Abstract
Inducing macromolecular interactions with small molecules to activate cellular signaling is a challenging goal. PROTACs (proteolysis-targeting chimaeras) are bifunctional molecules that recruit a target protein in proximity to an E3 ubiquitin ligase to trigger protein degradation. Structural elucidation of the key ternary ligase:PROTAC:target species and how this impacts target degradation selectivity remains elusive. We solved the crystal structure of Brd4-degrader MZ1 in complex with human VHL and the Brd4 bromodomain (Brd4BD2). The ligand folds into itself to allow formation of specific intermolecular interactions in the ternary complex. Isothermal titration calorimetry studies, supported by surface mutagenesis and proximity assays, are consistent with pronounced cooperative formation of ternary complexes with Brd4BD2. Structure-based-designed compound AT1 exhibits highly selective depletion of Brd4 in cells. Our results elucidate how PROTAC-induced de novo contacts dictate preferential recruitment of a target protein into a stable and cooperative complex with an E3 ligase for selective degradation.
Introduction
Regulating protein function through targeted degradation as opposed to more conventional target inhibition has emerged as a new modality of discovery chemistry with attractive potential both as tools for target validation and for the development of novel therapeutics 1–4. PROTACs (proteolysis-targeting chimaeras) are bifunctional molecules that bring a target protein into spatial proximity with an E3 ubiquitin ligase to trigger target ubiquitination and subsequent proteasomal degradation 5–11. Recent developments in the field have led to compounds being designed with increasingly “drug-like” molecular properties, and remarkable activities both in cells and in vivo 10. Furthermore, we and others have shown that target depletion selectivity by PROTACs can significantly exceed the binding selectivity of their constitutive warhead ligands 7,12. This realization provides proof-of-concept for turning non-selective or promiscuous ligands into more selective degraders, which can be highly desirable for both chemical probes and drug leads. A characteristic feature of PROTACs mode of action is their sub-stoichiometric catalytic activity that alleviates the requirement for target engagement and occupancy of traditional inhibitors 9. Effective redirection of ligase poly-ubiquitination activity toward a new substrate protein requires formation of a ligase:PROTAC:target ternary complex, an intermediate species that is crucial to the cellular activity of degrader molecules. However, structural elucidation of such a ternary species and its influence on selectivity of target degradation have remained elusive since the PROTAC concept was first incepted and demonstrated in 2001 (ref. 5).
In 2015 we and others reported the first examples of small-molecule PROTACs that target the bromo- and extra-terminal (BET) family proteins for degradation by recruiting substrate-recognition subunits von Hippel–Lindau protein (VHL) 7 and cereblon (CRBN) 6,8 of the respective cullin RING ligases (CRLs), CRL2VHL and CRL4CRBN. Compound MZ1 (ref. 7) conjugates the pan-BET inhibitor JQ1 (ref. 13) to VH032, a potent and specific VHL ligand 14,15, via a 3-unit PEG linker (Fig. 1a). MZ1 and its analogues, MZ2 and MZ3 (Supplementary Results, Supplementary Fig. 1), induce more effective depletion of a single BET member, Brd4 (a validated drug target against cancer and other diseases 16), over its family paralogues Brd2 and Brd3 (ref. 7). These observations led us to hypothesize a structural basis for target selectivity, imparted as a result of PROTAC-induced recruitment of the ligase and bromodomain together in a ternary complex. To reveal the molecular details of complex formation, we pursued the crystal structure of MZ1 in complex with VHL and a BET bromodomain. Here we present for the first time a structure of a PROTAC bound to both E3 ligase and target protein. The structure reveals MZ1 is “sandwiched” between the two proteins, inducing extensive new protein-protein and protein-ligand contacts of both hydrophobic and electrostatic nature. Biophysical binding studies in solution allowed measurement of full thermodynamic parameters of complex formation, which revealed marked isoform-specific cooperativity of ternary complexes. Surface mutagenesis swap and proximity binding assays data support the theory that the induced protein–protein interaction (PPI) contacts drive specificity of the cooperative recognition, impacting on the relative population of ternary complexes. Furthermore, new PROTAC molecules designed guided by the crystal structure showed exquisite selectivity for inducing cellular depletion of Brd4 over its BET family members Brd2 and Brd3.
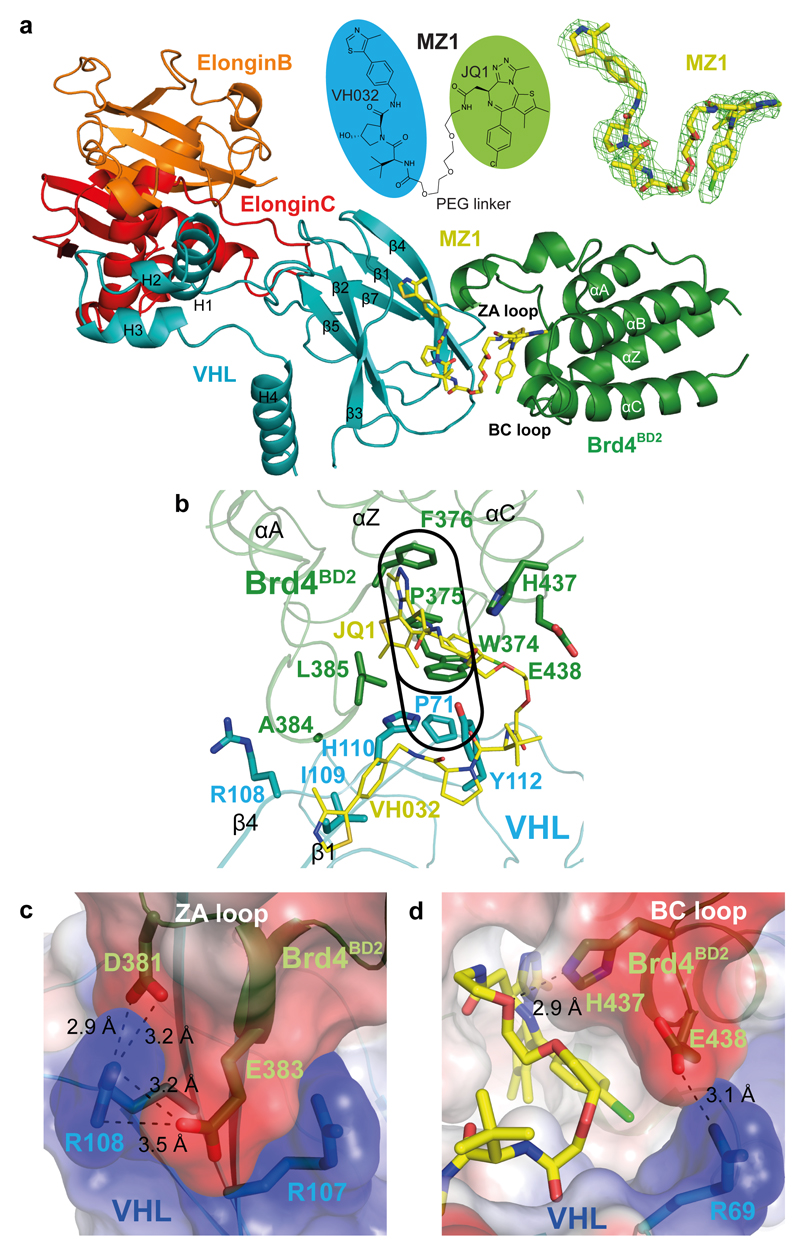
Figure 1. The crystal structure of the Brd4BD2:MZ1:VHL-ElonginC-ElonginB complex.
a, Overall structure of Brd4BD2:MZ1:VHL-ElonginC-ElonginB in ribbon representation. Top middle, chemical structure of bifunctional PROTAC molecule MZ1. Top right, Fo−Fc omit map generated prior to ligand modelling contoured at 3.0σ around bound MZ1. b, Key residues forming the hydrophobic “base” of the induced Brd4BD2:VHL interface. The "WPF" shelf of Brd4BD2 and extended "PWPF" stack are outlined in black. The JQ1 and VH032 elements of MZ1 are labelled in yellow. c, Electrostatic potential map showing the charged zipper contacts between Brd4BD2 residues D381 and E383 with VHL residues R107 and R108. d, Electrostatic potential map showing the interaction between Brd4BD2 residue E438 with VHL residue R69. The hydrogen bond between H437 of Brd4BD2 and the PEG linker of MZ1 is also shown. Dashed lines indicate hydrogen bonds with distances shown in angstroms (Å).
Results
Ternary complex crystal structure
To elucidate the structural details of PROTAC-induced substrate recruitment to an E3 ligase, we solved the crystal structure of MZ1 bound in a ternary complex with the second bromodomain (BD) of Brd4 (Brd4BD2) and VHL to 2.7 Å resolution (Fig. 1a, Supplementary Table 1). The asymmetric unit of the crystal contained two ternary Brd4BD2:MZ1:VCB (VHL, ElonginB and ElonginC) complexes of overall identical quaternary architecture (Supplementary Fig. 2a) and only minor deviations at either end when superposed over the central VHL subunit (Supplementary Fig. 2b,c). The first complex (chains A, B, C and D) had lower average B factors (Supplementary Fig. 2a) so we refer to this in all subsequent analyses. The electron density around MZ1 was fully defined (see inset panel in Fig 1a and Supplementary Fig. 2d,e for each protomer in the asymmetric unit). MZ1 is bound within a bowl-shaped interface formed by extensive protein-protein interactions (PPIs) between Brd4BD2 and VHL (Supplementary Fig. 3a). The bowl has a hydrophobic "base" which is formed by two key points of contact (Fig. 1b and Supplementary Fig. 3a). Firstly, Trp374 from the characteristic hydrophobic region named “WPF shelf” (ref. 17) of Brd4BD2 interacts with residues Arg69, Pro71 and Tyr112 of VHL (Fig. 1b). Pro71 provides an additional stack to the WPF, forming an extended “PWPF” shelf (Fig. 1b). Secondly, Ala384 and Leu385 from the second helical turn of the ZA loop of Brd4BD2 contact the hydrophobic side chains of Arg108, Ile109 and His110 in β4 of VHL (Fig. 1b). Two electrostatic "arms" complete the rim of the bowl. At one end, Asp381 and Glu383 in the ZA loop of Brd4BD2 form a tight zipper structure of complementary charges with Arg107 and Arg108 (Fig. 1c). At the opposite end Brd4BD2 residue Glu438, residing in the BC loop, contacts Arg69 from VHL (Fig. 1d). In the induced interface between the two proteins, Brd4BD2 recapitulates some of the interactions made by the HIF-1α CODD segment peptide with VHL 18,19, as the electrostatic zipper structure generated by Asp381 and Glu383 contacts the same VHL residues (Arg107 and Arg108) as Asp569 and Asp571 of HIF-1α (Fig. 1c and Supplementary Fig. 3b,c). In contrast, VHL does not contact the surface of Brd4 bromodomain bound to acetyllysine histone H4 peptide 20 (Supplementary Fig. 3d,e). In total the PPIs induced in the complex bury a surface area of 688 Å2 (Supplementary Table 2).
MZ1 is cupped within the bowl structure in such a way that its two heads recapitulate the binding modes of the respective ligands individually – JQ1 in the acetyllysine-binding pocket of Brd4BD2 13,20, and VH032 in the hydroxyproline-binding site of VHL 14 (Supplementary Fig. 3f,g). In addition to the expected binary protein-ligand interactions, MZ1 forms additional protein-ligand interactions within the ternary complex. The PEG linker makes van der Waals interactions with the BC loop of Brd4BD2 and a hydrogen bond between the ether oxygen adjacent to the amide linkage to JQ1 and the BD2-specific residue His437 (Fig. 1d). The same hydrogen bond between His437 and a PEG oxygen is also observed in a recent crystal structure of Brd4BD2 bound to MT1, a bivalent BET inhibitor comprised of two JQ1 moieties linked by PEG units in a similar fashion as in MZ1 (Supplementary Fig. 4) 21. VHL and Brd4BD2 come together to sandwich MZ1 against their respective binding surfaces, burying otherwise solvent-exposed regions of the JQ1 and VH032 ligands. Specifically, Ala384, Leu385 and Gly386 from the ZA loop of Brd4BD2 contact the –CH2–phenyl portion of VH032, whilst His110 and Tyr112 of β4 of VHL contact one of the two thiophene methyl groups and the para-chlorophenyl ring of JQ1 (Fig. 1b). In addition, MZ1 folds onto itself in such a way that its PEG linker is packed between the tert-butyl group of the VH032 moiety and the para-chlorophenyl ring of JQ1 (Fig. 1a,d). Throughout a 100 ns molecular dynamics simulation, favorable intermolecular contacts were observed from JQ1 atoms to VHL, from VH032 atoms to Brd4BD2, and from the PEG linker atoms to both VHL and Brd4BD2, as well as intramolecular contacts within MZ1 involving the PEG linker and the JQ1 and VH032 moieties (Fig. 2a and Supplementary Fig. 5). The surface area further buried by the ligand folding within the bowl-shaped interface was 1,933 Å2, resulting in a total extended buried surface area of 2,621 Å2 for the ternary complex (see Supplementary Table 2 for a comparison with cereblon:phthalimides:target ternary complexes 2,3).
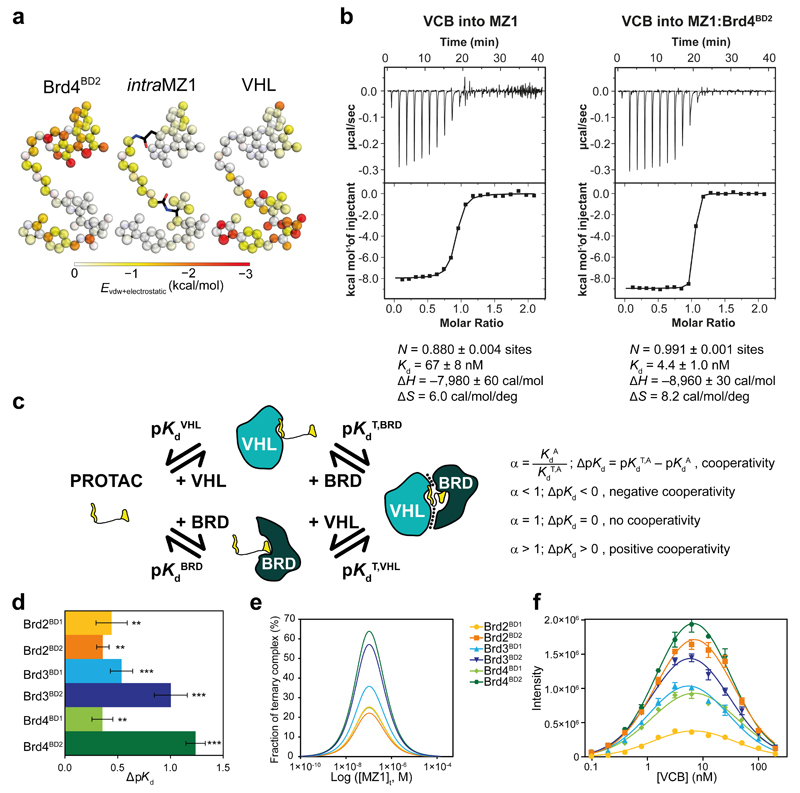
Figure 2. Brd4BD2 and VHL form a stable, cooperative complex in the presence of MZ1.
a, Novel ligand contacts are induced by ternary complex formation. Colour strength (from white to red) indicates the mean enthalpic energies of individual MZ1 atoms in contacting Brd4BD2 (left) or VHL (right), as well as intra-ligand contacts within MZ1 (centre) in a 100 ns MD simulation. b, Inverse ITC titrations of VCB into MZ1 (left, representative of eight replicates) and VCB into the pre-formed MZ1:Brd4BD2 (right, performed in duplicate) c, Ternary complex equilibria and definition of cooperativities. d, ΔpKd measured for VCB with MZ1 and the indicated BET-BDs, reported as difference (± uncertainty), from pKd values measured as mean (± 1 s.e.m.) as described in Online Methods. Statistical significance of pKd values for ternary titrations compared to the corresponding binary titrations was assessed by two-tailed t-test assuming equal variances, and is indicated as ** (p-value < 0.01) or *** (p-value < 0.001). e, Simulated fraction of ternary complexes based on mathematical model described in ref. 23. f, AlphaLISA intensity values titrating VCB against BET-BDs with MZ1. AlphaLISA intensities represent mean (± 1 s.d.) of intensity values from four technical replicates. The hook effect observed on these curves is due to biotinylated-VCB oversaturating the donor beads, resulting in a progressive decrease in signal.
Isoform-specific cooperativity of ternary complexes
The extensive new contacts observed in the crystal structures suggested the possibility that isoform-specific PPIs could play a role in the “cooperativity” of the ternary complex equilibria 22,23. To assess the thermodynamics of PROTAC-induced complex formation with VHL and BET BDs, we employed isothermal titration calorimetry (ITC) in solution (Table 1 and Supplementary Fig. 6). To disentangle contributions from binary and ternary complex formation equilibria we performed reverse titrations i.e. protein in syringe and ligand in cell. This experimental strategy avoids the characteristic “hook effect” observed with increasing concentrations of PROTACs, as formation of binary complexes competes with and eventually surpasses formation of ternary ones 23. First, we titrated a solution of BET BD against MZ1, ensuring no excess unbound PROTAC compound would be present at the end of the titration. This was followed by a titration of VCB into the saturated MZ1:BD complex, forming the ternary VCB:MZ1:BD complex (Fig. 2b, right panel). Titration of VCB into MZ1 alone (Fig. 2b, left panel) was then performed and used as reference, allowing the potential cooperativity of ternary complex formation to be accurately quantified. By definition, a ternary system is considered positively cooperative if interactions enhance formation of the ternary complex ( ΔpKd = pKd (ternary) − pKd (binary) >0). Conversely, a system is termed negatively cooperative when formation of the ternary complex is diminished (α < 1; ΔpKd < 0), for example because of repulsive interactions or steric hindrance between the two components in the ternary complex. Non-cooperative equilibria would instead show unchanged Kd for the two steps (α = 1; ΔpKd = 0), (Fig. 2c). With all BET BDs used, we observed significant positively cooperative ternary complex formation (see Table 1, and plots of ΔpKd in Fig. 2d). Strikingly, the strongest cooperativity was observed for Brd4BD2 (α = 18), followed by Brd3BD2 (α = 11; Table 1 and Fig. 2d). The large cooperativity observed led to a steep transition of the sigmoidal binding curve (Fig. 2b, right panel), suggesting that the fitted Kd value could potentially be underestimating the real binding affinity of this ternary complex. All BD1s also exhibited positive cooperativity, albeit to a much lesser extent (α between 2 and 3). Brd4BD2 and Brd3BD2 not only exhibited the greatest cooperativity amongst all BET BDs, they also formed the most stable ternary complexes overall (ΔG (binary + ternary) = –22.2 ± 0.1 and 22.0 ± 0.2 kcal/mol, respectively), ~2 kcal/mol more stable than e.g. Brd2BD1 (ΔG = −20.3 ± 0.2 kcal/mol).
Table 1. Thermodynamic parameters of formation of binary and ternary complexes between MZ1, VCB and BET bromodomains measured by isothermal titration calorimetry.
| Protein in syringe | Species in cell | Kd (nM) | ΔG (kcal × (mol-1) | ΔH (kcal × (mol-1) | −TΔS (kcal × (mol-1) | N | α | ΔpKd |
|---|---|---|---|---|---|---|---|---|
| Brd2BD1 | MZ1 | 62 ± 6 | −9.84 ± 0.06 | −12.8 ± 0.7 | 3.0 ± 0.8 | 1.1 ± 0.1 | ||
| Brd2BD2 | 60 ± 3 | −9.85 ± 0.03 | −9.8 ± 0.3 | −0.1 ± 0.3 | 1.2 ± 0.1 | |||
| Brd3BD1 | 21 ± 5 | −10.2 ± 0.1 | −14.7 ± 0.8 | 4.2 ± 0.9 | 1.1 ± 0.1 | |||
| Brd3BD2 | 13 ± 3 | −10.8 ± 0.1 | −14.0 ± 0.9 | 3.3 ± 0.7 | 1.05 ± 0.02 | |||
| Brd4BD1 | 39 ± 9 | −10.1 ± 0.1 | −14.7 ± 0.4 | 4.6 ± 0.5 | 0.95 ± 0.03 | |||
| Brd4BD2 | 15 ± 1 | −10.68 ± 0.04 | −10.9 ± 0.4 | 0.2 ± 0.4 | 1.08 ± 0.07 | |||
| Brd2BD1 KEA | 69 ± 9 | −9.78 ± 0.08 | −14 ± 1 | 4 ± 1 | 0.83 ± 0.08 | |||
| Brd4BD2 QVK | 22 ± 8 | −10.5 ± 0.2 | −12.4 ± 0.5 | 1.9 ± 0.2 | 1.0 ± 0.1 | |||
| VCBa | MZ1a | 66 ± 6 | −9.81 ± 0.05 | −7.7 ± 0.3 | −2.1 ± 0.3 | 0.93 ± 0.04 | ||
| VCB | MZ1:Brd2BD1 | 24 ± 8 | −10.4 ± 0.2 | −7.3 ± 0.2 | −3.1 ± 0.4 | 1.1 ± 0.2 | 2.9 | 0.4 ± 0.2 |
| MZ1:Brd2BD2 | 28 ± 3 | −10.3 ± 0.1 | −10.5 ± 0.1 | 0.2 ± 0.2 | 1.07 ± 0.02 | 2.3 | 0.36 ± 0.06 | |
| MZ1:Brd3BD1 | 19 ± 4 | −10.6 ± 0.1 | −8.8 ± 0.5 | −1.8 ± 0.7 | 1.01 ± 0.01 | 3.5 | 0.5 ± 0.1 | |
| MZ1:Brd3BD2 | 7 ± 2 | −11.2 ± 0.2 | −6.3 ± 0.1 | −4.9 ± 0.3 | 0.99 ± 0.04 | 10.7 | 1.0 ± 0.2 | |
| MZ1:Brd4BD1 | 28 ± 6 | −10.3 ± 0.1 | −9.1 ± 0.9 | −1 ± 1 | 0.97 ± 0.06 | 2.3 | 0.4 ± 0.1 | |
| MZ1:Brd4BD2 | 3.7 ± 0.7 | −11.5 ± 0.1 | −8.9 ± 0.1 | −2.6 ± 0.2 | 1.02 ± 0.02 | 17.6 | 1.24 ± 0.09 | |
| MZ1:Brd2BD1 KEA | 12 ± 7 | −10.9 ± 0.4 | −5.7 ± 0.2 | −5.2 ± 0.2 | 0.8 ± 0.1 | 7.9 | 0.8 ± 0.3 | |
| MZ1:Brd4BD2 QVK | 14.9 ± 0.1 | −10.68 ± 0.03 | −6.2 ± 0.3 | −4.5 ± 0.3 | 0.9 ± 0.1 | 4.2 | 0.62 ± 0.04 |
All ITC titrations were performed at 25 °C. Values reported are the mean ± S.E.M. from two independent measurements, except for VCB titration into MZ1 (line a) for which values reported are the mean ± S.E.M. from eight independent measurements.
To understand the impact of cooperativities of different BET BDs ternary complexes on their relative population, we applied a mathematical model of ternary equilibria 23. We simulated the fraction of ternary complex formation for VCB, MZ1 and six individual BET BDs, using our measured binary Kds and cooperativities α (Table 1) and protein concentrations of 40 nM (to be around the Kd values and to match the concentrations used later in AlphaLISA). An overlay of simulations showed that the relative populations of each ternary complex vary significantly, with Brd4BD2 being the most populated, and, as an example, ~2.5 fold greater than Brd2BD1 at any given concentration of PROTAC (Fig. 2e). To interrogate this relative trend experimentally, we employed a proximity AlphaLISA assay that can achieve high signal amplification in response to formation of ternary complexes over an energy transfer distance of up to 200 nm 24,25. At every fixed component concentration, the relative trend observed in AlphaLISA signal was broadly consistent with the cooperativity trends measured by ITC, with Brd4BD2 and Brd3BD2 giving greater signal, while Brd2BD1 giving the lowest response (Fig. 2f). A similar trend was observed with the analogous MZ2 (PEG4), MZ3 (PEG3-Phe) and MZ4 (PEG2) (Supplementary Fig. 7a–h). Taken together the data are consistent with target-specific cooperativities and stabilities of ternary complexes impacting on the relative population of this key intermediate species.
Specificity of MZ1-induced protein-protein interactions
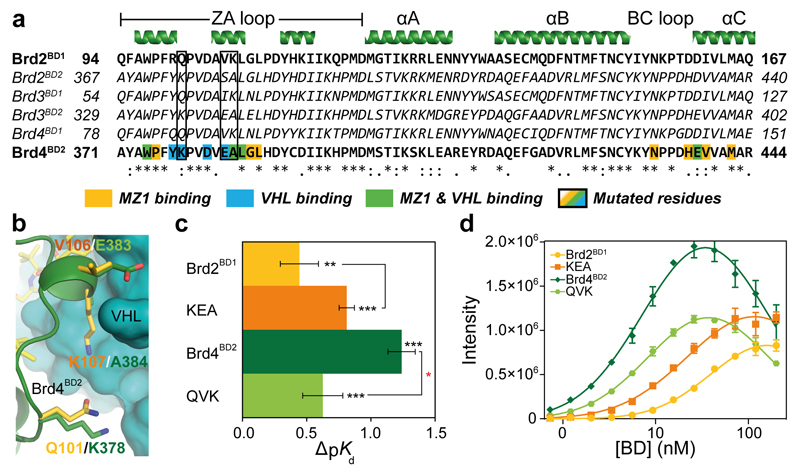
To evaluate to what extent the cooperativity of ternary complex formation is dictated by surface complementarity between VHL and the Brd4BD2 bromodomain, we mutated semi-conserved or non-conserved bromodomain residues forming key induced PPI contacts, but not directly involved in binding of MZ1 (Fig. 3a and Supplementary Fig. 5). Inspection of sequence alignments (Fig. 3a) and the crystal structure (Fig. 3b) guided us to select residues Glu383 and Ala384 in Brd4BD2 (the most cooperative of bromodomains), for site directed mutagenesis. These residues are Val106 and Lys107 in the corresponding positions in Brd2BD1, one of the least cooperative BET domains. In addition, the MD simulation evidenced extensive movement of loop 7 of VHL (ref. 26) bringing it in close contact with Lys378 of Brd4BD2 (Supplementary Fig. 5b–d), which corresponds to Gln101 in Brd2BD1. Based on these considerations, triple mutant Brd4BD2 K378Q/E383V/A384K (named QVK for simplicity) was designed. The mutations would make the PPI surface of Brd4BD2 closer to that of Brd2BD1, albeit with the caveat of introducing an extra charge overall. Conversely, a triple mutant of Brd2BD1 was designed in which the corresponding residues are switched to those of Brd4BD2 (Q101K/V106E/K107A, named KEA for simplicity). In ITC, the QVK mutant exhibited significantly weakened cooperative complex formation relative to Brd4BD2 wild-type (WT) (α = 4; Table 1 and ΔpKd = 0.64 ± 0.04, see plots in Fig. 3c). Conversely, the cooperativity of the KEA mutant increased relative to Brd2BD1 WT (α = 8; Table 1 and Fig. 3c). Crucially, these mutations did not affect the Kd of each domain for MZ1 (Table 1), suggesting that the switch of cooperativity is independent of binary target engagement and instead is dictated by the induced PPIs. Consistent with the cooperativity switch measured by ITC, QVK reduced ternary complex formation in AlphaLISA compared to WT, whereas KEA displayed the opposite effect (Fig. 3d with MZ1 and Supplementary Fig. 7i with MZ2). Taken together these data validate the ternary complex structure in solution and elucidate how specific PPIs influence cooperative recruitment of two target proteins to each other by a bifunctional molecule.
Figure 3. The molecular basis of MZ1-induced compact complex formation between Brd4BD2 and VHL.
a, Sequence alignment of BET bromodomains. Residues of Brd4BD2 in contact with MZ1 and/or VHL are highlighted. b, Structural alignment of Brd2BD1 (yellow) superposed on Brd4BD2 (dark green) in the ternary structure with MZ1 and VHL. Key, non-conserved interacting residues are shown in sticks. c, ΔpKd measured for VCB with MZ1 and the indicated BET-BDs, reported as difference (± uncertainty), from pKd values measured as mean (± 1 s.e.m.) as described in Online Methods. Statistical significance of pKd values for ternary titrations compared to the corresponding binary titrations (in black) and for ternary titrations of WT compared to the corresponding triple-mutant (in red) was assessed by two-tailed t-test assuming equal variances, and is indicated as * (p-value < 0.05), ** (p-value < 0.01) or *** (p-value < 0.001). d, AlphaLISA intensity values titrating Brd2BD1, Brd4BD2 and corresponding mutants against VCB with MZ1. AlphaLISA intensities are the mean (± 1 s.d.) of intensity values from four technical replicates.
Structure-designed AT1 is highly selective Brd4 degrader
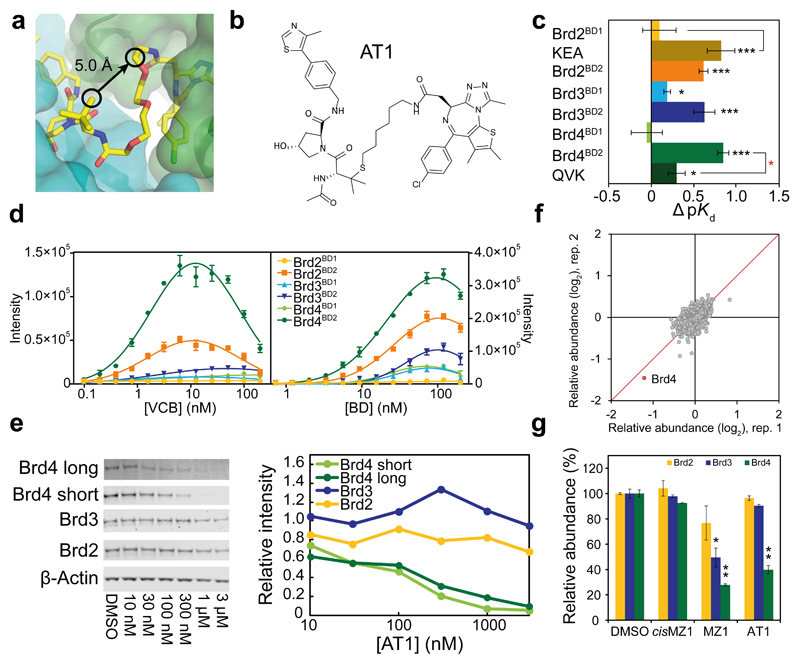
In chemical probe and drug development knowledge of ligand-bound structures can guide the design of next-generation compounds. We therefore sought to create new PROTACs based on our crystal structure that could exhibit enhanced target depletion selectivity in cells toward Brd4. We noted that the side chain of the key tert-Leu group of VH032 projected an attractive vector to link directly to the JQ1 moiety (Fig. 4a), which we hypothesized could better discriminate against the relative binding orientation observed in the crystal. We therefore replaced tert-Leu with penicillamine and synthesized 1 (AT1, Fig. 4b) and other analogues 2–6 (AT2–AT6) bearing thioether linkages of varying lengths to JQ1 (Supplementary Fig. 1 and Methods). The modified VHL ligand within AT1 retained binding to VHL (Kd 330 nM, Supplementary Table 3, and Supplementary Fig. 8), with a loss of potency of less than two-fold relative to VH032 (ref. 14) but approximately five-fold less compared to MZ1. ITC data comparing binary and ternary complexes revealed Brd4BD2 as the BET bromodomain forming the most cooperative (α = 7; ΔpKd = 0.84 ± 0.07) and most stable (ΔG = −20.2 ± 0.2 kcal/mol) of all ternary complexes with AT1 (Fig. 4c, Supplementary Table 3, and Supplementary Fig. 8). The same trend of cooperativity observed for MZ1 and the QVK and KEA mutants relative to WT was also observed with AT1 (Fig. 4c, Supplementary Table 3), suggesting AT1 recruits VHL and Brd4BD2 in the same relative orientation as does MZ1. We consistently observed by AlphaLISA preferential recruitment of Brd4BD2 over the other BDs by AT1–6 (Fig. 4d and Supplementary Fig. 9). We next tested the activity of the new structure-designed molecules to induce degradation of BET proteins in cells, and observed remarkable Brd4-selective depletion at all concentrations tested, with depletion of Brd4 after 24 h treatment with 1–3 μM of AT1 and negligible activity against Brd2 and Brd3 (Fig. 4e, see Supplementary Fig. 9 for AT2–6). Specificity for Brd4 degradation was not due to differences in protein synthesis rates, as shown by control treatments with cycloheximide, which blocks protein translation (Supplementary Fig. 10). To assess whether ubiquitination of lysine residues could play a role in the observed selectivity, we combined our crystal structure of the EloBC–VHL–MZ1–Brd4BD2 complex with existing whole CRL structural information into a model of the entire CRL2VHL–MZ1–bromodomain assembly (Supplementary Fig. 12a). Several lysine residues are surface exposed and accessible to the E2-Ub in this model at distances between 50–60 Å, consistent with known CRL substrates 27. Mapping MZ1-induced ubiquitination sites in vitro identified Lys346 on Brd4BD2 and several sites on the other BET-BDs (Supplementary Fig. 12b-d). Unbiased and quantitative isobaric tagging mass spectrometry proteomics confirmed Brd4 as the sole protein markedly depleted (to ~40%) upon treatment with AT1, amongst the 5,674 detected proteins that passed filtering criteria (Fig. 4f and Supplementary Data Set 1). Crucially, no effect on protein levels of Brd2 and Brd3 was observed with AT1, in contrast with MZ1 that exerted a broader and more profound effect across all BET proteins, albeit still preferential toward Brd4 (Fig. 4g). Together these data qualify AT1 as a new highly selective degrader of Brd4 in cells.
Figure 4. Structure-guided design and characterization of Brd4-selective degrader AT1.
a, A vector linking VH032 to JQ1 that maintains the relative binding orientation. b, Chemical structure of AT1. c, ΔpK d measured for VCB with AT1 and the indicated BET-BDs, reported as difference (± uncertainty), from pKd values measured as mean (± 1 s.e.m.). Statistical significance of pKd values for ternary titrations compared to corresponding binary (black) and for ternary WT compared to corresponding mutant (red) was assessed by two-tailed t-test assuming equal variances. d, AlphaLISA intensity values titrating VCB against BET-BDs with AT1 (left) and BET-BDs against VCB with AT1 (right). AlphaLISA intensities are the mean (± 1 s.d.) of intensity values from four technical replicates. e–g, Highly selective degradation of Brd4 by AT1 in HeLa cells after 24 h. e, Protein levels are shown from one representative of three biological replicates, visualized by immunoblot (left) and quantified relative to DMSO (right). Intensity values were measured as described in Online Methods. Full gels are provided in Supplementary Fig. 11. f, Impact of AT1 (1 µM, 24 h) on the cellular proteome. Data plotted as fold change (log2) of replicate 1 vs replicate 2, for a total of 5,674 proteins quantified (see Online Methods). g, Quantified levels of BET proteins shown are mean (± 1 s.e.m.) from two replicates relative to mean of vehicle. Statistical significance of relative protein abundance compared to DMSO was assessed by two-tailed t-test assuming equal variances. Statistical significance indicated as * (p-value < 0.05), ** (p-value < 0.01) or *** (p-value < 0.001).
Discussion
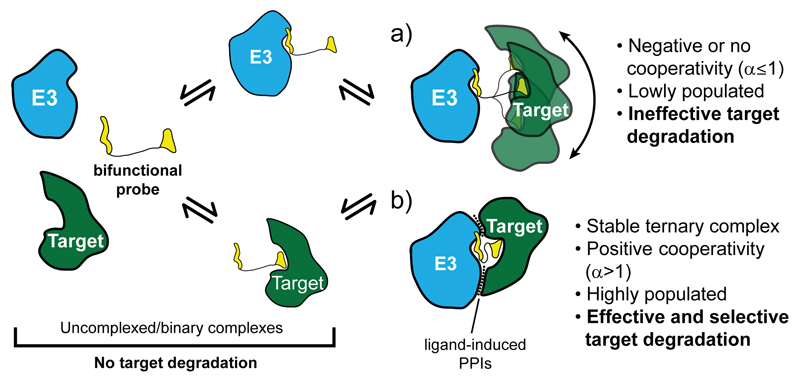
We put forth a model for how cooperative recruitment of a target close to an E3 ligase in a ternary complex by a PROTAC molecule can impact on the effectiveness and selectivity of target degradation (Fig. 5). Our work shines structural insights into how bifunctional molecules can induce target-specific interactions in the ‘enzyme-substrate’ ternary complex species key for PROTAC catalytic activity. These cooperative molecular recognition features contribute to how tightly and stably the ‘neo-substrate’ can be bound to the ligase, impacting on the relative population of the complex and consequently on the catalytic efficiency of the process. For homologous targets, as is the case with BET bromodomains, we show how these features add a level of target depletion selectivity independently of binary target engagement.
Figure 5. Schematic model of selective PROTAC-induced target degradation.
A target is preferentially recruited in a stable and positively cooperative ternary complex with the E3 ubiquitin ligase upon folding of the bifunctional probe to induce formation of specific PPIs.
We illustrate the relevance of measuring cooperativities of ternary complex formation in solution using ITC. One of the main advantages of the assay setup as described here is that it is designed to avoid issues associated with the hook effect. While the assay may be used to characterize in full a handful of compounds and systems, it requires large quantities of material and, notably, lacks throughput. Alternative bioassays to quantify formation of ternary complexes could circumvent this limitation, and we provide evidence that proximity AlphaLISA assay can be used for these purposes. However, AlphaLISA data should be interpreted with caution, even when comparing highly conserved domains as done here, because in this bead-based technology the multiplicity of binding sites and relative linkage and orientation of components immobilized to the beads may influence the measured signal 24,28. Moreover, it can be difficult to deconvolute individual binding parameters from dose–response curves monitoring ternary complex formation, because these are often bell–shaped curves complicated by the hook effect 23. We anticipate that future assay developments in this direction will help prioritize complexes for structural studies and to drive drug development programs. The results of our study are of particular relevance because VHL-based BET degraders similar to those described here have proven to be bioavailable and active in vivo 29 and could potentially enter clinical trials as early as 2017 (ref. 4).
Our study points to the importance that the bifunctional molecule “folds on itself” in such a way that its two heads can recruit the respective targets into productive proximity. The result of this process is the burial of extensive surface area, the formation of new PPIs, contributing to the high stability and cooperativity of ternary complexes. Our work has therefore major implications for future PROTAC drug design, which has so far been somewhat empirical and has largely adopted a combinatorial “plug-and-play” strategy 11. The proposed model suggests that bifunctional molecules should be rationally designed and prioritized based on their ability to induce favorable contacts and allow forming a stable complex between the E3 ligase and the target. While ligand-induced proximity is expected to strengthen potential PPIs because of the reduced entropic cost, we show that the level of surface complementarity between the two proteins in their relative orientation imposed by the bifunctional molecule dictates cooperative complexation. Although the exact relative orientation between VHL and Brd4 observed in the crystal may not be the only one that the system can adopt in solution once free from potential constraints of crystal packing, our data suggests that it captures a significant species underlying MZ1 function. Maximizing the diversity of E3s recruited 30, and linking positions and vectors from the E3 and target ligands, will thus be important to achieve target-specific degraders.
In an example of first layer of this rational design, we show how new PROTACs designed based on our ternary structure can lead to enhanced selectivity of depletion in cells for the crystallized target Brd4. The efficiency and selectivity of cellular protein knockdown will inevitably depend on other factors, including compound permeability and stability, the expression level of the hijacked CRL and its relative activity and flexibility 27,31, as well as target abundance and re-synthesis rates. Differing ubiquitination rates could also in principle influence target degradation selectivity. To this end, in addition to increasing their relative population, cooperative and stable complexes would be expected to exhibit slower dissociation rates and longer half-lives, potentially aiding the efficiency of target ubiquitination by the hijacked ligase. Differing availability and access of surface lysine residues between alternate substrates could also play a role. However, based on our data, the presence of many surface Lys on BET-BDs, and the flexibility and large ubiquitination zone of CRLs 2,27, we view it unlikely that target ubiquitination plays a role in the observed selectivity of Brd4 degradation. Obtaining a more detailed biochemical picture of target ubiquitination in a cellular context will be of clear importance for future investigation.
For targeted protein degradation, converting a pan-selective or promiscuous probe ligand into a more selective degrader probe provides new opportunities to improve target validation and could minimize off-target effects. In addition to dictating selectivity of target degradation, highly cooperative ternary E3:PROTAC:target systems would be anticipated to unlock the possibility to effectively degrade hitherto “undruggable” targets using ligands with inherently weak binary binding affinities. A more general implication of this study is the feasibility to induce de novo protein-protein interactions, or stabilize weakened ones, using bifunctional small molecules, a feature previously established with mono-functional ‘molecular glues’ 32 such as the plant hormones auxin 33 and jasmonate 34, the phthalimide immunomodulatory drugs (IMiDs) 2,3,31,35–38, and macrocyclic natural products such as rapamycin and cyclosporine 39,40. We envision that extensions of PPI-stabilizing capabilities to hetero- or homo-bifunctional small molecules 21,41,42 beyond PROTACs as highlighted here could expand the target spectrum accessible to PPI stabilizers, and provide a new paradigm of selective chemical intervention for structural chemical biology and drug discovery.
Online Methods
Chemical synthesis
Synthesis of compounds described in this paper and their intermediates are described in the Supplementary Note.
Constructs, protein expression and purification
Wild-type and mutant versions of human proteins VHL (UniProt accession number: P40337), ElonginC (Q15369), ElonginB (Q15370), Brd2 (P25440), Brd3 (Q15059) and Brd4 (O60885) were used for all protein expression. For expression of VBC, N-terminally His6-tagged VHL (54–213), ElonginC (17–112) and ElonginB (1–104) were co-expressed in Escherichia coli BL21(DE3) at 24 °C for 16 h using 3 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). E. coli cells were lysed using a pressure cell homogenizer (Stansted Fluid Power) and lysate clarified by centrifugation. His6-tagged VCB was purified on a HisTrapFF affinity column (GE Healthcare) by elution with an imidazole gradient. The His6 tag was removed using TEV protease and the untagged complex dialysed into low concentration imidazole buffer. VCB was then flowed through the HisTrapFF column a second time, allowing impurities to bind as the complex eluted without binding. VCB was then additionally purified by anion exchange and size-exclusion chromatography using MonoQ and Superdex-75 columns (GE Healthcare), respectively. The final purified complex was stored in 20 mM Bis Tris, pH 7, 150 mM sodium chloride and 1 mM dithiothreitol (DTT). Brd2BD1 (71–194), Brd2BD2 (344–455), Brd3BD1 (24–146), Brd3BD2 (306–416), Brd4BD1 (44–178) and Brd4BD2 (333–460) as well as equivalent mutant constructs were expressed with an N-terminal His6 tag in E. coli BL21(DE3) at 18 °C for 20 h using 0.2 mM IPTG. His6-tagged BDs were purified on nickel Sepharose™ 6 fast flow beads (GE Healthcare) by elution with increasing concentrations of imidazole. For crystallography the His6-tagged BD was cleaved with TEV protease and dialysed into low concentration imidazole buffer. The BD was then flowed over the nickel beads a second time to remove impurities and protease. BDs were then additionally purified by size-exclusion chromatography using a Superdex-75 column. For AlphaLISA, ITC and ubiquitination reactions, following elution of His6-tagged BDs from the nickel beads, the BDs were purified by size-exclusion chromatography using a Superdex-75 column. The final purified proteins were stored in 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.5, 150 mM sodium chloride and 1 mM DTT. All chromatography purification steps were performed using Äkta FPLC purification systems (GE Healthcare) or glass econo-columns (Bio-Rad) at room temperature.
Crystallography
VCB, MZ1 and Brd4BD2 were mixed as a 1:1:1 stoichiometric ternary complex with a final concentration of 10 mg/mL. Drops of the ternary complex were mixed 1:1 with 13% (w/v) PEG 8000 and 0.1 M sodium citrate (pH 6.3) in the hanging-drop vapour diffusion format. Crystals appeared within minutes and were fully grown after one day. A crystal was dehydrated in a solution containing 18% (w/v) PEG 8000 for a few minutes and flash-cooled in liquid nitrogen using 20% 2-methyl-2,4-pentanediol in liquor solution as a cryoprotectant. Diffraction data were collected at Diamond Light Source beamline I04-1 using a Pilatus 6M-F detector at a wavelength of 0.92819 Å. Indexing and integration of reflections was performed using XDS with the XDSGUI interface43, and scaling and merging with AIMLESS44 in CCP4i45. The Wilson B factor was estimated at 47.2 Å2. To solve the phase problem the molecular replacement method was used with the program PHASER46 using search models derived from the coordinates of VCB (PDB entry 1VCB26) and Brd4BD2 (PDB entry 2OUO20). Two instances of the ternary complex were found in the asymmetric unit, indicating a final solvent content of 68% as calculated from the Matthews coefficient. The initial model was refined iteratively using REFMAC47 and COOT48. Ligand structures and restraints were generated using the PRODRG server49. The MOLPROBITY server50 was used to validate the geometry and steric clashes in the structures; the distribution of backbone torsion angles in the Ramachandran plot are 98.3% in the favored region and 1.7% in the allowed region. The structure has been deposited in the protein data bank (PDB) with accession code 5T35 and data collection and refinement statistics are presented in Supplementary Table 1. Interfaces and contacts observed in the crystal structure were calculated with PISA51 and LIGPLOT52. All figures were generated using PyMOL.
Molecular dynamics simulations
MD simulations were carried out using the NAMD program53 and the CHARMM 36 force field54. We attempted to derive ab initio topology and parameter files for MZ1 using Jaguar v. 9.0 (Schrödinger Inc., LLC, New York, NY, US). However, characterization of the minimized structure as a minimum by vibrational analysis proved unsuccessful (number of imaginary frequencies > 0) using several approaches and initial structures, probably due to the large number of atoms. Therefore topology and parameter files were generated using the CGenFF server55.
To simulate the Brd4BD2:MZ1:VHL ternary complex in solution, the coordinates of the X-ray crystal structure of the complex (chains A and D) were used as starting structure for simulation. ElonginB and ElonginC, which are sufficiently far from the hydroxyproline recognition site of VHL (> 20 Å), were excluded to increase computational efficiency throughout the simulation. The model was solvated in a TIP3P water box with a padding of 12 Å from the edge of the box to any protein atom. The system charges were neutralized with sodium or chloride ions as appropriate. The solvated system was minimized for 3,000 steps with all protein and MZ1 atoms restrained to eliminate residual unfavorable interactions between each other and the solvent, followed by another 5,000 steps with all atoms free to move. Heating of the system from 0 to 300 K was achieved in 100 ps (time step of 1 fs), with fixed protein backbone atoms to allow relaxation of the solvent. The system was subsequently equilibrated for 600 ps (time step of 2 fs) with all atoms free to move. The NPT ensemble was used during production simulation of 100 ns (time step of 2 fs). The temperature was controlled with a Langevin thermostat at 300 K, and the pressure with a Nose-Hoover Langevin piston barostat at 1 bar. A SHAKE constraint was applied to all bonds containing hydrogen atoms. Short-range nonbonded interactions were switched at 10 Å and cut off at 12 Å, and particle mesh Ewald summation was employed for long-range non-bonded interactions.
The trajectory was analysed using VMD v. 1.9.256 and taking snapshots every 10 ps of simulation, unless otherwise stated. To calculate root-mean-square deviations (RMSD) throughout the simulation, ternary complexes were superposed to the crystallographic complex using an in-house PyMOL script considering only Cα atoms of the VHL protein within a shell of 10 Å from MZ1. This was implemented in order to diminish the effect of structural rearrangements occurring far from the hydroxyproline recognition site of VHL during the simulation arising from the absence of ElonginB and ElonginC. Radius of gyration (Rg) of the ternary complex, i.e. the radius of a sphere with equivalent moment of inertia, was computed using Carma57 at each snapshot considering the protein backbone. Buried surface area (BSA) upon complex formation, i.e. the difference in surface-accessible surface area (SASA) between the formed complex and the unbound partners in the system, was computed considering all protein atoms and a spherical probe of radius 1.4 Å. Intermolecular contacts, i.e. pair-to-pair contacts between an amino acid in Brd4BD2 and VHL, were considered formed if more than five atoms of the amino acid were at a distance closer than 4.0 Å from the partner protein. Intermolecular contacts were computed using the Timeline plugin v. 2.3 as implemented in VMD. Per-residue and per-atom inter- and intramolecular interaction energies (EvdW + electrostatic) were computed using an in-house automated routine of the NAMD Energy plugin v. 1.4 as implemented in VMD. Interaction energies were estimated by adding the pair-wise van der Waals and electrostatic contributions between individual amino acids or atoms and the corresponding partner. In the case of per-atom calculations, the interaction energies of hydrogen atoms were added to their corresponding heavy atom. For intramolecular interactions analysis, MZ1 was divided into three sections (JQ1, PEG linker, and VH032) and pair-wise energetic contributions from the atoms of each section to the rest of the molecule (excluding 1–4 bonded atoms) were calculated. In order to obtain comparable and interpretable results, the following scaling factor and cutoff value were applied to the electrostatic contribution:
Isothermal titration calorimetry (ITC)
Titrations were performed on an ITC200 micro-calorimeter (GE Healthcare). The titrations were all performed as reverse mode (protein in syringe, ligand in cell) and consisted of 19 injections of 2 µL protein solution (20 mM Bis-tris propane, 150 mM NaCl, 1 mM tris(2-carboxyethyl)phosphine (TCEP), 0.02 % DMSO, pH 7.4 ) at a rate of 2 sec/µL at 120 s time intervals. An initial injection of protein (0.4 µL) was made and discarded during data analysis. All experiments were performed at 25 ºC, whilst stirring at 600 rpm. PROTACs (MZ1 or AT1) were diluted from a 10 mM DMSO stock solution to 20 µM in buffer containing 20 mM Bis-tris propane, 150 mM NaCl, 1 mM tris(2-carboxyethyl)phosphine (TCEP), pH 7.4. The final DMSO concentration was 0.01 or 0.02 %. Bromodomain (100 or 200 µM, in the syringe) was titrated into the PROTAC (10 or 20 µM, in the cell). At the end of the titration, the excess of solution was removed from the cell, the syringe was washed and dried, VCB complex (84 or 168 µM, in the same buffer) was loaded in the syringe and titrated into the complex PROTAC:bromodomain. The concentration of the complex in the cell (C) after the first titration (8.4 or 16.8 µM), was calculated as follow:
where: C0 is the initial concentration of the PROTAC in the cell (20 µM), Vcell is the volume of the sample cell (200.12 µL) and Vinj is the volume of titrant injected during the first titration (38.4 µL). Titrations for the binary complex PROTAC:VCB were performed as follow: to the solution of PROTAC (10 or 20 µM, in the cell), buffer (38.4 µL) was added by means of a single ITC injection. The excess of solution was removed from the cell, the syringe was washed and dried, VCB complex (84 or 168 µM, in the same buffer) was loaded in the syringe and titrated into the diluted PROTAC solution. The data were fitted to a single binding site model to obtain the stoichiometry n, the dissociation constant Kd and the enthalpy of binding ΔH using the Microcal LLC ITC200 Origin software provided by the manufacturer. The reported values are the mean ± s.e.m. from independent measurements (eight for VCB into MZ1; seven for VCB into AT1; two for each BD into VCB:PROTAC).
Simulations of ternary complex fractions
Fractions of ternary complexes were calculated by applying the ternary equilibria model in the excel spreadsheet provided in ref. 23. Input parameters were [VHL]t = [BD]t = 40 nM; Kd (VHL) = 66 nM; Kd and α (BD) were as measured by ITC (Table 1).
Biotinylation of VCB
To biotinylate VCB the complex was mixed in a 1:1 stoichiometry with EZ-Link NHS-PEG4-Biotin (Thermo Scientific) and incubated at room temperature for 1 h. To remove any unreacted NHS-biotin the sample was run over a PD-10 desalting column (GE Healthcare) into 20 mM HEPES, pH 7.5, 150 mM sodium chloride and 1 mM DTT.
AlphaLISA assays
All assays were performed at room temperature and plates sealed with transparent film between addition of reagents. All reagents were diluted in 50 mM HEPES, pH 7.5, 100 mM NaCl, 0.1% (w/v) bovine serum albumin and 0.02% (w/v) 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) and each solution was prepared as a 5× stock and mixed into a final volume of 25 μL per well. Each protein sample (biotinylated VCB and His6-BD) and PROTAC were mixed and incubated for 1 h. Ni-coated acceptor beads were added and plates incubated another 1 h. Streptavidin-coated donor beads were added and plates incubated for a final 1 h. Plates were read on a PHERAstar FS (BMG Labtech) using an optic module with an excitation wavelength of 680 nm and emission wavelength of 615 nm. Within each read there was a settling time of 0.1 s, an excitation time of 0.3 s and an integration time of 0.6 s. For BET-BD titration experiments, VCB and PROTACs were kept constant at a final concentration of 20 nM and His6-BD was serially diluted three-in-five from 200 nM. For VCB titration experiments, His6-BDs and PROTACs were kept constant at a final concentration of 40 nM and VCB was serially diluted one in-two from 200 nM. The intensity values were plotted with concentration values on a log10 scale.
Tissue culture
Human HeLa cell lines were obtained from ATCC and were kept in DMEM medium (Gibco) supplemented with 10% FBS (Gibco), L-glutamate (Gibco), penicillin and streptomycin. Cells were kept in an incubator at 37 °C, 5% CO2. All cell lines were tested for mycoplasma contamination every month using MycoAlert™Mycoplasma detection kit (Lonza).
Cell treatment and lysis
HeLa cells were seeded at 2.5×105 per well on a standard six-well plate. After a day, cells were treated with test compounds for the desired period of time. Cells were washed with PBS twice before lysis. Lysis was achieved by applying RIPA buffer (Sigma), supplemented with 1× protease inhibitor cocktail (Roche), Benzonase (Merck) and 0.5 mM MgCl2 to the cells on ice. Lysate was briefly sonicated and then centrifuged at 20,000 × g for 10 min at 4 °C. Supernatant was collected as cell extract and protein concentration was measured by BCA assay. The extract was snap frozen in liquid nitrogen for storage before being used for Western blot analysis. Cycloheximide (C7698, Sigma Aldrich) was used at 100 μg/mL for the indicated times.
Western blot
Blots were probed with antibodies for Brd4 (AbCam ab128874, 1:1,000 dilution), Brd3 (AbCam ab50818, 1:500 dilution), Brd2 (AbCam ab139690, 1:2,000 dilution), β-actin (AbCam ab8227, 1:2,000 dilution) and cMyc (AbCam ab32072, 1:1,000 dilution) antibodies. Blots were developed with anti-Mouse or anti-Rabbit IRDye® 800CW secondary antibody from Licor (1:10,000 dilution) and bands visualized using Licor Odyssey Sa imaging system. Image processing and band intensity quantification were done using Licor Image Studio software Version 5.2.5. Ubiquitination blots were probed with anti-6×His antibody (AbCam ab18184, 1:2,000 dilution) and then with anti-Mouse IgG, HRP-linked antibody (Cell Signaling Technology #7076, 1:2,000 dilution). Probed blots were visualised with ECL Western Blotting Substrate (Pierce #32106) on film.
Sample preparation for MS proteomics
HeLa cells were seeded at 2×106 on a 100-mm plate 24 h before treatment. To treat the cells, culture medium was replaced with 12 mL of medium containing the test compound. After 24 h, medium was removed and cells were washed with 12 mL of cold PBS twice. Samples were kept on ice from this point onwards. Cells were lysed in 0.5 mL of 100 mM Tris pH 8.0, 4% (w/v) SDS supplemented with protease inhibitor cocktail (Roche). The lysate was pulse sonicated briefly and then centrifuged at 17,000 × g for 15 min at 4 °C. The supernatant fraction of cell extract was snap-frozen in liquid nitrogen and stored in −80 °C freezer before further processing.
Sample preparation for MS proteomics
Samples were quantified using a micro BCA protein assay kit (Thermo Fisher Scientific) and 200μg of each sample was processed and digested using the Filter Aided Sample Preparation (FASP) method 58. The samples were then desalted using a 7 mm, 3 ml C18 SPE cartridge column (Empore™, 3M) and labelled with TMT 59 10plex™ Isobaric Label Reagent Set (Thermo Fisher Scientific) as per manufacturers instructions. After labelling, the peptides from the 10 samples were pooled together in equal proportion. The pooled sample was fractionated into 20 discrete fractions using high pH reverse phase chromatography 60 on a XBridge peptide BEH column (130 Å, 3.5 µm 2.1 X 150 mm, Waters) using an Ultimate 3000 HPLC system (Thermo Scientific/Dionex). Column temperature was set to 20 °C. The peptides were separated using a mix of buffers A (10 mM ammonium formate in water, pH 10) and B (10 mM ammonium formate in 90% CH3CN, pH 10). The peptides were eluted from the column using a flow rate of 200 μl/min and a linear gradient from 5% to 60% buffer B over 60 min. The peptides eluted from the column were separated into 40 fractions prior to concatenation into 20 fractions based on the UV signal of each fraction. All the fractions were dried in a speedvac concentrator and resuspended in 10 μl 5% formic acid, then diluted to 1% prior to MS analysis.
nLC-MS/MS analysis
The fractions were analysed sequentially on a Q Exactive™ HF Hybrid Quadrupole-Orbitrap™ Mass Spectrometer (Thermo Scientific) coupled to an Ultimate 3000 RSLC nano UHPLC system (Thermo Scientific) and Easyspray column (75 μm × 50 cm, PepMap RSLC C18 column, 2 μm, 100 Å, Thermo Scientific). The peptides from each fraction were separated using a mix of buffer A (0.1% formic acid in MS grade water) and B (0.08% formic acid in 80% MS grade CH3CN). The peptides from each fraction were eluted from the column using a flow rate of 300 nl/min and a linear gradient from 5% to 40% buffer B over 122 min. The column temperature was set at 50 °C. The Q Exactive™ HF Hybrid Quadrupole-Orbitrap™ Mass Spectrometer was operated in data dependent mode with a single MS survey scan followed by 10 sequential m/z dependant MS2 scans. The 10 most intense precursor ions were sequencially fragmented by Higher energy Collision Dissociation (HCD). The MS1 isolation window was set to 0.4 Da and the resolution set 120,000. MS2 resolution was set as 60,000. The maximum ion injection time for MS1 and MS2 were set at 50 msec and 200 msec, respectively.
Peptide and protein identification
The raw ms data files for all 20 fractions were merged and searched against the Sprot database with taxonomy set to Homo sapiens by Proteome Discoverer Version 1.4 (Thermo Scientific) using the Mascot v.2.4.1 (Matrix Science) search engine for protein identification and TMT reporter ion quantitation. The identification was based on the following database search criteria: enzyme used Trypsin/P; maximum number of missed cleavages equal to 2; precursor mass tolerance equal to 10 ppm; fragment mass tolerance equal to 0.06 Da; dynamic modifications: Oxidation (M), Dioxidation (M), Acetyl (N-term), Gln->pyro-Glu (N-term Q), Pro->Hyp (P), Deamidation(N,Q); static modifications: Carbamidomethyl (C), TMT10plex (K), TMT10plex (N-term). For protein identification the mascot ion score threshold was set at 30 and a minimum of 2 peptides was required.
Peptide and protein quantitation
The ratios of TMT reporter ion abundances in MS/MS spectra generated by HCD from raw data sets were used for TMT quantification. Isotopic correction factors were applied for the batch of TMT reagents used in this experiment as per manufacturers recommendation. A minimum of two unique peptides was used for quantitation and the resultant ratios were normalized based on protein median. Quantified proteins were filtered if the absolute fold change difference between the two DMSO replicates was ≥ 1.3.
Model construction of the multisubunit CRL2VHL–MZ1–Brd4 complex
A structural model of the CRL2VHL (VHL–EloC–EloB–Cul2–Rbx1) with bound MZ1–Brd4BD2 at one end and E2–Ubiquitin at the other end was constructed in PyMOL by aligning our Brd4BD2–MZ1–VHL–EloC–EloB on to the quaternary structure VHL–EloC–EloB–Cul2NTD (PDB entry 4WQO). Cul2NTD and Cul2CTD were modelled based on the structures of Cul5NTD (PDB entry 2WZK) and Cul1CTD–Rbx1 (PDB entry 3RTR) and superposed onto full-length Cul1 from PDB entry 1LDK. Finally, the Rbx1–E2–Ub arm was modelled based on the crystal structure of Rbx1–Ubc12~NEDD8–Cul1–Dcn1 (PDB entry 4P5O) superposed via the cullin subunit.
Recombinant ubiquitination experiments and ubiquitination site identification
His6-tagged BET-BDs (2 μM) were ubiquitinated in the presence of E1 Ube1 (19 nM), E2 Ube2d1 (145 nM), ubiquitin (Ubiquigent, 1 mg/mL), recombinant VHL-ElonginC-ElonginB-Cullin2-Rbx1 complex (330 nM) and MZ1 (2 μM) standing for 24 h at room temperature in a buffer of 25 mM HEPES, pH 7.5, 5 mM MgCl2, 100 mM NaCl, 2 mM ATP, 0.1 mg/mL BSA and 1 mM TCEP. Reactions were terminated by the addition of 1× NuPAGE LDS sample buffer (Invitrogen).
Sample preparation for MS
Samples were run 1–2 cm into a pre-cast 4–12% Bis-Tris NuPAGE gel and the entire protein content of each lane excised, washed and dried. Proteins were reduced with 10 mM DTT and 20 mM ammonium carbonate at 56 °C for 60 min and then alkylated with 50 mM N-ethylmaleimide and 20 mM ammonium carbonate at room temperature for 30 min. Proteins were trypsinized overnight at 30 °C and the resulting peptides extracted and dried down.
nLC-MS/MS analysis
Each peptide sample was reconstituted in 10 µl 5% formic acid then diluted to 1% prior to MS analysis. Peptide samples were injected onto a C18 PepMap 100 (300 μm x 5 mm, Thermo Scientific) trap column with buffer A (0.1% formic acid in MS grade water) using an Ultimate 3000 RSLC nano UHPLC system. After a 5 min wash at 5 µl/min the sample was then eluted onto an EasySpray PepMap RSLC C18 column (75 µm x 50 cm, Thermo Scientific) into a LTQ Orbitrap Velos Pro via an EasySpray ion source. The peptides were eluted from the column using a flow rate of 300 nl/min and a linear gradient from 2% to 40% buffer B (0.08% formic acid in 80% MS grade CH3CN) over 124 min. The column temperature was set at 50 °C. The Orbitrap Velos Pro ms system was operated in data dependant acquisition mode using a Top 15 method with Lockmass = 445.120024. A MS1 survey scan with a range of 335–1800 m/z and a resolution of 60,000 was followed by 15 sequential MS2 scans at the normal scan rate using the LTQ Velos ion trap. The FTMS and ITMS AGC targets were set to 1e6 ions and 5e3 ions respectively. The FTMS and ITMS maximum fill times were set to 500 msec and 100 msec respectively. ITMS isolation width was set at 2 Da with a normalised collision energy of 35, a default charge state of 2, an Activation Q of 0.250 and Activation Time of 10 msec.
Peptide and protein identification
The resultant raw data was searched against the Sprot database with a taxonomy filter set to H. sapiens using the Mascot v. 2.4.1 (Matrix Science) search engine to identify peptides containing Lysines with εN-linked di-glycine modifications. Peptide mass tolerance was set to 10 ppm and the fragment mass tolerance set to 0.6 Da. The number of maximum miss-cleavages was set to 2. The enzyme was set to Trypsin/P and the following variable modifications were considered: Acetyl (N-term), Deamidated (NQ), Dioxidation (M), Gln->pyro-Glu (N-term Q), GlyGly (K), Oxidation (M). A fixed modification for Cysteine was set to N-ethylmaleimide. A mascot ion score threshold was set at 37 to filter non-significant peptide identifications.
Statistical methods
No statistical methods were used to predetermine sample size. The experiments were not randomized, and the investigators were not blinded to allocation during experiments and outcome assessment. For all experiments, number of replicates (n), mean value, error value and P value cutoffs are described in the respective figure legends. Error bars are shown for all data points with replicates as a measure of variation with the group. All t-tests performed were two-tailed t-tests assuming equal variances.
Supplementary Material
Acknowledgements
This work was supported by the European Research Council (ERC-2012-StG-311460 DrugE3CRLs Starting Grant to A.C.); the UK Biotechnology and Biological Sciences Research Council (BBSRC grant BB/J001201/2 to A.C.); the European Commission (H2020-MSCA-IF-2014-655516 Marie Skłodowska-Curie Actions Individual Fellowship to K-H.C., and H2020-MSCA-IF-2015-806323 Marie Skłodowska-Curie Actions Individual Fellowship to X.L.); and the Wellcome Trust (Strategic Awards 100476/Z/12/Z for biophysics and drug discovery and 094090/Z/10/Z for structural biology and X-ray crystallography to the Division of Biological Chemistry and Drug Discovery). We are thankful to P. Fyfe for support with the in-house X-ray facility; L. Finn for support with tissue culture facility (MRC-PPU); the Ferguson lab for access to LI-COR equipment; T. Cardote for the gift of full-length Cul2-Rbx1 and A. Knebel (MRC-PPU/DSTT) for the gift of E1 and E2 enzymes; the Division of Computational Biology for support with computational cluster; and to Diamond Light Source for beamtime (BAG proposal MX10071) and beamline support at beamline I04-1.
Footnotes
Accession codes
Atomic coordinates and structure factors for hsBrd4BD2–MZ1–hsVHL–hsEloC–hsEloB have been deposited in the Protein Data Bank (PDB) under accession number 5T35.
Data availability
Any supplementary information, chemical compound information and source data are available in the online version of the paper.
Author Contributions
A.C. conceived the idea and directed the project.
M.S.G., X.L., A.T., K.-H.C. and A.C. designed the experiments and interpreted results
M.S.G., X.L., A.T., and K.-H.C. performed experiments
A.T. and M.Z. contributed to compound design and synthesized compounds
W.C. performed MS proteomics experiments under the supervision of D.J.L.
M.S.G., X.L. and A.C. wrote the manuscript with input from all other authors.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Huang X, Dixit VM. Drugging the undruggables: exploring the ubiquitin system for drug development. Cell Res. 2016;26:484–498. doi: 10.1038/cr.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petzold G, Fischer ES, Thomä NH. Structural basis of lenalidomide-induced CK1α degradation by the CRL4(CRBN) ubiquitin ligase. Nature. 2016;532:127–130. doi: 10.1038/nature16979. [DOI] [PubMed] [Google Scholar]
- 3.Matyskiela ME, et al. A novel cereblon modulator recruits GSPT1 to the CRL4(CRBN) ubiquitin ligase. Nature. 2016;535:252–257. doi: 10.1038/nature18611. [DOI] [PubMed] [Google Scholar]
- 4.Lai AC, Crews CM. Induced protein degradation: an emerging drug discovery paradigm. Nat Rev Drug Discov. 2016 doi: 10.1038/nrd.2016.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakamoto KM, et al. Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. P Natl Acad Sci Usa. 2001;98:8554–8559. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winter GE, et al. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348:1376–1381. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zengerle M, Chan K-H, Ciulli A. Selective Small Molecule Induced Degradation of the BET Bromodomain Protein BRD4. ACS Chem Biol. 2015;10:1770–1777. doi: 10.1021/acschembio.5b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu J, et al. Hijacking the E3 Ubiquitin Ligase Cereblon to Efficiently Target BRD4. Chem Biol. 2015;22:755–763. doi: 10.1016/j.chembiol.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bondeson DP, et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat Chem Biol. 2015;11:611–617. doi: 10.1038/nchembio.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deshaies RJ. Protein degradation: Prime time for PROTACs. Nat Chem Biol. 2015;11:634–635. doi: 10.1038/nchembio.1887. [DOI] [PubMed] [Google Scholar]
- 11.Toure M, Crews CM. Small-Molecule PROTACS: New Approaches to Protein Degradation. Angew Chem Int Ed Engl. 2016;55:1966–1973. doi: 10.1002/anie.201507978. [DOI] [PubMed] [Google Scholar]
- 12.Lai AC, et al. Modular PROTAC Design for the Degradation of Oncogenic BCR-ABL. Angew Chem Int Ed Engl. 2016;55:807–810. doi: 10.1002/anie.201507634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galdeano C, et al. Structure-guided design and optimization of small molecules targeting the protein-protein interaction between the von Hippel-Lindau (VHL) E3 ubiquitin ligase and the hypoxia inducible factor (HIF) alpha subunit with in vitro nanomolar affinities. J Med Chem. 2014;57:8657–8663. doi: 10.1021/jm5011258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frost J, et al. Potent and selective chemical probe of hypoxic signalling downstream of HIF-α hydroxylation via VHL inhibition. Nat Commun. 2016;7:13312. doi: 10.1038/ncomms13312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicodeme E, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hon W-C, et al. Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL. Nature. 2002;417:975–978. doi: 10.1038/nature00767. [DOI] [PubMed] [Google Scholar]
- 19.Min J-H, et al. Structure of an HIF-1alpha -pVHL complex: hydroxyproline recognition in signaling. Science. 2002;296:1886–1889. doi: 10.1126/science.1073440. [DOI] [PubMed] [Google Scholar]
- 20.Filippakopoulos P, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka M, et al. Design and characterization of bivalent BET inhibitors. Nat Chem Biol. 2016;12:1089–1096. doi: 10.1038/nchembio.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitty A. Cooperativity and biological complexity. Nat Chem Biol. 2008;4:435–439. doi: 10.1038/nchembio0808-435. [DOI] [PubMed] [Google Scholar]
- 23.Douglass EF, Miller CJ, Sparer G, Shapiro H, Spiegel DA. A comprehensive mathematical model for three-body binding equilibria. J Am Chem Soc. 2013;135:6092–6099. doi: 10.1021/ja311795d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eglen RM, et al. The use of AlphaScreen technology in HTS: current status. Curr Chem Genomics. 2008;1:2–10. doi: 10.2174/1875397300801010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts JM, Bradner JE. A Bead-Based Proximity Assay for BRD4 Ligand Discovery. Curr Protoc Chem Biol. 2015;7:263–278. doi: 10.1002/9780470559277.ch150024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stebbins CE, Kaelin WG, Pavletich NP. Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function. Science. 1999;284:455–461. doi: 10.1126/science.284.5413.455. [DOI] [PubMed] [Google Scholar]
- 27.Duda DM, et al. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou M, Li Q, Wang R. Current Experimental Methods for Characterizing Protein-Protein Interactions. ChemMedChem. 2016;11:738–756. doi: 10.1002/cmdc.201500495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raina K, et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. P Natl Acad Sci Usa. 2016;113:7124–7129. doi: 10.1073/pnas.1521738113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bulatov E, Ciulli A. Targeting Cullin-RING E3 ubiquitin ligases for drug discovery: structure, assembly and small-molecule modulation. Biochem J. 2015;467:365–386. doi: 10.1042/BJ20141450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fischer ES, et al. Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature. 2014;512:49–53. doi: 10.1038/nature13527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischer ES, Park E, Eck MJ, Thomä NH. SPLINTS: small-molecule protein ligand interface stabilizers. Curr Opin Struct Biol. 2016;37:115–122. doi: 10.1016/j.sbi.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan X, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 34.Sheard LB, et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature. 2010;468:400–405. doi: 10.1038/nature09430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ito T, et al. Identification of a primary target of thalidomide teratogenicity. Science. 2010;327:1345–1350. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- 36.Lu G, et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science. 2014;343:305–309. doi: 10.1126/science.1244917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krönke J, et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343:301–305. doi: 10.1126/science.1244851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chamberlain PP, et al. Structure of the human Cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs. Nat Struct Mol Biol. 2014;21:803–809. doi: 10.1038/nsmb.2874. [DOI] [PubMed] [Google Scholar]
- 39.Pommier Y, Marchand C. Interfacial inhibitors: targeting macromolecular complexes. Nat Rev Drug Discov. 2012;11:25–36. doi: 10.1038/nrd3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thiel P, Kaiser M, Ottmann C. Small-Molecule Stabilization of Protein-Protein Interactions: An Underestimated Concept in Drug Discovery? Angew Chem Int Ed Engl. 2012;51:2012–2018. doi: 10.1002/anie.201107616. [DOI] [PubMed] [Google Scholar]
- 41.Illendula A, et al. A small-molecule inhibitor of the aberrant transcription factor CBFβ-SMMHC delays leukemia in mice. Science. 2015;347:779–784. doi: 10.1126/science.aaa0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waring MJ, et al. Potent and selective bivalent inhibitors of BET bromodomains. Nat Chem Biol. 2016;12:1097–1104. doi: 10.1038/nchembio.2210. [DOI] [PubMed] [Google Scholar]
- 43.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schüttelkopf AW, van Aalten DMF. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 50.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 52.Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 53.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brooks BR, et al. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vanommeslaeghe K, Mackerell AD. Automation of the CHARMM General Force Field (CGenFF) I: bond perception and atom typing. J Chem Inf Model. 2012;52:3144–3154. doi: 10.1021/ci300363c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graphics. 1996;14:33–8. 27–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 57.Glykos NM. Software news and updates. Carma: a molecular dynamics analysis program. J Comput Chem. 2006;27:1765–1768. doi: 10.1002/jcc.20482. [DOI] [PubMed] [Google Scholar]
- 58.Manza LL, Stamer SL, Ham A-JL, Codreanu SG, Liebler DC. Sample preparation and digestion for proteomic analyses using spin filters. Proteomics. 2005;5:1742–1745. doi: 10.1002/pmic.200401063. [DOI] [PubMed] [Google Scholar]
- 59.Thompson A, et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 60.Gilar M, Olivova P, Daly AE, Gebler JC. Orthogonality of separation in two-dimensional liquid chromatography. Anal Chem. 2005;77:6426–6434. doi: 10.1021/ac050923i. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.