC substrate (29 nt) : 5' GAGGGGGCGCGGCGCUCAUUCCUGCGAAA These were made by chemical synthesis, incorporating modified nucleosides as required. Cleavage kinetics were studied under single-turnover conditions. Ribozyme and radioactively [5'-32P]-labeled substrate were annealed in 100 mM KCl by rapid cooling (0.2 deg. per second) from 80°C to 25°C in a thermal cycler. Aliquots of annealed TS-4 were equilibrated to 25°C and the reaction initiated by adding an equal volume of equilibrated 2x reaction buffer. The final reaction contained 1 µM ribozyme and 10 nM substrate in 30 mM HEPES (pH 7.0), 200 µM MgCl2, 100 mM KCl. In some experiments the MgCl2 was replaced with the same concentration of the chloride salt of other divalent cations or with 1 mM Co(NH3)6Cl3, as detailed in the text. Activity in monovalent ions was investigated using 25 mM HEPES (pH 7.0), 1.0 M LiCl, 100 mM EDTA, the latter to sequester any divalent ions that may be present. Slow reactions requiring long incubations were carried out under mineral oil to prevent evaporation. 2 µl aliquots were removed at intervals and the reaction terminated by addition to 13 µl of a mixture containing 95% (v/v) formamide, 50 mM EDTA and electrophoresis dyes. Substrate and product were separated by electrophoresis in 20% polyacrylamide gels containing 7 M urea and quantified by phosphorimaging. Progress curves were fitted by non-linear regression analysis to single or double exponential functions using Kaleidagraph (Abelbeck Software).
Data Availability
Structural data have been deposited in the Protein Data Bank (PDB) with coordinate accession number 5T5A. All other data generated or analysed during this study are included in this published article (and its supplementary information files) or are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgements
We thank Saira Ashraf for expert synthesis of RNA, the CRUK for program support A18604 (to DMJL), the Wellcome Trust for the in-house diffractometer and ESRF for synchrotron beam time.
Footnotes
Author Contributions
Y.L. performed crystallography, T.J.W. performed mechanistic investigations and Y.L., T.J.W. and D.M.J.L. designed the research, analyzed data and wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests
References
- 1.Lilley DMJ, Eckstein F, editors. Ribozymes and RNA Catalysis. Royal Soc Chemistry; Cambridge: 2008. pp. 1–318. [Google Scholar]
- 2.Wilson TJ, Lilley DMJ. The evolution of ribozyme chemistry. Science. 2009;323:1436–1438. doi: 10.1126/science.1169231. [DOI] [PubMed] [Google Scholar]
- 3.Fedor MJ. Tertiary structure stabilization promotes hairpin ribozyme ligation. Biochemistry. 1999;38:11040–11050. doi: 10.1021/bi991069q. [DOI] [PubMed] [Google Scholar]
- 4.McLeod AC, Lilley DM. Efficient, pH-dependent RNA ligation by the VS ribozyme in trans. Biochemistry. 2004;43:1118–1125. doi: 10.1021/bi035790e. [DOI] [PubMed] [Google Scholar]
- 5.Nahas MK, et al. Observation of internal cleavage and ligation reactions of a ribozyme. Nature structural & molecular biology. 2004;11:1107–1113. doi: 10.1038/nsmb842. [DOI] [PubMed] [Google Scholar]
- 6.Canny MD, Jucker FM, Pardi A. Efficient ligation of the Schistosoma hammerhead ribozyme. Biochemistry. 2007;46:3826–3834. doi: 10.1021/bi062077r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakano S, Proctor DJ, Bevilacqua PC. Mechanistic characterization of the HDV genomic ribozyme: assessing the catalytic and structural contributions of divalent metal ions within a multichannel reaction mechanism. Biochemistry. 2001;40:12022–12038. doi: 10.1021/bi011253n. [DOI] [PubMed] [Google Scholar]
- 8.Ke A, Zhou K, Ding F, Cate JH, Doudna JA. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature. 2004;429:201–205. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- 9.Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas JM, Perrin DM. Probing general acid catalysis in the hammerhead ribozyme. J Amer Chem Soc. 2009;131:1135–1143. doi: 10.1021/ja807790e. [DOI] [PubMed] [Google Scholar]
- 11.Klein DJ, Ferré-D'Amaré AR. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- 12.Cochrane JC, Lipchock SV, Strobel SA. Structural investigation of the GlmS ribozyme bound to its catalytic cofactor. Chem Biol. 2007;14:97–105. doi: 10.1016/j.chembiol.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cochrane JC, Lipchock SV, Smith KD, Strobel SA. Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme. Biochemistry. 2009;48:3239–3246. doi: 10.1021/bi802069p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson TJ, Lilley DMJ. Do the hairpin and VS ribozymes share a common catalytic mechanism based on general acid-base catalysis? A critical assessment of available experimental data. RNA. 2011;17:213–221. doi: 10.1261/rna.2473711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 16.Przybilski R, et al. Functional hammerhead ribozymes naturally encoded in the genome of Arabidopsis thaliana. Plant Cell. 2005;17:1877–1885. doi: 10.1105/tpc.105.032730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salehi-Ashtiani K, Luptak A, Litovchick A, Szostak JW. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science. 2006;313:1788–1792. doi: 10.1126/science.1129308. [DOI] [PubMed] [Google Scholar]
- 18.Martick M, Horan LH, Noller HF, Scott WG. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature. 2008;454:899–902. doi: 10.1038/nature07117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Webb CH, Riccitelli NJ, Ruminski DJ, Luptak A. Widespread occurrence of self-cleaving ribozymes. Science. 2009;326:953. doi: 10.1126/science.1178084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roth A, et al. A widespread self-cleaving ribozyme class is revealed by bioinformatics. Nature chemical biology. 2014;10:56–60. doi: 10.1038/nchembio.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weinberg Z, et al. New classes of self-cleaving ribozymes revealed by comparative genomics analysis. Nature Chem Biol. 2015;11:606–610. doi: 10.1038/nchembio.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Wilson TJ, McPhee SA, Lilley DM. Crystal structure and mechanistic investigation of the twister ribozyme. Nature Chem Biol. 2014;10:739–744. doi: 10.1038/nchembio.1587. [DOI] [PubMed] [Google Scholar]
- 23.Eiler D, Wang J, Steitz TA. Structural basis for the fast self-cleavage reaction catalyzed by the twister ribozyme. Proc Natl Acad Sci USA. 2014;111:13028–13033. doi: 10.1073/pnas.1414571111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ren A, et al. In-line alignment and Mg2+ coordination at the cleavage site of the env22 twister ribozyme. Nature Comm. 2014;5:5534. doi: 10.1038/ncomms6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lilley DMJ, et al. Nomenclature Committee of the International Union of Biochemistry: A nomenclature of junctions and branchpoints in nucleic acids. Recommendations 1994. Eur J Biochem. 1995;230:1–2. doi: 10.1111/j.1432-1033.1995.tb20526.x. [DOI] [PubMed] [Google Scholar]
- 26.Weinberg Z, et al. New classes of self-cleaving ribozymes revealed by comparative genomics analysis. Nature Chem Biol. 2015;11:606–610. doi: 10.1038/nchembio.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murray JB, Seyhan AA, Walter NG, Burke JM, Scott WG. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem & Biol. 1998;5:587–595. doi: 10.1016/s1074-5521(98)90116-8. [DOI] [PubMed] [Google Scholar]
- 28.Das SR, Piccirilli JA. General acid catalysis by the hepatitis delta virus ribozyme. Nature Chem Biol. 2005;1:45–52. doi: 10.1038/nchembio703. [DOI] [PubMed] [Google Scholar]
- 29.Chen JH, et al. A 1.9 A crystal structure of the HDV ribozyme precleavage suggests both Lewis acid and general acid mechanisms contribute to phosphodiester cleavage. Biochemistry. 2010;49:6508–6518. doi: 10.1021/bi100670p. [DOI] [PubMed] [Google Scholar]
- 30.Chen J, et al. Identification of the catalytic Mg2+ ion in the hepatitis delta virus ribozyme. Biochemistry. 2013;52:557–567. doi: 10.1021/bi3013092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boots JL, Canny MD, Azimi E, Pardi A. Metal ion specificities for folding and cleavage activity in the Schistosoma hammerhead ribozyme. RNA. 2008;14:2212–2222. doi: 10.1261/rna.1010808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee TS, et al. Role of Mg2+ in hammerhead ribozyme catalysis from molecular simulation. J Amer Chem Soc. 2008;130:3053–3064. doi: 10.1021/ja076529e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson TJ, Liu Y, Domnick C, Kath-Schorr S, Lilley DM. The novel chemical mechanism of the twister ribozyme. J Amer Chem Soc. 2016;138:6151–6162. doi: 10.1021/jacs.5b11791. [DOI] [PubMed] [Google Scholar]
- 34.Wilson TJ, Lilley DMJ. The evolution of ribozyme chemistry. Science. 2009;323:1436–1438. doi: 10.1126/science.1169231. [DOI] [PubMed] [Google Scholar]
- 35.Shan S, Kravchuk AV, Piccirilli JA, Herschlag D. Defining the catalytic metal ion interactions in the Tetrahymena ribozyme reaction. Biochemistry. 2001;40:5161–5171. doi: 10.1021/bi002887h. [DOI] [PubMed] [Google Scholar]
- 36.Stahley MR, Strobel SA. Structural evidence for a two-metal-ion mechanism of group I intron splicing. Science. 2005;309:1587–1590. doi: 10.1126/science.1114994. [DOI] [PubMed] [Google Scholar]
- 37.Forconi M, Lee J, Lee JK, Piccirilli JA, Herschlag D. Functional identification of ligands for a catalytic metal ion in group I introns. Biochemistry. 2008;47:6883–6894. doi: 10.1021/bi800519a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frederiksen JK, Li NS, Das R, Herschlag D, Piccirilli JA. Metal-ion rescue revisited: biochemical detection of site-bound metal ions important for RNA folding. RNA. 2012;18:1123–1141. doi: 10.1261/rna.028738.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crary SM, Kurz JC, Fierke CA. Specific phosphorothioate substitutions probe the active site of Bacillus subtilis ribonuclease P. RNA. 2002;8:933–947. doi: 10.1017/s1355838202025025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reiter NJ, et al. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature. 2010;468:784–789. doi: 10.1038/nature09516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pinard R, et al. Functional involvement of G8 in the hairpin ribozyme cleavage mechanism. EMBO J. 2001;20:6434–6442. doi: 10.1093/emboj/20.22.6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson TJ, Zhao ZY, Maxwell K, Kontogiannis L, Lilley DM. Importance of specific nucleotides in the folding of the natural form of the hairpin ribozyme. Biochemistry. 2001;40:2291–2302. doi: 10.1021/bi002644p. [DOI] [PubMed] [Google Scholar]
- 43.Bevilacqua PC. Mechanistic considerations for general acid-base catalysis by RNA: revisiting the mechanism of the hairpin ribozyme. Biochemistry. 2003;42:2259–2265. doi: 10.1021/bi027273m. [DOI] [PubMed] [Google Scholar]
- 44.Kuzmin YI, Da Costa CP, Fedor MJ. Role of an active site guanine in hairpin ribozyme catalysis probed by exogenous nucleobase rescue. J Molec Biol. 2004;340:233–251. doi: 10.1016/j.jmb.2004.04.067. [DOI] [PubMed] [Google Scholar]
- 45.Han J, Burke JM. Model for general acid-base catalysis by the hammerhead ribozyme: pH-activity relationships of G8 and G12 variants at the putative active site. Biochemistry. 2005;44:7864–7870. doi: 10.1021/bi047941z. [DOI] [PubMed] [Google Scholar]
- 46.Klein DJ, Been MD, Ferré-D'Amaré AR. Essential role of an active-site guanine in glmS ribozyme catalysis. J Amer Chem Soc. 2007;129:14858–14859. doi: 10.1021/ja0768441. [DOI] [PubMed] [Google Scholar]
- 47.Wilson TJ, McLeod AC, Lilley DMJ. A guanine nucleobase important for catalysis by the VS ribozyme. EMBO J. 2007;26:2489–2500. doi: 10.1038/sj.emboj.7601698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson TJ, et al. Nucleobase-mediated general acid-base catalysis in the Varkud satellite ribozyme. Proc Natl Acad Sci USA. 2010;107:11751–11756. doi: 10.1073/pnas.1004255107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kath-Schorr S, et al. General acid-base catalysis mediated by nucleobases in the hairpin ribozyme. J Amer Chem Soc. 2012;134:16717–16724. doi: 10.1021/ja3067429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beaucage SL, Caruthers MH. Deoxynucleoside phosphoramidites - a new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Letters. 1981;22:1859–1862. [Google Scholar]
- 51.Wilson TJ, Zhao Z-Y, Maxwell K, Kontogiannis L, Lilley DMJ. Importance of specific nucleotides in the folding of the natural form of the hairpin ribozyme. Biochemistry. 2001;40:2291–2302. doi: 10.1021/bi002644p. [DOI] [PubMed] [Google Scholar]
- 52.Hakimelahi GH, Proba ZA, Ogilvie KK. High yield selective 3'-silylation of ribonucleosides. Tetrahedron let. 1981;22:5243–5246. [Google Scholar]
- 53.Perreault J-P, Wu T, Cousineau B, Ogilvie KK, Cedergren R. Mixed deoxyribo- and ribooligonucleotides with catalytic activity. Nature. 1990;344:565–567. doi: 10.1038/344565a0. [DOI] [PubMed] [Google Scholar]
- 54.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Cryst D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCoy AJ, et al. Phaser crystallographic software. Journal of applied crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Cryst D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feig AL, Uhlenbeck OC. In: The RNA World. Gesteland RF, Cech TR, Atkins JF, editors. Cold Spring Harbor Laboratory Press; 1999. pp. 287–319. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structural data have been deposited in the Protein Data Bank (PDB) with coordinate accession number 5T5A. All other data generated or analysed during this study are included in this published article (and its supplementary information files) or are available from the corresponding author on reasonable request.