Abstract
Escherichia coli clones ST131, ST69, ST95 and ST73 are frequent causes of urinary tract infections (UTI) and bloodstream infections. Specific clones and virulence profiles of E. coli causing UTI in men has been rarely described. The aim of this study was to characterize patient and clonal characteristics of community-acquired UTI caused by E. coli in men (n=12) and women (n=127) in Rio de Janeiro, Brazil, complementing a previous work. We characterized isolates in phylogenetic groups, ERIC2-PCR and PFGE types, MLST, genome similarity and virulence gene-profiles. UTI from men were more frequently caused by phylogenetic group B2 isolates (83% vs 42%, respectively, p 0.01), a group with significantly higher virulence scores compared with women. ST73 was the predominant clone in men (50%) and the second most frequent in women (12%), with the highest virulence score (mean and median=9) among other clones. ST73 gnomes formed at least six clusters. E. coli from men carried significantly higher numbers of virulence genes, such as sfa/focDE (67% vs 27%), hlyA (58% vs 24%), cnf 1 (58% vs 16%), fyuA (100% vs 82%) and MalX (92% vs 44%), compared with isolates from women. These data suggest the predominance and spread of ST73 isolates likely relates to an abundance of virulence determinants.
Keywords: Uropathogenic Escherichia coli, ST73, virulence factors, urinary tract infection
1 Introduction
Successful clones of uropathogenic Escherichia coli (UPEC), determined by multilocus sequence typing (MLST), have been described in various parts of the world. Some of these clones, such as ST131, ST69, ST95 and ST73, predominate as cause of community-acquired urinary tract infections (UTI) and blood stream infections (BSI) (Riley 2014; Miajlovic et al. 2015). The reasons for such predominance are unclear; however, the pathogenic potential and resistance to antimicrobials are frequently described explanations. ST131 is a classic UPEC strain associated with extended spectrum β-lactamase (ESBL) production and fluoroquinolone resistance (Johnson et al. 2009; Fam et al. 2011; Kudinha et al. 2013; Nicolas-Chanoine et al. 2014). ST69 usually shows a multidrug-resistant (MDR) profile (Manges et al. 2001; Colgan et al. 2008). ST95 is associated with neonatal meningitis (Alkeskas et al. 2015), and ST73, also a virulent clone, has been described with plasmid-encoded CTX-M genes (Alhashash et al. 2016).
UTI in men is uncommon, except when abnormalities or invasive procedures in the urinary tract are present (Lipsky 1989; Raynor and Carson 2011). Nevertheless, isolates of the more pathogenic UPEC phylogenetic group B2 are able to cause infection in men without urinary abnormalities (Johnson et al. 2005). A single study specified clones and respective virulence genes affecting men with prostatitis, and showed predominance of B2 isolates carrying at least one toxin-encoding gene (Krieger et al. 2011). Another similar study defined only the presence of ST131, which as B2 isolates, were predominant in pyelonephritis (75%) and cystitis (68%), but scanter among fecal isolates (34%) among E. coli isolates from previously healthy men (Kudinha et al. 2013). Finally, in another study of note, shared strains in a cohort of couples of men with febrile community-acquired UTI and their female sexual partners (Ulleryd et al. 2015) were more likely to belong to MLST clonal complexes (CC)73 and CC127, and carried several virulence genes (Ulleryd et al. 2015). Thus, the specific virulence traits of clones affecting men are poorly described.
In a previous study performed in 2005–2006, we described ST69 as the predominant cause of community-acquired UTI in women in Rio of Janeiro, Brazil (Dias et al. 2009). The second predominant clone was ST73, affecting 12% of the women (Dias et al. 2009) and the most common affecting men, as shown in the present study. While ST69 isolates were significantly more resistant to trimethoprim-sulfamethoxazole and ampicillin, ST73 isolates were more susceptible to trimethoprim-sulfamethoxazole, compared to all other isolates combined. We hypothesized ST69 and ST73 could carry additional virulence genes potentially favoring uropathogenesis and predominance. The aim of this study was to characterize patient and clonal characteristics of community-acquired UTI caused by E. coli in men (n=12) and women (n=127) in Rio de Janeiro, Brazil, complementing a previous work. (Dias et al. 2009).
2 Methods
2.1 Study design and patient population
Patients with symptomatic community-acquired UTI attending one walk-in clinic in Rio of Janeiro from March to November 2005 and from March to November 2006 were included in a previous study (Dias et al. 2009). To define UTI precisely, patients were inquired about UTI symptoms and baseline diseases. The patient population included 139 patients (127 women and 12 men) with UTI. The study was approved by the ethics committee of the institution.
2.2 Characterization of isolates
All isolates were identified by standard microbiological analysis, antimicrobial susceptibility by disk diffusion (CLSI 2016), phylogenetic grouping (A, B1, B2 and D) by triplex PCR and strain typed by ERIC2-PCR, PFGE and MLST (Dias et al. 2009). Whole genome sequences of ST73 isolates were obtained by MiSeq Illumina V3 (2x300pb) in a paired-end library at the University of California Sequencing Facility, Berkeley, CA. DNA was extracted with DNeasy boold and tissue kit (Quiagen, Valencia, CA). The geographic distribution of strain types was visualized with BioNumerics software (version 7.1, Applied Maths, Kortrijk, Belgica). For all isolates, virulence factor coding genes were detected by multiplex-PCR, including papA, papG alleles I, II and III; fimH; afa/draBC and sfa/focDE; hlyA and cnf 1; iutA and fyuA; kpsMT II; traT; ibeA and malX (Johnson and Stell 2000). Control isolates were E. coli ATCC BAA-457, E.coli KS52, E. coli J96, E. coli RS 218, E. coli 20001. For each isolate, we calculated a score for the sum of positive virulence traits. For pap gene positive isolates, only one unit was counted for the presence of either papA and/or papG allele I, II and/or III (Moreno et al. 2005).
2.3 Dendrogram of MLST concatenates, virulence profiles and phylogenetic tree
We obtained a dendrogram using the MLST concatenated data using UPGMA method and the 15 virulence genes converted into binary data, with BioNumerics software (version 7.1, Applied Maths, Kortrijk, Belgium). Draft genome sequence alignments were performed with Progressive Mauve (Darling et al. 2004) and a phylogenetic guide tree was built with Neighbor Joining, visualized with the software Geneious version 10.0 created by Biomatters (Kearse et al. 2012). Reference strains, and their respective accession #, included Nissle1917, CP007799; ABU83972, CP001671; and CFT073, AEO14075.
2.4 Statistical methods
Data was analyzed by OpenEpi (Dean, Sullivan, Soe, 2011). Differences between categorical variables were analyzed by chi-square or Fisher's exact test; continuous variables were compared with Student's t-test. Two-tailed p-value ≤ 0.05 was defined as statistically significant.
3 Results
3.1 Men with UTI
The 12 patients had characteristic UTI symptoms, including dysuria, frequency, urgency, fever, and lumbar pain. Eight individuals had no underlying diseases or urinary abnormalities. Two had prostate increase; one each had bowel cancer and kidney stone. No patients had previous hospitalizations or invasive procedures.
3.2 UPEC antimicrobial susceptibility, clonal groups and virulence factors
Antimicrobial susceptibility prevalences of isolates from men did not differed significantly from isolates from women, as shown in Table 1. Among the 12 study isolates from men, 10 (83%) belonged to phylogenetic group B2, in a frequency significantly higher than in the collection from women (42%, p 0.013), as shown in Table 2. Both patients with UTI by non-group B2 isolates had an underlying urinary tract disease, while only two of the 10 patients with B2 had such conditions (p 0.18). The analysis of virulence genes in these isolates (Table 3) revealed a virulence score significantly higher for group B2 compared to D, A and B1 phylogenetic groups (p < 0.001).
Table 1.
Distribution of antimicrobial resistance prevalences in study isolates by gender, ST69 and ST 73
| Antimicrobial | Number and (%) of resistant antimicrobial isolates | Number and (%) in ST | |||
|---|---|---|---|---|---|
|
| |||||
| Total N=139 | Women N=127 | Men N=12 | ST69 N=15 | ST73 N=21 | |
| Amikacin | 3 (2) | 3 (2) | 0 | 0 | 1 (5) |
| Amoxicillin-clavulanic acid | 28 (20) | 24 (19) | 4 (33) | 3 (20) | 6 (29) |
| Ampicillin | 101 (73) | 91 (72) | 10 (83) | 14 (93) | 14 (67) |
| Cefotaxime | 4 (3) | 2 (1.5) | 2 (17) | 0 | 0 |
| Cefoxitin | 12 (9) | 10 (0.8) | 2 (17) | 2 (13) | 0 |
| Ceftazidime | 1 (0.7) | 0 | 1 (8) | 0 | 0 |
| Cefuroxime | 8 (6) | 6 (5) | 2 (17) | 2 (13) | 0 |
| Cephalothin | 107 (77) | 98 (77) | 9 (75) | 14 (93) | 17 (81) |
| Ciprofloxacin | 9 (7) | 8 (6) | 1 (8) | 0 | 0 |
| Fosfomycin | 4 (3) | 4 (3) | 0 | 1(7) | 0 |
| Gentamicin | 3 (2) | 3 (2) | 0 | 0 | 0 |
| Imipenem | 0 | 0 | 0 | 0 | 0 |
| Levofloxacin | 9 (7) | 8 (6) | 1(8) | 0 | 0 |
| Meropenem | 0 | 0 | 0 | 0 | 0 |
| Nalidixic acid | 14 (10) | 13 (10) | 1 (8) | 1 (7) | 0 |
| Nitrofurantoin | 4 (3) | 4 (3) | 0 | 0 | 0 |
| Norfloxacin | 9 (7) | 8 (6) | 1(8) | 0 | 0 |
| Trimethoprim/Sulfamethoxazole | 63 (45) | 59 (46) | 4 (33) | 14 (93) | 2 (10)a |
p <0.001 for comparison with ST69. All other comparisons are p >0.05.
Table 2.
Distribution of 139 E. coli study isolates by phylogenetic group, gender, and sequence types
| Phylogenetic group and ST | Number and (%) of
isolates in group |
||
|---|---|---|---|
| Female | Male | Total | |
| B2 | 53 (42) | 10 (83)a | 63 (45) |
| ST73b | 15 | 6 | 21 |
| ST14 | 2 | 1 | 3 |
| ST131 | 2 | 1 | 3 |
| ST706 | 2 | 1 | 3 |
| ST699 | 0 | 1 | 1 |
| D | 46 (36) | 1 (8) | 47 (34) |
| ST405 b | 3 | 1 | 4 |
| A | 18 (14) | 0 | 18 (13) |
| B1 | 10 (8) | 1 (8) | 11 (8) |
| ST90 b | 1 | 1 | 2 |
| Total | 127 (100) | 12 (100) | 139 (100) |
p 0.013, other comparisons are p >0.05.
ST identified in patients of male gender with underlying disease of the urinary tract.
Table 3.
Distribution of virulence factors in study isolates by phylogenetic group
| Virulence factor/Gene | Number and (%) of positive isolates | p value | ||||||
|---|---|---|---|---|---|---|---|---|
| B2 (n=63) | D (n=47) | A (n=18) | B1 (n=11) | Total (n=139) | B2 vs D | B2 vs A | B2 vs B1 | |
| Adhesin | ||||||||
| papA | 39 (62) | 26 (55) | 0 | 0 | 65 (45) | NS | <0.001 | <0.001 |
| papG I | 0 | 0 | 0 | 0 | 0 | |||
| papG II | 29 (46) | 33 (70) | 1 (6) | 0 | 63 (45) | 0.02 | <0.01 | <0.01 |
| papG III | 12 (19) | 4 (9) | 1 (6) | 0 | 17 (12) | 0.02 | NS | NS |
| fimH | 63 (100) | 37 (79) | 13 (72) | 10 (91) | 123 (88) | <0.001 | <0.001 | NS |
| afa/draBC | 4 (6) | 7 (23) | 1 (6) | 0 | 12 (9) | NS | NS | NS |
| sfa/focDE | 41 (65) | 1 (2) | 0 | 0 | 42 (30) | <0.001 | <0.001 | <0.001 |
| Toxins | ||||||||
| hlyA | 37 (59) | 0 | 0 | 0 | 37 (27) | <0.001 | <0.001 | <0.001 |
| cnf1 | 27 (43) | 0 | 0 | 0 | 27 (19) | <0.001 | <0.001 | <0.01 |
| Siderophore | ||||||||
| iutA | 45 (71) | 44 (94) | 12 (67) | 7 (64) | 108 (78) | <0.01 | NS | NS |
| fyuA | 59 (94) | 43 (91) | 12 (67) | 2 (18) | 116 (84) | NS | 0.01 | <0.001 |
| Invasin | ||||||||
| kpsMT II | 53 (84) | 39 (83) | 9 (50) | 1 (9) | 102 (73) | NS | <0.01 | <0.001 |
| ibeA | 11 (17) | 2 (4) | 0 | 0 | 13 (9) | NS | NS | NS |
| traT | 30 (48) | 31 (66) | 4 (22) | 6 (55) | 71 (51) | NS | NS | NS |
| Pathogenicity island | ||||||||
| MalX | 55 (87) | 11(23) | 1 (6) | 0 | 67 (48) | <0.001 | <0.001 | <0.001 |
|
Virulence Scorea mean; median |
7; 7 | 5; 5 | 3; 3 | 2; 3 | 6; 6 | <0.001 | <0.001 | <0.001 |
The sum of positive virulence traits, proposed by Moreno (2005)
NS: non-significant difference (p > 0.05)
Comparison of D with B1 and A groups revealed differences for genes papG II, fyuA and kpsMT II (D vs B1, p < 0.001); papA, papG II (D vs A, p < 0.001) and traT (D vs A, p < 0.003)
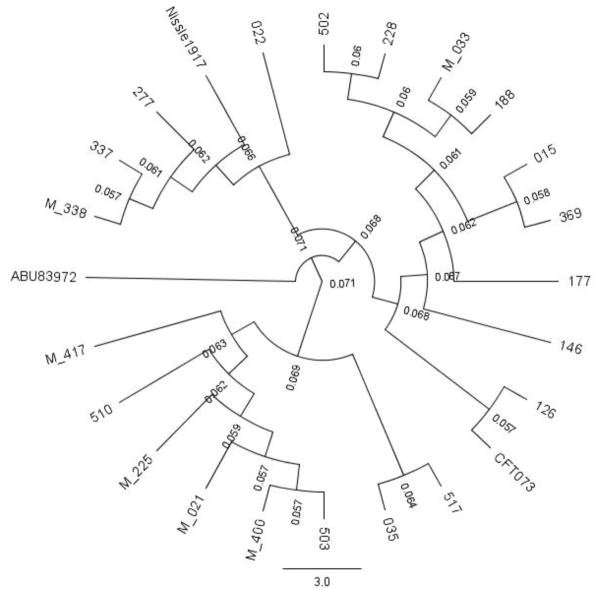
ST73 was the predominant clone; other clones included only a few isolates (Table 3). From the six men with UTI by ST73, four had no underlying disease or urinary abnormality. To explore the relationship among isolates and three other ST73 reference strains, Nissle 1917, ABU83972 and CFT073 (Vejborg et al. 2010), we built a phylogenetic guide tree, shown in Fig. 1. Isolates formed at least six clusters with >0.068 substitutions per site. Four of the six isolates from men remained in the same cluster.
Fig 1.
Phylogenetic guide tree performed by Progressive MAUVE with Neighbor Joining. Tree was visualized with Geneious version 10.0 created by Biomatters. Isolate codes are at the branch tips. Codes from male patients start with M. Values in smaller caption are numbers of substitutions per site.
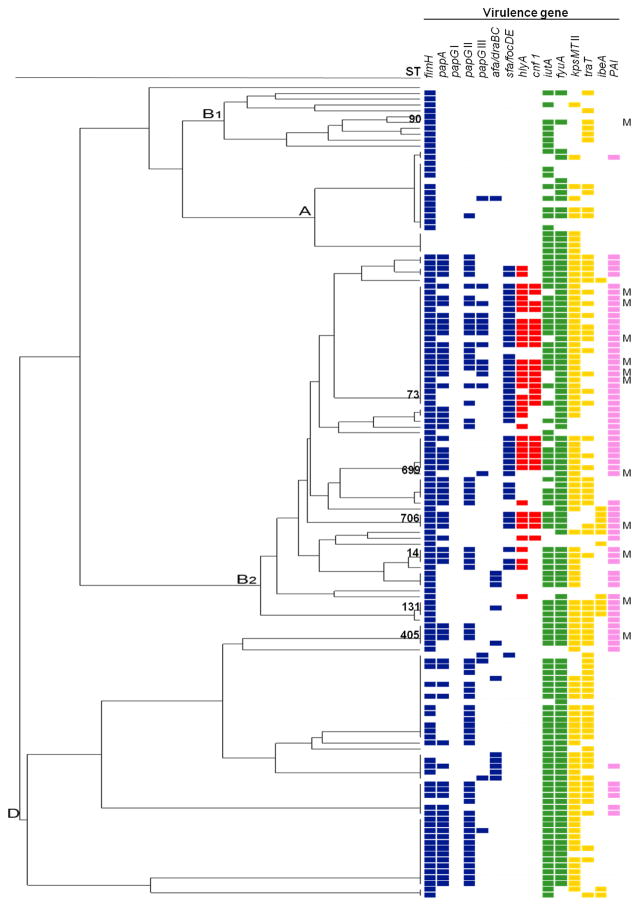
Comparison of antimicrobial susceptibility prevalences of all ST69 and ST73 isolates revealed resistance to trimethoprim/sulfamethoxazole was significantly higher in ST69 (p <0.001). The presence of virulence genes in each isolate shown in a dendrogram with the MLST concatenate revealed a clear accumulation of genes in B2 group (Fig. 2). Isolates from men carried significantly higher numbers of virulence genes such as sfa/focDE (67% vs 27%), hlyA (58% vs 24%), cnf 1 (58% vs 16%), fyuA (100% vs 82%) and MalX (92% vs 44%) compared to women (Table 4). In fact, differences between ST73 and ST69 accounted for this finding for adhesin sfa/focDE, toxins hlyA and cnf1, and pathogenicity island MalX. ST73 isolated from men and women had a similar virulence profile; only iutA gene was more common in women (p=0.06).
Fig. 2.
Dendrogram shows virulence genotypes of 139 UPEC isolates obtained from community acquired UTI. Colored squares indicate the presence of the gene. Color codes: blue, adhesins; red, toxins; green, siderophore receptors; yellow: invasins; pink: miscellaneous virulence genes. Sequence types (ST) and phylogenetic groups are indicated for all isolates. Isolates for men are indicated by letter M. The order of ST for each phylogenetic group is described respectively, B1: ST398, ST453, ST708, ST336, ST707, ST295, ST90, ST155, ST58, ST443, ST1136; A: ST697, ST10, ST93; B2: ST95, ST421, ST420, ST73, ST827, ST698, ST12, ST452, ST127, ST699, ST144, ST703, ST372, ST706, ST135, ST141, ST583, ST14, ST704, ST550, ST700, ST702, ST131, ST705; D: ST405, ST701, ST69, ST393, STS94, ST38, ST59, ST62, ST354.
Table 4.
Distribution of virulence genes in study isolates by gender, ST69, and ST found in men
| Virulence factor/Gene | Number and(%)of
positive isolates |
pa | Number and (%) in ST
|
pb | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total N=139 | Women N=127 | Men N=12 | ST69 N=15 | ST73 N=21 | ST405 N=4 | ST131 N=3 | ST706 N=3 | ST14 N= 3 | ST699 N=1 | ST90 N=1 | |||
| Adhesin | |||||||||||||
| papA | 65 (45) | 59 (46) | 6 (50) | NS | 4 (27) | 14 (67) | 3 (75) | 0 | 3 | 3 | 0 | 0 | 0.041 |
| papG I | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| papG II | 63 (45) | 59 (46) | 4 (33) | NS | 12 (80) | 14 (67) | 3 (75) | 0 | 0 | 3 | 0 | 0 | NS |
| papG III | 17 (12) | 13 (10) | 4 (33) | NS | 2 (13) | 11 (52) | 0 | 0 | 0 | 0 | 1 | 0 | 0.04 |
| fimH | 123 (88) | 111 (87) | 12 (100) | 15 (100) | 21 (100) | 4 (100) | 3 | 3 | 3 | 1 | 1 | NS | |
| afa/draBC | 12 (9) | 12 (9) | 0 | NS | 1 (7) | 0 | 1 (25) | 1 | 0 | 0 | 0 | 0 | NS |
| sfa/focDE | 42 (30) | 34 (27) | 8 (67) | 0.02 | 1 (7) | 20 (95) | 0 | 0 | 3 | 2 | 1 | 0 | <0.001 |
| Toxins | |||||||||||||
| hlyA | 37 (27) | 30 (24) | 7 (58) | 0.03 | 0 | 16 (76) | 0 | 0 | 3 | 2 | 0 | 0 | <0.001 |
| cnf 1 | 27 (19) | 20 (16) | 7 (58) | 0.004 | 0 | 17 (81) | 0 | 0 | 0 | 0 | 0 | 0 | <0.001 |
| Siderophore | |||||||||||||
| iutA | 108 (78) | 101 (80) | 7 (58) | NS | 13 (87) | 15 (71) | 3 | 3 | 3 | 3 | 0 | 1 | NS |
| fyuA | 116 (84) | 104 (82) | 12 (100) | <0.001 | 14 (93) | 21 (100) | 3 | 3 | 3 | 3 | 1 | 1 | NS |
| Invasin | |||||||||||||
| kpsMT II | 102 (73) | 92 (72) | 10 (83) | NS | 10 (67) | 21 (100) | 3 | 3 | 0 | 3 | 1 | 0 | 0.02 |
| traT | 13 (9) | 11 (9) | 2 (17) | NS | 14 (93) | 10 (48) | 3 | 3 | 1 | 1 | 0 | 1 | 0.01 |
| ibeA | 71 (51) | 62 (49) | 9 (75) | NS | 0 | 0 | 3 | 3 | 3 | 0 | 0 | 0 | 0 |
| Pathogenicity island | |||||||||||||
| MalX | 67 (48) | 56 (44) | 11 (92) | 0.003 | 0 | 21 (100) | 3 | 3 | 3 | 3 | 1 | 0 | <0.001 |
| Virulence Score: mean; median | 6; 6 | 7; 7 | 8; 8 | NS | 5; 6 | 9; 9 | 7; 7 | 7; 7 | 9; 9 | 8; 8 | 6; 6 | 4; 4 | <0.001 |
p for comparison between women vs men;
p for comparison between ST69 and ST73.
p < 0.05 are shown in bold; NS: no significant difference (p > 0.05) ST: sequence type. ST69: isolates identified only in women. ST73, ST405, ST131, ST706, ST14; ST699 and ST90: isolates identified in women and men Virulence score: the sum of positive virulence traits, proposed by Moreno (2005)
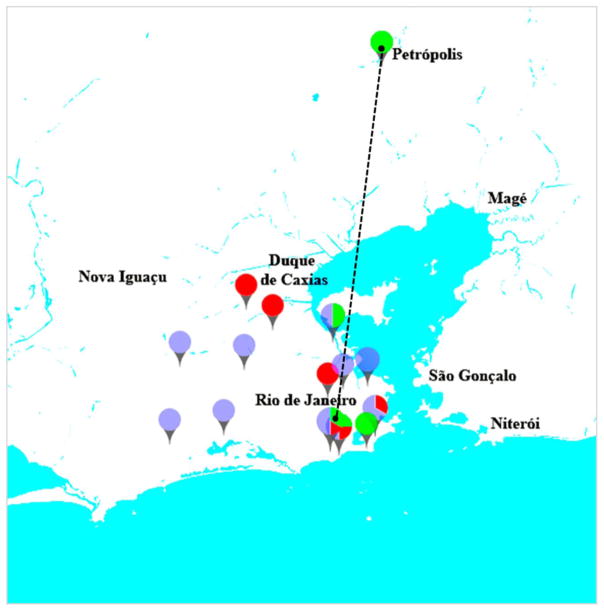
3.3 Geographical distribution of ST in men
ST73 isolates were widely distributed in the city of Rio de Janeiro (Fig. 3), affecting male patients who lived at distances 54 Km apart. Review of patients’ appointment dates showed each subject with ST73 UTI visited the clinic on a different day.
Fig. 3.
Geographic distribution of ST73 isolates identified in men (green) and women (purple), and other ST (red) detected in men. Names refer to municipalities. Dashed line marks 54 Km distance.
4 Discussion
In the present study, analysis of UPEC isolates showed most of them belonged to phylogenetic group B2, but such predominance was higher among men with UTI. Similar results were observed by others (Johnson et al. 2005; Piatti et al. 2008; Salvador et al. 2012). Although a sample size with 12 cases seems low, this sample came from a cohort prospective study, and reflects all cases occurring over 18 months sampling. Group B2 UPEC isolates are more likely to colonize the host and break through barrier defenses to cause UTI. It is plausible that isolates with increased capacity to cause disease, as in group B2, are more likely to cause infection in men. Indeed, isolates in group B2 were the most virulent. The comparison of virulence gene frequencies among the two most common phylogenetic groups B2 and D showed, as expected, increased frequencies of adhesins papGIII, fimH and sfa/focDE, toxins hlyA, cnf 1 and pathogenicity island MalX, and a higher virulence score in group B2. Similar differences were described by others (Johnson and Stell 2000; Moreno et al. 2005; Piatti et al. 2008; Srivastava et al. 2014).The predominant clones causing UTI in the present study, ST73, ST69, ST10, ST62, ST127 and ST59, are quite similar to those described by others (Gibreel et al. 2012; Banerjee et al. 2013; Horner et al. 2014). ST73, one of the major clones described by others, caused half of the infections in men in this study (Salvador et al. 2012; Banerjee et al. 2013; Manges et al. 2015). These data show ST73 may have increased virulence to humans; in fact, it had the highest virulence score in the study (mean 9), together with ST706 (a clone with only three isolates). However, the small sample size of isolates from men was a limitation of this study; a bigger sample size could have pointed additional important clones affecting men.
ST73 may include a variety of strain types, including the non-pathogenic fecal isolate Nissle 1917, the asymptomatic bacteriuria isolate ABU83972, and the highly virulent pyelonephritis strain CFT073 (Vejborg et al. 2010). Indeed, a cladrogram built with these prototypes and ST73 study isolates formed various clusters, leaving ABU83972 in a monophyletic branch, as described by others (Vejborg et al. 2010).
Comparison of virulence factors of ST73 and ST69 revealed increased frequencies of several genes in ST73. However, traT was significantly higher in ST69 (93% vs. 48%, p: 0.01), as already described (Blanco et al. 2011; Gibreel et al. 2012). This gene encodes serum resistance capacity and may confer additional advantage to ST69 isolates in women. The success of ST69 can also be due to the presence of antimicrobial resistance, especially to trimethoprim-sulfamethoxazole, selected by the high prescription of this drug, freely available in Brazilian pharmacies by the time the study was performed. The presence of several adhesins, might have also favored adaptation of ST69 isolates to uroepithelium. The presence of invasins also supports the virulence of this clone to the host. Finally, in the present study, the presence of toxins was significantly higher in isolates from men compared to women, and in isolates of ST73 compared to ST69. A high frequency of toxin producing in UTI isolates from men was also noticed by others (Andreu et al. 1997).
In a large study of E. coli from food animals and humans, ST73 was exclusive of humans, showing food sources are unlikely (Manges et al. 2015). This clone is probably a true human pathogen, circulating unnoticed in human microbiota and causing UTI in those with unidentified predisposing conditions.
Highlights.
We provide key characteristic of Escherichia coli from UTI in men.
The characteristics presented may help understand UTI pathogenesis.
ST73 was a predominant clone.
ST73 had the highest virulence score compared to the other isolates.
Acknowledgments
This study was supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ). This work used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 Instrumentation Grants S10RR029668 and S10RR027303.
Footnotes
Conflict of interest: The authors declare that they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alhashash F, Wang X, Paszkiewicz K, Diggle M, Zong Z, McNally A. Increase in bacteraemia cases in the East Midlands region of the UK due to MDR Escherichia coli ST73: high levels of genomic and plasmid diversity in causative isolates. J Antimicrob Chemother. 2016 Feb;71(2):339–43. doi: 10.1093/jac/dkv365. [DOI] [PubMed] [Google Scholar]
- Alkeskas A, Ogrodzki P, Saad M, Masood N, Rhoma NR, Moore K, et al. The molecular characterisation of Escherichia coli K1 isolated from neonatal nasogastric feeding tubes. BMC Infect Dis. 2015;15:449. doi: 10.1186/s12879-015-1210-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreu A, Stapleton AE, Fennell C, Lockman HA, Xercavins M, Fernandez F, et al. Urovirulence determinants in Escherichia coli strains causing prostatitis. J Infect Dis. 1997;176(2):464–469. doi: 10.1086/514065. [DOI] [PubMed] [Google Scholar]
- Banerjee R, Johnston B, Lohse C, Chattopadhyay S, Tchesnokova V, Sokurenko EV, et al. The clonal distribution and diversity of extraintestinal Escherichia coli isolates vary according to patient characteristics. Antimicrob Agents Chemother. 2013 Dec;57(12):5912–7. doi: 10.1128/AAC.01065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco J, Mora A, Mamani R, Lopez C, Blanco M, Dahbi G, et al. National survey of Escherichia coli causing extraintestinal infections reveals the spread of drug-resistant clonal groups O25b:H4-B2-ST131, O15:H1-D-ST393 and CGA-D-ST69 with high virulence gene content in Spain. J Antimicrob Chemother. 2011 Sep 1;66(9):2011–21. doi: 10.1093/jac/dkr235. [DOI] [PubMed] [Google Scholar]
- Colgan R, Johnson JR, Kuskowski M, Gupta K. Risk Factors for Trimethoprim-Sulfamethoxazole Resistance in Patients with Acute Uncomplicated Cystitis. Antimicrob Agents Chemother. 2008 Mar 1;52(3):846–51. doi: 10.1128/AAC.01200-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004 Jul;14(7):1394–403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias RCS, Marangoni DV, Smith SP, Alves EM, Pellegrino FLPC, Riley LW, et al. Clonal composition of Escherichia coli causing community-acquired urinary tract infections in the State of Rio de Janeiro, Brazil. Microb Drug Resist Larchmt N. 2009 Dec;15(4):303–8. doi: 10.1089/mdr.2009.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fam N, Leflon-Guibout V, Fouad S, Aboul-Fadl L, Marcon E, Desouky D, et al. CTX-M-15-Producing Escherichia coli Clinical Isolates in Cairo (Egypt), Including Isolates of Clonal Complex ST10 and Clones ST131, ST73, and ST405 in Both Community and Hospital Settings. Microb Drug Resist. 2011 Mar;17(1):67–73. doi: 10.1089/mdr.2010.0063. [DOI] [PubMed] [Google Scholar]
- Gibreel TM, Dodgson AR, Cheesbrough J, Fox AJ, Bolton FJ, Upton M. Population structure, virulence potential and antibiotic susceptibility of uropathogenic Escherichia coli from Northwest England. J Antimicrob Chemother. 2012 Feb 1;67(2):346–56. doi: 10.1093/jac/dkr451. [DOI] [PubMed] [Google Scholar]
- Horner C, Fawley W, Morris K, Parnell P, Denton M, Wilcox M. Escherichia coli bacteraemia: 2 years of prospective regional surveillance (2010–12) J Antimicrob Chemother. 2014 Jan 1;69(1):91–100. doi: 10.1093/jac/dkt333. [DOI] [PubMed] [Google Scholar]
- Johnson JR, Menard M, Johnston B, Kuskowski MA, Nichol K, Zhanel GG. Epidemic Clonal Groups of Escherichia coli as a Cause of Antimicrobial-Resistant Urinary Tract Infections in Canada, 2002 to 2004. Antimicrob Agents Chemother. 2009 Jul 1;53(7):2733–9. doi: 10.1128/AAC.00297-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JR, Scheutz F, Ulleryd P, Kuskowski MA, O’Bryan TT, Sandberg T. Host-pathogen relationships among Escherichia coli isolates recovered from men with febrile urinary tract infection. Clin Infect Dis. 2005;40(6):813–822. doi: 10.1086/428048. [DOI] [PubMed] [Google Scholar]
- Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis. 2000;181(1):261–272. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinforma Oxf Engl. 2012 Jun 15;28(12):1647–9. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger JN, Dobrindt U, Riley DE, Oswald E. Acute Escherichia coli Prostatitis in Previously Health Young Men: Bacterial Virulence Factors, Antimicrobial Resistance, and Clinical Outcomes. Urology. 2011 Jun;77(6):1420–5. doi: 10.1016/j.urology.2010.12.059. [DOI] [PubMed] [Google Scholar]
- Kudinha T, Johnson JR, Andrew SD, Kong F, Anderson P, Gilbert GL. Distribution of phylogenetic groups, sequence type ST131, and virulence-associated traits among Escherichia coli isolates from men with pyelonephritis or cystitis and healthy controls. Clin Microbiol Infect. 2013 Apr;19(4):E173–80. doi: 10.1111/1469-0691.12123. [DOI] [PubMed] [Google Scholar]
- Lipsky BA. Urinary tract infections in men. Epidemiology, pathophysiology, diagnosis, and treatment. Ann Intern Med. 1989 Jan 15;110(2):138–50. doi: 10.7326/0003-4819-110-2-138. [DOI] [PubMed] [Google Scholar]
- Manges AR, Harel J, Masson L, Edens TJ, Portt A, Reid-Smith RJ, et al. Multilocus sequence typing and virulence gene profiles associated with Escherichia coli from human and animal sources. Foodborne Pathog Dis. 2015 Apr;12(4):302–10. doi: 10.1089/fpd.2014.1860. [DOI] [PubMed] [Google Scholar]
- Manges AR, Johnson JR, Foxman B, O’Bryan TT, Fullerton KE, Riley LW. Widespread distribution of urinary tract infections caused by a multidrug-resistant Escherichia coli clonal group. N Engl J Med. 2001;345(14):1007–1013. doi: 10.1056/NEJMoa011265. [DOI] [PubMed] [Google Scholar]
- Miajlovic H, Mac Aogáin M, Collins CJ, Rogers TR, Smith SG. Characterization of Escherichia coli bloodstream isolates associated with mortality. J Med Microbiol. 2015 Oct 30; doi: 10.1099/jmm.0.000200. [DOI] [PubMed]
- Moreno E, Planells I, Prats G, Planes AM, Moreno G, Andreu A. Comparative study of Escherichia coli virulence determinants in strains causing urinary tract bacteremia versus strains causing pyelonephritis and other sources of bacteremia. Diagn Microbiol Infect Dis. 2005 Oct;53(2):93–9. doi: 10.1016/j.diagmicrobio.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Nicolas-Chanoine M-H, Bertrand X, Madec J-Y. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev. 2014 Jul;27(3):543–74. doi: 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti G, Mannini A, Balistreri M, Schito AM. Virulence Factors in Urinary Escherichia coli Strains: Phylogenetic Background and Quinolone and Fluoroquinolone Resistance. J Clin Microbiol. 2008 Feb 1;46(2):480–7. doi: 10.1128/JCM.01488-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynor MC, Carson CC. Urinary Infections in Men. Med Clin North Am. 2011 Jan;95(1):43–54. doi: 10.1016/j.mcna.2010.08.015. [DOI] [PubMed] [Google Scholar]
- Riley LW. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin Microbiol Infect. 2014 May;20(5):380–90. doi: 10.1111/1469-0691.12646. [DOI] [PubMed] [Google Scholar]
- Salvador E, Wagenlehner F, Kohler C-D, Mellmann A, Hacker J, Svanborg C, et al. Comparison of Asymptomatic Bacteriuria Escherichia coli Isolates from Healthy Individuals versus Those from Hospital Patients Shows that Long-Term Bladder Colonization Selects for Attenuated Virulence Phenotypes. Infect Immun. 2012 Feb 1;80(2):668–78. doi: 10.1128/IAI.06191-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R, Agarwal J, Srivastava S, Mishra B. Role of special pathogenicity versus prevalence theory in pathogenesis of acute cystitis caused by Escherichia coli. J Med Microbiol. 2014 Aug 1;63(Pt_8):1038–43. doi: 10.1099/jmm.0.073270-0. [DOI] [PubMed] [Google Scholar]
- Ulleryd P, Sandberg T, Scheutz F, Clabots C, Johnston BD, Thuras P, et al. Colonization with Escherichia coli Strains among Female Sex Partners of Men with Febrile Urinary Tract Infection. In: Munson E, editor. J Clin Microbiol. 6. Vol. 53. 2015. Jun, pp. 1947–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vejborg RM, Friis C, Hancock V, Schembri MA, Klemm P. A virulent parent with probiotic progeny: comparative genomics of Escherichia coli strains CFT073, Nissle 1917 and ABU 83972. Mol Genet Genomics MGG. 2010 May;283(5):469–84. doi: 10.1007/s00438-010-0532-9. [DOI] [PubMed] [Google Scholar]