Abstract
The ErbB3 receptor binding protein EBP1 encodes two alternatively spliced isoforms p48 and p42. While there is evidence of differential roles for these isoforms in tumorigenesis, little is known about their underlying mechanisms. Here we demonstrate that EBP1 isoforms interact with the SCF-type ubiquitin ligase FBXW7 in distinct ways to exert opposing roles in tumorigenesis. EBP1 p48 bound to the WD domain of FBXW7 as an oncogenic substrate of FBXW7. EBP1 p48 binding sequestered FBXW7α to the cytosol, modulating its role in protein degradation and attenuating its tumor suppressor function. In contrast, EBP1 p42 bound to both the F-box domain of FBXW7 as well as FBXW7 substrates. This adapter function of EBP1 p42 stabilized the interaction of FBXW7 with its substrates and promoted FBXW7-mediated degradation of oncogenic targets, enhancing its overall tumor suppressing function. Overall, our results establish distinct physical and functional interactions between FBXW7 and EBP1 isoforms which yield their mechanistically unique isoform-specific functions of EBP1 in cancer.
Keywords: Colorectal cancer, FBXW7, Protein isoform, EBP1 P48, EBP1 P42
Introduction
The ErbB3 binding protein EBP1 is a member of the proliferation-associated 2G4 protein family and ubiquitously expressed in all human tissues and involved in the regulation of cell growth and differentiation (1, 2). EBP1 encodes two different protein isoforms, the long form P48 and short form P42, due to alternative splicing of pre-mRNA, which are believed to have different cellular activities (3–5). The predominant P48 isoform can promote cell proliferation, differentiation or survival including cancer cells (4, 6), and localizes to both cytoplasm and nucleus (5). Moreover, correlation of high expression level of P48 with poor clinical outcomes in pancreatic ductal adenocarcinoma (6), brain tumor (4), cervical (7) and prostate cancer patients (8) suggests P48 is closely related with cancer progression. In contrast, the P42 isoform which lacks the N-terminal 54 amino acids is considered to be a potent tumor suppressor because of its growth inhibitory function (3, 5, 9). Although it is reported that P48 has an oncogenic activity and P42 plays a suppressive role in various cancer cells, the underlying mechanism regarding the distinctive functions of the two isoforms of EBP1 in cancer remains largely unclear.
FBXW7 (F-box and WD40 domain protein 7) functions as a substrate recognition subunit of the SCF (SKP1/CUL1/F-box protein) E3 ubiquitin ligase complex (10) to regulate a network of proteins with central roles in cell division, growth and differentiation (11–13). FBXW7 contains two important functional domains: the F-box domain which interacts directly with SKP1 to recruit ubiquitin-conjugating enzymes, and the WD40 domain which binds to a consensus phosphor-binding motif called the CDC4 phosphodegron (CPD) in its substrates (14, 15). FBXW7 has been characterized as a general tumor suppressor. FBXW7 mutation or deletion is often observed in multiple human cancers including colorectal cancer (16, 17) and loss of FBXW7 function results in tumorigenesis (18–23). Accumulating data indicate that FBXW7 exerts its anti-tumor function mainly through targeting multiple oncoproteins for ubiquitination and proteasomal degradation, such as cyclin E (24), Notch (25), mTOR (26), Aurora-A (27) and c-Myc (28). However, a detailed understanding of the full set of FBXW7 substrates and the mechanisms that link FBXW7 deficiency to tumorigenesis are still lacking.
In this study, we used proteomics approach to globally screen FBXW7-regulated proteins in colorectal cancer cells and first identified that EBP1 P48 expression was dramatically up-regulated in FBXW7 deficient cells. Further mechanism study showed that FBXW7 physically binds to EBP1 P48 and facilitate the ubiquitination and degradation of P48 through the proteasome pathway in a GSK3β-dependent manner. Interestingly, we also found that FBXW7 is able to interact with EBP1 P42 but fail to result in the degradation of P42. On the contrary, the tumor inhibitory function of FBXW7 was enhanced by P42 but attenuated by P48. The underlying mechanical protein interaction among the three proteins was further investigated. Therefore, our data demonstrate a physical and functional protein interaction network among FBXW7 and EBP1 isoforms which provide novel mechanistic insights into the distinctive functions and regulations of EBP1 isoforms.
Materials and Methods
Cell lines and Cell culture
HCT116 (FBXW7+/+ and FBXW7−/−) and DLD-1 (FBXW7+/+ and FBXW7−/−) cell lines were kind gifts from B. Vogelstein and cultured in McCoy’s 5A supplemented with 10% fetal bovine serum (FBS). MDA-MB-468, PC3, DU145 and HEK293T cells were purchased from the American Type Culture Collection (Manassas, VA, USA), where they were characterized by DNA-fingerprinting and isozyme detection. MDA-MB-468 and HEK293T cells were maintained in DME medium with 10% FBS and PC3 and DU145 cells in RPMI-1640 medium with 10% FBS. All cells were grown at 37 °C with 5% CO2/95% air atmosphere and were revived every 3 to 4 months.
Expression plasmids
Wild type HA-FBXW7α, FBXW7α mutants (ΔWD, ΔF,), shFBXW7 A and shFBXW7 B plasmids were previously constructed by our lab (Supplementary Figure 1C) (21, 26, 29). The shRNA sequences for FBXW7 are as follows: shFBXW7 A: GGCAACAACGACGCCGAAT; shFBXW7 B: GAGTTGGCACTCTATGTGC.EBP1 P48 and P42 human cDNA were subcloned using the PrimeSTAR Max DNA polymerase (TaKaRa, Japan) into the Myc-CMV10 vector and Flag-CMV10 vector. FBXW7αNΔF were subcloned into the pCGN-HA vector. Flag-EBP1 P48 S40A, Flag-EBP1 P48 S44A, HA-FBXW7α mutants (R465C, R465H, R479H, R505C) and HA-FBXW7αΔFR465C were generated using PCR-directed mutagenesis method. Ubiquitin gene was amplified and subcloned into Myc-CMV10 vector and pCGN-HA vector. The shRNA plasmids specific for EBP1 P48 were purchased from Genechem (Shanghai, China) with the following target sequences: shP48.1: GAGCAACAGGAGCAAACTA; shP48.3: GGTGGAAGCATCTAGCTCA.
Antibodies and Reagents
The antibodies used in our experiments are listed in supplementary table 1. CHX, MG132, GSK3β inhibitor VIII and protein G/A were from Calbiochem (Darmstadt, Germany). Protease inhibitor cocktail was from Roche (Basel, Switzerland). Trizol and lipo-2000 were from Invitrogen (Carlsbad, CA, USA). RIPA was from Beyotime Biotechnology (Jiangsu, China).
Immunoblots and Immunoprecipitation
Whole cell lysates were prepared using RIPA lysis buffer supplemented with protease inhibitors. Nuclear and cytoplasmic extracts were prepared using a nuclear and cytoplasmic protein extraction kit (Beyotime Biotechnology, Jiangsu, China) according to the manufacturer’s instructions. Western blotting and immunoprecipitation were performed as described before (30).
RNA isolation and real-time RT-PCR
RNA was isolated using the Trizol Reagent. The reverse transcription reaction was performed using the RevertAid First Strand cDNA synthesis Kit (Thermo, Waltham, MA, USA). qRT-PCR was performed using the SYBR Green PCR Master Mix (Thermo, Waltham, MA, USA) and the ABI PRISM 7900HT Real-time PCR detection system (Eppendorf, Hamburg, Germany).
Immunohistochemistry and Scoring
50 samples of colorectal adenocarcinoma tissues and corresponding adjacent non-cancerous tissues were obtained from patients undergoing surgical excision of tumors in Qilu hospital of Shandong University (Jinan, China). The samples were fixed with 10% formalin and embedded in paraffin, sliced into 5μm sections. The sections were processed and stained as described previously (30). Antibodies against N-EBP1 P48 and FBXW7 were diluted 1:300. Staining was observed in 5 randomly selected high-power fields. The staining intensity was based on the average percentage of positive cells. The scoring results were analyzed by two investigators.
Cell proliferation and Motility assay
MTT and colony formation assays were performed to assess cell proliferation, Wound healing and invasion assays were used to examine cell motility as described (30). Cells for motility assays were cultured in a low serum concentration (0.2%) to exclude the affection of proliferation on cell motility.
Nude mice xenograft model
Nude mice (6 weeks old) were purchased from Shanghai Slac Laboratory Animal Co. Ltd. and maintained in microisolator cages. All animals were used in accordance with institutional guidelines and the current experiments were approved by the Use Committee for Animal Care of the institute. For metastasis assays, HCT116 FBXW7+/+, FBXW7−/− and FBXW7−/− shEBP1 P48.1 cells (3×106) were injected into the tail veins. After 40 days, the mice were sacrificed by anaesthesia with chloral hydrate. The lung and liver were dissected out for Hematoxylin and Eosin staining.
Statistical analysis
Results are expressed as mean ± standard deviation (SD) from at least three independent experiments. SPSS17.0 statistical software package (SPSS Inc.) was used for statistical analysis. Statistical differences between groups were assessed using Student t-test. Association between FBXW7 and EBP1 P48 expression in colorectal cancer tissue was evaluated by spearman rank correlation test. P < 0.05 was considered statistically significant.
Results
EBP1 isoforms are differentially regulated by FBXW7
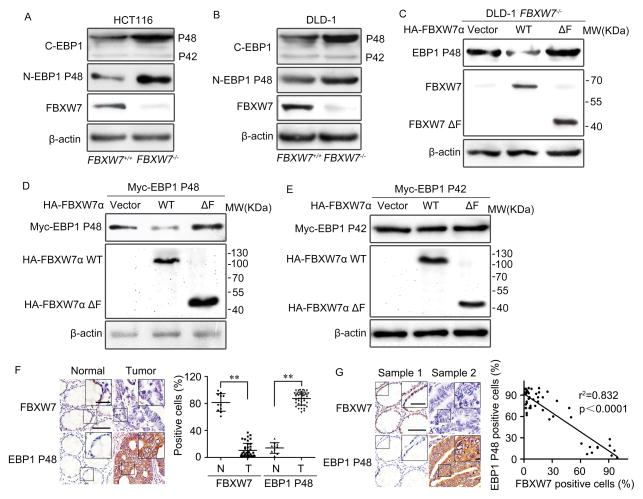
To screen new substrates recognized by FBXW7, we performed Two-dimensional Gel Electrophoresis (2-DE) and Mass Spectrometry (MS) in human colorectal cancer cell line HCT116 with homozygous deletion of the FBXW7 gene (HCT116 FBXW7−/−) (31). Several differentially expressed proteins were separated and identified in HCT116 FBXW7−/− and FBXW7+/+ cells, of which EBP1 protein was most significantly elevated in HCT116 FBXW7−/− cells (Supplementary Figure 1A, B). The altered expression of EBP1 was validated in HCT116 and DLD-1 FBXW7−/− cells by Western blotting using two EBP1 antibodies, anti-C-EBP1 to detect both EBP1 isoforms, and anti-N-EBP1 P48 to specific P48. Interestingly, EBP1 P48 but not P42 is significantly elevated in the FBXW7-deficient cells (Figure 1A, B). Restoration of FBXW7α wild type (WT) reduces the endogenous P48 level, which was not found in the cells restored with truncated dominant negative form (FBXW7ΔF) (Figure 1C, the diagrammatic drawings of FBXW7α mutants were shown in Supplementary Figure 1C). Moreover, elevated P48 protein levels were also observed in other cancer cell lines such as MDA-MB-468, DU145 and PC3 cells when silencing FBXW7 expression by two different FBXW7 shRNA constructs (shFBXW7A and B) (Supplementary Figure 1D–F). To confirm the differential regulation of FBXW7 on EBP1 isoforms, Myc-tagged P48 (Myc-P48) or P42 (Myc-P42) were co-transfected into HEK293T cells along with HA-tagged FBXW7α or FBXW7ΔF. FBXW7α dramatically reduced while FBXW7ΔF increased the protein level of Myc-P48 compared with the empty vector (Figure 1D). In contrast, neither FBXW7α nor FBXW7ΔF had influence on the protein levels of Myc-P42 (Figure 1E). No change in P48 mRNA level was observed after depleting or silencing FBXW7 expression in the cell lines used (Supplementary Figure 1G–J). These results indicated that FBXW7 downregulates P48 expression at post-transcriptional level.
Figure 1. FBXW7 down-regulates EBP1 P48 protein level.
A and B. Western blotting on the whole cell lysates of HCT116 (A) and DLD1 (B) cells to confirm the protein levels of EBP1 isoforms with or without FBXW7 depletion. C. DLD1 FBXW7−/− cells transfected with HA-tagged FBXW7α or HA- tagged FBXW7ΔF. Western blotting confirmed that FBXW7 reduces endogenous EBP1 P48 level. D and E. HEK293T cells were co-transfected with HA-tagged FBXW7α or HA-tagged FBXW7ΔF and Myc-tagged EBP1 P48 (D) or Myc-tagged EBP1 P42 (E). Western blotting to assess the regulation of exogenous EBP1 isoforms by FBXW7. F. Immunohistochemistry staining of human colon tumor sections for EBP1 P48 and FBXW7 to show their expression differences in adjacent normal colon tissues and tumor tissues. Left: Representative images of FBXW7 and EBP1 P48 staining; Right: scatter diagram of relative expression of FBXW7 and EBP1 P48, Data as mean ± SD, Normal: adjacent normal colon tissues, Tumor: tumor tissue. N = 11 for normal tissues and 50 for colon tissues. G. EBP1 P48 expression was negatively correlated with FBXW7 expression in human colon cancer tissues. N = 50 for colon tissues. The scale bars in (F, G) represent 50μm and in the inserts of (F, G) represent 20μm. The results from (A–E) are repeated at least three times. ** P<0.01 based on Student t-test.
To recognize any clinical correlation of FBXW7 and EBP1 P48 expression, immunohistochemistry was performed on 50 colorectal cancer tissue samples (11 of them with paired adjacent non-tumorous tissues) to analyze FBXW7 and P48 expression. As expected, FBXW7 expression was significantly down-regulated and P48 was highly expressed in colon cancer tissues compared with their adjacent normal colon tissues (Figure 1F). Statistical analysis showed that P48 expression was negatively correlated with FBXW7 level in colon cancer tissues (Figure 1G).
FBXW7 promotes proteasomal degradation of EBP1 P48 in a GSK3β dependent manner
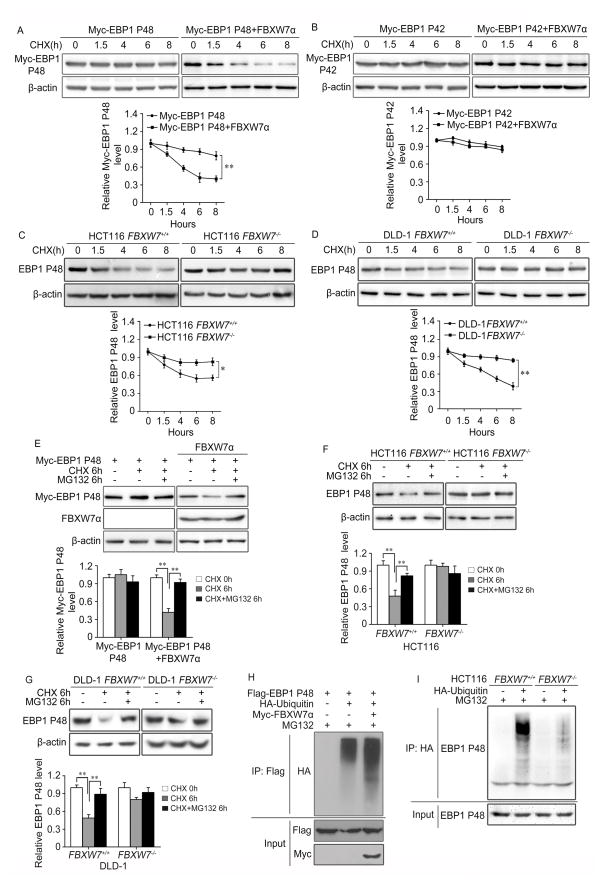
To evaluate whether FBXW7 affects the protein stability of EBP1, we overexpressed FBXW7α and Myc-tagged P48 or Myc-tagged P42 in HEK293T cells and measured the exogenous P48 and P42 protein degradation by cycloheximide (CHX) chase assay. We found that Myc-P48 but not Myc-P42 was more quickly degraded when FBXW7 was present (Figure 2A, B). The effect of FBXW7 on the turnover of endogenous P48 protein was verified in HCT116 and DLD-1 cells (Figure 2C, D). These results indicate that FBXW7 facilitates P48 protein degradation.
Figure 2. FBXW7 facilitates EBP1 P48 turnover in ubiquitin-dependent proteasome pathway.
A and B. HEK293T cells co-transfected with Myc-tagged EBP1 P48 (A) or Myc-tagged EBP1 P42 (B) and HA-tagged FBXW7α followed by treatment with 50μg/ml cycloheximide (CHX) for 0, 1.5, 4, 6 and 8 hours (h). Western blotting shows that FBXW7 affects EBP1 P48 protein degradation but not EBP1 P42. The graphs show quantitative analysis of CHX chase data. C and D. HCT116 FBXW7+/+ and FBXW7−/− cells (C) or DLD1 FBXW7+/+ and FBXW7−/− cells (D) were treated with 50μg/ml CHX for 0, 1.5, 4, 6 and 8 hours (h). Western blotting detected the effects of FBXW7 on the degradation of endogenous EBP1 P48. The graphs show quantitative analysis of CHX chase data. E. HEK293T cells co-transfected with Myc-tagged EBP1 P48 and HA-tagged FBXW7α followed by treatment with 50μg/ml CHX alone or plus 10μM MG132 for 6h. Western blotting shows FBXW7 promotes proteasomal degradation of exogenous EBP1 P48. The graph shows quantitative analysis. F and G. HCT116 FBXW7+/+ and FBXW7−/− cells (F) or DLD1 FBXW7+/+ and FBXW7−/− cells (G) treated with 50μg/ml CHX alone or plus 10μM MG132 for 6 hours (h). Western blotting analysis showing FBXW7 promotes proteasomal degradation of endogenous EBP1 P48. The graphs show quantitative analysis. H. HEK293T cells were co-transfected with Myc-tagged EBP1 P48, HA-tagged FBXW7α and HA-ub. Immunoprecipitation with anti-Flag antibody followed by immunoblotting with an anti-HA antibody to show FBXW7 promotes exogenous EBP1 P48 ubiquitination. I. HCT116 FBXW7+/+ and FBXW7−/− cells were transfected with HA-ub. Immunoprecipitated with anti-HA antibody followed by immunoblotting with anti-EBP1 P48 antibody showing FBXW7 promotes endogenous EBP1 P48 ubiquitination. All results were repeated at least three times. * P<0.05; ** P<0.01 based on Student t-test.
Next, we determined whether the degradation of P48 by FBXW7 is proteasome-dependent. HEK293T, HCT116 and DLD1 cells were treated with proteasome inhibitor MG132. Proteasome inhibition not only increased exogenous P48 protein levels (Figure 2E) but also increased endogenous P48 protein levels in a FBXW7-dependent manner (Figure 2F, G). Proteins targeted for proteasome destruction are usually polyubiquitinated. To examine whether FBXW7 influence the ubiquitination status of P48, Flag-tagged P48 was transfected into HEK293T cells with HA-tagged ubiquitin (HA-ub) or with HA-ub and Myc-tagged FBXW7α. Immunoprecipitation with Flag-EBP1 P48 followed by HA immunoblotting showed overexpression of FBXW7 increased the ubiquitination of Flag-P48 (Figure 2H). Additionally, we compared the P48 ubiquitination in HCT116 FBXW7+/+ and FBXW7−/− cells and found that the P48 ubiquitination is dramatically decreased in HCT116 FBXW7−/− cells (Figure 2I).
Since FBXW7 mediates the degradation of most substrates in a GSK3β-dependent manner, we then examined whether FBXW7 targets P48 in a GSK3β-dependent manner. GSK3β was found to co-immunoprecipitate with Flag-tagged P48 (Supplementary Figure 2A). Moreover, GSK3β inhibitor VIII reversed the reduced expression of ectopic and endogenous P48 protein mediated by FBXW7 (Supplementary Figure 2B, C). We then investigated whether GSK3β inhibitor treatment could inhibit the ubiquitination of P48. HEK293T cells were transfected with HA-ub and treated with or without GSK3β inhibitor. Immunoprecipitation of HA followed by immunoblot analysis of P48 showed that P48 ubiquitination was dramatically reduced in the cells following GSK3β inhibitor treatment (Supplementary Figure 2D). Taken together, these results indicate that FBXW7 promotes the ubiquitination and proteasomal degradation of P48 in a GSK3β-dependent manner.
FBXW7 distinctively interacts with EBP1 isoform P48 and P42
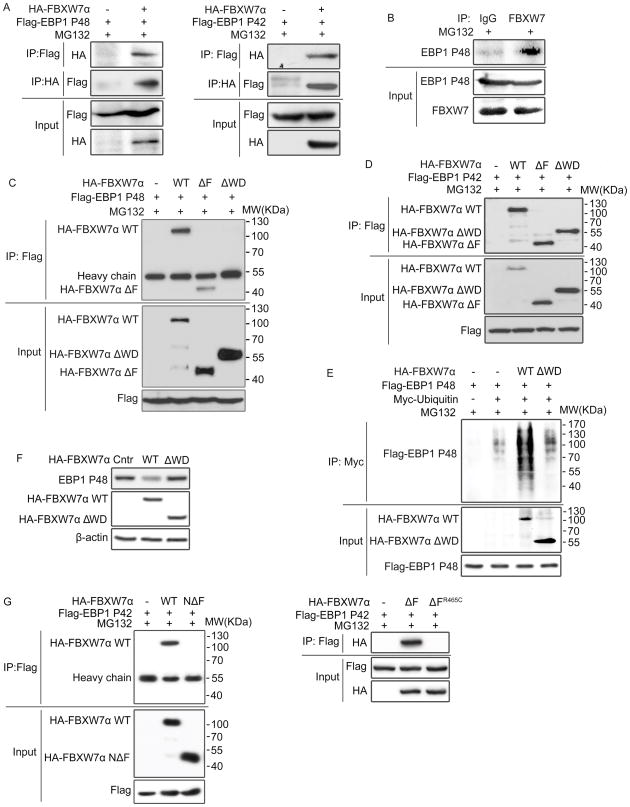
To test if FBXW7 physically interacts with EBP1 isoforms, HEK293T cells were co-transfected with HA-tagged FBXW7α and Flag-tagged P48 or Flag-tagged P42 respectively. Immunoprecipitation and subsequent immunoblot analyzes showed that FBXW7 co-precipitated with both P48 and P42 (Figure 3A). Moreover, endogenous FBXW7 co-precipitated with endogenous P48 (Figure 3B). To identify the sites of FBXW7 that interacts with P48 and P42, HEK293T cells were co-transfected with Flag-tagged EBP1 isoforms and HA-tagged FBXW7α, FBXW7ΔF (F-box deletion), FBXW7ΔWD (WD domain deletion) (Supplementary Figure 1C) or WD mutants in the residues 465, 479 and 505 of FBXW7α (R465C, R465H, R479H and R505C) which are essential for the substrate interaction (32). Co-immunoprecipitation analysis showed that both FBXW7 and FBXW7ΔF could precipitate with P48 and P42. However, FBXW7ΔWD and WD mutants failed to bind to P48 but still had the binding activity to P42 (Figure 3C, D, Supplementary Figure 3A–C). Consistently, functional analysis revealed that neither FBXW7ΔWD nor WD mutants could increase the ubiquitination of P48 (Figure 3E) and had effects on the protein level of P48 (Figure 3F, Supplementary Figure 3D). These indicate that FBXW7 binds to P48 solely through its WD domain but to P42 in a more complex mechanism.
Figure 3. FBXW7 differentially interacts with EBP1 isoforms.
A. HEK293T cells were co-transfected with Flag-tagged EBP1 P48 or Flag-tagged EBP1 P42 and HA-tagged FBXW7α. Immunoprecipitation and subsequent immunoblotting shows the binding between FBXW7 and EBP1 P48 or EBP1 P42. B. HCT116 cell lysates were immunoprecipitated with FBXW7 antibody followed by immunoblotting with anti-N-EBP1 P48 antibody to show the interaction between endogenous FBXW7 and EBP1 P48. C and D. HEK293T cells co-transfected with HA-tagged FBXW7α, ΔF and ΔWD mutants and Flag-tagged EBP1 P48 (C) or Flag-tagged EBP1 P42 (D). Cell lysates were immunoprecipitated with anti-Flag antibody followed by immunoblotting with anti-HA antibody. E. HEK293T cells co-transfected with Flag-tagged EBP1 P48, Myc-tagged ubiquitin and HA-tagged FBXW7α, ΔWD mutant. Immunoprecipitation followed by immunoblotting showing EBP1 P48 ubiquitination is FBXW7 WD domain dependent. F. HEK293T cells were transfected with HA-tagged FBXW7α or ΔWD respectively. Western blotting analysis was done using anti-N-EBP1 P48 antibody. G. HEK293T cells were co-transfected with Flag-tagged EBP1 P42 and HA-tagged FBXW7α, FBXW7αNΔF, FBXW7αΔF or FBXW7αΔFR465C, cell lysates were immunoprecipitated with anti-Flag antibody to detect the binding site of FBXW7 which bind to EBP1 P42. All results were repeated at least three times.
To further investigate the functional region of FBXW7 to bind with P42, we co-transfected HEK293T cells with Flag-tagged P42 and HA-tagged FBXW7NΔF with the deletion of both F-box and WD domain (Supplementary Figure 1C) or HA-tagged FBXW7ΔFR465C with F-box domain deletion and a point mutation in WD domain which lose its binding ability to its substrates. Subsequent co-immunoprecipitation showed that neither FBXW7NΔF nor FBXW7ΔFR465C could precipitate with EBP1 P42 (Figure 3G). Taken together, these data indicate that FBXW7 differentially interacts with EBP1 isoforms, FBXW7 binds to P48 through its WD domain and promotes P48 degradation, but P42 binds to both FBXW7 F-box domain directly and to WD domain mediated by FBXW7 substrates.
EBP1 P48 S44 and S40 sites are responsible for its binding to FBXW7
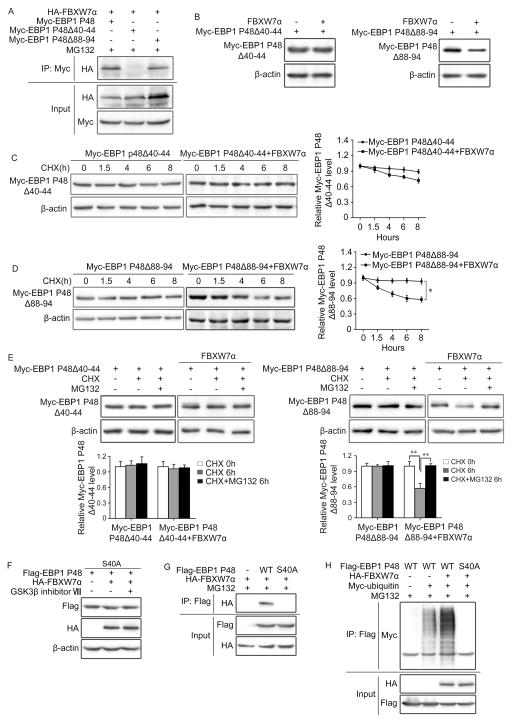
FBXW7 is known to target substrates containing a consensus motif termed CDC4 phosphodegron (CPD) comprising residues that are often phosphorylated by GSK3β. Using the PhosphoMotif Finder software, we found the N-terminal region of P48 harbours two potential CPDs at residues 40–44 and 88–94. To test if these motifs in P48 are critical for its regulation by FBXW7, Myc-tagged P48Δ40–44 and Myc-tagged P48Δ88–94 constructs were generated and co-transfected with HA-tagged FBXW7α into HEK293T cells to examine the binding activity. Both full length P48 and P48Δ88–94 co-precipitated with FBXW7, in contrast, P48Δ40–44 did not form a detectable complex with FBXW7 (Figure 4A). Consistent with these findings, we found that overexpression of FBXW7α significantly decreased the protein level and accelerated the degradation of P48Δ88–94 but had no effect on those of P48Δ40–44 (Figure 4B–D). Additionally, in the presence of proteasome inhibitor MG132, a significant increase in P48Δ88–94 protein level occurred, while EBP1 P48Δ40–44 protein level did not change (Figure 4E). Together, these results suggest that the motif at residues 40–44 is a functional FBXW7 degradation signal.
Figure 4. EBP1 P48 S40 is responsible for the binding to FBXW7.
A and B. HEK293T cells were co-transfected with HA-tagged FBXW7α and Myc-tagged EBP1 P48, Δ40–44 or Δ88–94 mutants. Immunoprecipitation followed by immunoblotting show that EBP1 P48 binds to FBXW7 through AA 40–44 (A) Western blotting show the degradation of EBP1 P48 by FBXW7 is AA 40–44 dependent (B). C and D. HEK293T cells were co-transfected with HA-tagged FBXW7α and Myc-tagged EBP1 P48Δ40–44 (C) or Myc-tagged EBP1 P48Δ88–94 (D), followed by treatment with 50μg/ml CHX for 0, 1.5, 4, 6 and 8 hours (h). Western blotting shows the degradation of EBP1 P48 mutants by FBXW7. E. HEK293T cells transfected as in (C, D) and treated with 50μg/ml CHX alone or plus 10μM MG132 for 6 hours (h). Western blotting shows the proteasomal degradation of EBP1 P48 is dependent on its AA 40–44. F. HEK293T cells were transfected with HA-tagged FBXW7α and Flag-tagged EBP1 P48S40A, Western blotting analysed whether EBP1 P48 S40A affected degradation by FBXW7 or sensitivity to the GSK3β inhibitor. G. HEK293T cells co-transfected with HA-tagged FBXW7α and Flag-tagged EBP1 P48 or P48 S40A. Immunoprecipitation followed by immunoblotting shows EBP1 P48 S40 is responsible for the binding to FBXW7. H. HEK293T cells co-transfected with HA-tagged FBXW7, Myc-tagged ubiquitin and Flag-tagged EBP1 P48 or EBP1 P48 S40A. Immunoprecipitation with Flag antibody followed by immunoblotting with Myc antibody confirmed that EBP1 P48S40 is responsible for the ubiquitination of EBP1 P48. All results are repeated at least three times. * P<0.05; ** P<0.01 based on Student t-test.
We than investigated the significance of individual site for regulation by FBXW7. As the sequence of EBP1 P48 Ser40 to Ser44, S40SGVS44, is highly similar to the putative GSK3β conserved phosphorylation sequence (Ser/Thr-X-X-X-Ser/Thr), we hypothesized that P48 Ser40 and Ser44 are crucial for its GSK3β-dependent degradation by FBXW7. We generated the mutant EBP1 P48S40A where serine at 40 was substituted by alanine. As expected, FBXW7 as well as GSK3β inhibitor VIII cannot affect the protein level of EBP1 P48S40A (Figure 4F). Co-immunoprecipitation showed EBP1 P48S40A mutant completely lost the capacity to bind to FBXW7 (Figure 4G), and in turn the ubiquitination of EBP1 P48S40A mutant was dramatically reduced compared with that of wild-type EBP1 P48 (Figure 4H). As GSK3β usually requires priming phosphorylation on Ser or Thr at position +4 (33, 34), we then constructed the mutant EBP1 P48S44A where serine at 44 was substituted by alanine. Co-immunoprecipitation showed that the mutant EBP1 P48S44A cannot bind to FBXW7 (Supplementary Figure 4A) or GSK3β (Supplementary Figure 2A). Moreover, EBP1 P48S44A cannot be efficiently ubiquitinated by FBXW7 (Supplementary Figure 4B). These results suggested that EBP1 P48S44 may serve as a priming phosphorylation site to recruit GSK3β and then S40 is phosphorylated by GSK3β. Taken all together, we concluded that the residual S40 and S44 sites of P48 are responsible for the physical interaction with FBXW7 and for the FBXW7-mediated proteasomal degradation of EBP1 P48.
EBP1 P48 mediates tumor phenotypes resulted from FBXW7 deficiency
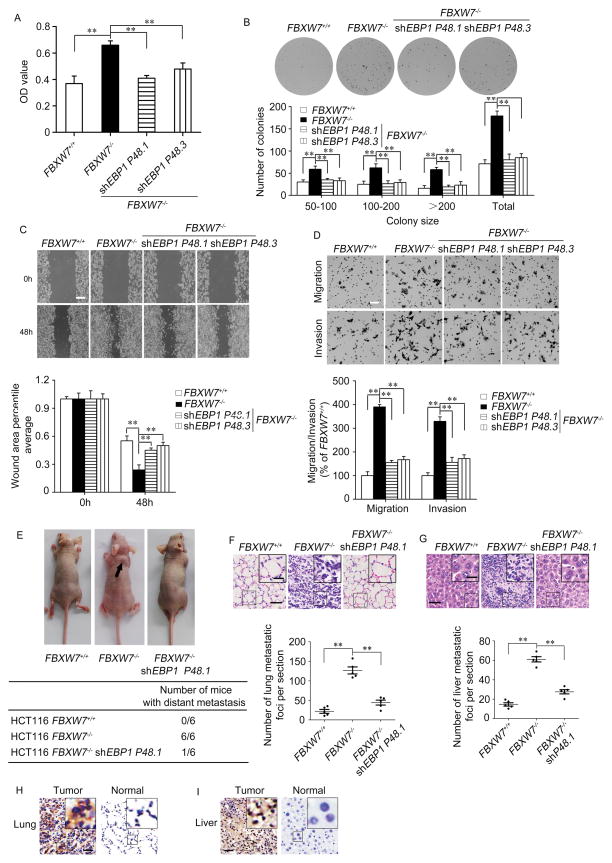
Since EBP1 P48 is identified as a target of FBXW7, we questioned whether P48 is responsible for the cell proliferation and motility phenotypes induced by FBXW7 deficiency. Depletion of P48 expression in HCT116 FBXW7−/− cells by specific P48 shRNA (Supplementary Figure 5A) abolished FBXW7 loss-induced cell proliferation using MTT and colony formation assays (Figure 5A, B). In addition, knockdown of P48 significantly alleviated the increase of cell migration and invasion induced by FBXW7 deficiency in HCT116 cells (Figure 5C, D). Similar results were observed in DLD-1 FBXW7−/− cells and prostate cancer DU145-shFBXW7 cells when silencing P48 expression (Supplementary Figure 5B–G). These data collectively showed that knockdown of P48 effectively suppressed FBXW7 loss-driven cell proliferation, migration and invasion, indicating that FBXW7 executes tumor suppression function at least partly through regulating P48 level.
Figure 5. EBP1 P48 mediates the tumor suppressing function of FBXW7.
A and B. MTT (A) and colony formation (B) analyzes were used to detect the influence of EBP1 P48 knockdown on FBXW7 loss-induced cell proliferation of HCT116 cells. The graph shows quantitative analysis. C and D. The influence of EBP1 P48 knockdown on FBXW7 loss-induced migration and invasion ability of HCT116 cells was determined by wound healing (C), uncoated or Matrigel-coated transwell assay (D). The graphs show quantitative analysis. E. The total numbers of nude mice with distant metastasis at 40–50 days after injection of HCT116 FBXW7+/+, HCT116 FBXW7−/− or HCT116 FBXW7−/− cells with silent EBP1 P48 into tail vein. F and G. The representative HE staining and numbers of metastatic foci per section in lung (F) and liver (G) of individual mouse with injection of indicated cells. The scale bars in (C) represents 500μm, in (D, F, G) represent 50μm and in the inserts of (F, G) represent 20μm. H and I. Anti-human GAPDH antibody which does not react with mouse GAPDH was used to distinguish the colonized cell population between human and mouse by IHC. The results from (A–E) are repeated at least three times. ** P<0.01 based on Student t-test.
To verify whether P48 mediates FBXW7 loss-induced metastasis in vivo, HCT116 FBXW7−/− cells with or without silencing P48 expression were injected into nude mice through the tail vein. Silencing P48 not only significantly decreased the number of mice with distant metastasis (Figure 5E), but also dramatically decreased the number of metastatic tumors in both lung and liver of each mouse (Figure 5F, G). In order to distinguish the colonized cell population between human and mouse, anti-human GAPDH antibody which does not react with mouse GAPDH was used to detect the lung and liver tissues of mice by IHC. The results showed that the colonized cell populations were from human colon cancer cells (Figure 5H, I). Therefore, the in vivo results further demonstrated the critical role of P48 in the colon cancer cell metastatic behaviour induced by loss of FBXW7.
EBP1 P48 relocates FBXW7α into cytoplasm
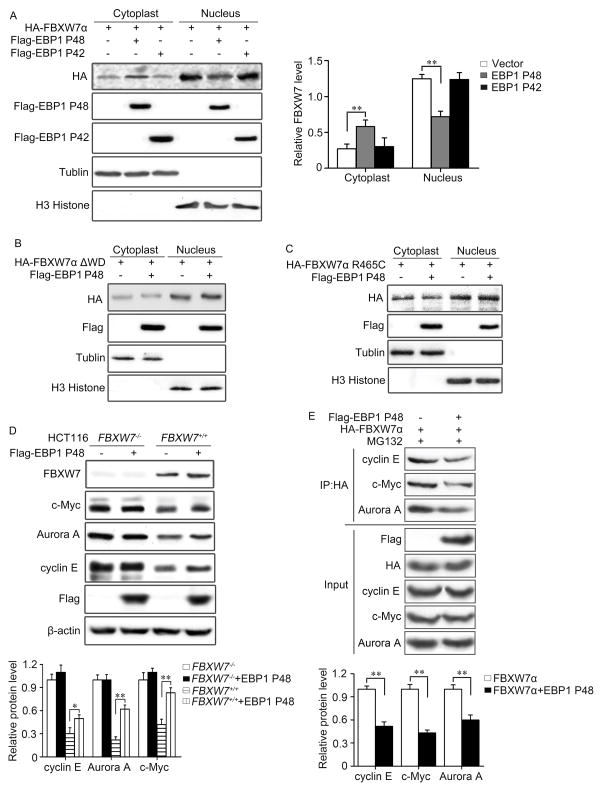
In our efforts to explore the modulation of FBXW7 function, EBP1 P48 was observed to be able to influence the activity of FBXW7. We first found that overexpression of P48 in HEK293T cells inhibited the nuclear localization of FBXW7α (Figure 6A, Supplementary Figure 6A), but had no effect on ΔWD or WD domain mutant forms of FBXW7 (Figure 6B, C, Supplementary Figure 6B, C). As nuclear localization of FBXW7α is essential for nuclear oncoprotein degradation, and FBXW7α mislocalization is of pathological significance (35), we then subsequently examined the protein levels of FBXW7α nuclear substrates, including c-Myc, cyclin E and Aurora-A. These protein levels were increased after ectopic expression of P48 in a FBXW7-dependent manner (Figure 6D). Further co-immunoprecipitation in HEK293T cells showed that EBP1 P48 could decrease the binding of FBXW7 with its substrates, including c-Myc, Aurora-A and cyclin E (Figure 6E). mRNA levels of these analyzed FBXW7 substrates were not affected by ectopic expression of P48 (Supplementary Figure 6D). Collectively, we concluded that P48 may suppress FBXW7-mediated substrates degradation through relocalizing FBXW7α to the cytosol. Thus, FBXW7α and P48 form a negative regulatory loop, FBXW7 induces P48 degradation and meanwhile P48 partially inhibits the degradation of nuclear target proteins of FBXW7α.
Figure 6. EBP1 P48 inhibits FBXW7-mediated substrate-degradation.
A–C. HEK293T cells co-transfected with Flag-tagged EBP1 P48 or Flag-tagged EBP1 P42 and HA-tagged FBXW7α (A), ΔWD (B) or WD point mutant (C). The effect of EBP1 P48 on the subcellular distribution of FBXW7α was analyzed using Western blotting analysis. D. HCT116 FBXW7+/+ and HCT116 FBXW7−/− cells transfected with Flag-tagged EBP1 P48 respectively. The protein levels of indicated substrates of FBXW7 were analyzed by Western blotting. E. HEK293T cells were transfected with HA-tagged FBXW7α with or without Flag-tagged EBP1 P48. Immunoprecipitation with HA antibody followed by immunoblotting show that EBP1 P48 could decrease the binding of FBXW7 with its substrates, including cyclin E, c-Myc and Aurora A. All results are repeated at least three times. * P<0.05; ** P<0.01 based on Student t-test.
Next, we transfected EBP1 P48 into HCT116 FBXW7+/+ and HCT116 FBXW7−/− cells respectively and detected the cell proliferation and motility phenotypes induced by EBP1 P48. Resultantly, overexpression of EBP1 P48 enhanced the proliferation and migration of both HCT116 FBXW7+/+ and HCT116 FBXW7−/− cells. But the promoting effect of EBP1 P48 was more significant in HCT116 FBXW7+/+ cells, indicating that P48 executes its oncogenic activity at least partly dependent on its interaction with FBXW7 (Supplementary Figure 6 E, F).
EBP1 P42 promotes FBXW7-mediated substrate degradation and enhances the tumor suppressing function of FBXW7
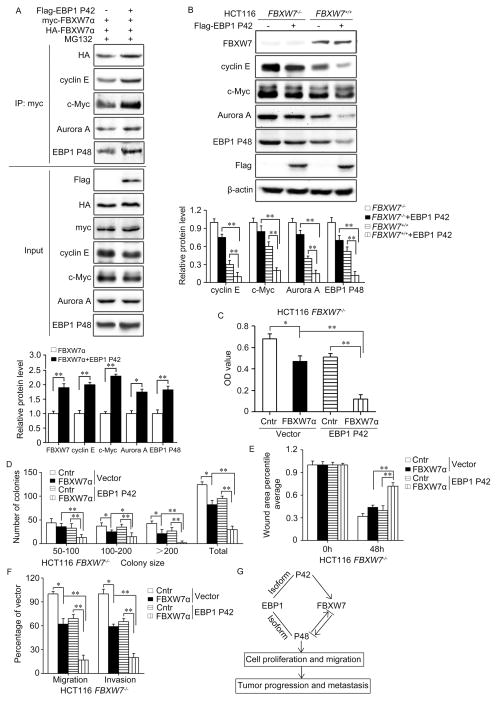
As shown above that EBP1 P42 interacts with FBXW7 F-box domain and FBXW7-substrate complex, we questioned the possibility that P42 functions as a bridge to enhance the binding of substrates to the WD domain of FBXW7. Consistently, when HEK293T were co-transfected with Myc-tagged and HA-tagged FBXW7α and Flag-tagged P42, co-immunoprecipitation results confirmed that P42 did enhance the binding of FBXW7 with its substrates such as c-Myc, Aurora-A, cyclin E and P48 (Figure 7A). We also examined if P42 has any influence of the dimerization of FBXW7 as the dimerization regulates its interaction with substrates and the robustness of substrate degradation (36, 37). Co-immunoprecipitation results showed that P42 enhanced the binding between Myc-tagged and HA-tagged FBXW7α (Figure 7A), which may also contribute to the enhanced binding between FBXW7α and its substrates.
Figure 7. EBP1 P42 promotes FBXW7-mediated substrate degradation and enhances the tumor suppressing function of FBXW7.
A. HEK293T cells were co-transfected with Flag-tagged EBP1 P42, myc-tagged FBXW7α and HA-tagged FBXW7α, Immunoprecipitation and subsequent immunoblotting shows that EBP1 P42 not only enhanced the dimerization of HA-tagged and Myc-tagged FBXW7α, but also increased the binding of FBXW7 with its substrates. B. HCT116 FBXW7+/+ and HCT116 FBXW7−/− cells were transfected with Flag-tagged EBP1 P42, the protein levels of indicated substrates of FBXW7 were measured by Western blotting. C and D. HCT116 FBXW7 −/− cells were transfected FBXW7α and/or EBP1 P42. MTT analysis (C) and colony formation analysis (D) were used to detect the influence of EBP1 P42 on FBXW7-induced inhibitory effect on cell proliferation. The graphs show quantitative analysis. E and F. The influence of EBP1 P42 on FBXW7-induced inhibitory effect on cell migration and invasion of HCT116 FBXW7−/− cells were determined by wound healing (E), uncoated or Matrigel-coated transwell assay (F). The graphs show quantitative analysis. G. Schematic illustration of the proposed interaction among EBP1 isoforms and FBXW7 in cancer progression and metastasis. All results are repeated at least three times. * P<0.05; ** P<0.01 based on Student t-test.
We then investigated the biological significance of this interaction between P42 and FBXW7. Although overexpression of P42 had no significant effects on FBXW7 protein level and localization (Figure 6A), overexpression of wild type P42 led to a more significant decrease of several FBXW7 substrates including c-Myc, cyclin E, Aurora-A and P48 in FBXW7+/+ HCT116 cells compared with that in FBXW7−/− HCT116 cells without affecting the mRNA levels of these substrate (Figure 7B, Supplementary Figure 7), suggesting that P42 can promote FBXW7-mediated substrate degradation.
To verify whether P42 has any effect on the tumor suppressive function of FBXW7, combinations of P42 and FBXW7 expression were constructed in HCT116 FBXW7−/− cells. Overexpression of P42 dramatically enhanced FBXW7-induced inhibition of cell proliferation using MTT and colony formation assays (Figure 7C, D). In addition, scratch healing and Boyden chamber assays showed that overexpression of P42 promotes the inhibitory effect of FBXW7 on cell migration and invasion (Figure 7E, F). These data illustrated that P42 could enhance the tumor suppressing role of FBXW7.
Furthermore, although overexpression of P42 led to a significant decrease in the cell growth, migration and invasion activity of HCT116 FBXW7+/+ cells but to a less extent in that of HCT116 FBXW7−/− cells (Supplementary Figure 8), suggesting that P42 plays its tumor suppressive activity partially through enhancing the tumor suppressive role of FBXW7.
Together, our data demonstrated that FBXW7 differentially regulates EBP1 isoforms. In turn, the activity of FBXW7 is differently regulated by EBP1 isoforms, which was attenuated by P48 but enhanced by P42. The mutual interactions among FBXW7, P48 and P42 may play important roles in tumor initiation and progression (Figure 7G).
Discussion
EBP1 was originally identified as an ErbB3 binding protein which was considered as an oncogene or tumor suppressor in different circumstances (38–41) and has prognostic significance in a panel of cancers such as cervical (7, 42), pancreatic (6), hepatocellular (43), brain (44) and other tumors. Recent study has clarified that EBP1 possesses two isoforms, the longer form P48 and the shorter form P42, which distinctively regulate cell survival and differentiation (5). The isoform P48 with the oncogenic potential promotes cell proliferation and facilitates cancer development (4, 45), whereas the isoform P42 is considered as a tumor suppressor by inhibiting cell proliferation and suppresses tumorigenesis (3, 9, 46). However, the molecular mechanism(s) underlying these differential functions of the two isoforms especially in cancers remain largely unknown. It was previous reported that P48 and P42 bind to ErbB3 with different affinities, thus differentially regulate AKT and PI3K activities to regulate cell proliferation and differentiation (5), and interact differentially with Nucleophosmin/B23 to regulate cell proliferation and apoptosis in PC12 cells (2). In this study, through searching novel targets of FBXW7, we discovered the mechanism by which EBP1 isoforms execute differential functions in tumor development through distinctive interactions with FBXW7.
We first identified that the oncogenic isoform P48 is a substrate of FBXW7. Our results show that FBXW7 binds to the P48 via its WD40 domains, and mediates its ubiquitination and subsequent proteasomal degradation in GSK3β-dependent manner, which is consistent with the stereotyped pattern of a vast majority of characterized substrates recognized by FBXW7 (47–49). In addition, depletion of P48 alleviated FBXW7 loss-induced motility and invasiveness in colon cancer cells in vitro and in vivo. These experimental findings provide a mechanistic framework to explain the inverse correlation between FBXW7 and P48 abundance in colon cancer clinical samples.
Besides the reported mechanistically oncogenic functions of P48 (4), we found P48 promotes FBXW7α nuclear export and the relocalization of FBXW7α has been reported to be of pathological significance because it is intimately linked to its role of substrate down-regulation (35). Consistent with this finding, the levels of some nuclear substrates of FBXW7 are accumulated in the cells, and subsequently the inhibitory effects of FBXW7 on cell migration were attenuated. Our findings further support the oncogenic role of P48 in cancer development. FBXW7α contains two nuclear localization signals (NLSs), one in the isoform-specific N-terminus and another in the shared WD40 domain. Our preliminary data showed that P48 can regulate nuclear localization of FBXW7 through binding to its WD40 domain, raising a possibility that P48 may mask the NLS in WD40 domain thereby influencing its localization. Thus, this study reveals a mechanism by which P48 provokes cancer through regulating FBXW7α activities.
We have identified the residues S40 and S44within the N-terminus of P48 are critical for binding to FBXW7 and ubiquitination mediated by FBXW7. P42 lacks N-terminal 54 amino acids, but is still able to bind to FBXW7 and cannot be degraded by FBXW7. The lack of N-terminal 54 amino acids causes P42 to have different crystal structure from P48 (50), thus P48 and P42 bind to different domains of FBXW7 with different motifs. The deletion of 40–44 aa or mutation of key amino acid of Ser40 or Ser44 in P48 results in destroy of the structure of P48 which necessary for binding to FBXW7 WD domain. However, P42 without the N terminal 54 amino acids of P48 still remains its own structure and binding mode with FBXW7. This is also consistent with previous studies that P42 shows different binding partners to P48 (2, 5).
Detailed analysis of protein interaction revealed that P42 binds to FBXW7 F-box directly and to WD40 domain in a substrate binding-dependent manner. Such interactions potentiate the binding between FBXW7 and its substrates and subsequently lead to reduced levels of these substrates. Our findings truly indicate that P42 interact with FBXW7-substrate complex and function as a bridge to enhance the binding of substrates to the WD40 domain of FBXW7. Moreover, our further mechanism study demonstrates that P42 fortify the dimerization of FBXW7. Emerging evidences have suggested that FBXW7 dimerization plays a critical role in regulating FBXW7 stability and its function on substrate degradation (36, 37). As a result, P42 promotes FBXW7-mediated substrate degradation and enhances the tumor suppressing role of FBXW7 in a FBXW7 dependent manner. Our study reveals a mechanism by which P42 suppresses tumor development by promoting FBXW7 activities.
In conclusion, our study uncovers a physical and functional relationship among FBXW7 and EBP1 isoforms (P48 and P42). Our results demonstrate that two EBP1 isoforms distinctively modulate the role of FBXW7 by different and independent mechanisms. The knowledge of the role of FBXW7 and EBP1 isoforms in the molecular mechanisms governing tumorigenesis shed new lights on improving current anti-cancer therapies.
Supplementary Material
Acknowledgments
Financial support:
Financial support: This work was supported by National Natural Science Foundation of China No. 31271461, 81472583, 81528017 and the Taishan Scholar Program of Shandong Province (G. Wei); by the National Institutes of Health, National Cancer Institute R01 CA116481, and the Low Dose Scientific Focus Area, Office of Biological & Environmental Research, US Department of Energy (DE-AC02-05CH11231) (J. Mao); by National Natural Science Foundation of China No.81470127, the Natural Science Foundation of Shandong Province No. ZR2014HM032, China Postdoctoral Science Foundation Funded Project No.2011M501136, 2012T50616 (P. Zhang); by the China Postdoctoral International Exchange Program 2015, National Natural Science Foundation of China No. 81402193, Postdoctoral Innovation Project of Shandong Province No.147751, and Postdoctoral Science Foundation of China No.2015M570597 (Y. Wang).
Footnotes
Disclosure of potential conflicts of interest: The authors disclose no potential conflicts of interest.
Authors’ Contributions: Y.L.W., P.J.Z., P.P.Z., C.Y.L, and Y.S.W. performed all the experiments and the data analysis. G.W.W and J-H.M. designed the project and supervised the study. Y.L.W., P.J.Z., G.W.W and J-H.M wrote the manuscript.
References
- 1.Figeac N, Serralbo O, Marcelle C, Zammit PS. ErbB3 binding protein-1 (Ebp1) controls proliferation and myogenic differentiation of muscle stem cells. Developmental biology. 2014;386:135–51. doi: 10.1016/j.ydbio.2013.11.017. [DOI] [PubMed] [Google Scholar]
- 2.Okada M, Jang SW, Ye K. Ebp1 association with nucleophosmin/B23 is essential for regulating cell proliferation and suppressing apoptosis. The Journal of biological chemistry. 2007;282:36744–54. doi: 10.1074/jbc.M706169200. [DOI] [PubMed] [Google Scholar]
- 3.Ko HR, Nguyen TL, Kim CK, Park Y, Lee KH, Ahn JY. P42 Ebp1 functions as a tumor suppressor in non-small cell lung cancer. BMB reports. 2015;48:159–65. doi: 10.5483/BMBRep.2015.48.3.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim CK, Nguyen TL, Joo KM, Nam DH, Park J, Lee KH, et al. Negative regulation of p53 by the long isoform of ErbB3 binding protein Ebp1 in brain tumors. Cancer research. 2010;70:9730–41. doi: 10.1158/0008-5472.CAN-10-1882. [DOI] [PubMed] [Google Scholar]
- 5.Liu Z, Ahn JY, Liu X, Ye K. Ebp1 isoforms distinctively regulate cell survival and differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10917–22. doi: 10.1073/pnas.0602923103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gong C, Zhang Y, Chen Y, Zhang H, Liu X, Xue H, et al. High expression of ErbB3 binding protein 1 (EBP1) predicts poor prognosis of pancreatic ductal adenocarcinoma (PDAC) Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36:9189–99. doi: 10.1007/s13277-015-3625-6. [DOI] [PubMed] [Google Scholar]
- 7.Liu L, Li XD, Chen HY, Cui JS, Xu DY. Significance of Ebp1 and p53 protein expression in cervical cancer. Genetics and molecular research: GMR. 2015;14:11860–6. doi: 10.4238/2015.October.2.19. [DOI] [PubMed] [Google Scholar]
- 8.Gannon PO, Koumakpayi IH, Le Page C, Karakiewicz PI, Mes-Masson AM, Saad F. Ebp1 expression in benign and malignant prostate. Cancer cell international. 2008;8:18. doi: 10.1186/1475-2867-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ko HR, Kim CK, Lee SB, Song J, Lee KH, Kim KK, et al. P42 Ebp1 regulates the proteasomal degradation of the p85 regulatory subunit of PI3K by recruiting a chaperone-E3 ligase complex HSP70/CHIP. Cell death & disease. 2014;5:e1131. doi: 10.1038/cddis.2014.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–19. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 11.Davis RJ, Welcker M, Clurman BE. Tumor suppression by the Fbw7 ubiquitin ligase: mechanisms and opportunities. Cancer Cell. 2014;26:455–64. doi: 10.1016/j.ccell.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Q, Karnak D, Tan M, Lawrence TS, Morgan MA, Sun Y. FBXW7 Facilitates Nonhomologous End-Joining via K63-Linked Polyubiquitylation of XRCC4. Mol Cell. 2016;61:419–33. doi: 10.1016/j.molcel.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, Inuzuka H, Zhong J, Wan L, Fukushima H, Sarkar FH, et al. Tumor suppressor functions of FBW7 in cancer development and progression. FEBS Lett. 2012;586:1409–18. doi: 10.1016/j.febslet.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243–56. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 16.Grim JE. Fbxw7 hotspot mutations and human colon cancer: mechanistic insights from new mouse models. Gut. 2014;63:707–9. doi: 10.1136/gutjnl-2013-305144. [DOI] [PubMed] [Google Scholar]
- 17.Bai J, Gao J, Mao Z, Wang J, Li J, Li W, et al. Genetic mutations in human rectal cancers detected by targeted sequencing. J Hum Genet. 2015;60:589–96. doi: 10.1038/jhg.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Liu Y, Lu J, Zhang P, Wang Y, Xu Y, et al. Rapamycin inhibits FBXW7 loss-induced epithelial-mesenchymal transition and cancer stem cell-like characteristics in colorectal cancer cells. Biochem Biophys Res Commun. 2013;434:352–6. doi: 10.1016/j.bbrc.2013.03.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis H, Lewis A, Behrens A, Tomlinson I. Investigation of the atypical FBXW7 mutation spectrum in human tumours by conditional expression of a heterozygous propellor tip missense allele in the mouse intestines. Gut. 2014;63:792–9. doi: 10.1136/gutjnl-2013-304719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Babaei-Jadidi R, Li N, Saadeddin A, Spencer-Dene B, Jandke A, Muhammad B, et al. FBXW7 influences murine intestinal homeostasis and cancer, targeting Notch, Jun, and DEK for degradation. J Exp Med. 2011;208:295–312. doi: 10.1084/jem.20100830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang H, Lu X, Liu Z, Chen L, Xu Y, Wang Y, et al. FBXW7 suppresses epithelial-mesenchymal transition, stemness and metastatic potential of cholangiocarcinoma cells. Oncotarget. 2015;6:6310–25. doi: 10.18632/oncotarget.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yumimoto K, Akiyoshi S, Ueo H, Sagara Y, Onoyama I, Ueo H, et al. F-box protein FBXW7 inhibits cancer metastasis in a non-cell-autonomous manner. J Clin Invest. 2015;125:621–35. doi: 10.1172/JCI78782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao JH, Perez-Losada J, Wu D, Delrosario R, Tsunematsu R, Nakayama KI, et al. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature. 2004;432:775–9. doi: 10.1038/nature03155. [DOI] [PubMed] [Google Scholar]
- 24.Strohmaier H, Spruck CH, Kaiser P, Won KA, Sangfelt O, Reed SI. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413:316–322. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- 25.Tetzlaff MT, Yu W, Li M, Zhang P, Finegold M, Mahon K, et al. Defective cardiovascular development and elevated cyclin E and Notch proteins in mice lacking the Fbw7 F-box protein. Proc Natl Acad Sci U S A. 2004;101:3338–45. doi: 10.1073/pnas.0307875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mao JH, Kim IJ, Wu D, Climent J, Kang HC, DelRosario R, et al. FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science. 2008;321:1499–502. doi: 10.1126/science.1162981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwon YW, Kim IJ, Wu D, Lu J, Stock WA, Jr, Liu Y, et al. Pten regulates Aurora-A and cooperates with Fbxw7 in modulating radiation-induced tumor development. Mol Cancer Res. 2012;10:834–44. doi: 10.1158/1541-7786.MCR-12-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. Embo Journal. 2004;23:2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rocher-Ros V, Marco S, Mao JH, Gines S, Metzger D, Chambon P, et al. Presenilin modulates EGFR signaling and cell transformation by regulating the ubiquitin ligase Fbw7. Oncogene. 2010;29:2950–61. doi: 10.1038/onc.2010.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Wen M, Kwon Y, Xu Y, Liu Y, Zhang P, et al. CUL4A induces epithelial-mesenchymal transition and promotes cancer metastasis by regulating ZEB1 expression. Cancer Res. 2014;74:520–31. doi: 10.1158/0008-5472.CAN-13-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rajagopalan H, Jallepalli PV, Rago C, Velculescu VE, Kinzler KW, Vogelstein B, et al. Inactivation of hCDC4 can cause chromosomal instability. Nature. 2004;428:77–81. doi: 10.1038/nature02313. [DOI] [PubMed] [Google Scholar]
- 32.Malyukova A, Dohda T, von der Lehr N, Akhondi S, Corcoran M, Heyman M, et al. The tumor suppressor gene hCDC4 is frequently mutated in human T-cell acute lymphoblastic leukemia with functional consequences for Notch signaling. Cancer Research. 2007;67:5611–5616. doi: 10.1158/0008-5472.CAN-06-4381. [DOI] [PubMed] [Google Scholar]
- 33.Luan Y, Wang P. FBW7-mediated ubiquitination and degradation of KLF5. World J Biol Chem. 2014;5:216–23. doi: 10.4331/wjbc.v5.i2.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, et al. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–21. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- 35.Welcker M, Larimore EA, Frappier L, Clurman BE. Nucleolar targeting of the fbw7 ubiquitin ligase by a pseudosubstrate and glycogen synthase kinase 3. Mol Cell Biol. 2011;31:1214–24. doi: 10.1128/MCB.01347-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Welcker M, Larimore EA, Swanger J, Bengoechea-Alonso MT, Grim JE, Ericsson J, et al. Fbw7 dimerization determines the specificity and robustness of substrate degradation. Genes Dev. 2013;27:2531–6. doi: 10.1101/gad.229195.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Welcker M, Clurman BE. Fbw7/hCDC4 dimerization regulates its substrate interactions. Cell Div. 2007;2:7. doi: 10.1186/1747-1028-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen LX, Zhu L, Lee Y, Ta L, Mitchell BS. Expression and Role of the ErbB3-Binding Protein 1 in Acute Myelogenous Leukemic Cells. Clin Cancer Res. 2016 doi: 10.1158/1078-0432.CCR-15-2282. [DOI] [PubMed] [Google Scholar]
- 39.Ahn JY, Liu X, Liu Z, Pereira L, Cheng D, Peng J, et al. Nuclear Akt associates with PKC-phosphorylated Ebp1, preventing DNA fragmentation by inhibition of caspase-activated DNase. The EMBO journal. 2006;25:2083–95. doi: 10.1038/sj.emboj.7601111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lessor TJ, Yoo JY, Xia X, Woodford N, Hamburger AW. Ectopic expression of the ErbB-3 binding protein ebp1 inhibits growth and induces differentiation of human breast cancer cell lines. Journal of cellular physiology. 2000;183:321–9. doi: 10.1002/(SICI)1097-4652(200006)183:3<321::AID-JCP4>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Wang XW, Jelovac D, Nakanishi T, Yu MH, Akinmade D, et al. The ErbB3-binding protein Ebp1 suppresses androgen receptor-mediated gene transcription and tumorigenesis of prostate cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:9890–5. doi: 10.1073/pnas.0503829102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu L, Xu DY, Yang SS, Li XD. Ebp1 protein expression in cervical cancer tissue and its significance. Genetics and molecular research: GMR. 2015;14:5496–500. doi: 10.4238/2015.May.22.20. [DOI] [PubMed] [Google Scholar]
- 43.Hu B, Xiong Y, Ni R, Wei L, Jiang D, Wang G, et al. The downregulation of ErbB3 binding protein 1 (EBP1) is associated with poor prognosis and enhanced cell proliferation in hepatocellular carcinoma. Molecular and cellular biochemistry. 2014;396:175–85. doi: 10.1007/s11010-014-2153-9. [DOI] [PubMed] [Google Scholar]
- 44.Kwon IS, Ahn JY. p48 Ebp1 acts as a downstream mediator of Trk signaling in neurons, contributing neuronal differentiation. Neurochemistry international. 2011;58:215–23. doi: 10.1016/j.neuint.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 45.Ko HR, Kim CK, Ahn JY. Phosphorylation of the N-terminal domain of p48 Ebp1 by CDK2 is required for tumorigenic function of p48. Molecular carcinogenesis. 2015;54:1283–91. doi: 10.1002/mc.22203. [DOI] [PubMed] [Google Scholar]
- 46.Oh SM, Liu Z, Okada M, Jang SW, Liu X, Chan CB, et al. Ebp1 sumoylation, regulated by TLS/FUS E3 ligase, is required for its anti-proliferative activity. Oncogene. 2010;29:1017–30. doi: 10.1038/onc.2009.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao D, Zheng HQ, Zhou ZM, Chen CS. The Fbw7 Tumor Suppressor Targets KLF5 for Ubiquitin-Mediated Degradation and Suppresses Breast Cell Proliferation. Cancer Research. 2010;70:4728–4738. doi: 10.1158/0008-5472.CAN-10-0040. [DOI] [PubMed] [Google Scholar]
- 48.Busino L, Millman SE, Scotto L, Kyratsous CA, Basrur V, O’Connor O, et al. Fbxw7 alpha- and GSK3-mediated degradation of p100 is a pro-survival mechanism in multiple myeloma. Nature Cell Biology. 2012;14:375. doi: 10.1038/ncb2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren H, Koo JH, Guan BX, Yue P, Deng XM, Chen MW, et al. The E3 ubiquitin ligases beta-TrCP and FBXW7 cooperatively mediates GSK3-dependent Mcl-1 degradation induced by the Akt inhibitor API-1, resulting in apoptosis. Molecular Cancer. 2013:12. doi: 10.1186/1476-4598-12-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Monie TP, Perrin AJ, Birtley JR, Sweeney TR, Karakasiliotis I, Chaudhry Y, et al. Structural insights into the transcriptional and translational roles of Ebp1. The EMBO journal. 2007;26:3936–44. doi: 10.1038/sj.emboj.7601817. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.