Summary
Global transcriptome reprogramming during spermatogenesis ensures timely expression of factors in each phase of male germ cell differentiation. Spermatocytes and spermatids require particularly extensive reprogramming of gene expression to switch from mitosis to meiosis and to support gamete morphogenesis. Here, we uncovered an extensive alternative splicing program during this transmeiotic differentiation. Notably, intron retention was largely the most enriched pattern, with spermatocytes showing generally higher levels of retention compared with spermatids. Retained introns are characterized by weak splice sites and are enriched in genes with strong relevance for gamete function. Meiotic intron-retaining transcripts (IRTs) were exclusively localized in the nucleus. However, differently from other developmentally regulated IRTs, they are stable RNAs, showing longer half-life than properly spliced transcripts. Strikingly, fate-mapping experiments revealed that IRTs are recruited onto polyribosomes days after synthesis. These studies reveal an unexpected function for regulated intron retention in modulation of the timely expression of select transcripts during spermatogenesis.
Keywords: transcriptome profiling, alternative splicing, intron retention, mRNA stability, germ cell differentiation, spermatogenesis
Highlights
-
•
Alternative splicing in meiotic spermatocytes favors intron retention
-
•
Intron retention stabilizes nuclear transcripts in meiotic spermatocytes
-
•
A high transcription rate outcompetes the spliceosome from weak introns in meiosis
-
•
Long-lived intron-retaining mRNAs encode key proteins for male gamete function
Transcription during mammalian spermatogenesis is discontinuous and ceases in late post-meiotic stages. Naro et al. show that regulated intron retention in male meiotic germ cells generates long-lived intron-retaining transcripts that are preserved for days after their synthesis and can be timely translated by transcriptionally inactive post-meiotic germ cells.
Introduction
Spermatogenesis is a remarkable cell differentiation process that yields the male gamete and remains active throughout the adult life of mammals (Griswold, 2016). It comprises three main phases: a “mitotic” phase during which spermatogonia stem cells divide either to self-renew or to originate successive generations of mitotic spermatogonial cells; a “meiotic” phase during which primary spermatocytes engage two consecutive divisions that increase genetic variability and half genome content; and “spermiogenesis” when haploid spermatids undergo dramatic morphological changes, such as DNA compaction and development of a motile flagellum, which endow the mature spermatozoa with features essential for fertilization (Hermo et al., 2010a, Hermo et al., 2010b).
Each stage of spermatogenesis requires a specific repertoire of factors to accomplish the profound genomic and morphological modifications that characterize male germ cell differentiation. Proper expression of these factors is dynamically controlled at transcriptional and post-transcriptional levels (Paronetto and Sette, 2010, Chalmel and Rolland, 2015). Testicular paralogs of core transcriptional components, such as TAF7L (Zhou et al., 2013), or testis-specific transcription factors, such as the cyclic AMP-responsive factor CREM (Nantel and Sassone-Corsi, 1996), together with stage-specific epigenetic modifications (Godmann et al., 2007, Soumillon et al., 2013, Hammoud et al., 2014), contribute to orchestrate gene expression programs during spermatogenesis. Moreover, since transcription is not continuously active during male germ cell differentiation (Monesi, 1964), translational regulation of stored mRNAs ensures timely expression of proteins in the transcriptionally silent stages of spermatogenesis (Paronetto and Sette, 2010).
High-throughput transcriptome analyses identified testis among the tissues displaying the highest proportion of splice variants (Kan et al., 2005), with germ cells providing the largest contribution to this transcriptome complexity (Soumillon et al., 2013). Moreover, profiling of whole testis transcriptome during the first wave of mouse spermatogenesis highlighted some alternative splicing (AS) events that are timely regulated during this process (Schmid et al., 2013, Margolin et al., 2014). AS of exons and introns in virtually every mammalian gene yields multiple transcripts that often display different coding properties and/or patterns of spatial/temporal expression (Barbosa-Morais et al., 2012, Yang et al., 2016). In this way, AS represents a powerful resource to amplify the complexity and flexibility of the genome and to precisely modulate gene expression during cell differentiation and development (Kalsotra and Cooper, 2011). Thus, AS modulation likely contributes to fine-tune the germ cell differentiation program.
To date, no study has investigated whether global splicing changes occur during the transmeiotic phases of spermatogenesis, when unique processes take place. Indeed, meiotic spermatocytes undergo homologous recombination and chromosome segregation, whereas post-meiotic spermatids need to differentiate into highly specialized, motile spermatozoa (Griswold, 2016, Hermo et al., 2010a, Hermo et al., 2010b), and all of these events require specific factors that are often only expressed in germ cells. In this study, by performing high-throughput transcriptome profiling of purified male germ cells we uncovered a robust intron retention (IR) program in meiotic spermatocytes. Intron-retaining genes were strongly enriched in functional categories related to differentiation and properties of the male gamete. Mechanistically, we found that weak splice sites coupled with high RNA polymerase II (RNAPII) activity contribute to IR in spermatocytes. Reducing the transcriptional burden by inhibiting RNAPII phosphorylation rescued splicing, suggesting that competition for the spliceosome selects weak introns for retention. Although IR was generally shown to cause transcript instability in mammals, thus contributing to the quenching of gene expression during cell differentiation or in response to stress (Yap et al., 2012, Wong et al., 2013, Shalgi et al., 2014, Edwards et al., 2016, Pimentel et al., 2016, Ni et al., 2016), we found that meiotic intron-retaining transcripts (IRTs) are long-lived mRNAs, which are preserved until they need to be translated. Thus, our study reveals an unexpected physiological role for IR in ensuring proper and timely control of gene expression during male germ cell differentiation.
Results
A Regulated Alternative Splicing Program Characterizes the Meiotic Transition of Male Germ Cells
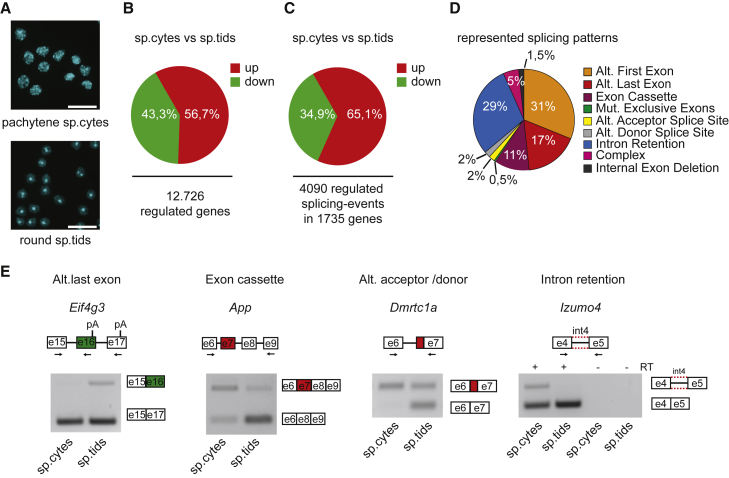
To achieve a comprehensive characterization of the splicing changes occurring during transmeiotic differentiation of male germ cells, we performed high-throughput RNA sequencing (RNA-seq) analyses of poly(A)+ RNA isolated from highly purified populations of meiotic spermatocytes (n = 2) and post-meiotic spermatids (n = 3) (Figure 1A). Gene expression analyses revealed a remarkable and global reprogramming of the transcriptome across meiosis, with 12,726 genes (corresponding to nearly 60% of the expressed genes; data not shown) differentially expressed between spermatocytes and spermatids (Figure 1B). As expected, transcripts for the spermatid-specific nucleoproteins (i.e., protamines and transition proteins) (Dadoune, 2003) and the sperm-specific calcium channels (i.e., CatSper proteins) (Singh and Rajender, 2015) were strongly upregulated in spermatids (Figure S1A). By contrast, expression of the Sycp2 and Sycp3 genes, encoding synaptonemal proteins (Wang et al., 2005), and of Mlh3 and Spo11, encoding homologous recombination proteins (Romanienko and Camerini-Otero, 2000, Santucci-Darmanin et al., 2002), were upregulated in spermatocytes (Figure S1A). These observations confirmed the purity of the germ cell populations and the reliability of the bioinformatics analysis.
Figure 1.
An Extensive Alternative Splicing Program Characterizes the Meiotic Transition of Male Germ Cells
(A) Representative images of Hoechst nuclear staining of purified cellular populations of meiotic pachytene spermatocytes (sp.cytes, upper panel) and post-meiotic round spermatids (sp.tids, lower panel). Scale bar, 25 μm.
(B and C) Pie charts showing percentages of expression-regulated genes (B) and splicing-regulated exons (C) identified in sp.cytes compared with sp.tids (fold change ≥2; p ≤ 0.05).
(D) Pie chart representing distribution of regulated splicing events in sp.cytes versus sp.tids among different splicing patterns.
(E) Representative images of RT-PCR analyses for indicated AS events differentially regulated between sp.cytes and sp.tids. Schematic representation for each event analyzed is depicted above the representative agarose gel. Red and green boxes indicate respectively up- and downregulated events in sp.cytes compared with sp.tids. Black arrows in the scheme indicate primers used for the PCR analysis.
See also Figure S1.
By using the FAST-DB splicing patterns annotation tool, we also identified 4,090 AS events in 1,753 genes that are modulated across meiosis in male germ cells, with the majority (65.1%) being upregulated in spermatocytes (Figure 1C and Table S1). Classification of the events corresponding to annotated splice variants in the FAST-DB reference database highlighted selection of alternative first and last exons and IR as the most regulated splicing modalities (Figure 1D and Table S2). RT-PCR analysis of randomly selected events yielded a high validation rate in all splicing patterns (80%; 12/15 events tested) (Figures 1E and S1B). Moreover, comparison of our dataset with a previous RNA-seq analysis of spermatocyte and spermatid transcriptomes (GSE43717; Soumillon et al., 2013) revealed a highly significant overlap (p = 0; modified Fisher's test) in splicing-regulated genes (Figure S1C) and a very similar distribution of splicing patterns (Figure S1D). Although many splicing-regulated genes (44.8%) were also regulated at the expression level (Figure S1E), with almost equal frequency in all splicing patterns (Figure S1F), they were underrepresented when compared with the total of genes differentially expressed in the two cell types (60% of expressed genes). These results uncover an extended transcriptome reprogramming across meiosis and highlight a specific splicing program activated during this crucial phase of spermatogenesis.
Intron Retention Is a Specific Feature of the Meiotic Transcriptome
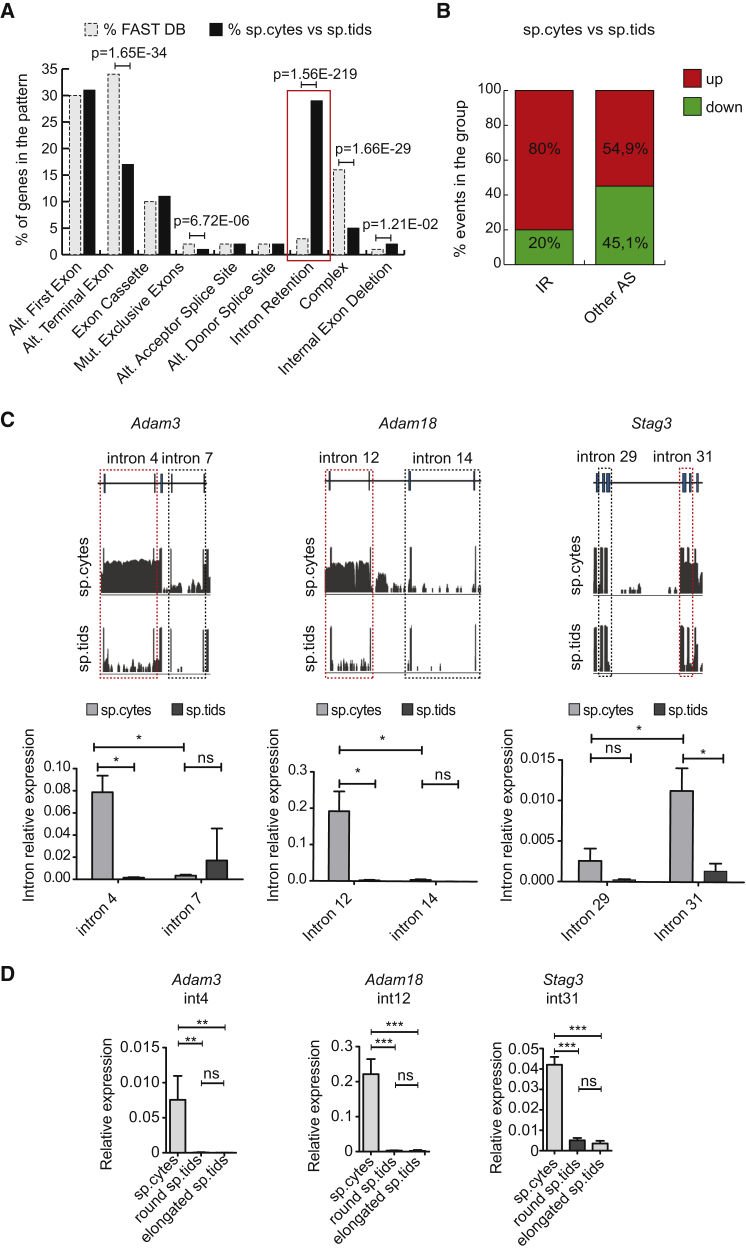
An unexpected finding of our analysis was the large extent of IR detected in male germ cells (Figure 1D). Indeed, unlike other splicing patterns, IR events were strikingly over-represented with respect to their expected frequency in the FAST-DB reference database (Figure 2A). To test whether this enrichment was due to underestimation of IR events in the FAST-DB, we analyzed RNA-seq data for other developing/differentiation systems using the same approach. Notably, in the comparison between the transcriptome of round spermatids and somatic Sertoli cells of the testis (GSE43717; Soumillon et al., 2013), IR accounted for 10% of the regulated splicing events, whereas this percentage was even lower (6%) when we compared newborn with adult cardiomyocytes (GSE49906; Giudice et al., 2014) (Figures S2A and S2B). By contrast, a similar percentage of IR events (22% compared with 29% in our condition) was regulated between spermatocyte and spermatid transcriptomes from an independent source (Soumillon et al., 2013) (Figures S1C and S2B). Thus, IR regulation is a specific feature of the transmeiotic differentiation of male germ cells.
Figure 2.
Intron Retention Is a Key Feature of the Male Meiotic Transcriptome
(A) Bar graph showing percentages of events annotated in FAST-DB (dashed, light gray columns) and of those differentially regulated between spermatocytes (sp.cytes) and spermatids (sp.tids) (black columns) within each AS pattern. p Values above the graph indicate a significant enrichment for regulated events within specific AS pattern in our dataset compared with the reference database (modified Fisher's test). Red box highlights the IR pattern.
(B) Bar graph representing percentages of up- and downregulated events among exonic and intronic splicing events. Enrichment in upregulated events in IR with respect to all other AS events was statistically significant (p = 4.9 × 10−61; χ2 test).
(C) (Top) Visualization of the RNA-seq reads profile of the intron-retaining genes Adam3, Adam18, and Stag3 in sp.cytes (upper graph) and sp.tids (lower graph). Sequence reads (vertical gray lines), exons (blue boxes), and introns (horizontal lines) are shown. (Bottom) Bar graphs showing results of qPCR analyses for the expression of the retained and non-retained introns (red and black dashed boxes, respectively) relative to their flanking exons in sp.cytes and sp.tids (mean ± SD, n = 3; ∗p ≤ 0.05, ns = not significant; one-way ANOVA).
(D) Bar graphs showing results of qPCR analyses for the expression of indicated introns relative to spliced product of their flanking exons in sp.cytes, and round and elongated sp.tids (mean ± SD, n = 4; ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001; ns, not significant; one-way ANOVA).
See also Figures S2 and S3.
Further classification of all splicing events differentially regulated between spermatocytes and spermatids (thus including novel splice variants besides the annotated ones of Figure 1C) showed that IR accounts for 40.9% of the regulated splicing events (Figure S2C). Notably, 80% of IR events were upregulated in spermatocytes compared with spermatids (Figure 2B). This trend was statistically significant and specific, as all other splicing types were more equally distributed among up- and downregulated events (Figure 2B). Moreover, analysis of IR regulation from an independent RNA-seq analysis of mouse spermatocytes and spermatids (Soumillon et al., 2013) yielded highly significant overlap with our results (Figures S2D–S2F). These analyses strongly suggest that IR represents a prominent modality of splicing regulation in meiotic spermatocytes. Time-course analysis of representative IR events in whole testis RNAs confirmed that introns are spliced more efficiently in adult testis (60 days post partum [dpp]), when the majority of germ cells are at post-meiotic stages, than in juvenile testis (12–20 dpp) (Figures S3A and S3B), when the organ is enriched in meiotic cells (Griswold, 2016).
To validate these observations, we selected a subset of IR events predicted by the RNA-seq analysis. In all 20 cases tested, real-time PCR analysis using primers spanning exon-intron regions and exon-exon junctions (spliced introns) confirmed much higher expression levels of IRTs in spermatocytes compared with both early-stage (round) and late-stage (elongated) spermatids (Figures 2C and S3C–S3E). IR events were specific, as neighboring introns not detected in the RNA-seq analysis were not significantly retained in spermatocytes (Figure 2C). Likewise, five genes that resulted properly spliced by RNA-seq analysis did not display any significant accumulation of introns (Figure S3F). These results confirmed that IR is a prominent pattern in mouse spermatocytes, which affects a large proportion of their splicing signature.
Meiotic Intron-Retaining mRNAs Are Stable Nuclear Transcripts
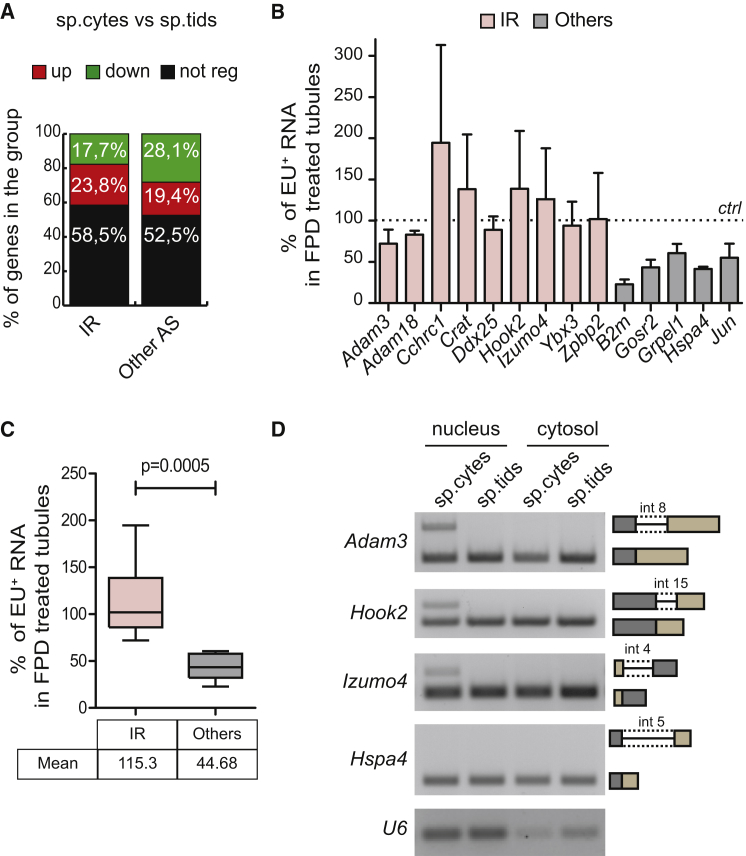
Generation of IRTs in mammalian cells has been mainly described as a device utilized to downregulate expression of genes that are no longer required during stress or development (Wong et al., 2016). In these transcripts, IR often introduces premature termination codons (PTCs), thus targeting them to degradation by the non-sense-mediated decay (NMD) pathway or by the nuclear RNA quality-control machinery (Yap et al., 2012, Wong et al., 2013, Shalgi et al., 2014, Edwards et al., 2016, Pimentel et al., 2016, Ni et al., 2016). However, we observed no enrichment for downregulated genes within those affected by IR compared with other splicing-regulated genes (Figure 3A). On the contrary, IRTs are slightly enriched for unaffected and upregulated genes (Figure 3A). Furthermore, among the 542 IR events introducing PTCs that we predicted from in silico analysis, we did not observe enrichment for downregulated genes (Figure S4). These observations suggest that IR does not target transcripts for degradation in meiosis.
Figure 3.
IRTs Are Highly Stable mRNAs Retained in the Nucleus of Meiotic Cells
(A) Bar graph showing percentages of expression-regulated genes within the groups of intron-retention and other splicing-regulated genes. A significant association was found between the gene regulation and the type of event (p = 0.0156; χ2 test).
(B) Bar graph showing qPCR analysis using primers spanning exon-intron regions for EU-labeled RNA transcripts of indicated genes isolated from total RNA of seminiferous tubules of 20-dpp (days post-partum) mice treated or not for 1 hr with 1 μM FPD. Results are expressed as percentage of the EU-labeled RNA pulled down relative to control condition, arbitrarily set to 100% (dashed line in the graph, mean ± SD, n = 3).
(C) Box plot showing the distribution of the mean percentages of EU-labeled RNA transcripts pulled down as described in (B) in the IR-regulated genes and other expressed genes. Whiskers indicate 1.5 interquartile range. p Value indicates a significant difference between means of the two groups (Welch's t test).
(D) PCR analysis of the subcellular localization of the IRTs of Adam3, Hook2, and Izumo4 genes. The properly spliced genes Hspa4 and U6 were evaluated as control.
See also Figure S4.
To directly test whether IR affects the stability of meiotic transcripts, we analyzed their decay after transcription inhibition. Fragments of testicular tubules were cultured for 4 hr in the presence of 5-Ethynyl uridine (EU), a nucleoside analog of uracil, to label nascent transcripts (Jao and Salic, 2008). After EU removal, transcription was inhibited by treatment with flavopiridol (FPD) and EU-labeled RNAs were isolated. Interestingly, FPD treatment did not cause a detectable reduction in the expression levels of IRTs (Figure 3B). By contrast, expression of properly spliced genes, such as Gosr2, B2m, Grpel1, or Hspa4, was reduced after transcriptional inhibition, similarly to what was observed for the high-turnover Jun transcript (Figure 3B). Moreover, by comparing transcript levels in FPD versus vehicle-treated tubules, we observed a significantly higher stability for IRTs with respect to properly spliced transcripts (Figure 3C).
To further investigate the regulation of IR-containing transcripts, we checked their localization. Cellular fractionation followed by conventional PCR analysis demonstrated that IRTs (i.e., Adam3, Izumo4, and Hook2 transcripts) were specifically localized in the nucleus of meiotic spermatocytes, whereas no signal for IRTs was detected in the nuclear fraction of spermatids (Figure 3D). These observations further confirmed the meiosis-specific nature of the IR phenomenon in male germ cells and suggest that nuclear localization of these transcripts may prevent their degradation. Thus, unlike what was observed in other differentiation processes, IR during spermatogenesis is not functional in dampening the regulated transcripts.
Transcriptional Overload Negatively Selects Splicing of Weak Introns in Meiosis
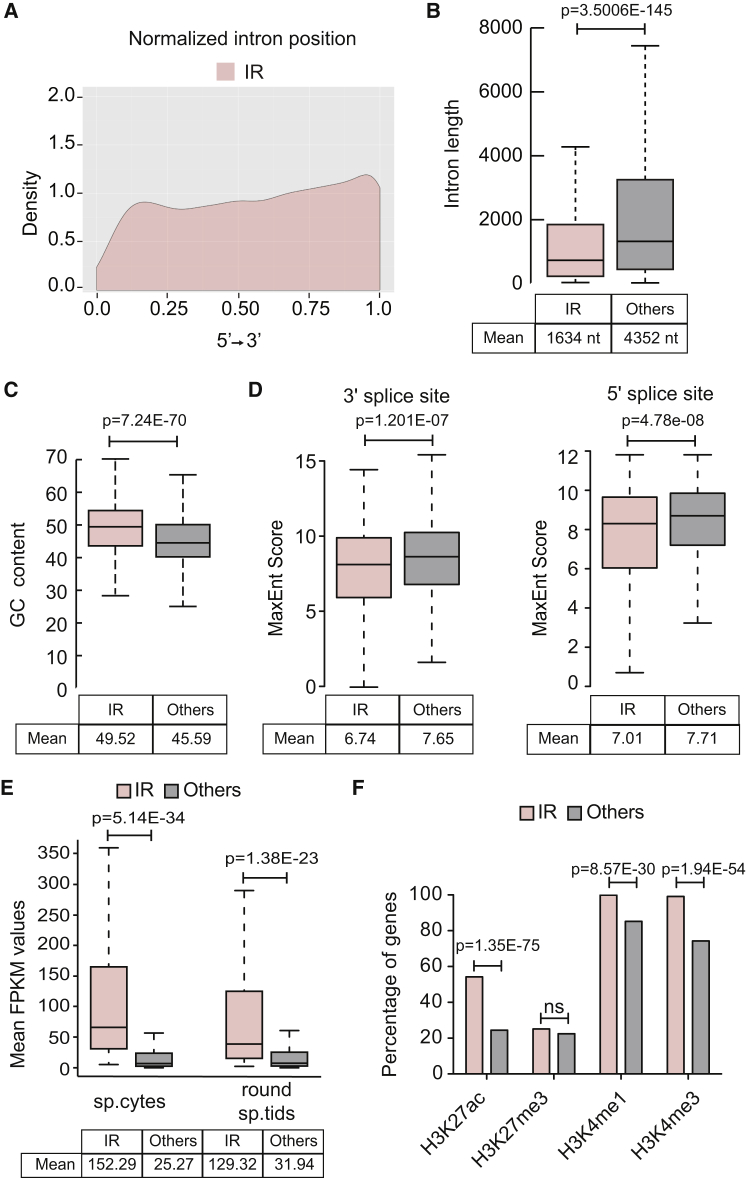
To gain insight into the nature of the IRTs enriched in meiosis, we explored whether retained introns displayed specific structural features that may distinguish them from properly spliced introns. Bioinformatics analysis documented that retained introns are preferentially localized toward the 3′ end of genes (Figure 4A). Similarly to what was previously reported for other introns affected by developmentally or physiologically regulated retention (Braunschweig et al., 2014, Boutz et al., 2015), meiotically retained introns are significantly shorter in length and display a higher GC content (Figures 4B and 4C). Furthermore, they are flanked by weaker 5′ and 3′ splice sites when compared with all other introns (Figure 4D). These results suggest that meiotically retained introns represent a distinct class characterized by specific features, which might contribute to the mechanism(s) underlying their retention. Intriguingly, by evaluating the mean FPKM (fragments per kilobase of transcript per million mapped reads) value as estimate of the gene expression levels (Trapnell et al., 2010), we observed significantly higher expression in the group of IR genes compared with all the other transcribed genes (Figure 4E). Moreover, comparison of our dataset with chromatin immunoprecipitation sequencing (ChIP-seq) analyses of histone marks in spermatocytes available from the GEO database (GSE49624; Hammoud et al., 2014) highlighted a significant enrichment of epigenetic marks of active transcription (i.e., acetylated H3K27, trimethylated H3K4) in the 5′ region of IR genes with respect to all other genes, whereas repressive marks (i.e., trimethylated H3K27) were not affected (Figure 4F). These results suggest that the high transcriptional level of IR genes combined with the specific features of their sequence negatively selects retained introns from recognition and excision by the spliceosome in meiosis.
Figure 4.
Specific cis-Acting Elements and Elevated Transcription Rate Feature Meiotic IRTs
(A) Graphic representation of retained introns location within gene body.
(B–D) Box plots representing comparison between retained introns and other introns for their length (B), GC content (C), and splice-site strength (D). Whiskers indicate 1.5 interquartile range. p Values indicate a significant difference between means of the two populations (Welch's t test).
(E) Box plots showing distribution of FPKM values for IR and other expressed genes in spermatocytes (sp.cytes) and spermatids (sp.tids). p Values indicate a significant difference between means of the two groups in the two cell types (Welch's t test).
(F) Bar graph showing percentages of intron-retaining and other expressed genes enriched for indicated histone marks in sp.cytes. p Values indicate a significant difference in the enrichment for different histone marks between the two groups (χ2 test).
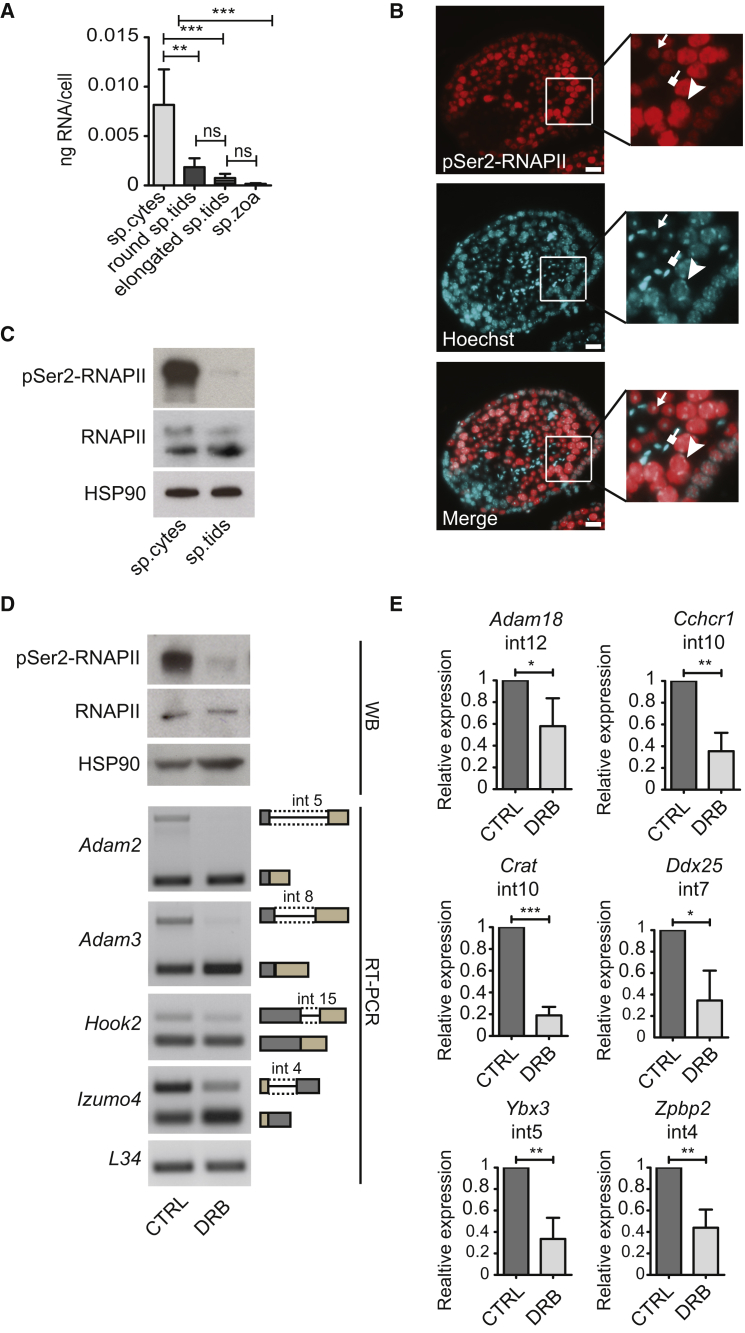
Next, we asked why meiotically retained introns were properly spliced in spermatids. Transcription is discontinuous during spermatogenesis, with three active bursts in mitotic spermatogonia, meiotic spermatocytes, and haploid early spermatids being followed by transcriptionally inactive phases (Paronetto and Sette, 2010). Measurement of total RNA per cell showed much higher levels in spermatocytes than in later stages of male germ cell differentiation (Figure 5A). Fast transcription elongation rates correlate with increased phosphorylation of RNAPII in serine 2 (Ser-2) (Phatnani and Greenleaf, 2006). Immunofluorescence analysis of seminiferous tubules and western blot analysis of isolated germ cells revealed strong Ser-2 phosphorylation of RNAPII in spermatocytes and much lower signal in round spermatids (Figures 5B and 5C), while no staining was detected in elongated spermatids (Figure 5B). Moreover, a short pulse of EU labeling showed higher levels of nascent transcripts in meiotic spermatocytes than in post-meiotic spermatids, which positively correlated with Ser-2 RNAPII staining (Figure S5). These observations indicate that spermatocytes display higher transcriptional activity than spermatids.
Figure 5.
The Elevated Transcriptional Activity of Meiotic Spermatocytes Modulates IR Events
(A) Histogram showing the total RNA per cell synthesized by spermatocytes (sp.cytes), round spermatids (sp.tids), elongated sp.tids, and spermatozoa (sp.zoa) (mean ± SD, n = 4, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001; ns, not significant; one-way ANOVA test).
(B) Representative images of immunofluorescence analysis of seminiferous tubule cross-sections from adult mice using anti-pSer2-RNAPII (H5) antibody. Hoechst staining was used to identify nuclear morphology. Insets show magnified images of meiotic spermatocytes (arrowhead), post-meiotic round spermatids (arrow), and elongated spermatids (square-headed arrow). Scale bar, 25 μm.
(C) Representative western blot analysis for RNAPII and its Ser2-CTD phosphorylated form (pSer2-RNAPII) in nuclear-enriched extracts of sp.cytes and round sp.tids. HSP90 protein level was evaluated as loading control.
(D) Upper panels show representative western blot analysis for total and pSer2-RNAPII in nuclear-enriched extracts obtained from seminiferous tubules treated or not for 24 hr with 10 μg/mL DRB. HSP90 was evaluated as loading control. Lower panels show representative images of agarose gel analysis for RT-PCR products of indicated IR events under the same experimental conditions.
(E) Bar graphs showing results of qPCR analyses for indicated retained introns relative to their flanking exons in seminiferous tubules treated for 24 hr with 10 μg/mL DRB compared with the control condition, arbitrarily set to 1 (mean ± SD, n = 4; ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001; one-way ANOVA).
See also Figure S5.
We hypothesized that the high levels of pre-mRNA synthesis in spermatocytes could impose competition for spliceosome recruitment, thus causing retention of introns with weak features (Figure 4D) in highly expressed transcripts such as IRTs (Figure 4E). To test whether release of this transcriptional burden allows splicing of retained introns, we treated fragments of testicular tubules with 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB) to reduce Ser-2 phosphorylation (Figure 5D), thus decreasing the RNAPII elongation rate (Ip et al., 2011). Strikingly, DRB treatment was sufficient to enhance splicing of retained introns in IRTs (Figures 5D and 5E). These results show that reducing the transcription rate of mouse spermatocytes is sufficient to rescue recognition and splicing of retained introns by the spliceosome.
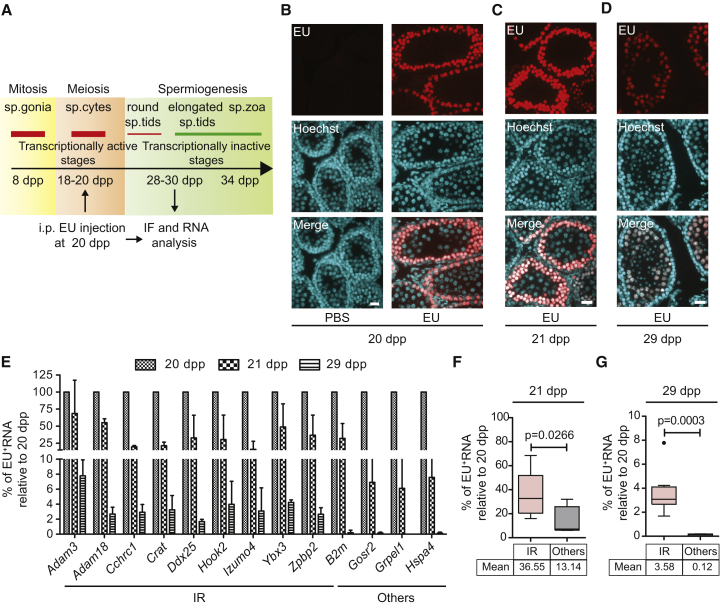
Meiotic Intron-Retaining Transcripts Are Preserved until Late Phases of Spermatogenesis and Encode for Key Spermatogenic Proteins
It was previously reported that a large amount of poly(A)+ mRNAs is accumulated in the nucleus of spermatocytes, suggesting nuclear RNA storage at this stage of germ cell differentiation (Morales and Hecht, 1994). To examine whether mRNAs are actually stably stored in the nucleus of meiotic cells, we performed in vivo EU labeling of nascent RNAs and followed their fate upon germ cell differentiation (Figure 6A). Immunofluorescence analysis of testis 5 hr after EU injection indicated meiotic germ cells as the preferential site of accumulation of nascent RNAs in testis (Figure 6B). Importantly, these transcripts were stable for days in the nuclei of differentiating germ cells, as they were still detected in more luminal late meiotic (Figures 6C and 6D) and early post-meiotic germ cells (Figure S6A) even 9 days after EU injection. Stability of nuclear transcripts was a specific feature of germ cells, because it was not detected in the nuclei of the somatic Sertoli cells within the seminiferous tubules nor in the peritubular and interstitial cells of testis (Figure S6B). Furthermore, while EU-positive cells could be readily detected in kidney 5 hr after injection, no staining was present after 24 hr (Figure S6C).
Figure 6.
IRTs Are Stable during Spermatogenesis
(A) Schematic representation of the timeline of the key phases of murine spermatogenesis and their transcriptional activity. Time points of the in vivo RNA labeling experiments with EU are depicted below.
(B–D) EU staining with Alexa 594-azide of testicular paraffin-embedded cross-sections from EU-injected and PBS-injected (as control) mice harvested at 20 dpp (B), 21 dpp (C), and 29 dpp (D). Hoechst nuclear staining was performed to discriminate the different cellular types within the seminiferous tubules. Scale bar, 25 μm.
(E) Bar graph showing qPCR analysis for EU-labeled RNAs pulled down at indicated time points. Results are expressed as percentage of the EU-labeled RNA captured 5 hr after injection at 20 dpp, arbitrarily set to 100% (mean ± SD, n = 3).
(F and G) Box plot showing the distribution of the mean percentages of EU-labeled RNAs pulled down as described in (E) in the IR-regulated genes and other expressed genes. Whiskers indicate 1.5 interquartile range. p Values indicate a significant difference between means of the two groups (Welch's t test).
See also Figure S6.
Next, we asked whether IR promoted the accumulation of stable transcripts in spermatocyte nuclei. To this end, EU-labeled mRNAs were isolated by pull-down and analyzed by qPCR. We found that IRTs were stable in germ cells for up to 9 days after their synthesis, whereas the turnover of properly spliced mRNAs was much faster (Figure 6E). Indeed, ∼50% of EU-labeled IRTs were still present after 24 hr and ∼3%–10% of them were stored up to 9 days after labeling. By contrast, less than 10% of properly spliced mRNAs were detected 24 hr after labeling whereas they were undetectable after 9 days (Figure 6E). The higher stability in vivo of IRTs with respect to properly spliced transcripts was statistically significant at both times (Figures 6F and 6G) and not influenced by levels of initial labeling, as IRTs did not show higher labeling efficiency (percentage of total RNA) 5 hr after EU injection (Figure S6D). These observations indicate that IR promotes nuclear accumulation and storage of a specific subset of transcripts in meiosis.
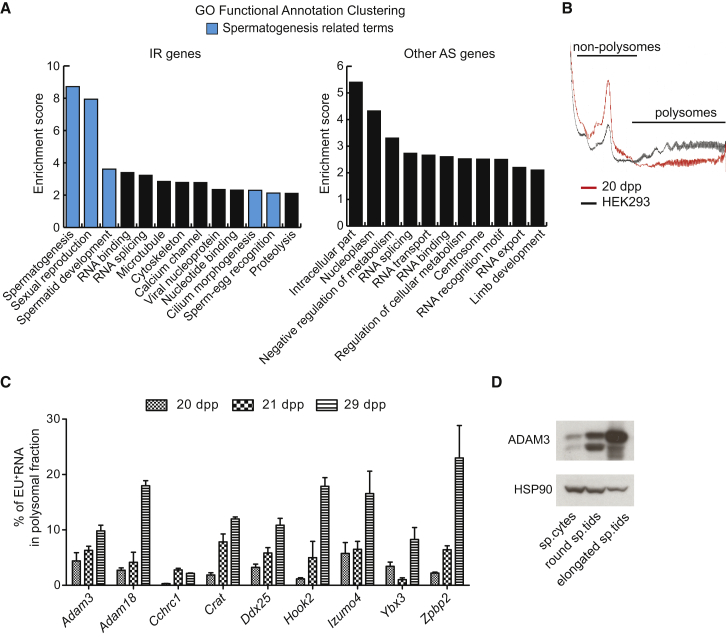
Post-transcriptional regulation of gene expression plays a key role during spermatogenesis (Paronetto and Sette, 2010), and some meiotic mRNAs are preserved and translated at later stages (Monesi, 1964, Geremia et al., 1977, Iguchi et al., 2006). To investigate whether IR genes displayed distinct functional properties with respect to all other AS-regulated genes, we performed gene ontology (GO) analysis. Notably, IR genes were specifically enriched in functional categories with strong relevance for spermatogenesis, and in particular for the late phase of this process when de novo mRNA synthesis is abolished (Monesi, 1964, Geremia et al., 1977) (Figure 7A). To test whether usage of IRTs is delayed during germ cell differentiation, we performed polysome profiling of testicular extracts at different times from EU injection. As previously suggested (Messina et al., 2010), translation efficiency was very low in testis (Figures 7B, S7A, and S7B), with transcripts being enriched in the translationally inactive “non-polysome” fractions (Figure S7C). However, we found that EU-labeled transcripts encoded by IR-regulated genes displayed a time-dependent shift to translationally active polysomes at times (9 days) (Figure 7C) when properly spliced transcripts have already disappeared (Figure 6E). This clear relative increase in transcript recruitment to the polysomes, however, may not result in more protein, as the total amount of transcripts decreases between day 21 and day 29 (Figure 6E). To test the possibility that proteins encoded by IR-regulated genes accumulate in late germ cells, we analyzed the expression of ADAM3 as a representative example. In line with the polysomal recruitment at later stages, ADAM3 protein levels steadily increased from spermatocytes to elongated spermatids (Figure 7D). These results suggest that IR is a mechanism designed to accumulate and preserve transcripts with strong relevance for gamete differentiation, thus prolonging their use until later stages of spermatogenesis.
Figure 7.
Meiotic IRTs Are Enriched in GO Terms Relative to Spermatogenesis and Are Actively Translated Several Days after Their Synthesis
(A) Bar graphs showing the enrichment score of the GO functional clusters enriched in the groups of IR-regulated and other AS-regulated genes (p ≤ 0.05, high stringency).
(B) Absorbance profile (OD = 260 nm) of sucrose gradient sedimentation of extracts from mitotic HEK293 cells (black line) and testes of 20-dpp mice (red line).
(C) Bar graphs showing results of qPCR analysis for EU-labeled RNAs of indicated genes within the polysomal fraction derived from sucrose gradient fractionation of whole testes harvested at indicated time points after EU injection at 20 dpp. Results are expressed as percentage of the total EU-labeled RNA in all the fractions of the gradient (mean ± SD, n = 3).
(D) Representative images of western blot analysis for ADAM3 in total extracts of spermatocytes (sp.cytes), and round and elongated spermatids (sp.tids). HSP90 was evaluated as loading control.
See also Figure S7.
Discussion
Testis displays the highest transcriptome complexity among mammalian tissues, and most of this complexity is due to germ cells undergoing differentiation (Soumillon et al., 2013). Although AS is known to amplify transcriptome diversity (Fu and Ares, 2014) and testis is characterized by a high content of tissue-specific splice variants (Kan et al., 2005, Schmid et al., 2013), scant information is available on the global splicing changes occurring during germ cell differentiation. Here we uncovered a widespread IR program in meiosis, which stabilizes highly expressed transcripts with strong relevance for spermatogenesis. IRTs are retained in the nucleus of meiotic cells for days and can be recruited onto the polysomes for translation long after their synthesis. Thus our results indicate that, in addition to being a device to destabilize transcripts at specific developmental transitions (Yap et al., 2012, Wong et al., 2013, Shalgi et al., 2014, Edwards et al., 2016, Pimentel et al., 2016, Ni et al., 2016), IR can also favor accumulation, storage, and timely usage of specific transcripts during highly organized cell differentiation programs, such as spermatogenesis. A similar “positive” role for IR was also described in embryonic stem cells in response to stress (Boutz et al., 2015). However, the peculiarity of our finding is that IRTs can be maintained for days after synthesis and that IR regulation timely determines the usage of transcripts when transcription is repressed. This observation suggests that post-transcriptional splicing is particularly suited to temporally control gene expression in cells undergoing transcriptional silencing, such as germ cells at specific stages of their differentiation (Paronetto and Sette, 2010).
Meiotic spermatocytes and post-meiotic spermatids were selected for analyses because these cells undergo profound and unique morphogenetic changes, such as homologous recombination and spermiogenesis (Griswold, 2016), which likely require a global reprogramming of gene expression. Accordingly, we found that ∼60% of the transcribed genome is differentially regulated between these cell types, indicating a widespread reorganization of the transcriptome across meiosis. In addition to changes in transcript levels, our analyses also highlighted an extensive AS program regulated during transmeiotic differentiation of germ cells. We focused on IR because it was the most represented pattern, far beyond its expected frequency, and because intron-retaining genes were enriched in functional categories strongly relevant for spermatogenesis with respect to all other AS-regulated genes. Most IR events were detected in meiotic spermatocytes, whereas these introns were spliced in post-meiotic cells. These observations suggested that temporal regulation of IR plays an important role in male germ cell differentiation. One question arising from these results is why retained introns are less efficiently spliced in meiotic cells. Interestingly, bioinformatics analyses identified sequence features (i.e., weak splice sites, short length, high GC content) that were also found in retained introns in other cell types and tissues (Braunschweig et al., 2014). In addition, we found that IRTs are expressed at significantly higher level than all other transcripts in the germ cell transcriptome. In line with the observations that spermatocytes display higher transcriptional rates (Monesi, 1964, Geremia et al., 1977, Paronetto et al., 2011) and RNAPII phosphorylated levels (Ser2) than post-meiotic spermatids (this work), we hypothesized that meiotically retained introns could be outcompeted by stronger introns during co-transcriptional splicing. In support of this hypothesis, we found that relieving this “transcriptional burden” by reducing RNAPII activity rescued splicing of retained introns in meiotic cells, thus recapitulating the pattern observed in spermatids. Notably, a similar developmentally modulated competition for the spliceosome was also described in yeast (Munding et al., 2013). In that case, however, high transcription of ribosomal genes titrated the splicing machinery away from weak introns and favored their retention in vegetative cells. When ribosomal genes are turned off in meiosis, splicing efficiency is restored (Munding et al., 2013), indicating that pre-mRNAs compete for limiting factors during nuclear processing. Thus, modulation of IR by competition for the spliceosome could represent a conserved mechanism of regulation for genes involved in specialized developmental programs in eukaryotes.
One key feature of mammalian spermatogenesis is the discontinuous nature of transcription. In particular, de novo synthesis of mRNAs is ceased in post-meiotic cells due to exchange of histones with protamines and tight compaction of the chromatin (Paronetto and Sette, 2010, Hermo et al., 2010b). Thus, transcripts required for the late phases of differentiation must be synthesized prematurely and stored until needed. While much of this regulation was attributed to translational control in the cytoplasm (Paronetto and Sette, 2010, Kleene, 2013), it was also observed that a large fraction of poly(A)+ transcripts accumulated in the nucleus of meiotic spermatocytes (Morales and Hecht, 1994). Here we found that IRTs are highly expressed polyadenylated transcripts exclusively localized in the meiotic nuclei, which may account for this earlier observation. Moreover, fate-mapping experiments demonstrated that IRTs are particularly stable transcripts, suggesting that they might be stored in the nuclei to be utilized at later stages. Indeed, although we cannot formally prove that the spliced mRNAs recruited for translation directly derive from IRT molecules, polysome profiling indicated that their translational efficiency increases with time, likely as a reflection of their slow and delayed splicing. Since functional categories related to mature gamete properties, such as spermatid development and sperm-egg recognition, are significantly enriched in IR-regulated genes, we suggest that IR allows their accumulation during the transcriptional burst of meiotic cells, whereas the slower transcriptional rate of post-meiotic cells favors spliceosome recruitment, splicing, and translation. As a proof of principle of this hypothesis, we show that accumulation of ADAM3 protein parallels the delay in pre-mRNA processing and polysome recruitment of its transcript. Thus, IR might represent a device to temporally uncouple transcription and translation of abundant mRNAs that are required for proper sperm function.
In conclusion, our study expands the role of IR in the regulation of gene expression in mammals. While it was previously appreciated that IR promotes elimination of unwanted transcripts at developmental or differentiation transitions, we now show that it can also play a positive effect by ensuring timely expression of selected genes during highly specialized developmental programs.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat anti-RNA polymerase II subunit B1 (phospho CTD Ser-2) clone 3E1 | Millipore | Cat # 04-1571; RRID: AB_10627998 |
| Rabbit polyclonal anti HSP 90α/β (H-114) | Santa Cruz | Cat # sc-7947; RRID: AB_2121235 |
| Rabbit polyclonal anti-RNA polymerase II (N-20) | Santa Cruz | Cat # sc-899; RRID: AB_632359 |
| Mouse monoclonal anti-ADAM3 | Santa Cruz | Cat # sc-365288; RRID: AB_10846942 |
| Mouse monoclonal [H5] to RNAPII H5 | Abcam | Cat # ab24758; RRID: AB_2167352 |
| Donkey polyclonal Cy3 conjugated Anti-Mouse IgM | Jackson Immunoresearch | Cat # 715-165-020; RRID: AB_2340811 |
| Donkey whole antibody anti Rabbit IgG HRP-linked | GE Healthcare | Cat # NA934; RRID: AB_772206 |
| Sheep HRP-linked F(ab')2 fragment anti mouse IgG | GE Healthcare | Cat # NA9310; RRID: AB_772193 |
| Goat anti-rat IgG HRP-linked | Santa Cruz | Cat # sc-2032; RRID: AB_631755 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Collagenase from Clostridium histolyticum | Sigma Aldrich | Cat # C7657; CAS 9001-12-1 |
| Dnase I from bovine pancreas | Sigma Aldrich | Cat # DN25; CAS 9003-98-9 |
| Trypsin from bovine pancreas | Sigma Aldrich | Cat # T9201; CAS 9002-07-7 |
| 5,6-Dichlorobenzimidazole 1-β-D-ribofuranoside (DRB) | Sigma Aldrich | Cat # D1916; CAS 53-85-0 |
| Flavopiridol hydrochloride hydrate (FPD) | Sigma Aldrich | Cat # F3055 |
| EU | Life Technologies | Cat # E10345 |
| Critical Commercial Assays | ||
| miRNEasy minikit | Qiagen | Cat # 217004 |
| TruSeq Stranded mRNA Library Prep Kit | Illumina | Cat # RS-122-2101 |
| RNase-free DNase | Roche | Cat # 04 716 728 001 |
| M-MLV reverse transcriptase | Promega | Cat # M170B |
| GoTaq | Promega | Cat # M3005 |
| LightCycler 480 SYBR Green I Master | Roche | Cat # 04 887 352 001 |
| RNasin Ribonuclease Inhibitor | Promega | Cat # N251B |
| Immunocruz Western Blotting Luminol Reagent | Santa Cruz | Cat # sc-2048 |
| Click-IT RNA Imaging kit | Life Technologies | Cat # C10330 |
| Click-IT Nascent RNA Capture kit | Life Technologies | Cat # C10365 |
| SuperScript VILO cDNA Synthesis Kit | Life Technologies | Cat #11754-050 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GSE95138 |
| Raw data | Soumillon et al., 2013 | GSE43717 |
| Raw data | Giudice et al., 2014 | GSE49906 |
| Raw data | Hammoud et al., 2014 | GSE49624 |
| Mouse reference genome assembly mm10, GRCm38 | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/grc/mouse |
| Mouse FAST DB v2013_2 | GenoSplice technology | http://www.easana.com |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6 | Animal facility of the University of Rome Tor Vergata | N/A |
| Oligonucleotides | ||
| See Table S3 for primer sequences used for conventional and qPCR analysis | This paper | N/A |
|
Software and Algorithms | ||
| FastQC V0.11.2 | http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ | |
| Picard-Tools V1.119 | https://broadinstitute.github.io/picard/ | |
| Bedtools V2-2.20.1 | http://bedtools.readthedocs.io/en/latest/ | |
| Rseqc V2.3.9 | http://rseqc.sourceforge.net/ | |
| STARv2.4.0 | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| DAVID Functional annotation Tool (v6.8) | Huang da et al., 2009 | https://david.ncifcrf.gov/ |
| MaxEntScan tool | Yeo and Burge, 2004 | http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html |
| Python V2.7.6 | https://www.python.org/ | |
| R V3.2.5 | https://www.r-project.org | |
| GraphPad Prism 5.0 | http://www.graphpad.com/ | |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Claudio Sette (claudio.sette@uniroma2.it).
Experimental Model and Subject Details
Mouse Husbandry and Male Germ Cells Isolation
C57BL/6 mice were maintained on a normal 12 hr light/dark cycle in the animal facility of the University of Rome Tor Vergata, according to the Guideline of the Italian Institute of Health (protocol n. 1088/2016-PR). Spermatocytes and spermatids were collected from 12-week-old mice by gravimetric decantation (Sta-put) (for RNA-seq experiments) or by centrifugal elutriation of testicular cellular suspension as described (Geremia et al., 1976, Paronetto et al., 2006). Testicular cellular suspensions were prepared from single animal for the Sta-put method or by pooling germ cells collected from 3 age-matched animals for the centrifugal elutriation. Briefly, testes were dissected from the albuginea membrane and mildly digested for 15 minutes in 0,25 mg/ml collagenase (Sigma Aldrich); 0,05 mg/ml DNase I (Sigma Aldrich) in EKRB (120,1 mM NaCl; 4,8mM KCl; 25,2 mM NaHCO3; 1,2 mM KH2PO4; 1,2 mM MgSO4; 1,3 mM CaCl2; 11 mM glucose) at 32°C with constant shaking. Digestion was followed by two washes in EKRB and by further digestion in EKRB containing 1 mg/ml trypsin; 0,05 mg/ml DNase I (Sigma Aldrich) for 30 min at 30 °C. Digestion was stopped by adding 10% fetal calf serum and the released germ cells were collected after sedimentation of tissue debris. Germ cells were centrifuged for 10 min at 1500 rpm at 4 °C, the pellet resuspended in 20 ml of EKRB supplemented with 0,2% bovine serum albumin (BSA) and finally injected in the sample chamber of the Sta-put apparatus or of the elutriator. Homogeneity of cell populations was 80-85% (pachytene spermatocytes and elongated sp.tids), 95% (round spermatids) and was monitored by analysis by Hoechst-staining of nuclear-morphology. For spermatozoa collection, cauda of epidydimis of adult mice were dissected, placed in a petri dish containing PBS and incised in order to allow the sperm to swim out for 5 min into the medium (Sette et al., 1997). Sperm suspension was then collected and further processed for subsequent analyses.
Seminiferous Tubules Culture
For seminiferous tubules culture, testes collected from 20 dpp or adult mice were dissected from the albuginea membrane and mildly digested for 15 minutes in 0,25 mg/ml collagenase (Sigma Aldrich) at 32°C with constant shaking. Following two washes in MEM (Sigma Aldrich), released seminiferous tubules were cultured in MEM, supplemented with 0,5% BSA, 1 mM sodium pyruvate, 2 mM sodium lactate, non-essential aminoacids (Gibco), at 32°C in a humified atmosphere containing 5% CO2. For 5,6-Dichlorobenzimidazole 1-β-D-ribofuranoside (DRB) treatment experiments seminiferous tubules were treated with 10 μg/ml DRB (Sigma) or vehicle alone (DMSO) for 24 hr.
Method Details
RNA Extraction, Library Preparation and RNA-Seq Data Analysis
For RNA-seq analysis, purified populations of meiotic spermatocytes (n=2) and post-meiotic round spermatids (n=3) were fractionated by Sta-put and total RNA was extracted and DNAse treated using the miRNEasy extraction kit (Qiagen) according to manufacturer’s instruction. PolyA plus RNA-seq libraries were constructed according to Illumina’s protocol and sequenced using a 100 bp single-end format on an Illumina HiSeq 2000. RNA-Seq data analysis was performed by GenoSplice technology (www.genopslice.com). Sequencing, data quality, reads repartition (e.g., for potential ribosomal contamination), and insert size estimation were performed using FastQC, Picard-Tools, Samtools and rseqc. Reads were mapped using STARv2.4.0 (Dobin et al., 2013). Gene expression regulation study was performed as already described (Noli et al., 2015) using Mouse FAST DB v2013_2 annotations. Only genes expressed in at least sp.tids or sp.cytes were further analyzed. Genes were considered as expressed if their rpkm value was greater than 97,5% of the background rpkm value based on intergenic regions. Results were considered statistically significant for p-values ≤ 0.05 and fold-changes ≥ 1.5. Analysis at the splicing level was performed as already described (Traunmüller et al., 2016, Furney et al., 2013): first, by taking into account only exon reads and flanking exon-exon junction reads (“exon” analysis) in order to potentially detect new alternative events that could be differentially regulated (i.e., without taking into account known alternative events); then, by taking into account known patterns (“pattern” analysis) using the FAST DB splicing patterns annotation (i.e., for each gene, all possible splicing patterns were defined by comparing exon content of transcripts). All types of alternative events can be analyzed: Alternative first exons, alternative terminal exons, cassette exon, mutually exclusive exons, alternative 5' donor splice site, alternative 3' acceptor splice sites, intron retention, internal exon deletion and complex events corresponding to mix of several alternative event categories). “exon” and “pattern” analyses were based on the splicing-index calculation as previously described (Gandoura et al., 2013, Wang et al., 2012). Results were considered statistically significant for p-values ≤ 0.05 and fold-changes ≥1.5 for “PATTERN” and p-values ≤ 0.05 and fold-changes ≥ 2.0 for “EXON”. Same procedures were carried out for analysis of downloaded published datasets (GSE43717, Soumillon et al., 2013; GSE49906, Giudice et al., 2014). Analysis for enriched GO functional clusters among IR and other AS regulated genes was performed using DAVID Functional annotation Tool (v6.8), using the expressed genes as background (Huang da et al., 2009). Ontologies were considered as enriched if fold enrichment ≥ 2.0 and p-value ≤ 0.05.
Extraction of RNA, RT-PCR and Real-Time PCR Analysis
Total RNA was extracted using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. Isolation of nuclear and cytoplasmic RNA was performed as described (Rio et al., 2010). Briefly, isolated germ cells were washed three times with ice-cold PBS by centrifuging at 1000 g for 5 min and then lysed with three volumes of cold buffer A (10 mM KCl; 1,5 mM MgCl2; 20 mM TrisHCl; 1 mM DTT). Cellular lysates were incubated for 10 min on ice and then homogenized in a Dounce homogenizer by applying 15 strokes with a type B pestle. After addition of Triton X-100 to a final concentration of 0,1%, homogenates were centrifuged at 1500 g for 5 min at 4°C. Trizol was immediately added to the pellet nuclei, while supernatant corresponding to the cytosolic fraction was transferred in a new tube and three volumes of Trizol added. After digestion with RNase-free DNase (Roche), 250 ng - 1 μg of total RNA was retro-transcribed with oligo-dT primers, using M-MLV reverse transcriptase (Promega). cDNA was used as template for PCR (GoTaq, Promega) and reactions were analyzed on agarose or acrylamide gels. PCR analysis for validation of AS events was performed in triplicate. Quantitative real-time PCRs (RT-qPCR) were performed using LightCycler 480 SYBR Green I Master and the LightCycler 480 System (Roche), according to the manufacturer’s instructions. Control reactions omitting M-MLV reverse transcriptase were also carried out. All primers used are listed in Table S3.
Analysis of Cis-Acting Features of Retained Introns
To visualize the preferential location of retained introns, the number of retained intron was normalized with the total number of introns in the corresponding gene. 3' and 5' splice-sites strengths of retained introns and other introns were scored using MaxEntScan tool (Yeo and Burge, 2004) and GC content for each intron sequence was obtained by weighing the GC count with its length.
PTC and NMD Prediction
The representative RefSeq transcript was selected for each of the 768 genes with intron retention event. In case no RefSeq transcript existed, the GenBank transcript with a higher number of exons was selected. Sequence of retained intron was then included in transcript sequence. Open Reading Frame (ORF) predictions were performed using “find_orfs_with_trans” from Python module Bio.SeqIO (Python V2.7.6) on these new transcript sequences. Only ORFs from frame +1, +2 and +3 and with defined stop were selected. If the coding sequence (CDS) start site was already known in the original transcript sequence, ORF with the same start was selected. If no CDS was already known, the longest ORF was selected. If distance between the predicted CDS stop and the last exon-exon junction was greater than 50 nucleotides, the corresponding transcript was predicted to be targeted by NMD.
Comparison with ChIP-Seq Data
For comparison with ChIP-seq dataset for histone modifications, bedgraph files from GSE49624 (Hammoud et al., 2014) were annotated after changing the mm9 coordinates into the corresponding mm10 coordinates. The proportion of genes from “IR” and “Others” groups that were present in the GSE49624 data and had histone marks (score>=10) were plotted using R. These proportions were compared using Chi2 test, or Fisher's test when Chi2 conditions were not met (i.e. when expected values of the number of sample observations were < 5).
Protein Extracts and Western Blot Analysis
Nuclear-enriched cellular extracts were prepared by resuspending isolated germ cells or seminiferous tubules in RSB100 buffer [10 mM Tris HCl pH 7.4; 100 mM NaCl; 2,5 mM MgCl2; 0,5 % (v/v) Triton X-100; 15 mM β-glycerophosphate; 1 mM DTT; 0,5 mM NaVO3; protease inhibitor cocktail (Sigma Aldrich); RNasin Ribonuclease inhibitor 40U/ml (Promega)]. After incubation on ice for 15 min, samples were sonicated, stratified on 30% sucrose cushion and centrifuged for 15 min at 7000 g at 4°C. For total cellular extracts, isolated germ cells were resuspended in RIPA buffer [50 mM Tris pH 7.4; 1% NP-40; 0,5% Na deoxycholate; 0,1% SDS; 150 mM NaCl; 1 mM EDTA; 1 mM DTT; 0,5 mM NaVO3; protease inhibitor cocktail (Sigma Aldrich)], incubated on ice for 20 min, briefly sonicated, and centrifuged for 20 min at 12000 g at 4°C. Protein extracts were then analyzed by Western Blot using the following primary antibodies: rat anti-RNA polymerase II subunit B1 (phospho CTD Ser-2) clone 3E1 (Millipore; dilution 1:1000); rabbit anti-HSP90α/β (dilution 1:1000), anti- RNA polymerase II N-20 (dilution 1:1000) and mouse anti-ADAM3 (dilution 1:400) (Santa Cruz). Anti-rabbit, anti-mouse (GE Healthcare) and anti-rat (Santa Cruz) HRP-linked secondary antibodies were all used at 1:10000 dilution and ECL signal developed using Immunocruz Western Blotting Luminol Reagent (Santa Cruz).
Immunofluorescence Analysis of Nascent RNAs
Nascent RNAs were analyzed by immunofluorescence using the Click-IT RNA Imaging kit (Life Technologies). Seminiferous tubules from adult mice were grown for 2 hr 30 min in presence of 1 mM EU (Life Technologies) and then collected for analysis. For in vivo labeling, 20 dpp mice were intraperitoneally injected with 300 μg/gr EU (Life Technologies) or PBS, as control, and organs collected 5 hr, 24 hr (21 dpp), and 9 days (29 dpp) after injection. Samples were formalin-fixed, paraffin embedded and EU-staining performed according to manufacturer’s instructions. Briefly, 5-μm paraffin sections were mounted on polylysine-coated slides, dewaxed and rehydrated as previously described (Muciaccia et al., 2013). After permeabilization in 0,5% Triton X-100 in PBS for 20 min, sections were stained with Alexa Fluor 594 or Alexa Fluor 488 azide and nuclei counterstained with Hoechst 33342. Images were taken using a DMI6000B inverted microscope (LEICA Geosystems) equipped with a Pan-Neofluar 40X /0.75 objective lens and elaborated with Photoshop (Adobe) for composing panels.
Pull-Down Assay of Nascent RNAs
Seminiferous tubules from 20 dpp mice were grown for 4 hr in presence of 1 mM EU (Life Technologies) or PBS as control. Following two washes in MEM, tubules were released for 1 hr in fresh medium added or not with 1 μM flavopiridol (FPD) (Sigma-Aldrich) and then harvested for analysis. For in vivo labeling, 20 dpp mice were intraperitoneally injected with 300 μg/gr EU (Life Technologies) or PBS as control and testes collected 5 hr, 24 hr (21 dpp), and 9 days (29 dpp) after injection. Samples were collected in Trizol or snap-frozen for polysome fractionation, performed as previously described (Paronetto et al., 2006, Paronetto et al., 2009). Briefly, testes were homogenized in lysis buffer [100 mM NaCl; 10 mM MgCl2; 30 mM Tris-HCl pH 7.5; 1% Triton X-100; 1 mM DTT; 0,5 mM NaVO3; protease inhibitor cocktail (Sigma Aldrich); RNasin Ribonuclease inhibitor 40U/ml (Promega)]. After 15 min of incubation on ice, lysates were centrifuged for 10 min at 12000 g at 4°C. Supernatants were loaded on a 15-50% (w/v) sucrose gradients and sedimented by ultracentrifugation for 2 hr at 37000 rpm in a Beckman SW41 rotor. UV-absorption (A260) profiles of polysome gradients were measured by UV detector (UVis-920; GE Healthcare) and each gradient was collected in 10 fractions of 1 ml each. Fractions 1–5 and 6-10 were pooled to generate polysomal and non-polysomal fractions respectively and RNA was isolated by phenol:chloroform extraction. Isolated total or fractionated EU-labeled RNA was biotinylated and captured using the Click-IT Nascent RNA Capture kit (Life Technologies) according to manufacturer’s instructions. Captured nascent RNAs were retrotranscribed using the SuperScript VILO cDNA Synthesis Kit (Life Technologies) followed by RT-qPCR analysis performed as aforementioned.
Immunofluorescence Analysis
5-μm sections from paraffin embedded seminiferous tubules cross-sections were processed for immunofluorescence analysis as follows: sections were serially collected and mounted on polylysine-coated slides. Dewaxed and rehydrated sections were incubated in 10 mM sodium citrate, 0,05% Tween 20 pH 6.0 in a microwave oven for antigen retrieval at 750 W three times for 5 min. After permeabilization in 0,5% Triton X-100 in PBS for 15 min, sections were incubated with glycine 1 M in PBS for quenching autofluorescence and blocked 1 hr in a 0,1% Triton, 1% BSA, 5% donkey serum PBS solution. Sections were then incubated 2 hr at room temperature with primary antibody RNAPII H5 (1:200, Abcam) and then 1 hr with Cy3- conjugated anti-mouse IgM (1:500, Jackson Immunoresearch). Nuclei were counterstained with Hoechst 33342. Images were taken using a DMI6000B inverted microscope (LEICA Geosystems) equipped with a Pan-Neofluar 40X /0.75 objective lens and elaborated with Photoshop (Adobe) for composing panels.
Quantification and Statistical Analysis
Statistical analyses for differential gene expression, splicing changes, comparison of different datasets, analysis of cis-acting sequence features of introns, were performed in R according the statistical tests described in the figure legends. Statistical analyses for qPCR analysis were performed in GraphPad Prism according to the statistical tests described in the figure legends. Number of different cellular preparations or of animals independently analyzed is indicated by the “n” in each figure legend. In all figures, if p-value is not stated: ∗, p-value ≤ 0.05; ∗∗, p-value ≤ 0.01; ∗∗∗, p-value ≤ 0.001.
Data and Software Availability
The RNA-seq data have been deposited in the GEO database under ID GEO: GSE95138.
Author Contributions
Conceptualization, C.N and C.S.; Methodology, C.N., C.S., N.S., A.J.A., E.V., and R.G.; Software, Formal Analysis, and Data Curation, A.J. and P.D.l.G.; Investigation, C.N., S.D.P., N.S., and P.B.; Writing – Original Draft and Review & Editing, C.N. and C.S.; Visualization, C.N. and C.S.; Supervision, Project Administration, and Funding Acquisition, C.S.
Acknowledgments
We thank Dr. Eleonora Cesari and Livia Pellegrini for technical assistance. This work was supported by grants from Telethon (GGP 12189; GGP 14095), Associazione Italiana Ricerca sul Cancro (AIRC; IG14581), and Ministry of Health “Ricerca Corrente” and “5x1000 Anno 2014” to Fondazione Santa Lucia.
Published: March 30, 2017
Footnotes
Supplemental Information includes seven figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.devcel.2017.03.003.
Supplemental Information
References
- Barbosa-Morais N.L., Irimia M., Pan Q., Xiong H.Y., Gueroussov S., Lee L.J., Slobodeniuc V., Kutter C., Watt S., Colak R. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- Boutz P.L., Bhutkar A., Sharp P.A. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev. 2015;29:63–80. doi: 10.1101/gad.247361.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig U., Barbosa-Morais N.L., Pan Q., Nachman E.N., Alipanahi B., Gonatopoulos-Pournatzis T., Frey B., Irimia M., Blencowe B.J. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 2014;24:1774–1786. doi: 10.1101/gr.177790.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmel F., Rolland A.D. Linking transcriptomics and proteomics in spermatogenesis. Reproduction. 2015;150:R149–R157. doi: 10.1530/REP-15-0073. [DOI] [PubMed] [Google Scholar]
- Dadoune J.P. Expression of mammalian spermatozoal nucleoproteins. Microsc. Res. Tech. 2003;61:56–75. doi: 10.1002/jemt.10317. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards C.R., Ritchie W., Wong J.J., Schmitz U., Middleton R., An X., Mohandas N., Rasko J.E., Blobel G.A. A dynamic intron retention program in the mammalian megakaryocyte and erythrocyte lineages. Blood. 2016 doi: 10.1182/blood-2016-01-692764. pii:blood-2016-01-692764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X.D., Ares M., Jr. Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 2014;15:689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furney S.J., Pedersen M., Gentien D., Dumont A.G., Rapinat A., Desjardins L., Turajlic S., Piperno-Neumann S., de la Grange P., Roman-Roman S. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov. 2013;3:1122–1129. doi: 10.1158/2159-8290.CD-13-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandoura S., Weiss E., Rautou P.E., Fasseu M., Gustot T., Lemoine F., Hurtado-Nedelec M., Hego C., Vadrot N., Elkrief L. Gene- and exon-expression profiling reveals an extensive LPS-induced response in immune cells in patients with cirrhosis. J. Hepatol. 2013;58:936–948. doi: 10.1016/j.jhep.2012.12.025. [DOI] [PubMed] [Google Scholar]
- Geremia R., Goldberg R.B., Bruce W.R. Kinetics of histone and protamine synthesis during meiosis and spermiogenesis in the mouse. Andrologia. 1976;8:147–156. doi: 10.1111/j.1439-0272.1976.tb02124.x. [DOI] [PubMed] [Google Scholar]
- Geremia R., Boitani C., Conti M., Monesi V. RNA synthesis in spermatocytes and spermatids and preservation of meiotic RNA during spermiogenesis in the mouse. Cell Differ. 1977;5:343–355. doi: 10.1016/0045-6039(77)90072-0. [DOI] [PubMed] [Google Scholar]
- Giudice J., Xia Z., Wang E.T., Scavuzzo M.A., Ward A.J., Kalsotra A., Wang W., Wehrens X.H., Burge C.B., Li W. Alternative splicing regulates vesicular trafficking genes in cardiomyocytes during postnatal heart development. Nat. Commun. 2014;5:3603. doi: 10.1038/ncomms4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godmann M., Auger V., Ferraroni-Aguiar V., Di Sauro A., Sette C., Behr R., Kimmins S. Dynamic regulation of histone H3 methylation at lysine 4 in mammalian spermatogenesis. Biol. Reprod. 2007;77:754–764. doi: 10.1095/biolreprod.107.062265. [DOI] [PubMed] [Google Scholar]
- Griswold M.D. Spermatogenesis: the commitment to meiosis. Physiol. Rev. 2016;96:1–17. doi: 10.1152/physrev.00013.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud S.S., Low D.H., Yi C., Carrell D.T., Guccione E., Cairns B.R. Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell. 2014;15:239–253. doi: 10.1016/j.stem.2014.04.006. [DOI] [PubMed] [Google Scholar]
- Hermo L., Pelletier R.M., Cyr D.G., Smith C.E. Surfing the wave, cycle, life history, and genes/proteins expressed by testicular germ cells. Part 1: background to spermatogenesis, spermatogonia, and spermatocytes. Microsc. Res. Tech. 2010;73:241–278. doi: 10.1002/jemt.20783. [DOI] [PubMed] [Google Scholar]
- Hermo L., Pelletier R.M., Cyr D.G., Smith C.E. Surfing the wave, cycle, life history, and genes/proteins expressed by testicular germ cells. Part 2: changes in spermatid organelles associated with development of spermatozoa. Microsc. Res. Tech. 2010;73:279–319. doi: 10.1002/jemt.20787. [DOI] [PubMed] [Google Scholar]
- Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Iguchi N., Tobias J.W., Hecht N.B. Expression profiling reveals meiotic male germ cell mRNAs that are translationally up- and down-regulated. Proc. Natl. Acad. Sci. USA. 2006;103:7712–7717. doi: 10.1073/pnas.0510999103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip J.Y., Schmidt D., Pan Q., Ramani A.K., Fraser A.G., Odom D.T., Blencowe B.J. Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res. 2011;21:390–401. doi: 10.1101/gr.111070.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao C.Y., Salic A. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl. Acad. Sci. USA. 2008;105:15779–15784. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsotra A., Cooper T.A. Functional consequences of developmentally regulated alternative splicing. Nat. Rev. Genet. 2011;12:715–729. doi: 10.1038/nrg3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan Z., Garrett-Engele P.W., Johnson J.M., Castle J.C. Evolutionarily conserved and diverged alternative splicing events show different expression and functional profiles. Nucleic Acids Res. 2005;33:5659–5666. doi: 10.1093/nar/gki834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleene K.C. Connecting cis-elements and trans-factors with mechanisms of developmental regulation of mRNA translation in meiotic and haploid mammalian spermatogenic cells. Reproduction. 2013;146:R1–R19. doi: 10.1530/REP-12-0362. [DOI] [PubMed] [Google Scholar]
- Margolin G., Khil P.P., Kim J., Bellani M.A., Camerini-Otero R.D. Integrated transcriptome analysis of mouse spermatogenesis. BMC Genomics. 2014;15:39. doi: 10.1186/1471-2164-15-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina V., Di Sauro A., Pedrotti S., Adesso L., Latina A., Geremia R., Rossi P., Sette C. Differential contribution of the MTOR and MNK pathways to the regulation of mRNA translation in meiotic and postmeiotic mouse male germ cells. Biol. Reprod. 2010;83:607–615. doi: 10.1095/biolreprod.110.085050. [DOI] [PubMed] [Google Scholar]
- Monesi V. Ribonucleic acid synthesis during mitosis and meiosis in the mouse testis. J. Cell Biol. 1964;22:521–532. doi: 10.1083/jcb.22.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales C.R., Hecht N.B. Poly(A)+ ribonucleic acids are enriched in spermatocyte nuclei but not in chromatoid bodies in the rat testis. Biol. Reprod. 1994;50:309–319. doi: 10.1095/biolreprod50.2.309. [DOI] [PubMed] [Google Scholar]
- Muciaccia B., Boitani C., Berloco B.P., Nudo F., Spadetta G., Stefanini M., de Rooij D.G., Vicini E. Novel stage classification of human spermatogenesis based on acrosome development. Biol. Reprod. 2013;89:60. doi: 10.1095/biolreprod.113.111682. [DOI] [PubMed] [Google Scholar]
- Munding E.M., Shiue L., Katzman S., Donohue J.P., Ares M., Jr. Competition between pre-mRNAs for the splicing machinery drives global regulation of splicing. Mol. Cell. 2013;51:338–348. doi: 10.1016/j.molcel.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nantel F., Sassone-Corsi P. CREM: a transcriptional master switch during the spermatogenesis differentiation program. Front. Biosci. 1996;1:d266–d269. doi: 10.2741/a131. [DOI] [PubMed] [Google Scholar]
- Ni T., Yang W., Han M., Zhang Y., Shen T., Nie H., Zhou Z., Dai Y., Yang Y., Liu P. Global intron retention mediated gene regulation during CD4+ T cell activation. Nucleic Acids Res. 2016;44:6817–6829. doi: 10.1093/nar/gkw591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noli L., Capalbo A., Ogilvie C., Khalaf Y., Ilic D. Discordant growth of monozygotic twins starts at the blastocyst stage: a case study. Stem Cell Rep. 2015;5:946–953. doi: 10.1016/j.stemcr.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paronetto M.P., Sette C. Role of RNA-binding proteins in mammalian spermatogenesis. Int. J. Androl. 2010;33:2–12. doi: 10.1111/j.1365-2605.2009.00959.x. [DOI] [PubMed] [Google Scholar]
- Paronetto M.P., Zalfa F., Botti F., Geremia R., Bagni C., Sette C. The nuclear RNA-binding protein Sam68 translocates to the cytoplasm and associates with the polysomes in mouse spermatocytes. Mol. Biol. Cell. 2006;17:14–24. doi: 10.1091/mbc.E05-06-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paronetto M.P., Messina V., Bianchi E., Barchi M., Vogel G., Moretti C., Palombi F., Stefanini M., Geremia R., Richard S. Sam68 regulates translation of target mRNAs in male germ cells, necessary for mouse spermatogenesis. J. Cell Biol. 2009;185:235–249. doi: 10.1083/jcb.200811138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paronetto M.P., Messina V., Barchi M., Geremia R., Richard S., Sette C. Sam68 marks the transcriptionally active stages of spermatogenesis and modulates alternative splicing in male germ cells. Nucleic Acids Res. 2011;39:4961–4974. doi: 10.1093/nar/gkr085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phatnani H.P., Greenleaf A.L. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- Pimentel H., Parra M., Gee S.L., Mohandas N., Pachter L., Conboy J.G. A dynamic intron retention program enriched in RNA processing genes regulates gene expression during terminal erythropoiesis. Nucleic Acids Res. 2016;44:838–851. doi: 10.1093/nar/gkv1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio D.C., Ares M., Jr., Hannon G.J., Nilsen T.W. Preparation of cytoplasmic and nuclear RNA from tissue culture cells. Cold Spring Harb. Protoc. 2010;2010 doi: 10.1101/pdb.prot5441. pdb.prot5441. [DOI] [PubMed] [Google Scholar]
- Romanienko P.J., Camerini-Otero R.D. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell. 2000;6:975–987. doi: 10.1016/s1097-2765(00)00097-6. [DOI] [PubMed] [Google Scholar]
- Santucci-Darmanin S., Neyton S., Lespinasse F., Saunières A., Gaudray P., Paquis-Flucklinger V. The DNA mismatch-repair MLH3 protein interacts with MSH4 in meiotic cells, supporting a role for this MutL homolog in mammalian meiotic recombination. Hum. Mol. Genet. 2002;11:1697–1706. doi: 10.1093/hmg/11.15.1697. [DOI] [PubMed] [Google Scholar]
- Schmid R., Grellscheid S.N., Ehrmann I., Dalgliesh C., Danilenko M., Paronetto M.P., Pedrotti S., Grellscheid D., Dixon R.J., Sette C. The splicing landscape is globally reprogrammed during male meiosis. Nucleic Acids Res. 2013;41:10170–10184. doi: 10.1093/nar/gkt811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette C., Bevilacqua A., Bianchini A., Mangia F., Geremia R., Rossi P. Parthenogenetic activation of mouse eggs by microinjection of a truncated c-kit tyrosine kinase present in spermatozoa. Development. 1997;124:2267–2274. doi: 10.1242/dev.124.11.2267. [DOI] [PubMed] [Google Scholar]
- Shalgi R., Hurt J.A., Lindquist S., Burge C.B. Widespread inhibition of posttranscriptional splicing shapes the cellular transcriptome following heat shock. Cell Rep. 2014;7:1362–1370. doi: 10.1016/j.celrep.2014.04.044. [DOI] [PubMed] [Google Scholar]
- Singh A.P., Rajender S. CatSper channel, sperm function and male fertility. Reprod. Biomed. Online. 2015;30:28–38. doi: 10.1016/j.rbmo.2014.09.014. [DOI] [PubMed] [Google Scholar]
- Soumillon M., Necsulea A., Weier M., Brawand D., Zhang X., Gu H., Barthès P., Kokkinaki M., Nef S., Gnirke A. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep. 2013;3:2179–2190. doi: 10.1016/j.celrep.2013.05.031. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traunmüller L., Gomez A.M., Nguyen T.M., Scheiffele P. Control of neuronal synapse specification by a highly dedicated alternative splicing program. Science. 2016;352:982–986. doi: 10.1126/science.aaf2397. [DOI] [PubMed] [Google Scholar]
- Wang P.J., Page D.C., McCarrey J.R. Differential expression of sex-linked and autosomal germ-cell-specific genes during spermatogenesis in the mouse. Hum. Mol. Genet. 2005;14:2911–2918. doi: 10.1093/hmg/ddi322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E., Aslanzadeh V., Papa F., Zhu H., de la Grange P., Cambi F. Global profiling of alternative splicing events and gene expression regulated by hnRNPH/F. PLoS One. 2012;7:e51266. doi: 10.1371/journal.pone.0051266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J.J., Ritchie W., Ebner O.A., Selbach M., Wong J.W., Huang Y., Gao D., Pinello N., Gonzalez M., Baidya K. Orchestrated intron retention regulates normal granulocyte differentiation. Cell. 2013;154:583–595. doi: 10.1016/j.cell.2013.06.052. [DOI] [PubMed] [Google Scholar]
- Wong J.J., Au A.Y., Ritchie W., Rasko J.E. Intron retention in mRNA: no longer nonsense: known and putative roles of intron retention in normal and disease biology. Bioessays. 2016;38:41–49. doi: 10.1002/bies.201500117. [DOI] [PubMed] [Google Scholar]
- Yang X., Coulombe-Huntington J., Kang S., Sheynkman G.M., Hao T., Richardson A., Sun S., Yang F., Shen Y.A., Murray R.R. Widespread expansion of protein interaction capabilities by alternative splicing. Cell. 2016;164:805–817. doi: 10.1016/j.cell.2016.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap K., Lim Z.Q., Khandelia P., Friedman B., Makeyev E.V. Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes Dev. 2012;26:1209–1223. doi: 10.1101/gad.188037.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G., Burge C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- Zhou H., Grubisic I., Zheng K., He Y., Wang P.J., Kaplan T., Tjian R. Taf7l cooperates with Trf2 to regulate spermiogenesis. Proc. Natl. Acad. Sci. USA. 2013;110:16886–16891. doi: 10.1073/pnas.1317034110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.