Abstract
Objective
To examine the use of the Prostate Health Index (phi)* as a continuous variable in multivariable risk assessment for aggressive prostate cancer in a large multicenter US study.
Materials and Methods
The study population included 728 men with PSA levels of 2-10 ng/mL and negative digital rectal examination enrolled in a prospective, multi-site early detection trial. The primary endpoint was aggressive prostate cancer, defined as biopsy Gleason score ≥7. First, we evaluated whether the addition of phi improves the performance of currently available risk calculators (PCPT and ERSPC). We also designed and internally validated a new phi-based multivariable predictive model, and created a nomogram.
Results
Of 728 men undergoing biopsy, 118 (16.2%) had aggressive prostate cancer. Phi predicted the risk of aggressive prostate cancer across the spectrum of values. Adding phi significantly improved the predictive accuracy of the PCPT and ERSPC risk calculators for aggressive disease. A new model was created using age, prior biopsy, prostate volume, PSA, and phi with an AUC of 0.746. The bootstrap-corrected model showed good calibration with observed risk for aggressive prostate cancer and had net benefit on decision curve analysis.
Conclusion
Using phi as part of multivariable risk assessment leads to a significant improvement in the detection of aggressive prostate cancer, potentially reducing harms from unnecessary prostate biopsy and overdiagnosis.
Introduction
The Prostate Health Index (phi)* is a mathematical formula derived from the relative concentrations of 3 different PSA forms: total PSA, free PSA and [−2]proPSA. The assay for [−2]proPSA and the clinical utility for phi were approved by the FDA in 2012 for the early detection of prostate cancer.
Several large prospective studies from the United States (US) and Europe have demonstrated that phi outperforms both total and percent free PSA for the prediction of prostate biopsy outcome.[1] Our group recently reported that among US men with PSA levels of 4-10 ng/mL and negative DRE, phi outperformed total and percent free PSA for the identification of clinically significant prostate cancer on biopsy.[2] Another recent study from the Early Disease Research Network showed that phi had an AUC of 0.82 for identifying aggressive prostate cancer.[3] In that population, deferring prostate biopsy for men with a phi <24 would have avoided 36-41% of unnecessary biopsies. Several studies also have shown the ability of phi to predict biopsy reclassification among US and Asian men enrolled in active surveillance.[4, 5]
Currently, phi test results were reported in categories of 4 distinct “risk bins”: 0-24.9, 25.0-34.9, 35.0-54.9, and >55.0. These have a probability of detecting prostate cancer on biopsy of 11.0%, 18.1%, 32.7%, and 52.1%, respectively. Although phi classified in this way results in improved performance compared with total and percent free PSA, an individualized approach using continuous risk assessment is desirable to predict aggressive disease for an individual patient. Furthermore, since no single marker is perfect, a multivariable risk-adapted strategy has been advocated.[6] Previous studies from Europe suggest that phi is a useful addition to multivariable risk stratification. The objective of this study was to evaluate whether the inclusion of phi improves the performance of risk calculators for the prediction of aggressive prostate cancer in a large multicenter cohort of US men with PSA values in the gray zone of 2-10ng/ml and to design an optimized phi-based predictive model.
Methods
From 2003 to 2009, we performed a prospective multicenter US clinical trial of phi, as previously described.[1] At 8 centers, men age ≥50 years were enrolled who met the following criteria: (1) total PSA level between 2 and 10 ng/mL, (2) negative digital rectal examination (DRE), and (3) prostate biopsy with ≥6 cores within 6 months after blood draw providing a histological diagnosis. Men with previous prostate surgery, active urinary tract infection or those using medications that affect PSA levels (e.g., 5 alpha reductase inhibitors) were excluded.
The Beckman Coulter Access 2 Immunoassay Analyzer was used to measure PSA, free PSA and [−2] proPSA. Samples were processed within 8 hours of collection and then were stored at ≤ −70°C before testing (≤5 years from the date of blood draw) at one of 3 laboratories. Phi was calculated using the following formula: [−2]proPSA/fPSA × √PSA.
The prospective multicenter study ultimately included 892 men, of which 79.2% were undergoing initial biopsy, 17.8% had previously undergone biopsy and 3% had unknown biopsy history.[1] Both participants and investigators were blinded to phi results, and all men underwent biopsy irrespective of the phi value. Of these, men with missing data for prior biopsy (n = 27; 3.0%), prostate volume (n = 136; 15.3%), and biopsy Gleason score (n = 1; 0.1%) were excluded from the current analysis.
The primary endpoint for this study was detecting aggressive prostate cancer, defined as biopsy Gleason score ≥7. The secondary endpoint was overall prostate cancer detection on biopsy. Separate models were performed for the overall population with PSA levels from 2 to 10 ng/mL, and for the subset with PSA levels of 4 to 10 ng/mL that currently is the range approved by the FDA. Subset analysis was also performed for men undergoing initial prostate biopsy (n=611), since this represented the majority of the cohort.
First, we used a univariate logistic regression model to determine the probability of aggressive prostate cancer at each value of phi from 1-100. We also calculated the risks of prostate cancer and aggressive prostate cancer for our study population using published coefficients for the modified Prostate Cancer Prevention Trial Risk Calculator (PCPTRC) 2.0 [7] and the European Study of Screening for Prostate Cancer (ERSPC) Risk Calculator 4 + DRE.[8, 9] For PCPTRC, we incorporated age, race, DRE, PSA, and prior biopsy, but not family history, as it was not available in our data set. For the ERSPC risk calculator, we used DRE, prior biopsy, PSA, and prostate volume (categorized into <30 cm3, 30 - 49 cm3, and ≥ 50 cm3, as described by Roobol et al.). The DRE variable was set to 0 for both models, as our dataset only included patients with normal DRE. We assessed whether phi increases the performance of each model by comparing the AUCs of each model with and without phi, using the nonparametric method.[10]
Next, multivariable logistic regression models were fitted using backwards stepwise approach for variable selection. Log-transformed values of PSA and phi were included in the models. Area under the curve (AUC) was used to examine the discrimination of the model, and calibration plots were used to compare agreement between the model predictions with the observed risk of Gleason ≥7 prostate cancer on biopsy. The final model was assessed for the presence of multicollinearity between independent variables by calculating the tolerance statistic.[11]
Decision curve analysis was used to determine the net benefit of the competing models with and without phi compared to strategies of biopsying all men (typical practice) or biopsying none of the men.[12] We adjusted for the oversampling of cancer patients in our study cohort (cancer prevalence of 45% by design) by adding a constant to the linear predictor derived from the model. This adjustment factor was calculated based on the proportion of patients with Gleason ≥7 that would be expected in a patient population with a prostate cancer prevalence of 25%, which resulted in 8.8% prevalence of aggressive cancer. Bootstrap-based internal validation was performed by estimating model performance on 500 resampled datasets.[13, 14] SAS version 9.3 (SAS Institute, Cary, North Carolina) was used for all analyses, and statistical significance was defined as p<0.05.
Results
Overall, 728 men with complete data were included in the analysis. Of these, 118 (16.2%) had aggressive prostate cancer. Table 1 shows the demographics of the study population.
Table 1.
Demographics and clinical characteristics of the study population
| Characteristic | Total n=728 | Aggressive n=118 | Not aggressive/benign n=610 | P value | |
|---|---|---|---|---|---|
| Age | Mean ± SD | 62.8 ± 6.9 | 63.8 ± 6.7 | 62.6 ± 7.0 | 0.069 |
| Race | Black | 39 (5.4) | 6 (5.1) | 33 (5.4) | 0.058 |
| Caucasian | 606 (83.2) | 106 (89.8) | 500 (82.0) | ||
| Other/Unknown | 83 (11.4) | 6 (5.1) | 77 (12.6) | ||
| Prior Biopsy | # (%) | 117 (16.1) | 8 (6.8) | 109 (17.9) | 0.003 |
| PSA (ng/mL) | Mean ± SD | 5.4 ± 1.9 | 5.7 ± 2.0 | 5.3 ± 1.9 | 0.050 |
| Median (Range) | 5.2 (2.0-10.0) | 5.5 (2.2-9.8) | 5.1 (2.0-10.0) | ||
| %fPSA | Mean ± SD | 19.0 ± 8.3 | 15.6 ± 6.8 | 19.6 ± 8.4 | <0.001 |
| Median (Range) | 17.7 (3.1-53.2) | 14.6 (3.7-35.0) | 18.3 (3.1-53.2) | ||
| [−2]proPSA (pg/mL) | Mean ± SD | 15.2 ± 8.6 | 17.4 ± 12.5 | 14.8 ± 7.5 | 0.057 |
| Median (Range) | 13.2 (2.9-90.8) | 14.1 (5.5-90.8) | 13.0 (2.9-51.1) | ||
| phi | Mean ± SD | 38.4 ± 21.1 | 51.5 ± 34.1 | 35.9 ± 16.4 | <0.001 |
| Median (Range) | 33.8 (13.7-325.8) | 43.7 (15.8-325.8) | 32.0 (13.7-144.9) | ||
| Prostate Volume (cc) | Mean ± SD | 50.1 ± 22.2 | 43.1 ± 19.3 | 51.5 ± 22.4 | <0.001 |
| Median (Range) | 45.0 (14.0-209.0) | 39.0 (15.0-120.0) | 47.0 (14.0-209.0) | ||
| Positive biopsy | # (%) | 334 (45.9) | 118 (100) | 216 (35.4) | <0.001 |
| Gleason Score | 6 | 216 (64.7) | 216 (100) | <0.001 | |
| 3 + 4 | 83 (24.9) | 83 (70.3) | |||
| 4 + 3 | 17 (5.1) | 17 (14.4) | |||
| 8 | 7 (2.1) | 7 (5.9) | |||
| 9 | 11 (3.3) | 11 (9.3) |
The median value of phi was 43.7 in the men with aggressive prostate cancer, compared to 32.0 in men with low-grade cancer or a negative biopsy (p<0.001). As shown in Supplemental Table 1, phi predicted the risk of aggressive prostate cancer across the spectrum of values. Table 2 shows the performance characteristics using phi cutoffs of 15-35 for overall and aggressive prostate cancer.
Table 2.
Performance of Prostate Health Index thresholds of 15 to 35
|
phi cutoff |
Probability of prostate cancer |
Sensitivity, PCa |
Specificity, PCa |
NPV, PCa |
Probability of aggressive cancer |
Sensitivity, Aggressive |
Specificity, Aggressive |
NPV, Aggressive |
|---|---|---|---|---|---|---|---|---|
| 15 | 0.072 | 0.997 | 0.020 | 0.953 | 0.020 | 0.997 | 0.015 | 0.981 |
| 16 | 0.080 | 0.994 | 0.036 | 0.947 | 0.022 | 0.992 | 0.025 | 0.968 |
| 17 | 0.088 | 0.991 | 0.053 | 0.947 | 0.024 | 0.983 | 0.036 | 0.957 |
| 18 | 0.097 | 0.982 | 0.081 | 0.931 | 0.026 | 0.975 | 0.057 | 0.959 |
| 19 | 0.105 | 0.976 | 0.099 | 0.925 | 0.029 | 0.975 | 0.072 | 0.967 |
| 20 | 0.114 | 0.967 | 0.137 | 0.926 | 0.031 | 0.975 | 0.102 | 0.976 |
| 21 | 0.122 | 0.955 | 0.155 | 0.912 | 0.034 | 0.966 | 0.118 | 0.973 |
| 22 | 0.131 | 0.928 | 0.206 | 0.896 | 0.037 | 0.949 | 0.162 | 0.971 |
| 23 | 0.140 | 0.910 | 0.251 | 0.894 | 0.039 | 0.941 | 0.200 | 0.972 |
| 24 | 0.149 | 0.895 | 0.292 | 0.893 | 0.042 | 0.932 | 0.233 | 0.973 |
| 25 | 0.158 | 0.880 | 0.322 | 0.890 | 0.045 | 0.907 | 0.256 | 0.966 |
| 26 | 0.167 | 0.862 | 0.358 | 0.886 | 0.048 | 0.898 | 0.287 | 0.967 |
| 27 | 0.177 | 0.844 | 0.414 | 0.889 | 0.051 | 0.890 | 0.331 | 0.969 |
| 28 | 0.186 | 0.814 | 0.447 | 0.878 | 0.054 | 0.873 | 0.366 | 0.968 |
| 29 | 0.195 | 0.784 | 0.482 | 0.870 | 0.057 | 0.831 | 0.397 | 0.960 |
| 30 | 0.205 | 0.757 | 0.518 | 0.865 | 0.060 | 0.805 | 0.430 | 0.958 |
| 31 | 0.214 | 0.725 | 0.556 | 0.858 | 0.064 | 0.780 | 0.467 | 0.956 |
| 32 | 0.223 | 0.692 | 0.586 | 0.851 | 0.067 | 0.754 | 0.500 | 0.955 |
| 33 | 0.233 | 0.677 | 0.609 | 0.850 | 0.070 | 0.754 | 0.523 | 0.957 |
| 34 | 0.242 | 0.650 | 0.647 | 0.847 | 0.073 | 0.737 | 0.559 | 0.957 |
| 35 | 0.251 | 0.608 | 0.678 | 0.838 | 0.077 | 0.686 | 0.592 | 0.951 |
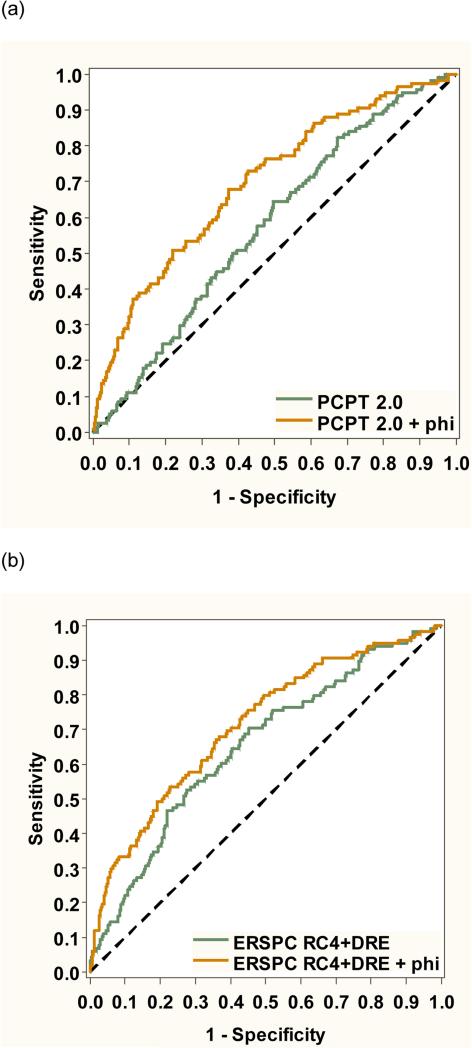
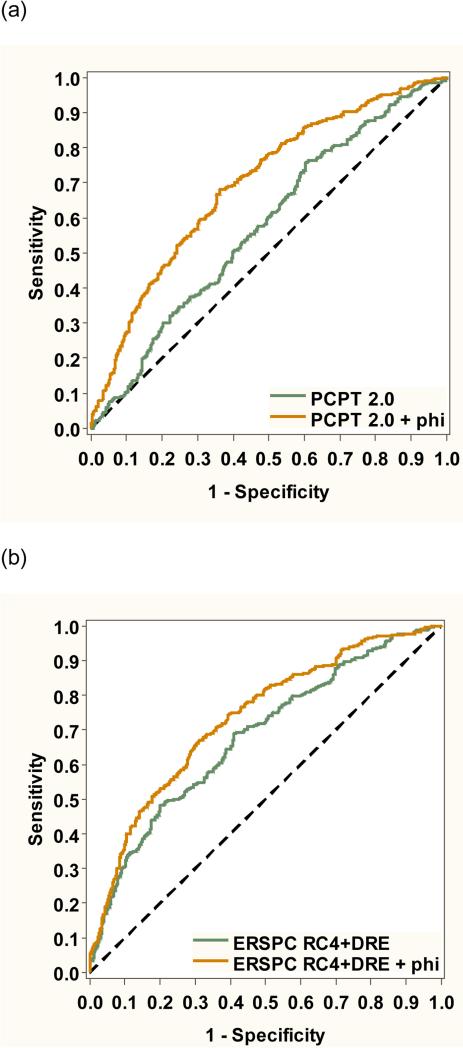
As shown in Figure 1, adding phi to the PCPT risk calculator significantly improved discrimination of aggressive disease (from AUC 0.577 to 0.697, p<0.001). Predictive accuracy for aggressive disease was also improved adding phi to the ERSPC risk calculator (from AUC 0.650 to 0.711, p=0.014). Figure 2 shows the receiver operating characteristic analysis for overall prostate cancer. Adding phi improved discrimination using the PCPT risk calculator (from AUC 0.575 to 0.696, p<0.001) and ERSPC risk calculator (from AUC 0.680 to 0.733, p<0.001).
Figure 1.
Receiver operating characteristic analysis showing the improvement in predictive accuracy for aggressive prostate cancer by adding phi to the (a) PCPT risk calculator and (b) ERSPC risk calculator.
Figure 2.
Receiver operating characteristic analysis showing the improvement in predictive accuracy for overall prostate cancer by adding phi to the (a) PCPT risk calculator and (b) ERSPC risk calculator.
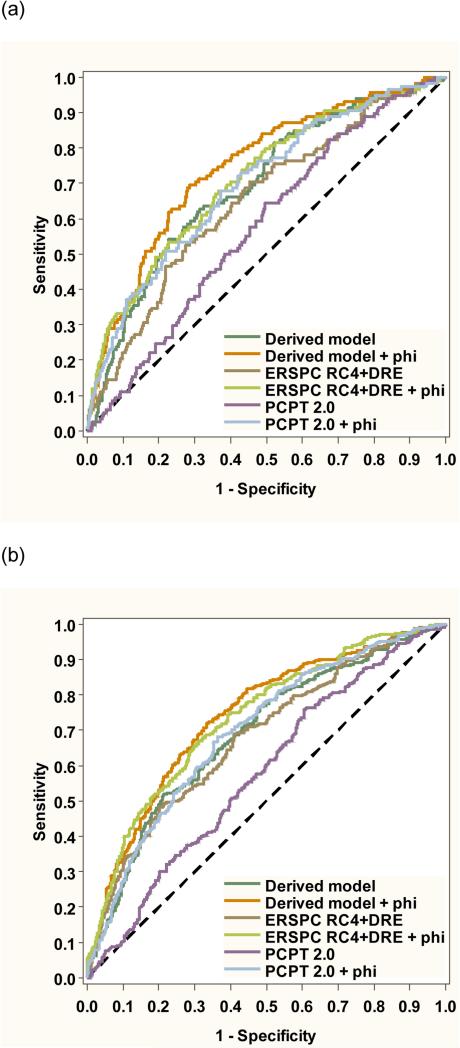
Next, we designed a new model including age, prior biopsy, prostate volume, PSA, and phi for the primary endpoint of aggressive prostate cancer. We found no evidence of multicollinearity in the model (tolerance >0.84 for all independent variables). As shown in Figure 3a, the inclusion of phi improved the model AUC from 0.695 to 0.746 (P = 0.005). The model with phi offered significantly better discrimination than the modified PCPT risk calculator (AUC 0.577, p=<0.001) or ERSPC risk calculator (AUC 0.650, p<0.001). In a separate model additionally including percent free PSA, the addition of phi also significantly improved predictive accuracy (AUC 0.714 to 0.747, p=0.028). In the subset with PSA levels of 4-10 ng/mL, the inclusion of phi improved the model AUC from 0.714 to 0.766 (p=0.018) for aggressive prostate cancer. Among men undergoing initial prostate biopsy, the inclusion of phi improved the model AUC from 0.670 to 0.723 (p=0.006).
Figure 3.
Combined receiver operating characteristic plot comparing the new derived model to the PCPT and ERSPC risk calculators for (a) aggressive and (b) overall prostate cancer detection.
Figure 3b shows the ROC analysis for the secondary endpoint of overall prostate cancer detection on biopsy. The addition of phi significantly improved the AUC for total prostate cancer detection compared to the base model (0.739 vs. 0.688, p < 0.001) among men with PSA levels from 2-10 ng/mL. The phi-based model had significantly better performance for overall prostate cancer compared to the modified PCPT risk calculator (0.739 vs. 0.575, p<0.001) and ERSPC risk calculator (0.739 vs. 0.680, p<0.001). Phi also improved discrimination for total prostate cancer detection beyond the base model in the subset with PSA levels of 4 to 10 ng/mL (0.742 vs 0.696, p=0.004) and in the subset undergoing initial biopsy, the (0.729 vs 0.629, p=<0.001).
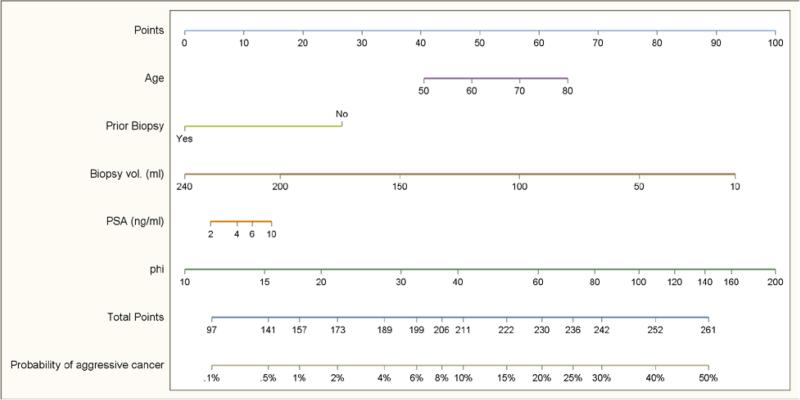
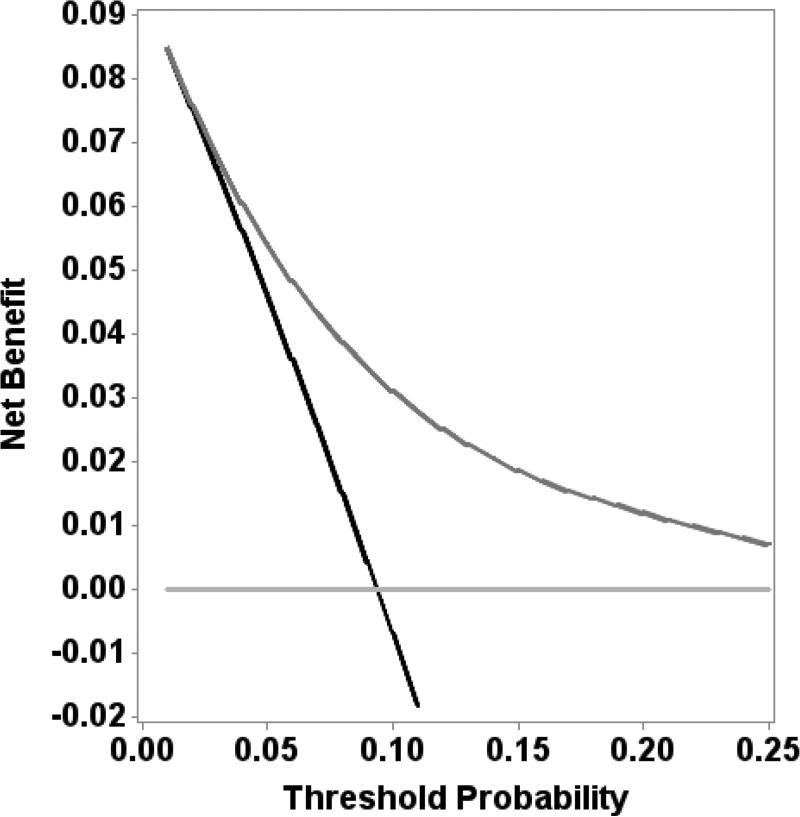
Figure 4 shows a nomogram for aggressive prostate cancer based on our final model incorporating phi with clinical variables. The final model showed good calibration with observed risk for aggressive prostate cancer (Supplemental Figure 1). The bootstrap-corrected AUC was 0.736. Decision curve analysis revealed a net benefit for the model including phi at threshold values of greater than 3% (Figure 5).
Figure 4.
Nomogram with phi and other variables to predict aggressive prostate cancer.
Based on adjusted model for patient population with 25% prevalence of prostate cancer and 8.8% prevalence of aggressive cancer.
Figure 5.
Decision curve analysis comparing final model with phi to biopsy-all and biopsy-none strategies.
Based on adjusted model for patient population with 25% prevalence of prostate cancer and 8.8% prevalence of aggressive cancer.
Discussion
Our results show that phi can be used in a continuous fashion to predict the risk of high-grade prostate cancer on biopsy. Adding phi to currently available risk prediction tools from the PCPT and ERSPC significantly improved the prediction of aggressive prostate cancer in a US population, as in prior studies from Europe.[15] We also designed and internally validated a new phi-based multivariable model, with the greatest overall discrimination for aggressive prostate cancer (AUC 0.746). The model was well-calibrated and led to a net benefit at threshold probabilities of aggressive cancer > 3%.
In the past, a one-size-fits-all approach was used for early prostate cancer detection, wherein a single PSA threshold was used to determine the need for prostate biopsy in all men. However, there is no PSA value at which prostate cancer can be excluded.[16] Rather, PSA and its derivatives are continuous variables reflecting the spectrum of prostate cancer risk. Although phi is currently reported in 4 distinct risk bins, the present results confirm its utility as a continuous variable. In this manuscript, we provide a tabulation of the probability of aggressive cancer (Gleason score ≥7) at each value of phi from 1 −100 estimated using a univariate logistic regression prediction model (Supplemental Table 1) and a nomogram for estimating the probability of aggressive prostate cancer on biopsy based upon patient age, prior prostate biopsy history, prostate volume, PSA, and phi score (Figure 4).
The clinical paradigm has now shifted toward a more personalized approach to prostate biopsy decisions, considering the PSA value along with other risk factors. For example, the European Association of Urology guidelines recommend that multivariable clinical risk-prediction tools should be incorporated into the decision-making process.[17] The Melbourne Consensus statement also recommends that “PSA testing should not be considered on its own, but rather as part of a multivariable approach to early prostate cancer detection.”[6] To this end, we created a multivariable model using continuous phi values along with other well-documented risk factors with improved performance for the identification of clinically-significant prostate cancer. Previous studies have documented high levels of compliance by patients and physicians with recommendations about prostate biopsy from the ERSPC risk calculator, suggesting that multivariable tools provide useful support for clinical decision-making.[18]
A limitation of our study is that the prospective trial did not include men with positive digital rectal examination; therefore, it was not possible to evaluate this factor in our model. However, a previous study by Lughezzani et al. showed that phi significantly improved performance of a multivariable predictive model including digital rectal examination findings.[19] This model was externally validated in men from a multicentric European population in which 17% of men had positive DRE.[20] Nevertheless, men with suspicious nodules on DRE are recommended to undergo prostate biopsy irrespective of the values for PSA and its derivatives. Therefore, it is more clinically relevant to determine a set of factors to aid in biopsy decisions for men with a negative DRE.
Our study also excluded men taking finasteride; however, emerging data suggests that phi may also work in this population.[21] In addition, only 5.4% of the study population was African American, and data on family history were not available; therefore, these factors should be incorporated into future refinements of the model. Finally, we used biopsy endpoints to determine disease aggressiveness, since biopsy results are used to make initial treatment decisions and are therefore useful endpoints for biomarker studies. Nevertheless, our results are consistent with several other studies showing that phi also predicts adverse pathology in the radical prostatectomy specimen.[22-24]
It is noteworthy that several other serum and urine markers are currently commercially available to aid in prostate biopsy decisions, such as the 4K score and PCA3.[7] Data on these markers were not available in the current study population to perform a comparative analysis, and it is not possible to compare AUC's across studies due to differences in the underlying population. However, previous head-to-head comparisons within the same patient population have suggested similar performance of phi and 4K score for predicting high-grade disease on biopsy,[25] and that phi outperforms PCA3 for the identification of clinically significant prostate cancer.[26] There are also logistical differences in that PCA3 requires vigorous digital rectal examination first while phi and 4K score are blood tests. Multiparametric magnetic resonance imaging (mpMRI) also being used increasingly in prostate cancer detection and risk assessment; thus, additional studies are warranted to evaluate a strategy combining phi with mpMRI to reduce unnecessary biopsies. Finally, although the results of internal validation were favorable, external validation of the continuous phi-based model for aggressive prostate cancer is necessary. That notwithstanding, we also show the ability of phi to improve the predictive accuracy of existing clinical tools such as the PCPT and ERSPC risk calculators.
Strengths of our study include the multicenter, prospective study design in which all participants underwent a prostate biopsy for histological evaluation. Unlike several previous studies[19], we focused specifically on men in the “gray zone” of PSA (2-10 ng/ml) with negative DRE where a nomogram is most clinically useful to help decide on biopsy, since men with PSA levels >10 ng/mL or suspicious nodule on DRE are likely to proceed to biopsy regardless of other factors.
Conclusion
Using continuous values of phi as part of multivariable model improves the prediction of aggressive prostate cancer among individual patients with PSA between 2 to 10 ng/mL and benign digital rectal examination. We present a nomogram for estimating the probability of Gleason 7 prostate cancer on biopsy based upon patient age, prior prostate biopsy history, prostate volume, PSA, and phi score.
Supplementary Material
Acknowledgments
Source of Funding: This work was funded by Beckman Coulter Incorporated, Chaska, Minnesota; and supported in part by the Laura and Isaac Perlmutter Cancer Center at New York University to SL, the Louis Feil Charitable Lead Trust to SL; the National Institutes of Health/National Cancer Institute (NIH/NCI) Johns Hopkins Prostate SPORE Grant #P50CA58236, the Early Detection Research Network NIH/NCI Grant #U01-CA86323, and NIH/NCI U01 CA86323 to AWP; NIH/NCI U24 CA115102 to DWC; NIH/NCI U01CA113913 to MS; the Urological Research Foundation, Northwestern-University of Chicago-NorthShore University Prostate SPORE grant (NIH/NCI P50 CA90386-05S2), the Robert H. Lurie Comprehensive Cancer Center grant (NIH/NCI P30 CA60553), and Beckman Coulter Incorporated to WJC.
Footnotes
Phi is a combination of the Beckman Coulter Access Hybritech PSA, free PSA, and p2PSA assays. This manuscript is not intended as off-label promotion of any Beckman Coulter, Inc. product. All trademarks are the property of their respective owners.
References
- 1.Catalona WJ, Partin AW, Sanda MG, Wei JT, Klee GG, Bangma CH, Slawin KM, Marks LS, Loeb S, Broyles DL, Shin SS, Crus AB, Chan DW, Sokoll LJ, Roberts WL, van Schaik RHN, Mizrahi IA. A Multicenter Study of [−2] Pro-Prostate Specific Antigen Combined With Prostate Specific Antigen and Free Prostate Specific Antigen for Prostate Cancer Detection in the 2.0 to 10.0 ng/ml Prostate Specific Antigen Range. J Urol. 2011;185:1650–5. doi: 10.1016/j.juro.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loeb S, Sanda MG, Broyles DL, Shin SS, Bangma CH, Wei JT, et al. The Prostate Health Index Selectively Identifies Clinically Significant Prostate Cancer. J Urol. 2014 Nov 15; doi: 10.1016/j.juro.2014.10.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de la Calle C, Patil D, Wei JT, Scherr DS, Sokoll L, Chan DW, et al. Multicenter Evaluation of the Prostate Health Index (PHI) for Detection of Aggressive Prostate Cancer in Biopsy-Naive Men. J Urol. 2015 Jan 27; doi: 10.1016/j.juro.2015.01.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tosoian JJ, Loeb S, Feng Z, Isharwal S, Landis P, Elliot DJ, et al. Association of [−2]proPSA with Biopsy Reclassification During Active Surveillance for Prostate Cancer. The Journal of urology. 2012 Oct;188:1131–6. doi: 10.1016/j.juro.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirama H, Sugimoto M, Ito K, Shiraishi T, Kakehi Y. The impact of baseline [−2]proPSA-related indices on the prediction of pathological reclassification at 1 year during active surveillance for low-risk prostate cancer: the Japanese multicenter study cohort. Journal of cancer research and clinical oncology. 2014 Feb;140:257–63. doi: 10.1007/s00432-013-1566-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murphy DG, Ahlering T, Catalona WJ, Crowe H, Crowe J, Clarke N, et al. The Melbourne Consensus Statement on the Early Detection of Prostate Cancer. BJU international. 2013 Nov 8; doi: 10.1111/bju.12556. [DOI] [PubMed] [Google Scholar]
- 7.Parekh DJ, Punnen S, Sjoberg DD, Asroff SW, Bailen JL, Cochran JS, et al. A Multi-institutional Prospective Trial in the USA Confirms that the 4Kscore Accurately Identifies Men with High-grade Prostate Cancer. Eur Urol. 2014 Oct 27; doi: 10.1016/j.eururo.2014.10.021. [DOI] [PubMed] [Google Scholar]
- 8.Roobol MJ, van Vugt HA, Loeb S, Zhu X, Bul M, Bangma CH, et al. Prediction of prostate cancer risk: the role of prostate volume and digital rectal examination in the ERSPC risk calculators. Eur Urol. 2012 Mar;61:577–83. doi: 10.1016/j.eururo.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 9.Roobol MJ, Schroder FH, Hugosson J, Jones JS, Kattan MW, Klein EA, et al. Importance of prostate volume in the European Randomised Study of Screening for Prostate Cancer (ERSPC) risk calculators: results from the prostate biopsy collaborative group. World J Urol. 2012 Apr;30:149–55. doi: 10.1007/s00345-011-0804-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988 Sep;44:837–45. [PubMed] [Google Scholar]
- 11.Kleinbaum DG, Kupper LL, Nizam A, Muller KE. Applied Regression Analysis and Other Multivariable Methods (Duxbury Applied) 4th Edition Duxbury Press; North Scituate, MA: 2008. [Google Scholar]
- 12.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006 Nov-Dec;26:565–74. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steyerberg EW, Eijkemans MJ, Harrell FE, Jr., Habbema JD. Prognostic modeling with logistic regression analysis: in search of a sensible strategy in small data sets. Med Decis Making. 2001 Jan-Feb;21:45–56. doi: 10.1177/0272989X0102100106. [DOI] [PubMed] [Google Scholar]
- 14.Nguyen CT, Kattan MW. How to tell if a new marker improves prediction. Eur Urol. 2011 Aug;60:226–8. doi: 10.1016/j.eururo.2011.04.029. discussion 8-30. [DOI] [PubMed] [Google Scholar]
- 15.Foley RW, Gorman L, Sharifi N, Murphy K, Moore H, Tuzova AV, Perry AS, Murphy TB, Lundon DJ, Watson WG. Improving Multivariable Prostate Cancer Risk Assessment Using the Prostate Health Index. BJU Int. 2015 doi: 10.1111/bju.13143. [DOI] [PubMed] [Google Scholar]
- 16.Thompson IM, Pauler DK, Goodman PJ, Tangen CM, Lucia MS, Parnes HL, et al. Prevalence of prostate cancer among men with a prostate-specific antigen level < or =4.0 ng per milliliter. N Engl J Med. 2004 May;27350:2239–46. doi: 10.1056/NEJMoa031918. [DOI] [PubMed] [Google Scholar]
- 17.Heidenreich A, Abrahamsson PA, Artibani W, Catto J, Montorsi F, Van Poppel H, et al. Early detection of prostate cancer: European Association of Urology recommendation. European urology. 2013 Sep;64:347–54. doi: 10.1016/j.eururo.2013.06.051. [DOI] [PubMed] [Google Scholar]
- 18.van Vugt HA, Roobol MJ, Busstra M, Kil P, Oomens EH, de Jong IJ, et al. Compliance with biopsy recommendations of a prostate cancer risk calculator. BJU Int. 2012 May;109:1480–8. doi: 10.1111/j.1464-410X.2011.10611.x. [DOI] [PubMed] [Google Scholar]
- 19.Lughezzani G, Lazzeri M, Larcher A, Lista G, Scattoni V, Cestari A, et al. Development and internal validation of a Prostate Health Index based nomogram for predicting prostate cancer at extended biopsy. The Journal of urology. 2012 Oct;188:1144–50. doi: 10.1016/j.juro.2012.06.025. [DOI] [PubMed] [Google Scholar]
- 20.Lughezzani G, Lazzeri M, Haese A, McNicholas T, de la Taille A, Buffi NM, et al. Multicenter European External Validation of a Prostate Health Index-based Nomogram for Predicting Prostate Cancer at Extended Biopsy. Eur Urol. 2013 Dec 16; doi: 10.1016/j.eururo.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 21.Crawford ED, Arangua P, Jones C, Poage W, Stone N, La Rosa FG, Loeb S, Werahera PN. Prostate Health Index Predicts Upgrading of Men on 5-alpha Reductase Inhibitors. American Urological Association; New Orleans, LA: 2015. [Google Scholar]
- 22.Ferro M, Lucarelli G, Bruzzese D, Perdona S, Mazzarella C, Perruolo G, et al. Improving the prediction of pathologic outcomes in patients undergoing radical prostatectomy: the value of prostate cancer antigen 3 (PCA3), prostate health index (phi) and sarcosine. Anticancer Res. 2015 Feb;35:1017–23. [PubMed] [Google Scholar]
- 23.Tallon L, Luangphakdy D, Ruffion A, Colombel M, Devonec M, Champetier D, et al. Comparative evaluation of urinary PCA3 and TMPRSS2: ERG scores and serum PHI in predicting prostate cancer aggressiveness. International journal of molecular sciences. 2014;15:13299–316. doi: 10.3390/ijms150813299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwen ZR, Tosoian JJ, Sokoll LJ, Mangold L, Humphreys E, Schaeffer EM, et al. Prostate Health Index (PHI) Predicts High-Stage Pathology in African-American Men. Urology. 2015 Dec 10; doi: 10.1016/j.urology.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 25.Nordstrom T, Vickers A, Assel M, Lilja H, Gronberg H, Eklund M. Comparison Between the Four-kallikrein Panel and Prostate Health Index for Predicting Prostate Cancer. Eur Urol. 2015 Jul;68:139–46. doi: 10.1016/j.eururo.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seisen T, Roupret M, Brault D, Leon P, Cancel-Tassin G, Comperat E, et al. Accuracy of the prostate health index versus the urinary prostate cancer antigen 3 score to predict overall and significant prostate cancer at initial biopsy. Prostate. 2015 Jan;75:103–11. doi: 10.1002/pros.22898. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.