Abstract
Ferroptosis is a form of regulated cell death driven by oxidative injury promoting lipid peroxidation, although detailed molecular regulators are largely unknown. Here we show that heat shock 70kDa protein 5 (HSPA5) negatively regulates ferroptosis in human pancreatic ductal adenocarcinoma (PDAC) cells. Mechanistically, activating transcription factor 4 (ATF4) resulted in the induction of HSPA5, which in turn bound glutathione peroxidase 4 (GPX4) and protected against GPX4 protein degradation and subsequent lipid peroxidation. Importantly, the HSPA5-GPX4 pathway mediated ferroptosis resistance, limiting the anticancer activity of gemcitabine. Genetic or pharmacological inhibition of the HSPA5-GPX4 pathway enhanced gemcitabine sensitivity by disinhibiting ferroptosis in vitro and in both subcutaneous and orthotopic animal models of PDAC. Collectively, these findings identify a novel role of HSPA5 in ferroptosis and suggest a potential therapeutic strategy for overcoming gemcitabine resistance.
Introduction
The therapeutic goal of all cancer treatment has been to trigger tumor-selective cell death and spare normal tissues. Acquisition of various genetic or epigenetic alterations in cancer cells may result in drug resistance by activating stress-adaptation responses such as the heat shock response or the unfolded protein response. Understanding the molecular mechanisms of these protective responses in the setting of cell injury and death may help overcome drug resistance in cancer cells.
Ferroptosis is a form of regulated cell death identified by Dr. Brent Stockwell’s laboratory in 2012 (1). Different from apoptosis and necroptosis, activation of caspase and receptor-interacting protein kinase is not required for induction of ferroptosis (1). In contrast, lipid peroxidation plays a key role in mediating ferroptosis (2). Recently, the discovery and characterization of ferroptosis regulators has begun to unravel the molecular mechanisms of ferroptosis (3). Among them, glutathione peroxidase 4 (GPX4, an antioxidant enzyme) inhibits ferroptosis (4). Genetic or pharmacological inhibition of GPX4 increases lipid peroxidation, which contributes to ferroptotic cancer death (4). Conditional knockout of GPX4 in mice enhances ferroptosis in vivo (5-7). In addition to inhibition of GPX4 activity, increased GPX4 degradation also contributes to ferroptotic cancer cell death (8, 9).
The heat shock 70kDa protein 5 (HSPA5, also termed GRP78 or BIP) is a member of the molecular chaperones expressed primarily in the endoplasmic reticulum (ER) (10). As an important component of the unfolded protein response, HSPA5 promotes cell survival under conditions of ER stress (11). The role of HSPA5 in ferroptosis has not been explored, although ferroptosis itself is associated with ER stress (12).
Pancreatic ductal adenocarcinoma (PDAC) is an extremely lethal cancer with limited treatment options. In this study, we provide the first evidence that increased HSPA5 expression suppresses ferroptosis by direct inhibition of GPX4 protein degradation in human PDAC cells. Furthermore, we demonstrate that the HSPA5-GPX4 pathway mediates ferroptosis resistance, limiting the anticancer activity of gemcitabine (the first-line drug used alone or in combination for the treatment of patients with advanced PDAC). These findings not only identify a novel role of HSPA5 in ferroptosis, but also suggest a potential therapeutic strategy for overcoming gemcitabine resistance in PDAC cells by triggering ferroptosis.
Materials and Methods
Antibodies and reagents
The antibodies to HSPA5 (#3177), CANX (#2679), EIF2AK3 (#3192), ACTB (#3700), glyceraldehyde 3-phosphate dehydrogenase (GAPDH, #5174), ATF4 (#11815), HSP90 (#4877), SLC7A11 (#12691), and Myc-tag (#2278) were obtained from Cell Signaling Technology (Danvers, MA, USA). The antibody to GPX4 (#ab125066), p-EIF2AK3 at T982 (#ab192591), and p-ERN1 at S724 (#ab124945) were obtained from Abcam (Cambridge, MA, USA). Z-VAD-FMK (#V116); staurosporine (#S4400), rapamycin (#R0395), H2O2 (#216763), EGCG (#E4143), cycloheximide (#C7698), sulfasalazine (#S0883), and gemcitabine (#G6423) were obtained from Sigma (St. Louis, MO, USA). Necrosulfonamide (#480073) was obtained from EMD Millipore Corporation (Darmstadt, Germany). TNFα (#210-TA-005) was obtained from R&D Systems (Minneapolis, MN, USA). Erastin (#E7781), ferrostatin-1 (#S7243), and liproxstatin-1 (#S7699) were obtained from Selleck Chemicals (Houston, TX, USA).
Cell culture
PANC1, CFPAC1, MiaPaCa2, Panc2.03, and Panc02 cells were obtained from American Type Culture Collection (ATCC, USA) or the National Cancer Institute (NCI, USA). GPX4−/− cells were a gift from Dr. Marcus Conrad (Institute of Developmental Genetics; Helmholtz Zentrum München; München, Germany) (7). These cells were grown in Dulbecco's Modified Eagle's Medium or RPMI-1640 Medium with 10% fetal bovine serum, 2 mM L-glutamine, and 100 U/ml of penicillin and streptomycin. All cells were mycoplasma free and authenticated by Short Tandem Repeat DNA Profiling Analysis.
Cell viability assay
Cell viability was evaluated using the Cell Counting Kit-8 (CCK-8) (#96992, Sigma) according to the manufacturer’s instructions. The assay is based on utilizing the highly water-soluble tetrazolium salt WST-8 [2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium,monosodium salt] to produce a water-soluble formazan dye upon reduction in the presence of an electron carrier. Absorbance at 450 nm is proportional to the number of living cells in the culture.
Clonogenic cell survival assay
Long-term cell survival was monitored in a colony formation assay. In brief, 1,000 cells were plated into 24-well plates after treatment with indicated drugs for 24 hours. The cells were allowed to grow for the next 10 to 12 days to allow colony formation and the colonies were visualized using crystal violet staining.
GPX4 activity assay
The activity of GPX4 was determined using l-α-phosphatidylcholine hydroperoxide substrate and a coupled enzymatic assay as previously described (13, 14). It was based on the oxidation of glutathione (GSH) to oxidized glutathione disulfide (GSSG) catalyzed by GPX4, which was then coupled to the recycling of GSSG back to GSH utilizing glutathione reductase and reduced β-nicotinamide adenine dinucleotide phosphate, reduced (NADPH). The decrease in NADPH absorbance measured at 340 nm during the oxidation of NADPH to NADP+ indicated GPX4 activity.
Malondialdehyde (MDA) assay
The relative MDA concentration in cell lysates was assessed using a Lipid Peroxidation Assay Kit (#ab118970, Abcam) according to the manufacturer’s instructions. Briefly, the MDA in the sample reacted with thiobarbituric acid (TBA) to generate a MDA-TBA adduct. The MDA-TBA adduct were quantified colorimetrically (OD = 532 nm) or fluorometrically (Ex/Em = 532/553 nm).
GSH assay
The relative GSH concentration in cell lysates was assessed using a kit from Sigma (#CS0260) according to the manufacturer’s instructions. The measurement of GSH used a kinetic assay in which catalytic amounts (nmoles) of GSH caused a continuous reduction of 5,5’-dithiobis (2-nitrobenzoic acid) to 5-thio-2-nitrobenzoic acid and the GSSG formed was recycled by glutathione reductase and NADPH. The reaction rate was proportional to the concentration of glutathione up to 2 mM. The yellow product (5-thio-2-nitrobenzoic acid) was measured spectrophotometrically at 412 nm.
GSSG assay
The relative GSSG concentration in cell lysates was assessed using a kit from Cayman (#703002) according to the manufacturer’s instructions. The quantification of GSSG, exclusive of GSH, was accomplished by first derivatizing GSH with 2-vinylpyridine. In brief, 1 μl of 1 M 2-vinylpyridine in absolute ethanol were added to 99 μl of cell homogenate. This suspension was incubated at room temperature for 60 minutes to block the thiol group of the GSH already present. NADPH (95 μl of 2 mg/ml) in nanopure water and 5 μl of 2 units/ml glutathione reductase were added to reduce GSSG.
Western blot analysis
Proteins in the cell lysate or supernatants were resolved on 4%-12% Criterion XT Bis-Tris gels (Bio-Rad, Hercules, CA, USA) and transferred to a nitrocellulose membrane. After blocking with 5% milk, the membrane was incubated for two hours at 25? or overnight at 4? with various primary antibodies. After incubation with peroxidase-conjugated secondary antibodies for one hour at routine temperature, the signals were visualized via using enhanced or super chemiluminescence (Pierce, Rockford, IL, USA) and by exposure to X-ray films. The relative band intensity was quantified using the Gel-pro Analyzer® software (Media Cybernetics, Bethesda, MD, USA).
Immunoprecipitation analysis
Cells were lysed at 4°C in ice-cold radioimmunoprecipitation assay buffer (Millipore, USA), and cell lysates were cleared by a brief centrifugation (12,000 g, 10 min). Concentrations of proteins in the supernatant were determined by BCA assay. Prior to immunoprecipitation, samples containing equal amount of proteins were pre-cleared with protein A or protein G agarose/sepharose (Millipore, USA) (4°C, 3 h), and subsequently incubated with various irrelevant IgG or specific antibodies (5 μg/mL) in the presence of protein A or G agarose/sepharose beads for 2 h or overnight at 4°C with gentle shaking. Following incubation, agarose/sepharose beads were washed extensively with phosphate buffered saline and proteins were eluted by boiling in 2 × sodium dodecyl sulfate sample buffer before sodium dodecyl sulfate polyacrylamide gel electrophoresis.
RNAi and gene transfection
The YY1-shRNA-1 (Sequence: CCGGGCCTCTCCTTTGTATATTATTCTCGAGAATAATATACAAAGGAGAGGCTTTTT); YY1-shRNA-2 (Sequence: CCGGGACGACGACTACATTGAACAACTCGAGTTGTTCAATGTAGTCGTCGTCTTTTT); GTF2I-shRNA-1 (Sequence: CCGGTGCTGACAGGTCAATACTATCCTCGAGGATAGTATTGACCTGTCAGCATTTTTG); GTF2I-shRNA-2 (Sequence: CCGGATGGCAGCTGTGACAGTAAAGCTCGAGCTTTACTGTCACAGCTGCCATTTTTTG); ATF4-shRNA-1 (Sequence: CCGGGCCTAGGTCTCTTAGATGATTCTCGAGAATCATCTAAGAGACCTAGGCTTTTT); ATF4-shRNA-2 (Sequence: CCGGGCCAAGCACTTCAAACCTCATCTCGAGATGAGGTTTGAAGTGCTTGGCTTTTT); ATF6-shRNA-1 (Sequence: CCGGGCAGCAACCAATTATCAGTTTCTCGAGAAACTGATAATTGGTTGCTGCTTTTT); ATF6-shRNA-2 (Sequence: CCGGCCCAGAAGTTATCAAGACTTTCTCGAGAAAGTCTTGATAACTTCTGGGTTTTT); HSPA5-shRNA-1 (Sequence: GTACCGGAGATTCAGCAACTGGTTAAAGCTCGAGCTTTAACCAGTTGCTGAATCTTTTTTTG); HSPA5-shRNA-2 (Sequence: CCGGGAAATCGAAAGGATGGTTAATCTCGAGATTAACCATCCTTTCGATTTCTTTTTG); SLC7A11-shRNA (Sequence: CCGGCCCTGGAGTTATGCAGCTAATCTCGAGATTAGCTGCATAACTCCAGGGTTTTTG); and GPX4-shRNA (Sequence: CCGGGTGGATGAAGATCCAACCCAACTCGAGTTGGGTTGGATCTTCATCCACTTTTTG) were obtained from Sigma. pcDNA3.1-HSPA5 (#32701) and pCMV-Myc-HSPA5 (#27164) were obtained from Addgene (Cambridge, MA, USA). pCMV6-GPX4 (#RC208065) was obtained from OriGene Technologies (Rockville, MD, USA). pDRIVE-HA-GPX4-U46C and pDRIVE-HA-GPX4-U46S were developed as described previously (15). Transfections were performed with Lipofectamine™ 3000 (#L3000008, Invitrogen) according to the manufacturer’s instructions.
Quantitative real time polymerase chain reaction (Q-PCR) analysis
First-strand cDNA synthesis was carried out by using a Reverse Transcription System Kit according to the manufacturer’s instructions (#11801-025, OriGene Technologies). cDNA from various cell samples was amplified with specific primers (HSPA5: 5’-CTGTCCAGGCTGGTGTGCTCT-3’ and 5’-CTTGGTAGGCACCACTGTGTTC-3’; CANX: 5’-GCTGGTTAGATGATGAGCCTGAG-3’ and 5’-ACACCACATCCAGGAGCTGACT-3’; EIF2AK3: 5’-GTCCCAAGGCTTTGGAATCTGTC-3’ and 5’-CCTACCAAGACAGGAGTTCTGG-3’; YY1: 5’-GGAGGAATACCTGGCATTGACC-3’ and 5’-CCCTGAACATCTTTGTGCAGCC-3’; GTF2I: 5’-CGACTTTTGGCATTCCGAGGCT-3’ and 5’-GTGCTCTCCTTAATCGCCGTCT-3’; ATF4: 5’-TTCTCCAGCGACAAGGCTAAGG-3’ and 5’-CTCCAACATCCAATCTGTCCCG-3’; ATF6: 5’-CAGACAGTACCAACGCTTATGCC-3’ and 5’-GCAGAACTCCAGGTGCTTGAAG-3’; GPX4: 5’-ACAAGAACGGCTGCGTGGTGAA-3’ and 5’-GCCACACACTTGTGGAGCTAGA-3’) and the data was normalized to actin RNA (5’-CACCATTGGCAATGAGCGGTTC-3’ and 5’-AGGTCTTTGCGGATGTCCACGT-3’).
Animal models
All animal experiments were approved by the Institutional Animal Care and Use Committees and performed in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care guidelines (http://www.aaalac.org).
To generate murine subcutaneous tumors, 2×106 PANC1 cells were injected subcutaneously to the right of the dorsal midline in nude mice. Once the tumors reached ~50 mm3 at day seven, mice were randomly allocated into groups and treated with chemotherapy for two weeks (n=5 mice/group). Tumors were measured twice weekly and volumes were calculated using the formula length×width2×π/6.
To generate orthotopic tumors, B6 mice were surgically implanted with 1× 106 Panc02 into the tail of the pancreas. Two weeks after implantation, mice were randomly allocated into groups and treated with chemotherapy for three weeks (n=6 mice/group). Animal survival was monitored every week for three months.
Statistical analysis
Data are expressed as means ± SD of three independent experiments. Unpaired Student’s t tests were used to compare the means of two groups. One-way Analysis of Variance (ANOVA) was used for comparison among the different groups. When ANOVA was significant, post hoc testing of differences between groups was performed using the Least Significant Difference (LSD) test. The Kaplan-Meier method was used to compare differences in mortality rates between groups. A p-value < 0.05 was considered statistically significant.
Results
ATF4 promotes HSPA5 expression in ferroptosis
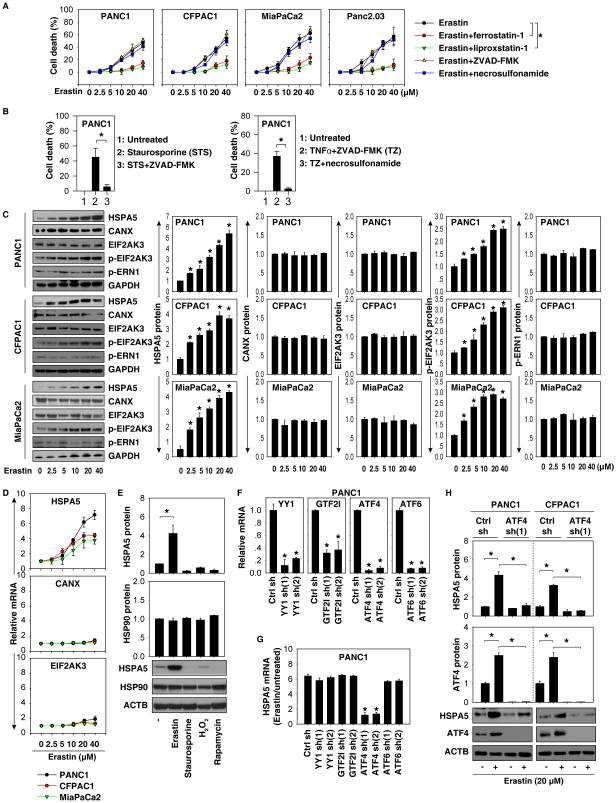
Erastin is a classical inducer of ferroptosis that was originally identified in a screening for small molecules that are selectively lethal to oncogenic RAS mutant cells (16). Given that PDAC is initiated almost exclusively by the expression of mutant K-RAS, we analyzed the anticancer activity of erastin in several human PDAC cell lines (e.g., PANC1, CFPAC1, MiaPaCa2, and Panc2.03) harboring K-RAS mutation using CCK8 cell viability assay kit. Erastin-dose dependently induced cell death in PDAC cells; this process was reversed by the ferroptosis inhibitors ferrostatin-1 and liproxstatin-1 (Fig. 1A). In contrast, the apoptosis inhibitor (e.g., ZVAD-FMK) and necroptosis inhibitor (e.g., necrosulfonamide) had no effect on erastin-induced cell death (Fig. 1A). Clonogenic cell survival assay confirmed the results from the CCK8 cell viability assay in PANC1 and CFPAC1 cells (Fig. S1). As expected, ZVAD-FMK limited apoptotic stimuli staurosporine-induced cell death and necrosulfonamide blocked necroptotic stimuli (tumor necrosis factor α [TNFα]+ZVAD-FMK)-induced cell death in PANC1 cells (Fig. 1B).These observations indicate that the anticancer activity of erastin in PDAC cells depends on the induction of ferroptosis (1).
Figure 1. ATF4 regulates HSPA5 expression in ferroptosis.
(A) Indicated human PDAC cells were treated with erastin (2.5-40 μM) with or without a cell death inhibitor (ferrostatin-1, 1 μM; liprostatin-1, 1 μM; ZVAD-FMK, 10 μM; necrosulfonamide, 0.5 μM) for 24 hours. Cell death was assayed using a CCK8 kit (n=3, *p < 0.05). (B) ZVAD-FMK (10 μM) limited apoptotic stimuli staurosporine (0.5 μM)-induced cell death, whereas necrosulfonamide (0.5 μM) blocked necroptotic stimuli (ZVAD-FMK [10 μM]/TNFα (10 ng/ml))-induced cell death at 24 hours in PANC1 cells. (C) Western blot analysis indicated protein expression in PDAC cells following treatment with erastin (2.5-40 μM) for 24 hours (n=3, *p < 0.05 versus untreated group). (D) In parallel, the mRNA levels of HSPA5, CANX, and EIF2AK3 were assayed using Q-PCR. (E) Western blot analysis indicated protein expression in PANC1 cells following treatment with erastin (20 μM), staurosporine (0.5 μM), H2O2 (1 mM), and rapamycin (1 μM) for 24 hours (n=3, *p < 0.05). (F-H) Knockdown of ATF4 (but not YY1, GTF2I, or ATF6) by shRNA (F) inhibited erastin (20 μM, 24 hours)-induced HSPA5 expression at mRNA (G) and protein levels (H) (n=3, *p < 0.05 versus control shRNA group).
Erastin triggers the heat shock response and ER stress during ferroptosis in cancer cells (10, 17). HSPA5 is not only a heat shock protein, but also an ER chaperone. We therefore examined HSPA5 expression in PDAC cells following erastin treatment. Erastin dose-dependently induced HSPA5 expression at the protein (Fig. 1C) and mRNA (Fig. 1D) levels in human PDAC cells. In contrast, protein and mRNA expression of other ER stress-associated proteins such as CANX (also termed calnexin) and eukaryotic translation initiation factor 2 alpha kinase 3 (EIF2AK3, also termed PERK) were not significantly affected by erastin (Fig. 1C and 1D). Western blot analysis also showed that phosphorylated EIF2AK3 (p-EIF2AK3) was increased by erastin (Fig. 1C). In contrast, phosphorylation of endoplasmic reticulum to nucleus signaling 1 (ERN1, also termed IRE1) was not significantly changed by erastin (Fig. 1C). Additionally, erastin-induced HSPA5 protein upregulation was not observed in PANC1 cells following induction of apoptosis with staurosporine, initiation of autophagy with rapamycin, or enhancement of necrosis with H2O2 (Fig. 1E). These findings suggest that HSPA5 is significantly upregulated during the induction of ferroptosis in PDAC cells.
Several transactional factors including Yin Yang 1 (YY1) (18), general transcription factor IIi (GTF2I, also termed TFII-I) (19), activating transcription factor (ATF)-4 (20), and ATF6 (21) have been reported to regulate HSPA5 expression in individual cell lines in response to ER stress. We examined the effects of knockdown of these transactional factors using two individual shRNAs (Fig. 1F) on HSPA5 expression in ferroptosis. Notably, suppression of ATF4 (but not YY1, GTF2I, and ATF6) inhibited erastin-induced HSPA5 mRNA expression in PANC1 cells (Fig. 1G). Knockdown of ATF4 also inhibited erastin-induced HSPA5 protein expression in PANC1 and CFPAC1 cells (Fig. 1H). Collectively, these findings indicate that ATF4 mediates HSPA5 expression in ferroptosis.
HSPA5 negatively regulates ferroptosis
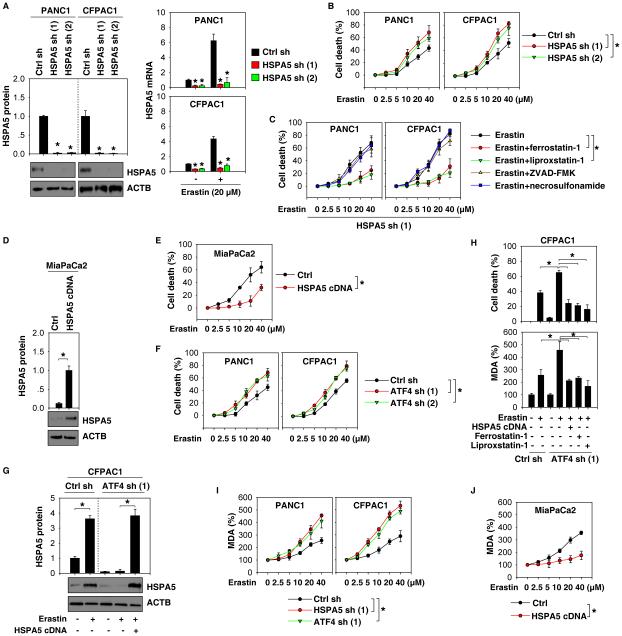
To determine whether increased HSPA5 expression affects ferroptosis, we knocked down HSPA5 using two specific shRNAs in PDAC cell lines (e.g., PANC1 and CFPAC1), which had relatively high expression of HSPA5 at baseline (Fig. 1C). Suppression of HSPA5 expression by RNAi (Fig. 2A) increased erastin-induced death in PANC1 and CFPAC1 by CCK8 cell viability assay (Fig. 2B) and clonogenic cell survival assay (Fig. S2). This process can be reversed by ferrostatin-1 and liproxstatin-1 (but not ZVAD-FMK and necrosulfonamide) (Fig. 2C and Fig. S3), suggesting that loss of HSPA5 enhances erastin-induced cell death in a ferroptosis-dependent manner. To further confirm the role of HSPA5 in ferroptosis, we overexpressed HSPA5 by gene transfection in PDAC cell lines (e.g., MiaPaCa2), which had a relatively low expression of HSPA5 at baseline (Fig. 1C). Overexpressed HSPA5 (Fig. 2D) inhibited erastin-induced ferroptotic cell death by CCK8 cell viability assay (Fig. 2E) and clonogenic cell survival assay (Fig. S4). These findings suggest that HSPA5 may be a negative regulator of ferroptosis. To test this notion further, we determined whether ATF4-mediated HSPA5 expression regulates ferroptosis. Knockdown of HSPA5 (Fig. 2B), as well as inhibition of ATF4 expression by RNAi, increased erastin-induced cell death in PDAC cells by CCK8 cell viability assay (Fig. 2F) and clonogenic cell survival assay (Fig. S5). Forced overexpression of HSPA5 (Fig. 2G) or pharmacological inhibition of ferroptosis using ferrostatin-1 and liproxstatin-1 reduced erastin-induced cell death in ATF4-knockdown CFPAC1 cells by CCK8 cell viability assay (Fig. 2H) and clonogenic cell survival assay (Fig. S6). These findings indicate that ATF4-regulated HSPA5 expression may facilitate ferroptosis resistance in PDAC cells.
Figure 2. HSPA5 negatively regulates ferroptosis.
(A, B) Knockdown of HSPA5 by shRNA enhanced erastin-induced cell death in indicated PDAC cells at 24 hours (n=3, *p < 0.05 versus control shRNA group). (C) Indicated HSPA5 knockdown PDAC cells were treated with erastin (2.5-40 μM) with or without indicated inhibitors (ferrostatin-1, 1 μM; liprostatin-1, 1 μM; ZVAD-FMK, 10 μM; necrosulfonamide, 0.5 μM) for 24 hours. Cell death was assayed using a CCK8 kit (n=3, *p < 0.05). (D, E) Overexpression of HSPA5 by gene transfection of pcDNA3.1-HSPA5 cDNA inhibited erastin-induced cell death in MiaPaCa2 cells at 24 hours (n=3, *p < 0.05). (F) Knockdown of ATF4 by shRNA enhanced erastin-induced cell death in indicated PDAC cells at 24 hours (n=3, *p < 0.05). (G, H) Overexpression of HSPA5 (pcDNA3.1-HSPA5 cDNA) or treatment with ferroptosis inhibitors (ferrostatin-1, 1 μM; liprostatin-1, 1 μM) reversed erastin (20 μM, 24 hours)-induced cell death and MDA production in ATF4 knockdown CFPAC1 cells (n=3, *p < 0.05). (I, J) Knockdown of HSPA5 or ATF4 increased erastin-induced MDA production in PANC1 and CFPAC1 cells (I), whereas overexpression of HSPA5 (pcDNA3.1-HSPA5 cDNA) limited erastin-induced MDA production at 24 hours in MiaPaCa2 cells (J) (n=3, *p < 0.05).
Given that ferroptosis is characterized by lipid peroxidation (2), we next investigated whether ATF4-mediated HSPA5 expression affects malondialdehyde (MDA, an end product of lipid peroxidation) production in erastin-induced ferroptosis. Knockdown of HSPA5 or ATF4 increased erastin-induced MDA production in PANC1 and CFPAC1 cells (Fig. 2I), whereas overexpression of HSPA5 inhibited erastin-induced MDA production in MiaPaCa2 cells (Fig. 2J). Overexpression of HSPA5 or treatment with ferrostatin-1 and liproxstatin-1 also inhibited erastin-induced MDA production in ATF4-knockdown CFPAC1 cells (Fig. 2H). These findings suggest that ATF4-dependent HSPA5 expression blocks ferroptosis through inhibition of lipid peroxidation.
HSPA5 inhibits GPX4 degradation in ferroptosis
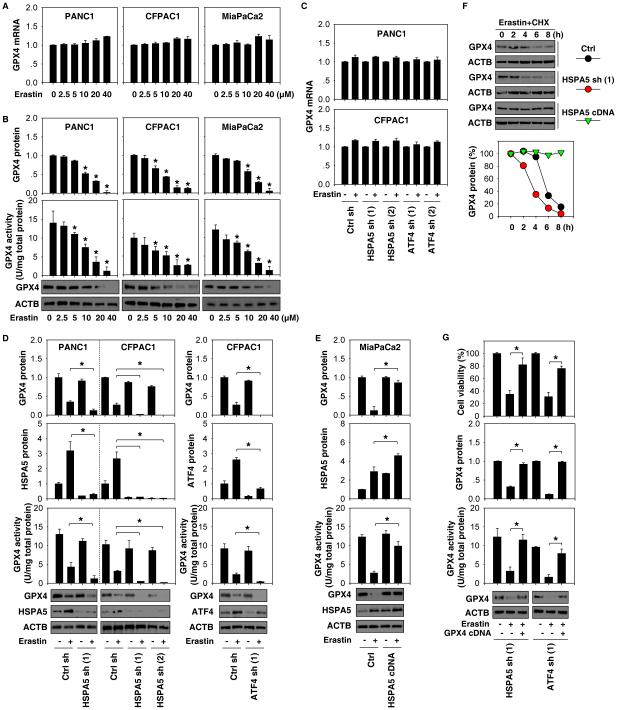
GPX4 is the critical enzyme that can reduce lipid peroxidation in ferroptosis (4). We next determined whether HSPA5 regulates lipid peroxidation through affecting GPX4 expression and activity. Erastin did not significantly affect GPX4 mRNA expression (Fig. 3A), whereas it remarkably suppressed GPX4 protein expression and activity (Fig. 3B) in PANC1 and CFPAC1 cells. These data suggest that erastin may result in GPX4 protein degradation during ferroptosis. Knockdown of HSPA5 or ATF4 expression had no influence on GPX4 mRNA levels in CFPAC1 and PANC1 cells with or without erastin treatment (Fig. 3C). In contrast, knockdown of HSPA5 or ATF4 promoted erastin-induced GPX4 protein degradation in CFPAC1 or PANC1 cells (Fig. 3D), whereas overexpression of HSPA5 inhibited erastin-induced GPX4 protein degradation in MiaPaCa2 cells (Fig. 3E). As expected, GPX4 activity was inhibited after knockdown of HSPA5 or ATF4 (Fig. 3D). In contrast, GPX4 activity was increased after overexpression of HSPA5 in erastin-induced ferroptosis (Fig. 3E). These findings suggest a potential role of HSPA5 in the regulation of GPX4 protein levels and subsequent activity. To further test whether HSPA5 affects GPX4 protein degradation, we analyzed the half-life of GPX4 protein using the cycloheximide chase assay. We observed that knockdown of HSPA5 shortened, whereas overexpression of HSPA5 prolonged GPX4 protein stabilization in PANC1 cells following erastin treatment (Fig. 3F), suggesting that HSPA5 protects against erastin-induced GPX4 protein degradation in ferroptosis.
Figure 3. HSPA5 inhibits GPX4 degradation in ferroptosis.
(A, B) Indicated PDAC cells were treated with erastin (2.5-40μM) for 24 hours. The expression of GPX4 at mRNA (A) and protein (B) levels, as well as GPX4 activity (B), were assayed (n=3, *p < 0.05 versus untreated group). (C, D) The effects of knockdown of HSPA5 or ATF4 on the expression of GPX4 at mRNA (C) and protein (D) levels, as well as GPX4 activity (D) in indicated PDAC cells following treatment with erastin (20 μM) for 24 hours (n=3, *p < 0.05). (E) The effects of overexpression of HSPA5 by gene transfection of pcDNA3.1-HSPA5 cDNA on the expression of GPX4 protein as well as GPX4 activity in MiaPaCa2 cells following treatment with erastin (20 μM) for 24 hours (n=3, *p < 0.05). (F) HSPA5 knockdown or overexpression PANC1 cells were treated with 20 μM erastin and 20 μg/ml cycloheximide (CHX) for indicated time points. Cell lysates were subjected to western blot analysis with anti-GPX4 and anti-ACTB antibodies. (G) Overexpression of GPX4 by gene transfection of pCMV6-GPX4 cDNA restored GPX4 expression, GPX4 activity, and cell viability in HSPA5- or ATF4-knockdown PANC1 cells following treatment with erastin (20 μM) for 24 hours (n=3, *p < 0.05).
We next addressed whether GPX4 degradation contributes to ferroptosis when the ATF4-HSPA5 pathway is deficient. Overexpression of GPX4 by gene transfection restored GPX4 expression (Fig. 3G), GPX4 activity (Fig. 3G), and erastin resistance (Fig. 3G and Fig. S7) in HSPA5- or ATF4-knockdown PANC1 cells. Collectively, these findings suggest that HSPA5 inhibits ferroptosis through protecting against GPX4 protein degradation.
Interaction between HSPA5 and GPX4 in ferroptosis
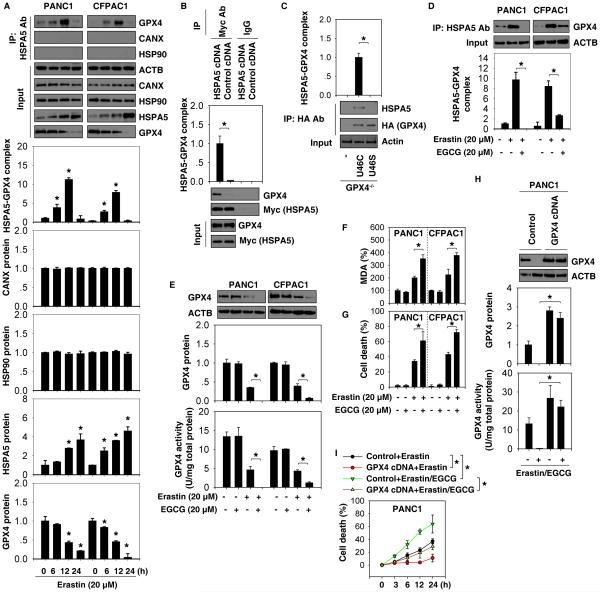
With respect to the molecular chaperone activity of HSPA5, we investigated whether HSPA5 directly binds to GPX4 to protect against GPX4 degradation in ferroptosis. Immunoprecipitation analysis showed that the interaction between GPX4 and HSPA5 was increased in PANC1 and CFPAC1 cells following erastin treatment at six to 12 hours and was then decreased at 24 hours (Fig. 4A). In contrast, there was no interaction between GPX4 and other ER stress-associated proteins (e.g., HSP90 and CANX) (Fig. 4A). Moreover, transfection with exogenous Myc-tagged HSPA5 cDNA increased the interaction between GPX4 and HSPA5 in PANC1 cells (Fig. 4B). In contrast, transfection with an empty vector did not increase the interaction between GPX4 and HSPA5 in PANC1 cells (Fig. 4B). GPX4 is a selenocysteine (Sec)-containing protein. The anti-oxidative activity of GPX4 depends on a selenocysteine residue at 46 (U46) (22). Previous studies have shown that re-expression of Sec/Cys (U46C) mutant into GPX4−/− cells inhibited cell death, whereas re-expression of the Sec/Ser (U46S) mutant failed (7, 15). We found that re-expression of U46C mutant (but not U46S mutant) in GPX4−/− cells restored the interaction between GPX4 and HSPA5 (Fig. 4C). These findings suggest that U46 is important for regulation of the binding of GPX4 to HSPA5.
Figure 4. Interaction between HSPA5 and GPX4 is observed in ferroptosis.
(A) Immunoprecipitation (IP) analysis of the interaction between HSPA5 and GPX4 in indicated PDAC cells following treatment with erastin for six to 24 hours (n=3, *p < 0.05 versus untreated group). (B) IP analysis of the interaction between HSPA5 and GPX4 in PANC1 cells after transfection with pCMV-Myc-HSPA5 cDNA (n=3, *p < 0.05). (C) IP analysis of the interaction between HSPA5 and GPX4 in GPX4−/− cells after transfection with U46C mutant or U46S mutant (n=3, *p < 0.05). (D) IP analysis of the interaction between HSPA5 and GPX4 in indicated PDAC cells after treatment with erastin in the absence or presence of EGCG for 12 hours. (E) Analysis of GPX4 expression and activity in indicated PDAC cells after treatment with erastin in the absence or presence of EGCG for 12 hours (n=3, *p < 0.05). (F-G) Intracellular MDA levels and cell death were assayed in indicated PDAC cells after treatment with erastin in the absence or presence of EGCG for 24 hours (n=3, *p < 0.05). (H-I) Overexpression of GPX4 by gene transfection with pCMV6-GPX4 cDNA limited the anticancer activity of erastin (20 μM) in combination with EGCG (20 μM) in PANC1 cells (n=3, *p < 0.05).
Epigallocatechine gallate (EGCG) is an inhibitor of HSPA5 function in cancer cells (23). We further examined the influence of pharmacologic inhibition of HSPA5 by EGCG in ferroptosis. EGCG inhibited the interaction between HSPA5 and GPX4 (Fig. 4D) and promoted GPX4 protein degradation (Fig. 4E) and decreased GPX4 activity (Fig. 4E) in PANC1 and CFPAC1 cells following erastin treatment. EGCG alone did not affect MDA production (Fig. 4F) and cell viability (Fig. 4G). However, EGCG enhanced erastin-induced MDA production (Fig. 4F) and cell death (Fig. 4G and Fig. S8) in these PDAC cells. In contrast, overexpression of GPX4 by gene transfection with GPX4 cDNA restored GPX4 activity (Fig. 4H) and reduced the anticancer activity of erastin in combination with EGCG in PANC1 cells by CCK8 cell viability assay (Fig. 4I) and clonogenic cell survival assay (Fig. S9). Collectively, our findings suggest that pharmacologic inhibition of HSPA5 by EGCG increases GPX4 protein degradation and subsequent erastin-induced ferroptosis in PDAC cells.
Inhibition of system Xc− is not required for HSPA5 expression and GPX4 degradation in ferroptosis
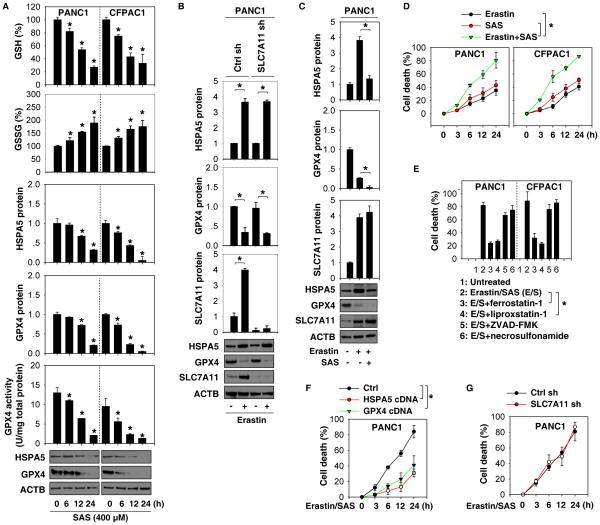
System Xc− is an amino acid antiporter that typically mediates the exchange of extracellular cystine and intracellular glutamate across the cellular plasma. In addition to erastin, sulfasalazine induces ferroptotic cancer death by inhibition of system Xc−, which leads to glutathione (GSH) depletion (1, 12). We therefore asked whether system Xc− activity regulates HSPA5 expression and GPX4 degradation in ferroptosis. Unlike erastin-induced HSPA5 expression (Fig. 1C and 1D), sulfasalazine suppressed HSPA5 expression with GSH depletion in PDAC cells (Fig. 5A). Moreover, GPX4 expression and activity was inhibited, whereas the oxidized form glutathione disulfide (GSSG) was increased by sulfasalazine (Fig. 5A). Interestingly, knockdown of solute carrier family 7 member 11 (SLC7A11, a core component of system Xc−) by shRNA had no effect on erastin-induced HSPA5 expression and GPX4 degradation in PANC1 cells (Fig. 5B). In contrast, sulfasalazine blocked erastin-induced HSPA5 upregulation and increased GPX4 degradation in PANC1 cells (Fig. 5C). These findings suggest that inhibition of system Xc− may be not required for erastin-induced HSPA5 upregulation and GPX4 degradation.
Figure 5. Inhibition of system Xc− is not required for HSPA5 expression or GPX4 degradation in ferroptosis.
(A) Indicated PDAC cells were treated with sulfasalazine (SAS) for six to 24 hours. The levels of GSH, GSSG, indicated protein, and GPX4 activity were assayed (n=3, *p < 0.05 versus untreated group). (B) Knockdown of SLC7A11 by shRNA did not affect erastin (20μM)-induced HSPA5 expression and GPX4 degradation at 24 hours in PANC1 cells (n=3, *p < 0.05). (C) SAS (400μM) inhibited erastin (20μM)-induced HSPA5 expression and GPX4 degradation at 24 hours in PANC1 cells (n=3, *p < 0.05). (D) Indicated PDAC cells were treated with erastin (20μM) with or without SAS (400μM) for three to 24 hours and cell death was assayed (n=3, *p < 0.05). (E) Indicated PDAC cells were treated with erastin (20 μM) /SAS (400 μM) with or without indicated inhibitors (ferrostatin-1, 1 μM; liprostatin-1, 1 μM; ZVAD-FMK,10 μM; necrosulfonamide, 0.5 μM) for 24 hours. Cell death was assayed using a CCK8 kit (n=3, *p < 0.05). (F, G) Indicated PANC1 cells were treated with erastin (20 μM) in combination with SAS (400 μM) for three to 24 hours. Cell death was assayed with a CCK8 kit (n=3, *p < 0.05).
We next addressed whether sulfasalazine-mediated HSPA5 degradation enhances the anticancer activity of erastin. The synergistic anticancer effect of erastin combined with sulfasalazine causes cell death in PANC1 and CFPAC1 cells by CCK8 cell viability assay (Fig. 5D) and clonogenic cell survival assay (Fig. S10). This process was blocked by ferroptosis inhibitors (e.g., ferrostatin-1 and liproxstatin-1), but not the apoptosis inhibitor (e.g., ZVAD-FMK) or necroptosis inhibitor (e.g., necrosulfonamide) (Fig. 5E). Furthermore, overexpression of HSPA5 or GPX4 by gene transfection reduced the anticancer activity of erastin in combination with sulfasalazine in PDAC1 cells by CCK8 cell viability assay (Fig. 5F) and clonogenic cell survival assay (Fig. S11). In contrast, knockdown of SLC7A11 did not affect the anticancer activity of erastin in combination with sulfasalazine (Fig. 5G). These findings suggest that activation of the HSPA5-GPX4 pathway promotes ferroptosis resistance and that this process is likely SLC7A11-independent.
Inhibition of the HSPA5-GPX4 pathway enhances gemcitabine sensitivity in vitro
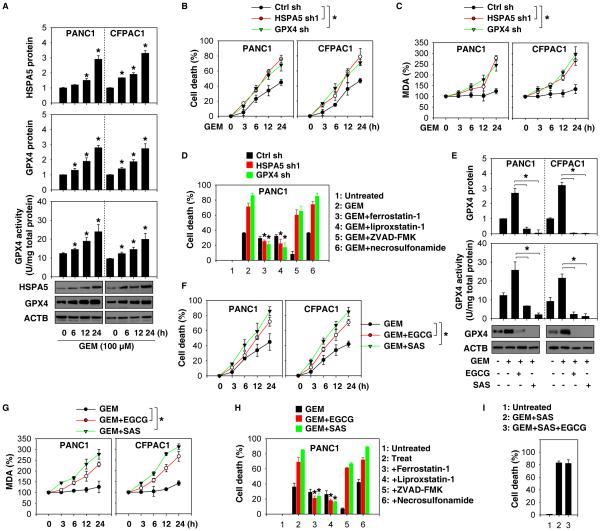
We next investigated whether gemcitabine affects HSPA5 and GPX4 expression in PDAC cells. Gemcitabine time-dependently increased HSPA5 and GPX4 protein expression, as well as GPX4 activity, in PANC1 or CFPAC1 cells (Fig. 6A), suggesting a potential role of the HSPA5-GPX4 pathway in the regulation of the anticancer activity of gemcitabine. Indeed, knockdown of HSPA5 and GPX4 increased gemcitabine-induced cell death (Fig. 6B and Fig. S12) with increased MDA production (Fig. 6C) in PANC1 or CFPAC1 cells. Ferroptosis inhibitors (e.g., ferrostatin-1 and liproxstatin-1), but not apoptosis inhibitor (e.g., ZVAD-FMK) or necroptosis inhibitor (e.g., necrosulfonamide), reversed gemcitabine-induced cell death by CCK8 cell viability assay (Fig. 6D) and clonogenic cell survival assay (Fig. S13) in HSPA5- or GPX4-knockdown PANC1 cells. Thus, genetic inhibition of the HSPA5-GPX4 pathway enhances the anticancer activity of gemcitabine by induction of ferroptosis.
Figure 6. Inhibition of the HSPA5-GPX4 pathway enhances gemcitabine sensitivity in vitro.
(A) Indicated PDAC cells were treated with gemcitabine (GEM) for six to 24 hours and then protein levels and GPX4 activity were assayed (n=3, *p < 0.05 versus untreated group). (B, C) Knockdown of HSPA5 or GPX4 by shRNA in PDAC cells increased GEM (100 μM)-induced cell death and MDA production (n=3, *p < 0.05). (D) Indicated PANC1 cells were treated with GEM (100 μM) with or without indicated inhibitors (ferrostatin-1, 1 μM; liprostatin-1, 1 μM; ZVAD-FMK,10 μM; necrosulfonamide, 0.5 μM) for 24 hours. Cell death was assayed with a CCK8 kit (n=3, *p < 0.05 versus GEM group). (E) Indicated PDAC cells were treated with GEM (100 μM) in the absence or presence of SAS (400 μM) or EGCG (20 μM) for 24 hours and then protein levels and GPX4 activity were assayed (n=3, *p < 0.05). (F-G) In parallel, cell death and MDA levels were assayed. (H) Ferroptosis inhibitors (e.g., ferrostatin-1 and liproxstatin-1), but not apoptosis inhibitor (e.g., ZVAD-FMK) or necroptosis inhibitor (e.g., necrosulfonamide) reversed GEM/EGCG or GEM/SAS-induced cell death in PANC1 cells (n=3, *p < 0.05). (I) PANC1 cells were treated with GEM (100 μM)+SAS (400 μM) or GEM (100 μM)+SAS (400 μM)+EGCG (20 μM) for 24 hours; cell death was then assayed.
We further addressed whether pharmacologic inhibition of HSPA5 by EGCG or sulfasalazine enhances the anticancer activity of gemcitabine. Indeed, EGCG or sulfasalazine blocked gemcitabine-induced GPX4 upregulation and promoted GPX4 degradation and inhibited GPX4 activity in PANC1 and CFPAC1 cells (Fig. 6E), confirming that HSPA5 is an important regulator of GPX4 protein levels. As a result, EGCG or sulfasalazine promoted gemcitabine-induced cell death (Fig. 6F and Fig. S14) with increased MDA production (Fig. 6G) in PANC1 and CFPAC1 cells. This process was reversed by ferrostatin-1 and liproxstatin-1 (but not ZVAD-FMK nor necrosulfonamide) in PDAC cells by CCK8 cell viability assay (Fig. 6H) and clonogenic cell survival assay (Fig. S15). Of note, there was no significant difference between the anticancer effects of gemcitabine/sulfasalazine and gemcitabine/sulfasalazine/EGCG (Fig. 6I). Collectively, these findings suggest that pharmacologic inhibition of the HSPA5-GPX4 pathway may enhance the anticancer activity of gemcitabine by inducing effective ferroptosis in PDAC cells.
Inhibition of the HSPA5-GPX4 pathway enhances gemcitabine sensitivity in vivo
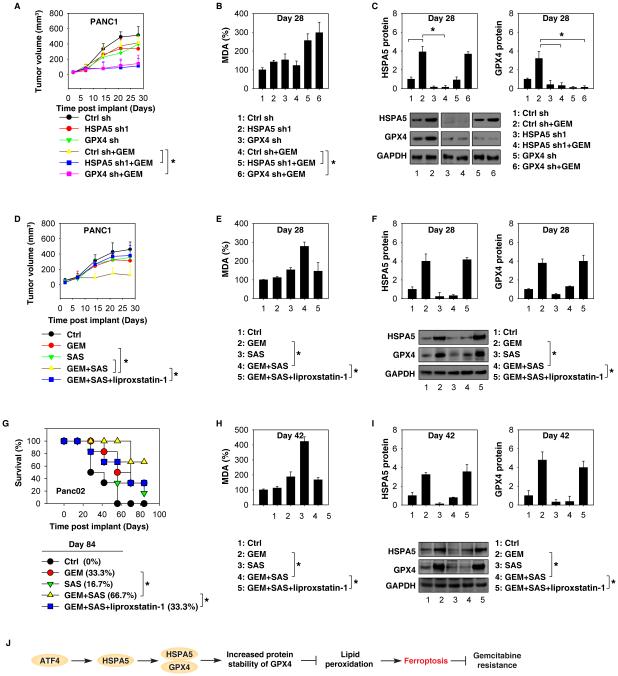
To determine whether suppression of the HSPA5-GPX4 pathway enhances the anticancer activity of gemcitabine in vivo, HSPA5- or GPX4-knockdown PANC1 cells were implanted into the subcutaneous space of the right flank of nude mice. Beginning at day seven, these mice were treated with gemcitabine. Compared with the control shRNA group, gemcitabine treatment effectively reduced the size of tumors formed (Fig. 7A) with increased MDA levels (Fig. 7B) by HSPA5- or GPX4-knockdown cells (Fig. 7C) observed within the tumor. Similarly, sulfasalazine also enhanced the anticancer activity of gemcitabine in subcutaneous tumor models (Fig. 7D), which was associated with increased MDA levels (Fig. 7E) and decreased HSPA5 and GPX4 expression (Fig. 7F) within tumors. In contrast, the ferroptosis inhibitor liproxstatin-1 suppressed MDA production in tumor (Fig. 7E) and the anticancer activity of gemcitabine combined with sulfasalazine (Fig. 7D) and restored the expression of HSPA5 and GPX4 (Fig. 7F). These findings demonstrate that inhibition of the HSPA5-GPX4 pathway enhances the anticancer activity of gemcitabine by induction of ferroptosis in a subcutaneous PDAC mouse model.
Figure 7. Inhibition of the HSPA5-GPX4 pathway enhances gemcitabine sensitivity in vivo.
(A-C) Nude mice were injected subcutaneously with indicated PANC1 cells (2 × 106 cells/mouse) and intraperitoneally (i.p.) treated with GEM (20 mg/kg once every other day) at day seven for two weeks (n=5 mice/group). Tumor volume was calculated weekly (A) and the MDA levels (B) and the protein levels of HSPA5 and GPX4 (C) in isolated tumor at day 28 were assayed (* p < 0.05). (D-F) Nude mice were injected subcutaneously with PANC1 cells (2 × 106 cells/mouse) and treated with GEM (20 mg/kg/i.p., once every other day), SAS (100 mg/kg/i.p., once every other day), GEM+SAS, or GEM+SAS+liproxstatin-1 (10 mg/kg/i.p., once every other day) at day seven for two weeks (n=5 mice/group). Tumor volume was calculated weekly (D) and MDA levels (E) and protein levels of HSPA5 and GPX4 (F) in isolated tumor at day 28 were assayed (* p < 0.05). (G-I) B6 mice were surgically implanted with 1× 106 Panc02 cells into the tail of the pancreas. Two weeks after implantation, mice were randomized (six mice/group) to receive either: 1) GEM (20 mg/kg/i.p., once every other day); 2) SAS (100 mg/kg/i.p., once every other day); 3) GEM+SAS; or 4) GEM+SAS+liproxstatin-1 (10 mg/kg/i.p., once every other day) for three weeks. Animal survival was calculated weekly (G) and MDA levels (H) and the protein levels of HSPA5 and GPX4 (I) in isolated tumor at day 42 were assayed (* p < 0.05). (J) Schematic depicting HSPA5-mediated ferroptosis resistance in the regulation of gemcitabine sensitivity in PDAC cells.
Orthotopic tumor models are considered more clinically relevant and more predictive of drug efficacy than standard subcutaneous models. We next determined whether induction of ferroptosis by sulfasalazine also enhances the anticancer activity of gemcitabine in orthotopic tumor models with established mouse Panc02 cells in B6 mice. Indeed, gemcitabine combined with sulfasalazine significantly prolonged animal survival (Fig. 7G) with increased MDA levels (Fig. 7H) and decreased HSPA5 and GPX4 expression (Fig. 7I) in tumor. In contrast, the ferroptosis inhibitor liproxstatin-1 inhibited MDA production in tumor (Fig. 7H) and decreased the anticancer activity of gemcitabine combined with sulfasalazine (Fig. 7G) and restored the expression of HSPA5 and GPX4 (Fig. 7I). These results support the idea that targeting ferroptotic repressor (e.g., HSPA5) improves the anticancer activity of gemcitabine for PDAC in vivo.
Discussion
Ferroptosis is a relatively recently identified means to induce non-apoptotic cell death in cancer cells (1, 4). The current results establish that upregulation of HSPA5 is a negative regulator of ferroptosis in PDAC cells. Increased HSPA5 expression inhibits lipid peroxidation in ferroptosis by directly protecting against GPX4 degradation (Fig. 7J). These findings also result in the significant improvement of anticancer activity of gemcitabine in combined with HSPA5 inhibitor in the treatment of PDAC by induction of ferroptosis.
Lipid peroxidation is a free radical chain reaction and generates a range of reactive carbonyl species in both pathological and physiological conditions (24). Accumulation of lipid peroxidation products is recognized as a central mediator of ferroptosis (2). We and others previously identified several molecules, including GPX4 (4), cysteinyl-tRNA synthetase (25), p53 (26), nuclear factor erythroid 2-related factor 2 (27, 28), and acyl-CoA synthetase long-chain family member 4 (29, 30) that regulate ferroptotic cancer death by directly or indirectly targeting lipid peroxidation in a transcription-independent or -dependent manner. The work presented here identifies an additional pathway upstream of GPX4 by HSPA5 to limit lipid peroxidation and underscores the importance of molecular chaperones in ferroptosis (12). Similar to previous studies conducted with endothelial (31) and glial cells (32), the current study demonstrates that HSPA5 plays a role in regulating lipid peroxidation in PDAC cells. Inhibition of lipid peroxidation by antioxidants (e.g., ferrostatin-1 (4) and liprostatin-1 (7)) diminishes erastin-induced ferroptosis in HSPA5-kncodown PDAC cells.
Our studies also provide the molecular mechanism that underlies the change between ER stress and ferroptosis (12). Potent triggers for ER stress include redox imbalances and lipid peroxidation (33). ER stress involves a wide range of enzymes and molecular chaperones depending on the biological contexts. Upregulation of ER stress markers such as ATF4 and phosphorylation of eIF2α have been observed in ferroptosis (12). The current study demonstrated a marked increase of HSPA5 protein expression by activation of ATF4 (but not other transcriptional factors) in ferroptosis. Further functional analysis documents that suppression of the ATF4-HSPA5 pathway by RNAi enhances erastin-induced ferroptosis, suggesting that induction of HSPA5 expression by ATF4 is a stress-adaptation response against ferroptosis. These findings are consistent with previous observations that ATF4 serves to enhance the transcriptional expression of genes involved in amino acid metabolism and resistance to oxidative stress (34, 35). In contrast, ATF4-mediated DNA damage inducible transcript 3 expression contributes to ER stress-induced apoptosis (36). Further studies will be required to elucidate the mechanism and context-dependent functions of ATF4 in various forms of regulated cell death.
Despite structural and catalytic similarities, GPX4 is unique among GPX isoforms, as it is the only enzyme capable of inhibiting lipid peroxidation by reducing phospholipid hydroperoxide in mammalian cells (37). Previous work has shown that ferroptosis inducer (e.g., erastin and FIN56) can cause GPX4 protein degradation in cancer cells (8, 9). We have now established that the interaction between HSPA5 and GPX4 protects against GPX4 protein degradation in ferroptosis. Thus, inhibition of HSPA5 can increase GPX4 protein degradation and subsequent lipid peroxidation. Like global knockout of HSPA5 (38), global knockout of GPX4 also can cause early embryonic lethality in mice (39), suggesting that ferroptosis plays an important role in early mouse embryonic development.
Pancreatic cancer is one of the most difficult malignancies to successfully treat, despite advances in modern medicine. Gemcitabine-based treatment is currently a standard first line therapy for patients with advanced PDAC; however overall survival remains poor and few options are available for patients in whom gemcitabine-based therapies fail. Elevated HSPA5 expression is associated with poor prognosis in patients with PDAC (40). Our results indicate that upregulation of the HSPA5-GPX4 pathway contributes to gemcitabine resistance. In contrast, genetic and pharmacological inhibition of the HSPA5-GPX4 pathway by RNAi, EGCG, or sulfasalazine enhances gemcitabine sensitivity in PDAC cells in vitro and in two animal tumor models.
EGCG, the most abundant catechin in tea, is a polyphenol under investigation for its ability to affect human health and diseases, including cancer. A molecular and structural pharmacology study established that EGCG is a direct inhibitor of HSPA5 by binding to the ATP-binding site of HSPA5 (23). Early studies indicate that EGCG prevents the formation of the anti-apoptotic HSPA5-caspase-7 complex, which leads to an increased etoposide-induced apoptosis in breast cancer cells (23). Our current study demonstrates that EGCG prevents the formation of the antiferroptotic HSPA5-GPX4 complex, which results in an increased erastin-induced ferroptosis in PDAC cells. Thus, EGCG has a great potential for PDAC treatment.
Importantly, we demonstrated that sulfasalazine not only inhibits system Xc−, but also promotes GPX4 degradation in ferroptosis. Inhibition of system Xc− is not required for sulfasalazine-mediated GPX4 degradation, although system Xc− also plays a role in pancreatic cancer growth and drug resistance (41). In contrast, sulfasalazine inhibits HSPA5 expression, which in turn promotes GPX4 degradation in ferroptosis. Sulfasalazine is an FDA-approved drug to treat patients with ulcerative colitis or rheumatoid arthritis (42). Our results highlight a potential PDAC treatment strategy by combining gemcitabine containing regimens with sulfasalazine to induce ferroptosis.
In summary, we provide the evidence of the involvement of HSPA5 in mediating ferroptosis resistance in PDAC cells. As a protein chaperone, HSPA5 forms complexes with GPX4 that inhibit GPX4 degradation. Inhibition of GPX4 activity and promotion of GPX4 degradation are responsible for lipid peroxidation in ferroptosis. Since human cancer cells are inherently heterologous, different types of cancer cells can use diverse signaling and defense mechanisms to acquire resistance to specific anticancer agents. In addition to mediating tumor therapy, excessive ferroptosis has been implicated in injury- and inflammation-associated diseases (7, 43). Further studies are needed to confirm whether increased HSPA5 expression contributes to ferroptosis resistance in other tumors and diseases.
Supplementary Material
Acknowledgments
We thank Christine Heiner (Department of Surgery, University of Pittsburgh) for her critical reading of the manuscript.
Financial Support: This work was supported by the National Institutes of Health (R01CA160417, R01GM115366, and R01CA181450), a Pancreatic Cancer Action Network-AACR Career Development Award (13-20-25-TANG), a Research Scholar Grant from the American Cancer Society (RSG-16-014-01-CDD), the National Natural Science Foundation of China (31671435), and the National Natural Science Foundation of Guangdong (2016A030308.). This project partly utilized University of Pittsburgh Cancer Institute shared resources supported by award P30CA047904.
Footnotes
Conflict of Interest: The authors declare no conflicts of interest.
References
- 1.Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–72. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang WS, Stockwell BR. Ferroptosis: Death by Lipid Peroxidation. Trends Cell Biol. 2016;26:165–76. doi: 10.1016/j.tcb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie Y, Hou W, Song X, Yu Y, Huang J, Sun X, et al. Ferroptosis: process and function. Cell Death Differ. 2016;23:369–79. doi: 10.1038/cdd.2015.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–31. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen L, Hambright WS, Na R, Ran Q. Ablation of the Ferroptosis Inhibitor Glutathione Peroxidase 4 in Neurons Results in Rapid Motor Neuron Degeneration and Paralysis. J Biol Chem. 2015;290:28097–106. doi: 10.1074/jbc.M115.680090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsushita M, Freigang S, Schneider C, Conrad M, Bornkamm GW, Kopf M. T cell lipid peroxidation induces ferroptosis and prevents immunity to infection. J Exp Med. 2015;212:555–68. doi: 10.1084/jem.20140857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedmann Angeli JP, Schneider M, Proneth B, Tyurina YY, Tyurin VA, Hammond VJ, et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol. 2014;16:1180–91. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu Y, Xie Y, Cao L, Yang L, Yang M, Lotze MT, et al. The ferroptosis inducer erastin enhances sensitivity of acute myeloid leukemia cells to chemotherapeutic agents. Mol Cell Oncol. 2015;2:e1054549. doi: 10.1080/23723556.2015.1054549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimada K, Skouta R, Kaplan A, Yang WS, Hayano M, Dixon SJ, et al. Global survey of cell death mechanisms reveals metabolic regulation of ferroptosis. Nat Chem Biol. 2016;12:497–503. doi: 10.1038/nchembio.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee AS. Glucose-regulated proteins in cancer: molecular mechanisms and therapeutic potential. Nat Rev Cancer. 2014;14:263–76. doi: 10.1038/nrc3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 12.Dixon SJ, Patel DN, Welsch M, Skouta R, Lee ED, Hayano M, et al. Pharmacological inhibition of cystine-glutamate exchange induces endoplasmic reticulum stress and ferroptosis. Elife. 2014;3:e02523. doi: 10.7554/eLife.02523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weitzel F, Ursini F, Wendel A. Phospholipid hydroperoxide glutathione peroxidase in various mouse organs during selenium deficiency and repletion. Biochim Biophys Acta. 1990;1036:88–94. doi: 10.1016/0304-4165(90)90018-r. [DOI] [PubMed] [Google Scholar]
- 14.Sneddon AA, Wu HC, Farquharson A, Grant I, Arthur JR, Rotondo D, et al. Regulation of selenoprotein GPx4 expression and activity in human endothelial cells by fatty acids, cytokines and antioxidants. Atherosclerosis. 2003;171:57–65. doi: 10.1016/j.atherosclerosis.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 15.Mannes AM, Seiler A, Bosello V, Maiorino M, Conrad M. Cysteine mutant of mammalian GPx4 rescues cell death induced by disruption of the wild-type selenoenzyme. FASEB J. 2011;25:2135–44. doi: 10.1096/fj.10-177147. [DOI] [PubMed] [Google Scholar]
- 16.Dolma S, Lessnick SL, Hahn WC, Stockwell BR. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell. 2003;3:285–96. doi: 10.1016/s1535-6108(03)00050-3. [DOI] [PubMed] [Google Scholar]
- 17.Sun X, Ou Z, Xie M, Kang R, Fan Y, Niu X, et al. HSPB1 as a novel regulator of ferroptotic cancer cell death. Oncogene. 2015;34:5617–25. doi: 10.1038/onc.2015.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baumeister P, Luo S, Skarnes WC, Sui G, Seto E, Shi Y, et al. Endoplasmic reticulum stress induction of the Grp78/BiP promoter: activating mechanisms mediated by YY1 and its interactive chromatin modifiers. Mol Cell Biol. 2005;25:4529–40. doi: 10.1128/MCB.25.11.4529-4540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong M, Lin MY, Huang JM, Baumeister P, Hakre S, Roy AL, et al. Transcriptional regulation of the Grp78 promoter by endoplasmic reticulum stress: role of TFII-I and its tyrosine phosphorylation. J Biol Chem. 2005;280:16821–8. doi: 10.1074/jbc.M413753200. [DOI] [PubMed] [Google Scholar]
- 20.Luo S, Baumeister P, Yang S, Abcouwer SF, Lee AS. Induction of Grp78/BiP by translational block: activation of the Grp78 promoter by ATF4 through and upstream ATF/CRE site independent of the endoplasmic reticulum stress elements. J Biol Chem. 2003;278:37375–85. doi: 10.1074/jbc.M303619200. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Shen J, Arenzana N, Tirasophon W, Kaufman RJ, Prywes R. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J Biol Chem. 2000;275:27013–20. doi: 10.1074/jbc.M003322200. [DOI] [PubMed] [Google Scholar]
- 22.Scheerer P, Borchert A, Krauss N, Wessner H, Gerth C, Hohne W, et al. Structural basis for catalytic activity and enzyme polymerization of phospholipid hydroperoxide glutathione peroxidase-4 (GPx4) Biochemistry. 2007;46:9041–9. doi: 10.1021/bi700840d. [DOI] [PubMed] [Google Scholar]
- 23.Ermakova SP, Kang BS, Choi BY, Choi HS, Schuster TF, Ma WY, et al. (−)-Epigallocatechin gallate overcomes resistance to etoposide-induced cell death by targeting the molecular chaperone glucose-regulated protein 78. Cancer Res. 2006;66:9260–9. doi: 10.1158/0008-5472.CAN-06-1586. [DOI] [PubMed] [Google Scholar]
- 24.Ayala A, Munoz MF, Arguelles S. Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid Med Cell Longev. 2014;2014:360438. doi: 10.1155/2014/360438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayano M, Yang WS, Corn CK, Pagano NC, Stockwell BR. Loss of cysteinyl-tRNA synthetase (CARS) induces the transsulfuration pathway and inhibits ferroptosis induced by cystine deprivation. Cell Death Differ. 2016;23:270–8. doi: 10.1038/cdd.2015.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang L, Kon N, Li T, Wang SJ, Su T, Hibshoosh H, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57–62. doi: 10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun X, Ou Z, Chen R, Niu X, Chen D, Kang R, et al. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology. 2016;63:173–84. doi: 10.1002/hep.28251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun X, Niu X, Chen R, He W, Chen, Kang R, et al. Metallothionein-1G Facilitates Sorafenib Resistance through Inhibition of Ferroptosis. Hepatology. 2016 doi: 10.1002/hep.28574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doll S, Proneth B, Tyurina YY, Panzilius E, Kobayashi S, Ingold I, et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat Chem Biol. 2017;13:91–8. doi: 10.1038/nchembio.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan H, Li X, Zhang X, Kang R, Tang D. Identification of ACSL4 as a biomarker and contributor of ferroptosis. Biochem Biophys Res Commun. 2016;478:1338–43. doi: 10.1016/j.bbrc.2016.08.124. [DOI] [PubMed] [Google Scholar]
- 31.Birukova AA, Singleton PA, Gawlak G, Tian X, Mirzapoiazova T, Mambetsariev B, et al. GRP78 is a novel receptor initiating a vascular barrier protective response to oxidized phospholipids. Mol Biol Cell. 2014;25:2006–16. doi: 10.1091/mbc.E13-12-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suyama K, Watanabe M, Sakabe K, Otomo A, Okada Y, Terayama H, et al. GRP78 suppresses lipid peroxidation and promotes cellular antioxidant levels in glial cells following hydrogen peroxide exposure. PLoS One. 2014;9:e86951. doi: 10.1371/journal.pone.0086951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vladykovskaya E, Sithu SD, Haberzettl P, Wickramasinghe NS, Merchant ML, Hill BG, et al. Lipid peroxidation product 4-hydroxy-trans-2-nonenal causes endothelial activation by inducing endoplasmic reticulum stress. J Biol Chem. 2012;287:11398–409. doi: 10.1074/jbc.M111.320416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roybal CN, Hunsaker LA, Barbash O, Vander Jagt DL, Abcouwer SF. The oxidative stressor arsenite activates vascular endothelial growth factor mRNA transcription by an ATF4-dependent mechanism. J Biol Chem. 2005;280:20331–9. doi: 10.1074/jbc.M411275200. [DOI] [PubMed] [Google Scholar]
- 35.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–33. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 36.Szegezdi E, Logue SE, Gorman AM, Samali A. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 2006;7:880–5. doi: 10.1038/sj.embor.7400779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Imai H, Nakagawa Y. Biological significance of phospholipid hydroperoxide glutathione peroxidase (PHGPx, GPx4) in mammalian cells. Free Radic Biol Med. 2003;34:145–69. doi: 10.1016/s0891-5849(02)01197-8. [DOI] [PubMed] [Google Scholar]
- 38.Luo S, Mao C, Lee B, Lee AS. GRP78/BiP is required for cell proliferation and protecting the inner cell mass from apoptosis during early mouse embryonic development. Mol Cell Biol. 2006;26:5688–97. doi: 10.1128/MCB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yant LJ, Ran Q, Rao L, Van Remmen H, Shibatani T, Belter JG, et al. The selenoprotein GPX4 is essential for mouse development and protects from radiation and oxidative damage insults. Free Radic Biol Med. 2003;34:496–502. doi: 10.1016/s0891-5849(02)01360-6. [DOI] [PubMed] [Google Scholar]
- 40.Niu Z, Wang M, Zhou L, Yao L, Liao Q, Zhao Y. Elevated GRP78 expression is associated with poor prognosis in patients with pancreatic cancer. Sci Rep. 2015;5:16067. doi: 10.1038/srep16067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lo M, Ling V, Wang YZ, Gout PW. The xc- cystine/glutamate antiporter: a mediator of pancreatic cancer growth with a role in drug resistance. Br J Cancer. 2008;99:464–72. doi: 10.1038/sj.bjc.6604485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plosker GL, Croom KF. Sulfasalazine: a review of its use in the management of rheumatoid arthritis. Drugs. 2005;65:1825–49. doi: 10.2165/00003495-200565130-00008. [DOI] [PubMed] [Google Scholar]
- 43.Gao M, Monian P, Quadri N, Ramasamy R, Jiang X. Glutaminolysis and Transferrin Regulate Ferroptosis. Mol Cell. 2015;59:298–308. doi: 10.1016/j.molcel.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.