Abstract
Diffuse intrinsic pontine glioma (DIPG) is a childhood brainstem tumor with a universally poor prognosis. Here, we characterize on a positron emission tomography (PET) probe for imaging DIPG in vivo. In human histological tissues, the probes target, poly(ADP)ribose polymerase 1 (PARP1), was highly expressed in DIPG compared to normal brain. PET imaging allowed for the sensitive detection of DIPG in a genetically engineered mouse model (GEMM), and probe uptake correlated to histologically determined tumor infiltration. Imaging with the sister fluorescence agent revealed that uptake was confined to proliferating, PARP1 expressing cells. Comparison to other imaging technologies revealed remarkable accuracy of our biomarker approach. We subsequently demonstrated that serial imaging of DIPG in mouse models enables monitoring of tumor growth, as shown in modeling of tumor progression. Overall, this validated method for quantifying DIPG burden would serve useful in monitoring treatment response in early phase clinical trials.
Keywords: PARP1, Molecular Imaging, DIPG, PET Imaging, Optical Imaging
INTRODUCTION
Brain and central nervous system (CNS) tumors are the most common solid tumor and the second-leading cause of cancer-related death in individuals 0 to 19 years of age in the United States and Canada (1, 2). While the treatment and survival rates for more common forms of pediatric CNS tumors have improved (3), survival significantly depend on the location and histologic subtype of the individual CNS lesions (4, 5). Certain types of brainstem tumors, particularly the diffuse intrinsic pontine glioma (DIPG), face a grim prognosis. Dozens of clinical trials have been conducted over several decades, evaluating a wide range of therapeutic strategies, but none of them could demonstrate a survival benefit beyond traditional external beam radiation therapy (6–12). DIPG remains an incurable disease, with the mean survival being less than a year from diagnosis (13). One unique feature of this tumor is the infiltrative and nonfocal growth pattern and the sparse uptake of MR contrast agents. Critically, while MRI can be used for initially diagnosing DIPG, this approach has proven imperfect for defining the true tumor burden and monitoring of treatment response (14, 15).
As a result, established methods used for response monitoring in other CNS tumors are of limited value in DIPG (16). Length of survival thus serves as the conventional measure of therapeutic efficacy. Intuitively, this falls short of being able to accurately monitor response to experimental therapeutics in early stages of clinical development, which therefore hinders the effective and hypothesis-driven discovery of therapeutic strategies. Accordingly, central nervous system tumors that share an infiltrative and non-enhancing morphology would benefit enormously from new imaging technologies that can accurately diagnose and delineate tumor tissue.
One near-ideal biomarker for DIPG imaging is PARP1. The enzyme is overexpressed in many malignancies (17–24), including DIPG (25–27), and is rooted in its key role during tumor cell proliferation and DNA damage repair (26, 28). The high expression levels of PARP1 in DIPG have attracted considerable interest in using PARP1 as a therapeutic option to control this malignancy. This is in addition to the finding that the expression levels of PARP1 are in fact prognostic for overall survival (29–31).
Particularly for brainstem diseases such as DIPG, where obtaining ex vivo materials for aiding diagnostic decision-making is not only challenging, but also not feasible on a longitudinal basis, we recognized that a non-invasive imaging biomarker has enormous value. We have used optically (PARPi-FL) and isotopically [18F]PARPi) labeled versions of PARP1-targeted molecules previously for imaging of solid tumors and have shown their capability to image PARP1 expression in tumors with high specificity and sensitivity (32–37). Using the 18F-labeled PARP1-targeted small molecule, [18F]PARPi, and its fluorescent sister agent PARPi-FL, we aim to explore three clinically relevant questions: (I) How does PARP1 expression in DIPG compare to healthy tissue, and do genetically engineered mouse models (GEMMs) accurately reflect this? (II) Do 18F-labeled PARP1 agents allow non-invasive delineation of DIPG in a pediatric DIPG mouse model? (III) How accurately does [18F]PARPi reflect the tumor burden and tumor growth?
We believe that a quickly and selectively distributing PARP1-targeted marker that can be injected intravenously can overcome many hurdles currently hampering the development of efficient DIPG therapeutics. Here we test one such PET tracer, validate its high sensitivity and accuracy in pediatric GEMMs, and demonstrate its ability to delineate and monitor DIPG in vivo.
MATERIALS & METHODS
Human biospecimens
Formalin fixed paraffin embedded tissue (FFPE) (n=4 surgical biopsies and n=1 autopsy specimen of diffuse midline glioma) were obtained from the tissue archives at Weill Cornell Medicine. The use of archival tissues was approved by the Institutional Review Board (IRB) at Weill Cornell Medical College and consent was waived as by the IRB approval.
In vivo tumor generation
DIPG development was modeled in p53 deficient nestin/tv-a mice (ntv-a/p53fl/fl mice) as described in the Supplementary Methods using a glioma model based on the RCAS/tv-a system (38). Tumor development was initiated by intracranial injection of immortalized chicken fibroblasts (DF1) either at postnatal day 2–4 (juvenile model) or at an age of 4–6 weeks (adult model). DF1 cells were obtained from ATCC in 2014 (CRL12203™) and were transfected with oncogene-expressing RCAS vectors (RCAS-PDGFB and RCAS-Cre) as described previously (39). Mice started showing symptoms around 4–5 weeks post-inoculation. All animal experiments were performed in accordance with institutional guidelines and approved by the IACUC of MSK and WCMC, and followed NIH guidelines for animal welfare.
Immunohistochemical staining
Staining was carried out using primary antibodies against PARP1 (sc-7150, Santa Cruz Biotechnology), Ki67 (proliferation; AB16667, Abcam), and CD31 (endothelial cells; DIA-310, Dianova) using formalin-fixed, paraffin-embedded sections (FFPE) or frozen sections (indicated in the respective results section). For more details on the staining protocols, see the Supplementary Material. Adjacent sections were H&E stained for morphological evaluation.
Multicolor immunofluorescence staining
To evaluate co-localization of PARPi-FL, PARP1, CD31, and Ki-67, multicolor immunofluorescence (IF) staining of all markers on the same cryosection was conducted as described in the Supplementary Methods.
Synthesis of imaging agents
We followed established workflows to realize the synthesis of the imaging agents PARPi-FL (33), [18F]PARPi (36), [11C]Choline (40, 41) and [18F]FLT (42, 43). For detailed information, refer to Supplementary Methods.
PARPi-FL detection in DIPG brain stem model
We determined penetration and localization of PARPi-FL in the brain of tumor-bearing mice. Juvenile mice that showed symptoms of tumor development (gait instability, weight loss, crouching, head swelling) received an intravenous injection of 22.4 nmol PARPi-FL. Brains were extracted 2 h p.i. following intracardiac perfusion with 4% Paraformaldehyde (PFA) and fixation in 4% PFA was continued for 4–24 h at 4 °C. Subsequently, brains were transferred to a 30% sucrose solution for 24–72 h at 4 °C, followed by embedding in OCT compound (Sakura Finetech) and freezing on dry ice. Cryosections of the frozen brains were obtained using a cryostat (Vibratome Ultra Pro 5000).
PARPi-FL was imaged using confocal microscopy after counterstaining slides with Hoechst DNA stain and mounting with Mowiol mounting medium (Sigma-Alrich). On adjacent sections, H&E staining for morphological analysis and PARP1 IHC were carried out (see previous method) to determine the relation between tumor presence, PARP1 expression, and PARPi-FL localization.
Autoradiography
To determine the localization of [18F]PARPi in the brain, we conducted autoradiography on tumor-bearing (n=4) and healthy (n=2) juvenile ntv-a;p53fl/fl mice. For more detailed methodology see Supplementary Methods. Briefly, brains were extracted 2 h p.i. of 150–170 μCi [18F]PARPi and nine coronal cryosections per brain were exposed to a storage phosphor autoradiography plate (Fujifilm, BAS-MS2325, Fiji Photo Film) overnight at −20 °C for radiotracer localization. Adjacent sections were H&E stained for morphological correlation. Relative count intensities were determined using ImageJ 1.47u processing software (source: http://rsbweb.nih.gov/ij/).
PET/CT imaging - general
PET/CT images were acquired on an Inveon PET/CT (Siemens) and reconstructed scans were analyzed using the Inveon Research Workplace Software. To acquire PET/CT images, animals were anesthetized with 2% isoflurane and positioned on the scanner bed. For [18F]PARPi imaging, animals were intravenously injected with 120–180 μCi of the tracer 2 h prior to PET/CT imaging, if not indicated otherwise. Typically, PET data were collected for 5–10 min, followed by CT. Correction for partial volume effects was not carried out, due to the lack of availability of a reference standard for tumor volume.
Specificity of [18F]PARPi accumulation in vivo
To determine specificity of in vivo accumulation of [18F]PARPi, juvenile ntv-a;p53fl/fl mice with tumors located in the brainstem (n=6) or control mice without tumors (n=3) were divided into three groups. Tumor-bearing and healthy (=control) animals were injected with [18F]PARPi (n=3/group) or tumor-bearing animals were injected with 1 mg olaparib 30 min before injection of [18F]PARPi (n=3) to occupy specific binding sites (“Block”). We calculated the % injected dose/gram (%ID/g) for the entire brain or the pons region of all three groups. Therefore, we created three-dimensional volumes of interest (VOI) using the CT data acquired together with the PET images.
Subsequently, animals were submitted to intracardiac perfusion and preservation of brains for histology as described above. H&E staining and PARP1 IHC (methodology described above) were used to compare localization of [18F]PARPi to the presence of tumors.
Comparison of [18F]PARPi, [18F]FLT, and [11C]Choline in PET/CT using adult mice
The PET contrast agents [18F]FLT and [11C]Choline, which are clinically used for imaging malignancies of the brain, as well as MRI, were compared to [18F]PARPi imaging. Here, we used ntv-a;p53fl/fl mice injected with DF1 cells in the right hemisphere at 4–6 weeks old (n=5). Each animal underwent all four imaging procedures. Five weeks after tumor inoculation, we first imaged [11C]-Choline and [18F]PARPi on the same day, followed by [18F]FLT and MR imaging 48 h later.
For [11C]Choline imaging, we injected 100–900 μCi per animal. Imaging was started 10 min post i.v. injection of [11C]Choline for 10 min, followed by CT. Five hours later, 100–200 μCi [18F]PARPi was injected into the same animals and imaged 2 h p.i. using a 5-min scan protocol followed by CT. Two days later, we imaged the same animals using [18F]FLT after injection of 120–180 μCi. Images were acquired at 2 h p.i. using a 10-min scan followed by CT. Later on the same day, we acquired MR images of the same animals.
PET/CT imaging in adult mice using [18F]PARPi to follow tumor growth
To monitor tumor growth rate and determine minimum required tumor volumes for imaging, we conducted an imaging study over the course of 6 weeks using ntv-a;p53fl/fl mice injected with DF1 cells in the right hemisphere at 4–6 weeks old (n=4). At 3 weeks after tumor inoculation, we started a weekly imaging schedule. Every 7 days, we injected 100–200 μCi [18F]PARPi and conducted PET/CT imaging 2 h p.i. using a 5-min PET scan, followed by CT. At the end of the study, we conducted an MRI. Intracardiac perfusion was carried out as described above and brains were submitted for H&E and PARP1 histology. Quantification of the %ID/g was carried out retrospectively. VOIs were created after tumor development was confirmed using the PET/CT and MRI images from week 6 and placed in the same brain areas in the PET/CT images of the previous weeks. Furthermore, the %ID/g of the entire brain, independent of tumor presence or size, was analyzed.
Magnetic resonance imaging
MRI was performed on a 1.05 T small animal imaging system (NanoScan, Mediso Inc.). The imaging protocol included localizer gradient-echo images used to plan sequential high-resolution T2-weighted, pre-contrast T1-weighted image, and post-contrast T1-weighted images. The axial slice FOV was 32 x 32 mm, 2 mm slice thickness (no slice gap), with 8 slices sufficient for full brain coverage. The matrix size was 128 x 128. A NEX of 4 provided sufficient SNR. For the T2-weighted images, TR was 4377 ms, effective TE was 90 ms, and ETL was 32.
Statistical analysis
For histological analysis, we evaluated at least 3 different stained samples with more than 5 analyzed fields-of-view before statistical analysis was carried out. In vivo, a minimal animal number per group of 3 mice was required for statistical analysis and was extended to 5 where possible. Unless otherwise stated, data points represent mean values and error bars represent standard deviations of biological replicates. Statistical analysis was carried out using Prism 6.0c (GraphPad Software, La Jolla, CA) and R. To evaluate differences between groups we used the unpaired Student’s t-test, assuming populations with an equal SD, unless otherwise stated. If more than two groups were compared, we corrected for multiple comparisons using the Holm-Sidak method. Cutoffs for statistically significant differences between groups were chosen at p-values smaller than 0.05 and discrete p-values are reported in the respective locations.
RESULTS
DIPG biospecimens show PARP1 overexpression
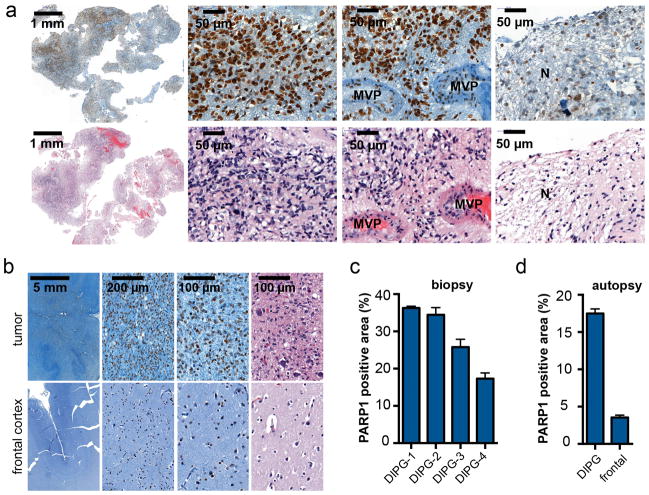
In order to probe the clinical relevance of PARP1 in DIPG, we obtained both biopsy tissue specimens as well as autopsy tissue from patients with histopathologically confirmed DIPG. In the examined infiltrative malignant brainstem glioma biopsies (n=4), more than 90% of observed cell nuclei were positive for PARP1. PARP1 expression was present throughout the tumors, with the exception of necrotic tissue and areas with proliferative vasculature (Fig. 1a). Similarly high PARP1 expression was observed in the autopsy specimen, where we were able to compare the tumor region to non-neoplastic neocortex in the same patient, which showed no tumor involvement. As opposed to the tumor, in the frontal cortex only, a subset of cells expressed PARP1, such as nuclei of oligodendritic cells and astrocytes, as well as nucleolei of a subset of neurons (Fig. 1b, Supplementary Fig. 1a). PARP1 abundance in DIPG was further amplified by the much higher cellularity of high-grade DIPG tissue compared to healthy tissue (Fig. 1b). In grade IV biopsy and autopsy tissue, the total area covered by PARP1-positive nuclei was high (28.5 ± 8.7% PARP1-positive area in biopsy and 17.5 ± 2.9% in autopsy tissue vs. 3.5 ± 0.9% in the frontal cortex of the same brain Fig. 1c–d). The PARP1-positive area, together with the associated high target abundance, is fundamental for sufficient uptake of a molecularly targeted tracer, and to achieve a high tumor-to-background contrast.
Figure 1. PARP1 expression in human biospecimens of DIPG.
(a) Immunohistochemical staining of DIPG biopsy tissue for PARP1 protein and H&E. From left to right: low magnification overview, viable tumor tissue, tumor area with microvascular proliferation (MVP), and necrosis (N). (b) PARP1 staining of a DIPG autopsy specimen shows strong infiltrative growth of PARP1-positive tumor cells into the dentate nucleus (first row), while PARP1 expression and cellular density in the frontal cortex of the same brain (second row) is low (H&E in Supplementary Fig. 1a). (c) Quantification of PARP1 in biopsy and (d) autopsy specimens based on histological staining (brown stained area (=PARP1) vs. whole tissue area). N=4 histopathologically confirmed cases of DIPG; n=1 DIPG autopsy brain.
PARP1 overexpression in mouse models of DIPG is comparable to biospecimens
Encouraged by the robustness of PARP1 expression in human DIPG, we turned to a mouse model with the aim of determining whether the high levels of PARP1 expression found in human tissue of DIPG is reflected in GEMMs of this malignancy. Specifically, we quantified and correlated PARP1 overexpression in a previously established DIPG mouse model of ntv-a/p53fl/fl, where tumors are formed after somatic gene transfer of oncogenes via retroviral delivery (38). For this study, we used two variants of the model, where cells are either injected into the pons of 2- to 4-day-old mice (juvenile model) or into the right hemisphere of 4- to 6-week-old mice (adult model). Upon occurrence of symptoms, which appear in both models within 4–6 weeks after inoculation, brains were processed for histology and stained for PARP1 expression, proliferation index (Ki67), and morphological features (H&E).
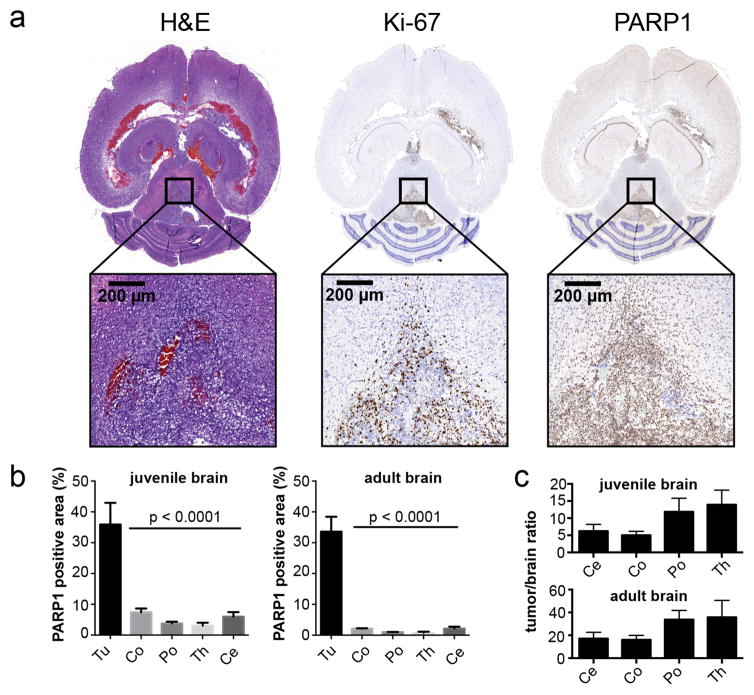
In the juvenile model, tumors were located primarily in the pons, but also showed spread to the third ventricle and parts of the hippocampus (Fig. 2a). All tumor areas showed a strong proliferative activity, particularly toward the infiltrative edges, while the central regions of tumors contained a mix of Ki67-positive and Ki67-negative cells. All other regions of the brain showed almost no Ki67 expression at all (Fig. 2a). Analogous to what was found for Ki67, PARP1 expression was also significantly higher within areas of infiltration as compared to healthy, non-tumorous tissues (Fig. 2a). Interestingly, PARP1 expression was rather homogeneous throughout the tumor, with the exception of necrotic areas. Regarding non-tumorous brain areas, we detected slightly higher PARP1 expression in the cortex and the cerebellum compared to pons and thalamus. While lower than what we found for tumor tissues, physiological PARP1 expression was observed in the Purkinje cell layer in the cerebellum and the dentate gyrus (Supplementary Fig. 1b). When we quantified the PARP1-positive area of the regions of interest (ROI) in tumor and non-tumor affected brain areas of healthy animals in the juvenile model, we found a PARP1-positive area of 35.9 ± 7.1% in the tumor, compared to 7.4 ± 1.2% in the cortex, 3.8 ± 1.1% in the pons, 3.1 ± 0.9% in the thalamus, and 5.9 ± 1.5% in the cerebellum (p<0.01, t-test, vs. cortex, pons, thalamus, cerebellum) (Fig. 2b). When tumors were induced in the cerebral hemisphere of adult mice, very similar PARP1 staining (Supplementary Fig. 1c) and expression values were found (33.6 ± 4.8% PARP1-positive area). Expression in different brain areas was significantly lower (2.1 ± 0.15% in the cortex, 1.0 ± 0.1% in the pons, 0.8 ± 0.3% in the thalamus, and 2.1 ± 0.6% in the cerebellum (p<0.01, t-test, vs. cortex, pons, thalamus, cerebellum; Fig. 2b). Ratios of PARP1 expression in tumor tissue compared to healthy brain areas were generally high. In the juvenile model they ranged from 5.0 ± 0.6 to 13.8 ± 2.4 (tumor/cortex and tumor/thalamus, respectively) and in the adult model from 16.1 ± 2.2 to 35.8 ± 10.4 (tumor/cortex and tumor/thalamus, respectively) (Fig. 2c). Overall, PARP1 expression levels in tumors were found to be statistically indistinguishable between the juvenile and adult model applying a random effects model and using a Likelihood Ratio Test (X2 = 1.57, p-value = 0.21). Our GEMM PARP1 expression data corresponds to PARP1 expression in human DIPG biospecimens that showed grade IV histopathology (Fig. 1).
Figure 2. PARP1 is overexpressed in RCAS-based murine DIPG mouse models.
(a) Immunohistochemical staining of formalin-fixed, paraffin-embedded tumor-bearing brains following tumor generation in ntv-a/p53fl/fl mice injected with DF1 cells (transfected with RCAS-PDGFB, RCAS-Cre) in the brainstem at 2–4 days of age or right hemisphere at 4–6 weeks of age (Supplementary Fig. 1c). Anti-PARP1, anti-Ki67 (proliferation), and H&E staining for morphological evaluation were carried out on adjacent sections. (b) Quantification of PARP1 staining in juvenile and adult tumor-bearing mice in order to calculate the relative PARP1-positive area in relation to the entire tissue area. ROIs were placed on tumor (Tu) regions and anatomically defined brain regions (Co=cortex, Po=pons, Ce=cerebellum, Th=thalamus; n≥3 animals/4–7 ROIs per region; data represented as mean ± standard error). Statistical significance was determined using an unpaired Student’s t-test, corrected for multiple comparisons by the Holm-Sidak method with an alpha of 0.01. Automated analysis based on color thresholding was carried out using Metamorph Software. (c) Tumor-to-brain-area ratios are displayed as mean values ± standard error. N=4 animals for the juvenile brain; n=3 for the adult brain.
The fluorescently labeled PARP1 inhibitor PARPi-FL shows intratumoral accumulation in vivo after intravenous injection
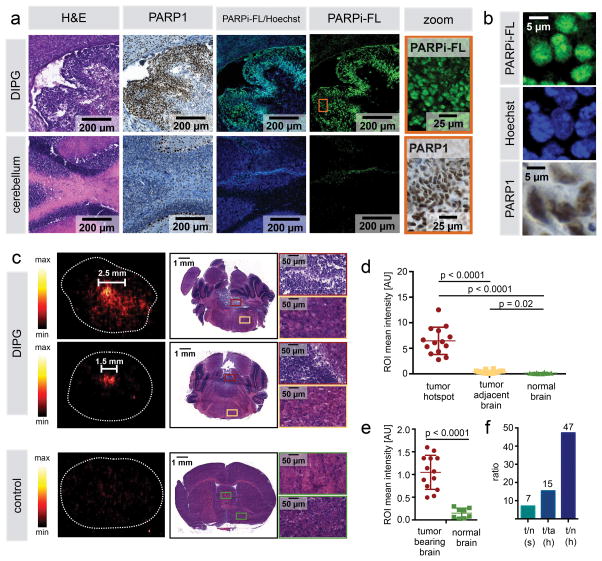
Next, we determined if a PARP1-targeted imaging agent accumulates in the experimental DIPG and if its uptake is specific to PARP1 expression. We injected the fluorescent PARP1-targeted tracer PARPi-FL (32, 34) (Supplementary Fig. 2) into tumor-bearing mice (juvenile mice showing symptoms of tumor development) and determined its cellular localization at 2 hours post-injection (p.i.). We were able to detect fluorescence emission originating from PARPi-FL areas of the brain that spatially correlated with histological features of tumor tissue on H&E stained sections (Fig. 3a). Simultaneously, these areas showed a strongly increased PARP1 expression when compared to surrounding normal brain. The PARPi-FL signal co-localized with PARP1 protein expression in cell nuclei. Confinement of the signal to the nuclei could was shown by co-localization with Hoechst nuclear stain (Fig. 3b), while PARPi-FL was not detected in the cytoplasmatic and extracellular space. This confirms not just delivery of the imaging probe into the intracellular space, but also efficient washout of the excess non-specifically bound agent.
Figure 3. Intratumoral accumulation of PARPi-FL and [18F]PARPi in mouse model of DIPG after intravenous injection.
(a) Juvenile brainstem tumor-bearing ntv-a/p53fl/fl mice were intravenously injected with 22.4 nmol PARPi-FL upon occurrence of symptoms of tumor growth (gait instability, weight loss, crouching, head swelling). Brains were processed for cryoconservation 2 h p.i. of PARPi-FL. Cryosections were co-stained with Hoechst DNA stain and evaluated for presence of PARPi-FL (green fluorescence) using confocal microscopy. Adjacent sections were stained for PARP1 expression and H&E. Representative staining from n=5. (b) High magnification images from (a) showing nuclear localization of PARPi-FL, Hoechst, and PARP1. (c) [18F]PARPi accumulation in DIPG and healthy control mice after intravenous injection. 150–170 μCi [18F]PARPi were injected in juvenile brainstem tumor-bearing ntv-a/p53fl/fl mice (n=4) or healthy ntv-a/p53fl/fl control mice (n=2). For autoradiography, animals were sacrificed 2 h p.i., brains were extracted and flash-frozen, and 20 μm coronal cryosections were cut. Sections were exposed to a storage phosphor autoradiography plate overnight at −20 °C and read the following day. Adjacent sections were submitted for H&E staining for morphological confirmation that activity hotspots represent tumor tissue. (d) For signal quantification, ROIs were placed on activity hotspots, tumor-adjacent brain, and corresponding sections in healthy brains (n=3–8 ROIs per specimen) and results were pooled. (e) To evaluate the difference between tumor-bearing and non-tumor-bearing brains, ROIs were placed over entire tumor-bearing cross-sections, independent of size and number of activity hotspots and intensities were compared to non-tumor-bearing brains. (f) Calculation of signal ratios using mean values of ROI categories; t=tumor, n=normal brain, ta=tumor adjacent, h=hotspot, s=section. Statistical significance was determined using an unpaired student’s t-test, assuming equal SD of populations.
Within the same animals, and depending on the location within the brain and overall stage of a given tumor, we observed areas that histopathologically corresponded to lower- and high-grade malignant gliomas (44). High-grade patterns showed a high cellular density, strong dedifferentiation, and pseudopalisading necrosis, while lower-grade malignancy showed less cellularity and exhibited no signs of necrosis (Supplementary Fig. 3a). Ki67 and CD31 expression showed different features of alterations in low-grade and high-grade malignant areas compared to normal brain tissue (Supplementary Fig. 3b). PARP1 expression and PARPi-FL uptake were clearly present and correlated in both low-grade and high-grade areas, but expression and uptake were higher in high-grade areas. In normal brain tissue, we detected very little expression of both Ki67 and PARP1 and consequently, no retained PARPi-FL was detected (Supplementary Fig. 3b).
Cerebral uptake of [18F]PARPi is localized to DIPG-like brainstem tumors
As a next step, and analogous to what we have seen for the fluorescent PARP1 tracer PARPi-FL, we tested whether the fluorescent agent’s PET active counterpart [18F]PARPi could be used for non-invasive DIPG imaging. We injected the radiolabeled imaging agent intravenously in tumor bearing juvenile ntv-a/p53fl/fl mice. Correlation of autoradiography images with H&E histology showed that at 2 h p.i. of [18F]PARPi, the PET signal originated from areas infiltrated by tumor cells (Fig. 3c), identifying tumor activity hotspots, and even tumors of less than 1.5 mm diameter showed uptake of [18F]PARPi with clear contrast to background. In mice that did not receive an injection of virus carrying DF1 cells, no activity hotspots or histological signs of malignant growths were found, indicating that the uptake is connected to PARP1 expression and, consequently, tumor-specific (Fig. 3c). Quantification of the signal intensity showed clearly higher signal in DIPG than in normal brain of tumor bearing and control animals (Fig. 3d). When size and number of hotspots was not considered and average signal intensity of tumor-bearing brains was compared to normal brains, there was also a clear, statistically significant cutoff between the groups with no overlap (Fig. 3e; p<0.0001, student’s unpaired t-test). To evaluate the strength of contrast ratios between the different groups, we calculated mean values for each group. The average signal in entire sections containing tumor hotspots was 7 times higher than healthy brain sections. The average signal in ROIs focusing on tumor hotspots was 15-fold higher than in tumor-adjacent areas of the same sections and 47-fold higher than randomly assigned areas of the same size in healthy brains (Fig. 3f).
Specificity of [18F]PARPi tumor uptake was confirmed by blocking with Olaparib
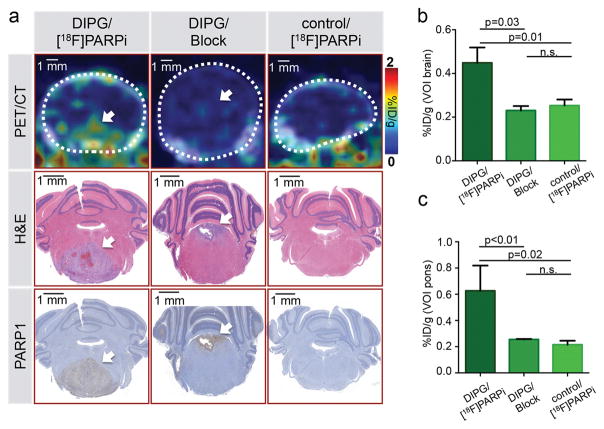
Encouraged by the robustness of our PET imaging tracer, we decided to use the juvenile DIPG mouse model to non-invasively probe the specificity of [18F]PARPi accumulation in vivo, using both animals that showed symptoms of tumor presence and age-matched healthy controls. For tumor-bearing mice, PET/CT images 2 h p.i. revealed an accumulation of [18F]PARPi in the pons of animals that could be correlated to the presence of tumor on H&E and PARP1 histological slides (Fig. 4a). Probing the selectivity of [18F]PARPi, we injected 1 mg olaparib prior to injection of the tracer. This was done in an attempt to saturate available binding sites, and consequently block the uptake of the tracer. For this cohort of mice, no accumulation of the probe could be observed, irrespective of tumor tissue being present in the pons, as confirmed by H&E and PARP1 stained slides (Fig. 4a). [18F]PARPi PET/CT imaging of healthy mice looked similarly negative. For quantification, volumes of interest (VOIs) were created to encompass either the entire brain or the pons of each animal using the CT images for anatomical reference. Calculation of the % injected dose per gram (%ID/g) revealed that the average %ID/g was significantly increased to 0.45 ± 0.07 %ID/g in tumor-bearing brains injected with [18F]PARPi, compared to healthy animals and animals receiving a pre-injection of Olaparib (0.25 ± 0.03 %ID/g and 0.23 ± 0.02 %ID/g, respectively; p<0.05; Fig. 4b). The differences were even more pronounced when only the pons was considered (0.63 ± 0.19 %ID/g vs. 0.25 ± 0.01 and 0.22 ± 0.03 %ID/g, respectively; p<0.05; Fig. 4c). Hence, binding specificity could be confirmed by statistically significant reduction of [18F]PARPi uptake by pre-injection of excess PARP1 inhibitor Olaparib. No statistically significant differences were found between healthy animals and animals receiving Olaparib (p = 0.3 when considering the whole brain and p = 0.1 when considering the pons; student’s unpaired t-test).
Figure 4. Specificity of [18F]PARPi tumor uptake was confirmed by blocking with Olaparib.
In vivo PET/CT imaging and quantification of [18F]PARPi in juvenile brainstem tumor-bearing mice (ntv-a/p53fl/fl mice injected with transfected DF1 cells RCAS-PDGFB and RCAS-Cre in the brainstem at 2–4 days of age). [18F]PARPi was intravenously injected in tumor-bearing (DIPG) or healthy mice (control); to control for specificity, 1 mg olaparib was injected 30 min prior to [18F]PARPi (Block) to occupy specific binding sites (n=3/group). After imaging, intracardiac perfusion with 4% PFA was carried out and brains were conserved for histology. (a) Representative PET/CT images of the brainstem region compared to H&E and PARP1 staining of the same animals. (b) Quantification of %ID/g of the entire brain was derived from the PET/CT data set using the CT as reference for creating VOIs. (c) Similarly, the %ID/g was analyzed for the brainstem region using the CT as reference for creating VOIs. Statistical significance was determined using an unpaired student’s t-test assuming equal SD. White arrow points at tumor location.
[18F]PARPi localization in comparison with MRI, CT, and histology
Next, we compared the ability of different in vivo imaging modalities to spatially resolve the growth pattern of DIPG, using histology as a gold standard. We could confirm that the location and extent of the tumor as determined using [18F]PARPi PET accurately represents the tumor volume and the area of increased PARP1 expression (white arrow, Fig. 5). In the adult model of DIPG, we were also able to delineate tumors in T2-weighted MRI. Increased interstitial edema caused a strong MRI signal change, which was not seen in [18F]PARPi-PET images (orange arrow, Fig. 5). Using CT alone, tumors did not show contrast due to absence of soft tissue contrast in small animal CT. Outside of the brain, [18F]PARPi generated a strong signal in the gut, due to its hepatobiliary excretion (Supplementary Fig. 4). Beyond the gut, it specifically localizes only to PARP1-expressing tissues, which can be seen by comparing PARP1 IHC of a range of tissues and organs (Supplementary Fig. 5) to biodistribution data of [18F]PARPi with and without blocking using Olaparib (Supplementary Fig. 6), revealing the spleen, lymph nodes, salivary gland, and pancreas as tissues with PARP1 expression.
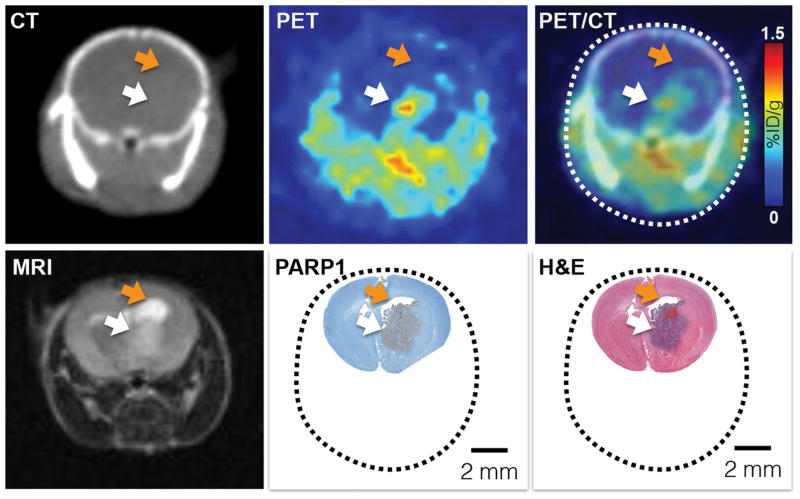
Figure 5. [18F]PARPi accurately delineates tumors in vivo.
To evaluate the ability and quality of [18F]PARPi to delineate brain tumors in vivo, we compared [18F]PARPi PET/CT imaging to MRI and histology. First row (left to right): CT, PET, and PET/CT 1 h p.i. of [18F]PARPi. Second row (left to right): T2-weighted 1.05T MRI image, PARP1 IHC (brown staining) and H&E. All images are from the same animal. PET/CT and MRI were conducted on the same day and the animal was sacrificed immediately after MRI to preserve the brain for histology. White arrow indicates tumor location. Orange arrow points at an accumulation of cerebrospinal fluid (CSF), which causes a strong MRI signal, but is not seen in [18F]PARPi imaging.
Comparison to other PET brain tracers
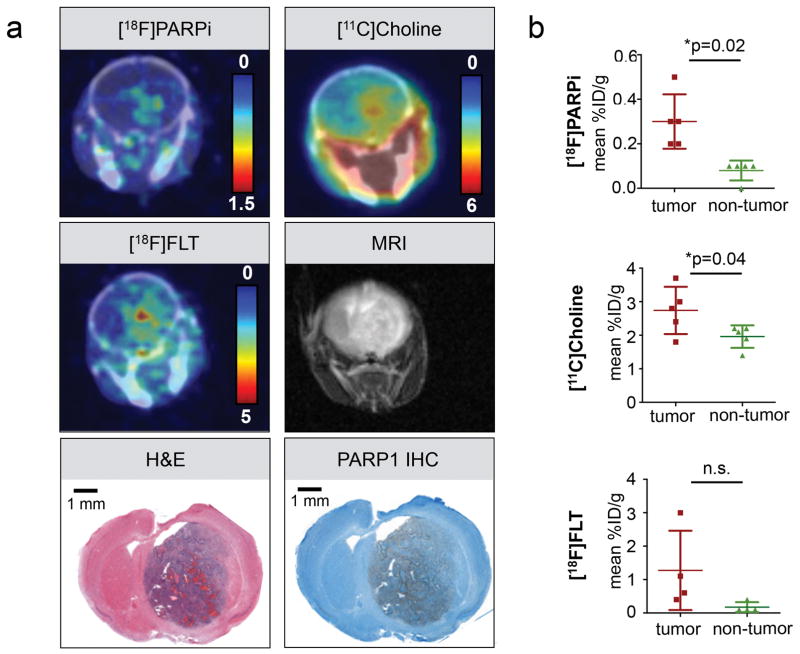
In order to determine whether PARP1 expression and [18F]PARPi uptake in the brain coincides with the elevated metabolism and DNA synthesis rate typically seen for aggressively growing malignant cells, we subjected tumor-bearing animals to [11C]Choline and [18F]FLT PET/CT in addition to [18F]PARPi PET/CT. All animals (n = 5) also received non-contrast-enhanced MRI scans (Fig. 6a). Unequivocal tumor detection with [11C]Choline imaging (10 min post injection of 378 ± 343 μCi) led to a visible tumor to background contrast in 3/5 animals. The mean %ID/g in tumor was 2.74 ± 0.71 and in non-tumor tissue 1.96 ± 0.34 %ID/g). [18F]PARPi and [18F]FLT were both conducted at 2 h p.i. of comparable amounts of activity (162 ± 22 μCi and 147 ± 19 μCi for [18F]PARPi and [18F]FLT, respectively). The mean uptake in tumor was 0.3 ± 0.12 %ID/g for [18F]PARPi and 1.27 ± 1.19 %ID/g for [18F]FLT, showing higher overall uptake, but also higher variability in probe uptake, yielding less consistent uptake values (Fig. 6b). The background uptake in non-tumor brain tissue was lower for [18F]PARPi than [18F]FLT (0.08 ± 0.04 %ID/g vs. 0.18 ± 0.15 %ID/g, respectively. While both [18F]PARPi and [11C]Choline showed statistically significant differences in mean %ID/g comparing tumor and non-tumor areas (p=0.02 and p=0.04, respectively, paired two-tailed t-test), [18F]FLT did not (p=0.1). However, we observed a high variability of the signal and the number of animals studies was rather small. Tumors were also visible in T2-weighted MRI images acquired on a 1.05T MRI; however, strong variations of the T2 signal within animals was observed. Histological evaluation of all tumors confirmed presence of PARP1 overexpression within tumor tissue (Fig. 6a). We confirmed that [18F]PARPi is able to delineate brain tumors with a clear contrast to normal brain tissue, and uptake location is in agreement with metabolic PET tracers. While these, together with MRI, are also able to delineate tumors, they cannot provide information on PARP1 expression, target engagement, and treatment eligibility monitoring, which is a unique feature of [18F]PARPi.
Figure 6. Comparison of [18F]PARPi to [11C]Choline and [18F]FLT imaging.
(a) Representative PET/CT images of the same tumor-bearing animal (adult model, right hemisphere) using [18F]PARPi (2 h p.i.), [11C]Choline (5 min p.i.), [18F]FLT (2 h p.i.), and MRI (T2 weighted) compared to PARP1 IHC and H&E histology. [18F]FLT and MRI were conducted 48 h after the [18F]PARPi and [11C]Choline imaging. (b) Quantification of the mean %ID/g of a VOI in the tumor area compared to a control area in the back region of the brain. Statistical significance was determined using a paired t-test.
Observation of tumor growth and tumor volume delineation
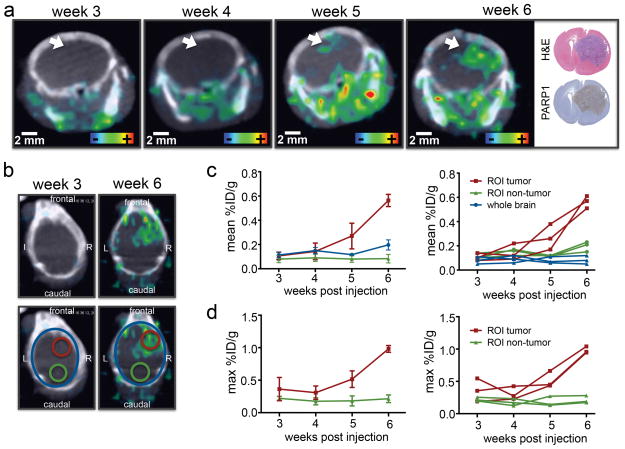
We further evaluated the capability of [18F]PARPi to monitor tumor growth by performing weekly imaging using ntv-a/p53fl/fl mice injected with DF1 cells in the right hemisphere at 4–6 weeks of age. We were able to detect focal accumulation of [18F]PARPi in week 5 p.i., showing hotspots of activity of 1.5–2.0 mm in size. These hotspots showed considerably increased mean and max. %ID/g within one week (Fig. 7a, Supplementary Fig. 7a). The hotspots in the PET/CT imaging could be confirmed to correlate with tumor tissue using histology. The [18F]PARPi retention was quantified using a retrospective approach, where the confirmed tumor location in week 6 was used to draw a VOI in the same region of all scans recorded in previous weeks (Fig. 7b). Quantification of mean and maximum %ID/g in the tumor area showed that an increase in uptake was observed in week 5, which strongly increased in week 6, while the uptake in non-tumorous tissue and the entire brain remained unchanged over the course of the experiment (Fig. 7c and 7d). Tumor-bearing animals began showing symptoms such as weight loss, crouching, gait-instability and scruffy fur in week 6 of the experiment and had to be sacrificed by week 7. One animal showed no uptake of [18F]PARPi over 7 weeks and was histologically confirmed to have no tumor growth (Supplementary Fig. 7b).
Figure 7. Tumor growth monitoring is feasible using [18F]PARPi.
(a) Tumor development was followed over the course of 6 weeks using ntv-a;p53fl/fl mice injected with DF1 cells in the right hemisphere at 4–6 weeks old (n=4 total, n=3 that showed tumor growth displayed in figure). In the displayed example the white arrow indicates tumor location. For weekly PET/CT imaging animals were injected with 100–200 μCi [18F]PARPi 2 h prior to PET/CT imaging. H&E and PARP1 histology were conducted after the last imaging time point. (b) To quantify uptake, VOIs were created in the tumor region and control region, using the PET/CT and histologically confirmed tumor location. These were then applied to earlier imaging time points. Mean (c) and max (d) %ID/g were quantified between week 3 and 6 post-tumor inoculation. One animal showed no tumor development and was histologically confirmed to have no tumor (Supplementary Fig. 7b).
DISCUSSION
We have shown that our 18F-labeled tracer [18F]PARPi accurately delineates and quantifies tumor tissue in a genetically engineered mouse model of DIPG. DIPG is a childhood brainstem tumor without curative treatment options and a mean survival of less than one year. Only recently, insights into the etiology and molecular characteristics have been made (45–48), alongside the isolation and development of cell lines and animal models (38, 49). These disease models enabled us to explore and validate whether [18F]PARPi, a radiolabeled PARP1-targeted tracer, is capable of molecularly and non-invasively quantifying PARP1 expression in DIPG. Importantly, biopsy confirmation of DIPG has been abandoned, because the inherently risky procedure does not significantly impact disease management or outcome (50). Lack of biopsy derived tissue has minimized not only the availability of tissue for clinical research, and more fundamentally, precludes the validation of molecular biomarkers prior to therapy, which is vital for the development of targeted and personalized treatment approaches. A unique value of [18F]PARPi imaging is therefore that it does not only help diagnose DIPG, but its uptake also correlates with the expression of a druggable target, which has been explored in combination treatment approaches, e.g., with external beam radiation and DNA-damaging chemotherapeutics (51). This sets [18F]PARPi apart from metabolic markers and anatomic non-invasive imaging, which ultimately might be capable of detecting DIPG as well (Fig. 7), but otherwise do not allow physicians to predict target engagement and, consequently, predict treatment response.
Furthermore, [18F]PARPi can not only demonstrate the presence, but also the accessibility of PARP1 for PARP inhibitor therapy. Previous studies revealed widespread perturbations in DNA repair pathways in DIPG including non-homologous end-joining (NHEJ), homologous recombination (HR), BRCA1 and BRCA2 (25), as well as PARP1 overexpression, implying potentially high sensitivity of DIPG to PARP1 inhibition, which was indeed found in a recent study (52). In the setting of PARP inhibitor therapy, [18F]PARPi could be particularly valuable for measuring target engagement of the drug by imaging the displacement of [18F]PARPi by the PARP inhibitor. This would be feasible without impacting the treatment itself, because while [18]PARPi and olaparib have comparable inhibitory profiles (36), the doses needed for imaging are 800- to 1600-fold lower than a typical daily dose of the PARP1 inhibitor Olaparib.
Importantly, the current standard of care for monitoring DIPG is MRI, which is used both for diagnosis and treatment monitoring. However, both the initial MRI scan as well as a potential volume reduction in a post-irradiation follow-up scan have very limited prognostic value (14, 15), partly due to the influence of edema on the assessed tumor volume (53). Additionally, volume alone does not necessarily reflect grade and aggressiveness, as has been shown in a study where patients with the largest pre-radiotherapy tumor volumes showed the longest progression-free survival (54). Instinctively, volumetric MRI measurement seems to be inadequate as a response criterion in DIPG since tumor volume measurement might not reflect tumor burden to the patient, while the value of metabolic evaluation by 1H magnetic resonance spectroscopy remains to be determined (55). The use of targeted molecular imaging methods in DIPG management, however, could play a decisive role in characterizing tumors to personalize treatment strategies.
In summary, our radiolabeled small molecule [18F]PARPi efficiently targets PARP1, in vitro and in vivo, a biomarker that is highly expressed in DIPG. To date, no biomarkers are used as the standard of care in diagnosis or treatment of DIPG. Our results indicate that not only does [18F]PARPi allow serial imaging, but histological slides also show that PARP1 expression might correlate to different types of CNS neoplasms (including benign lesions), inflammation, and radiation-induced necrosis. Notably, quantification of [18F]PARPi in the brain indicates that it might be possible to determine the presence of tumor by looking at the average total brain uptake. Since PET imaging only requires miniscule amounts of radiolabeled tracer, the imaging technology we present here would not interfere with therapeutic treatment protocols and might provide robust treatment criteria for patients — even before the progression/regression of tumor tissue is observed. In conclusion, our preclinical results indicate a strong rationale for further investigating [18F]PARPi, and clinical translation of our small molecule is within reach.
Supplementary Material
Acknowledgments
Grant support. This work was supported by National Institutes of Health grants NIH 1 R01 HL125703 (T.R.), R01CA204441 (T.R.), R21CA191679 (T.R. and W.W.), K25 EB016673 (T.R.), and P30 CA008748-50. For financial support, the authors also thank the Center for Molecular Imaging and Nanotechnology (CMINT, T.R.), the Tow Fellowship Program in Molecular Imaging and Nanotechnology (B.C., S.K.), the MSKCC Radiology Development Project Grant (V.M.) the National Science Foundation’s Integrative Graduate Education and Research Traineeship (IGERT 0965983 for B.C.) and the German Research Foundation (S.K.).
The authors thank the Small Animal Imaging Core (V. Longo, P. Zanzonico), the Radiochemistry and Molecular Imaging Probes Core (J. Lewis, S. Lyashenko, P. DeNoble), and the Molecular Cytology Core (N. Fang, M. Turkekul, D. Yarilin) at Memorial Sloan Kettering Cancer Center for support. Furthermore, we thank Christian Brand for supplying PARPi-FL and Ranjodh Singh for his help with mouse DIPG tissue procurement. We thank Kristen Cunanan and Mithat Gonen for their advice on statistical analysis and Leah Bassity for editing the manuscript.
Footnotes
Competing financial interests. The authors declare no potential conflicts of interest.
References
- 1.Kohler BA, Ward E, McCarthy BJ, Schymura MJ, Ries LA, Eheman C, et al. Annual report to the nation on the status of cancer, 1975–2007, featuring tumors of the brain and other nervous system. Journal of the National Cancer Institute. 2011;103:714–36. doi: 10.1093/jnci/djr077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ostrom QT, Gittleman H, Fulop J, Liu M, Blanda R, Kromer C, et al. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2008–2012. Neuro-oncology. 2015;17(Suppl 4):iv1–iv62. doi: 10.1093/neuonc/nov189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith MA, Seibel NL, Altekruse SF, Ries LA, Melbert DL, O’Leary M, et al. Outcomes for children and adolescents with cancer: challenges for the twenty-first century. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:2625–34. doi: 10.1200/JCO.2009.27.0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel S, Bhatnagar A, Wear C, Osiro S, Gabriel A, Kimball D, et al. Are pediatric brain tumors on the rise in the USA? Significant incidence and survival findings from the SEER database analysis. Child Nerv Syst. 2014;30:147–54. doi: 10.1007/s00381-013-2307-1. [DOI] [PubMed] [Google Scholar]
- 5.Deorah S, Lynch CF, Sibenaller ZA, Ryken TC. Trends in brain cancer incidence and survival in the United States: Surveillance, Epidemiology, and End Results Program, 1973 to 2001. Neurosurgical focus. 2006;20:E1. doi: 10.3171/foc.2006.20.4.E1. [DOI] [PubMed] [Google Scholar]
- 6.Cohen KJ, Heideman RL, Zhou T, Holmes EJ, Lavey RS, Bouffet E, et al. Temozolomide in the treatment of children with newly diagnosed diffuse intrinsic pontine gliomas: a report from the Children’s Oncology Group. Neuro-oncology. 2011;13:410–6. doi: 10.1093/neuonc/noq205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haas-Kogan DA, Banerjee A, Poussaint TY, Kocak M, Prados MD, Geyer JR, et al. Phase II trial of tipifarnib and radiation in children with newly diagnosed diffuse intrinsic pontine gliomas. Neuro-oncology. 2011;13:298–306. doi: 10.1093/neuonc/noq202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jalali R, Raut N, Arora B, Gupta T, Dutta D, Munshi A, et al. Prospective evaluation of radiotherapy with concurrent and adjuvant temozolomide in children with newly diagnosed diffuse intrinsic pontine glioma. International journal of radiation oncology, biology, physics. 2010;77:113–8. doi: 10.1016/j.ijrobp.2009.04.031. [DOI] [PubMed] [Google Scholar]
- 9.Korones DN, Fisher PG, Kretschmar C, Zhou T, Chen Z, Kepner J, et al. Treatment of children with diffuse intrinsic brain stem glioma with radiotherapy, vincristine and oral VP-16: a Children’s Oncology Group phase II study. Pediatric blood & cancer. 2008;50:227–30. doi: 10.1002/pbc.21154. [DOI] [PubMed] [Google Scholar]
- 10.Sharp JR, Bouffet E, Stempak D, Gammon J, Stephens D, Johnston DL, et al. A multi-centre Canadian pilot study of metronomic temozolomide combined with radiotherapy for newly diagnosed paediatric brainstem glioma. European journal of cancer. 2010;46:3271–9. doi: 10.1016/j.ejca.2010.06.115. [DOI] [PubMed] [Google Scholar]
- 11.Michalski A, Bouffet E, Taylor RE, Hargrave D, Walker D, Picton S, et al. The addition of high-dose tamoxifen to standard radiotherapy does not improve the survival of patients with diffuse intrinsic pontine glioma. Journal of neuro-oncology. 2010;100:81–8. doi: 10.1007/s11060-010-0141-9. [DOI] [PubMed] [Google Scholar]
- 12.Kretschmar CS, Tarbell NJ, Barnes PD, Krischer JP, Burger PC, Kun L. Preirradation Chemotherapy and Hyperfractionated Radiation-Therapy 66 Gy for Children with Brain-Stem Tumors - a Phase-Ii Study of the Pediatric-Oncology-Group, Protocol 8833. Cancer. 1993;72:1404–13. doi: 10.1002/1097-0142(19930815)72:4<1404::aid-cncr2820720441>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 13.Hargrave D, Bartels U, Bouffet E. Diffuse brainstem glioma in children: critical review of clinical trials. The Lancet Oncology. 2006;7:241–8. doi: 10.1016/S1470-2045(06)70615-5. [DOI] [PubMed] [Google Scholar]
- 14.Liu AK, Brandon J, Foreman NK, Fenton LZ. Conventional MRI at presentation does not predict clinical response to radiation therapy in children with diffuse pontine glioma. Pediatric radiology. 2009;39:1317–20. doi: 10.1007/s00247-009-1368-5. [DOI] [PubMed] [Google Scholar]
- 15.Hargrave D, Chuang N, Bouffet E. Conventional MRI cannot predict survival in childhood diffuse intrinsic pontine glioma. Journal of neuro-oncology. 2008;86:313–9. doi: 10.1007/s11060-007-9473-5. [DOI] [PubMed] [Google Scholar]
- 16.Wen PY, Macdonald DR, Reardon DA, Cloughesy TF, Sorensen AG, Galanis E, et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:1963–72. doi: 10.1200/JCO.2009.26.3541. [DOI] [PubMed] [Google Scholar]
- 17.Aleskandarany M, Caracappa D, Nolan CC, Macmillan RD, Ellis IO, Rakha EA, et al. DNA damage response markers are differentially expressed in BRCA-mutated breast cancers. Breast cancer research and treatment. 2015;150:81–90. doi: 10.1007/s10549-015-3306-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bieche I, de Murcia G, Lidereau R. Poly(ADP-ribose) polymerase gene expression status and genomic instability in human breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 1996;2:1163–7. [PubMed] [Google Scholar]
- 19.Chow JP, Man WY, Mao M, Chen H, Cheung F, Nicholls J, et al. PARP1 is overexpressed in nasopharyngeal carcinoma and its inhibition enhances radiotherapy. Molecular cancer therapeutics. 2013;12:2517–28. doi: 10.1158/1535-7163.MCT-13-0010. [DOI] [PubMed] [Google Scholar]
- 20.Galia A, Calogero AE, Condorelli R, Fraggetta F, La Corte A, Ridolfo F, et al. PARP-1 protein expression in glioblastoma multiforme. European journal of histochemistry : EJH. 2012;56:e9. doi: 10.4081/ejh.2012.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green AR, Caracappa D, Benhasouna AA, Alshareeda A, Nolan CC, Macmillan RD, et al. Biological and clinical significance of PARP1 protein expression in breast cancer. Breast cancer research and treatment. 2015;149:353–62. doi: 10.1007/s10549-014-3230-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nosho K, Yamamoto H, Mikami M, Taniguchi H, Takahashi T, Adachi Y, et al. Overexpression of poly(ADP-ribose) polymerase-1 (PARP-1) in the early stage of colorectal carcinogenesis. European journal of cancer. 2006;42:2374–81. doi: 10.1016/j.ejca.2006.01.061. [DOI] [PubMed] [Google Scholar]
- 23.Ossovskaya V, Koo IC, Kaldjian EP, Alvares C, Sherman BM. Upregulation of Poly (ADP-Ribose) Polymerase-1 (PARP1) in Triple-Negative Breast Cancer and Other Primary Human Tumor Types. Genes & cancer. 2010;1:812–21. doi: 10.1177/1947601910383418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Staibano S, Pepe S, Lo Muzio L, Somma P, Mascolo M, Argenziano G, et al. Poly(adenosine diphosphate-ribose) polymerase 1 expression in malignant melanomas from photoexposed areas of the head and neck region. Human pathology. 2005;36:724–31. doi: 10.1016/j.humpath.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 25.Buczkowicz P, Hawkins C. Pathology, Molecular Genetics, and Epigenetics of Diffuse Intrinsic Pontine Glioma. Frontiers in oncology. 2015;5:147. doi: 10.3389/fonc.2015.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.deMurcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:7303–7. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith SJ, Long A, Barrow JH, Macarthur DC, Coyle B, Grundy RG, et al. Pediatric high-grade glioma: identification of poly(ADP-ribose) polymerase as a potential therapeutic target. Neuro-oncology. 2011;13:1171–7. doi: 10.1093/neuonc/nor115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouchard VJ, Rouleau M, Poirier GG. PARP-1, a determinant of cell survival in response to DNA damage. Experimental Hematology. 2003;31:446–54. doi: 10.1016/s0301-472x(03)00083-3. [DOI] [PubMed] [Google Scholar]
- 29.Rojo F, Garcia-Parra J, Zazo S, Tusquets I, Ferrer-Lozano J, Menendez S, et al. Nuclear PARP-1 protein overexpression is associated with poor overall survival in early breast cancer. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2012;23:1156–64. doi: 10.1093/annonc/mdr361. [DOI] [PubMed] [Google Scholar]
- 30.Michels J, Adam J, Goubar A, Obrist F, Damotte D, Robin A, et al. Negative prognostic value of high levels of intracellular poly(ADP-ribose) in non-small cell lung cancer. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2015;26:2470–7. doi: 10.1093/annonc/mdv393. [DOI] [PubMed] [Google Scholar]
- 31.Park SH, Noh SJ, Kim KM, Bae JS, Kwon KS, Jung SH, et al. Expression of DNA Damage Response Molecules PARP1, gamma H2AX, BRCA1, and BRCA2 Predicts Poor Survival of Breast Carcinoma Patients. Translational oncology. 2015;8:239–49. doi: 10.1016/j.tranon.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reiner T, Lacy J, Keliher EJ, Yang KS, Ullal A, Kohler RH, et al. Imaging Therapeutic PARP Inhibition In Vivo through Bioorthogonally Developed Companion Imaging Agents. Neoplasia. 2012;14 doi: 10.1593/neo.12414. 169–IN3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thurber GM, Yang KS, Reiner T, Kohler RH, Sorger P, Mitchison T, et al. Single-cell and subcellular pharmacokinetic imaging allows insight into drug action in vivo. Nature communications. 2013;4:1504. doi: 10.1038/ncomms2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thurber GM, Reiner T, Yang KS, Kohler RH, Weissleder R. Effect of small-molecule modification on single-cell pharmacokinetics of PARP inhibitors. Molecular cancer therapeutics. 2014;13:986–95. doi: 10.1158/1535-7163.MCT-13-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Irwin CP, Portorreal Y, Brand C, Zhang Y, Desai P, Salinas B, et al. PARPi-FL--a fluorescent PARP1 inhibitor for glioblastoma imaging. Neoplasia. 2014;16:432–40. doi: 10.1016/j.neo.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carney B, Carlucci G, Salinas B, Di Gialleonardo V, Kossatz S, Vansteene A, et al. Non-invasive PET Imaging of PARP1 Expression in Glioblastoma Models. Molecular imaging and biology : MIB : the official publication of the Academy of Molecular Imaging. 2015 doi: 10.1007/s11307-015-0904-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kossatz S, Brand C, Gutiontov S, Liu JT, Lee NY, Gonen M, et al. Detection and delineation of oral cancer with a PARP1 targeted optical imaging agent. Scientific reports. 2016;6:21371. doi: 10.1038/srep21371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Becher OJ, Hambardzumyan D, Walker TR, Helmy K, Nazarian J, Albrecht S, et al. Preclinical evaluation of radiation and perifosine in a genetically and histologically accurate model of brainstem glioma. Cancer research. 2010;70:2548–57. doi: 10.1158/0008-5472.CAN-09-2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hambardzumyan D, Amankulor NM, Helmy KY, Becher OJ, Holland EC. Modeling Adult Gliomas Using RCAS/t-va Technology. Translational oncology. 2009;2:89–95. doi: 10.1593/tlo.09100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sutinen E, Nurmi M, Roivainen A, Varpula M, Tolvanen T, Lehikoinen P, et al. Kinetics of [(11)C]choline uptake in prostate cancer: a PET study. European journal of nuclear medicine and molecular imaging. 2004;31:317–24. doi: 10.1007/s00259-003-1377-9. [DOI] [PubMed] [Google Scholar]
- 41.Tolvanen T, Yli-Kerttula T, Ujula T, Autio A, Lehikoinen P, Minn H, et al. Biodistribution and radiation dosimetry of [(11)C]choline: a comparison between rat and human data. European journal of nuclear medicine and molecular imaging. 2010;37:874–83. doi: 10.1007/s00259-009-1346-z. [DOI] [PubMed] [Google Scholar]
- 42.Mosdzianowski C, Eisenhut M, Martin J, Eisenbarth J, Korenjak C, Nader M. Automated FLT synthesis using 3-N-Boc-1-(5-O-(4,4′-dimethoxytrityl)-3-O-nosyl-2-deoxy-beta-D-lyxofuranosyl)thymine as precursor. Eur J Nucl Med. 2001;28:1228. [Google Scholar]
- 43.Martin SJ, Eisenbarth JA, Wagner-Utermann U, Mier W, Henze M, Pritzkow H, et al. A new precursor for the radiosynthesis of [F-18]FLT. Nuclear medicine and biology. 2002;29:263–73. doi: 10.1016/s0969-8051(01)00289-x. [DOI] [PubMed] [Google Scholar]
- 44.Buczkowicz P, Bartels U, Bouffet E, Becher O, Hawkins C. Histopathological spectrum of paediatric diffuse intrinsic pontine glioma: diagnostic and therapeutic implications. Acta neuropathologica. 2014;128:573–81. doi: 10.1007/s00401-014-1319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nikbakht H, Panditharatna E, Mikael LG, Li R, Gayden T, Osmond M, et al. Spatial and temporal homogeneity of driver mutations in diffuse intrinsic pontine glioma. Nature communications. 2016;7:11185. doi: 10.1038/ncomms11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grasso CS, Tang Y, Truffaux N, Berlow NE, Liu L, Debily MA, et al. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nature medicine. 2015;21:827. doi: 10.1038/nm0715-827a. [DOI] [PubMed] [Google Scholar]
- 47.Warren KE. Diffuse intrinsic pontine glioma: poised for progress. Frontiers in oncology. 2012;2:205. doi: 10.3389/fonc.2012.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khuong-Quang DA, Buczkowicz P, Rakopoulos P, Liu XY, Fontebasso AM, Bouffet E, et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta neuropathologica. 2012;124:439–47. doi: 10.1007/s00401-012-0998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hashizume R, Smirnov I, Liu S, Phillips JJ, Hyer J, McKnight TR, et al. Characterization of a diffuse intrinsic pontine glioma cell line: implications for future investigations and treatment. Journal of neuro-oncology. 2012;110:305–13. doi: 10.1007/s11060-012-0973-6. [DOI] [PubMed] [Google Scholar]
- 50.Albright AL, Packer RJ, Zimmerman R, Rorke LB, Boyett J, Hammond GD, et al. Magnetic-Resonance Scans Should Replace Biopsies for the Diagnosis of Diffuse Brain-Stem Gliomas - a Report from the Childrens Cancer Group. Neurosurgery. 1993;33:1026–30. doi: 10.1227/00006123-199312000-00010. [DOI] [PubMed] [Google Scholar]
- 51.Scott CL, Swisher EM, Kaufmann SH. Poly (ADP-Ribose) Polymerase Inhibitors: Recent Advances and Future Development. Journal of Clinical Oncology. 2015;33:1397. doi: 10.1200/JCO.2014.58.8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chornenkyy Y, Agnihotri S, Yu M, Buczkowicz P, Rakopoulos P, Golbourn B, et al. Poly-ADP-Ribose Polymerase as a Therapeutic Target in Pediatric Diffuse Intrinsic Pontine Glioma and Pediatric High-Grade Astrocytoma. Molecular cancer therapeutics. 2015;14:2560–8. doi: 10.1158/1535-7163.MCT-15-0282. [DOI] [PubMed] [Google Scholar]
- 53.Oh J, Cha S, Aiken AH, Han ET, Crane JC, Stainsby JA, et al. Quantitative apparent diffusion coefficients and T2 relaxation times in characterizing contrast enhancing brain tumors and regions of peritumoral edema. Journal of magnetic resonance imaging : JMRI. 2005;21:701–8. doi: 10.1002/jmri.20335. [DOI] [PubMed] [Google Scholar]
- 54.Poussaint TY, Kocak M, Vajapeyam S, Packer RI, Robertson RL, Geyer R, et al. MRI as a central component of clinical trials analysis in brainstem glioma: a report from the Pediatric Brain Tumor Consortium (PBTC) Neuro-oncology. 2011;13:417–27. doi: 10.1093/neuonc/noq200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lobel U, Hwang S, Edwards A, Li Y, Li X, Broniscer A, et al. Discrepant longitudinal volumetric and metabolic evolution of diffuse intrinsic Pontine gliomas during treatment: implications for current response assessment strategies. Neuroradiology. 2016;58:1027–34. doi: 10.1007/s00234-016-1724-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.