Abstract
Inactivation of the tumor suppressor neurofibromin 1 (NF1) presents a newly characterized melanoma subtype, for which currently no targeted therapies are clinically available. Pre-clinical studies suggest that ERK inhibitors are likely to provide benefit, albeit with limited efficacy as single agent; therefore, there is a need for rationally designed combination therapies. Here, we evaluate the combination of the ERK inhibitor SCH772984 and the biguanide phenformin. Combination of both compounds showed potent synergy in cell viability assays and cooperatively induced apoptosis. Treatment with both drugs was required to fully suppress mTOR signaling, a known effector of NF1 loss. Mechanistically, SCH772984 increased the oxygen consumption rate (OCR), indicating that these cells relied more on oxidative phosphorylation upon treatment. Consistently, SCH772984 increased expression of the mitochondrial transcriptional co-activator PGC1α. In contrast, co-treatment with phenformin, an inhibitor of complex I of the respiratory chain, decreased the OCR. SCH772984 also promoted the expansion of the H3K4 demethylase KDM5B (also known as JARID1B)-positive subpopulation of melanoma cells, which are slow-cycling and treatment-resistant. Importantly, phenformin suppressed this KDM5B-positive population, which reduced the emergence of SCH772984-resistant clones in long-term cultures. Our results warrant the clinical investigation of this combination therapy in patients with NF1 mutant melanoma.
Keywords: AMPK, JARID1B, KDM5B, SCH772984, drug resistance
INTRODUCTION
Cutaneous melanoma can be classified according to its genetic landscape into four subcategories: BRAF-mutant (~52% of cases), NRAS-mutant (~28%), NF1-mutant (~14%) and triple-wild-type (~6%) (Akbani et al. 2015). Mutations in BRAF, NRAS and NF1 lead to constitutive activation of the RAS/RAF/MEK/ERK signaling pathway, resulting in uncontrolled proliferation and tumor growth. Therefore, small-molecule inhibitors against several targets in this pathway have been developed, including the BRAF inhibitors (BRAFi) vemurafenib and dabrafenib; MEK inhibitors (MEKi) trametinib and cobimetinib; and other compounds undergoing clinical evaluation. While BRAF and MEK inhibitors are approved by the FDA for the treatment of BRAF-mutant melanoma, targeted therapies for NF1-mutant melanoma are currently unavailable.
NF1 is a tumor suppressor that belongs to the family of RAS GTPase-activating proteins (GAP) and functions to negatively regulate RAS (Martin et al. 1990). RAS proteins are activated when bound to GTP; conversely, hydrolysis of GTP to GDP, which is accelerated by GAPs, inactivates RAS (Ratner and Miller 2015). Loss-of-function mutations in NF1 consequently activate the RAS/RAF/MEK/ERK signaling pathway. Therefore, MEKi and ERK inhibitors (ERKi) have been evaluated in preclinical studies of this melanoma subtype. While sensitivities as single agents are variable, NF1-mutant melanoma cells more consistently respond to ERKi compared to MEKi (Krauthammer et al. 2015). Rational combination therapies may further enhance the limited efficacy of ERKi and turn it into a promising treatment option for the NF1 subtype of melanoma (Morris et al. 2013).
We have recently shown that the anti-diabetes biguanide drug and AMP-activated kinase (AMPK) activator phenformin, enhances the antitumor activity of BRAFi in cultured cells, xenografts, and genetically engineered mouse models (Yuan et al. 2013). Phenformin and its analog metformin target complex I of the respiratory chain and subsequently activate AMPK and suppress mTOR signaling (Pollak 2013). This acts as an energy break and reprograms proliferative cancer metabolism to catabolism. In addition, metformin and MEKi were shown to synergistically reduce cell viability and tumor growth in NRAS-mutant melanoma (Vujic et al. 2014). We therefore sought to investigate the potential benefit of combining the ERKi SCH772984 with phenformin in NF1-mutant melanoma cells. In this study we show that the combination of SCH772984 with phenformin provides a therapeutic advantage over ERKi treatment alone by synergistically blocking melanoma cell proliferation and enhancing the induction of apoptosis. The combination cooperatively inhibited mTOR signaling, a known effector of NF1-deficient tumors. Importantly, phenformin suppressed the ERKi-resistant, KDM5B-positive subpopulation of melanoma cells and inhibited the emergence of resistant clones in long-term culture.
RESULTS
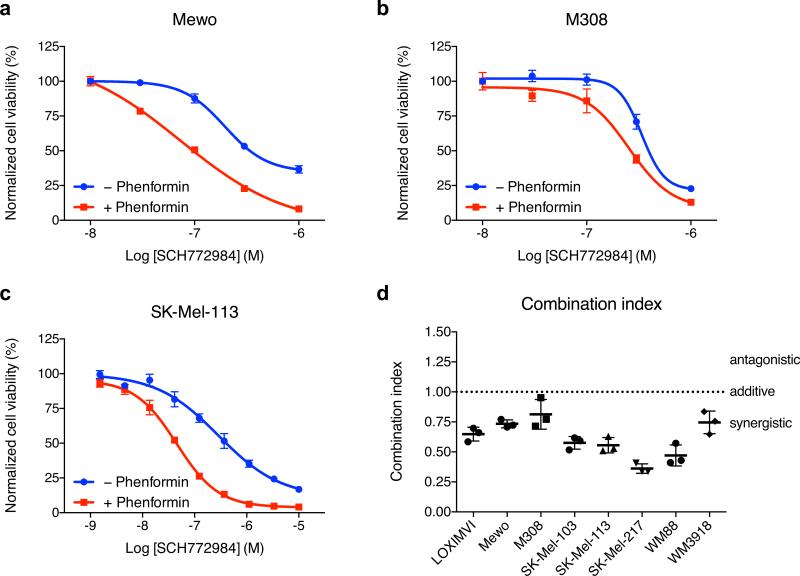
We first examined the antiproliferative activity of phenformin in combination with ERKi SCH772984 by MTS viability assays in various melanoma cells with inactivated NF1 (see Supplementary Table 1 for mutation status). Co-treatment with phenformin enhanced the antiproliferative activity of SCH772984 in Mewo, M308 and SK-Mel-113 cells, compared with SCH772984 treatment alone as measured by MTS viability assay (Figure 1a-c). All three of these cell lines harbor loss-of-function mutations in NF1. While Mewo and SK-Mel-113 cells are wild type for BRAF and NRAS, M308 additionally has a BRAFV600E mutation and is resistant to BRAFi (Søndergaard et al. 2010). To exclude the possibility that phenformin treatment, by targeting mitochondrial metabolism, was interfering with the MTS viability assay and confounding our results, we independently confirmed these results in M308 and SK-Mel-103 cells using the DNA content-based CyQUANT assay (Supplementary Figure 1). We then quantitated the potency of the drug combination by calculating the combination index (CI) using the Chou-Talalay method (Chou 2006). Drug interactions with a combination index < 1 are defined as synergistic, ~1 as additive and > 1 as antagonistic. The combinatory effect in all eight tested cell lines was confirmed to be synergistic as CI values ranged between 0.4 and 0.8 (Figure 1d).
Figure 1. Phenformin and SCH772984 synergistically inhibit proliferation in NF1-mutant cancer cells.
(a) Mewo, (b) M308 and (c) SK-Mel-113 cells were treated for 72 hours with ERKi SCH772984 in absence or presence of 1 mM phenformin and cell viability measured by MTS assay. One representative experiment of three is shown with n = 3 ± S.D. for each data point. (d) Synergy studies of SCH772984 and phenformin were carried out in various NF1-deficient cell lines according to the Chou-Talalay method and combination index (CI) calculated by CompuSyn. CI < 1 demonstrates synergism, CI = 1 additive effect and CI > 1 antagonism. Mean of n = 3 ± S.D.
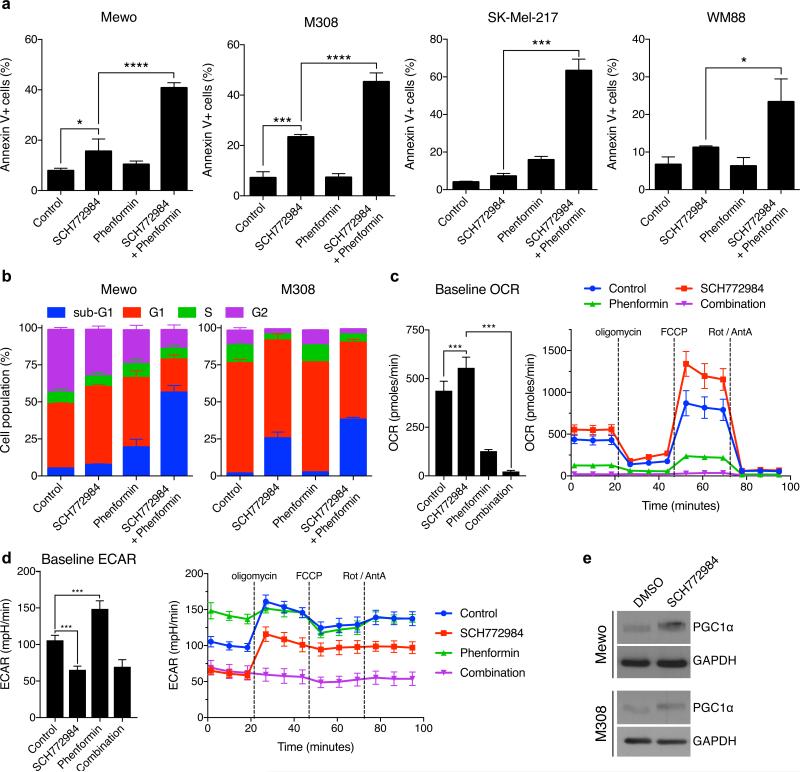
To assess whether the combination increases the induction of apoptosis or inhibition of cell cycle progression, we performed FACS-based Annexin V and cell cycle analyses. SCH772984 or phenformin alone only modestly induced the Annexin V-positive, apoptotic cell population (1.7-3.3-fold increase in all tested cell lines for SCH772984 and 0.9-3.9-fold increase with phenformin treatment; Figure 2a). In contrast, the combination synergistically increased the apoptotic cell populations up to 15.7-fold. Notably, in all cell lines, this increase was greater than the sum of the individual drug treatments. Furthermore, while SCH772984 arrested cells in the G1 phase of the cell cycle, combined treatment with phenformin potently increased the sub-G1 population of cells in Mewo and M308 cells (Figure 2b).
Figure 2. Effect of combined SCH772984 and phenformin treatment on apoptosis, cell cycle and mitochondrial metabolism.
(a) Annexin V staining and (b) cell cycle analysis after treatment with SCH772984 (0.3 μM for both cell lines) and phenformin (Mewo, 1 mM; M308, 0.3 mM) for 48 hours. Mean of n = 3 ± S.D. OCR (c) and ECAR (d) in Mewo cells upon pre-treatment with 0.3 μM SCH772984 and 30 μM phenformin for 24 hours and exposure to mitochondrial stressors oligomycin, FCCP and rotenone/anitmycin A (Rot/AntA). Mean of n = 5 ± S.D. (e) PGC1α expression after treatment with inhibitors in Mewo (0.1 μM SCH772984) and M308 (1 μM SCH772984) cells for 24 hours.
We then explored the molecular mechanism underlying the synergy of phenformin and SCH772984. As phenformin targets complex I of the respiratory chain and therefore directly impacts oxidative phosphorylation (Pollak 2013), we measured the effect of these treatments on the oxygen consumption rate (OCR), which is an indicator of mitochondrial respiration (Brand and Nicholls 2011). Treatment with SCH772984 increased the OCR in Mewo cells, which suggests that the surviving cells relied more on oxidative phosphorylation (Figure 2c). As expected, phenformin inhibited oxidative phosphorylation and made cells more glycolytic, as shown by a reduced OCR and increased ECAR (Figure 2c-d). Combination of both inhibitors further reduced the OCR while cells maintained a low ECAR (Figure 2c-d). Upon treatment with the combination of SCH772984 and phenformin, cells were not able to respond to the mitochondrial stressors oligomycin, a Fo-ATPase inhibitor of Complex V, or the uncoupler FCCP. Therefore, the drug combination inhibited both oxidative phosphorylation and glycolysis, which ultimately abolishes the ability to sustain ATP production. Consistently, SCH772984 treatment induced expression of PGC1α, a key regulator in mitochondrial biogenesis (Figure 2e).
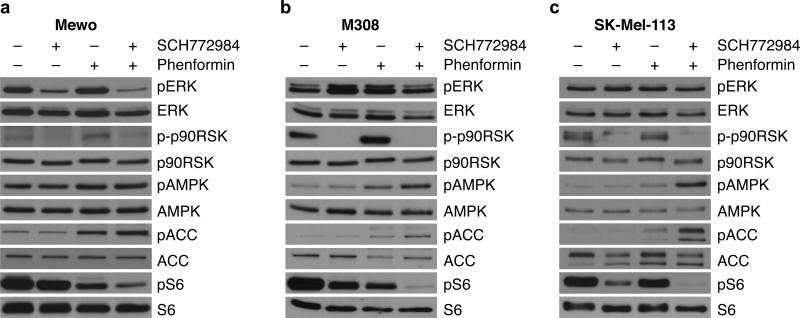
We then performed western blot analyses for key signaling pathways in NF1-mutant melanoma cells treated with each drug individually and in combination. As shown in Figure 3, 24-hour treatment with SCH772984 did not alter ERK1/2 phosphorylation in the tested NF1-deficient cell lines, but potently suppressed activation of the ERK downstream target p90-RSK. Phenformin, as expected, activated AMPK, as seen by increases in AMPK phosphorylation. Notably, phosphorylation of acetyl-CoA carboxylase (ACC), an AMPK substrate, was further increased by the drug combination. As both the RAS/RAF/MEK/ERK and AMPK signaling pathways can intersect with mTOR signaling, we investigated the molecular effects on S6 phosphorylation, a well-established mTORC1 downstream target. While SCH772984 and phenformin alone only moderately decreased S6 phosphorylation, the combination essentially abolished S6 activation. Taken together, these results suggest that phenformin and ERKi cooperatively inhibit mTOR signaling, which may underlie the synergistic effect of this combination on cell viability and apoptosis.
Figure 3. mTOR signaling is cooperatively suppressed by SCH772984 and phenformin.
(a) Mewo, (b) M308 and (c) SK-Mel-113 cells were treated with indicated doses of inhibitors (Mewo, 0.1 μM SCH772984; 0.3 mM phenformin; M308 and SK-Mel-113, 1 μM SCH772984; 1 mM phenformin) for 24 hours and subjected to Western blotting with indicated antibodies.
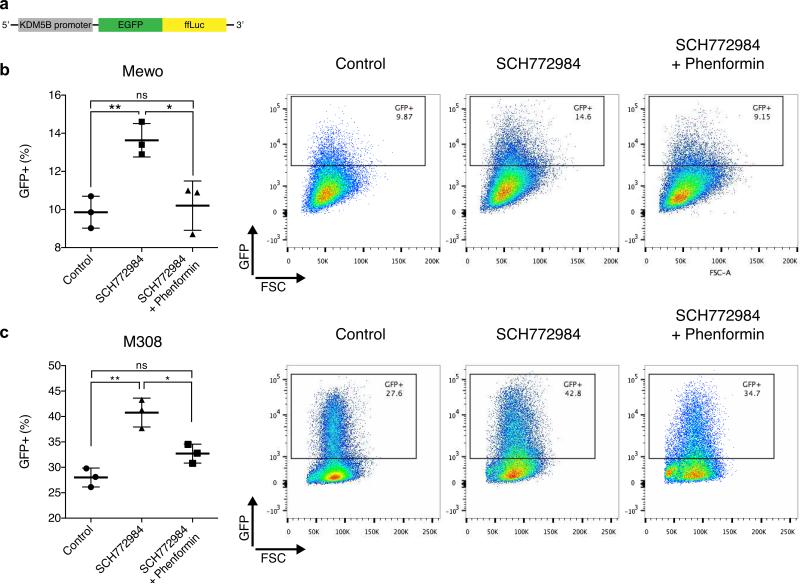
We then investigated if the SCH772984-phenformin combination impacts the distribution of KDM5B-positive cells in NF1-deficient melanoma. KDM5B encodes for a histone 3 lysine 4 demethylase, which is predominantly expressed in slow-cycling melanoma cells that are critical for long-term tumor maintenance (Roesch et al. 2010). KDM5B-positive cells are more resistant to targeted therapies and chemotherapy (Roesch et al. 2013; Yuan et al. 2013; Zhang et al. 2016). We and others have previously shown that BRAFi increase the KDM5B-positive population, likely as a survival mechanism to evade treatment response and lead to emergence of drug resistance (Roesch et al. 2013; Yuan et al. 2013). In contrast, phenformin preferentially inhibits the proliferation of KDM5B-positive BRAF-mutant melanoma cells (Roesch et al. 2013; Yuan et al. 2013). We therefore sought to investigate the effects of ERKi and phenformin on the KDM5B-positive population in NF1-deficient melanoma. To capture the dynamic nature of KDM5B expression in response to treatment, we developed a KDM5B-eGFP-ffLuc reporter construct (Figure 4a), where the promoter region of KDM5B drives expression of an eGFP-firefly luciferase fusion construct. The promoter region of KDM5B used here is identical to a published reporter driving eGFP (JARID1Bprom-EGFP (Roesch et al. 2013)). The slow-cycling nature of KDM5B-positive cells was confirmed, as GFP expression in KDM5B reporter-positive cells was mutually exclusive with expression of the proliferation marker Ki67 by FACS (Supplementary Figure 2a).
Figure 4. Phenformin inhibits expansion of KDM5B-positive cells upon SCH772984 treatment.
(a) Scheme of KDM5B-reporter construct. (b) Mewo and (c) M308 cells stably transduced with the KDM5B reporter were treated with 0.3 μM SCH772984 in absence or presence of 1 mM (Mewo) or 0.3 mM (M308) phenformin for 72 hours and distribution of KDM5B-positive cells compared to vehicle-treated controls. Mean of n = 3 ± S.D. Representative FACS plots are shown.
Treatment with ERKi SCH772984 significantly increased the proportion of KDM5B-positive cells in both Mewo and M308 cells stably expressing the KDM5B reporter (Figure 4b-c). This effect was also seen with the MEKi trametinib in Mewo cells and in NRAS-mutant, NF1-wildtype SK-Mel-2 cells (Supplementary Figure 2b-c), suggesting that inhibition of RAF-MEK-ERK at various levels leads to similar effects on the KDM5B-subpopulation of melanoma cells across different subtypes. Importantly, phenformin potently prevented the induction of the KDM5B-positive subpopulation of cells upon SCH772984 treatment (Figure 4b-c), suggesting that in NF1-loss melanomas, phenformin may prevent the emergence of KDM5B-positive, drug-resistant cells.
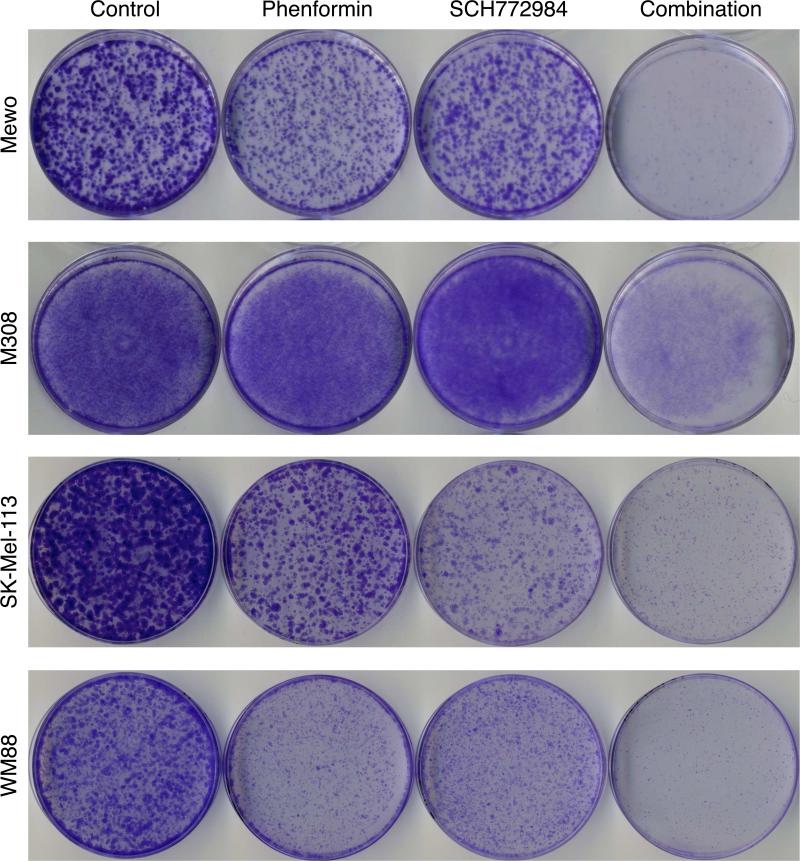
Finally, we sought to assess whether the synergistic interaction of phenformin with SCH772984 might therefore also affect the emergence of SCH772984-resistant melanoma clones in long-term culture. We addressed this possibility by employing clonogenic growth assays. Mewo, M308, SK-Mel-113 and WM88 cells seeded at single-cell density were exposed to phenformin and SCH772984 alone or in combination and the presence of resistant clones was identified by crystal violet staining. The combination strikingly reduced the formation of colonies compared to either treatment alone in these cells (Figure 5), supporting that phenformin delays the development of ERKi resistance in NF1 mutant melanoma.
Figure 5. Phenformin and SCH772984 synergistically inhibit emergence of resistant clones in NF1-deficient cancer cells.
Clonogenic growth assays were carried out in Mewo (0.3 μM SCH772984, 0.3 mM phenformin for 11 days), M308 (0.3 μM SCH772984, 30 μM phenformin for 14 days), SK-Mel-113 (1 μM SCH772984, 0.3 mM phenformin for 11 days) and WM88 (0.03 μM SCH772984, 0.5 mM phenformin for 11 days) cells.
DISCUSSION
Recent advances in the genomic classification of melanoma have deepened our disease understanding, but also underscored the importance to develop new targeted therapies for advanced melanomas, especially ones that do not harbor BRAF mutations. Inactivating mutations in NF1 define such a sub-class and we have shown here that combined treatment with the ERKi SCH772984 and phenformin could provide an attractive new treatment option.
Clinical trials assessing the efficacy of MEKi and ERKi in patients with BRAF WT melanomas, including those harboring inactivated NF1 are currently planned or ongoing (Sullivan 2016). Pre-clinical studies of RAF, MEK and ERK inhibitors in NF1-mutant melanoma models, showed mixed results in cell line models. NF1 mutations have been proposed as a potential resistance mechanism to BRAFi and MEKi in BRAF-mutant melanoma cells, as knockdown of NF1 by shRNA in these cells decreases their sensitivity to BRAFi PLX4720 and MEKi AZD6244 (Whittaker et al. 2013). Another study, however, demonstrated that NF1-deficient cells are sensitive to MEKi trametinib (Nissan et al. 2014). Two recent studies addressed this controversy by directly comparing the sensitivities of MEK inhibition in the context of NF1 loss and found that there was no difference between NF1-wildtype and NF1-mutant cells with respect to their sensitivity to MEKi trametinib (Ranzani et al. 2015) or selumetinib (Krauthammer et al. 2015). Different binding modes of MEKi could account for mixed responses (Nissan et al. 2014) as well as the genetic make-up of the NF1 melanoma type. This group of cancers is genetically heterogeneous and often co-occurs with second-hit mutations in so-called “RASopathy genes” (Krauthammer et al. 2015). Compared to MEKi, ERKi consistently showed higher response rates across various NF1-mutant cell line models in these recent studies. For example, one study reported that 5 of 7 (70%) NF1-mutant cell lines displayed high sensitivity to ERKi SCH772984 (Krauthammer et al. 2015), in line with previous results showing that NF1 knockdown cells retained sensitivity to ERKi VTX-11e despite induced resistance to BRAFi PLX4720 and MEKi selumetinib (Whittaker et al. 2013). We have confirmed this observation, as six out of eight tested NF1-deficient melanoma cell lines had sub-micromolar IC50 values against SCH772984. These data together suggest that ERKi may show higher efficacy than MEKi and could emerge as the inhibitor of choice in NF1-inactivated melanoma.
Responses to targeted therapies in melanoma are usually not durable; therefore, rationally designed combination strategies are required to defer acquired drug resistance and improve clinical outcomes. We have recently shown that phenformin synergistically enhances the antiproliferative activity of BRAFi in several BRAF-mutant cancer cells and mouse models (Yuan et al. 2013) and therefore investigated in this study its suitability as a combination partner with SCH772984 in NF1-inactivaed melanoma. SCH772984 treatment induced an oxidative metabolic program as shown by increased OCR and elevated PGC1α expression. Comparably, inhibition of the RAS/RAF/MEK/ERK pathway further upstream using BRAFi in BRAF-mutant melanoma has been shown to trigger a PGC1α-driven transcriptional program that facilitates mitochondrial biogenesis and enhances oxidative phosphorylation, which improves the resilience to oxidative stress (Haq et al. 2013; Vazquez et al. 2013). Elevated PGC1α expression has further been observed in patients treated with BRAFi and/or MEKi (Gopal et al. 2014). Interestingly, increased oxidative phosphorylation and expression of PGC1α is also a phenotype observed in cells surviving oncogene ablation in a pancreatic cancer mouse model (Viale et al. 2014). We demonstrate that combined treatment of SCH772984 and phenformin, which targets oxidative phosphorylation, profoundly inhibited the viability of NF1-mutant melanoma cells by altering the cellular metabolism and limiting the energy production.
Additionally, synergy of phenformin and ERKi was accompanied by profound suppression of mTOR signaling, as shown by almost complete abrogation of ribosomal protein S6 phosphorylation. Activation of mTOR is critical for tumorigenesis driven by NF1 loss in genetically engineered neurofibromatosis mouse models. NF1 knockout leads to hyperactivation of mTOR signaling (Dasgupta et al. 2005; Johannessen et al. 2005), which sensitizes these tumors to mTOR inhibition by rapamycin (Johannessen et al. 2008). However, mTOR inhibition by rapamycin has proven to be less effective in NF1-mutant melanoma as compared to malignant peripheral nerve sheath tumors (MPNST), the most common malignancy of neurofibromatosis 1 (Nissan et al. 2014). Sustained and potent suppression of S6 phosphorylation is required for clinical responses to RAF and MEK pathway inhibition in melanoma patients (Corcoran et al. 2013). Our data indicate that SCH772984 or phenformin alone can only partially suppress S6 phosphorylation, but combination of both drugs cooperatively dephosphorylates S6 to a greater extent. This might contribute to the synergy of these drugs in NF1-mutant melanoma cells and supports their combined use as a melanoma therapeutic.
We have previously shown that vemurafenib and phenformin synergistically kill BRAF-mutant melanoma cells and delay the development of resistance. A clinical trial based on these findings will be accruing shortly. Melanomas are characterized by a high degree of intratumoral heterogeneity and the histone H3K4 demethylase KDM5B (also known as JARID1B) has been proposed as a marker for a subset of slow-cycling melanoma cells, which are critical for long-term tumor maintenance (Roesch et al. 2010). Heterogeneous KDM5B expression has very recently been confirmed by single-cell RNA-sequencing in a large-scale melanoma patient cohort (Tirosh et al. 2016). While in BRAF-mutant melanoma BRAFi target the KDM5B-negative bulk of the tumor, phenformin kills the otherwise unblemished KDM5B-positive population. We show that the relevance of the KDM5B-positive population appears to be a more general phenomenon in melanoma, extending beyond BRAF-mutant cases, namely to NF1-deficient and NRAS-mutant melanoma. Furthermore, inhibition of the RAS/RAF/MEK/ERK pathway at multiple targets increases the KDM5B-positive population. More importantly, phenformin would target KDM5B-positive cells in NF1-mutant melanoma, similar to BRAF-mutant melanoma models. How exactly phenformin inhibits this subpopulation is currently unknown. It is possible that the KDM5B-positive population has metabolic features distinct from the KDM5B-negative population, and such metabolic heterogeneity dictates the selective effects of phenformin, a mitochondrial complex I inhibitor, on KDM5B-positive cells. The notion that different metabolic programs co-exist in tumors has been recently suggested in a pancreatic ductal adenocarcinoma mouse models (Viale et al. 2014; Elgogary et al. 2016) and lung cancer patients (Hensley et al. 2016). Understanding the establishment, maintenance and biological consequence of intra-tumor metabolic heterogeneity in solid tumors will help the design of strategies to target cell subpopulations with distinct metabolic states and improve therapeutic efficacy. Our previous findings in BRAF mutant melanoma (Yuan et al. 2013) and results from this study in NF1 mutant melanoma suggest that targeting oxidative phosphorylation with phenformin is critical to limit the emergence of drug-resistant clones in melanoma regardless of driver mutation and therefore could contribute to more durable clinically responses to targeted therapies. Taken together, our study reveals a potent combination therapy of ERKi and phenformin for NF1-mutant melanoma that warrants further clinical investigation.
MATERIALS & METHODS
Reagents and cell culture
SCH772984 and trametinib were purchased form Selleck (Houston, TX) and phenformin from Toronto Research Chemicals (Toronto, ON, Canada). KDM5B reporter was generated by subcloning of the human KDM5B promoter (from −228 to + 21) into pFuGW, a lentiviral based expression vector with the ffLuc2-eGFP expression cassette (obtained from Dr. Dominic Esposito at Frederick National Laboratory for Cancer Research).
NF1, BRAF and NRAS mutation status of all cell lines is shown in Supplementary Table 1. Mewo and WM88 were obtained from Lynda Chin and M308 from Antoni Ribas, respectively. SK-Mel-103, SK-Mel-113, SK-Mel-217, WM3918 were obtained from Memorial Sloan Kettering Cancer Center and LOXIMVI from National Cancer Institute. WM115 cells were purchased from ATCC (Manassas, VA). All cells were cultured in RPMI 1640 containing 10% fetal bovine serum and penicillin/streptomycin/glutamine. Cells were cultured at 37°C in humidified atmosphere containing 5% CO2. Transfection and lentiviral infection were performed as previously described (Yuan et al. 2013) and stable clones selected with puromycin.
Cell viability assays
For the cell viability analysis, cells were seeded in a 96-well plates and drug treatment was started the following day. After 72-hour incubation, MTS (Promega, Madison, WI) or CyQUANT assays (Thermo Scientific, Waltham, MA) were performed according to manufacturer's instructions (Promega, Madison, WI) and normalized cell viability curves fitted using sigmoidal dose-response with variable slope algorithm in GraphPad Prism software (version 6). Combination index was calculated with CompuSyn software according to manufacturer's instructions.
Apoptosis and cell cycle analysis
Cells were treated with indicated doses of SCH772984 and phenformin for 48 hours. Analysis of apoptotic cells by annexin V staining (Annexin V FITC and PI kit, Thermo Scientific) was done according to manufacturer's instructions. Cell cycle analysis was performed after fixation in 70% ethanol and incubation with PBS containing 0.5 μg/mL RNaseA (Sigma-Aldrich) and 50 μg/mL propidium iodide (PI; Sigma-Aldrich, St. Louis, MO). Data was acquired on a Canto Flow Cytometer (Becton Dickinson, Franklin Lakes, NJ) and analyzed using FlowJo software (Treestar, Ashland, OR).
Measurement of mitochondrial bioenergetics using Seahorse XFe24 bioanalyzer
Oxygen consumption rate was measured using an XFe24 bioanalyzer (Seahorse Bioscience, Billerica, MA) as previously described (Parmenter et al. 2014). In brief, 5 × 104 cells were seeded into Seahorse plates. The following day, cells were treated with vehicle, 0.3 μM SCH772984 or 30 μM phenformin for 24 hours. Prior to the assay, cells were washed twice with assay media (unbuffered RPMI supplemented with 5 mM glucose, 1 mM glutamine, 1 mM sodium pyruvate). Cells were resuspended in assay media containing SCH772984 and phenformin. The assay comprised of 3-minute mix, 2-minute wait and 3-minute measurement cycles with injection of ATP synthase inhibitor oligomycin (1 μM), uncoupler FCCP (0.5 μM) and rotenone / antimycin A (0.5 μM). After the experiment, cells were trypsinized and OCR and ECAR were normalized to cell number.
FACS
Melanoma cells stably expressing the KDM5B reporter were treated with indicated doses of inhibitors for 72 hours. Cells were trypsinized, washed in PBS and resuspended in PBS containing 0.5 g/mL live cell stain 7-AAD (Biolegend, San Diego, CA) for FACS analysis using a Canto Flow Cytometer. Upon exclusion of doublets and dead cells, the KDM5BGFP-positive population was determined by comparison to parental, untransduced cells.
For antibody staining, KDM5B reporter cells were permeabilized with Foxp3 Fixation/Permeabilization kit (eBioscience, San Diego, CA) and stained with Ki67 antibody (Vector Laboratories, Burlingame, CA; product number VP-K451). Anti-rabbit AlexaFluor 594 (ThermoScientific) was used as secondary antibody. Cells were acquired on a Canto Flow Cytometer and data was analyzed using FlowJo software.
Western blotting
Western blotting was performed as previously described (Zheng et al. 2009) using the following antibodies: pACC S79 (product number 3661), ACC (3676), pAMPK T172 (2535), AMPK (2603), pERK T202/Y204 (9101), ERK (4695), GAPDH (2118), p-p90RSK Thr573 (9346), p90RSK1/2/3 (9355), pS6 (5364), S6 (2217), all purchased from Cell Signaling Technologies, Danvers, MA; or PGC1a (sc-13067; Santa Cruz Biotechnology, Dallas, TX).
Clonogenic assay
Cells were seeded at single-cell density (Mewo, 1 × 104 cells; M308, 6 × 104 cells) in 6-cm dishes and the following day indicated treatments commenced. Media containing inhibitors was changed every 2-3 days. After 11-14 days of drug treatment, cells were washed with PBS, fixed with 10% formalin for 5 min and stained with 1% crystal violet (Sigma-Aldrich) solution for 1 hour. Dishes were washed in water and dried.
Statistical analysis
Quantitative data is presented as mean of n = 3 ± standard deviation (S.D.). Two groups were compared by Student's t test and multiple groups were compared by unmatched, one-way ANOVA with Tukey correction by comparing the mean of each column with the mean of every other column in GraphPad Prism (version 6; La Jolla, CA).
Supplementary Material
ACKNOWLEDGEMENT
We thank Michael Evers, Jung Hyun Lee, Nunciada Salma and Che-Hung Shen for technical assistance.
GRANT SUPPORT
This work is supported by Melanoma Research Alliance Team Science Award (B.Z., J.Z. L.C.C.); National Institutes of Health (NIH) grants R01 CA166717 (B.Z.), K08 CA 160657 (J.Z.) and P01 CA120964 (L.C.C.); Clinique Clinical Scholar Award (J.Z.); the Harry J. Lloyd Charitable Trust (B.Z.); China Scholarship Council grant (S.C., C.M., Y.R.); American Skin Association award (F.M.S.).
Abbreviations
- AMPK
AMP-activated kinase
- BRAFi
BRAF inhibitor
- ERKi
ERK inhibitor
- MEKi
MEK inhibitor
- mTOR
mechanistic target of rapamycin
- NF1
neurofibromin 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
L.C.C. serves on the Board of Directors of Agios Pharmaceuticals and the Scientific Advisory Board of Enlibrium, companies developing drugs that target cancer metabolism. The authors declare that they have no other conflicts of interest.
REFERENCES
- Akbani R, Akdemir KC, Aksoy BA, Albert M, Ally A, Amin SB, et al. Genomic Classification of Cutaneous Melanoma. Cell. 2015;161(7):1681–96. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand MD, Nicholls DG. Assessing mitochondrial dysfunction in cells. Biochem J. 2011;435(2):297–312. doi: 10.1042/BJ20110162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T-C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58(3):621–81. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- Corcoran RB, Rothenberg SM, Hata AN, Faber AC, Piris A, Nazarian RM, et al. TORC1 Suppression Predicts Responsiveness to RAF and MEK Inhibition in BRAF-Mutant Melanoma. Sci Transl Med. 2013;5(196):196ra98–8. doi: 10.1126/scitranslmed.3005753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta B, Yi Y, Chen DY, Weber JD, Gutmann DH. Proteomic Analysis Reveals Hyperactivation of the Mammalian Target of Rapamycin Pathway in Neurofibromatosis 1–Associated Human and Mouse Brain Tumors. Cancer Res. 2005;65(7):2755–60. doi: 10.1158/0008-5472.CAN-04-4058. [DOI] [PubMed] [Google Scholar]
- Elgogary A, Xu Q, Poore B, Alt J, Zimmermann SC, Zhao L, et al. Combination therapy with BPTES nanoparticles and metformin targets the metabolic heterogeneity of pancreatic cancer. Proc Natl Acad Sci U S A. 2016;113(36):E5328–36. doi: 10.1073/pnas.1611406113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal YNV, Rizos H, Chen G, Deng W, Frederick DT, Cooper ZA, et al. Inhibition of mTORC1/2 overcomes resistance to MAPK pathway inhibitors mediated by PGC1α and oxidative phosphorylation in melanoma. Cancer Res. 2014;74(23):7037–47. doi: 10.1158/0008-5472.CAN-14-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haq R, Shoag J, Andreu-Perez P, Yokoyama S, Edelman H, Rowe GC, et al. Oncogenic BRAF Regulates Oxidative Metabolism via PGC1α and MITF. Cancer Cell. 2013;23(3):302–15. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley CT, Faubert B, Yuan Q, Lev-Cohain N, Jin E, Kim J, et al. Metabolic Heterogeneity in Human Lung Tumors. Cell. 2016;164(4):681–94. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen CM, Johnson BW, Williams SMG, Chan AW, Reczek EE, Lynch RC, et al. TORC1 Is Essential for NF1-Associated Malignancies. Curr Biol. 2008;18(1):56–62. doi: 10.1016/j.cub.2007.11.066. [DOI] [PubMed] [Google Scholar]
- Johannessen CM, Reczek EE, James MF, Brems H, Legius E, Cichowski K. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proc Natl Acad Sci U S A. 2005;102(24):8573–8. doi: 10.1073/pnas.0503224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauthammer M, Kong Y, Bacchiocchi A, Evans P, Pornputtapong N, Wu C, et al. Exome sequencing identifies recurrent mutations in NF1 and RASopathy genes in sun-exposed melanomas. Nat Genet. 2015;47(9):996–1002. doi: 10.1038/ng.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GA, Viskoohil D, Bollag G, McCabe PC, Crosier WJ, Haubruck H, et al. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell. 1990;63(4):843–9. doi: 10.1016/0092-8674(90)90150-d. [DOI] [PubMed] [Google Scholar]
- Morris EJ, Jha S, Restaino CR, Dayananth P, Zhu H, Cooper A, et al. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov. 2013;3(7):742–50. doi: 10.1158/2159-8290.CD-13-0070. [DOI] [PubMed] [Google Scholar]
- Nissan MH, Pratilas CA, Jones AM, Ramirez R, Won H, Liu C, et al. Loss of NF1 in Cutaneous Melanoma Is Associated with RAS Activation and MEK Dependence. Cancer Res. 2014;74(8):2340–50. doi: 10.1158/0008-5472.CAN-13-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmenter TJ, Kleinschmidt M, Kinross KM, Bond ST, Li J, Kaadige MR, et al. Response of BRAF-mutant melanoma to BRAF inhibition is mediated by a network of transcriptional regulators of glycolysis. Cancer Discov. 2014;4(4):423–33. doi: 10.1158/2159-8290.CD-13-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak M. Potential applications for biguanides in oncology. J Clin Invest. 2013;123(9):3693–700. doi: 10.1172/JCI67232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranzani M, Alifrangis C, Perna D, Dutton-Regester K, Pritchard A, Wong K, et al. BRAF/NRAS wild-type melanoma, NF1 status and sensitivity to trametinib. Pigment Cell Melanoma Res. 2015;28(1):117–9. doi: 10.1111/pcmr.12316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner N, Miller SJ. A RASopathy gene commonly mutated in cancer: the neurofibromatosis type 1 tumour suppressor. Nat Rev Cancer. 2015;15(5):290–301. doi: 10.1038/nrc3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, et al. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141(4):583–94. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch A, Vultur A, Bogeski I, Wang H, Zimmermann KM, Speicher D, et al. Overcoming Intrinsic Multidrug Resistance in Melanoma by Blocking the Mitochondrial Respiratory Chain of Slow-Cycling JARID1Bhigh Cells. Cancer Cell. 2013;23(6):811–25. doi: 10.1016/j.ccr.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan RJ. The role of mitogen-activated protein targeting in melanoma beyond BRAFV600. Curr Opin Oncol. 2016;28(2):185–91. doi: 10.1097/CCO.0000000000000271. [DOI] [PubMed] [Google Scholar]
- Søndergaard JN, Nazarian R, Wang Q, Guo D, Hsueh T, Mok S, et al. Differential sensitivity of melanoma cell lines with BRAF V600E mutation to the specific Raf inhibitor PLX4032. J Transl Med. 2010;20(8(1)):1. doi: 10.1186/1479-5876-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Izar B, Prakadan SM, Wadsworth MH, Treacy D, Trombetta JJ, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352(6282):189–96. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Lim JH, Chim H, Bhalla K, Girnun G, Pierce K, et al. PGC1α Expression Defines a Subset of Human Melanoma Tumors with Increased Mitochondrial Capacity and Resistance to Oxidative Stress. Cancer Cell. 2013;23(3):287–301. doi: 10.1016/j.ccr.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sanchez N, Marchesini M, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514(7524):628–32. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vujic I, Sanlorenzo M, Posch C, Esteve-Puig R, Yen AJ, Kwong A, et al. Metformin and trametinib have synergistic effects on cell viability and tumor growth in NRAS mutant cancer. Oncotarget. 2014;6(2):969–78. doi: 10.18632/oncotarget.2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker SR, Theurillat JP, Van Allen E, Wagle N, Hsiao J, Cowley GS, et al. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Discov. 2013;3(3):350–62. doi: 10.1158/2159-8290.CD-12-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, Yuan P, Ito K, Ito K, Perez-Lorenzo R, Perez-Lorenzo R, et al. Phenformin enhances the therapeutic benefit of BRAF(V600E) inhibition in melanoma. Proc Natl Acad Sci U S A. 2013;110(45):18226–31. doi: 10.1073/pnas.1317577110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Wong P, Salvaggio C, Salhi A, Osman I, Bedogni B. Synchronized Targeting of Notch and ERBB Signaling Suppresses Melanoma Tumor Growth through Inhibition of Notch1 and ERBB3. J Invest Dermatol. Feb. 2016;136(2):464–72. doi: 10.1016/j.jid.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Jeong JH, Asara JM, Yuan Y-Y, Granter SR, Chin L, et al. Oncogenic B-RAF negatively regulates the tumor suppressor LKB1 to promote melanoma cell proliferation. Mol Cell. 2009;33(2):237–47. doi: 10.1016/j.molcel.2008.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.