Abstract
Humans consider themselves discrete autonomous organisms, but recent research is rapidly strengthening the appreciation that associated microorganisms make essential contributions to human health and well-being. Each person is inhabited and also surrounded by his/her own signature microbial cloud. A low diversity of microorganisms is associated with a plethora of diseases including allergy, diabetes, obesity, arthritis, inflammatory bowel diseases and even neuropsychiatric disorders. Thus, an interaction of microorganisms with the host immune system is required for a healthy body. Exposure to microorganisms from the moment we are born and appropriate microbiome assembly during childhood are essential for establishing an active immune system necessary to prevent disease later in life. Exposure to microorganisms educates the immune system, induces adaptive immunity and initiates memory B and T cells that are essential to combat various pathogens. The correct microbial-based education of immune cells may be critical in preventing the development of autoimmune diseases and cancer. This review provides a broad overview of the importance of the host microbiome and accumulating knowledge of how it regulates and maintains a healthy human system.
Keywords: Microbiome, host, human health, disease, infant, cancer
Role of the microbiome in maintaining host health
At all stages of life, humans are associated with microorganisms and their products. Humans co-evolved with microbes in the environment, and each body habitat has a unique set of microorganisms in its microbiota (1). The microbiome (term coined by Joshua Lederberg) consists of the ecological community of commensal, symbiotic, and pathogenic microorganisms that share our body (2). The host organism together with its microbiome constitutes the “holobiont” (Greek, holos, whole/entire), and the totality of the genome is the “hologenome” (3,4). Changes in the holobiont may impact the complex signaling network thereby influencing the hologenome leading to health or disease.
The human body is estimated to be composed of 3 × 1013 eukaryotic cells and 3.9 × 1013 colonizing microorganisms, such that host cells and microbiota are of nearly the same number in an individual (5). The largest concentrations of microbes occupy the gut, skin and oral cavity. The microbiota of our system is well tolerated by our immune system due to the co-evolution of these microorganisms over time. The overwhelming majority of gut microbiota are Eubacteria. The collective genome, or metagenome, of the enteric microbiota contains over 100 times the number of genes in the human genome, and there are approximately 10-fold more genes in each of our microbiomes than in each of us, encoding the greatest source of potential antigens with which the immune system must cope (6). The microbiome in humans significantly enriches the metabolism of glycans, amino acids and xenobiotics. It is also responsible for the synthesis of vitamins, isoprenoids and other nutrients making humans “super organisms” whose metabolism represents an amalgamation of microbial and human attributes (7).
Each individual emits a distinct and personalized cloud of microorganisms into his or her surroundings (8). The microbiome in humans is also not constant during lifespan, but rather changes with age. Culture and location also have a profound impact on the microbiome (9). Health status is yet another factor influencing the microbiome compositional status. In one study, the growth dynamics of gut microbiota in health and disease have been inferred from a single metagenomic sample (10). These authors copy the number and ratio at origin and terminus to detect the actively growing species in a microbiome. In this way, they showed differences between virulent and avirulent strains, population diurnal oscillations and bacterial species predominant during disease and diet changes.
Diet is a particularly important factor in determining the microbiota composition of the gut (11). Thus vegans, vegetarians and omnivores have distinct microbiomes. Total counts of Bacteroides spp., Bifidobacterium spp., Escherichia coli and Enterobacteriaceae spp. were significantly lower in vegan samples than in controls. In contrast, total counts of Klebsiella spp., Enterobacter spp., other Enterobacteriaceae, Enterococcus spp., Lactobacillus spp., Citrobacter spp. and Clostridium spp. were similar in people with different diets. Subjects on a vegetarian diet ranked between vegans and omnivores. The total microbial count did not differ between the dietary groups (12). Notably, the microbiome of a person can be altered rapidly by changes in dietary patterns. It has been demonstrated by David et al. (13) that short-term consumption of diets composed entirely of animal or plant products can alter the microbial community structure. An animal-based diet increased the abundance of bile-tolerant microorganisms, including Alistipes, Bilophila and Bacteroides and decreased the levels of Firmicutes that metabolize dietary plant polysaccharides (Roseburia spp, Eubacterium rectale and Ruminococcus bromii). Thus, the gut microbiome rapidly responds to diet.
Microbiome taxonomy and its future
The taxonomy of microbiomes reflects their complexity and the challenges encountered in their understanding. Microbiomes include species across all major kingdoms, including viruses as well as Archaea, bacteria and microbial eukaryotes. Our present depth of knowledge is associated with different methods of investigation, targeted surveys and scope of studies conducted. To date, the most comprehensively investigated phylogenic group in health and disease has been bacteria.
Prokaryotes
List of Prokaryotic Names with Standing in Nomenclature (LPSN; http://www.bacterio.net) includes two prokaryotic domains (or empires), subdivided into 30 phyla in the domain Bacteria and 5 phyla in the domain Archaea. Together these 35 phyla encompass about 2,400 genera and 12,400 species (14). This list is based on strict requirements, including the availability of reference strains, and does not include all available reference strains deposited in culture collections, including the ones for which genome sequences are already available (15-17). The addition of whole-genome phylogenetic analysis allows a refined positioning in the phylogenetic hierarchy as new tools are being developed (18-21). This approach brings some conflicts with the present classification, as has happened when the 16S rRNA gene phylogenetic classification competed with the phenotypic classification (22-24). Additionally the ability to target and obtain the sequence of the genes used for phylogenetic classification (16S rRNA, recA, rpoB, gyrB etc.) using culture-independent methods also adds to the known diversity. Specialized curated databases that allow the propagation of this knowledge include SILVA, Ribosomal Database Project and the Human Oral Microbiome Database (23,25,26). This culture-independent approach raised to 46 the number of phyla (23). How much of this diversity is in the human microbiome is unclear. However, it is already clear that organisms known to be environmental are also associated with health, and that at least 30 prokaryotic phyla and 950 genera are associated with the human microbiome (27,28).
Microbial eukaryotes
The microbial eukaryotes are extremely diverse and do not fit under a single keyword. While accurate, the eukaryotic supergroups defined by phylogenomics (Opisthokonta, Amoebozoa, Excavata, Archaeplastida and SAR [Stramenopiles+Alveolates+Rhizaria]), are unfortunately uninformative compared to previous classification methods used in the literature. From the clinician to the lay person, terms such as fungi, protists, parasites, protozoa and amoebae are much more familiar. In this area, present knowledge is based mostly on their roles as causative agents of disease; few studies have focused on healthy individuals or within a defined illness in a restricted number of individuals (29-33). Until recently, the focus on single disease agents also meant neglecting the reminder of the eukaryotic microbiome (34,35).
The human eukaryotic microbiome includes pathogens, commensals and beneficial organisms. The fungi (Opisthokonta) harbor a wide diversity of organisms, with an overlap for the skin with the local environment (35). The fungi include the Ascomycota (Candida albicans), Basidiomycota (Cryptococcus neoformans), Microsporida (Encephalitozoon intestinalis), Zygomycota (Rhizopus microsporus). As fungi are part of the environment and human alimentation, it may be difficult to differentiate between transient and commensal organisms without a longitudinal study, unless a disease or an opportunistic infection occurs (36,37). The Acanthocephala, most closely related to the rotifers, include Macracanthorhynchus hirudinaceus. The helminths (Opisthokonta), which are classified as part of the animals, include the cestodes (tapeworms: Taenia saginata), trematodes (flukes: Schistosoma mansoni), and nematodes (roundworms: Enterobius vermicularis). While the majority of the helminths cause illness in millions of people worldwide, a few helminth species have been used in therapy (38,39). The protozoa include the Amoebozoa [Amoebozoa] (amoeba: Entamoeba histolytica), Metamonada [Excavata] (flagellates: Giardia intestinalis), Parabasilia [Excavata] (Dientamoeba fragilis), Ciliophora [SAR] (ciliates: Ballentidium coli), Apicomplexa [SAR] (Cryptosporidium parvum) and Stramenopile [SAR] (Blastocystis hominis). These protozoa are all medically important even though not all carriers are symptomatic (30,35). The Archaeplastida (including green and red algae) can be present in the microbiome of the skin and digestive tract. Additional sequences available through the sequencing of targeted genes including via metagenomics has expanded this knowledge and is maintained and curated in databases such as SILVA (23). A current view of the tree of life, encompassing the total diversity represented by sequenced genomes was published recently by Hug et al. (40).
Viruses
The gut microbiome includes bacteriophages that influence the bacterial hosts. The bacteriophage in the human gut are of three classes: a set of core bacteriophages shared among more than half of the human population, a common set of bacteriophages found in 20%–50% of individuals, and a set of bacteriophages that are unique to a person. Healthy gut phageome (aggregate of bacteriophage in the gut) is significantly decreased in individuals with gastrointestinal diseases (41). The International Committee on Taxonomy of Viruses in its 2014 release listed 104 families, 505 genera and 3,186 species of all known viruses (42). The human virome overlaps with other animal viromes. These dsDNA, ssDNA, dsRNA, ssRNA- ssRNA+ viruses and dsDNA and ssRNA retroviruses can affect any of the tissues within the body. Human protists (non-fungal microbial eukaryotes) have their own viral challenges which are being uncovered within the human virome (43). Much more is known about mycoviruses and their intracellular transmission during cell division and sporogenesis and it is recognized that their life cycles generally lack an extracellular phase. Most known mycoviruses have dsRNA genome, but an increasing number of positive- or negative-strand ssRNA and ssDNA viruses have been isolated and characterized (44). Most of the archaeal viruses have been isolated from members of the Euryarchaeota and Crenarchaeota with broader morphological characteristics than their bacterial counterpart (45). Little is known of the impact of archaeal viruses on the human virome. The dsDNA, ssDNA, dsRNA, ssRNA bacteriophages have a great impact on prokaryotic ecology through their ability to modify population structure. A list of viral pathogens is maintained by the ViPR resource, while prophages are available at PHAST (46,47).
Future of taxonomy
An open challenge in taxonomy is to refine the classifications to be more compatible with the emerging methods of molecular bio-surveillance and detection requiring targets associated to an outcome or being able to identify strains at multiple body-sites across the domains of life (21,48). This work is dependent on a greater understanding of the true diversity in the population with the direct sequencing of large sample sets and/or large cohorts such as the Human Microbiome Project, MetaHIT, BioMarks, and future large scale projects (49-51). New resources are now attempting to bridge the different approaches and topics of detection across domains, such as the Human Pan-Microbe Community database (HPMCD) (34). The ability to isolate and sequence single cells offers the opportunity to deepen both our understanding of the genomic composition of taxonomic diversity, as well to put this diversity in context of its environment, microbial partners, biogeography, and host physiologic status both at the local and systemic levels (52-54).
Beyond the present time taxonomy and the species level classification focused on vertical transmission of conserved information to descendant cells. The strain definition is associated with gene composition and gene modification including mutations and antigenic variation following homologous DNA recombination, CRISPR system (Clustered Regularly Interspaced Short Palindromic Repeats), gene transfer, mobile elements, epigenetics, etc. Moving from targeted-gene phylogeny, to whole-genome comparison has its own limitations that can be complemented by the inclusion of other omics as once metabolic panels, protein and DNA finger-printing profiles were used. This polyphasic analysis allows understanding genetic relatedness and phylogenic relationship in the context of disease, reservoir, niche transmission within a single individual, propagation within a population and dissemination among environments. An increasing number of gene transfer events among domains are being documented as well as across ecological niches (55-57). The genetic modification can be due to increases in genetic content but genetic loss also has critical consequences in competitiveness or niche settlement (58,59). As no clear general phylogenetic definition of strain has emerged in this era of genomics, efforts are to differentiate the different isolates with markers not yet found in other genomes and/or single nucleotide polymorphism (20,21,60,61).
The infant microbiome and transgeneration effects
Until recently the placenta was considered a “sterile” intrauterine environment. Aagaard et al. (62) reported that the placental microbiome is consistently different from other parts of the body, including the skin and urogenital tract. Interestingly, the placental microbiome is most similar to that of the oral cavity. Thus, these authors suggested that microbes travel to the placenta from the mouth via the blood. The results reinforce data that have suggested a link between periodontal disease in the mother and the risk of preterm birth.
Infant-associated microbial communities initially possess high concentrations of facultative anaerobes such as Escherichia coli and Streptococcus spp., but these populations are replaced by strict anaerobes coinciding with a reduction in oxygen tension (63-65). In addition to environmental routes of inoculation, the specific mode by which the infant is delivered is now known to influence the early gut microbiome structure and trajectory.
Although there is emerging evidence that the fetus encounters placental and amniotic bacteria in utero, it is clear that parturition contributes to the infant's first major inoculation of colonizing microbes (63,66,67). Microbial communities colonize external surfaces of the infant immediately following birth. This includes various microbial populations that are established and maintained along the gastrointestinal tract (GIT). The percentage of babies delivered through cesarean section has risen in many countries. While a number of cesarean section deliveries are performed for obstetrical indications, a large proportion is not medically indicated and may be due to maternal request and may incur several risks for the child (68). Obstetricians in many medical settings are paid more for cesarean delivery and it is well known that private hospitals and practitioners encourage cesarean delivery (69). However, recent studies demonstrated that babies born vaginally are healthier compared to babies born by cesarean delivery. As such, infants delivered vaginally tend to harbor microbiota that are typically encountered in the female reproductive tract, such as Lactobacillus. In contrast, cesarean delivery is typically associated with Staphylococcus spp. and other bacteria that are associated with the mother's skin and hospital environment (65,70,71). Children delivered by cesarean section have significantly increased risk of asthma, systemic connective tissue disorders, juvenile arthritis, inflammatory bowel disease, immune deficiencies, leukemia (72) and Crohn's disease (73). Although there are some indications that infants born via cesarean section may be more susceptible to colonization by Clostridium difficile or methicillin-resistant Staphylococcus aureus (MRSA), and may be at an increased risk for pathologies later in life (71,72), additional mechanistic studies are required to conclude causal relationships in this regard.
The infant microbiome exhibits several shared attributes regardless of birth method. In general, the infant microbiome is often dominated by the genera Bifidobacterium, Bacteroidetes, and members of clostridial taxa (74). In the seminal 2012 study conducted by Yatsunenko et al. (9) gut microbiomes were characterized from individuals located in three distinct geographic locations (i.e., United States, rural Malawi, and rural Venezuela). Regardless of the host's location microbial populations converge toward an adult community by three years of age (9). Furthermore, microbial community diversity increased as the host aged across all populations (9). Infant microbiomes exhibit hallmarks of functional redundancy, in that inter-individual taxonomic variation is common despite sharing a stable and uniform metabolic potential (75). This functional redundancy during neonatal development may contribute to metabolic, digestive and immune system homeostasis (74,76). During infancy, the impact of alterations in community assembly on function has been linked to outcomes such as malnourishment, C. difficile-associated diarrhea, and necrotizing enterocolitis (74,76,77). Early life microbiome disruption may potentially increase risk for developing celiac disease, asthma, type-1 diabetes, and obesity (70,72,78-81). These conditions could have long-term medical implications that interact reciprocally with the gut microbiome.
Perturbations or durable disruptions of the infant microbiome may proceed via several paths, with hospitalization and antibiotic use considered to be primary causes. Pre-term and term infants who are hospitalized early in life are at a greater risk for nosocomial C. difficile infection (74,82). Thus the hospital environment is a reservoir for infectious agents that may be deleterious for at-risk populations such as pre-term infants with underdeveloped immunological function. As antibiotics select for resistant and resilient strains, indiscriminant usage of antimicrobials may drive gut dysbiosis in certain instances. Several studies have characterized the influence of antibiotic usage on restricting gut microbiota diversity, potentially increasing susceptibility to aggressive bacterial infections like C. difficile and vancomycin-resistant Enterococcus bacteremia (82-84).
As it does in adults, diet exerts a strong influence on the structural composition of infant-hosted microbiomes. Culture-dependent and independent studies indicate that breastfed infants often possess a significantly different and less diverse gut microbiome relative to formula-fed infants (9,64,85-87). Accordingly, human milk incorporates several bioactive compounds important for infant nutrition, including lipids, proteins and lactose. In addition several milk molecules enhance immunological and neurological development (88-90). Escaping digestion by host glycosyl hydrolases, soluble milk glycans are transferred to the distal colon where they are exposed to the gut microbiota of the infant. Thus these human milk oligosaccharides (HMO) are available to guide the establishment and function of the infant microbiome. HMOs are heterogeneous carbohydrate polymers that are the third most abundant milk component at several grams per liter (91). HMO structures incorporate the monosaccharides D-glucose, D-galactose, N-acetylglucosamine, L-fucose and N-acetylneuraminic acid, with over 200 unique HMO isomers composed of these components identified to date (91,92).
Breastfeeding infants often display a microbiome enriched for commensal Bifidobacteria that can utilize HMOs. Bifidobacteria are Gram-positive anaerobes of the phylum Actinobacteria which typically colonize infants and adults to lesser degree (93-95). Accordingly, Bifidobacterium longum subsp. infantis is a major commensal of breastfed infants with this lineage possessing a large genomic cluster that enables HMOs utilization (95). That unique gene assemblage permits the catabolism of specific small mass HMOs that other Bifidobacteria cannot process. For example, in comparison to other Bifidobacteria, B. longum subsp. infantis flourishes in the presence of milk that contains α1,2-fucosylated HMOs (96). An individual's complement of HMO structures is somewhat dependent on the mother's genotype and may vary by gestational age and stage of lactation. The relative concentrations of α1,2-linked fucosyl moieties depend on the fucosyltransferase 2 (FUT2) allele (96). Women with a functional copy of this gene, termed secretors, may confer certain health benefits to their infant such as a decreased risk for diarrheal diseases (96-98). HMOs can decrease the presence of gastrointestinal pathogens using two primary mechanisms. HMOs themselves mimic pathogen binding sites of receptors that decorate the surface of host cells (99-101). Studies have documented this effect using Vibrio cholerae, Streptococcus pnemoniae and Escherichia coli (102-104). In addition, high levels of Bifidobacteria are correlated with lower incidence of potentially dangerous neonatal infections, potentially due to competitive exclusion (105,106). Gut microbiota development during infancy can have long-lasting effects on the individual's future health. Colonization of fucosyllactose (FL)-utilizing bifidobacteria is due to an ABC transporter that acts as a key genetic factor for FL utilization (107).
Human milk is generally accepted as the best nutrition for newborns and has been shown to support the optimal growth and development of infants (108). Human milk also provides bioactive components that are important to optimize gut microbial colonization, immune maturation, metabolic development and even cognitive development. Breast milk has a low buffering capacity which would make the gut more susceptible to a lowering of pH due to acid production from bacterial fermentation in the colon. The fecal pH of the breast-fed infant is between 5 and 6 dominated by Bifidobacteria, whereas formula-fed infants have a fecal pH in the range of 8–9. The acetic acid in the gut of breast-fed infants is frequently present as an acetate buffer. This effect was not observed in formula-fed infants. The lower pH in the gut is an important factor in restricting the growth of Enterobacteria, Clostridia and the Bacteroides, and favors the proliferation of the acid-tolerant Bifidobacteria and Lactobacilli (109). Human milk also contains many antimicrobial factors, such as partially digested or fermented peptides, milk-borne fatty acids, human lactoferrin, lysozyme, and secretory IgA (sIgA). These factors may decrease the prevalence of pathogens in the gut's ecosystem in infants. The broad range of nondigestible oligosaccharides specifically found in human milk but not in other mammals' milk (108) is a major factor in the prevention of pathogen growth in the gastrointestinal tract. Stunted infants fed poorly have low amounts of sialylated HMOs in the gut. These oligosaccharides are not used by the body, but rather used by the gut microbes. Charbonneau et al. (110) colonized germ-free mice with a consortium of bacterial strains cultured from the fecal microbiota of a 6-month-old stunted infant and fed recipient animals with normal diet with or without purified sialylated bovine milk oligosaccharides (S-BMO). S-BMO produced a microbiota-dependent body weight gain indicating growth promotion in the presence of gut microbiota. However control animals that were germ free did not increase body weight, demonstrating some bacteria in the gut are involved in weight gain.
Infant formula is often based on bovine milk unless it is plant derived. Fluid dairy milk contains oligosaccharides with similar structure to HMOs, which may suggest similar functionality despite being incorporated at relatively low concentrations (108,111-113). At the moment, there are efforts to supplement infant formula with oligosaccharides although HMO structures are difficult to synthesize and may not be commercially viable (114). However, oligosaccharides from other sources may increase bifidobacterial concentration as a preferred endpoint, including galacto-oligosaccharides and fructo-oligosaccharides.
Use of oral probiotics by the mother during pregnancy is thought to help the developing baby. Microbes in placenta or amniotic fluid affect fetal innate immune gene expression during late pregnancy. Maternal probiotic supplementation significantly modulated the expression of TLR-related genes both in the placenta and in the fetal gut. Thus, fetal and placental immune physiology may be modulated by maternal dietary interventions including using specific probiotics (115,116). It has also been shown that maternal probiotic supplementation during pregnancy and breastfeeding reduces the risk of eczema in the infant (116). Probiotic supplements continue to impact infants in their early years. It has been shown that infant formula supplemented with the probiotics Lactobacillus rhamnosus GG and Bifidobacterium lactis Bb-12 offer a safe means of reducing the risk of early acute otitis media and antibiotic use and the risk of recurrent respiratory infections during the first year of life (117). Probiotics enhance gut-specific IgA responses, which are frequently defective in children with food allergy (118). Kainonen et al. (119) have demonstrated that exclusive breast-feeding promotes an anti-inflammatory cytokine milieu, which is maintained throughout infancy. Such an immunological environment limits hyper-responsiveness and promotes tolerization, thereby prohibiting the onset of allergic disease.
Infantile colic (excessive crying), is a common problem in about 20% of healthy thriving infants in the first three months of life (120). The risk factors associated with the development of infantile colic include, maternal smoking, increased maternal age and firstborn status. Infantile colic could also be related to cow's milk protein allergy and atopy (121). Several studies have demonstrated that administration of probiotics containing Lactobacillus reuteri DSM 17938 significantly improved colic symptoms by reducing crying and fussing times in breastfed infants with colic (122,123). Treatment of colic with L. reuteri did not affect the global composition of the microbiota. The decrease in colicky symptoms was linked to changes in the microbiota, with a relative increased abundance of the phyla Bacteroidetes and genus Bacteroides after treatment with L. reuteri (124).
Microbiome and aging
As humans age, the composition of the microbiome also changes (9). Aging is accompanied by the onset of a myriad of clinical changes, including a basal proinflammatory state (“inflamm-aging”) that directly interfaces with the microbiota of older adults and enhances their susceptibility to diseases that accompany aging. Studies in older adults demonstrate that the gut microbiota correlates with diet, basal level of inflammation and location of residence (e.g., community dwelling, long-term care settings) (125-127). Links between the microbiota and a variety of clinical problems plaguing older adults have been made, including physical frailty, Clostridium difficile, colitis, vulvovaginal atrophy, colorectal carcinoma and atherosclerotic disease (128).
The most drastic change associated with the aging gut is a change in the relative proportion of organisms, e.g., Firmicutes dominate in the young and Bacteroidetes in the elderly. Reduction in the diversity of bacteria comprising subpopulations is seen in individuals with high frailty, although living in a community undermines this alteration (125,129).
Aging-associated oxidative stress induces aggressive potential and virulence factors of anaerobic bacteria, thereby causing morphologic alterations of bacterial cells that could impact the host. The microbiota may also influence host gene expression by regulating microRNAs (130). Analysis of the network functions revealed that differentially regulated miRNAs between infants and adults and miRNAs that decreased during aging shared two network functions: inflammatory disease and inflammatory response. miRNAs promote aging by modulating their targets to drive cell senescence and aging in different tissues or organs. There is significant variation in the expression of miRNA during aging. Genome-wide assessment of miRNA expression revealed that the majority of miRNAs decreased with age (131,132). Interestingly, host-derived miRNAs may also influence the composition of the gut microbiome (133).
It has been documented that calorie restriction can increase the life span of model organisms (134). Notably, Zhang et al. (135) demonstrated that calorie restriction enriches bacterial phylotypes positively correlated with lifespan. Bacteria of the genus Lactobacillus have been shown to increase in animals on low-fat diet, and this environment reduces phylotypes which are negatively correlated with lifespan. Caloric restriction-induced changes in gut microbiota occur concomitantly with a significant reduction in serum levels of lipopolysaccharide-binding protein (LPS), suggesting that animals undergoing calorie restriction establish a structurally balanced architecture of gut microbiota that exerts a health benefit through the reduction of antigen load from the gut. Strikingly, dietary changes can detectibly influence host environment in as little as 24 hours, with longer term changes correlating with novel enterotype clustering in the host (136).
Multiple studies in centenarians indicate extreme aging is characterized by microbial changes deemed unique from other age groups, with emphasis placed on organismal composition and increased inflammatory effects (137,138). Fecal sampling by Rampelli et al. (138) revealed a distinctive functional profile with a decrease in short chain fatty acid production and saccharolytic potential but an increase in proteolytic potential. One hundred and sixteen microbial genes were found to be correlated with aging, including those essential to the metabolism of tryptophan, phenylalanine, tyrosine, valine, and lysine. Implications of such variability include changes in wellbeing, aging, and disease susceptibility. This was accompanied by an increase in occurrence of pathobionts, bacteria usually present in low numbers in the adult. Pro-inflammatory effects of the pathobionts are exaggerated by a decrease in Faecalibacterium prausnitzii, a species associated with anti-inflammatory influences (137).
The relationship between aging and the microbiome is not strictly one-sided; it has been demonstrated that host aging can actually be impacted by interspecies communication. Animal fecundity, development time and lifespan were all dependent on the amount and type of bacteria they were fed. There are multiple lines of evidence demonstrating the ability of microbes to substantially change host physiology as it pertains to these parameters (139). Accordingly, manipulating the microbiome of older adults holds promise as an innovative strategy to positively influence the development of co-morbidities associated with aging (128).
Rozsa et al. (140) recently proposed the ‘microbiome mutiny hypothesis’, whereby some microorganisms of the microbiome could switch to higher virulence (microbiome mutiny) in old or seriously ill people, to optimize their transmission under the conditions of increased background mortality. This proposed virulence shift might contribute to the death of old or seriously ill people even in the absence of apparent disease.
In the central nervous system (CNS), polyphenols present in many edible plants exert anti-inflammatory effects (141) and act on the brain in several ways. Like antioxidant vitamins, dietary polyphenols contribute to the regulation of oxidative stress and improve vascular health. Notably, intestinal microbiota convert dietary polyphenols to phenolic acids, stimulating proliferation of Bifidobacteria and decreasing the ratio of Firmicutes to Bacteroidetes, relative to controls. Polyphenols also stimulate short chain fatty acid production by bacteria (142). Wang et al. (143) recently reported that the microbiome can convert grape-derived polyphenol to the phenolic acids 3-hydroxybenzoic acid and 3-(3′-hydroxyphenyl)propionic acid, which interfere with the assembly of β-amyloid peptides into neurotoxic β-amyloid aggregates that play key roles in the pathogenesis of Alzheimer's disease (AD). Thus, in the brain and other tissues, many healthful effects of polyphenols may relate to their conversion to various metabolic derivatives by the gut microbiome while aging.
Microorganisms and immune function
Through co-evolution with their hosts, microbes exert a major influence in shaping the development of the immune system, putting it under selective pressure to develop the capability to discern between invasive pathogens that it is imperative to control and commensal resident microbes that are beneficial to tolerate (144,145). Many developmental aspects of the adaptive immune system are influenced by the composition of bacterial colonization of the gut. Thus, the mammalian immune system, which is tasked with the duty of controlling microorganisms, is in turn fundamentally shaped by microorganisms (146). For example, it has been demonstrated that changes to the symbiotic microbiota early in life, or the absence of it, can lead to exacerbated type 2 immunity and allergies due to aberrant immune functionality. The microbiota is a strong inducer of proinflammatory T helper 17 (TH17) cells and regulatory T cells (Treg) in the intestine. The microbiota-induced Treg express the nuclear hormone receptor RORγt and differentiate along a pathway that also leads to TH17 cells. In the absence of RORγt+ Treg, TH2-driven defense against helminths is more efficient, whereas TH2-associated pathology is exacerbated. Thus, the microbiota regulate type 2 responses through induction of type 3 RORγt+ Treg and TH17 cells, thereby balancing immune responses at mucosal surfaces (147). Exercise can also influence the immune system and how they modulate microorganisms (148,149). Intense exercise causes immunosuppression, while moderate intensity exercise improves immune function and potentially reduces risk and severity of respiratory viral infection by increasing stress hormones, reduce excessive local inflammation and skew the immune response to a Th2 phenotype (148). Similarly, exercise can also influence bacterial infections. Pape et al. (150) demonstrated a reduction of bacterial infection in people with physical activity compared to those that maintain a sedentary lifestyle.
Similar to adaptive immunity, the innate immunity is also influenced by the microbiome. One example of this is neutrophil aging. Aged neutrophils exhibit impaired migration and reduced pro-inflammatory properties. Microbiota influence neutrophil aging via Toll-like receptor and MyD88-mediated signaling pathways. Depletion of the microbiota significantly reduces circulating numbers of aged neutrophils and improves pathogenesis and inflammation-related organ damage in models of sickle-cell disease or endotoxin-induced septic shock. Thus, host microbiota may play a role in regulating a disease-promoting neutrophil subset that promotes tissue injury in various inflammatory diseases (151).
While active immunity is essential to combat infection, inadequate control over immune responsiveness due to the inability to establish immune tolerance can also have dire consequences, regardless of whether the response is directed against a foreign pathogen or self. Meanwhile, one of the major benefits of immune tolerance is the ability to maintain a commensal microbiome consisting of a multitude of foreign microorganisms. Thus, the mechanisms for establishing tolerance are a vital aspect of the immune regulatory framework. One crucial element in instructing the immune system to be self-tolerant is the education of thymus T cells during development. In the thymus, self-reactive cells are either eliminated or differentiated into tolerogenic Foxp3(+) regulatory T cells (Treg) (152). Apart from the thymus, the immune system is also educated in the gut where it has been shown that the interaction of T cells with commensal microbiota results in the peripheral generation of Treg rather than pathogenic effector cells. Failure of this tolerogenic process can lead to the development of autoimmune diseases including colitis (152).
Microorganisms encountered early in life prevent autoimmunity and allergy
The human microbiome is important for human health, behavior and disease, yet its function and dynamics during healthy and disease states are not fully understood (153). It is also not fully understood how the microbiome interacts with the host immunity thereby preventing autoimmunity. The hygiene hypothesis first put forward by Strachan (154) postulates that the lack of early exposure to microorganisms (either beneficial or pathogenic) would lead to the poor development of the immune system. The leading idea is that some microorganisms that co-evolved with us are able to protect against a large spectrum of immune-related disorders (155). While the hygiene hypothesis is not universally applicable, it offers some explanative power to interpret the effects of microorganism exposure in early life on preventing autoimmunity and allergy.
Children growing up on dairy farms are protected from allergy, hay fever, and asthma (156,157). Asthma is a chronic inflammatory disease triggered by acute inflammation of the bronchial tube leading to production of extra mucus. This can make breathing difficult and trigger coughing, wheezing and shortness of breath. The number of asthma cases is increasing all over the world, but the causes remain obscure. It has been hypothesized that increased cleanliness, reduced family size and subsequent decreased microbial exposure could explain the increases in global asthma prevalence (158). Evidence from bronchial brushings implicates phyla present in healthy individuals with variation present in disease conditions such as cystic fibrosis, chronic obstructive airways disease and asthma. The microbiome can exacerbate the phenotypes seen in the condition, as well as explain the variability in phenotypes observed (159). Many cytokines and chemokines are involved in the pathophysiology of asthma. Th2 cytokines may play an important role in the pathophysiology of asthma. The Th1 cells secrete IL-2 and interferon-γ, whereas the cytokines, IL-4, IL-5, IL-9 and IL-13, are derived from T helper type 2 (Th2) cells, although they may also be derived from other cell types. The distinction between Th1 and Th2 cells is not as distinct in humans as in mice (160,161). According to the hygiene hypothesis, the lack of infection and exposure to environmental endotoxins may alter the balance between Th1 and Th2 cells.
Though children on farms are much less likely to get asthma the underpinnings of protection is not clearly understood. Early-life contact with livestock and their fodder, and consumption of unprocessed cow's milk have been identified as the most effective protective exposures. Studies of the immunobiology of farm living point to activation and modulation of innate and adaptive immune responses by intense microbial exposures (162). Schuijs et al. (157) demonstrated that chronic exposure to low-dose endotoxin or farm dust protects mice from developing house dust mite (HDM)-induced asthma. Endotoxin reduced epithelial cell cytokines that activate dendritic cells (DCs), thus suppressing type 2 immunity to HDM. Loss of the ubiquitin-modifying enzyme A20 in lung epithelium abolished the protective effect. A single-nucleotide polymorphism in the gene encoding A20 has been associated with allergy and asthma risk in children growing up on farms. Thus, the farming environment protects from allergy by modifying the communication between barrier epithelial cells and dendritic cells through A20 induction.
From delivery, the microbiome assembly might influence asthma. Babies born via cesarean section, who experience an altered trajectory of microbiome assembly, are more prone to asthma than those born vaginally. Similarly, children treated with antibiotics are also more prone to asthma attack (163). Lif Holgerson et al. (164) analyzed the oral biofilm in healthy three-month-old infants born by cesarian section or delivered vaginally. Among over 300 bacterial taxa analyzed, Slackia exigua was detected only in infants delivered by C-section. Further, significantly more bacterial taxa were detected in the infants delivered vaginally (79 species/species clusters) compared with infants delivered by cesarean section (54 species/species clusters). Overall, the vaginally delivered infants had a higher number of bacterial taxa. A higher prevalence of salivary Streptococcus salivarius, Lactobacillus curvata, Lactobacillus salivarius, and Lactobacuillus casei was detected in infants delivered vaginally (165).
A longitudinal human study by Arrieta et al. (166) reported that infants at risk of asthma have transient gut microbial dysbiosis during the first 100 days of life. The authors collected stool and urine samples from more than 300 babies at 3 months and 1 year-old, as well as information on their health at 1, 3, and 5 years. Then, they analyzed levels of gut microbes in each stool sample. Babies that had low or undetectable levels of four bacteria—Lachnospira, Veillonella, Faecalibacterium, and Rothia—at 3 months old all went on to show early signs of asthma—wheezing and skin allergies—at 1 year old. The babies that did not develop these symptoms invariably had high levels of the four microbes in their 3-month stool samples. The authors also used the stool samples from the asthma-prone 3-month-olds to colonize the guts of mice that had been raised in a bacteria-free environment. The animals developed inflamed lungs indicative of asthma. However, upon inoculating the four missing microbes to the digestive tracts of these mice along with the feces, they no longer had a heightened risk of developing asthma. The studies demonstrated that certain bacterial species that are encountered early in life could train the immune system to prevent asthma.
Microbial dysbiosis in early life can alter the trajectory of immune development and provide the setting for allergic disorders in later life (167). Dysbiosis may trigger autoimmune diseases via inappropriate post-translational modification of host proteins (168). Endogenous and microbial enzymes have the capacity of intestinal enzymatic neo-antigen generation by post-translational modification of host proteins. The hygiene hypothesis stipulates that microbial exposure during early life induces immunologic tolerance via immune stimulation, and hence reduces the risk of allergy development. Several common lifestyle factors and household practices, such as dishwashing methods, may increase microbial exposure. Hesselmar et al. (169) investigated whether lifestyle factors are associated with allergy prevalence. The authors demonstrated that in families that used hand dishwashing, allergic diseases in children are less common than in children from families who use machine dishwashing. The authors were of the opinion that a less-efficient dishwashing method may induce immune tolerance via increased microbial exposure.
Autoimmunity is more prevalent in the population of some northern European countries such as Finland and Estonia when compared to Russia. It was found that Bacteroides species are less abundant in Russians but dominate in Finnish and Estonian infants. Their lipopolysaccharide (LPS) exposures arose primarily from Bacteroides rather than from Escherichia coli, which is a potent innate immune activator. The Bacteroides LPS was found to be structurally distinct from E. coli LPS and inhibits innate immune signaling and endotoxin tolerance. It was observed that unlike LPS from E. coli, Bacteroides dorei LPS does not decrease incidence of autoimmune diabetes in non-obese diabetic mice. Early colonization by immunologically silencing microbiota may thus preclude aspects of immune education (170).
Rheumatoid arthritis (RA) is an autoimmune disease where the immune system attacks the joints leading to swollen and painful joints. The mucosal surfaces are sites of RA initiation. The common occurrence of periodontal dysbiosis in RA suggests that oral pathogens may trigger the production of disease-specific autoantibodies and arthritis in susceptible individuals. Periodontitis is characterized by the presence of citrullinated autoantigens that are primary immune targets in RA. The citrullinome in periodontitis is similar to the hypercitrullination observed in the rheumatoid joint, implicating this mucosal site in RA pathogenesis. Recent studies identified the periodontal pathogen Aggregatibacter actinomycetemcomitans as a bacterial trigger of autoimmunity in RA by inducing hypercitrullination in host neutrophils. The pore-forming toxin leukotoxin A secreted by A. actinomycetemcomitans triggers autoantigen citrullination in the RA joint (171). Zhang et al. (172) reported alterations in the gut, dental or saliva microbiome that distinguished patients with RA from healthy controls. Individuals with RA had low numbers of Haemophilus spp., whereas Lactobacillus salivarius was very high in these patients. It has been reported in experiments in mice that inoculation with Bifidobacterium adolescentis exacerbated autoimmune arthritis. B. adolescentis is known to induce Th17 cells in the intestine (173). Interestingly, the frequencies of Th17 cells and levels of interleukin-17 strongly correlated with systemic disease activity at both the onset and the progression of RA (174).
Role of microbiome in obesity
Obesity results from an imbalance of food intake, basal metabolism, digestive tract microbial composition and energy expenditure (175,176). According to Turnbaugh et al. (177) the gut microbiome should be considered as a set of genetic factors that together with host genotype and lifestyle contribute to the pathophysiology of obesity. It is observed that the intestinal bacteria in obese humans and mice differ from those in lean individuals. Obese mice microbiota was found to be rich in Firmicutes compared to the lean mice microbiota, which was abundant in Bacteroidetes (177). Strikingly, colonization of germ-free mice with microbiota from obese mice was sufficient to cause a significant increase in total body fat, as compared to colonization with microbiota from lean mice (177). The obese microbiome has an increased capacity to harvest energy from the diet, thereby increasing weight gain in the host (177,178). Colonization of adult germ-free mice with a gut microbial community harvested from conventionally raised mice increased body fat within 10–14 days, despite an associated decrease in food consumption. This change involves several linked mechanisms: microbial fermentation of dietary polysaccharides that cannot be digested by the host; subsequent intestinal absorption of monosaccharides and short-chain fatty acids; their conversion to more complex lipids in the liver; and microbial regulation of host genes that promote deposition of the lipids in adipocytes (179).
Transfer of human microbiota to mice can phenocopy such effects, as shown by Ridaura et al. (180). Cohabitation of mice harboring an obese microbiota with mice containing the lean microbiota prevented the development of increased body mass and obesity-associated metabolic phenotypes in obese cage mates. Rescue correlated with invasion of specific members of Bacteroidetes from the lean microbiota into obese microbiota and was diet-dependent. The study confirmed that specific bacteria along with diet could induce obesity.
Childhood obesity is considered one of the most serious global health issues in our society. Obese children are more likely to be obese in adulthood and are at greater risk of premature death and adverse health outcomes in later life (181). Administration of three or more courses of antibiotics before children reach age 2 years is associated with an increased risk of early childhood obesity (182). When given early in life antibiotics that disrupt microbiota composition, and consequently the metabolic activity of the microbiota, can affect the body mass of the host by either promoting weight gain or stunting growth (183). The correlation of antibiotics to obesity has been earlier shown in animal models (184).
Food is broken up into components that tend beneficial microorganisms. Bacterial fermentation of a diet rich in fibers leads to production of short chain fatty acids (SCFA), which as noted above includes acetate, propionate, and butyrate in the gut (185,186). Interestingly, butyrate promotes colonic health and helps prevent cancer (185,187,188). The consumption of high fat and high calorie foods negatively impacts the beneficial microbes, which are believed in turn to promote obesity. Notably, obese people have lower Bacteroidetes and more Firmicutes in their distal gut than lean subjects and the introduction of low fat and carbohydrate diets increased the proportion of Bacteroidetes (175,189).
Obesity is also known to impair cognition and produces atrophy of brain regions associated with learning and memory. In animal studies it has been shown that, even before the onset of diabetes or metabolic syndrome, early stage obesity produced deficits on cognitive tasks that require the prefrontal cortex. Impaired cognition was associated with synapse loss, including reduced numbers of dendritic spines and expression of synaptic proteins, as well as structural alterations in the microglia. Thus, obesity must be considered as a contributing factor to brain dysfunction mediated through the gut-brain axis (190,191).
It has been demonstrated recently that some bacterial species are beneficial to the host by preventing obesity. In animal models and humans the abundance of Akkermansia muciniphila is decreased in obese and type 2 diabetic mice (192) and use of the bacterium as a probiotic is beneficial to the host. Interestingly, whole bacterium is not essential to prevent obesity. Intake of the membrane protein of the bacterium per se could be beneficial to the host. Amuc_1100, a specific protein isolated from the outer membrane of A. muciniphila, interacts with Toll-like receptor 2, is stable at temperatures used for pasteurization, improves the gut barrier and recapitulates the beneficial effects of the bacterium (193).
Metformin is a well-established drug in the management of type II diabetes and obesity. Recent studies suggest that the microorganisms are involved in mediating the beneficial effects of metformin on glucose metabolism. Metformin shifts gut microbiota composition through the enrichment of mucin-degrading A. muciniphila as well as several short- chain fatty acid-producing microbiota including Butyrivibrio, Bifidobacterium bifidum, Megasphaera, and Prevotella (194). Overall, there is substantial evidence of the key role of microbiota in obesity and its adverse effects.
Microbiome and cardiovascular diseases
The gut microbes produce a large range of metabolites which act not only in the gut, but also systemically, and this large pool of known and unknown metabolites is not fully understood (195). The metabolite trimethylamine N-oxide (TMAO) is the first potentially direct link between the gut microbiota and atherosclerotic heart disease. Trimethylamine (TMA) is produced by the gut microbiota from nutrients containing l-carnitine, choline, and phosphatidylcholine, and is subsequently oxidized by hepatic flavin-containing monooxygenases to TMAO. TMAO has been proposed to interfere with cholesterol transportation, and TMAO precursors promote foam cell formation and atherosclerosis in animal models, but not in the presence of antibiotics to the drinking water, suggesting a microbiota-dependent mechanism (195).
Hypertension is a risk factor for coronary heart disease, yet whether gut microbiota dysbiosis is involved in the development of hypertension remains largely unknown. In a recent study it was observed that the bacterial genus Prevotella and Klebsiella were overrepresented in individuals with hypertension. Fecal transplantation from hypertensive human donors to germ-free mice increased blood pressure in animals thereby demonstrating the direct influence of gut microbiota on high blood pressure (196).
Beneficial microorganisms are known to protect against atherosclerosis. The lack of gut microbiota in germ-free apolipoprotein E (ApoE)-null mice, an experimental model of human atherosclerosis, was found to induce the development of atherosclerotic plaques even when animals were fed a standard low-cholesterol diet. Colonization with normal human microbiota prevented atherogenesis in germ-free ApoE-null mice fed a standard low-cholesterol diet but not a diet with high cholesterol content (197). The bacterial genera Eubacteria, Anaeroplasma, Roseburia, Oscillospira and Dehalobacteria appeared to be protective against atherosclerosis and showed significant negative correlation with atherosclerotic plaque size and plasma adipocyte - fatty acid binding protein (A-FABP) and cholesterol (198). A. muciniphila is also beneficial to the heart. The bacteria attenuate atherosclerotic lesions by ameliorating metabolic endotoxemia-induced inflammation through restoration of the gut barrier (199).
Microbiome and behavior
The exponential growth in our collective knowledge of the human microbiome has seen the study of gut microorganisms move beyond the traditional preserve of strictly microbiological disciplines. As our appreciation of the structure and dynamics of the gut microbiome has flourished, so too has our grasp of the implications for host physiology in health and disease. Perhaps one of the more surprising aspects of this host-microbe dialogue is the complex interactions that manifest as alterations in brain and behavior. Moreover, the bidirectional nature of this conversation needs to be considered in the context that disruptions to CNS function can be expressed distally as alterations in microbiome composition and function in the gastrointestinal tract. These aspects of host-microbe dialogue are generally important to medicine, due to the impact of behavioral states that widely impact and/or reflect the operation and progression of pathogenic processes and their treatment (e.g., the negative impact of depression on general therapeutic compliance).
These complex reciprocal interactions are facilitated by the microbiome-gut-brain axis, which incorporates the gut microbiome as a critical node of the communication network encompassing the CNS, the neuroendocrine and neuroimmune systems, the sympathetic and parasympathetic arms of the autonomic nervous system and the enteric nervous system (200). The focus on the gut microbiome has proved to be a surprisingly fertile ground, and the evidence garnered from a variety of preclinical approaches has converged to illuminate how the gut microbiome regulates multiple behaviors, physiological readouts and indeed many fundamental aspects of brain function.
In this regard, surveys of microbiota-deficient germ-free animals have proved particularly informative. The use of these animals in general is not new, but their application to CNS-directed queries has been a notable feature of recent research efforts (201,202). From a behavioral perspective, these animals display a less-anxious phenotype (203-206) and this atypical performance can be normalized if the animals are colonized post-weaning (205). Remarkably, it has also been demonstrated using both the germ-free paradigm and an antibiotic-induced microbiota deficiency that anxiety-like behaviors can be transferred via the gut microbiota by means of a fecal transplant (207,208). Germ-free animals also display alterations in social behaviors (209,210) and, insofar as it has been logistically possible to address in detail in this paradigm, aspects of cognitive function (211). Gut microbiota depletion using a cocktail of antibiotics from early adolescence in mice replicates many of the behavioral characteristics of germ-free mice, including reduced anxiety-like behaviors and impaired cognitive performance (212).
Other approaches have both largely supported and extended the behavioral picture painted by microbiota-deficient animals. For example, administration of a probiotic Lactobacillus rhamnosus strain reduced anxiety and depression-related behaviors (213), while alternative candidate probiotics, including a Bifidobacterium longum strain, exerted a beneficial impact on cognitive processes (214). Prebiotics (eg. fiber rich foods that can influence the microbiome) can also exert anxiolytic effects (215), while bacterial infection with an enteric pathogen can impact both learning and memory (211) and modulate pain behaviors (216).
Physiologically, germ-free animals also exhibit profound differences with conventionally colonized controls. These differences include a defective immune system and exaggerated corticosterone outputs to acute stressors (205,217,218). The microbiome is also required for the development of microglia, cells that defend the central nervous system. Microglia from germ-free mice had altered gene expression that influenced its development (219).
An increased availability of tryptophan - the amino acid precursor to neuroactives such as serotonin and kynurenine pathway catabolites, as generated respectively by IDO1 or TPH - is one feature of the germ-free state (205,220,221). Many aberrant physiological features can be rescued if the animals are colonized with a normal microbiota, especially if this intervention takes place during specific time windows post weaning (205,217). As is the case for behavior, other microbiota manipulations such as rendering mice microbiota deficient or probiotic ingestion by rodents can also impact parameters such as corticosterone outputs or tryptophan availability (212,213,222).
The reciprocal interaction between stress and the microbiome is a particularly interesting facet of this bidirectional relationship. As indicated above, the gut microbiota exert an influence on the hypothalamic-pituitary adrenal axis, the main host stress response system and this can be captured by measuring cortisol in humans or corticosterone in rodents (223). There are now studies showing that the opposite is also true and that a variety of stressful insults which are linked to psychopathology in adulthood can alter the composition of the gut microbiome in animal models. This is reflected in studies that have examined early-life stress (224,225), prenatal stress (226,227) and psychosocial stress (228-230). Interestingly, the gut microbiota seems necessary for the expression of some of the pathological behavioral features induced by maternal separation (231), a well validated early life stress based model of gut-brain axis dysfunction (232). In the clinical setting, maternal prenatal stress is also associated with alterations in the infant gut microbiome (233). Another example of such feedback loop is stress-related microbiome-gut-brain axis dysfunction in irritable bowel syndrome (234).
Growing up, germ-free influence biological function such as blood-brain barrier integrity (235), transcriptional regulation (203,236), neurogenesis (237) microglial function (238) and myelination (239). Recently, it has been demonstrated that a germ-free mouse model of Alzheimer's disease displayed a marked reduction of cerebral amyloid pathology and that colonization of these mice with the gut microbiota of their conventionally-colonised counterparts reinstated the amyloid pathology (240). Although the behavioral implications of these altered amyloid phenotypes requires elaboration, this intriguing study does provide support for the hypothesized role of the gut microbiota in neurodegenerative disorders (241).
The mechanisms underpinning influence of the gut microbiome on brain and behavior are under investigation. The gut microbiota are required for motor deficits, microglia activation, and α-synuclein pathology. Colonization of microbiota from Parkinson's disease-affected patients enhances physical impairments compared to microbiota transplants from healthy human donors. Thus the gut bacteria is involved in movement disorders and alterations in the human microbiome represent a risk factor for Parkinson's disease (242). Recently the role of the vagus nerve (the main neural communication highway between the gut and the brain) has attracted much attention (243). It has been demonstrated, for example, that the beneficial CNS impact of a probiotic was abolished in vagotomised mice (213) while the anxiety-like behaviors that emerge in a colitis model were absent in previously vagotomised mice (207). The vagus nerve is not the sole conduit linking the gut and the brain (244) and a variety of alternatives have been considered. These include microbial regulation of tryptophan metabolism (245), microbial metabolites such as short chain fatty acids (SCFA) (246) or indoles derived from tryptophan (247), neuropeptide production (248) as well as immunomodulation (249). The important role of the gut microbiota in maintaining intestinal barrier integrity also needs to be taken in account (250,251).
The landscape for manipulating the microbiome is broad and increasingly financed (252). Consideration are being given to priming interventions which promote assembly of the infant microbiome (65,253,254), sustain the gut microbiota in healthy aging (125,126), more radical options such as fecal microbiota transplantation (255) as well as less controversial options such as psychobiotics (256), exercise (257,258) and diet-based manipulations (259,260). Indeed, a number of small studies using healthy volunteers have now demonstrated that ingestion of certain cocktails of probiotics, a fermented milk product with probiotic or prebiotics can impact on the CNS (261-264). Autism Spectrum Disorders (ASD) are complex neurobiological disorders characterized by impairment in social interaction and communication and restricted, repetitive, and stereotyped patterns of behavior, interests, and activities. Autistic children suffer from gastrointestinal disorders. Autistic children have less diverse microbial population in the gut and significantly lower abundances of the genera Prevotella, Coprococcus, and Veillonellaceae involved in carbohydrate metabolism (265).
Future directions will likely see further elaboration of the role of the gut microbiome in sleep (266) and circadian rhythms (267-269). Our awareness of the interface between natural and built environments, the gut microbiota and human behavior is also growing (270-272). Of course, a key caveat is to what degree this promising but largely preclinical body of research will effectively impact the clinical setting. Moving from mouse to man, be it in the context of CNS-directed or gastrointestinal-focused research, is of course complicated for stress-related neuropsychiatric and other heterogeneous disorders associated with the gut microbiome (273,274). Of equal importance is the necessity to address the issue of whether the correlations that have been noted thus far between multiple disorders and the gut microbiota alterations are in fact causal relationships. Such obstacles are not insurmountable with due diligence and the necessary multidisciplinary expertise required to exploit the considerable opportunities presented by host-microbe interactions.
Beneficial microorganisms restrict the outgrowth of pathogens in the gut
The human microbiota encompasses all the microorganisms that reside on the skin and in all other tissues and organs including the gastrointestinal tracts. Of these body sites, the gastrointestinal (GI) tract is the most densely colonized organ. The microbiome includes bacteria, fungi, and archaea (275). There are approximately 1000 species of microbes colonizing the gut, with densities of 104 to 105 bacteria per millimeter of digestive effluent in the proximal small intestine and 1011 bacteria per gram of luminal content in the colon (276). The physicochemical conditions in the gut influence the composition of the intestinal microbiota (277). The GI tract harbors many distinct niches, each containing a different microbial ecosystem that varies according to the location within the GI tract. The microbial density increases along the GI tract with101–104 microbial cells in the stomach and duodenum, 104 to 108 cells in the jejunum and ileum, to 1010 to 1012 cells per gram in the colon and feces (277-279).
The majority of all microorganisms in the human digestive tract are bacteria and belong to two phyla, the Bacteroidetes and the Firmicutes (280). In addition, the other significant phyla occupying the digestive tract includes Proteobacteria, Actinobacteria, Fusobacteria, Spirochaetes, Verrucomicrobia and Lentisphaerae (281,282). The methanogens, Methanobrevibacter and Methanosphaera are the most dominant archaeal groups (7,283). The two common fungal phyla in the gut include Ascomycota (which includes the genera Candida and Saccharomyces) and Basidiomycota (30,284).
Intestinal microbiota play a central role in the metabolic, nutritional, physiological and immunological processes of the human body, processing indigestible dietary polysaccharides including resistant starch and dietary fibers thereby leading to the production of important nutrients, such as SCFAs, vitamins (vitamin K, vitamin B12, folic acid) and certain amino acids that humans are unable to synthesize themselves (279,285,286). The plant polysaccharides in our diet are rich in xylan-, pectin-, and arabinose-containing carbohydrate structures. The human genome lacks most of the enzymes required for degrading these glycans. Nevertheless, the distal gut microbiome provides us with this capacity to process these polysaccharides. The human gut microbiome is enriched for genes involved in glucose, galactose, fructose, arabinose, mannose, and xylose, starch and sucrose metabolism. Our microbiome also has significantly enriched metabolism of glycans, amino acids, and xenobiotics; methanogenesis; and 2-methyl-d-erythritol 4-phosphate pathway-mediated biosynthesis of vitamins and isoprenoids (7). The intestinal microbiota also participates in the defense against pathogens by mechanisms such as colonization resistance and production of antimicrobial compounds. Furthermore, the intestinal microbiota is involved in the development, maturation and maintenance of the GI sensory and motoric functions, the intestinal barrier and the mucosal immune system (279). The microbiota of the intestine is also involved in promoting bone formation as well as resorption leading to skeletal growth. Microbiota induces the hormone insulin-like growth factor 1 (IGF-1), promoting bone growth and remodeling. When the microbiota ferment fiber short-chain fatty acids (SCFAs) are produced leading to induction of IGF-1 that promote bone growth (287).
The very high microbial content of the large intestine poses a major challenge to the mucosal immune system, as it needs to tolerate commensal microbiota and dietary antigens while maintaining the ability to eliminate pathogens. Induction of colonic Treg is crucial in fostering this immune homeostasis (288). CD4+CD25+FOXP3+ Treg are of two types: thymus-derived Treg (tTreg) and peripherally derived Treg (pTreg). Although it is difficult to distinguish these types phenotypically, both are thought to have an essential role in immune regulation (288). While tTreg develop in the thymus, the major site for pTreg development is the colon, resulting in a large population of regulatory cells that have a distinct TCR repertoire and are critical for intestinal homeostasis (152). Notably, the development of pTreg requires microbiota to be present in the colon. Though the mechanism of induction of colonic pTreg is not understood, several microbial components have been found to enhance their expansion and function, including SCFAs and polysaccharide A of Bacteroides fragilis (288). Acetate, propionate, and butyrate are the three main SCFAs, and butyrate has been found to be the most potent inducer of colonic Treg.
The newborn infant is colonized at birth with microbes from the mother's vaginal and fecal microbiota as well as with other environmental microbes encountered in the first days of life (289). The mode of delivery influences the microbial composition in man. A paper by Penders et al. (93) demonstrated that the important determinants of the gut microbiome composition in infants were the mode of delivery, type of infant feeding, gestational age, infant hospitalization and antibiotic use in the infant. Term infants born vaginally and exclusively breastfed had the most “beneficial” gut microbiota (had the highest numbers of Bifidobacteria and lowest numbers of C. difficile and E. coli). In contrast, infants born through C-section had lower numbers of Bifidobacteria and Bacteroides, and they were more often colonized with C. difficile, as compared to vaginally born infants. Exclusively formula-fed infants were more often colonized with E. coli, C. difficile, Bacteroides and Lactobacilli, as compared with breastfed infants. Hospitalization and prematurity were associated with higher prevalence and counts of C. difficile. Administration of antibiotics to infants was associated with decreased numbers of Bifidobacteria and Bacteroides. Infants with older siblings had slightly higher numbers of Bifidobacteria, compared with infants without siblings (93,290).
Clostridium difficile is an opportunistic, anaerobic gram-positive, spore-forming, toxin-producing bacillus that is transmitted among humans through the fecal–oral route. Notably, the pathogen is generally present in the human gut but it does not cause any disease under normal conditions. Abuse/misuse of antibiotics destroys beneficial microbiota that enables the proliferation of C. difficile leading to pathogenic conditions. Ampicillin, amoxicillin, cephalosporins, clindamycin, and fluoroquinolones are the antibiotics most frequently associated with disease, but almost all antibiotics have been associated with increased rates of opportunistic infection. C. difficile colonizes the large intestine and releases the exotoxins TcdA and TcdB that cause colitis in susceptible persons (291,292). C. difficile has emerged as a major enteric pathogen with worldwide distribution. In the United States, C. difficile is the most frequently reported nosocomial (i.e., hospital-acquired) pathogen. A recent surveillance study identified 453,000 cases of C. difficile infection and 29,000 deaths associated with C. difficile infection; approximately a quarter of those infections were community-acquired (293). The antibiotics prescribed to control C. difficile include metronidazole, vancomycin, fidaxomicin or surgery in extreme cases of infection (294). Fecal transplant is emerging as an alternative strategy for treating recurrent C. difficile infections (295). Use of probiotics (such as beneficial bacteria and yeast), which help restore a healthy balance to the intestinal tract are safe and effective for preventing C. difficile-associated diarrhea (296). Thus, beneficial microorganisms are essential to maintain the human gut immune homeostasis thereby preventing pathogenic infections. Further contributing to such defenses, beneficial microorganisms also modulate epithelial cell proliferation, villus architecture and angiogenesis within the intestine, along with xenobiotic metabolism, bone mineral density, behavior and key metabolic functions (297,298). Tipping the balance favoring the expansion of enterobacteria is one of the causes of several inflammatory bowel diseases. However, it is not known how the favorable bacteria prevent dysbiosis. A recent study demonstrated that microcins, the small proteins secreted by several favorable bacteria could limit the expansion of competing Enterobacteriaceae (299).
Microbiome effects on intestinal barrier function and inflammatory bowel disease
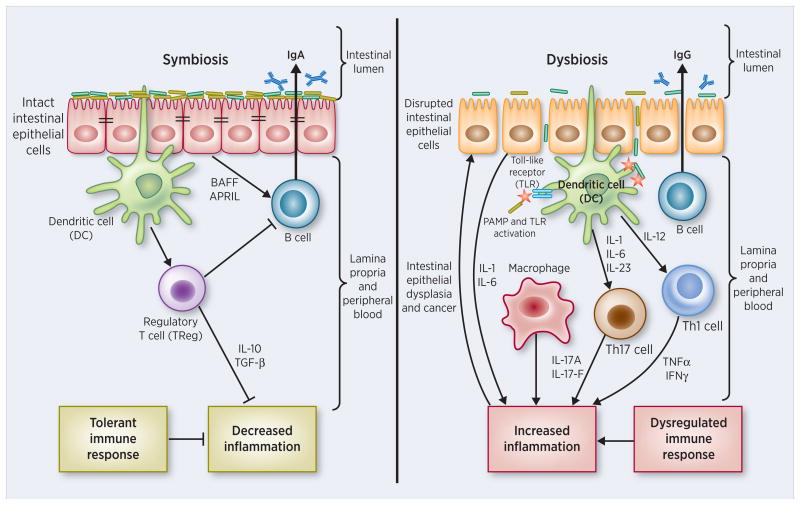
Mammals, including humans, support one of the most complex microbial ecosystems. Although the immune system is classically thought to have evolved to provide protection from infection by microbial pathogens, animals peacefully coexist with a vast and complex microbiota, which extensively interacts with the immune system. It has recently been proposed that the total information encoded by the mammalian genome is not sufficient to carry out all functions that are required to maintain health and that products of our microbiome are crucial for protection from various diseases (300). It is possible that alterations in the development or composition of the microbiota (dysbiosis) disturb the partnership between the microbiota and the human immune system, ultimately leading to altered immune responses that may underlie various human inflammatory disorders (146).
In inflammatory bowel disease (IBD), the role and interplay of the microbiome (301-303) with a GI barrier compromise (304-306) has been the subject of extensive review (307-311). GI barrier function is not governed solely by the tight junctional (TJ) complex, although this focus has certainly attracted the greatest basic research interest. TJs form the continuous intercellular barrier between epithelial cells, which is required to separate tissue spaces and regulate selective movement of solutes across the epithelium (312). From a wider perspective, GI barrier function also may be compromised by an impaired mucus layer over the epithelium (a topic reviewed nicely by others recently [Chen et al., (313)]), as well as by cell death in the epithelium or an epithelial-mesenchymal transition leading to impaired cell adhesion/attachment (to other cells and substratum). Leak at sites of compromised TJ may be quite distinct in nature from leak at sites of cell death (314). Likewise the remediation of leak is very different in these cases – repair of leak from impaired TJ may be a purely transcription/translation/phosphorylation-based process, whereas remediation of leak due to cell death/dedifferentiation/detachment could also require a careful orchestration of cell motility and cell replication. In these different cases, the microbiome may exert control over very different processes. It is worth considering that any given specific case of IBD likely involves gastrointestinal leak from all of these causes, and therefore alleviation of such leak – and the inflammatory cascades it gives rise to – is a quite complex task. Further research is required to determine whether microbiome may be better at repair of one type of leak than the other.
There is no doubt that IBD is in part driven by a breakdown or compromise of the gastrointestinal epithelial barrier, and many reviews on IBD have dealt with this feature as mentioned above. There is some controversy concerning whether a compromise of barrier function is the initial causation of the disease. The findings that asymptomatic, first degree relatives of IBD patients in fact harbor molecular-level leak in their GI tract mucosa has traditionally been powerful evidence tilting the argument toward causality (315,316). The question of course that leaps to the fore then is what induces the leak in the first place? The very fact that a genetic element exists in IBD (e.g. in first degree relatives) indicates a role for genetics in the disease, but equally obvious is that genetics is probably a necessary but insufficient condition.
Studies focusing on this involvement of gastrointestinal microbiome in IBD take two forms: 1) whether the microbiome is abnormal in IBD and possibly playing a role in etiology; 2) whether a microbiome modification can be designed as a therapeutic option in the disease. The second possibility appears achievable: some of the best clinical evidence – on the basis of its very applicability – is the success of “fecal-transplant” procedures in achieving therapeutic efficacy in IBD (317,318). Prior to the recent advent of these protocols there was the use of butyrate enemas to achieve therapeutic relief (286,319,320). The therapeutic efficacy of luminal administration of butyrate is cogent testimony to the positive role played by the normal microbiome in maintaining a functional epithelial barrier in the GI tract, as well as to the utility of targeting the microbiome as a viable clinical approach to IBD. Butyrate is a significant metabolite of dietary fiber by the normal gastrointestinal microbiota (321), with butyrate levels in the GI lumen being the highest in the body. Butyrate has been found in many recent in vitro studies to be highly effective in inducing structural changes to the epithelial TJ complex resulting in improved epithelial barrier function (322,323). A similar literature also exists for the GI microbiota metabolite, indole, a product of tryptophan metabolism by commensal bacteria (324,325). In combination, the fecal transplant and butyrate enema clinical studies, along with the in vitro studies of TJ modification and enhancement by butyrate and indole, provide a very powerful argument of not only maintenance and modification of the GI barrier by the microbiota, but also for targeting the microbiota as a viable, effective therapeutic strategy.
Better delineated proof of the ability of the microbiota to both positively and negatively affect the GI barrier comes out of animal model and epithelial cell culture studies. For example, the probiotic bacterium, Lactobacillus casei, both strengthened barrier function and decreased proinflammatory cytokine content in BALB/c mice (326). In addition, such treatment also modified the GI microbiota overall. The probiotic and commensal bacteria Lactobacillus rhamnosus and Faecalibacterium prausnitzii have also improved barrier function, as demonstrated in studies with CACO-2 cell culture models and experimentally induced colitis in C57BL/6 mice (327,328). The microbiome is also known to be involved in the wound healing of the mucosa of the gut (329). In mice it has been demonstrated that mucosal injury leads to increase in the expression of formyl peptide receptor 1 (FPR1) and neutrophilic NADPH oxidase (NOX2) that causes depletion of oxygen resulting in the enrichment of anaerobic bacteria. The anaerobic, mucinophilic gut symbiont, Akkermansia muciniphila, stimulated proliferation and migration of enterocytes adjacent to the colonic wounds mediated through FPR1 and intestinal epithelial-cell-specific, NOX1-dependent redox signaling, thereby leading to wound healing of the mucosa. These findings highlight a very important consideration in studies of the microbiota, barrier function and IBD – namely that not only can microbiota affect barrier function, but modification of barrier function (good and bad) may well affect microbiota composition – a “research road less traveled.” This less investigated area is well illustrated by the finding that anti-TNF immunologic medications – which reduce proinflammatory cytokines and allow for barrier repair –also result in changes in microbiota composition (330).
It is worth noting too that a factor as omnipresent as diet can affect both barrier function and microbiota, while also simultaneously affecting cytokine production. Administering a high-fat “Western” diet to CEABAC10 mice induced deleterious changes in GI microbiota (e.g. increased content of adherent-invasive E. coli), decreased mucus layer protection, and led to GI barrier compromise (331). Pathogenic E. coli have been implicated in Crohn's disease, in part due to an ability to not produce cell death while inducing synthesis of copious amounts of TNF (332). The dietary connection to an altered microbiota in terms of pathogenic bacteria is also apparent in the finding that vitamin D deficiency enables the barrier disruptive effects of pathogenic E. coli to be manifested (333). Even more fascinating and less intuitive is an effect of the environment at large on barrier function, microbiota and inflammatory status, as Kish et al. (334) show for particulate air pollutants. It will be instructive to discover in future research whether the principal actions of diet/nutrition/environment are on microbiota directly and barrier function indirectly, or vice versa.
As more and more studies reveal the intricate interplay between GI barrier function, GI microbiota and degree of inflammation, it is worth considering that the effects of microbiota on barrier function - and barrier function on microbiota - will in large degree derive from actions of protein kinases in signaling pathway transduction systems. This has been recently very well reviewed by Yang and Yan (335). One would caution however against taking an aggressively reductionist approach in dealing with the interplay of microbiota, barrier function and inflammation: The complexity of the gut microbiome in the GI tract, the complexity of the signaling pathways known to regulate barrier function, and the complexity of cytokine interactions all suggest strongly that one needs to tread carefully in what may well be overly ambitious undertakings to find and utilize specific molecular mechanisms involved in this intricate relationship. A properly functioning gastrointestinal mucosa is in essence a symphony scored by microbiota “strings,” barrier function “woodwinds,” and immune regulatory “brass.” To search for an all-pivotal kinase or phosphatase responsible for GI mucosal homeostasis may be analogous to trying to claim a single flute or viola as the fulcrum of Beethoven's Ninth – a fool's errand that ignores the extreme complexity and subtlety of the GI environment.
Because of the still singular importance of the studies showing epithelial barrier compromise in asymptomatic first-degree relatives of IBD patients to the field of IBD research, it will be very interesting to observe the outcome of studies yet to be performed on the microbiome of first degree relatives of IBD patients. It is likely that those results could be just as pivotal to the future understanding of IBD as were the now long ago studies of May and Hollander on barrier function and IBD (315,316).
Increased risk of developing IBD may be due to improved hygiene practices. Ramanan et al. (336) showed that intestinal helminth infection, caused by parasitic worms, protects IBD-susceptible mice from developing the disease. The infection of parasitic worms increased specific protective species and limited other inflammatory members of the microbiota. People from helminth-endemic regions harbored a similar protective microbiota, and their de-worming led to an increase in inflammatory Bacteroidales, as observed in the mice. Thus, a changing microbial environment may shape susceptibility to inflammatory disease.
Though we currently know at least a partial membership of the human microbiome, we are yet to fully understand how these microorganisms are contained in the intestine. Anatomy of the colon by light microscope reveals a mucus layer, mucosa, submucosa, gut associated lymphoid follicle and muscularis. The intestinal/colonic mucus is an efficient system for protecting the epithelium from bacteria by promoting their clearance and separating them from the epithelial cells, thereby inhibiting inflammation and infection (337). Colonic mucus is produced by the goblet cell. The main mucus component in the intestine is MUC2 mucin, a large and heavily O-glycosylated gel-forming mucin that forms enormous polymeric nets by C-terminal dimerization and N-terminal trimerization. Upon secretion from the goblet cells, the mucus expands rapidly and builds a stratified dense layer that is attached to the epithelium. Normal human sigmoid colon has an inner mucus layer that is impenetrable to bacteria. At a distance, far from the epithelial surface, the inner mucus is transformed into a soluble and less organized outer mucus layer that by proteolytic expansion, generates the preferred habitat for the commensal microbes (338).
IBDs are characterized by aberrant innate and adaptive immune responses to commensal luminal bacteria (339). Ulcerative colitis (UC) is thought to be caused by some strains of E. coli (340). In cell culture models it has been showed that UC-associated E. coli producing α-hemolysin can cause rapid loss of TJ integrity (341). The human intestinal epithelium is formed by a single layer of epithelial cells that separates the intestinal lumen from the underlying lamina propria and the space between these cells is sealed by TJ, which regulate the permeability of the intestinal barrier (342). TJ complexes allow passive absorption of small hydrophilic molecules (nutrients and ions), but they restrict passage of large molecules and infectious microbes. UC is characterized by a leaky intestinal barrier due to defective TJ. Our group had reported that attenuation of the Bin1 gene in a mouse model would protect against experimental colitis (343). Based on the study we recently demonstrated that treatment of experimental colitis with Bin1 monoclonal antibody would support mucosal barrier function by inducing the expression of TJ proteins thereby protecting the integrity of the lymphoid follicle. The therapy may be a novel strategy to treat UC and possibly limit risks of colorectal cancer (344). Thus lowering Bin1 levels may be a strategy that would lead to enhanced TJ proteins that, in turn, protects against pathogenic microorganisms from crossing the epithelial cells.
Other strategies may be used by the epithelial cells to protect against pathogenic microbes from entering the tissues. Recently while working on subconfluent CACO-2 cells (derived from the intestine) we observed that EEA1 endosomes (early endosome marker) were confined more toward the periphery of CACO-2 cell monolayers. To confirm whether these endosomes are present in the colon tissues, we stained for EEA1 in mouse colonic tissues. We observed EEA1 endosomes in the peripheral mucosa as well as in the muscularis. Insofar as endosomes are traditionally thought to protect against foreign bodies including microorganisms, one might speculate that early endosomes help protect against bacteria crossing the mucus. Microbes that overcome this barrier to cross the mucus layer might be destroyed within endosomes, with further protection afforded by the endosomes lining the muscularis (Fig. 1).
Figure 1. Early endosomes localize mainly to the periphery of colon mucosa and muscularis.
(A) Peripheral localization in a colony of human colonic Caco-2 cells. Cells were stained with the early endosomal marker EEA1 (green) or DAPI to visualize cell nuclei (blue) and processed for immunofluorescence microscopy. (B) Peripheral localization in murine colon mucosa processed as above. (C). Peripheral localization in murine colon muscular processed as above.
Additional endosomal strategies may be used to protect colonic tissues against the entry of pathogenic microbes. Endosomes are traditionally thought to protect colonic epithelial cell layers against microorganisms, possibly helping eliminate bacteria that cross the mucus layer, a strategy that also may be extended into the subordinate muscularis layer (Fig. 1). In subconfluent human colonic cells, which form island-like colonies in monolayer culture, the early endosome marker EEA1 can be seen to preferentially localize to the colony periphery. Similarly, in colon tissues early endosomes display the same localization with EEA1 staining in the periphery of the mucosa and muscularis. Beclin1-dependent autophagy associated with the endosome pathway also has been implicated in the bacterial and viral pathogen elimination (345), which downregulate Beclin1 to promote virulence and infection. Beclin1 associates with endosomes and regulates EEA1/early endosome localization and late endosome formation (346). Upon TLR signaling, Beclin1 rapidly translocates to the phagosome and mediates efficient phagosome-lysosome fusion to ensure rapid acidification and efficient destruction of the pathogen (345). In human colonic cells, Beclin1 staining occurs throughout the colon tissue at endosomes (Fig. 2), possibly helping direct pathogenic cargo to lysosomes and thereby restricting microbiome ecology to the gut lumen.
Figure 2. Late endosomes locate throughout colon mucosa and muscularis.
(A) Punctate cytosolic localization in a colony of human colonic Caco-2 cells. Cells were stained with Beclin1, an autophagic regulator associated with late endosomes (green), or DAPI to visualize cell nuclei (blue) and processed for immunofluorescence microscopy. (B) Cytosolic localization in murine colon mucosa processed as above. (C). Cytosolic localization in murine colon muscularis processed as above.
Indoleamine 2, 3-dioxygenase (IDO1) and the microbiome-host interaction
As noted above, establishing and maintaining the symbiotic mutualism that exists between the microbiome and its mammalian host necessitates the engagement of mechanisms of acquired immune tolerance as these microorganisms represent the epitome of non-self. Indoleamine 2,3-dioxygenase-1 (IDO1) is a metabolic enzyme that has gained recognition as an important mediator of acquired immune tolerance. IDO1 catalyzes the rate-limiting first step in the degradation of the essential amino acid tryptophan, but is not involved in maintaining tryptophan homeostasis, which instead is the role of the distinct liver enzyme TDO2 (tryptophan dioxygenase) (347). The concept of IDO1 as an immune regulator emerged from findings that tryptophan catabolism could suppress cytotoxic T cell activation (348,349). The demonstration that the IDO pathway inhibitor 1-methyl-tryptophan (1MT) could elicit T cell-dependent rejection of allogeneic mouse concepti (350,351) established the physiological relevance of tryptophan catabolism as a mediator of acquired immune tolerance. Subsequent findings linking attenuation of the tumor suppressor gene Bin1 to IDO1 dysregulation and tumoral immune escape (352) provided experimental substantiation for the corollary proposition that tumor cells might, by inducing IDO1, appropriate this mechanism of protection for the ‘foreign’ fetus to overcome immunosurveillance. Within the complex inflammatory milieu of the tumor microenvironment, IDO1 induction is not necessarily restricted to tumor cells, and non-malignant stromal cells expressing IDO1 can promote tumoral immune escape as well (353). In particular, IDO1 induction in antigen-presenting cells, such as dendritic cells and macrophages, has been implicated in promoting immune tolerance by suppressing effector cytotoxic T lymphocytes (CTL), converting naïve T lymphocytes to FoxP3+ Treg cells and elevating the suppressive activity of “natural” Treg (354).
A great deal of attention is now focused on the therapeutic potential of small molecule inhibitors of IDO1 for treating cancer patients (355), particularly in combination with cancer chemotherapy or “immune checkpoint” antibodies (352,356). It is not yet clear how IDO1 may influence host interactions with the microbiome, but there has been much attention to the related topic of its role in the host response to infection by various pathogens, which has been a topic of interest for a number of years. Indeed, well prior to findings of its involvement in immune modulation, it was noted that intraperitoneal administration of bacterial LPS could induce IDO1 activity in the lungs of mice by 30- to 50-fold (357). This initial indication that IDO1 induction might be associated with the inflammatory response to microbial infection was followed by reports of pulmonary IDO1 induction in response to virus infection (358) and IFNγ (359).
Since IFNγ plays a major role in controlling a variety of infections, the finding that IDO1 is highly responsive to IFNγ spurred investigations addressing whether IDO1 might have a downstream anti-microbial effector role. In 1984, Pfefferkorn and colleagues reported that tryptophan degradation was responsible for the IFNγ-mediated restriction of the growth of the obligate intracellular protozoan Toxoplasma gondii in human fibroblasts (359). IFNγ-mediated restriction of the growth of the obligate, intracellular, gram-negative bacterium Chlamydophila psittaci was likewise linked to tryptophan deprivation (360). These studies focused attention on IFNγ- elicited tryptophan deprivation resulting from the induction of IDO1 as mediating the anti-proliferative effect on these intracellular pathogens. This assessment, that IDO1 provides a beneficial effect in combating infections, was complicated by additional studies demonstrating the activation of genes for Chlamydia persistence triggered by IDO1 mediated tryptophan depletion. Because the bacterium is sensitive to the antibiotic treatment only when it is metabolically active, IDO1 activity in this context was detrimental to clearing infections leading to the suggestion that tryptophan supplementation might help overcome antibiotic resistance (361). Furthermore, while the effects of IDO1 on microorganisms were initially attributed to depletion of tryptophan, evidence of microbial effects produced by downstream tryptophan metabolites, from what is collectively referred to as the kynurenine pathway, were also reported (362,363). In particular, 3-hydroxykynurenine was shown to suppress the proliferation of Staphylococcus aureus in vascular allografts (364), and both 3-hydroxykynurenine and 3-hydroxyanthranilic acid were found to contribute to controlling the replication of Trypanosoma cruzi in mice (365). In a single report, treatment with 1MT produced three different outcomes depending on the nature of the infection, exacerbating T. gondii toxoplasmosis, restraining L. major leishmaniasis, and having no apparent effect on HSV-1 replication or latency (366). In aggregate, the implication from these studies of infectious pathogens is that the overall impact of IDO1 activity, both in terms of tryptophan depletion and the production of various metabolites, on the diverse ecology of the commensal microbiome is likely to be complex and contextual.
Perhaps even more consequential than the direct effects of IDO1 activity on particular microbes are the effects that IDO1 can exert on the overall inflammatory environment. In accord with its ability to elicit T cell suppression, the general assumption has been that IDO1 should act in an immunosuppressive manner to limit the severity of inflammation. Data supporting this interpretation have been reported in a mouse model of chronic granulomatous disease in which defective IDO1 function in mice lacking an essential component of NADPH oxidase, p47phox, was implicated in the exaggerated inflammatory response to infection with Aspergillus fumigatus (367). The more severe illness was associated with higher numbers of IL17-producing γδT cells and fewer IL10 producing αβTreg, which could be reversed by the provision of exogenous kynurenine. However, as with the effects of tryptophan catabolism on microorganisms, the categorization of IDO1 as strictly immunosuppressive may be an oversimplification (368). In a mouse model of chemical carcinogenesis, genetic loss of IDO1 did not exacerbate inflammation in response to phorbol ester-elicited tumor promotion, as would be expected if it were broadly immunosuppressive, but did result in a dramatically reduced incidence of premalignant lesions (369). Perhaps even more strikingly, in a mouse model of rheumatoid arthritis (RA), 1MT treatment suppressed rather than exacerbated joint inflammation (370), while in a contact hypersensitivity model, genetic loss of IDO1 resulted in diminished ear swelling (371). Why IDO1 has such varied effects on the inflammatory response remains to be fully elucidated, but suggests that the outcome of the interactions with the complex microbiome may be contextual and difficult to predict.
The study in the RA model noted above also highlights a particular complication with interpreting results of the many studies that have relied on the use of the compound 1MT to inhibit IDO1 activity. Biochemical and pharmacological evaluation of this compound clearly indicates that it is not directly inhibiting the enzyme at the dose ranges administered in vivo, and it is able to signal as a mimetic for tryptophan sufficiency and thereby interfere with activation of downstream response pathways (372). However, a tryptophan deficiency signal can be provided by any of the enzymes that catalyze tryptophan degradation (IDO1, IDO2, TDO2 as well as TPH, the latter of which initiates an alternate pathway of tryptophan catabolism to serotonin). Therefore, 1MT is not a valid tool to discriminate which of these particular enzymes is involved. Indeed, in the RA model, genetic analysis revealed that the recently identified paralog IDO2, and not IDO1, is likely to be responsible for the effect of 1MT on joint inflammation, as the effect of 1MT administration was phenocopied in mice lacking IDO2 but not IDO1 (370,373). Studies using later generation, direct enzyme inhibitors (355), coupled with studies in genetically modified animals (371,374,375) can overcome ambiguities in data interpretation associated with 1MT treatment. Indeed, a recent study of LPS responses utilizing the IDO1, IDO2 and Tdo2 gene deletion mouse strains provides confirmatory evidence that these three genes have distinct, non-overlapping roles in the host immune response to this microbial signal (376).
While the regional microbiome present at all barrier surfaces is likely to influence immunity locally, the microbiome of the gastrointestinal tract is of particular interest because of the broader role it has been found to play in shaping systemic immune homeostasis (377). Correspondingly, current microbiome research is largely focused on the gastrointestinal tract, where commensal microorganisms have been found to contribute to host defense by limiting the growth of enteric pathogens and producing symbiosis factors that control intestinal inflammation and pathology (377). Evidence implicating IDO1 in this process has come from a study of the protective capacity of Lactobacillus salivarius, which is abolished in mice lacking the gene encoding NOD2, an intracellular pattern-recognition molecule that regulates inflammatory pathways in response to detection of bacterial peptidoglycans. In this model, IDO1 upregulation was found to correlate with NOD2-dependent protection (378). The increased regulatory complexity imposed by the gut microbiome may help also explain counterintuitive findings associated with IDO1 in this tissue. In a dextran sodium sulfate/1,2-dimethylhydrazine (DSS/DMH) elicited model of colon carcinogenesis, genetic loss of IDO1 resulted in increased tumor frequency (379), unlike other organ systems in which IDO1 loss has been associated with resistance to carcinoma development (380,381). This outcome is similar to the atypical impact on tumor formation ascribed to Treg in the gut, where their presence appears to be protective against carcinogenesis (382), despite evidence that Treg are generally associated in other organs with tumor promotion. It has been proposed that the effect of immunosuppressive mechanisms on inflammatory pathology of the gut may be quite different depending on whether there is any initial involvement of tissue damage, as the resulting microbial translocation produces a severe tumor-promoting inflammation (383). Under these circumstances, dampening the inflammatory response via immune suppressive mechanisms may provide a more consequential benefit that overshadows any detrimental role in promoting immune escape. This interpretation is consistent with the findings of two otherwise apparently contradictory studies in colitis models, where IDO1 blockade resulted in augmented colitis induced by trinitrobenzene sulfonic acid (384), but diminished colitis induced by Citrobacter rodentium (385).
In conjunction with the complex immune regulatory effects attributed to IDO1 activity in the gut, tryptophan metabolites produced by the microbiota affect mucosal reactivity. When switching from sugar to tryptophan as an energy source, the highly adaptive lactobacilli in the gut expand and produce an aryl hydrocarbon receptor (AhR) ligand, indole-3-aldehyde, that contributes to AhR-dependent IL-22 transcription. The resulting IL-22-dependent mucosal response promotes the survival of mixed microbial communities, while providing colonization resistance to the fungus Candida albicans and mucosal protection from inflammation. This example of coevolutionary commensalism through the microbiota-AhR axis represents yet another way in which tryptophan catabolism appears to be involved in fine tuning host mucosal reactivity (386). As further investigations into the physiological and pathophysiological interactions between IDO1, the commensal microbiome and host immunity are conducted, the indications from these early studies are that IDO1 is likely to play an integral but contextual role at the interface between homeostasis and dysbiosis.
Microbiome and cancer
The interplay between microbes, cancer and the immune system is in no manner fully defined. However, accumulating evidence argues provocatively that microbes exert a variety of functions on host oncogenesis, tumor progression and response to immunotherapy. Thus, selectively manipulating the gut microbiome is a critical parameter to consider in the ongoing battle against established cancers.
The metabolic potential of the gut microbiota is now regarded as vital to the process of malignant transformation. Disruption of the intimate relationship between the host and intestinal bacteria, known as dysbiosis, can affect oncogenesis, tumor progression, and response to cancer therapy. Dysbiosis can occur for several reasons: 1) direct occupancy of unwanted, foreign microbes (as discussed above) that outcompete friendly gut flora, 2) a response to immunosenescence with aging, and 3) direct environmental insults such as antibiotics and smoking (387). In the setting of chronic autoimmune processes such as Crohn's disease and UC, the integrity of gut epithelial, myeloid, and lymphoid components are disrupted (Fig. 3). These chronic insults ultimately increase host's risk for neoplastic transformation (388). Indeed, several factors that favor carcinogenesis similarly recapitulate dysbiosis.
Figure 3. Dysbiosis: an immunocompromised state characterized by pathobiont colonization that leads to hyperinflammation, dysplasia and tumorigenesis.
(Left) Symbiosis: A symbiotic gut microbiota operates under a functional intestinal epithelial cell (IEC) barrier, with steady state proportions of mucus, pattern recognition receptors (PRRs), antimicrobial peptides, and secretory IgA, which in turn contain the microbiota in the intestinal lumen. Under tight control by IECs, the intestinal immune system within the gut lamina propria becomes largely tolerant to the resident commensals. Signaling cascades that occur downstream of toll-like receptors (TLRs) are used by IECs to detect microbes through PRRs. Upon lipopolysaccharide (LPS) stimulation of TLRs, the MYD88 protein is recruited, activating the NF-κB pathway, leading to production of antimicrobial proteins and proinflammatory cytokines. In a symbiotic gut, IECs are desensitized by repeated exposure to LPS or are attenuated by LPS-mediated downregulation of the IL-1 receptor–associated kinase 1 (IRAK1), an activator of the NF-κB cascade. Exposure to LPS induces epithelial cells to secrete TGF-β, B-cell-activating factor of the TNF family (BAFF), and a proliferation-inducing ligand (APRIL), all of which promote the development of tolerogenic responses to the microbiota. CD103+ dendritic cells (DCs) support the development of regulatory T (Treg) cells to secrete IL-10 and TGF-β, and together they stimulate the production of commensal-specific IgA. (Right) Dysbiosis: Increased intestinal exposure of diverse PAMPs, pro-inflammatory cytokines, apoptotic debris, and toxins leads to microbial dysbiosis and overgrowth of “pathobionts”, transformed symbiotic bacteria now under pathologic conditions. Pathobiont overgrowth leads to the loss of barrier integrity and a breach in the IEC barrier. Translocation of bacteria and bacterial components triggers the intestinal immune system through TLR activation, resulting in potentially harmful effector T cell responses set to clear invading bacteria. Ultimately, the secretion of IL-1 and IL-6 from IECs fuels a TH1 and TH17 response by DCs and macrophages and leads to higher levels of commensal-specific IgG by B cells.
One well-studied model of the dysbiosis/cancer connection is that of repeated intra-abdominal infections, the use of antibiotics, or both leading to an increased incidence of colorectal cancer (389). In several preclinical studies, interventions that abrogate or directly alter gut microbiome composition increase the incidence and progression of colorectal carcinoma in both genetic and carcinogen-induced models of tumorigenesis (390-392). Moreover, several by-products of the gut microbiota directly target intestinal epithelial cells and either facilitate oncogenesis (as reported for hydrogen sulfide and the Bacteroides fragilis toxin) or suppress tumorigenesis (in the case of SCFAs) (393). Intestinal microbes have been characterized to participate in more than just colorectal carcinogenesis. Experimental models of gut flora also elucidate the development of other extraintestinal cancers such as hepatocellular carcinoma (394,395), presumably through systemically disseminated metabolic networks. Helicobacter pylori is a Gram-negative bacterial pathogen that selectively colonizes the gastric epithelium. It is postulated that half of the world's population is infected with H. pylori, though colonization of the pathogenic bacteria does not cause any symptoms in majority of the population. Nevertheless, long-term carriage of H. pylori significantly increases the risk of developing diseases. Among infected individuals, approximately 10% develop peptic ulcer disease, 1% to 3% develop gastric adenocarcinoma, and <0.1% develop mucosa-associated lymphoid tissue (MALT) lymphoma. However, at initial stages, gastric MALT lymphoma can be cured completely by eradication of H. pylori with antibiotics (396).
Apart from antibiotics, probiotics may also inhibit tumorigenesis and cancer progression. Konishi et al. (397) reported that the culture supernatant of Lactobacillus casei has tumor-suppressive effect on colon cancer cells. The authors reported that ferrichrome produced by L. casei is the molecule that provides tumor protection and is exerted via the JNK signaling pathway.
The etiology of breast cancer is still not understood, though it is believed the disease is due to a combination of both genetic and environmental factors. It is posited that environmental factors influence breast cancer as there is an increased incidence of breast cancer among migrants and their descendants after they move from a region of low breast cancer risk to a region of high risk. Bacterial communities within the host could be one such environmental factor that may influence breast cancer development. Different bacterial profiles in breast tissue exist between healthy women and those with breast cancer. Breast cancer patients had higher levels of Bacillus, Enterobacteriaceae and Staphylococcus. E.coli and Staphylococcus epidermidis isolated from breast cancer patients induced DNA double-stranded breaks in HeLa cells. There was also a decrease in some lactic acid bacteria, known for their beneficial health effects, including anticarcinogenic properties (398). It has been demonstrated that women who drink fermented milk products have a reduced risk of breast cancer development (399). Oral administration of Lactobacillus species has been shown to be protective in animal models of breast cancer (400).
Case studies back to the 1700s have recounted the development of bacterial infections in cancer patients that led to remissions of their malignant disease. One of the pioneers in this field, the U.S. surgeon William B. Coley, engaged in a lifelong study of this phenomenon after the loss of his very first patient in the late 1800s to a rapidly invasive sarcoma (401). Searching the literature available, Coley discovered records of another sarcoma patient with relentless sarcomatous recurrences following surgical resection and an ultimate wound infection (erysipelas) with Streptococcus pyogenes and high fever. To his surprise, after each attack of fever, the ulcer improved, the sarcoma shrank, and the lesion ultimately regressed completely. Coley suspected that in some manner the infection had induced tumor regression and began a series of trials to “cure” his cancer patients with pathogen inoculation. He infected his next 10 patients, but observed intrinsic variability in efficacy using this method(402). Due to this unpredictability, he elected to create a formulation containing two killed bacteria: Streptococcus pyogenes and Serratia marcescens. Under the form of an inactivated vaccine, he could simulate an infection (inflammation, chills, fever) without the actual risks of a life-threatening disease. This vaccine became known as “Coley toxins.” The relative success with Coley's vaccine was by no means limited to sarcomas. For decades, this vaccine form had been used by other contemporaries for carcinomas, lymphomas, melanomas and myelomas (403,404).
However, with the advent of radiotherapy and chemotherapy, and the empowerment of the U.S. FDA in 1964 that restricted clinical use of “Coley toxins,” the use of microbial toxins in oncology fell out of use. There were, however, rare instances in which this line of thinking endured and eventually received FDA approval. Perhaps the most prominent example is the use of Bacillus Calmette-Guerin (BCG) for the treatment for superficial bladder cancer (405). BCG is currently the only conventional bacterial vaccine in use for direct tumor killing. Unlike Coley toxins, BCG is not administered with the ultimate goal of induced fever. But similar to Coley's methods, the vaccine is applied directly to the tumor site with repeated courses following initial resection to prevent recurrence (406). After intravesicular administration of this vaccine, a wide range of cytokines become detectable in the urine, including interleukins-(IL)-1, IL-2, IL-6, IL-8, IL-10, IL-12, IL-18, interferon-γ, interferon-γ inducible protein-10, macrophage colony stimulating factor, and TNF-α (407-409). This inflammatory host response illustrates the point that individual immunomodulating cytokines are partial components of a much more complex immunological response to infection, and correspondingly, tumor regression. Some insights were gleaned from Coley's toxins and other historical reports on live or attenuated bacterial inoculations. One was that a local inoculation produces only a local response. Thus, BCG use is limited to superficial bladder cancer. The heat and immune activation associated with local inflammation are perceived to be a minimized febrile response, and correspondingly this local response is only effective in the immediate region where it occurs (401). Since Coley's passing, the field of tumor immunology has developed into a better founded and more sophisticated specialty, with investigators not only employing variety of basic immunologic principles (e.g., antitumor cytokines, cytotoxic T cells, immune-stimulatory antibodies, cell-based vaccines) but pivotal insights into the dominance of tumoral immune suppression in blunting the effectiveness of any immunotherapy. Overcoming this historical source of failure in the efficacy of cancer immunotherapy is empowering this field anew today.
There are more than a hundred chemotherapy drugs to treat many types of cancers. However, it is not fully understood the mechanism of some of these drugs. Cyclophosphamide is a clinically important chemotherapeutic cancer drug that stimulates anti-tumor immune responses. Viaud et al. (410) demonstrated that cyclophosphamide alters the composition of microbiota in the gut and induces the translocation of several Gram positive bacteria into mesenteric lymph nodes and spleen. In the lymphoid organs the Gram positive bacteria stimulated the generation of pathogenic T helper 17 (pTh17) cells and memory Th1 immune responses. Germ-free tumor-bearing mice or treated with antibiotics to kill Gram positive bacteria showed a reduction in T cell responses and their tumors were resistant to cyclophosphamide. Adoptive transfer of pTh17 cells restored the anti-tumor efficacy of cyclophosphamide. Overall, the study suggested that the gut microbiota help shape the anticancer immune response.
Turning the tables: using engineered microbes to attack cancer
Over the past century, knowledge gained on how selective microbes either facilitate the growth of cancer or alternatively act as tumoricidal agents permits us open access to utilize this double-edged sword to our advantage. Such interventions have already begun and span several modalities, including but not limited to the use of helper peptide sequences from bacterial subunits, bacterial toxin-fusion proteins, oncolytic viral vaccines, and, as recent studies elucidate, leveraging the metabolism of host flora to potentiate immune-modulators.
Broadly defined, an immunotherapeutic is any modality that manipulates the immune system for enhanced therapeutic outcome. These include non-specific activation of the immune system with microbial components or cytokines; antigen-specific adoptive immunotherapy with antibodies or lymphocyte transfers; and active immunotherapy by direct vaccination against tumor-specific proteins, or antigens. We traditionally regard vaccines as educators for the naïve immune system that are administered prophylactically in anticipation of any infection. However, in the setting of aggressive cancers, the use of cellular vaccines to mount reactive immune responses is now being employed with promising results.
A variety of cancer vaccines are currently under investigation, but perhaps the most widely investigated to date are 1) cellular vaccines composed of APC loaded with tumor antigen (411-414) and 2) peptide vaccines (415,416). Peptide vaccines are comprised of 8–25 amino acids that encompass an epitope - a recognizable sequence coding for an antigen. The transient nature and low magnitude of responses in many cancer patients has elucidated that tumors themselves are inherently proficient at downplaying immune responses as well as escaping antigen recognition altogether. Thus, there is an urgent need for improving vaccine immunogenicity and for ensuring that cancer antigens are sufficiently immunogenic. To enhance peptide vaccine immunogenicity, these small peptides are often conjugated to a carrier protein, such as keyhole limpet hemocyanin (417,418) and tetanus toxoid (419,420). These helper proteins enable recognition by and activation of the immune system with great potency and generate complimentary bystander activation with cytokine release and maturation of effector cell phenotype. Peptide vaccines are appealing in cancer therapy because they are relatively easy to manufacture and store, and they do not require laborious preparations. Due to their “off-the-shelf” feature, repeated boosting for enhanced immune activation also distinguishes peptide cancer vaccines as an expandable modality (421).
Adoptive immunotherapy has also been utilized to eradicate established tumors (422). This process involves ex vivo activation of autologous immune cells, isolated from either peripheral blood or intratumoral lymphocytes (423,424), into lymphocyte-activated killer cells. Lymphocyte-activated killer cells are generated by culturing autologous peripheral lymphocytes with IL-2, a vital growth cytokine for generating T cells and NK cells. These killer cells are then returned to the patient intratumorally or intravenously, where they become activated by host APCs and exert their tumoricidal effects. Using the knowledge we have gained from microbial components, NK cells have been potentiated by leveraging the cytolytic capacity of microbial diphtheria toxin. One study demonstrated that haploidentical NK cells for relapsed and refractory acute myeloid leukemia could be augmented and improved with a lymphodepletive platform using diphtheria toxin conjugated to IL-2. Using the immunotoxin IL2DT to deplete immunosuppressive Treg, investigators appreciably improved rates of in vivo NK-cell expansion (10% versus 27%) and AML 28-day remission (53% vs 21%; P= 0.02) compared to the cohort without IL2DT (425).
Oncolytic viruses represent another immunotherapy modality that has gained recent traction in the field of tumor immunology. Use of these microbes was first based on early reports of spontaneous cancer remissions coincident with natural infection or upon the use of live attenuated vaccines (426). Since then, an improved understanding of the molecular basis for viral host cell tropism, cytotoxicity, and cell type-specificity has opened up avenues for the very selective design of virally-based anti-cancer strategies. To be efficacious and safe, an oncolytic virus must possess 1) an inherently low human pathogenic potential (i.e. the orphan reovirus [(427)]), 2) a veterinary pathogen with unknown human pathogenicity (i.e. vesicular stomatitis virus [(428)]), or 3) a human pathogen genetically engineered to selectively kill cancerous cells without collateral cytotoxicity in normal cells (i.e. herpes simplex virus-1 [(429)]). One such group is utilizing a prototype nonpathogenic poliovirus recombinant, known as PVSRIPO. Poliovirus naturally targets the vast majority of ectodermal/neuroectodermal cancers expressing its cellular receptor, CD155. Evidence from glioblastoma patients suggests that the CD155 receptor is ectopically upregulated on tumor cells. Preclinical studies have shown that treatment of glioma xenografts with intratumoral inoculation of PVS-RIPO produced rampant tumor cell death, potent host-mediated inflammatory reactions against infected tumors, and rapid tumor decline (430,431). The use of PVS-RIPO is now being evaluated in Phase I setting for recurrent glioblastoma (432).
As multiple studies are being carried out yearly, investigators continue to uncover key barriers to efficacy that further clarify our strategies. As with any antigen-specific immune response, several homeostatic mechanisms remain at play to prevent rampant damage to the host or autoimmune toxicity. One of the most exciting regulatory axes to be studied recently is that of programmed death (PD)-1/PD-ligand-1 (PD-L1). PD-1 is a co-inhibitory receptor that is inducibly expressed by T and B cells upon activation. Antigen-specific T cells expressing the PD-1 receptor will engage with either of its ligands, PD-L1 or PD-L2 expressed on APC, which elicits an inhibitory cascade and subsequent inhibition of T cell receptor (TCR)-induced cytokine production and proliferation (433). PDL-1 is expressed in several other cell types, including tumor cells and some epithelial cells, lymphoid cells, and myeloid cells (434). Another axis involved in regulating self-recognition is that of cytotoxic T-lymphocyte antigen-4 (CTLA-4). Studies have demonstrated that tumor cells stimulate CTLA4, promoting a cascade of inhibitory immune processes and ultimate T cell inactivity against tumors themselves (435,436).
In cancer immunotherapy, monoclonal antibodies (mAb) against the immune checkpoints CTLA-4, PD-1 and its ligand PD-L1 have demonstrated high activity in melanoma and other tumors (437). Ipilimumab, an anti-CTLA-4 antibody, was the first approved “immune checkpoint inhibitor.” Although the response rate with ipilimumab is low (less than 20% of patients have objective responses), many of those positive responses were associated with long-term survival (438), with similar results in the first and second line settings. Nivolumab and pembrolizumab, both anti-PD-1 inhibitors, have now also been approved for the treatment of melanoma, with response rates of up to 40% and a demonstrated survival advantage in Phase III trials (434).
Strikingly, recent findings in preclinical models of cancer stress the importance of intact gut microbiota for effective immune checkpoint blockade. One recent study found that antitumor effects of CTLA-4 blockade depended on the presence of distinct Bacteroides species. In both mice and patients, T-cell responses specific for B. thetaiotaomicron or B. fragilis were associated with the efficacy of CTLA-4 blockade. Using antibiotic-treated as well as germ-free mice, tumors lacking these strains did not respond to CTLA blockade. This deficiency was rescued by B. fragilis gavage, by immunization with B. fragilis polysaccharides, or by adoptive transfer of B. fragilis–specific T cells. Ultimately, fecal microbial transplantation from humans to mice confirmed that treatment of melanoma patients with antibodies against CTLA-4 favored the outgrowth of B. fragilis with anticancer properties (439). A different preclinical study similarly elucidated that mice treated with gut commensals of Bifidobacterium displayed significantly improved suppression of melanoma growth in comparison with non-Bifidobacterium treated counterparts. These observed differences in spontaneous antitumor immunity were eliminated upon co-housing or after fecal transfer, eluding to the importance of shared bacterial colonization. Furthermore, administration of the bacteria to Bifidobacterium-naïve mice with established melanoma significantly enhanced tumor-specific immunity and response to anti-PD-L1 mAb therapy (440). Although the mechanisms from both these studies are not fully understood, they laid the foundation behind the requirement for an intact microbiome to enact antitumor responses. One key observation from both these studies was that they employed subcutaneous tumor models, meaning that intestinal microbiota exerted antitumor immunity in a systemic fashion. Both studies relied on the presence of CD8+ T cells. Secondly, both demonstrate that altered DC activation was a responsible intermediate event between the presence of gut microbiota and provision of checkpoint inhibitors (Fig. 4).
Figure 4. Gut microbiome directs the efficacy of immune checkpoint therapy.
Both anti-CTLA-4 and anti-PD-L1 therapies rely on gut microbiota for efficacy in immune activation. Anti-PD-L1 therapy has been shown to rely on the pre-existence of sufficient Bifidobacterium species, which are also thought to augment responses via PD-L1 binding on antigen-presenting cells such as DCs and macrophages. Subsequent ligation results in the prevention of suppressive signals to PD-1-expressing T cells. Similarly, anti-CTLA-4 indirectly alters the intestinal flora and enriches the Bacteroides species, possibly by promoting deterioration of the IEC barrier via activation of local lymphocytes. These bacteria then promote the activation of DCs, which present tumor antigens to prime and maintain anti-tumor T cell responses. Anti-CTLA-4 holds additional activation functions, including 1) preventing CTLA-4 from blocking activation of the co-stimulatory molecule CD28 on T cells and 2) blocking the immune-suppressive function of Tregs, which are required in deactivation of immune responses against tumors.
Conclusions
It is humbling to consider how much biomedical research has been conducted since the molecular biology revolution of the past century without appreciation of the importance of microbiomes in health and disease. Like all realms of biomedical investigation, the field of cancer research can no longer ignore the ‘other half’ of the organism; it must become as familiar with the genetics, biology, physiology and immunologic effects of host microorganisms as with the hosts themselves. In considering sources of experimental irreproducibility in biomedical publications that have been suggested recently to be disturbingly high, it seems likely that natural variations in microbiome infections present in experimental models and vivariums at different sites will provide one more challenge to the exquisitely difficult problem of how one defines a “molecular mechanism” in disease. More focus on practical applications (sought by most funding organizations) along with empirical explorations once traditional to biology may offer two paths forward, since, to paraphrase the pragmatic American philosopher Charles S. Peirce, “You know something if you can do something.” How the present obsession with molecular mechanism will change in the face of the challenge the microbiome poses to preclinical research is unclear. Nevertheless, as the molecular biology revolution continues to wash up on the shores of reductionism this century, it will be impossible not to re-embrace the roots of traditional biological thought, where ecology, evolution and a focus on emergent principles in complex organisms can help re-center the pursuit of new knowledge and its applications to improve disease management and healthful lifespans.
Acknowledgments
The laboratories of Jacques Izard are supported by National Institutes of Health (NIH) grants R01CA202704 and Nebraska Tobacco Settlement Biomedical Research Development Funds. The APC Microbiome Institute is supported by Science Foundation Ireland (SFI) (Grant Number SFI/12/RC/2273) and has conducted studies in collaboration with several companies including GSK, Pfizer, Wyeth and Mead Johnson. Dr Clarke's contribution to this review was specifically supported by the Irish Health Service Executive (Grant number HaPAI/2015/GC). Sunil Thomas, Alexander J. Muller, James M. Mullin, George C. Prendergast acknowledge support from the Lankenau Medical Center Foundation, the Women's Board of Lankenau Medical Center and Main Line Health. The content of this review was neither influenced nor constrained by funding from these Organizations.
Footnotes
Conflicts of interest: Conflicts of Interest: E. Walsh is an employee of Seres Therapeutics. D.A. Sela has ownership interest in a patent (2010/0113383 A1) and receives honoraria for speaking engagements. G.C. Prendergast has a commercial research grant from Janssen Pharmaceuticals, ownership interest in New Link Genetics Inc., Meditope Biosciences Inc., Dynamis Pharmaceuticals, and Man's Best Friend Therapeutics, and serves on consultant/advisory boards for New Link Genetics Inc., Ribonova Inc., Kyn Therapeutics Inc., Vitae Pharmaceuticals, Biogen Inc., OrbiMed Advisors LLC, and Guidepoint Global LLC. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science (New York, NY) 2009;326(5960):1694–7. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lederberg J, McCray AT. 'Ome Sweet 'Omics—a genealogical treasury of words. Scientist. 2001;15:8. [Google Scholar]
- 3.Margulis L. Symbiogenesis and symbionticism. In: Margulis L, Fester R, editors. Symbiosis as a source of evolutionary innovation: speciation and morphogenesis. MIT Press; Cambridge: 1991. pp. 1–14. [PubMed] [Google Scholar]
- 4.Bordenstein SR, Theis KR. Host Biology in Light of the Microbiome: Ten Principles of Holobionts and Hologenomes. PLoS biology. 2015;13(8):e1002226. doi: 10.1371/journal.pbio.1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sender R, Fuchs S, Milo R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans Cell. 2016;164(3):337–40. doi: 10.1016/j.cell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 6.Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489(7415):231–41. doi: 10.1038/nature11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. Science (New York, NY) 2006;312(5778):1355–9. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meadow JF, Altrichter AE, Bateman AC, Stenson J, Brown GZ, Green JL, et al. Humans differ in their personal microbial cloud. PeerJ. 2015;3:e1258. doi: 10.7717/peerj.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–7. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korem T, Zeevi D, Suez J, Weinberger A, Avnit-Sagi T, Pompan-Lotan M, et al. Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science (New York, NY) 2015;349(6252):1101–6. doi: 10.1126/science.aac4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graf D, Di Cagno R, Fak F, Flint HJ, Nyman M, Saarela M, et al. Contribution of diet to the composition of the human gut microbiota. Microbial ecology in health and disease. 2015;26:26164. doi: 10.3402/mehd.v26.26164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zimmer J, Lange B, Frick JS, Sauer H, Zimmermann K, Schwiertz A, et al. A vegan or vegetarian diet substantially alters the human colonic faecal microbiota. European journal of clinical nutrition. 2012;66(1):53–60. doi: 10.1038/ejcn.2011.141. [DOI] [PubMed] [Google Scholar]
- 13.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–63. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parte AC. LPSN--list of prokaryotic names with standing in nomenclature. Nucleic acids research. 2014;42(Database issue):D613–6. doi: 10.1093/nar/gkt1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. The Human Oral Microbiome. J Bacteriol. 2010;192(19):5002–17. doi: 10.1128/JB.00542-10. JB.00542-10 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson KE, Weinstock GM, Highlander SK, Worley KC, Creasy HH, Wortman JR, et al. A catalog of reference genomes from the human microbiome. Science (New York, NY) 2010;328(5981):994–9. doi: 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.A framework for human microbiome research. Nature. 2012;486(7402):215–21. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouckaert R, Heled J, Kuhnert D, Vaughan T, Wu CH, Xie D, et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS computational biology. 2014;10(4):e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang K, Brady A, Mahurkar A, White O, Gevers D, Huttenhower C, et al. MetaRef: a pan-genomic database for comparative and community microbial genomics. Nucleic acids research. 2014;42(Database issue):D617–24. doi: 10.1093/nar/gkt1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerepesi C, Banky D, Grolmusz V. AmphoraNet: the webserver implementation of the AMPHORA2 metagenomic workflow suite. Gene. 2014;533(2):538–40. doi: 10.1016/j.gene.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 21.Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, Pasolli E, et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nature methods. 2015;12(10):902–3. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 22.Stackebrandt E. Forces shaping bacterial systematics. Microbe. 2007:2. [Google Scholar]
- 23.Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, et al. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic acids research. 2014;42(Database issue):D643–8. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zuo G, Xu Z, Hao B. Phylogeny and Taxonomy of Archaea: A Comparison of the Whole-Genome-Based CVTree Approach with 16S rRNA Sequence Analysis. Life (Basel, Switzerland) 2015;5(1):949–68. doi: 10.3390/life5010949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database : the journal of biological databases and curation. 2010;2010 doi: 10.1093/database/baq013. baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic acids research. 2014;42(Database issue):D633–42. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome biology. 2012;13(6):R42. doi: 10.1186/gb-2012-13-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Y, Gao H, Mihindukulasuriya KA, La Rosa PS, Wylie KM, Vishnivetskaya T, et al. Biogeography of the ecosystems of the healthy human body. Genome biology. 2013;14(1):R1. doi: 10.1186/gb-2013-14-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agirbasli H, Ozcan SA, Gedikoglu G. Fecal fungal flora of pediatric healthy volunteers and immunosuppressed patients. Mycopathologia. 2005;159(4):515–20. doi: 10.1007/s11046-005-3451-2. [DOI] [PubMed] [Google Scholar]
- 30.Scanlan PD, Marchesi JR. Micro-eukaryotic diversity of the human distal gut microbiota: qualitative assessment using culture-dependent and -independent analysis of faeces. The ISME journal. 2008;2(12):1183–93. doi: 10.1038/ismej.2008.76. [DOI] [PubMed] [Google Scholar]
- 31.Chen Y, Chen Z, Guo R, Chen N, Lu H, Huang S, et al. Correlation between gastrointestinal fungi and varying degrees of chronic hepatitis B virus infection. Diagnostic microbiology and infectious disease. 2011;70(4):492–8. doi: 10.1016/j.diagmicrobio.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 32.Hamad I, Sokhna C, Raoult D, Bittar F. Molecular detection of eukaryotes in a single human stool sample from Senegal. PloS one. 2012;7(7):e40888. doi: 10.1371/journal.pone.0040888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gouba N, Raoult D, Drancourt M. Plant and fungal diversity in gut microbiota as revealed by molecular and culture investigations. PloS one. 2013;8(3):e59474. doi: 10.1371/journal.pone.0059474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamad I, Raoult D, Bittar F. Repertory of eukaryotes (eukaryome) in the human gastrointestinal tract: taxonomy and detection methods. Parasite immunology. 2016;38(1):12–36. doi: 10.1111/pim.12284. [DOI] [PubMed] [Google Scholar]
- 35.Parfrey LW, Walters WA, Lauber CL, Clemente JC, Berg-Lyons D, Teiling C, et al. Communities of microbial eukaryotes in the mammalian gut within the context of environmental eukaryotic diversity. Frontiers in microbiology. 2014;5:298. doi: 10.3389/fmicb.2014.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Irlinger F, Layec S, Helinck S, Dugat-Bony E. Cheese rind microbial communities: diversity, composition and origin. FEMS microbiology letters. 2015;362(2):1–11. doi: 10.1093/femsle/fnu015. [DOI] [PubMed] [Google Scholar]
- 37.Miceli MH, Diaz JA, Lee SA. Emerging opportunistic yeast infections. The Lancet Infectious diseases. 2011;11(2):142–51. doi: 10.1016/s1473-3099(10)70218-8. [DOI] [PubMed] [Google Scholar]
- 38.Summers RW, Elliott DE, Urban JF, Jr, Thompson RA, Weinstock JV. Trichuris suis therapy for active ulcerative colitis: a randomized controlled trial. Gastroenterology. 2005;128(4):825–32. doi: 10.1053/j.gastro.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 39.Wammes LJ, Mpairwe H, Elliott AM, Yazdanbakhsh M. Helminth therapy or elimination: epidemiological, immunological, and clinical considerations. The Lancet Infectious diseases. 2014;14(11):1150–62. doi: 10.1016/s1473-3099(14)70771-6. [DOI] [PubMed] [Google Scholar]
- 40.Hug LA, Baker BJ, Anantharaman K, Brown CT. A new view of the tree of life. 2016;1:16048. doi: 10.1038/nmicrobiol.2016.48. [DOI] [PubMed] [Google Scholar]
- 41.Manrique P, Bolduc B, Walk ST, van der Oost J, de Vos WM, Young MJ. Healthy human gut phageome. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(37):10400–5. doi: 10.1073/pnas.1601060113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.ICTV. The International Committee on Taxonomy of Viruses. 2015 http://wwwictvonlineorg.
- 43.Reteno DG, Benamar S, Khalil JB, Andreani J, Armstrong N, Klose T, et al. Faustovirus, an asfarvirus-related new lineage of giant viruses infecting amoebae. Journal of virology. 2015;89(13):6585–94. doi: 10.1128/jvi.00115-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghabrial SA, Caston JR, Jiang D, Nibert ML, Suzuki N. 50-plus years of fungal viruses. Virology. 2015;479-480:356–68. doi: 10.1016/j.virol.2015.02.034. [DOI] [PubMed] [Google Scholar]
- 45.Snyder JC, Bolduc B, Young MJ. 40 Years of archaeal virology: Expanding viral diversity. Virology. 2015;479-480:369–78. doi: 10.1016/j.virol.2015.03.031. [DOI] [PubMed] [Google Scholar]
- 46.Pickett BE, Sadat EL, Zhang Y, Noronha JM, Squires RB, Hunt V, et al. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic acids research. 2012;40(Database issue):D593–8. doi: 10.1093/nar/gkr859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: a fast phage search tool. Nucleic acids research. 2011;39(Web Server issue):W347–52. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gardy J, Loman NJ, Rambaut A. Real-time digital pathogen surveillance - the time is now. Genome biology. 2015;16(1):155. doi: 10.1186/s13059-015-0726-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–80. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Consortium HMP. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Massana R, Gobet A, Audic S, Bass D, Bittner L, Boutte C, et al. Marine protist diversity in European coastal waters and sediments as revealed by high-throughput sequencing. Environmental microbiology. 2015;17(10):4035–49. doi: 10.1111/1462-2920.12955. [DOI] [PubMed] [Google Scholar]
- 52.Beall CJ, Campbell AG, Dayeh DM, Griffen AL, Podar M, Leys EJ. Single cell genomics of uncultured, health-associated Tannerella BU063 (Oral Taxon 286) and comparison to the closely related pathogen Tannerella forsythia. PloS one. 2014;9(2):e89398. doi: 10.1371/journal.pone.0089398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mason OU, Hazen TC, Borglin S, Chain PS, Dubinsky EA, Fortney JL, et al. Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to Deepwater Horizon oil spill. The ISME journal. 2012;6(9):1715–27. doi: 10.1038/ismej.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoon HS, Price DC, Stepanauskas R, Rajah VD, Sieracki ME, Wilson WH, et al. Single-cell genomics reveals organismal interactions in uncultivated marine protists. Science (New York, NY) 2011;332(6030):714–7. doi: 10.1126/science.1203163. [DOI] [PubMed] [Google Scholar]
- 55.Hehemann JH, Correc G, Barbeyron T, Helbert W, Czjzek M, Michel G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464(7290):908–12. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- 56.Manna S, Harman A. Horizontal gene transfer of a Chlamydial tRNA-guanine transglycosylase gene to eukaryotic microbes. Molecular phylogenetics and evolution. 2016;94(Pt A):392–6. doi: 10.1016/j.ympev.2015.09.022. [DOI] [PubMed] [Google Scholar]
- 57.Smillie CS, Smith MB, Friedman J, Cordero OX, David LA, Alm EJ. Ecology drives a global network of gene exchange connecting the human microbiome. Nature. 2011;480(7376):241–4. doi: 10.1038/nature10571. [DOI] [PubMed] [Google Scholar]
- 58.Cisse OH, Pagni M, Hauser PM. Comparative genomics suggests that the human pathogenic fungus Pneumocystis jirovecii acquired obligate biotrophy through gene loss. Genome biology and evolution. 2014;6(8):1938–48. doi: 10.1093/gbe/evu155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Elias AF, Stewart PE, Grimm D, Caimano MJ, Eggers CH, Tilly K, et al. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infection and immunity. 2002;70(4):2139–50. doi: 10.1128/IAI.70.4.2139-2150.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rawat A, Engelthaler DM, Driebe EM, Keim P, Foster JT. MetaGeniE: characterizing human clinical samples using deep metagenomic sequencing. PloS one. 2014;9(11):e110915. doi: 10.1371/journal.pone.0110915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schloissnig S, Arumugam M, Sunagawa S, Mitreva M, Tap J, Zhu A, et al. Genomic variation landscape of the human gut microbiome. Nature. 2013;493(7430):45–50. doi: 10.1038/nature11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Science translational medicine. 2014;6(237) doi: 10.1126/scitranslmed.3008599. 237ra65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scholtens PA, Oozeer R, Martin R, Amor KB, Knol J. The early settlers: intestinal microbiology in early life. Annual review of food science and technology. 2012;3:425–47. doi: 10.1146/annurev-food-022811-101120. [DOI] [PubMed] [Google Scholar]
- 64.Underwood MA, German JB, Lebrilla CB, Mills DA. Bifidobacterium longum subspecies infantis: champion colonizer of the infant gut. Pediatric research. 2015;77(1-2):229–35. doi: 10.1038/pr.2014.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mueller NT, Bakacs E, Combellick J, Grigoryan Z, Dominguez-Bello MG. The infant microbiome development: mom matters. Trends in molecular medicine. 2015;21(2):109–17. doi: 10.1016/j.molmed.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Braundmeier AG, Lenz KM, Inman KS, Chia N, Jeraldo P, Walther-Antonio MR, et al. Individualized medicine and the microbiome in reproductive tract. Frontiers in physiology. 2015;6:97. doi: 10.3389/fphys.2015.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ardissone AN, de la Cruz DM, Davis-Richardson AG, Rechcigl KT, Li N, Drew JC, et al. Meconium microbiome analysis identifies bacteria correlated with premature birth. PloS one. 2014;9(3):e90784. doi: 10.1371/journal.pone.0090784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Neu J, Rushing J. Cesarean versus vaginal delivery: long-term infant outcomes and the hygiene hypothesis. Clinics in perinatology. 2011;38(2):321–31. doi: 10.1016/j.clp.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rehavi MMJE. Physicians treating physicians: Information and incentives in childbirth. NBER Working Paper. 2013 No. w19242. [Google Scholar]
- 70.Mueller NT, Whyatt R, Hoepner L, Oberfield S, Dominguez-Bello MG, Widen EM, et al. Prenatal exposure to antibiotics, cesarean section and risk of childhood obesity. International journal of obesity (2005) 2015;39(4):665–70. doi: 10.1038/ijo.2014.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(26):11971–5. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sevelsted A, Stokholm J, Bonnelykke K, Bisgaard H. Cesarean section and chronic immune disorders. Pediatrics. 2015;135(1):e92–8. doi: 10.1542/peds.2014-0596. [DOI] [PubMed] [Google Scholar]
- 73.Malmborg P, Bahmanyar S, Grahnquist L, Hildebrand H, Montgomery S. Cesarean section and the risk of pediatric Crohn's disease. Inflammatory bowel diseases. 2012;18(4):703–8. doi: 10.1002/ibd.21741. [DOI] [PubMed] [Google Scholar]
- 74.Johnson CL, Versalovic J. The human microbiome and its potential importance to pediatrics. Pediatrics. 2012;129(5):950–60. doi: 10.1542/peds.2011-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kostic AD, Gevers D, Siljander H, Vatanen T, Hyotylainen T, Hamalainen AM, et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell host & microbe. 2015;17(2):260–73. doi: 10.1016/j.chom.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Murgas Torrazza R, Neu J. The developing intestinal microbiome and its relationship to health and disease in the neonate. Journal of perinatology : official journal of the California Perinatal Association. 2011;31(Suppl 1):S29–34. doi: 10.1038/jp.2010.172. [DOI] [PubMed] [Google Scholar]
- 77.Rousseau C, Levenez F, Fouqueray C, Dore J, Collignon A, Lepage P. Clostridium difficile colonization in early infancy is accompanied by changes in intestinal microbiota composition. Journal of clinical microbiology. 2011;49(3):858–65. doi: 10.1128/jcm.01507-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McLoughlin RM, Mills KH. Influence of gastrointestinal commensal bacteria on the immune responses that mediate allergy and asthma. The Journal of allergy and clinical immunology. 2011;127(5):1097–107. doi: 10.1016/j.jaci.2011.02.012. quiz 108-9. [DOI] [PubMed] [Google Scholar]
- 79.Ajslev TA, Andersen CS, Gamborg M, Sorensen TI, Jess T. Childhood overweight after establishment of the gut microbiota: the role of delivery mode, pre-pregnancy weight and early administration of antibiotics. International journal of obesity (2005) 2011;35(4):522–9. doi: 10.1038/ijo.2011.27. [DOI] [PubMed] [Google Scholar]
- 80.Cheng J, Palva AM, de Vos WM, Satokari R. Contribution of the intestinal microbiota to human health: from birth to 100 years of age. Current topics in microbiology and immunology. 2013;358:323–46. doi: 10.1007/82_2011_189. [DOI] [PubMed] [Google Scholar]
- 81.Decker E, Hornef M, Stockinger S. Cesarean delivery is associated with celiac disease but not inflammatory bowel disease in children. Gut microbes. 2011;2(2):91–8. doi: 10.4161/gmic.2.2.15414. [DOI] [PubMed] [Google Scholar]
- 82.Hunter PA, Dawson S, French GL, Goossens H, Hawkey PM, Kuijper EJ, et al. Antimicrobial-resistant pathogens in animals and man: prescribing, practices and policies. The Journal of antimicrobial chemotherapy. 2010;65(Suppl 1):i3–17. doi: 10.1093/jac/dkp433. [DOI] [PubMed] [Google Scholar]
- 83.Stewardson AJ, Huttner B, Harbarth S. At least it won't hurt: the personal risks of antibiotic exposure. Current opinion in pharmacology. 2011;11(5):446–52. doi: 10.1016/j.coph.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 84.Blaser MJ. Who are we? Indigenous microbes and the ecology of human diseases. EMBO reports. 2006;7(10):956–60. doi: 10.1038/sj.embor.7400812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schwartz S, Friedberg I, Ivanov IV, Davidson LA, Goldsby JS, Dahl DB, et al. A metagenomic study of diet-dependent interaction between gut microbiota and host in infants reveals differences in immune response. Genome biology. 2012;13(4):r32. doi: 10.1186/gb-2012-13-4-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stark PL, Lee A. The microbial ecology of the large bowel of breast-fed and formula-fed infants during the first year of life. Journal of medical microbiology. 1982;15(2):189–203. doi: 10.1099/00222615-15-2-189. [DOI] [PubMed] [Google Scholar]
- 87.Harmsen HJ, Wildeboer-Veloo AC, Raangs GC, Wagendorp AA, Klijn N, Bindels JG, et al. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. Journal of pediatric gastroenterology and nutrition. 2000;30(1):61–7. doi: 10.1097/00005176-200001000-00019. [DOI] [PubMed] [Google Scholar]
- 88.Ballard O, Morrow AL. Human milk composition: nutrients and bioactive factors. Pediatric clinics of North America. 2013;60(1):49–74. doi: 10.1016/j.pcl.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Eidelman AI. Breastfeeding and the use of human milk: an analysis of the American Academy of Pediatrics 2012 Breastfeeding Policy Statement. Breastfeeding medicine : the official journal of the Academy of Breastfeeding Medicine. 2012;7(5):323–4. doi: 10.1089/bfm.2012.0067. [DOI] [PubMed] [Google Scholar]
- 90.Isaacs EB, Fischl BR, Quinn BT, Chong WK, Gadian DG, Lucas A. Impact of breast milk on intelligence quotient, brain size, and white matter development. Pediatric research. 2010;67(4):357–62. doi: 10.1203/PDR.0b013e3181d026da. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zivkovic AM, German JB, Lebrilla CB, Mills DA. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4653–8. doi: 10.1073/pnas.1000083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kunz C, Rudloff S, Baier W, Klein N, Strobel S. Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annual review of nutrition. 2000;20:699–722. doi: 10.1146/annurev.nutr.20.1.699. [DOI] [PubMed] [Google Scholar]
- 93.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118(2):511–21. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 94.Sela DA. Bifidobacterial utilization of human milk oligosaccharides. International journal of food microbiology. 2011;149(1):58–64. doi: 10.1016/j.ijfoodmicro.2011.01.025. [DOI] [PubMed] [Google Scholar]
- 95.Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(48):18964–9. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lewis ZT, Totten SM, Smilowitz JT, Popovic M, Parker E, Lemay DG, et al. Maternal fucosyltransferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome. 2015;3:13. doi: 10.1186/s40168-015-0071-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Davidson B, Meinzen-Derr JK, Wagner CL, Newburg DS, Morrow AL. Fucosylated oligosaccharides in human milk in relation to gestational age and stage of lactation. Advances in experimental medicine and biology. 2004;554:427–30. doi: 10.1007/978-1-4757-4242-8_56. [DOI] [PubMed] [Google Scholar]
- 98.Newburg DS, Ruiz-Palacios GM, Altaye M, Chaturvedi P, Meinzen-Derr J, Guerrero Mde L, et al. Innate protection conferred by fucosylated oligosaccharides of human milk against diarrhea in breastfed infants. Glycobiology. 2004;14(3):253–63. doi: 10.1093/glycob/cwh020. [DOI] [PubMed] [Google Scholar]
- 99.Newburg DS, Ruiz-Palacios GM, Morrow AL. Human milk glycans protect infants against enteric pathogens. Annual review of nutrition. 2005;25:37–58. doi: 10.1146/annurev.nutr.25.050304.092553. [DOI] [PubMed] [Google Scholar]
- 100.Newburg DS. Oligosaccharides in human milk and bacterial colonization. Journal of pediatric gastroenterology and nutrition. 2000;30(Suppl 2):S8–17. [PubMed] [Google Scholar]
- 101.Morrow AL, Ruiz-Palacios GM, Jiang X, Newburg DS. Human-milk glycans that inhibit pathogen binding protect breast-feeding infants against infectious diarrhea. The Journal of nutrition. 2005;135(5):1304–7. doi: 10.1093/jn/135.5.1304. [DOI] [PubMed] [Google Scholar]
- 102.Idota T, Kawakami H, Murakami Y, Sugawara M. Inhibition of cholera toxin by human milk fractions and sialyllactose. Bioscience, biotechnology, and biochemistry. 1995;59(3):417–9. doi: 10.1271/bbb.59.417. [DOI] [PubMed] [Google Scholar]
- 103.Andersson B, Porras O, Hanson LA, Lagergard T, Svanborg-Eden C. Inhibition of attachment of Streptococcus pneumoniae and Haemophilus influenzae by human milk and receptor oligosaccharides. The Journal of infectious diseases. 1986;153(2):232–7. doi: 10.1093/infdis/153.2.232. [DOI] [PubMed] [Google Scholar]
- 104.Newburg DS, Pickering LK, McCluer RH, Cleary TG. Fucosylated oligosaccharides of human milk protect suckling mice from heat-stabile enterotoxin of Escherichia coli. The Journal of infectious diseases. 1990;162(5):1075–80. doi: 10.1093/infdis/162.5.1075. [DOI] [PubMed] [Google Scholar]
- 105.Duijts L, Jaddoe VW, Hofman A, Moll HA. Prolonged and exclusive breastfeeding reduces the risk of infectious diseases in infancy. Pediatrics. 2010;126(1):e18–25. doi: 10.1542/peds.2008-3256. [DOI] [PubMed] [Google Scholar]
- 106.Abt MC, Pamer EG. Commensal bacteria mediated defenses against pathogens. Current opinion in immunology. 2014;29:16–22. doi: 10.1016/j.coi.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Matsuki T, Yahagi K, Mori H, Matsumoto H, Hara T, Tajima S, et al. A key genetic factor for fucosyllactose utilization affects infant gut microbiota development. 2016;7:11939. doi: 10.1038/ncomms11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Oozeer R, van Limpt K, Ludwig T, Ben Amor K, Martin R, Wind RD, et al. Intestinal microbiology in early life: specific prebiotics can have similar functionalities as human-milk oligosaccharides. The American journal of clinical nutrition. 2013;98(2):561s–71s. doi: 10.3945/ajcn.112.038893. [DOI] [PubMed] [Google Scholar]
- 109.Heavey PMRI. The Gut Microflora of the Developing Infant: Microbiology and Metabolism. Microbial ecology in health and disease. 1999;11:75–83. [Google Scholar]
- 110.Charbonneau MR, O'Donnell D, Blanton LV, Totten SM, Davis JC, Barratt MJ, et al. Sialylated Milk Oligosaccharides Promote Microbiota-Dependent Growth in Models of Infant Undernutrition. Cell. 2016;164(5):859–71. doi: 10.1016/j.cell.2016.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zivkovic AM, Barile D. Bovine milk as a source of functional oligosaccharides for improving human health. Advances in nutrition (Bethesda, Md) 2011;2(3):284–9. doi: 10.3945/an.111.000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tao N, DePeters EJ, Freeman S, German JB, Grimm R, Lebrilla CB. Bovine milk glycome. Journal of dairy science. 2008;91(10):3768–78. doi: 10.3168/jds.2008-1305. [DOI] [PubMed] [Google Scholar]
- 113.Tao N, DePeters EJ, German JB, Grimm R, Lebrilla CB. Variations in bovine milk oligosaccharides during early and middle lactation stages analyzed by high-performance liquid chromatography-chip/mass spectrometry. Journal of dairy science. 2009;92(7):2991–3001. doi: 10.3168/jds.2008-1642. [DOI] [PubMed] [Google Scholar]
- 114.Bode L. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology. 2012;22(9):1147–62. doi: 10.1093/glycob/cws074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rautava S, Kalliomaki M, Isolauri E. Probiotics during pregnancy and breast-feeding might confer immunomodulatory protection against atopic disease in the infant. The Journal of allergy and clinical immunology. 2002;109(1):119–21. doi: 10.1067/mai.2002.120273. [DOI] [PubMed] [Google Scholar]
- 116.Rautava S, Kainonen E, Salminen S, Isolauri E. Maternal probiotic supplementation during pregnancy and breast-feeding reduces the risk of eczema in the infant. The Journal of allergy and clinical immunology. 2012;130(6):1355–60. doi: 10.1016/j.jaci.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 117.Rautava S, Salminen S, Isolauri E. Specific probiotics in reducing the risk of acute infections in infancy--a randomised, double-blind, placebo-controlled study. The British journal of nutrition. 2009;101(11):1722–6. doi: 10.1017/s0007114508116282. [DOI] [PubMed] [Google Scholar]
- 118.Isolauri E, Rautava S, Salminen S. Probiotics in the development and treatment of allergic disease. Gastroenterology clinics of North America. 2012;41(4):747–62. doi: 10.1016/j.gtc.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 119.Kainonen E, Rautava S, Isolauri E. Immunological programming by breast milk creates an anti-inflammatory cytokine milieu in breast-fed infants compared to formula-fed infants. The British journal of nutrition. 2013;109(11):1962–70. doi: 10.1017/s0007114512004229. [DOI] [PubMed] [Google Scholar]
- 120.Xu M, Wang J, Wang N, Sun F, Wang L, Liu XH. The Efficacy and Safety of the Probiotic Bacterium Lactobacillus reuteri DSM 17938 for Infantile Colic: A Meta-Analysis of Randomized Controlled Trials. PloS one. 2015;10(10):e0141445. doi: 10.1371/journal.pone.0141445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Savino F. Focus on infantile colic. Acta paediatrica (Oslo, Norway : 1992) 2007;96(9):1259–64. doi: 10.1111/j.1651-2227.2007.00428.x. [DOI] [PubMed] [Google Scholar]
- 122.Sung V, Collett S, de Gooyer T, Hiscock H, Tang M, Wake M. Probiotics to prevent or treat excessive infant crying: systematic review and meta-analysis. JAMA pediatrics. 2013;167(12):1150–7. doi: 10.1001/jamapediatrics.2013.2572. [DOI] [PubMed] [Google Scholar]
- 123.Chau K, Lau E, Greenberg S, Jacobson S, Yazdani-Brojeni P, Verma N, et al. Probiotics for infantile colic: a randomized, double-blind, placebo-controlled trial investigating Lactobacillus reuteri DSM 17938. The Journal of pediatrics. 2015;166(1):74–8. doi: 10.1016/j.jpeds.2014.09.020. [DOI] [PubMed] [Google Scholar]
- 124.Roos S, Dicksved J, Tarasco V, Locatelli E, Ricceri F, Grandin U, et al. 454 pyrosequencing analysis on faecal samples from a randomized DBPC trial of colicky infants treated with Lactobacillus reuteri DSM 17938. PloS one. 2013;8(2):e56710. doi: 10.1371/journal.pone.0056710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Claesson MJ, Cusack S, O'Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4586–91. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.O'Toole PW, Jeffery IB. Gut microbiota and aging. Science (New York, NY) 2015;350(6265):1214–5. doi: 10.1126/science.aac8469. [DOI] [PubMed] [Google Scholar]
- 127.Jackson MA, Jeffery IB, Beaumont M, Bell JT, Clark AG, Ley RE, et al. Signatures of early frailty in the gut microbiota. Genome medicine. 2016;8(1):8. doi: 10.1186/s13073-016-0262-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zapata HJ, Quagliarello VJ. The microbiota and microbiome in aging: potential implications in health and age-related diseases. Journal of the American Geriatrics Society. 2015;63(4):776–81. doi: 10.1111/jgs.13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Saraswati S, Sitaraman R. Aging and the human gut microbiota-from correlation to causality. Frontiers in microbiology. 2014;5:764. doi: 10.3389/fmicb.2014.00764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Patrignani P, Tacconelli S, Bruno A. Gut microbiota, host gene expression, and aging. Journal of clinical gastroenterology. 2014;48(Suppl 1):S28–31. doi: 10.1097/mcg.0000000000000229. [DOI] [PubMed] [Google Scholar]
- 131.Noren Hooten N, Abdelmohsen K, Gorospe M, Ejiogu N, Zonderman AB, Evans MK. microRNA expression patterns reveal differential expression of target genes with age. PloS one. 2010;5(5):e10724. doi: 10.1371/journal.pone.0010724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lai CY, Wu YT, Yu SL, Yu YH, Lee SY, Liu CM, et al. Modulated expression of human peripheral blood microRNAs from infancy to adulthood and its role in aging. Aging cell. 2014;13(4):679–89. doi: 10.1111/acel.12225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liu S, da Cunha AP, Rezende RM, Cialic R, Wei Z, Bry L, et al. The Host Shapes the Gut Microbiota via Fecal MicroRNA. Cell host & microbe. 2016;19(1):32–43. doi: 10.1016/j.chom.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Heilbronn LK, Ravussin E. Calorie restriction and aging: review of the literature and implications for studies in humans. The American journal of clinical nutrition. 2003;78(3):361–9. doi: 10.1093/ajcn/78.3.361. [DOI] [PubMed] [Google Scholar]
- 135.Zhang C, Li S, Yang L, Huang P, Li W, Wang S, et al. Structural modulation of gut microbiota in life-long calorie-restricted mice. Nature communications. 2013;4:2163. doi: 10.1038/ncomms3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wu Z, Song L, Liu SQ, Huang D. A high throughput screening assay for determination of chronological lifespan of yeast. Experimental gerontology. 2011;46(11):915–22. doi: 10.1016/j.exger.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 137.Biagi E, Candela M, Fairweather-Tait S, Franceschi C, Brigidi P. Aging of the human metaorganism: the microbial counterpart. Age (Dordr) 2012;34(1):247–67. doi: 10.1007/s11357-011-9217-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rampelli S, Candela M, Turroni S, Biagi E, Collino S, Franceschi C, et al. Functional metagenomic profiling of intestinal microbiome in extreme ageing. Aging. 2013;5(12):902–12. doi: 10.18632/aging.100623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Heintz C, Mair W. You are what you host: microbiome modulation of the aging process. Cell. 2014;156(3):408–11. doi: 10.1016/j.cell.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rozsa L, Apari P, Muller V. The microbiome mutiny hypothesis: can our microbiome turn against us when we are old or seriously ill? Biology direct. 2015;10:3. doi: 10.1186/s13062-014-0034-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Caracciolo B, Xu W, Collins S, Fratiglioni L. Cognitive decline, dietary factors and gut-brain interactions. Mechanisms of ageing and development. 2014;136-137:59–69. doi: 10.1016/j.mad.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 142.Parkar SG, Trower TM, Stevenson DE. Fecal microbial metabolism of polyphenols and its effects on human gut microbiota. Anaerobe. 2013;23:12–9. doi: 10.1016/j.anaerobe.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 143.Wang D, Ho L, Faith J, Ono K, Janle EM, Lachcik PJ, et al. Role of intestinal microbiota in the generation of polyphenol-derived phenolic acid mediated attenuation of Alzheimer's disease beta-amyloid oligomerization. Molecular nutrition & food research. 2015;59(6):1025–40. doi: 10.1002/mnfr.201400544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Raberg L, Sim D, Read AF. Disentangling genetic variation for resistance and tolerance to infectious diseases in animals. Science (New York, NY) 2007;318(5851):812–4. doi: 10.1126/science.1148526. [DOI] [PubMed] [Google Scholar]
- 145.Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39(2):372–85. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 146.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nature reviews Immunology. 2009;9(5):313–23. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ohnmacht C, Park JH, Cording S, Wing JB, Atarashi K, Obata Y, et al. MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORgammat(+) T cells. Science (New York, NY) 2015;349(6251):989–93. doi: 10.1126/science.aac4263. [DOI] [PubMed] [Google Scholar]
- 148.Martin SA, Pence BD, Woods JA. Exercise and respiratory tract viral infections. Exercise and sport sciences reviews. 2009;37(4):157–64. doi: 10.1097/JES.0b013e3181b7b57b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Simpson RJ, Kunz H, Agha N, Graff R. Exercise and the Regulation of Immune Functions. Progress in molecular biology and translational science. 2015;135:355–80. doi: 10.1016/bs.pmbts.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 150.Pape K, Ryttergaard L, Rotevatn TA, Nielsen BJ, Torp-Pedersen C, Overgaard C, et al. Leisure-Time Physical Activity and the Risk of Suspected Bacterial Infections. Medicine and science in sports and exercise. 2016;48(9):1737–44. doi: 10.1249/mss.0000000000000953. [DOI] [PubMed] [Google Scholar]
- 151.Zhang D, Chen G, Manwani D, Mortha A, Xu C, Faith JJ, et al. Neutrophil ageing is regulated by the microbiome. Nature. 2015;525(7570):528–32. doi: 10.1038/nature15367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478(7368):250–4. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.The Integrative Human Microbiome Project: dynamic analysis of microbiome-host omics profiles during periods of human health and disease. Cell host & microbe. 2014;16(3):276–89. doi: 10.1016/j.chom.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Strachan DP. Hay fever, hygiene, and household size. BMJ (Clinical research ed) 1989;299(6710):1259–60. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Okada H, Kuhn C, Feillet H, Bach JF. The ‘hygiene hypothesis’ for autoimmune and allergic diseases: an update. Clinical and experimental immunology. 2010;160(1):1–9. doi: 10.1111/j.1365-2249.2010.04139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ege MJ, Mayer M, Normand AC, Genuneit J, Cookson WO, Braun-Fahrlander C, et al. Exposure to environmental microorganisms and childhood asthma. The New England journal of medicine. 2011;364(8):701–9. doi: 10.1056/NEJMoa1007302. [DOI] [PubMed] [Google Scholar]
- 157.Schuijs MJ, Willart MA, Vergote K, Gras D, Deswarte K, Ege MJ, et al. Farm dust and endotoxin protect against allergy through A20 induction in lung epithelial cells. Science (New York, NY) 2015;349(6252):1106–10. doi: 10.1126/science.aac6623. [DOI] [PubMed] [Google Scholar]
- 158.Brooks C, Pearce N, Douwes J. The hygiene hypothesis in allergy and asthma: an update. Current opinion in allergy and clinical immunology. 2013;13(1):70–7. doi: 10.1097/ACI.0b013e32835ad0d2. [DOI] [PubMed] [Google Scholar]
- 159.Chotirmall SH, Burke CM. Aging and the microbiome: implications for asthma in the elderly? Expert review of respiratory medicine. 2015;9(2):125–8. doi: 10.1586/17476348.2015.1002473. [DOI] [PubMed] [Google Scholar]
- 160.Barnes PJ. Cytokine-directed therapies for asthma. The Journal of allergy and clinical immunology. 2001;108(2 Suppl):S72–6. doi: 10.1067/mai.2001.116435. [DOI] [PubMed] [Google Scholar]
- 161.Barnes PJ. Immunology of asthma and chronic obstructive pulmonary disease. Nature reviews Immunology. 2008;8(3):183–92. doi: 10.1038/nri2254. [DOI] [PubMed] [Google Scholar]
- 162.von Mutius E, Vercelli D. Farm living: effects on childhood asthma and allergy. Nature reviews Immunology. 2010;10(12):861–8. doi: 10.1038/nri2871. [DOI] [PubMed] [Google Scholar]
- 163.Couzin-Frankel J. Bacteria and asthma: untangling the links. Science (New York, NY) 2010;330(6008):1168–9. doi: 10.1126/science.330.6008.1168. [DOI] [PubMed] [Google Scholar]
- 164.Lif Holgerson P, Harnevik L, Hernell O, Tanner AC, Johansson I. Mode of birth delivery affects oral microbiota in infants. Journal of dental research. 2011;90(10):1183–8. doi: 10.1177/0022034511418973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Nelun Barfod M, Magnusson K, Lexner MO, Blomqvist S, Dahlen G, Twetman S. Oral microflora in infants delivered vaginally and by caesarean section. International journal of paediatric dentistry / the British Paedodontic Society [and] the International Association of Dentistry for Children. 2011;21(6):401–6. doi: 10.1111/j.1365-263X.2011.01136.x. [DOI] [PubMed] [Google Scholar]
- 166.Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Science translational medicine. 2015;7(307) doi: 10.1126/scitranslmed.aab2271. 307ra152. [DOI] [PubMed] [Google Scholar]
- 167.Marsland BJ, Salami O. Microbiome influences on allergy in mice and humans. Current opinion in immunology. 2015;36:94–100. doi: 10.1016/j.coi.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 168.Lerner A, Aminov R, Matthias T. Dysbiosis May Trigger Autoimmune Diseases via Inappropriate Post-Translational Modification of Host Proteins. Frontiers in microbiology. 2016;7:84. doi: 10.3389/fmicb.2016.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hesselmar B, Hicke-Roberts A, Wennergren G. Allergy in children in hand versus machine dishwashing. Pediatrics. 2015;135(3):e590–7. doi: 10.1542/peds.2014-2968. [DOI] [PubMed] [Google Scholar]
- 170.Vatanen T, Kostic AD, d'Hennezel E, Siljander H, Franzosa EA, Yassour M, et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell. 2016;165(6):1551. doi: 10.1016/j.cell.2016.05.056. [DOI] [PubMed] [Google Scholar]
- 171.Konig MF, Abusleme L, Reinholdt J, Palmer RJ, Teles RP, Sampson K, et al. Aggregatibacter actinomycetemcomitans–induced hypercitrullination links periodontal infection to autoimmunity in rheumatoid arthritis. Science translational medicine. 2016;8(369):369ra176–369ra176. doi: 10.1126/scitranslmed.aaj1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhang X, Zhang D, Jia H. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Medicine. 2015;21(8):895–905. doi: 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- 173.Tan TG, Sefik E, Geva-Zatorsky N, Kua L, Naskar D, Teng F, et al. Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proceedings of the National Academy of Sciences of the United States of America. 2016 doi: 10.1073/pnas.1617460113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Leipe J, Grunke M, Dechant C, Reindl C, Kerzendorf U, Schulze-Koops H, et al. Role of Th17 cells in human autoimmune arthritis. Arthritis and rheumatism. 2010;62(10):2876–85. doi: 10.1002/art.27622. [DOI] [PubMed] [Google Scholar]
- 175.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022–3. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 176.Wang CY, Liao JK. A mouse model of diet-induced obesity and insulin resistance. Methods in molecular biology (Clifton, NJ) 2012;821:421–33. doi: 10.1007/978-1-61779-430-8_27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 178.Kallus SJ, Brandt LJ. The intestinal microbiota and obesity. Journal of clinical gastroenterology. 2012;46(1):16–24. doi: 10.1097/MCG.0b013e31823711fd. [DOI] [PubMed] [Google Scholar]
- 179.Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, et al. The gut microbiota as an environmental factor that regulates fat storage. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(44):15718–23. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science (New York, NY) 2013;341(6150):1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Koleva PT, Bridgman SL, Kozyrskyj AL. The infant gut microbiome: evidence for obesity risk and dietary intervention. Nutrients. 2015;7(4):2237–60. doi: 10.3390/nu7042237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Scott FI, Horton DB, Mamtani R, Haynes K, Goldberg DS, Lee DY, et al. Administration of Antibiotics to Children Before Age 2 Years Increases Risk for Childhood Obesity. Gastroenterology. 2016;151(1):120–9 e5. doi: 10.1053/j.gastro.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Cox LM, Blaser MJ. Antibiotics in early life and obesity. Nature reviews Endocrinology. 2015;11(3):182–90. doi: 10.1038/nrendo.2014.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158(4):705–21. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.McOrist AL, Miller RB, Bird AR, Keogh JB, Noakes M, Topping DL, et al. Fecal butyrate levels vary widely among individuals but are usually increased by a diet high in resistant starch. The Journal of nutrition. 2011;141(5):883–9. doi: 10.3945/jn.110.128504. [DOI] [PubMed] [Google Scholar]
- 186.De Filippis F, Pellegrini N, Vannini L, Jeffery IB, La Storia A, Laghi L, et al. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut. 2015 doi: 10.1136/gutjnl-2015-309957. [DOI] [PubMed] [Google Scholar]
- 187.Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. The microbiology of butyrate formation in the human colon. FEMS microbiology letters. 2002;217(2):133–9. doi: 10.1111/j.1574-6968.2002.tb11467.x. [DOI] [PubMed] [Google Scholar]
- 188.McIntyre A, Gibson PR, Young GP. Butyrate production from dietary fibre and protection against large bowel cancer in a rat model. Gut. 1993;34(3):386–91. doi: 10.1136/gut.34.3.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Musso G, Gambino R, Cassader M. Obesity, diabetes, and gut microbiota: the hygiene hypothesis expanded? Diabetes care. 2010;33(10):2277–84. doi: 10.2337/dc10-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Bocarsly ME, Fasolino M, Kane GA, LaMarca EA, Kirschen GW, Karatsoreos IN, et al. Obesity diminishes synaptic markers, alters microglial morphology, and impairs cognitive function. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(51):15731–6. doi: 10.1073/pnas.1511593112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Burokas A, Moloney RD, Dinan TG, Cryan JF. Microbiota regulation of the Mammalian gut-brain axis. Advances in applied microbiology. 2015;91:1–62. doi: 10.1016/bs.aambs.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 192.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(22):9066–71. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. 2017;23(1):107–13. doi: 10.1038/nm.4236. [DOI] [PubMed] [Google Scholar]
- 194.de la Cuesta-Zuluaga J, Mueller NT, Corrales-Agudelo V, Velasquez-Mejia EP, Carmona JA, Abad JM, et al. Metformin Is Associated With Higher Relative Abundance of Mucin-Degrading Akkermansia muciniphila and Several Short-Chain Fatty Acid-Producing Microbiota in the Gut. 2017;40(1):54–62. doi: 10.2337/dc16-1324. [DOI] [PubMed] [Google Scholar]
- 195.Troseid M. Gut microbiota and acute coronary syndromes: ready for use in the emergency room? European heart journal. 2017 doi: 10.1093/eurheartj/ehx005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Li J, Zhao F, Wang Y, Chen J, Tao J, Tian G, et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome. 2017;5(1):14. doi: 10.1186/s40168-016-0222-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Stepankova R, Tonar Z, Bartova J, Nedorost L, Rossman P, Poledne R, et al. Absence of microbiota (germ-free conditions) accelerates the atherosclerosis in ApoE-deficient mice fed standard low cholesterol diet. Journal of atherosclerosis and thrombosis. 2010;17(8):796–804. doi: 10.5551/jat.3285. [DOI] [PubMed] [Google Scholar]
- 198.Chan YK, Brar MS, Kirjavainen PV, Chen Y, Peng J, Li D, et al. High fat diet induced atherosclerosis is accompanied with low colonic bacterial diversity and altered abundances that correlates with plaque size, plasma A-FABP and cholesterol: a pilot study of high fat diet and its intervention with Lactobacillus rhamnosus GG (LGG) or telmisartan in ApoE-/- mice. BMC microbiology. 2016;16(1):264. doi: 10.1186/s12866-016-0883-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Li J, Lin S, Vanhoutte PM, Woo CW, Xu A. Akkermansia Muciniphila Protects Against Atherosclerosis by Preventing Metabolic Endotoxemia-Induced Inflammation in Apoe-/- Mice. Circulation. 2016;133(24):2434–46. doi: 10.1161/circulationaha.115.019645. [DOI] [PubMed] [Google Scholar]
- 200.Grenham S, Clarke G, Cryan JF, Dinan TG. Brain-gut-microbe communication in health and disease. Frontiers in physiology. 2011;2:94. doi: 10.3389/fphys.2011.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Kirk RG. “Life in a germ-free world”: isolating life from the laboratory animal to the bubble boy. Bulletin of the history of medicine. 2012;86(2):237–75. doi: 10.1353/bhm.2012.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Williams SC. Gnotobiotics. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(5):1661. doi: 10.1073/pnas.1324049111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Diaz Heijtz R, Wang S, Anuar F, Qian Y, Bjorkholm B, Samuelsson A, et al. Normal gut microbiota modulates brain development and behavior. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(7):3047–52. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Neufeld KM, Kang N, Bienenstock J, Foster JA. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterology and motility : the official journal of the European Gastrointestinal Motility Society. 2011;23(3):255–64. doi: 10.1111/j.1365-2982.2010.01620.x. e119. [DOI] [PubMed] [Google Scholar]
- 205.Clarke G, Grenham S, Scully P, Fitzgerald P, Moloney RD, Shanahan F, et al. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Molecular psychiatry. 2013;18(6):666–73. doi: 10.1038/mp.2012.77. [DOI] [PubMed] [Google Scholar]
- 206.Crumeyrolle-Arias M, Jaglin M, Bruneau A, Vancassel S, Cardona A, Dauge V, et al. Absence of the gut microbiota enhances anxiety-like behavior and neuroendocrine response to acute stress in rats. Psychoneuroendocrinology. 2014;42:207–17. doi: 10.1016/j.psyneuen.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 207.Bercik P, Denou E, Collins J, Jackson W, Lu J, Jury J, et al. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology. 2011;141(2):599–609. e1–3. doi: 10.1053/j.gastro.2011.04.052. [DOI] [PubMed] [Google Scholar]
- 208.Bruce-Keller AJ, Salbaum JM, Luo M, Blanchard Et, Taylor CM, Welsh DA, et al. Obese-type gut microbiota induce neurobehavioral changes in the absence of obesity. Biological psychiatry. 2015;77(7):607–15. doi: 10.1016/j.biopsych.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Desbonnet L, Clarke G, Shanahan F, Dinan TG, Cryan JF. Microbiota is essential for social development in the mouse. Molecular psychiatry. 2014;19(2):146–8. doi: 10.1038/mp.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Arentsen T, Raith H, Qian Y, Forssberg H, Diaz Heijtz R. Host microbiota modulates development of social preference in mice. Microbial ecology in health and disease. 2015;26:29719. doi: 10.3402/mehd.v26.29719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Gareau MG, Wine E, Rodrigues DM, Cho JH, Whary MT, Philpott DJ, et al. Bacterial infection causes stress-induced memory dysfunction in mice. Gut. 2011;60(3):307–17. doi: 10.1136/gut.2009.202515. [DOI] [PubMed] [Google Scholar]
- 212.Desbonnet L, Clarke G, Traplin A, O'Sullivan O, Crispie F, Moloney RD, et al. Gut microbiota depletion from early adolescence in mice: Implications for brain and behaviour. Brain, behavior, and immunity. 2015;48:165–73. doi: 10.1016/j.bbi.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 213.Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, et al. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(38):16050–5. doi: 10.1073/pnas.1102999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Savignac HM, Tramullas M, Kiely B, Dinan TG, Cryan JF. Bifidobacteria modulate cognitive processes in an anxious mouse strain. Behavioural brain research. 2015;287:59–72. doi: 10.1016/j.bbr.2015.02.044. [DOI] [PubMed] [Google Scholar]
- 215.Savignac HM, Couch Y, Stratford M, Bannerman DM, Tzortzis G, Anthony DC, et al. Prebiotic administration normalizes lipopolysaccharide (LPS)-induced anxiety and cortical 5-HT2A receptor and IL1-beta levels in male mice. Brain, behavior, and immunity. 2016;52:120–31. doi: 10.1016/j.bbi.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Verdu EF, Bercik P, Verma-Gandhu M, Huang XX, Blennerhassett P, Jackson W, et al. Specific probiotic therapy attenuates antibiotic induced visceral hypersensitivity in mice. Gut. 2006;55(2):182–90. doi: 10.1136/gut.2005.066100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu XN, et al. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. The Journal of physiology. 2004;558(Pt 1):263–75. doi: 10.1113/jphysiol.2004.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.O'Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO reports. 2006;7(7):688–93. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Matcovitch-Natan O, Winter DR, Giladi A, Vargas Aguilar S, Spinrad A, Sarrazin S, et al. Microglia development follows a stepwise program to regulate brain homeostasis. Science (New York, NY) 2016;353(6301) doi: 10.1126/science.aad8670. aad8670. [DOI] [PubMed] [Google Scholar]
- 220.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(10):3698–703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Mardinoglu A, Shoaie S, Bergentall M, Ghaffari P, Zhang C, Larsson E, et al. The gut microbiota modulates host amino acid and glutathione metabolism in mice. Molecular systems biology. 2015;11(10):834. doi: 10.15252/msb.20156487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Desbonnet L, Garrett L, Clarke G, Bienenstock J, Dinan TG. The probiotic Bifidobacteria infantis: An assessment of potential antidepressant properties in the rat. Journal of psychiatric research. 2008;43(2):164–74. doi: 10.1016/j.jpsychires.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 223.Dinan TG, Cryan JF. Regulation of the stress response by the gut microbiota: implications for psychoneuroendocrinology. Psychoneuroendocrinology. 2012;37(9):1369–78. doi: 10.1016/j.psyneuen.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 224.Bailey MT, Coe CL. Maternal separation disrupts the integrity of the intestinal microflora in infant rhesus monkeys. Developmental psychobiology. 1999;35(2):146–55. [PubMed] [Google Scholar]
- 225.O'Mahony SM, Marchesi JR, Scully P, Codling C, Ceolho AM, Quigley EM, et al. Early life stress alters behavior, immunity, and microbiota in rats: implications for irritable bowel syndrome and psychiatric illnesses. Biological psychiatry. 2009;65(3):263–7. doi: 10.1016/j.biopsych.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 226.Golubeva AV, Crampton S, Desbonnet L, Edge D, O'Sullivan O, Lomasney KW, et al. Prenatal stress-induced alterations in major physiological systems correlate with gut microbiota composition in adulthood. Psychoneuroendocrinology. 2015;60:58–74. doi: 10.1016/j.psyneuen.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 227.Jasarevic E, Howerton CL, Howard CD, Bale TL. Alterations in the Vaginal Microbiome by Maternal Stress Are Associated With Metabolic Reprogramming of the Offspring Gut and Brain. Endocrinology. 2015;156(9):3265–76. doi: 10.1210/en.2015-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Bailey MT, Dowd SE, Galley JD, Hufnagle AR, Allen RG, Lyte M. Exposure to a social stressor alters the structure of the intestinal microbiota: implications for stressor-induced immunomodulation. Brain, behavior, and immunity. 2011;25(3):397–407. doi: 10.1016/j.bbi.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Galley JD, Nelson MC, Yu Z, Dowd SE, Walter J, Kumar PS, et al. Exposure to a social stressor disrupts the community structure of the colonic mucosa-associated microbiota. BMC microbiology. 2014;14:189. doi: 10.1186/1471-2180-14-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Bharwani A, Mian MF, Foster JA, Surette MG, Bienenstock J, Forsythe P. Structural & functional consequences of chronic psychosocial stress on the microbiome & host. Psychoneuroendocrinology. 2016;63:217–27. doi: 10.1016/j.psyneuen.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 231.De Palma G, Blennerhassett P, Lu J, Deng Y, Park AJ, Green W, et al. Microbiota and host determinants of behavioural phenotype in maternally separated mice. Nature communications. 2015;6:7735. doi: 10.1038/ncomms8735. [DOI] [PubMed] [Google Scholar]
- 232.O'Mahony SM, Hyland NP, Dinan TG, Cryan JF. Maternal separation as a model of brain-gut axis dysfunction. Psychopharmacology. 2011;214(1):71–88. doi: 10.1007/s00213-010-2010-9. [DOI] [PubMed] [Google Scholar]
- 233.Zijlmans MA, Korpela K, Riksen-Walraven JM, de Vos WM, de Weerth C. Maternal prenatal stress is associated with the infant intestinal microbiota. Psychoneuroendocrinology. 2015;53:233–45. doi: 10.1016/j.psyneuen.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 234.Kennedy PJ, Cryan JF, Dinan TG, Clarke G. Irritable bowel syndrome: a microbiome-gut-brain axis disorder? World journal of gastroenterology. 2014;20(39):14105–25. doi: 10.3748/wjg.v20.i39.14105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Braniste V, Al-Asmakh M, Kowal C, Anuar F, Abbaspour A, Toth M, et al. The gut microbiota influences blood-brain barrier permeability in mice. Science translational medicine. 2014;6(263) doi: 10.1126/scitranslmed.3009759. 263ra158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Stilling RM, Ryan FJ, Hoban AE, Shanahan F, Clarke G, Claesson MJ, et al. Microbes & neurodevelopment--Absence of microbiota during early life increases activity-related transcriptional pathways in the amygdala. Brain, behavior, and immunity. 2015;50:209–20. doi: 10.1016/j.bbi.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 237.Ogbonnaya ES, Clarke G, Shanahan F, Dinan TG, Cryan JF, O'Leary OF. Adult Hippocampal Neurogenesis Is Regulated by the Microbiome. Biological psychiatry. 2015;78(4):e7–9. doi: 10.1016/j.biopsych.2014.12.023. [DOI] [PubMed] [Google Scholar]
- 238.Erny D, Hrabe de Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, et al. Host microbiota constantly control maturation and function of microglia in the CNS. 2015;18(7):965–77. doi: 10.1038/nn.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Hoban AE, Stilling RM, Ryan FJ, Shanahan F, Dinan TG, Claesson MJ, et al. Regulation of prefrontal cortex myelination by the microbiota. Translational psychiatry. 2016;6:e774. doi: 10.1038/tp.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Harach TMN, Dutilleul N, Cheatham V, Mc Coy KD, Neher JJ, Jucker M, Fåk F, Lasser T, Bolmont T. Reduction of Alzheimer's disease beta-amyloid pathology in the absence of gut microbiota. ArXiv e-prints. 2015:1509. [Google Scholar]
- 241.Friedland RP. Mechanisms of molecular mimicry involving the microbiota in neurodegeneration. Journal of Alzheimer's disease : JAD. 2015;45(2):349–62. doi: 10.3233/jad-142841. [DOI] [PubMed] [Google Scholar]
- 242.Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, et al. Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson's Disease. Cell. 2016;167(6):1469–80.e12. doi: 10.1016/j.cell.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Forsythe P, Bienenstock J, Kunze WA. Vagal pathways for microbiome-brain-gut axis communication. Advances in experimental medicine and biology. 2014;817:115–33. doi: 10.1007/978-1-4939-0897-4_5. [DOI] [PubMed] [Google Scholar]
- 244.Bercik P, Verdu EF, Foster JA, Macri J, Potter M, Huang X, et al. Chronic gastrointestinal inflammation induces anxiety-like behavior and alters central nervous system biochemistry in mice. Gastroenterology. 2010;139(6):2102–12.e1. doi: 10.1053/j.gastro.2010.06.063. [DOI] [PubMed] [Google Scholar]
- 245.O'Mahony SM, Clarke G, Borre YE, Dinan TG, Cryan JF. Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behavioural brain research. 2015;277:32–48. doi: 10.1016/j.bbr.2014.07.027. [DOI] [PubMed] [Google Scholar]
- 246.De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156(1-2):84–96. doi: 10.1016/j.cell.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 247.Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161(2):264–76. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Holzer P, Farzi A. Neuropeptides and the microbiota-gut-brain axis. Advances in experimental medicine and biology. 2014;817:195–219. doi: 10.1007/978-1-4939-0897-4_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.El Aidy S, Dinan TG, Cryan JF. Gut Microbiota: The Conductor in the Orchestra of Immune-Neuroendocrine Communication. Clinical therapeutics. 2015;37(5):954–67. doi: 10.1016/j.clinthera.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 250.Kelly JR, Kennedy PJ, Cryan JF, Dinan TG, Clarke G, Hyland NP. Breaking down the barriers: the gut microbiome, intestinal permeability and stress-related psychiatric disorders. Frontiers in cellular neuroscience. 2015;9:392. doi: 10.3389/fncel.2015.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Quigley EM. Leaky gut - concept or clinical entity? Current opinion in gastroenterology. 2016;32(2):74–9. doi: 10.1097/mog.0000000000000243. [DOI] [PubMed] [Google Scholar]
- 252.Garber K. Drugging the gut microbiome. Nature biotechnology. 2015;33(3):228–31. doi: 10.1038/nbt.3161. [DOI] [PubMed] [Google Scholar]
- 253.Borre YE, O'Keeffe GW, Clarke G, Stanton C, Dinan TG, Cryan JF. Microbiota and neurodevelopmental windows: implications for brain disorders. Trends in molecular medicine. 2014;20(9):509–18. doi: 10.1016/j.molmed.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 254.Clarke G, O'Mahony SM, Dinan TG, Cryan JF. Priming for health: gut microbiota acquired in early life regulates physiology, brain and behaviour. Acta paediatrica (Oslo, Norway : 1992) 2014;103(8):812–9. doi: 10.1111/apa.12674. [DOI] [PubMed] [Google Scholar]
- 255.Kelly CR, Kahn S, Kashyap P, Laine L, Rubin D, Atreja A, et al. Update on Fecal Microbiota Transplantation 2015: Indications, Methodologies, Mechanisms, and Outlook. Gastroenterology. 2015;149(1):223–37. doi: 10.1053/j.gastro.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Dinan TG, Stanton C, Cryan JF. Psychobiotics: a novel class of psychotropic. Biological psychiatry. 2013;74(10):720–6. doi: 10.1016/j.biopsych.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 257.Clarke SF, Murphy EF, O'Sullivan O, Lucey AJ, Humphreys M, Hogan A, et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut. 2014;63(12):1913–20. doi: 10.1136/gutjnl-2013-306541. [DOI] [PubMed] [Google Scholar]
- 258.O'Sullivan O, Cronin O, Clarke SF, Murphy EF, Molloy MG, Shanahan F, et al. Exercise and the microbiota. Gut microbes. 2015;6(2):131–6. doi: 10.1080/19490976.2015.1011875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Dash S, Clarke G, Berk M, Jacka FN. The gut microbiome and diet in psychiatry: focus on depression. Current opinion in psychiatry. 2015;28(1):1–6. doi: 10.1097/yco.0000000000000117. [DOI] [PubMed] [Google Scholar]
- 260.Luna RA, Foster JA. Gut brain axis: diet microbiota interactions and implications for modulation of anxiety and depression. Current opinion in biotechnology. 2015;32:35–41. doi: 10.1016/j.copbio.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 261.Messaoudi M, Lalonde R, Violle N, Javelot H, Desor D, Nejdi A, et al. Assessment of psychotropic-like properties of a probiotic formulation (Lactobacillus helveticus R0052 and Bifidobacterium longum R0175) in rats and human subjects. The British journal of nutrition. 2011;105(5):755–64. doi: 10.1017/s0007114510004319. [DOI] [PubMed] [Google Scholar]
- 262.Tillisch K, Labus J, Kilpatrick L, Jiang Z, Stains J, Ebrat B, et al. Consumption of fermented milk product with probiotic modulates brain activity. Gastroenterology. 2013;144(7):1394–401. 401.e1–4. doi: 10.1053/j.gastro.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Schmidt K, Cowen PJ, Harmer CJ, Tzortzis G, Errington S, Burnet PW. Prebiotic intake reduces the waking cortisol response and alters emotional bias in healthy volunteers. Psychopharmacology. 2015;232(10):1793–801. doi: 10.1007/s00213-014-3810-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Steenbergen L, Sellaro R, van Hemert S, Bosch JA, Colzato LS. A randomized controlled trial to test the effect of multispecies probiotics on cognitive reactivity to sad mood. Brain, behavior, and immunity. 2015;48:258–64. doi: 10.1016/j.bbi.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 265.Kang DW, Park JG, Ilhan ZE, Wallstrom G, Labaer J, Adams JB, et al. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PloS one. 2013;8(7):e68322. doi: 10.1371/journal.pone.0068322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Opp MR, Krueger JM. Sleep and immunity: A growing field with clinical impact. Brain, behavior, and immunity. 2015;47:1–3. doi: 10.1016/j.bbi.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014;159(3):514–29. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 268.Voigt RM, Forsyth CB, Green SJ, Mutlu E, Engen P, Vitaterna MH, et al. Circadian disorganization alters intestinal microbiota. PloS one. 2014;9(5):e97500. doi: 10.1371/journal.pone.0097500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Forsyth CB, Voigt RM, Burgess HJ, Swanson GR, Keshavarzian A. Circadian rhythms, alcohol and gut interactions. Alcohol (Fayetteville, NY) 2015;49(4):389–98. doi: 10.1016/j.alcohol.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Rook GA. Regulation of the immune system by biodiversity from the natural environment: an ecosystem service essential to health. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(46):18360–7. doi: 10.1073/pnas.1313731110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Hoisington AJ, Brenner LA, Kinney KA, Postolache TT, Lowry CA. The microbiome of the built environment and mental health. Microbiome. 2015;3:60. doi: 10.1186/s40168-015-0127-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Logan AC. Dysbiotic drift: mental health, environmental grey space, and microbiota. Journal of physiological anthropology. 2015;34:23. doi: 10.1186/s40101-015-0061-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Slattery DA, Cryan JF. The ups and downs of modelling mood disorders in rodents. ILAR journal / National Research Council, Institute of Laboratory Animal Resources. 2014;55(2):297–309. doi: 10.1093/ilar/ilu026. [DOI] [PubMed] [Google Scholar]
- 274.Nguyen TL, Vieira-Silva S, Liston A, Raes J. How informative is the mouse for human gut microbiota research? Disease models & mechanisms. 2015;8(1):1–16. doi: 10.1242/dmm.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Savage DC. Microbial ecology of the gastrointestinal tract. Annual review of microbiology. 1977;31:107–33. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 276.Walter J, Ley R. The human gut microbiome: ecology and recent evolutionary changes. Annual review of microbiology. 2011;65:411–29. doi: 10.1146/annurev-micro-090110-102830. [DOI] [PubMed] [Google Scholar]
- 277.Booijink CC, Zoetendal EG, Kleerebezem M, de Vos WM. Microbial communities in the human small intestine: coupling diversity to metagenomics. Future microbiology. 2007;2(3):285–95. doi: 10.2217/17460913.2.3.285. [DOI] [PubMed] [Google Scholar]
- 278.Dethlefsen L, Eckburg PB, Bik EM, Relman DA. Assembly of the human intestinal microbiota. Trends in ecology & evolution. 2006;21(9):517–23. doi: 10.1016/j.tree.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 279.Gerritsen J, Smidt H, Rijkers GT, de Vos WM. Intestinal microbiota in human health and disease: the impact of probiotics. Genes & nutrition. 2011;6(3):209–40. doi: 10.1007/s12263-011-0229-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Mariat D, Firmesse O, Levenez F, Guimaraes V, Sokol H, Dore J, et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC microbiology. 2009;9:123. doi: 10.1186/1471-2180-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Rajilic-Stojanovic M, Smidt H, de Vos WM. Diversity of the human gastrointestinal tract microbiota revisited. Environmental microbiology. 2007;9(9):2125–36. doi: 10.1111/j.1462-2920.2007.01369.x. [DOI] [PubMed] [Google Scholar]
- 282.Zoetendal EG, Rajilic-Stojanovic M, de Vos WM. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut. 2008;57(11):1605–15. doi: 10.1136/gut.2007.133603. [DOI] [PubMed] [Google Scholar]
- 283.Mihajlovski A, Alric M, Brugere JF. A putative new order of methanogenic Archaea inhabiting the human gut, as revealed by molecular analyses of the mcrA gene. Research in microbiology. 2008;159(7-8):516–21. doi: 10.1016/j.resmic.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 284.Ott SJ, Kuhbacher T, Musfeldt M, Rosenstiel P, Hellmig S, Rehman A, et al. Fungi and inflammatory bowel diseases: Alterations of composition and diversity. Scandinavian journal of gastroenterology. 2008;43(7):831–41. doi: 10.1080/00365520801935434. [DOI] [PubMed] [Google Scholar]
- 285.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science (New York, NY) 2005;307(5717):1915–20. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 286.Hamer HM, Jonkers DM, Bast A, Vanhoutvin SA, Fischer MA, Kodde A, et al. Butyrate modulates oxidative stress in the colonic mucosa of healthy humans. Clinical nutrition (Edinburgh, Scotland) 2009;28(1):88–93. doi: 10.1016/j.clnu.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 287.Yan J, Herzog JW, Tsang K, Brennan CA, Bower MA, Garrett WS, et al. Gut microbiota induce IGF-1 and promote bone formation and growth. 2016;113(47):E7554–e63. doi: 10.1073/pnas.1607235113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Hoeppli RE, Wu D, Cook L, Levings MK. The environment of regulatory T cell biology: cytokines, metabolites, and the microbiome. Frontiers in immunology. 2015;6:61. doi: 10.3389/fimmu.2015.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Karlsson F, Tremaroli V, Nielsen J, Backhed F. Assessing the human gut microbiota in metabolic diseases. Diabetes. 2013;62(10):3341–9. doi: 10.2337/db13-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Ly NP, Litonjua A, Gold DR, Celedon JC. Gut microbiota, probiotics, and vitamin D: interrelated exposures influencing allergy, asthma, and obesity? The Journal of allergy and clinical immunology. 2011;127(5):1087–94. doi: 10.1016/j.jaci.2011.02.015. quiz 95-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Voth DE, Ballard JD. Clostridium difficile toxins: mechanism of action and role in disease. Clinical microbiology reviews. 2005;18(2):247–63. doi: 10.1128/cmr.18.2.247-263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Leffler DA, Lamont JT. Clostridium difficile Infection. The New England journal of medicine. 2015;373(3):287–8. doi: 10.1056/NEJMc1506004. [DOI] [PubMed] [Google Scholar]
- 293.Lessa FC, Winston LG, McDonald LC. Burden of Clostridium difficile infection in the United States. The New England journal of medicine. 2015;372(24):2369–70. doi: 10.1056/NEJMc1505190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Cornely OA, Nathwani D, Ivanescu C, Odufowora-Sita O, Retsa P, Odeyemi IA. Clinical efficacy of fidaxomicin compared with vancomycin and metronidazole in Clostridium difficile infections: a meta-analysis and indirect treatment comparison. The Journal of antimicrobial chemotherapy. 2014;69(11):2892–900. doi: 10.1093/jac/dku261. [DOI] [PubMed] [Google Scholar]
- 295.Burke KE, Lamont JT. Fecal transplantation for recurrent Clostridium difficile infection in older adults: a review. Journal of the American Geriatrics Society. 2013;61(8):1394–8. doi: 10.1111/jgs.12378. [DOI] [PubMed] [Google Scholar]
- 296.Goldenberg JZ, Ma SS, Saxton JD, Martzen MR, Vandvik PO, Thorlund K, et al. Probiotics for the prevention of Clostridium difficile-associated diarrhea in adults and children. The Cochrane database of systematic reviews. 2013;(5) doi: 10.1002/14651858.CD006095.pub3. Cd006095. [DOI] [PubMed] [Google Scholar]
- 297.Tremaroli V, Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489(7415):242–9. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 298.Sommer F, Backhed F. The gut microbiota--masters of host development and physiology. Nature reviews Microbiology. 2013;11(4):227–38. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 299.Sassone-Corsi M, Nuccio SP, Liu H, Hernandez D, Vu CT, Takahashi AA, et al. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature. 2016;540(7632):280–3. doi: 10.1038/nature20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Zaneveld J, Turnbaugh PJ, Lozupone C, Ley RE, Hamady M, Gordon JI, et al. Host-bacterial coevolution and the search for new drug targets. Current opinion in chemical biology. 2008;12(1):109–14. doi: 10.1016/j.cbpa.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Bejaoui M, Sokol H, Marteau P. Targeting the Microbiome in Inflammatory Bowel Disease: Critical Evaluation of Current Concepts and Moving to New Horizons. Digestive diseases (Basel, Switzerland) 2015;33(Suppl 1):105–12. doi: 10.1159/000437104. [DOI] [PubMed] [Google Scholar]
- 302.Ohkusa T, Koido S. Intestinal microbiota and ulcerative colitis. Journal of infection and chemotherapy : official journal of the Japan Society of Chemotherapy. 2015;21(11):761–8. doi: 10.1016/j.jiac.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 303.Serban DE. Microbiota in Inflammatory Bowel Disease Pathogenesis and Therapy: Is It All About Diet? Nutrition in clinical practice : official publication of the American Society for Parenteral and Enteral Nutrition. 2015;30(6):760–79. doi: 10.1177/0884533615606898. [DOI] [PubMed] [Google Scholar]
- 304.Klag T, Stange EF, Wehkamp J. Defective antibacterial barrier in inflammatory bowel disease. Digestive diseases (Basel, Switzerland) 2013;31(3-4):310–6. doi: 10.1159/000354858. [DOI] [PubMed] [Google Scholar]
- 305.Atreya R, Neurath MF. IBD pathogenesis in 2014: Molecular pathways controlling barrier function in IBD. Nature reviews Gastroenterology & hepatology. 2015;12(2):67–8. doi: 10.1038/nrgastro.2014.201. [DOI] [PubMed] [Google Scholar]
- 306.Lee SH. Intestinal permeability regulation by tight junction: implication on inflammatory bowel diseases. Intestinal research. 2015;13(1):11–8. doi: 10.5217/ir.2015.13.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Chichlowski M, Hale LP. Bacterial-mucosal interactions in inflammatory bowel disease: an alliance gone bad. American journal of physiology Gastrointestinal and liver physiology. 2008;295(6):G1139–49. doi: 10.1152/ajpgi.90516.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Michielan A, D'Inca R. Intestinal Permeability in Inflammatory Bowel Disease: Pathogenesis, Clinical Evaluation, and Therapy of Leaky Gut. Mediators of inflammation. 2015;2015:628157. doi: 10.1155/2015/628157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Tang Y, Forsyth CB, Keshavarzian A. New molecular insights into inflammatory bowel disease-induced diarrhea. Expert review of gastroenterology & hepatology. 2011;5(5):615–25. doi: 10.1586/egh.11.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Coskun M. Intestinal epithelium in inflammatory bowel disease. Frontiers in medicine. 2014;1:24. doi: 10.3389/fmed.2014.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Merga Y, Campbell BJ, Rhodes JM. Mucosal barrier, bacteria and inflammatory bowel disease: possibilities for therapy. Digestive diseases (Basel, Switzerland) 2014;32(4):475–83. doi: 10.1159/000358156. [DOI] [PubMed] [Google Scholar]
- 312.Anderson JM, Van Itallie CM. Physiology and function of the tight junction. Cold Spring Harbor perspectives in biology. 2009;1(2):a002584. doi: 10.1101/cshperspect.a002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Chen SJ, Liu XW, Liu JP, Yang XY, Lu FG. Ulcerative colitis as a polymicrobial infection characterized by sustained broken mucus barrier. World journal of gastroenterology. 2014;20(28):9468–75. doi: 10.3748/wjg.v20.i28.9468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.DiGuilio KM, Mercogliano CM, Born J, Ferraro B, To J, Mixson B, et al. Sieving characteristics of cytokine- and peroxide-induced epithelial barrier leak: Inhibition by berberine. World journal of gastrointestinal pathophysiology. 2016;7(2):223–34. doi: 10.4291/wjgp.v7.i2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Hollander D, Vadheim CM, Brettholz E, Petersen GM, Delahunty T, Rotter JI. Increased intestinal permeability in patients with Crohn's disease and their relatives. A possible etiologic factor Annals of internal medicine. 1986;105(6):883–5. doi: 10.7326/0003-4819-105-6-883. [DOI] [PubMed] [Google Scholar]
- 316.May GR, Sutherland LR, Meddings JB. Is small intestinal permeability really increased in relatives of patients with Crohn's disease? Gastroenterology. 1993;104(6):1627–32. doi: 10.1016/0016-5085(93)90638-s. [DOI] [PubMed] [Google Scholar]
- 317.Wei Y, Zhu W, Gong J, Guo D, Gu L, Li N, et al. Fecal Microbiota Transplantation Improves the Quality of Life in Patients with Inflammatory Bowel Disease. Gastroenterology research and practice. 2015;2015:517597. doi: 10.1155/2015/517597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Moayyedi P, Surette MG, Kim PT, Libertucci J, Wolfe M, Onischi C, et al. Fecal Microbiota Transplantation Induces Remission in Patients With Active Ulcerative Colitis in a Randomized Controlled Trial. Gastroenterology. 2015;149(1):102–9.e6. doi: 10.1053/j.gastro.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 319.Luhrs H, Gerke T, Muller JG, Melcher R, Schauber J, Boxberge F, et al. Butyrate inhibits NF-kappaB activation in lamina propria macrophages of patients with ulcerative colitis. Scandinavian journal of gastroenterology. 2002;37(4):458–66. doi: 10.1080/003655202317316105. [DOI] [PubMed] [Google Scholar]
- 320.Scheppach W, Sommer H, Kirchner T, Paganelli GM, Bartram P, Christl S, et al. Effect of butyrate enemas on the colonic mucosa in distal ulcerative colitis. Gastroenterology. 1992;103(1):51–6. doi: 10.1016/0016-5085(92)91094-k. [DOI] [PubMed] [Google Scholar]
- 321.Cummings JH. Short chain fatty acids in the human colon. Gut. 1981;22(9):763–79. doi: 10.1136/gut.22.9.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Valenzano MC, DiGuilio K, Mercado J, Teter M, To J, Ferraro B, et al. Remodeling of Tight Junctions and Enhancement of Barrier Integrity of the CACO-2 Intestinal Epithelial Cell Layer by Micronutrients. PloS one. 2015;10(7):e0133926. doi: 10.1371/journal.pone.0133926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Peng L, Li ZR, Green RS, Holzman IR, Lin J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. The Journal of nutrition. 2009;139(9):1619–25. doi: 10.3945/jn.109.104638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Bansal T, Alaniz RC, Wood TK, Jayaraman A. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(1):228–33. doi: 10.1073/pnas.0906112107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Shimada Y, Kinoshita M, Harada K, Mizutani M, Masahata K, Kayama H, et al. Commensal bacteria-dependent indole production enhances epithelial barrier function in the colon. PloS one. 2013;8(11):e80604. doi: 10.1371/journal.pone.0080604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Zakostelska Z, Kverka M, Klimesova K, Rossmann P, Mrazek J, Kopecny J, et al. Lysate of probiotic Lactobacillus casei DN-114 001 ameliorates colitis by strengthening the gut barrier function and changing the gut microenvironment. PloS one. 2011;6(11):e27961. doi: 10.1371/journal.pone.0027961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Laval L, Martin R, Natividad JN, Chain F, Miquel S, Desclee de Maredsous C, et al. Lactobacillus rhamnosus CNCM I-3690 and the commensal bacterium Faecalibacterium prausnitzii A2-165 exhibit similar protective effects to induced barrier hyper-permeability in mice. Gut microbes. 2015;6(1):1–9. doi: 10.4161/19490976.2014.990784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Carlsson AH, Yakymenko O, Olivier I, Hakansson F, Postma E, Keita AV, et al. Faecalibacterium prausnitzii supernatant improves intestinal barrier function in mice DSS colitis. Scandinavian journal of gastroenterology. 2013;48(10):1136–44. doi: 10.3109/00365521.2013.828773. [DOI] [PubMed] [Google Scholar]
- 329.Alam A, Leoni G, Quiros M, Wu H, Desai C, Nishio H. The microenvironment of injured murine gut elicits a local pro-restitutive microbiota. 2016;1:15021. doi: 10.1038/nmicrobiol.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Busquets D, Mas-de-Xaxars T, Lopez-Siles M, Martinez-Medina M, Bahi A, Sabat M, et al. Anti-tumour Necrosis Factor Treatment with Adalimumab Induces Changes in the Microbiota of Crohn's Disease. Journal of Crohn's & colitis. 2015;9(10):899–906. doi: 10.1093/ecco-jcc/jjv119. [DOI] [PubMed] [Google Scholar]
- 331.Martinez-Medina M, Denizot J, Dreux N, Robin F, Billard E, Bonnet R, et al. Western diet induces dysbiosis with increased E coli in CEABAC10 mice, alters host barrier function favouring AIEC colonisation. Gut. 2014;63(1):116–24. doi: 10.1136/gutjnl-2012-304119. [DOI] [PubMed] [Google Scholar]
- 332.Darfeuille-Michaud A. Adherent-invasive Escherichia coli: a putative new E. coli pathotype associated with Crohn's disease. International journal of medical microbiology : IJMM. 2002;292(3-4):185–93. doi: 10.1078/1438-4221-00201. [DOI] [PubMed] [Google Scholar]
- 333.Assa A, Vong L, Pinnell LJ, Rautava J, Avitzur N, Johnson-Henry KC, et al. Vitamin D deficiency predisposes to adherent-invasive Escherichia coli-induced barrier dysfunction and experimental colonic injury. Inflammatory bowel diseases. 2015;21(2):297–306. doi: 10.1097/mib.0000000000000282. [DOI] [PubMed] [Google Scholar]
- 334.Kish L, Hotte N, Kaplan GG, Vincent R, Tso R, Ganzle M, et al. Environmental particulate matter induces murine intestinal inflammatory responses and alters the gut microbiome. PloS one. 2013;8(4):e62220. doi: 10.1371/journal.pone.0062220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Yang L, Yan Y. Protein kinases are potential targets to treat inflammatory bowel disease. World journal of gastrointestinal pharmacology and therapeutics. 2014;5(4):209–17. doi: 10.4292/wjgpt.v5.i4.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Ramanan D, Bowcutt R, Lee SC, Tang MS, Kurtz ZD, Ding Y, et al. Helminth infection promotes colonization resistance via type 2 immunity. Science (New York, NY) 2016;352(6285):608–12. doi: 10.1126/science.aaf3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Hansson GC. Role of mucus layers in gut infection and inflammation. Current opinion in microbiology. 2012;15(1):57–62. doi: 10.1016/j.mib.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Johansson ME, Sjovall H, Hansson GC. The gastrointestinal mucus system in health and disease. Nature reviews Gastroenterology & hepatology. 2013;10(6):352–61. doi: 10.1038/nrgastro.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Ciorba MA, Bettonville EE, McDonald KG, Metz R, Prendergast GC, Newberry RD, et al. Induction of IDO-1 by immunostimulatory DNA limits severity of experimental colitis. Journal of immunology (Baltimore, Md : 1950) 2010;184(7):3907–16. doi: 10.4049/jimmunol.0900291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Campieri M, Gionchetti P. Bacteria as the cause of ulcerative colitis. Gut. 2001;48(1):132–5. doi: 10.1136/gut.48.1.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Mirsepasi-Lauridsen HC, Du Z, Struve C, Charbon G, Karczewski J, Krogfelt KA, et al. Secretion of Alpha-Hemolysin by Escherichia coli Disrupts Tight Junctions in Ulcerative Colitis Patients. Clinical and translational gastroenterology. 2016;7:e149. doi: 10.1038/ctg.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Ulluwishewa D, Anderson RC, McNabb WC, Moughan PJ, Wells JM, Roy NC. Regulation of tight junction permeability by intestinal bacteria and dietary components. The Journal of nutrition. 2011;141(5):769–76. doi: 10.3945/jn.110.135657. [DOI] [PubMed] [Google Scholar]
- 343.Chang MY, Boulden J, Valenzano MC, Soler AP, Muller AJ, Mullin JM, et al. Bin1 attenuation suppresses experimental colitis by enforcing intestinal barrier function. Digestive diseases and sciences. 2012;57(7):1813–21. doi: 10.1007/s10620-012-2147-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Thomas S, Mercado JM, DuHadaway J, DiGuilio K, Mullin JM, Prendergast GC. Novel Colitis Immunotherapy Targets Bin1 and Improves Colon Cell Barrier Function. Digestive diseases and sciences. 2016;61(2):423–32. doi: 10.1007/s10620-015-3804-8. [DOI] [PubMed] [Google Scholar]
- 345.Wirawan E, Lippens S, Vanden Berghe T, Romagnoli A, Fimia GM, Piacentini M, et al. Beclin1: a role in membrane dynamics and beyond. Autophagy. 2012;8(1):6–17. doi: 10.4161/auto.8.1.16645. [DOI] [PubMed] [Google Scholar]
- 346.McKnight NC, Zhong Y, Wold MS, Gong S, Phillips GR, Dou Z, et al. Beclin 1 is required for neuron viability and regulates endosome pathways via the UVRAG-VPS34 complex. PLoS genetics. 2014;10(10):e1004626. doi: 10.1371/journal.pgen.1004626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Hayaishi ORY, Takikawa O, Yasui H. Progress in Tryptophan and Serotonin Research. 1984:33–42. [Google Scholar]
- 348.Mellor AL, Munn DH. Tryptophan catabolism and T-cell tolerance: immunosuppression by starvation? Immunology today. 1999;20(10):469–73. doi: 10.1016/s0167-5699(99)01520-0. [DOI] [PubMed] [Google Scholar]
- 349.Munn DH, Shafizadeh E, Attwood JT, Bondarev I, Pashine A, Mellor AL. Inhibition of T cell proliferation by macrophage tryptophan catabolism. The Journal of experimental medicine. 1999;189(9):1363–72. doi: 10.1084/jem.189.9.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Munn DH, Zhou M, Attwood JT, Bondarev I, Conway SJ, Marshall B, et al. Prevention of allogeneic fetal rejection by tryptophan catabolism. Science (New York, NY) 1998;281(5380):1191–3. doi: 10.1126/science.281.5380.1191. [DOI] [PubMed] [Google Scholar]
- 351.Mellor AL, Sivakumar J, Chandler P, Smith K, Molina H, Mao D, et al. Prevention of T cell-driven complement activation and inflammation by tryptophan catabolism during pregnancy. Nature immunology. 2001;2(1):64–8. doi: 10.1038/83183. [DOI] [PubMed] [Google Scholar]
- 352.Muller AJ, DuHadaway JB, Donover PS, Sutanto-Ward E, Prendergast GC. Inhibition of indoleamine 2,3-dioxygenase, an immunoregulatory target of the cancer suppression gene Bin1, potentiates cancer chemotherapy. Nature medicine. 2005;11(3):312–9. doi: 10.1038/nm1196. [DOI] [PubMed] [Google Scholar]
- 353.Munn DH, Sharma MD, Hou D, Baban B, Lee JR, Antonia SJ, et al. Expression of indoleamine 2,3-dioxygenase by plasmacytoid dendritic cells in tumor-draining lymph nodes. The Journal of clinical investigation. 2004;114(2):280–90. doi: 10.1172/jci21583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Munn DH, Mellor AL. Indoleamine 2,3 dioxygenase and metabolic control of immune responses. Trends in immunology. 2013;34(3):137–43. doi: 10.1016/j.it.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Prendergast GC, Smith C, Thomas S, Mandik-Nayak L, Laury-Kleintop L, Metz R, et al. Indoleamine 2,3-dioxygenase pathways of pathogenic inflammation and immune escape in cancer. Cancer immunology, immunotherapy : CII. 2014;63(7):721–35. doi: 10.1007/s00262-014-1549-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Holmgaard RB, Zamarin D, Munn DH, Wolchok JD, Allison JP. Indoleamine 2,3-dioxygenase is a critical resistance mechanism in antitumor T cell immunotherapy targeting CTLA-4. The Journal of experimental medicine. 2013;210(7):1389–402. doi: 10.1084/jem.20130066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Yoshida R, Hayaishi O. Induction of pulmonary indoleamine 2,3-dioxygenase by intraperitoneal injection of bacterial lipopolysaccharide. Proceedings of the National Academy of Sciences of the United States of America. 1978;75(8):3998–4000. doi: 10.1073/pnas.75.8.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Yoshida R, Urade Y, Tokuda M, Hayaishi O. Induction of indoleamine 2,3-dioxygenase in mouse lung during virus infection. Proceedings of the National Academy of Sciences of the United States of America. 1979;76(8):4084–6. doi: 10.1073/pnas.76.8.4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Pfefferkorn ER. Interferon gamma blocks the growth of Toxoplasma gondii in human fibroblasts by inducing the host cells to degrade tryptophan. Proceedings of the National Academy of Sciences of the United States of America. 1984;81(3):908–12. doi: 10.1073/pnas.81.3.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Byrne GI, Lehmann LK, Landry GJ. Induction of tryptophan catabolism is the mechanism for gamma-interferon-mediated inhibition of intracellular Chlamydia psittaci replication in T24 cells. Infection and immunity. 1986;53(2):347–51. doi: 10.1128/iai.53.2.347-351.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Beatty WL, Belanger TA, Desai AA, Morrison RP, Byrne GI. Tryptophan depletion as a mechanism of gamma interferon-mediated chlamydial persistence. Infection and immunity. 1994;62(9):3705–11. doi: 10.1128/iai.62.9.3705-3711.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Bozza S, Fallarino F, Pitzurra L, Zelante T, Montagnoli C, Bellocchio S, et al. A crucial role for tryptophan catabolism at the host/Candida albicans interface. Journal of immunology (Baltimore, Md : 1950) 2005;174(5):2910–8. doi: 10.4049/jimmunol.174.5.2910. [DOI] [PubMed] [Google Scholar]
- 363.Silva NM, Rodrigues CV, Santoro MM, Reis LF, Alvarez-Leite JI, Gazzinelli RT. Expression of indoleamine 2,3-dioxygenase, tryptophan degradation, and kynurenine formation during in vivo infection with Toxoplasma gondii: induction by endogenous gamma interferon and requirement of interferon regulatory factor 1. Infection and immunity. 2002;70(2):859–68. doi: 10.1128/iai.70.2.859-868.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Saito A, Motomura N, Kakimi K, Narui K, Noguchi N, Sasatsu M, et al. Vascular allografts are resistant to methicillin-resistant Staphylococcus aureus through indoleamine 2,3-dioxygenase in a murine model. The Journal of thoracic and cardiovascular surgery. 2008;136(1):159–67. doi: 10.1016/j.jtcvs.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 365.Knubel CP, Martinez FF, Fretes RE, Diaz Lujan C, Theumer MG, Cervi L, et al. Indoleamine 2,3-dioxigenase (IDO) is critical for host resistance against Trypanosoma cruzi. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2010;24(8):2689–701. doi: 10.1096/fj.09-150920. [DOI] [PubMed] [Google Scholar]
- 366.Divanovic S, Sawtell NM, Trompette A, Warning JI, Dias A, Cooper AM, et al. Opposing biological functions of tryptophan catabolizing enzymes during intracellular infection. The Journal of infectious diseases. 2012;205(1):152–61. doi: 10.1093/infdis/jir621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Romani L, Fallarino F, De Luca A, Montagnoli C, D'Angelo C, Zelante T, et al. Defective tryptophan catabolism underlies inflammation in mouse chronic granulomatous disease. Nature. 2008;451(7175):211–5. doi: 10.1038/nature06471. [DOI] [PubMed] [Google Scholar]
- 368.Muller AJ, Mandik-Nayak L, Prendergast GC. Beyond immunosuppression: reconsidering indoleamine 2,3-dioxygenase as a pathogenic element of chronic inflammation. Immunotherapy. 2010;2(3):293–7. doi: 10.2217/imt.10.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Muller AJ, Sharma MD, Chandler PR, Duhadaway JB, Everhart ME, Johnson BA, 3rd, et al. Chronic inflammation that facilitates tumor progression creates local immune suppression by inducing indoleamine 2,3 dioxygenase. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(44):17073–8. doi: 10.1073/pnas.0806173105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Scott GN, DuHadaway J, Pigott E, Ridge N, Prendergast GC, Muller AJ, et al. The immunoregulatory enzyme IDO paradoxically drives B cell-mediated autoimmunity. Journal of immunology (Baltimore, Md : 1950) 2009;182(12):7509–17. doi: 10.4049/jimmunol.0804328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Metz R, Smith C, DuHadaway JB, Chandler P, Baban B, Merlo LM, et al. IDO2 is critical for IDO1-mediated T-cell regulation and exerts a non-redundant function in inflammation. International immunology. 2014;26(7):357–67. doi: 10.1093/intimm/dxt073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Metz R, Rust S, Duhadaway JB, Mautino MR, Munn DH, Vahanian NN, et al. IDO inhibits a tryptophan sufficiency signal that stimulates mTOR: A novel IDO effector pathway targeted by D-1-methyl-tryptophan. Oncoimmunology. 2012;1(9):1460–8. doi: 10.4161/onci.21716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Merlo LM, Pigott E, DuHadaway JB, Grabler S, Metz R, Prendergast GC, et al. IDO2 is a critical mediator of autoantibody production and inflammatory pathogenesis in a mouse model of autoimmune arthritis. Journal of immunology (Baltimore, Md : 1950) 2014;192(5):2082–90. doi: 10.4049/jimmunol.1303012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Baban B, Chandler P, McCool D, Marshall B, Munn DH, Mellor AL. Indoleamine 2,3-dioxygenase expression is restricted to fetal trophoblast giant cells during murine gestation and is maternal genome specific. Journal of reproductive immunology. 2004;61(2):67–77. doi: 10.1016/j.jri.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 375.Kanai M, Funakoshi H, Takahashi H, Hayakawa T, Mizuno S, Matsumoto K, et al. Tryptophan 2,3-dioxygenase is a key modulator of physiological neurogenesis and anxiety-related behavior in mice. Molecular brain. 2009;2:8. doi: 10.1186/1756-6606-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Bessede A, Gargaro M, Pallotta MT, Matino D, Servillo G, Brunacci C, et al. Aryl hydrocarbon receptor control of a disease tolerance defence pathway. Nature. 2014;511(7508):184–90. doi: 10.1038/nature13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Dzutsev A, Goldszmid RS, Viaud S, Zitvogel L, Trinchieri G. The role of the microbiota in inflammation, carcinogenesis, and cancer therapy. European journal of immunology. 2015;45(1):17–31. doi: 10.1002/eji.201444972. [DOI] [PubMed] [Google Scholar]
- 378.Macho Fernandez E, Valenti V, Rockel C, Hermann C, Pot B, Boneca IG, et al. Anti-inflammatory capacity of selected lactobacilli in experimental colitis is driven by NOD2-mediated recognition of a specific peptidoglycan-derived muropeptide. Gut. 2011;60(8):1050–9. doi: 10.1136/gut.2010.232918. [DOI] [PubMed] [Google Scholar]
- 379.Chang MY, Smith C, DuHadaway JB, Pyle JR, Boulden J, Soler AP, et al. Cardiac and gastrointestinal liabilities caused by deficiency in the immune modulatory enzyme indoleamine 2,3-dioxygenase. Cancer biology & therapy. 2011;12(12):1050–8. doi: 10.4161/cbt.12.12.18142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Muller AJ, DuHadaway JB, Chang MY, Ramalingam A, Sutanto-Ward E, Boulden J, et al. Non-hematopoietic expression of IDO is integrally required for inflammatory tumor promotion. Cancer immunology, immunotherapy : CII. 2010;59(11):1655–63. doi: 10.1007/s00262-010-0891-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Smith C, Chang MY, Parker KH, Beury DW, DuHadaway JB, Flick HE, et al. IDO is a nodal pathogenic driver of lung cancer and metastasis development. Cancer discovery. 2012;2(8):722–35. doi: 10.1158/2159-8290.cd-12-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Erdman SE, Rao VP, Poutahidis T, Ihrig MM, Ge Z, Feng Y, et al. CD4(+)CD25(+) regulatory lymphocytes require interleukin 10 to interrupt colon carcinogenesis in mice. Cancer research. 2003;63(18):6042–50. [PubMed] [Google Scholar]
- 383.Saleh M, Trinchieri G. Innate immune mechanisms of colitis and colitis-associated colorectal cancer. Nature reviews Immunology. 2011;11(1):9–20. doi: 10.1038/nri2891. [DOI] [PubMed] [Google Scholar]
- 384.Gurtner GJ, Newberry RD, Schloemann SR, McDonald KG, Stenson WF. Inhibition of indoleamine 2,3-dioxygenase augments trinitrobenzene sulfonic acid colitis in mice. Gastroenterology. 2003;125(6):1762–73. doi: 10.1053/j.gastro.2003.08.031. [DOI] [PubMed] [Google Scholar]
- 385.Harrington L, Srikanth CV, Antony R, Rhee SJ, Mellor AL, Shi HN, et al. Deficiency of indoleamine 2,3-dioxygenase enhances commensal-induced antibody responses and protects against Citrobacter rodentium-induced colitis. Infection and immunity. 2008;76(7):3045–53. doi: 10.1128/iai.00193-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Zelante T, Iannitti RG, Fallarino F, Gargaro M, De Luca A, Moretti S, et al. Tryptophan Feeding of the IDO1-AhR Axis in Host-Microbial Symbiosis. Frontiers in immunology. 2014;5:640. doi: 10.3389/fimmu.2014.00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Zitvogel L, Galluzzi L, Viaud S, Vetizou M, Daillere R, Merad M, et al. Cancer and the gut microbiota: an unexpected link. Science translational medicine. 2015;7(271) doi: 10.1126/scitranslmed.3010473. 271ps1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Kamada N, Seo SU, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nature reviews Immunology. 2013;13(5):321–35. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 389.Wang JL, Chang CH, Lin JW, Wu LC, Chuang LM, Lai MS. Infection, antibiotic therapy and risk of colorectal cancer: a nationwide nested case-control study in patients with Type 2 diabetes mellitus. International journal of cancer Journal international du cancer. 2014;135(4):956–67. doi: 10.1002/ijc.28738. [DOI] [PubMed] [Google Scholar]
- 390.Bonnet M, Buc E, Sauvanet P, Darcha C, Dubois D, Pereira B, et al. Colonization of the human gut by E. coli and colorectal cancer risk. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20(4):859–67. doi: 10.1158/1078-0432.ccr-13-1343. [DOI] [PubMed] [Google Scholar]
- 391.Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, Fan TJ, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science (New York, NY) 2012;338(6103):120–3. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Zhan Y, Chen PJ, Sadler WD, Wang F, Poe S, Nunez G, et al. Gut microbiota protects against gastrointestinal tumorigenesis caused by epithelial injury. Cancer research. 2013;73(24):7199–210. doi: 10.1158/0008-5472.can-13-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nature reviews Microbiology. 2014;12(10):661–72. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 394.Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499(7456):97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- 395.Dapito DH, Mencin A, Gwak GY, Pradere JP, Jang MK, Mederacke I, et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer cell. 2012;21(4):504–16. doi: 10.1016/j.ccr.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Wroblewski LE, Peek RM, Jr, Wilson KT. Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clinical microbiology reviews. 2010;23(4):713–39. doi: 10.1128/cmr.00011-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Konishi H, Fujiya M, Tanaka H, Ueno N, Moriichi K, Sasajima J, et al. Probiotic-derived ferrichrome inhibits colon cancer progression via JNK-mediated apoptosis. Nature communications. 2016;7:12365. doi: 10.1038/ncomms12365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Urbaniak C, Gloor GB, Brackstone M, Scott L, Tangney M, Reid G. The Microbiota of Breast Tissue and Its Association with Breast Cancer. Applied and environmental microbiology. 2016;82(16):5039–48. doi: 10.1128/aem.01235-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.van't Veer P, Dekker JM, Lamers JW, Kok FJ, Schouten EG, Brants HA, et al. Consumption of fermented milk products and breast cancer: a case-control study in The Netherlands. Cancer research. 1989;49(14):4020–3. [PubMed] [Google Scholar]
- 400.de Moreno de LeBlanc A, Matar C, Theriault C, Perdigon G. Effects of milk fermented by Lactobacillus helveticus R389 on immune cells associated to mammary glands in normal and a breast cancer model. Immunobiology. 2005;210(5):349–58. doi: 10.1016/j.imbio.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 401.Hoption Cann SA, van Netten JP, van Netten C. Dr William Coley and tumour regression: a place in history or in the future. Postgraduate medical journal. 2003;79(938):672–80. [PMC free article] [PubMed] [Google Scholar]
- 402.Coley W. The Treatment of Malignant Tumors by Repeated Innoculations of Erysipelas: With a Report of Ten Original Cases. American Journal of the Medical Sciences. 1893;10:487–511. [PubMed] [Google Scholar]
- 403.Nauts HC, Fowler GA, Bogatko FH. A review of the influence of bacterial infection and of bacterial products (Coley's toxins) on malignant tumors in man; a critical analysis of 30 inoperable cases treated by Coley's mixed toxins, in which diagnosis was confirmed by microscopic examination selected for special study. Acta medica Scandinavica Supplementum. 1953;276:1–103. [PubMed] [Google Scholar]
- 404.Hoption Cann SA, van Netten JP, van Netten C, Glover DW. Spontaneous regression: a hidden treasure buried in time. Medical hypotheses. 2002;58(2):115–9. doi: 10.1054/mehy.2001.1469. [DOI] [PubMed] [Google Scholar]
- 405.Bassi P. BCG (Bacillus of Calmette Guerin) therapy of high-risk superficial bladder cancer. Surgical oncology. 2002;11(1-2):77–83. doi: 10.1016/s0960-7404(02)00008-7. [DOI] [PubMed] [Google Scholar]
- 406.Bohle A, Jocham D, Bock PR. Intravesical bacillus Calmette-Guerin versus mitomycin C for superficial bladder cancer: a formal meta-analysis of comparative studies on recurrence and toxicity. The Journal of urology. 2003;169(1):90–5. doi: 10.1097/01.ju.0000039680.90768.b3. [DOI] [PubMed] [Google Scholar]
- 407.Bohle A, Nowc C, Ulmer AJ, Musehold J, Gerdes J, Hofstetter AG, et al. Elevations of cytokines interleukin-1, interleukin-2 and tumor necrosis factor in the urine of patients after intravesical bacillus Calmette-Guerin immunotherapy. The Journal of urology. 1990;144(1):59–64. doi: 10.1016/s0022-5347(17)39366-7. [DOI] [PubMed] [Google Scholar]
- 408.Fleischmann JD, Toossi Z, Ellner JJ, Wentworth DB, Ratliff TL, Imbembo AL. Urinary interleukins in patients receiving intravesical Bacillus Calmette-Guerin therapy for superficial bladder cancer. Cancer. 1989;64(7):1447–54. doi: 10.1002/1097-0142(19891001)64:7<1447::aid-cncr2820640715>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 409.Taniguchi K, Koga S, Nishikido M, Yamashita S, Sakuragi T, Kanetake H, et al. Systemic immune response after intravesical instillation of bacille Calmette-Guerin (BCG) for superficial bladder cancer. Clinical and experimental immunology. 1999;115(1):131–5. doi: 10.1046/j.1365-2249.1999.00756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillere R, Hannani D, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science (New York, NY) 2013;342(6161):971–6. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Liau LM, Black KL, Prins RM, Sykes SN, DiPatre PL, Cloughesy TF, et al. Treatment of intracranial gliomas with bone marrow-derived dendritic cells pulsed with tumor antigens. Journal of neurosurgery. 1999;90(6):1115–24. doi: 10.3171/jns.1999.90.6.1115. [DOI] [PubMed] [Google Scholar]
- 412.Liau LM, Black KL, Martin NA, Sykes SN, Bronstein JM, Jouben-Steele L, et al. Treatment of a patient by vaccination with autologous dendritic cells pulsed with allogeneic major histocompatibility complex class I-matched tumor peptides. Case Report Neurosurgical focus. 2000;9(6):e8. doi: 10.3171/foc.2000.9.6.9. [DOI] [PubMed] [Google Scholar]
- 413.Brossart P. Dendritic cells in vaccination therapies of malignant diseases. Transfusion and apheresis science : official journal of the World Apheresis Association : official journal of the European Society for Haemapheresis. 2002;27(2):183–6. doi: 10.1016/s1473-0502(02)00041-1. [DOI] [PubMed] [Google Scholar]
- 414.So-Rosillo R, Small EJ. Sipuleucel-T (APC8015) for prostate cancer. Expert review of anticancer therapy. 2006;6(9):1163–7. doi: 10.1586/14737140.6.9.1163. [DOI] [PubMed] [Google Scholar]
- 415.Speiser DE, Lienard D, Pittet MJ, Batard P, Rimoldi D, Guillaume P, et al. In vivo activation of melanoma-specific CD8(+) T cells by endogenous tumor antigen and peptide vaccines. A comparison to virus-specific T cells European journal of immunology. 2002;32(3):731–41. doi: 10.1002/1521-4141(200203)32:3<731∷aid-immu731>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 416.Sampson JH, Archer GE, Mitchell DA, Heimberger AB, Herndon JE, 2nd, Lally-Goss D, et al. An epidermal growth factor receptor variant III-targeted vaccine is safe and immunogenic in patients with glioblastoma multiforme. Molecular cancer therapeutics. 2009;8(10):2773–9. doi: 10.1158/1535-7163.mct-09-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Heimberger AB, Sampson JH. The PEPvIII-KLH (CDX-110) vaccine in glioblastoma multiforme patients. Expert opinion on biological therapy. 2009;9(8):1087–98. doi: 10.1517/14712590903124346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Ragupathi G, Cappello S, Yi SS, Canter D, Spassova M, Bornmann WG, et al. Comparison of antibody titers after immunization with monovalent or tetravalent KLH conjugate vaccines. Vaccine. 2002;20(7-8):1030–8. doi: 10.1016/s0264-410x(01)00451-0. [DOI] [PubMed] [Google Scholar]
- 419.Slingluff CL, Jr, Yamshchikov G, Neese P, Galavotti H, Eastham S, Engelhard VH, et al. Phase I trial of a melanoma vaccine with gp100(280-288) peptide and tetanus helper peptide in adjuvant: immunologic and clinical outcomes. Clinical cancer research : an official journal of the American Association for Cancer Research. 2001;7(10):3012–24. [PubMed] [Google Scholar]
- 420.La Rosa C, Wang Z, Brewer JC, Lacey SF, Villacres MC, Sharan R, et al. Preclinical development of an adjuvant-free peptide vaccine with activity against CMV pp65 in HLA transgenic mice. Blood. 2002;100(10):3681–9. doi: 10.1182/blood-2002-03-0926. [DOI] [PubMed] [Google Scholar]
- 421.Purcell AW, McCluskey J, Rossjohn J. More than one reason to rethink the use of peptides in vaccine design. Nature reviews Drug discovery. 2007;6(5):404–14. doi: 10.1038/nrd2224. [DOI] [PubMed] [Google Scholar]
- 422.Alexander RB, Rosenberg SA. Adoptively transferred tumor-infiltrating lymphocytes can cure established metastatic tumor in mice and persist long-term in vivo as functional memory T lymphocytes. Journal of immunotherapy : official journal of the Society for Biological Therapy. 1991;10(6):389–97. doi: 10.1097/00002371-199112000-00001. [DOI] [PubMed] [Google Scholar]
- 423.Bartels CJ, Rosenberg SA, Yang JC. Adoptive cellular immunotherapy of cancer in mice using allogeneic T-cells. Annals of surgical oncology. 1996;3(1):67–73. doi: 10.1007/BF02409054. [DOI] [PubMed] [Google Scholar]
- 424.Barth RJ, Jr, Bock SN, Mule JJ, Rosenberg SA. Unique murine tumor-associated antigens identified by tumor infiltrating lymphocytes. Journal of immunology (Baltimore, Md : 1950) 1990;144(4):1531–7. [PubMed] [Google Scholar]
- 425.Bachanova V, Cooley S, Defor TE, Verneris MR, Zhang B, McKenna DH, et al. Clearance of acute myeloid leukemia by haploidentical natural killer cells is improved using IL-2 diphtheria toxin fusion protein. Blood. 2014;123(25):3855–63. doi: 10.1182/blood-2013-10-532531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Bluming AZ, Ziegler JL. Regression of Burkitt's lymphoma in association with measles infection. Lancet (London, England) 1971;2(7715):105–6. doi: 10.1016/s0140-6736(71)92086-1. [DOI] [PubMed] [Google Scholar]
- 427.Coffey MC, Strong JE, Forsyth PA, Lee PW. Reovirus therapy of tumors with activated Ras pathway. Science (New York, NY) 1998;282(5392):1332–4. doi: 10.1126/science.282.5392.1332. [DOI] [PubMed] [Google Scholar]
- 428.Stojdl DF, Lichty B, Knowles S, Marius R, Atkins H, Sonenberg N, et al. Exploiting tumor-specific defects in the interferon pathway with a previously unknown oncolytic virus. Nature medicine. 2000;6(7):821–5. doi: 10.1038/77558. [DOI] [PubMed] [Google Scholar]
- 429.Martuza RL, Malick A, Markert JM, Ruffner KL, Coen DM. Experimental therapy of human glioma by means of a genetically engineered virus mutant. Science (New York, NY) 1991;252(5007):854–6. doi: 10.1126/science.1851332. [DOI] [PubMed] [Google Scholar]
- 430.Gromeier M, Lachmann S, Rosenfeld MR, Gutin PH, Wimmer E. Intergeneric poliovirus recombinants for the treatment of malignant glioma. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(12):6803–8. doi: 10.1073/pnas.97.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Dobrikova EY, Broadt T, Poiley-Nelson J, Yang X, Soman G, Giardina S, et al. Recombinant oncolytic poliovirus eliminates glioma in vivo without genetic adaptation to a pathogenic phenotype. Molecular therapy : the journal of the American Society of Gene Therapy. 2008;16(11):1865–72. doi: 10.1038/mt.2008.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Brown MC, Dobrikova EY, Dobrikov MI, Walton RW, Gemberling SL, Nair SK, et al. Oncolytic polio virotherapy of cancer. Cancer. 2014;120(21):3277–86. doi: 10.1002/cncr.28862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Hobo W, Maas F, Adisty N, de Witte T, Schaap N, van der Voort R, et al. siRNA silencing of PD-L1 and PD-L2 on dendritic cells augments expansion and function of minor histocompatibility antigen-specific CD8+ T cells. Blood. 2010;116(22):4501–11. doi: 10.1182/blood-2010-04-278739. [DOI] [PubMed] [Google Scholar]
- 434.Marquez-Rodas I, Cerezuela P, Soria A, Berrocal A, Riso A, Gonzalez-Cao M, et al. Immune checkpoint inhibitors: therapeutic advances in melanoma. Annals of translational medicine. 2015;3(18):267. doi: 10.3978/j.issn.2305-5839.2015.10.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Agarwala SS. Novel immunotherapies as potential therapeutic partners for traditional or targeted agents: cytotoxic T-lymphocyte antigen-4 blockade in advanced melanoma. Melanoma research. 2010;20(1):1–10. doi: 10.1097/CMR.0b013e328333bbc8. [DOI] [PubMed] [Google Scholar]
- 436.Ansell SM, Hurvitz SA, Koenig PA, LaPlant BR, Kabat BF, Fernando D, et al. Phase I study of ipilimumab, an anti-CTLA-4 monoclonal antibody, in patients with relapsed and refractory B-cell non-Hodgkin lymphoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15(20):6446–53. doi: 10.1158/1078-0432.ccr-09-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, et al. Nivolumab plus ipilimumab in advanced melanoma. The New England journal of medicine. 2013;369(2):122–33. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Robert C, Thomas L, Bondarenko I, O'Day S, Weber J, Garbe C, et al. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. The New England journal of medicine. 2011;364(26):2517–26. doi: 10.1056/NEJMoa1104621. [DOI] [PubMed] [Google Scholar]
- 439.Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science (New York, NY) 2015;350(6264):1079–84. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science (New York, NY) 2015;350(6264):1084–9. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]