Abstract
PUF60 encodes a nucleic acid-binding protein, a component of multimeric complexes regulating RNA splicing and transcription. In 2013, patients with microdeletions of chromosome 8q24.3 including PUF60 were found to have developmental delay, microcephaly, craniofacial, renal and cardiac defects. Very similar phenotypes have been described in six patients with variants in PUF60, suggesting that it underlies the syndrome. We report 12 additional patients with PUF60 variants who were ascertained using exome sequencing: six through the Deciphering Developmental Disorders Study and six through similar projects. Detailed phenotypic analysis of all patients was undertaken. All 12 patients had de novo heterozygous PUF60 variants on exome analysis, each confirmed by Sanger sequencing: four frameshift variants resulting in premature stop codons, three missense variants that clustered within the RNA recognition motif of PUF60 and five essential splice-site (ESS) variant. Analysis of cDNA from a fibroblast cell line derived from one of the patients with an ESS variants revealed aberrant splicing. The consistent feature was developmental delay and most patients had short stature. The phenotypic variability was striking; however, we observed similarities including spinal segmentation anomalies, congenital heart disease, ocular colobomata, hand anomalies and (in two patients) unilateral renal agenesis/horseshoe kidney. Characteristic facial features included micrognathia, a thin upper lip and long philtrum, narrow almond-shaped palpebral fissures, synophrys, flared eyebrows and facial hypertrichosis. Heterozygote loss-of-function variants in PUF60 cause a phenotype comprising growth/developmental delay and craniofacial, cardiac, renal, ocular and spinal anomalies, adding to disorders of human development resulting from aberrant RNA processing/spliceosomal function.
Introduction
In 2009, Verheij et al.1 reported overlapping interstitial microdeletions of chromosome 8q24 in two patients who had colobomata, congenital heart defects, limb abnormalities, developmental delay and seizures (MIM 615583). Dauber et al.2 later described five patients who also had microdeletions of 8q24.3 and similar phenotypes including ocular colobomata, microcephaly, developmental delay, short stature, craniofacial, cardiac and renal defects. All five patients had in common a 78-kb deleted region containing three genes: SCRIB, NRBP2 and PUF60. A further patient with a missense variant in PUF602 and a foetus with an 8q24.3 deletion encompassing PUF60 occurring in association with an atrioventricular septal defect, a hypoplastic aortic arch, facial dysmorphism and other anomalies3 strongly implicated this single gene as the cause of the phenotype.
The poly-U-binding splicing factor gene (PUF60) encodes a nucleic acid-binding protein, which through interaction with other proteins such as SF3B4, regulates pre-RNA splicing and transcription.2, 4 In 2013 a single patient with an intragenic variant in PUF60 was identified2 and recently five other cases have been reported.5 Here, we present clinical and molecular data from 12 additional patients with de novo variants in PUF60 identified via exome sequencing undertaken for undiagnosed developmental disorders. The clinical features we observed in these patients and those previously published suggest that there is an emerging PUF60-related phenotype, although this is variable and may be difficult to recognize.
Methods
Patient ascertainment
Six of the affected patients were recruited via the Deciphering Developmental Disorders (DDD) study6 (http://www.ddduk.org) open to the UK NHS Regional Genetics Services. Five patients were recruited via locally based exome sequencing services by Clinical Geneticists in Norway, France and the United States and one via an exome sequencing study for patients with Cornelia de Lange Syndrome (CdLS; MIM 122470).6 All patients were assessed by their Clinical Geneticist who assisted with systematic detailed phenotyping. Patient growth centiles and z-scores were calculated using the UK WHO data (http://www.rcpch.ac.uk/growthcharts).
Genomic analysis
Trio-based exome sequencing was undertaken for the six affected patients and their parents were identified via the DDD study.7 High-resolution analysis for copy number abnormalities using array-based comparative genomic hybridization was also performed. Putative de novo variants were identified from exome data using the DeNovoGear software8 and then validated by targeted Sanger sequencing. For patients 7–12, alternative but similar trio exome sequence approaches were used, for example, for patient 10 a proband-based exome sequencing as described previously,8 for patient 11 exome capture with Nextera Rapid Capture Exome Kit (Illumina Inc., San Diego, CA, USA) and sequencing with HiSeq4000 (Illumina). For patient 12, Agilent Clinical Research Exome Kit (Agilent Technologies, Santa Clara, CA, USA) identified exonic and periexonic fragments, followed by massive parallel next-generation sequencing. Analysis of exome data was undertaken with DeNovoGear9 or FILTUS,10 and for patient 11, raw data was processed using an end-to-end in-house database. Identified variants were annotated using standard databases and were filtered based on the established criteria. Detailed descriptions of the wet and dry laboratory pipelines are published elsewhere.11 For patient 12, analysis was performed with a custom-developed Xome Analyser (Gene Dx, Bethesda, MD, USA). Mean depth of coverage for the analysis was x121; and quality threshold (≥10x) was achieved for 95.5% of the target sequence. As for DDD patients, significant variants were confirmed by targeted Sanger sequencing. Variant data has been submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) – submission ID SUB2319251.
Analysis of aberrant PUF60 splicing
Dermal fibroblasts were obtained from patient 8 (3781–3781) by skin punch biopsy and cultured in amnioMAX C-100 complete medium (Life Technologies, Carlsbad, CA, USA) as described previously.12 RNA was extracted from primary skin-derived fibroblast cell lines from patient 8 (3781–3781) and two sex-matched controls using RNeasy Kit (Qiagen, Hilden, Germany) and treated with DNAseI to eliminate genomic DNA, according to the manufacturer's instructions. Complementary DNA (cDNA) synthesis was carried out using random oligomer primers and AMV Reverse Transcriptase (Roche, Penzburg, Germany). The cDNA samples were resolved on an E-Gel electrophoresis system and extracted according to the manufacturer's instructions (Thermo Fisher Scientific, Waltham, MA, USA) to sequence the amplicons corresponding to the normal and mutant alleles. Sequences of primers used for cDNA amplification and Sanger sequencing of PUF60 are available upon request.
Results
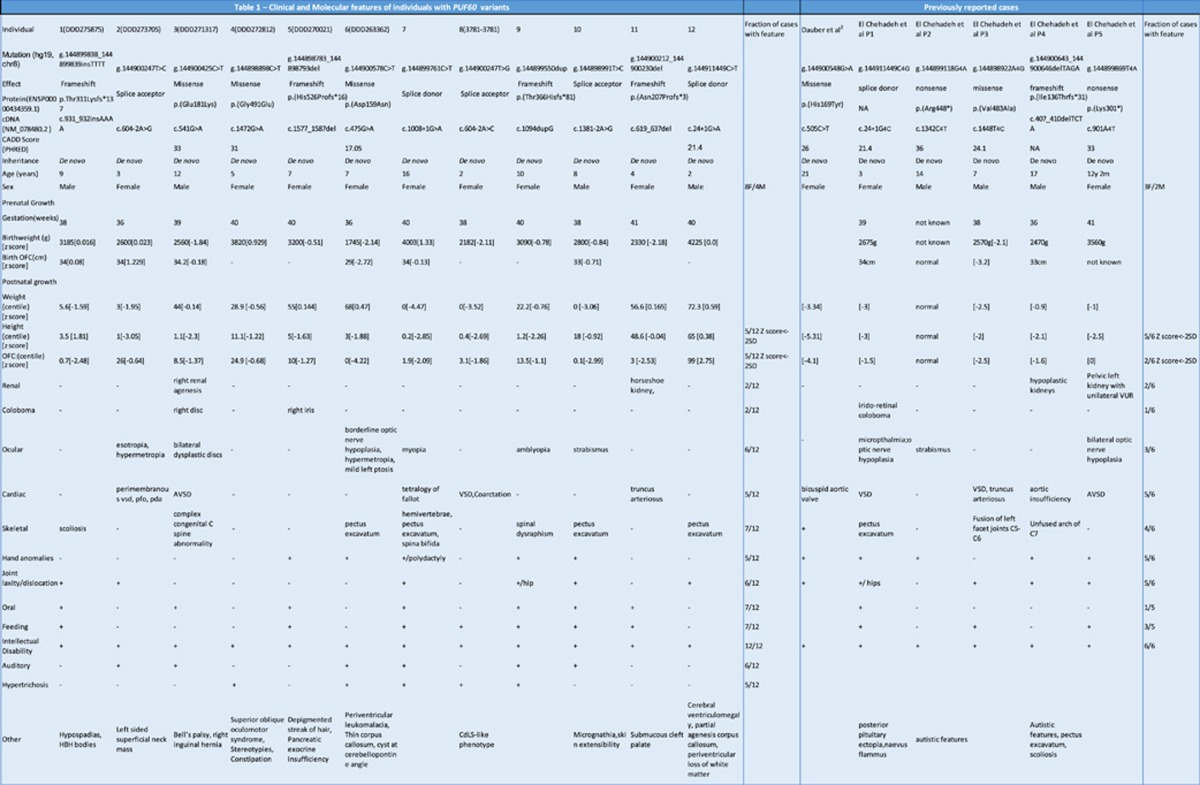
The clinical features and PUF60 variants in the 12 patients compared with the previously reported cases2, 5 are presented in Table 1.
Table 1. Clinical and genetic findings in patients with PUF60 variants.
Abbreviations: AVSD, atrioventricular septal defect; CADD, Combined Annotation Dependent Depletion; CdLS, Cornelia de Lange Syndrome; HBH, haemoglobin H; OFC, occipitofrontal circumference; PDA, patent ductus arteriosus; PFO, patent foramen ovale; VSD, ventricular septal defect.
Table lists presence of phenotypic features in each patient in our cohort, as well as in previously reported patients along with total frequency of each phenotypic feature. Mutation data including genomic location, predicted effect on protein are listed for all patients with CADD score14 calculations for all missense mutations and where previously reported. (CADD is a framework that integrates multiple existing annotation tools to give one scaled score of deleteriousness for a variant. A CADD score of 20 means that a variant is among the top 1% of deleterious variants in the human genome. A score of 30 means that the variant is in the top 0.1% and so forth.).
Variants
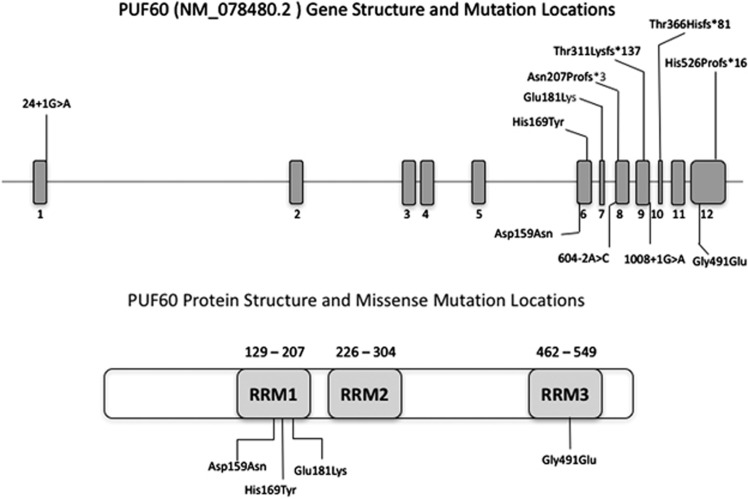
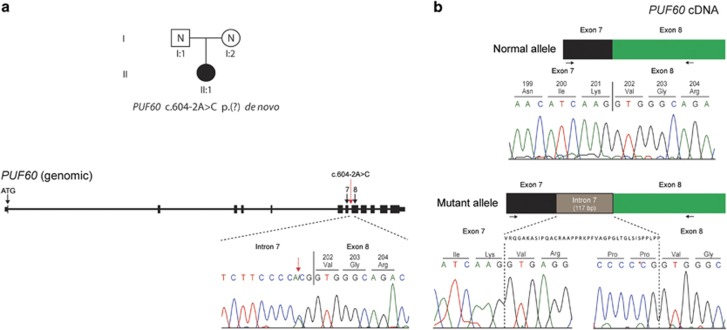
The distribution of the heterozygous de novo variants (DNV) is shown in context of the genomic structure of PUF60 in Figure 1. Nine of the DNV were predicted to cause loss-of function suggesting haploinsufficiency as the likely mutational mechanism. Four patients (1 (DDD275875),13 5 (DDD270021), 9 and 11) had frameshift variants resulting in early stop codons. Five patients (2 (DDD273705), 7 (who had the most complex phenotype), 8 (3781–3781), 10 and 12) had essential splice-site variants. In patient 8 (3781–3781), a de novo c.604−2A>C variant occurred in the exon 8 splice-acceptor site and was predicted to alter splicing of PUF60 mRNA. The effect of this variant was investigated as above: analysis of cDNA confirmed abnormal inclusion of the complete intron 7 in the mutant allele resulting in an apparent in-frame inclusion of 39 amino acids in the open reading frame of the transcript (Figure 2). It is thus not clear whether this essential splice-site variant will result in complete loss of function. Nonetheless, her phenotype is consistent with respect to developmental delay, short stature and cardiac involvement. Patient 12 was the only one to have a variant outside of the exon 6–12 region. The wild-type donor site is predicted to be disrupted resulting in skipping of exon 1; however, little is known about the exon 1 function (Human Splicing Finder score of −31.86 – probably affecting splicing; http://www.umd.be/HSF3/HSF.html).
Figure 1.
PUF60 gene diagram (above) indicating mutations found in our cohort which cluster in the second half of the coding region, apart from the single variant in exon 1. Note recurrent splicing variant in exon 8. Below is pictured the known protein structure and location of RRM domains in relation to the missense variants.
Figure 2.
A de novo variant in PUF60 results in aberrant splicing at exon 8 splice-acceptor site. (a) The heterozygous, de novo PUF60 variant, c.604−2A>C was identified in patient II:1 through trio-based exome sequencing of family 3781. (b) Sequencing of skin-derived cDNA from patient II:1 (family 3781) showed normal splicing of exons 7 and 8 from one allele (top) and aberrant splicing with inclusion of the complete intron 7 from the mutant allele (bottom). Black arrows show the position of the oligonucleotide primers used for cDNA amplification and sequencing. The genomic context of the PUF60 gene is shown, with exons indicated as black boxes. The location of the PUF60 c.604−2A>C variant in intron 7 is indicated by a dotted red arrow, with the Sanger sequence trace from patient II:1 (family 3781) presented underneath. Variant nomenclature, exon numbering and the PUF60 messenger RNA sequence are based on sequence accession numbers NM_078480.2 (mRNA) and NPs_510965.1 (protein) and GenBank accession number NG_033879.1.
Three patients in this study had missense variants located in regions encoding one of the three RNA recognition motif (RRM) domains of the protein (Alamut), c.541G>A(p.(Glu181Lys)) and c.475G>A(p.(Asp159Asn)) are located in RRM1 and c.1472G>A(p.(Gly491Glu) in RRM3. CADD scores14 are shown in Table 1 and support effect on function of all three variants.
None of the PUF60 variants we identified are listed in the Exome Variant Server, Exome Aggregation Consortium or the dbSNP population databases.
Growth
Birthweight was normal except for patients 6 (DDD263362; Z-score −2.14), 8 (3781–3781; Z-score −2.11) and 11 (Z-score −3.24). Body weight in childhood remained in the normal range in all patients apart from patients 7 and 8 (3781–3781; Z-scores −4.47 and −3.52 respectively) in both of whom other growth parameters were also significantly reduced. Nine patients had short stature defined as height below 5th centile and five had Z-scores below −2. Head circumference was proportionate to stature, apart from in five patients (1 (DDD275875), 6 (DDD263362), 7, 10 and 11) who had true microcephaly (Z-scores −2.48, −4.22, −2.09, −2.99, −2.53 respectively).
Musculoskeletal
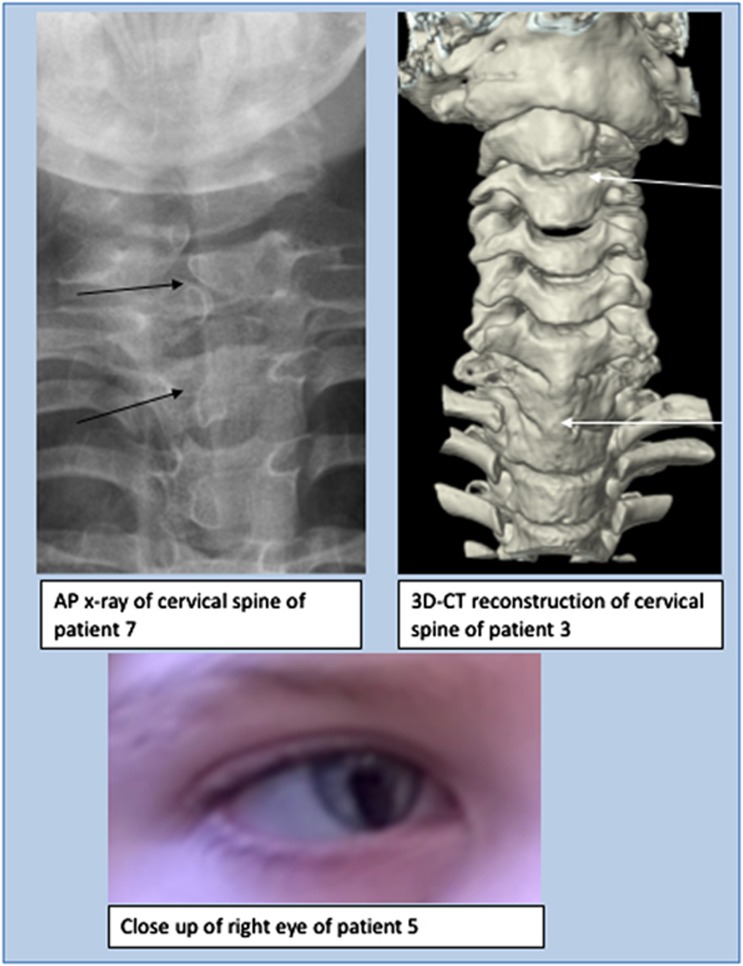
Skeletal abnormalities, especially in the spine, were seen in seven patients. In patient 3 (DDD271317), there was fusion of the whole vertebral bodies of C6 and C7 and the anterior part of C5 (Figure 3) and the posterior spinous processes of T6, T7 and T1 were unusually prominent. His cervical spine stability requires neurosurgical monitoring. Patient 7 had several cervical and thoracic hemivertebrae, spina bifida, thoracic kyphosis, bilateral rudimentary ribs at C7 and pectus excavatum (possibly exacerbated by sternotomy during cardiac surgery). In patient 9, the conus terminated at the L2-L3 disc space and the filum, 2 mm in width, extended from L3 to S2 associated with posterior osseous dysraphism of the sacrum. Patients 6 (DDD263362), 10 and 12 had pectus excavatum. Patient 1 (DDD275875) had a lower thoracic scoliosis and pes planus. He also had shoulder subluxation and generalized joint laxity, which was a common feature in the cohort. Patients 2 (DDD273705), 7, 9, 10 and 12 also had joint hypermobility; in one case this was associated with bilateral hip and interphalangeal joint dislocations. Digital anomalies in the group included unilateral preaxial polydactyly of the left hand with a broad proximal phalanx and duplicated distal phalanx (patient 7), short broad hands with bilateral fifth finger clinodactyly (patient 5 (DDD270021), left talipes, broad thumbs/halluces, fifth finger clinodactyly, 2–3 toe syndactyly with an overriding second toe on the left (patient 6 (DDD263362)) and bilateral hypoplasia of the fifth fingers (patient 9).
Figure 3.
Spinal features (above): AP x-ray of cervical spine of patient 7 demonstrating hemivertebrae (left) and 3D-CT reconstruction of cervical spine of patient 3 demonstrating abnormalities of articulation of atlas with C1, vertebral bodies of C2/C3 and fusion of bodies of C6/C7/T1 and ocular features (below): right iris coloboma in patient 5.
Auditory
Two patients had very narrow and easily occluded external auditory meatus, two had severe otitis media with episodic conductive hearing loss and two wear hearing aids for bilateral conductive hearing loss.
Development
All patients had mild to moderate global developmental delay. Most walked by 24 months and the latest did so by 30 months (patient 12 was not yet walking at 24 months). Most spoke their first word by 30 months. Three patients had difficulties forming a pincer grip and with fine motor coordination for activities such as handwriting. For those who are currently of school age, all receive additional educational support and four attend either a special needs school or class. Some episodes of difficult or immature behaviour including attention deficit disorder, head-banging, self-injury or temper tantrums were reported by parents of five patients.
Neurology
Patient 6 (DDD263362) had a stormy postnatal course during which bilateral intraventricular haemorrhages occurred, later associated with mild diplegia. An MRI brain scan showed periventricular leukomalacia, a thin corpus callosum and a cyst at the right cerebellopontine angle. Patient 7 had a cardiac catheterization at 9 months of age and following a right middle cerebral artery occlusion had a left-sided hemiparesis and right facial weakness. A brain MRI scan of patient 11 showed mild periventricular gliosis. Patient 12 had an MRI scan, which showed cerebral ventriculomegaly, partial agenesis corpus callosum and loss of white matter in the periventricular regions. Two patients have developed seizures.
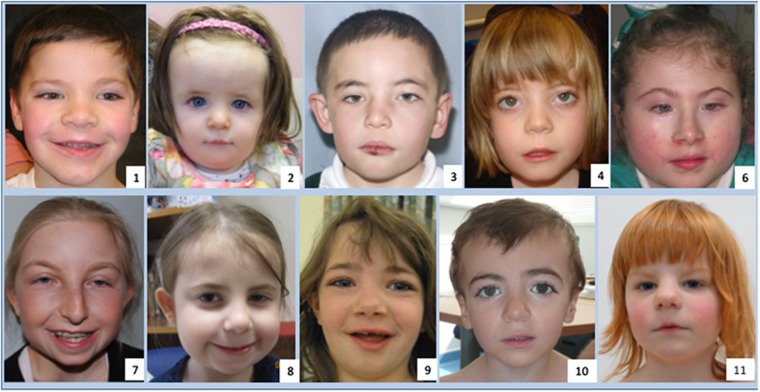
Facial features
The facial features (Figure 4) included a short neck in five >patients, a high forehead in six, micrognathia in four, a long philtrum in five, bushy eyebrows in four and a thin upper lip in four patients. One patient had synophrys and long eyelashes, another had bilateral preauricular pits and a third, a right accessory auricle and skin tag on the contralateral side of the neck. Two patients had facial features initially suggesting CdLS. Two other patients were tested for mutations in CREBBP and EP300 initially. One patient was tested for Coffin–Siris syndrome.
Figure 4.
Faces of patients (1, 2, 3, 4,6, 7, 8, 9, 10 and 11) with PUF60 variants, illustrating the thin upper lip, long philtrum, micrognathia and the flaring of eyebrows and narrow almond-shaped palpebral fissures, which are variably present. Patients 6 and 8 were assessed for CdLS and patients 7 and 10 had CREBBP analysis because of facial similarities with Rubinstein–Taybi syndrome. Patient 9 was investigated for Coffin–Siris syndrome.
Other
Recurrent ocular, cardiac and renal features occurred and are detailed in Table 1. Five patients had hypertrichosis of the face. Two patients had macrodontia, one with ectopic teeth and one early secondary dentition. Early feeding problems were very common: in one patient a gastrostomy was required from 9 months to 7 years of age and three patients were prone to aspiration episodes and complications in infancy, this resolved in two of them by late infancy.
Discussion
The clinical and molecular data we observed in 12 patients suggest that variants in PUF60 cause a syndrome characterized by short stature, developmental delay, dysmorphic facial features and structural malformations of the heart, eye and variably other organs. We are aware of 14 PUF60-related published cases: six with intragenic de novo missense variants2, 5 and eight with chromosome 8q24.3 deletions encompassing PUF60.1, 2, 3 The pattern of their congenital malformations and perturbation of growth and development shows some common themes.
Comparison of our patients with the six previously published for whom data are available (Table 1) suggests that short stature, relative microcephaly and developmental/cognitive delay are consistent findings. The only exception is patient 12 who had macrocephaly, hydrocephalus and normal stature, and in whom the variant was at the exon 1 boundary. A previously published patient had an identical variant5 but their phenotype was more typical, thus predictions on phenotype based on position of specific variants are not possible at this stage. We observed other similarities: a high proportion of cases overall had abnormal segmentation of the vertebrae, which occurred at different levels of the spine. One of our patients and two published patients5 had cervical vertebral fusion/abnormal cervical spines. Other authors found more distal lesions such as fusion of L5-S1, sacral dysplasia and agenesis of the coccyx,2 and other patients, including patients 7, had hemivertebrae in the thoracic and lumbar spine. We and others found minor developmental abnormalities of the distal limbs, including brachydactyly of the fifth fingers, clinodactyly and preaxial polydactyly. Ocular coloboma involving the anterior segment of the eye, choroid and/or the retina, were described in three previous as well as in two of our patients. Renal malformations such as ectopic fused kidneys, pelvic kidney, a unilateral polycystic kidney and renal hypoplasia or unilateral agenesis have been observed,2, 5 and in our cohort there was one case each of a unilateral kidney and a horseshoe kidney. Five of our patients had significant congenital heart defects and other authors5 have also reported ventricular septal defects in combination with truncus arteriosus coarctation or the aorta and bicuspid aortic valve2 and atrioventricular septal defects and hypoplastic aortic arch.3
While all 26 patients with deletions or variants of PUF60 had developmental delay and intellectual disability, those with deletions appear to be more severely affected.1, 2 The reported microdeletions of chromosome 8q24.3 may include other currently unknown genes that contribute to neurological and cognitive development. It is interesting that seizures were observed in our patients as well as in previous reports. Feeding difficulties and recurrent respiratory infections were common to some patients in both groups.
Previous authors commented on phenotypic similarity of PUF60-deleted patients with SF3BF4-related Nager syndrome (MIM 154400) and EFTUD2-related mandibulofacial dysostosis (MIM 610536) and suggested that PUF60 deficiency can be considered within the spectrum of craniofacial disorders resulting from spliceosome malfunction.15 However, coloboma, optic nerve hypoplasia and facial hirsutism have not hitherto been reported in syndromes ascribed to spliceosomal dysfunction. We and others noticed that some patients had features reminiscent of CdLS, particularly where facial hypertrichosis and prominent eyebrows were present. Comparison of the facial features in all patients with PUF60 deletions or variants where data were available indicated that micrognathia, a long philtrum and thin upper lip, synophrys, preauricular pits and cranial asymmetry were common findings. Additionally, two of our patients had very narrow external auditory canals, consistent with abnormal branchial arch development during embryogenesis, also thought to be a process disturbed in mandibulofacial dysostosis and Nager syndromes; however, the facial dysmorphology of patients with PUF60 variants is far more subtle than in EFTUD2 and SF3B4-related phenotypes and mandibulofacial dysostosis was not present in the patients we report. Furthermore, it is interesting to note that none of the patients were clinically suspected of a known spliceosomopathy. We conclude that the facial features associated with PUF60 might be recognized independently, but the diagnosis is more likely to be considered in a child with short stature, developmental delay and additional malformations described.
There is accumulating evidence from this and other phenotypic studies that PUF60 is important in human embryonic development. PUF60 belongs to the RRM half pint family and contains three RRM domains. Consistent with a developmental role, PUF60 encodes a DNA- and RNA-binding protein that is involved in diverse nuclear processes such as pre-mRNA splicing and regulation of transcription. During pre-mRNA splicing, PUF60 interacts with U2AF2 to promote splicing of an intron with a weak 3' splice site4 and has a role in alternative splicing.2 PUF60 is one of the non-core components that can ally with the U2 spliceosomal small nuclear ribonucleoprotein particles, or U2 spliceosome complex.16 Finally, PUF60 has been associated with the transcriptional repression of MYC through association with FUBP1. The mechanism by which reduced PUF60 expression affects cellular functions are not yet understood and beyond the scope of this study. It is an interesting though that the PUF60-related phenotype is more diverse than some others determined by spliceosomal genes, where very specific and recognizable malformations arise.17 Presumably, specific tissues are PUF60 dose-sensitive at specific times during early human development.
The similar phenotypes in 8q24.3 deletions encompassing the entire PUF60 gene and the point variants in our series, including both missense and null variants, suggest that haploinsufficiency is the common mechanism in all. Loss-of-function variants are predicted to result in altered dosage of different PUF60 isoforms and consequently abnormal splicing of target genes.2 Variants in this gene would therefore be expected to have widespread phenotypic effects. Previous researchers found that patient-derived cells demonstrated expression of a truncated specific isoform of PUF60.2 They also showed that suppression of puf60 in developing zebrafish resulted in reduced body length, microcephaly, craniofacial defects and cardiac anomalies. Additionally, they found evidence that suppression of the scrib gene (syntenic and adjacent to PUF60 in humans) caused defects such as colobomata and renal anomalies, and thus concluded that some features of the 8q24.3 deletion phenotype might be due to this contiguous gene. Our data together with the other six reported cases suggest that PUF60 deficiency itself could account for these features in humans. We anticipate that more patients with PUF60 variants will be described, which, in association with functional studies, will help to delineate the characteristics of this complex syndrome.
Conclusions
Chromosome 8q24.3 deletions have been associated with a phenotype encompassing microcephaly and short stature, developmental delay, colobomata, craniofacial, skeletal, cardiac and renal anomalies. Our findings demonstrate a very similar pattern of mild or moderate intellectual disability and physical characteristics observed in patients with variants in PUF60. We suggest that loss or reduction of expression of PUF60 results in a complex human phenotype with subtle facial features and a consistent pattern of congenital malformations, especially involving the heart and spine.
Acknowledgments
The DDD study presents independent research commissioned by the Health Innovation Challenge Fund (grant number HICF-1009-003), a parallel funding partnership between the Wellcome Trust and the Department of Health and the Wellcome Trust Sanger Institute (grant number WT098051). The views expressed in this publication are those of the author(s) and not necessarily those of the Wellcome Trust or the Department of Health. The study has UK Research Ethics Committee approval (10/H0305/83, granted by the Cambridge South REC, and GEN/284/12, granted by the Republic of Ireland REC). The research team acknowledges the support of the National Institute for Health Research, through the Comprehensive Clinical Research Network. The work of KKS and MDV was funded by the South-Eastern Norway Regional Health Authority (grant number 2012066). We thank Geoff Woodward and Marc Wadsley, bioinformaticians at the Bristol Genetics Laboratory, for their assistance with CADD scores. We would also like to thank all the local clinicians and health-care professionals involved with these children. We would especially like to thank the families of all the patients described here for working with us on this project and for consenting to this publication.
Author contributions
KJL: phenotypic analysis of patients, data collection and interpretation and first author of manuscript; MA: laboratory work and RNA work patient 8 and input to manuscript; RAJ: senior clinician patient 10 and review of manuscript; AC, clinician patient 1 and review of manuscript; SEC, referral of patient 10, input to manuscript; DRF: head of lab patient 8 and DDD senior collaborator; MG: lab collaborator, CADD score input, review of manuscript; AH: clinican patient 2, review of manuscript; JH: clinican patient 4, review of manuscript; KK: genetic counsellor patient 9, review of manuscript; PK: laboratory scientist patient 10, review of manuscript; TP: clinician patient 7, input to manuscript and review; FR: clinician patient 11, input to manuscript and review; KKS: laboratory scientist patient 7, review of manuscript; FS: clinician patient 5, review of manuscript; KT-B: clinician patient 8, review of manuscript; JT: laboratory scientist patient 10, review of manuscript; MDV: laboratory scientist patient 7, input to manuscript and review; JV: clinician patient 6, review of manuscript; JZ: clinician patient 9, review of manuscript; DDD study: exome analysis and reporting of all named DDD patients; SFS, clinican patient 3, phenotypic analysis, significant input in to manuscript and review, senior author and supervisor of project.
Footnotes
The authors declare no conflict of interest.
References
- Verheij JBGM, de Munnik SA, Dijkhuizen T et al: An 8.35 Mb overlapping interstitial deletion of 8q24 in two patients with coloboma, congenital heart defect, limb abnormalities, psychomotor retardation and convulsions. Eur J Med Genet 2009; 52: 353–357. [DOI] [PubMed] [Google Scholar]
- Dauber A, Golzio C, Guenot C et al: SCRIB and PUF60 are primary drivers of the multisystemic phenotypes of the 8q24.3 copy-number variant. Am J Hum Genet 2013; 93: 798–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells C, Spaggiari E, Malan V et al: First fetal case of the 8q24.3 contiguous genes syndrome. Am J Hum Genet 2016; 170A: 239–242. [DOI] [PubMed] [Google Scholar]
- Hastings ML, Allemand E, Duelli DM, Myers MP, Krainer AR: Control of pre-MRNA splicing by the general splicing factors PUF60 and U2AF(65). PLoS One 2, 2007; e538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Chehadeh S, Kerstjens-Frederikse WS, Thevenon J et al: Dominant variants in the splicing factor PUF60 cause a recognizable syndrome with intellectual disability, heart defects and short stature. Eur J Hum Genet 2016; 25: 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari M, Poke G, Ferry Q et al: Genetic heterogeneity in Cornelia de Lange syndrome (CdLS) and CdLS-like phenotypes with observed and predicted levels of mosaicism. J Med Genet 2014; 51: 659–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CF, Fitzgerald TW, Jones WD et al: Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet 2014; 385: 1305–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenon J, Duffourd Y, Masurel-Paulet A et al: Diagnostic odyssey in severe neurodevelopmental disorders: toward clinical whole-exome sequencing as a first-line diagnostic test. Clin Genet 2016; 89: 700–707. [DOI] [PubMed] [Google Scholar]
- Ramu A, Noordam MJ, Schwartz RS et al: DeNovoGear: de novo indel and point mutation discovery and phasing. Nat Methods 2013; 10: 985–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigeland MD, Gjøtterud KS, Selmer KK: FILTUS: a desktop GUI for fast and efficient detection of disease-causing variants, including a novel autozygosity detector. Bioinformatics 2016; 32: 1592–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillano D, Bertoli-Avella A, Kandaswamy K et al: Clinical exome sequencing – results from 2819 samples reflecting 1000 families. Eur J Hum Genet 2017; 25: 176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin CA, Ahmad I, Klingseisen A et al: Mutations in PLK4, encoding a master regulator of centriole biogenesis, cause microcephaly, growth failure and retinopathy. Nat Genet 2014; 46: 1283–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth HV, Richards SM, Bevan AP et al: DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources. Am J Hum Genet 2009; 84: 524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J: A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014; 46: 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehalle D, Wieczorek D, Zechi-Ceide RM et al: A review of craniofacial disorders caused by spliceosomal defects. Clin Genet 2015; 88: 405–415. [DOI] [PubMed] [Google Scholar]
- Patel SB, Bellini M: The assembly of a spliceosomal small nuclear ribonucleoprotein particle. Nucleic Acids Res 2008; 36: 6482–6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooley M, Lynch D, Bernier F: Cerebro-costo-mandibular syndrome: clinical, radiological and genetic findings. Am J Med Genet 2016; 170: 1115–1126. [DOI] [PubMed] [Google Scholar]