Abstract
Background
Modification of leaf traits in sugar beet requires a strong leaf specific promoter. With such a promoter, expression in taproots can be avoided which may otherwise take away available energy resources for sugar accumulation.
Results
Suppression Subtractive Hybridization (SSH) was utilized to generate an enriched and equalized cDNA library for leaf expressed genes from sugar beet. Fourteen cDNA fragments corresponding to thirteen different genes were isolated. Northern blot analysis indicates the desired tissue specificity of these genes. The promoters for two chlorophyll a/b binding protein genes (Bvcab11 and Bvcab12) were isolated, linked to reporter genes, and transformed into sugar beet using promoter reporter gene fusions. Transient and transgenic analysis indicate that both promoters direct leaf specific gene expression. A bioinformatic analysis revealed that the Bvcab11 promoter is void of G-box like regulatory elements with a palindromic ACGT core sequence. The data indicate that the presence of a G-box element is not a prerequisite for leaf specific and light induced gene expression in sugar beet.
Conclusions
This work shows that SSH can be successfully employed for the identification and subsequent isolation of tissue specific sugar beet promoters. These promoters are shown to drive strong leaf specific gene expression in transgenic sugar beet. The application of these promoters for expressing resistance improving genes against foliar diseases is discussed.
Background
Sugar beet (Beta vulgaris L.) is a biennial plant, a member of the Chenopodiaceae family [1]. In the first year after germination, a rosette of leaves develops while the taproot swells and accumulates sucrose. In the second year, flower initiation is induced after vernalization in the preceding winter. Beets are harvested at the end of the first year when sugar content of the taproot is high. Transgenic approaches towards modification of specific traits comprise the increase of pathogen resistance, the increase of sugar content and the improvement of sugar storage. These approaches require promoters that direct gene expression in a timely and spatial manner which is determined by the desired expression profile of the transgene.
In many cases improvement of transgenic traits in plants were achieved by using specific promoters [2,3]. Furthermore, to accomplish high tissue specific protein production in transgenic plants, often promoters from photosynthetic or storage specific genes are employed [4,5].
For the identification of desired promoters, a subtractive approach to enrich differentially expressed genes or a large scale approach to identify these genes in cDNA libraries may be employed prior to promoter isolation. One possible way to identify nonredundant clones in a cDNA library is the method of oligonucleotide fingerprinting (ofp) which was recently applied to sugar beet [6]. With this approach different cDNAs can be identified on a large scale basis within a cDNA library. While a large scale ofp approach is a feasible method to identify differentially expressed genes in different cDNA libraries, this method is very cost intensive and hence not applicable for many research groups.
A straight forward approach for the isolation of differentially expressed genes was achieved by the "Suppression Subtractive Hybridization" method (SSH) [7]. SSH is a cDNA- and a PCR-based technique that includes a step for the equalization of the abundance of different cDNA fragments during subtractive hybridization. Combined with suppression PCR, selective amplification of differentially expressed cDNA sequences was achieved without the application of physical separation methods [7]. This method was recently applied to isolate taproot expressed genes from sugar beet [8].
Here we have employed SSH for the isolation of leaf expressed sugar beet genes. Among the genes isolated was a cDNA fragment for a light-harvesting chlorophyll a/b binding protein (CAB). It is shown that sugar beet genotypes harbor either one or two cab genes that are both expressed. To investigate the use of the cab promoters for gene expression, the 5' regulatory sequences were isolated and linked to reporter genes. Transient reporter gene assays indicate that both promoters are expressed in sugar beet leaves. In transgenic sugar beet both promoters are expressed in green tissue. Sequence analysis revealed that the cab11 promoter, in contrast to the cab12 promoter, is void of G-box like regulatory sites with a palindromic ACGT core sequence. A leaf specific promoter in transgenic sugar beets can be employed for biotechnological applications.
Results
Identification of leaf expressed genes from sugar beet
To isolate cDNA fragments corresponding to leaf expressed genes, poly(A)+RNAs from leaf and taproot were isolated and subjected to cDNA synthesis. Suppression subtractive hybridization was performed as described in Methods. A total of 23 cDNA clones specific for the subtraction for leaf expressed genes were isolated and sequenced (data not shown). Fourteen out of 23 cDNAs were found to be different. Table 1 shows that 10 out of these 14 clones have homology mainly to nuclear encoded genes involved in the calvin cycle and in photosynthesis. Fragments L6 and L11 detect homology to the same gene but do not overlap. Four clones do not have any sequence homology to a known gene.
Table 1.
Homology of cDNA fragments enriched for leaf expressed genes.
| cDNA-fragment | Base pairs | Nearest homologa |
| L1 | 862 | Flaveria pringlei mRNA for glycine hydroxymethyltransferase (371 bp, 84 %) Acc. Z25859; [42] |
| L2 | 479 | Nicotiana tabacum mRNA for light-harvesting Chl a/b binding protein (337 bp, 86 %) Acc. X82497 |
| L3 | 424 | Spinacia oleracea psaH mRNA for photosystem I reaction centre subunit VI (361 bp, 90 %) Acc. X16858 [43] |
| L4 | 508 | unknown |
| L5 | 587 | Spinacia oleraceae ALDCHL mRNA for fructose-1,6-bisphosphate aldolase (495 bp, 87 %) Acc. X66814 [44] |
| L6 | 383 | Beta vulgaris clone RUB109UNI ribulose bisphosphate carboxylase, small subunit gene, partial sequence (139 bp, 98 %) Acc. AF173667 [45] |
| L7 | 451 | unknown |
| L8 | 221 | Arabidopsis thaliana mRNA for hydroxypyruvate reductase, complete cds (203 bp, 83 %) Acc. D85339 |
| L9 | 520 | Spinacia oleracea, ferredoxin-thioredoxin reductase A2 mRNA (151 bp, 82 %) Acc. X77163 [46] |
| L10 | 643 | unknown |
| L11 | 338 | Beta vulgaris clone RUB109UNI ribulose bisphosphate carboxylase, small subunit gene, partial sequence (206 bp, 93 %) Acc. AF173667 [45] |
| L12 | 532 | unknown |
| L13 | 265 | Spinacia oleraceae rubisco activase mRNA, complete cds (256 bp, 89 %) Acc. J03610 [47] |
| L14 | 540 | Spinacia oleraceae psaL mRNA for subunit XI of photosystem I reaction center (409 bp, 88 %) Acc. X64445 [48] |
aThe genes that gave the highest homology in a BLASTN gene bank comparison are listed. In parenthesis, the contiguous stretch of base pairs showing the indicated percent homology is given.
To further confirm that the enriched cDNA fragments correspond to tissue specific genes, Northern blot hybridizations to total RNA isolated from leaves (L), taproots (R), stems (S), and inflorescences (I) were performed with 7 cDNA fragments. Figure 1 shows that all but one of the cDNA fragments hybridize specifically or much stronger to RNA from leaves than to RNA from taproots. Clone L5 hybridizes with similar intensity to RNA from all four tissues.
Figure 1.
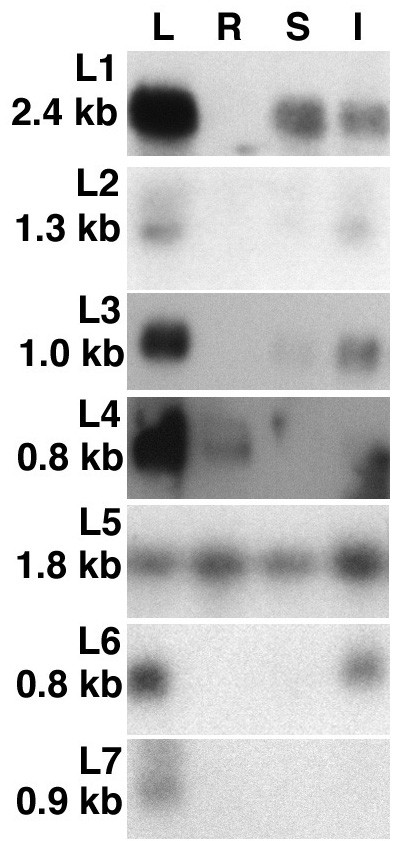
Northern blot analysis with cDNA fragments enriched for leaf expressed genes. Ten micrograms total RNA from leaves (L), taproots (R), stems (S), and inflorescences (I) were hybridized with the indicated cDNA fragments (Table 1). The sizes of the hybridizing transcripts are given in kilo base pairs (kb). The RNA gels were stained prior to blotting to confirm equal loading (data not shown).
The expression profile of the gene for SSH fragment L2 encoding a chlorophyll a/b binding protein (CAB) was analysed in more detail. RNA from different tissues and from different developmental stages were used in RNA blot experiments. RNA was isolated from different organs of sugar beets from a field 4, 6, 10, 12, 16 and 19 weeks after sowing. This interval covers the lifetime of a sugar beet in central Europe. Figure 2 shows that the gene is expressed only in above ground tissue like petiole, sink and source leaf regardless of the developmental stage. Since no expression is observed in below ground tissue like primary and lateral root the promoter of the cab gene was isolated and analysed with reporter gene fusions. Compared to the other genes, the expression profile of the gene for SSH fragment L2 showed the strongest tissue specificity during the different time points analysed (data not shown).
Figure 2.
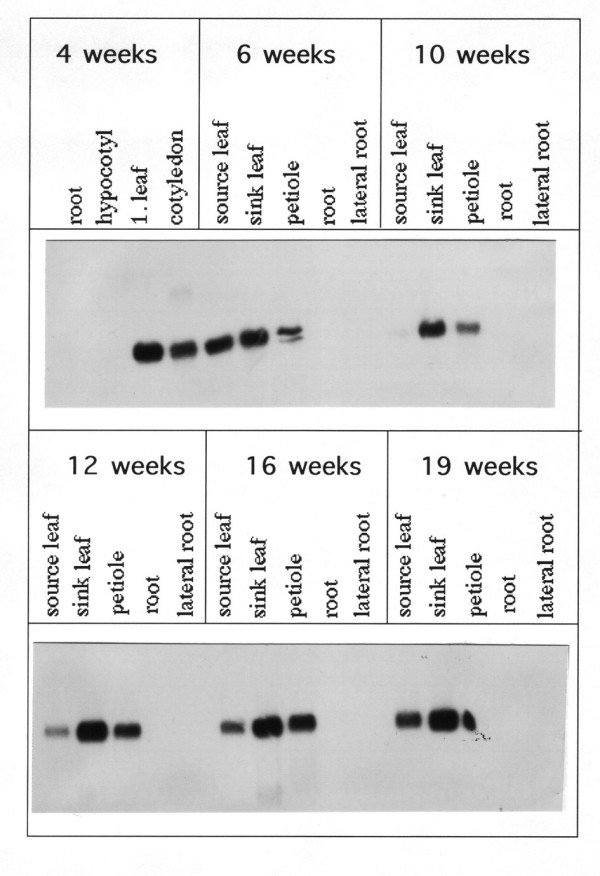
Organ specific expression of the cab gene in the above ground parts of sugar beet during development. RNA from root, lateral root, hypocotyl, cotyledon, petiole, sink and source leaf of sugar beets grown in the field was isolated 4, 6, 10, 12 16 and 19 weeks after sowing. Ten micrograms total RNA were separated in each lane and hybridized with the SSH fragment L2 (Table 1). The RNA gels were stained prior to blotting to confirm equal loading (data not shown).
Isolation of two promoters for the light-harvesting chlorophyll a/b binding protein
For the isolation of a sugar beet promoter corresponding to the cab gene, a complete cDNA clone was isolated (GenBank Acc. Nr. AJ579711, see Methods). A homology search with the encoded 252 amino acid long protein reveals a 87% identity to the cab11 and 85% identity to the cab12 gene from tomato encoding chlorophyll a/b binding protein [9].
Prior to the isolation of genomic clones, a gene copy number analysis was performed. DNA from sugar beet genotypes 1K0088 and 4B5421 was digested with different restriction enzymes and hybridized to the L2 fragment (Table 1). Figure 3 shows that in genomic DNA from genotype 1K0088 three (EcoRI, HindIII) and four (PstI) hybridizing fragments are detected, while only one (EcoRI) or two (PstI, HindIII) fragments are detected in genotype 4B5421. From this result it is concluded that genotype 4B5421 harbors one and genotype 1K0088 harbors two copies of the cab gene.
Figure 3.
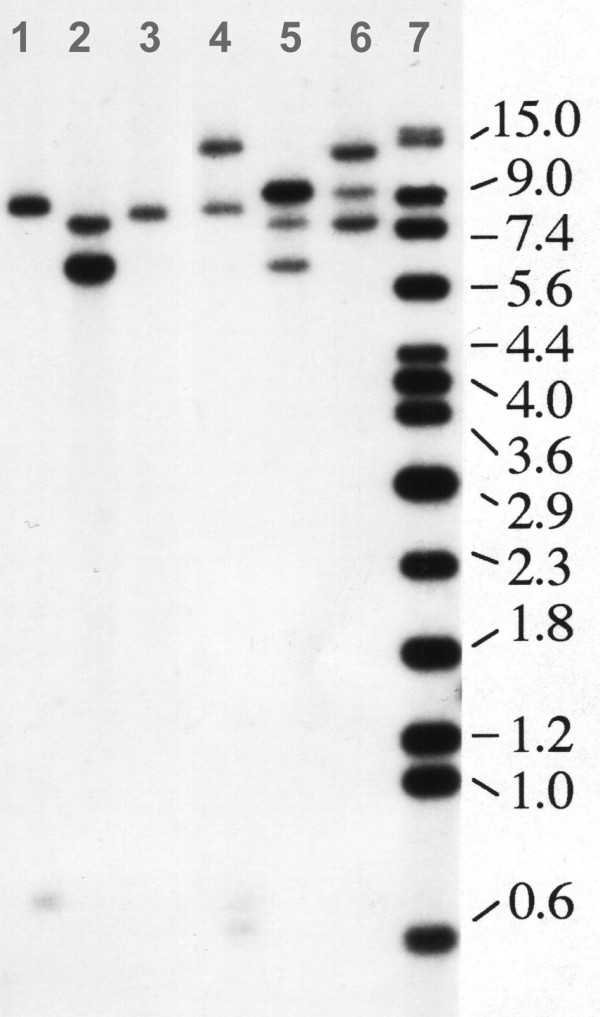
The cab gene is encoded by one or two copies in different sugar beet genotypes. Ten micrograms genomic DNA of genotype 4B5421 (lanes 1–3) and genotype 1K0088 (lanes 4–6) were restricted with different enzymes and hybridized with SSH fragment L2 (Table 1). Lane 1: PstI, lane 2: HindIII, lane 3: EcoRI, lane 4: PstI, lane 5: HindIII, lane 6: EcoRI, lane 7: size standard in kilo base pairs.
Genomic clones for the two different genes were isolated (Methods). Sequence comparison between the cDNA and both genomic clones indicate a very high degree of sequence identity within the coding region. The CAB11 and CAB12 amino acid sequence differ only in one position (data not shown).
From both genes the promoter regions were subcloned into plasmid vectors and sequenced (Methods). Sequence of 1148 and 3049 base pairs, respectively, containing most of the upstream region were deposited to GenBank (Acc. Nr. AX449166 and AX449167). The 1148 bp promoter fragment is designated cab11 promoter and the 3049 bp fragment cab12 promoter. Both fragments harbor 51 base pairs coding region of the CAB protein and 113 (cab11) and 70 (cab12) base pairs upstream untranslated sequence. Upstream of the untranslated region only about 300 bp are homologous between the two promoters while the rest of the sequence is highly divergent (data not shown). Because the cDNA clone isolated before originates from the genotype 4B5421 and corresponds to the cab11 gene, it was investigated if the second gene is also transcribed. Towards these ends 5' RACE amplifications were performed with RNA from genotype 1K0088 and sequenced. This analysis revealed that the cab12 gene is also transcribed (data not shown).
Transient expression assays in sugar beet leaves
To test whether the isolated promoters confer reporter gene expression in sugar beet leaves, a transient assay was performed. Towards these ends translational fusions of promoter fragments with the luciferase gene from Photinus pyralis in the vector pGEM-luc were constructed (see Methods). Table 2 summarizes the relative gene expression strength obtained with the different promoter reporter gene constructs when transformed into sugar beet leaves. Relative to the CaMV 35S promoter, the strength of all promoter fragments is approximately 20% and no significant drop in gene expression is observed between the largest and smallest promoter fragments. In summary, it can be concluded that the 1097 bp long fragment from the cab11 gene and the 342 bp fragment from the cab12 gene harbor all cis-regulatory sequences required for gene expression in sugar beet leaves at least under the conditions of the transient bioassay.
Table 2.
Transient gene expression analysis of two CAB promoters in sugar beet leaves. The plasmids harbor promoter fragments of cab11 (pC1L) and cab12 (pC2L).
| Promoter Reporter Gene Constructa | Relative expression level in sugar beet leavesb |
| pC1L-7126 | 7.0 +/- 3.4c |
| pC1L-1097 | 9.6 +/- 4.6 |
| pC2L-2998 | 8.0 +/- 0.2 |
| pC2L-1827 | 4.5 +/- 0 |
| pC2L-989 | 6.4 +/- 2.4 |
| pC2L-342 | 6.5 +/- 1.9 |
| CaMV 35S | 30.2 +/- 16.6 |
aThe number following the identifier of the promoter relates to fragment length
bThe relative expression level was calculated as:
(Photinus value(construct) - Photinus value (without DNA) / Renilla value (construct) - Renilla value (without DNA)) × 100.
cAverage of at least three assays +/- standard deviation
The promoter of two chlorophyll a/b binding protein genes confers leaf specific and light inducible gene expression in transgenic sugar beet plants
To investigate if the cab11 and cab12 promoters from sugar beet drive reporter gene expression in transgenic plants, promoter reporter gene fusions were introduced via Agrobacterium mediated transformation in sugar beet (see Methods). The length of the cab11 promoter fragment is 1097 bp and the length of the cab12 promoter fragment 2998 bp. Leaves of transgenic lines transferred to the greenhouse were analysed for reporter gene activity. The cab11 promoter of 12 independent transformants showed a specific β-glucuronidase (GUS) activity from 9 to 40599 pmol Mu × min-1 × mg-1, respectively (Fig. 4A). The cab12 promoter of 4 transformants showed a specific activity from 223 to 11656 pmol Mu × min-1 × mg-1, respectively (Fig. 4B). These results indicate that the 1097 bp cab11 promoter fragment and the 2998 bp cab12 promoter fragment are sufficient to confer promoter activity to transgenic sugar beet leaves. Furthermore, the cab11 promoter seems to be stronger than the cab12 promoter although more transgenic lines were analysed for cab11 than for cab12.
Figure 4.
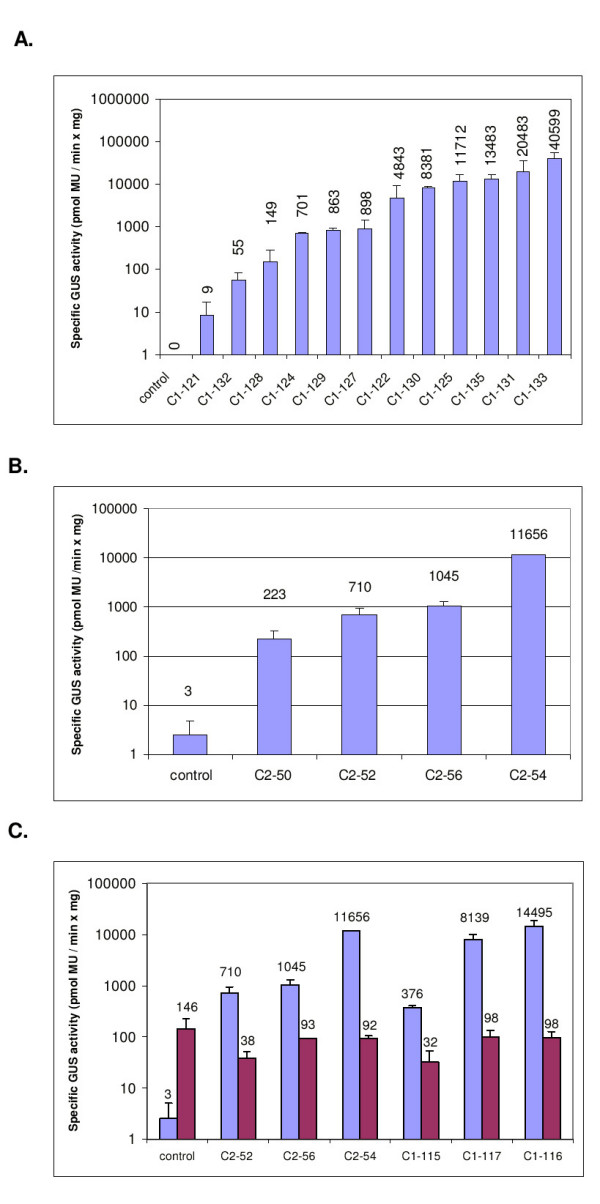
GUS activity of cab11 and cab12 promoter reporter gene constructs in leaves and roots of transgenic sugar beets. GUS activity was assayed using total protein prepared from the leaf or root. Each bar represents the activity of an individual transformant. The data are presented as the mean of three individual plants per transformant. The error bar describes the standard deviation of the mean. A. Activity of the cab11 promoter in leaves of transgenic sugar beet plants. B. Activity of the cab12 promoter in leaves of transgenic sugar beet plants. C. Comparison of the activity of the cab11 and cab12 promoter in leaves and roots of sugar beet. The GUS activity of the leaf extracts is shown by the left column and the activity of the root extracts by the right column.
In order to analyse if the cab11 and cab12 promoters confer tissue specific expression to sugar beet, the roots of three transgenic cab11 and three cab12 promoter lines were analysed. According to the strength of the cab11 and cab12 promoters in leaves (Fig. 4A and 4B) transgenic lines were selected which show low, moderate or high GUS activity in leaves. None of the lines showed GUS activity in the roots which was above the background level of nontransgenic control plants (Fig. 4C). Therefore the promoter activity of the cab11 and cab12 regulatory element is restricted to the above ground tissue of sugar beet and absent in roots. This result is consistent with the observations that transcripts of the cab genes are not detectable in the below ground tissue by Northern blot hybridization (Fig. 2).
To analyse cab11 and cab12 gene regulation in response to light, the reporter gene activtity of etiolated and green transgenic sugar beets was compared. In vitro shoots of the transgenic lines C1-121, C1-122, C2-50 and C2-52 were etiolated for 40 days in the dark. The GUS activity of the etiolated leaflets of one half of the plants was determined. Seven days after illumination the GUS activity of the remaining plants was determined after greening of the leaves. In two independent experiments the cab11 and cab12 promoter plants showed a 4,3 to 8,3 fold and a 95 to 118 fold induction during greening, respectively (Table 3). Although the GUS activity of the etiolated plants was comparable in the two assays, the greening plants showed a much stronger reporter gene activity in the second experiment. Apparently, different time points during the differentiation of etioplasts to chloroplasts were analysed. Time after illumination seems to have a strong influence on the level of promoter induction. Furthermore, gene expression is much stronger in these experiments compared to the analysis in transgenic plants (Table 3, Fig. 4). Finally, these results show that the cab11 and cab12 promoter are activated during the light induced plastid development.
Table 3.
Induction of two cab promoters seven days after illumination of etiolated sugar beet plants. The results for two cab11 (pC1) and two cab12 (pC2) promoter lines are shown.
| Experiment 1 | Experiment 2 | |||||
| Etiolated leaves, specific activitya | Green leaves, specific activity | Light induction | Etiolated leaves, specific activity | Green leaves, specific activity | Light induction | |
| non- transgenic | 10 | 0 | 0 | 16 | 45 | 2,8 |
| C1-121 | 155 | 1287 | 8,3 | 42 | 4126 | 98 |
| C1-122 | 36 | 153 | 4,3 | 50 | 5611 | 112 |
| C2-50 | 30 | 181 | 6 | 44 | 5207 | 118,4 |
| C2-52 | n.db | n.d | n.d | 43 | 4107 | 95,5 |
apmol Mu × min-1 × mg-1
bnot determined
The cab11 promoter lacks G-box elements with a palindromic ACGT core sequence
Gene expression is mainly regulated by the binding of transcription factors to specific cis-regulatory elements. Because the two sugar beet promoters show a similar expression profile, it was investigated if there are any differences or similarities in the composition of cis-regulatory sequences in both promoters. Towards these ends, a database-assisted approach was employed [10]. The Patch™ program [11] was used to inspect both promoter sequences for the occurence of plant transcription factor (TF) binding sites that are annotated to the TRANSFAC® database [11]. The results reveal a large number of putative TF binding sites in both promoters (data not shown). Upon closer inspection of the results it was striking that the cab11 promoter, in contrast to the cab12 promoter was completely void of putative G-box like binding sites that contain a conserved ACGT core sequence and are recognized by bZIP transcription factors. Using the program Patch™ and entering a lower score boundary of 100% to detect only experimentally verified binding sites, only two putative binding sites for bZIP transcription factors were found in the cab11 promoter. Figure 5 shows the positions of these two motifs that both lack the ACGT core sequence. The sequence motifs at position -749 and -489 relative to the translation start site were found in the rice glutelin-B1 promoter and are bound by the rice bZIP transcription factor family RISBZ [12]. Both sites were also found to be bound by the tobacco bZIP transcription factor TGA1a in a pea lectin promoter [13].
Figure 5.
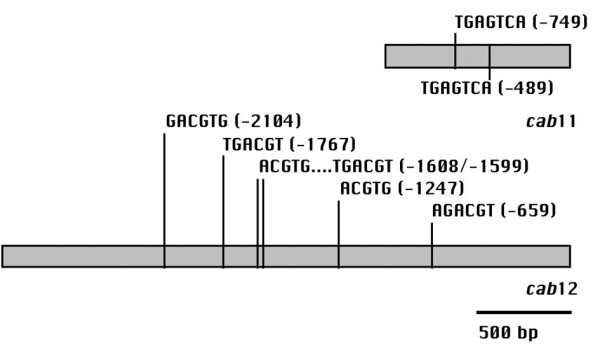
Schematic representation of the cab11 and cab 12 promoters with putative G-box elements. Positions of putative G-box like elements are given relative to the translation start site in the cab11 and cab12 promoters. All binding sites except the single one at position -489 in the cab11 promoter are detected in the upper strand.
Inspecting the sequence of the cab12 promoter for the ACGT core sequence of bZIP factor binding sites reveals 12 positions for this motif (data not shown). Using the program Patch™ six experimentally verified binding sites for bZIP factors were detected among these twelve sites that harbor the ACGT core in the cab12 promoter (Fig. 5). The motif at position -2104 is also present in the glutathione-S transferase 6 gene promoter of Arabidopsis where it is bound by the factor OBF4 [14]. The same site and the sites at position -1608 and -1247 occur in the embryonic abundant protein 1 promoter of rice and are recognized by the factors OSBZ8 and TRAB1 [15-17]. The sites at position -1767 and -1599 were recognized as bZIP binding sites in many other systems. The sequence TGACGT is part of the as-1 element of the CaMV 35S promoter that was shown to be bound by tobacco TGA1a, TGA1b, and TGA2.2 [18,19]. The site at position -659 is also present in the CaMV 35S promoter where it was shown to be bound by the wheat nuclear factor HBP-1 [20].
The observation that the cab11 promoter lacks G-box like elements with a conserved ACGT core sequence indicates that such sites are not required for leaf specific gene expression.
Discussion
The chlorophyll a/b binding proteins CAB11 and CAB12 from sugar beet belong to the light harvesting complex I – 730 (LHCI-730)
Subtractive hybridization was used to isolate leaf expressed genes from sugar beet. The goal was the identification of a promoter that drives leaf specific gene expression in transgenic sugar beet plants. Among seven analysed genes a cDNA fragment corresponding to a chlorophyll a/b binding protein gene was shown by RNA gel blot hybridization to be highly specific for green tissue (Fig. 1 and 2). Genomic DNA blot hybridizations indicate that the two sugar beet genotypes investigated harbor either one or two copies of the gene designated Bvcab11 and Bvcab12 (Fig. 3). A complete cDNA for the gene from genotype 4B5421 was isolated and encodes a protein of 252 amino acids that shows the highest homology (87%) to the cab11 gene from the light harvesting complex I (LHCI) in tomato [9]. This and homologies to other LHCI proteins indicate that the sugar beet gene belongs to the type IV LHCI complex [21]. Further support for this classification comes from the observation that the intron positions between cab11 from tomato and Bvcab11 from sugar beet are identical (data not shown).
LHCI can be subdivided into at least two different chlorophyll-protein complexes, one of which appears to be responsible for the 730 nm fluorescence of PSI (LHCI-730) and the other complex (LHCI-680) fluoresces at lower wavelength [21]. In barley the LHCI-730 complex was isolated as a heterodimer composed of the type I and type IV polypeptides [22]. Furthermore, tomato type I and type IV LHCI polypeptides (Lhca1/cab6a and Lhca4/cab11) expressed in E. coli form a heterodimer in vitro that closely resembles the native LHCI-730 dimer from tomato leaves [23]. Therefore, the sugar beet CAB11 and CAB12 proteins may be part of the LHCI-730 complex.
G-box like elements are not a prerequisite for leaf specific gene expression
The promoters for both sugar beet cab genes were isolated and linked to reporter genes. Transient gene expression studies in sugar beet indicated that 1097 bp upstream of the ATG from the Bvcab11 gene and 342 bp upstream of the ATG from the Bvcab12 gene are sufficient for leaf specific gene expression in sugar beet (Table 2).
Promoter reporter gene constructs for Bvcab11 and Bvcab12 were stably transformed into sugar beet (Beta vulgaris, var. VRB). In sugar beet both promoters are expressed in leaves (Fig. 4).
When the promoter sequences of both cab genes where analysed for putative transcription factor binding sites, a striking difference was observed. The Bvcab11 promoter lacks G-box like sequences with a palindromic ACGT core. Are G-boxes required for light or leaf specific gene expression? A 268 bp fragment of the wheat cab-1 promoter functions as a light responsive and organ specific enhancer in transgenic tobacco [24]. Most notably the three regions that interact with nuclear factors and that were able to enhance gene expression of a 90 bp CaMV 35S minimal promoter did not contain a G-box sequence [24]. The requirement of G-box sequences for light specific gene expression has also been analysed directly [25]. A trimer of the G-box motif found in the spinach ribulose-1,5-bisphosphate carboxylase small subunit-1 promoter was fused to a 90 bp CaMV 35S minimal promoter. While a mutant of this G-box did not confer gene expression to the minimal promoter in the dark and under different light conditions, the G-box increased reporter gene expression under these conditions [25]. Reporter gene expression in the dark was comparably higher than under different light conditions. This is similar to the finding that a G-box like sequence in the cab1R gene of rice is necessary for high level transient expression of a reporter gene in tobacco leaf tissue [26]. Taken together, this indicates that the presence of G-box sequences may have a quantitative effect but may not be a prerequisite for green tissue specific gene expression in sugar beet.
Biotechnological applications of leaf specific promoters in sugar beet
The major goal of this work was the isolation of a strong leaf specific sugar beet promoter that can be used for biotechnological applications. Disease control is one of the most important goals for biotechnological approaches towards improving sugar beet performance. There are many leaf spot diseases that are detrimental to the plant. For example, Cercospora leaf spot is one of the most widespread and destructive foliar diseases of sugar beet [27]. Expressing resistance improving genes in a strong and specific manner against pathogens causing foliar diseases may require a strong leaf specific promoter. With such a promoter, expression in taproots can be avoided which may otherwise take away available energy resources for sugar accumulation.
The work here shows that two promoters, Bvcab11 and Bvcab12, have been isolated that drive highly leaf specific gene expression in sugar beet (Fig. 4). No expression above background levels was detected for both promoters in sugar beet roots (Fig. 4C).
Based on the expression strength in transgenic plants, the Bvcab11 promoter may be suitable for biotechnological applications because it achieves a reporter gene activity comparable to the strong CaMV 35S promoter. CaMV 35S-mediated GUS activities in transgenic tobacco plants were reported as 113000 U (average of 10 plants, [28]) and 9000 U (average of 15 plants, [29]) in which 1 Unit refers to pmol 4-Mu produced min-1 × mg protein-1 [30].
The highest level of cab11 derived GUS expression is 40599 pmol Mu x min-1 × mg-1 which is comparable with the expression strength of the strong CaMV 35S promoter in tobacco.
Conclusions
In summary, this work presents the isolation and expression analysis of two cab promoters from sugar beet. Particularly, the Bvcab11 promoter may be useful to drive strong and specific gene expression in transgenic host plants. The lack of bZIP binding sites harboring the ACGT core sequence could also be advantageous for transient analysis of bZIP transcription factors when using a Bvcab11 reporter gene construct as a transformation control. Furthermore these promoters may be useful to express resistance improving genes against foliar diseases such as Cercospora leaf spot.
Methods
Preparation of RNA and genomic DNA
Two different methods for RNA preparation were employed. To isolate RNA for cDNA subtraction, the procedure described below was followed. For some of the Northern blot analyses a method described earlier was employed [31].
For RNA isolation plant material was homogenized in liquid nitrogen and resuspended in a solution containing 4 M guanidinthiocyanat, 25 mM Tris-HCl, pH8 und 100 mM β-mercaptoethanol. After centrifugation (4°C, 10 min. at 3300 rcf) nucleic acids in the supernatant were precipitated by addition of 0.03 volume sodium acetate (3 M, pH5) and 0.75 volume ethanol (100%) and incubation over night at -20°C. After centrifugation (4°C, 10000 g, 10 min.) the nucleic acid containing pellet was dissolved in 20 ml 100 mM NaCl, 10 mM EDTA pH8, 50 mM Tris-HCl pH8, and 0.2% SDS. Afterwards, a phenol:chloroform (1:1) and a chloroform:isoamylalcohol (24:1) extraction was performed. The pH of the aqueous solution was adjusted to about 5 with acidic acid and nucleic acids were precipitated by addition of 0.6 volume isopropanol and 0.05 volume 4 M NaCl and incubation for 2 hrs at -20°C. After centrifugation (20–30 min., 10000 g at 4°C) the nucleic acids containing pellet was resuspended in 10 ml H2ODEPC containing 0.1% SDS. Total RNA was precipitated by addition of 0.25 volume 8 M LiCl and incubation for at least 15 hrs at 4°C with subsequent centrifugation for 20 min at 4°C, 10000 g. Total RNA was resuspended in 400 μl H2ODEPC. After ethanol precipitation (addition of 0.1 volume sodium acetate, 3 M pH4.8, and 2.5 volume ethanol) total RNA was resuspended in a volume of 50–100 μl H2ODEPC. The isolation of poly(A)+ RNA was carried out with the Oligotex Kit according to the manufacturers protocol (Qiagen; Hilden, Germany). Measurements of RNA yield and electrophoretic separation on formaldehyde gels were done following standard protocols [[32], modified].
Genomic DNA was isolated from sugar beet genotypes 1K0088 and 4B5421 according to a previous published method [33]. For recombinant DNA work standard techniques were employed [32].
Suppression subtractive hybridization
The synthesis of cDNA was performed using the CLONTECH PCR-Select™ cDNA Subtraction Kit (Heidelberg, Germany). Each synthesis was carried out with 8 μg poly(A)+ RNA from sugar beet isolated either from leaves or taproots. Subtractive hybridization was done following the user manual (PT1117-1) of the CLONTECH PCR-Select™ cDNA Subtraction Kit. After the second PCR the amplified fragments from the forward and the reverse subtraction were cloned into the PCR cloning vector pCR®2.1. For each microgram PCR product approximately 300 recombinant plasmids were obtained. For the cloning of PCR products the Invitrogen T/A Cloning® Kit was employed (Karlsruhe, Germany). Prior to ligation into pCR®2.1 the subtracted PCR cDNA products were subjected to an additional incubation of 1 hour at 72°C with dATP and Taq polymerase (TaKaRa; Gennevilliers, France) to ensure that the majority of the PCR fragments contain "A-overhangs" for an efficient cloning into the T/A cloning vector.
DNA sequence analysis
The inserts of the plasmids were sequenced with fluorescently labeled M13 reverse and forward (-20) primers using the AutoRead Sequencing Kit (Pharmacia) and the Automated Laser Fluorescent A.L.F.™ DNA Sequencer from Pharmacia LKB (Freiburg, Germany). The DNA sequence analysis of the genomic and full-length cDNA clones was done by the custom sequencing service of MWG Biotech AG (Ebersberg, Germany). Sequences were subjected to data bank analysis using the BLAST algorithms [34] and analysed with the PILEUP programme of the GCG Wisconsin Analysis Package. For further promoter analysis the TRANSFAC® database was employed [11]. DNA sequences were also processed and analysed on a Macintosh computer using DNA Strider 1.3 [35] and a PC computer using Vector NTI Suite 8.0 (Informax).
Southern and Northern blot hybridizations
Radioactive probes were generated by the method of random hexamer priming with the Amersham Multiprime DNA Labelling System (Freiburg, Germany). Southern and Northern hybridizations were carried out following standard protocols [32,36].
For genomic Southern blot hybridizations 10 μg of DNA from sugar beet genotypes 1K0088 and 4B5421 was digested with different restriction enzymes. Electrophoretic separation, transfer to Hybond nylon membranes (Amersham Pharmacia Biotech, Freiburg), hybridization to radioactive probes, and exposure of the membrane to X-ray films were done according to standard protocols [32]. Radioactive probes were generated by labelling 20 ng of DNA with 50 μCi P32-dATP (6000 Ci/mMol, Amersham Pharmacia Biotech, Freiburg).
Isolation of cDNA and genomic clones
A leaf specific, directional cDNA library from sugar beet genotype 4B5421 was synthesized by the custom cDNA library service of GIBCO BRL (Rockville, USA) and cloned into the plasmid vector pCMV Sport 6.0. Screening of the library was done according to standard protocols [32]. Seven positive cab cDNA clones were identified after screening of 10000 clones using the SSH fragment L2 as a probe (Table 1). The longest cDNA is 1062 bp long, harbors a 114 bp non-translated leader, a 756 bp long reading frame, a 177 bp 3' nontranslated leader, and a 15 bp poly A tail (data not shown, GenBank Acc. Nr. AJ579711).
A genomic library from sugar beet genotype 1K0088 was generated in the lambda vector EMBL3 SP6/T7 and screened using standard protocols [32]. Genomic clones for two different cab loci were isolated.
The promoter for the gene cab11 is present on a ClaI fragment that was subcloned into a plasmid vector and completely sequenced. The fragment is 6294 bp long and contains 51 bp from the coding region of the gene. The resulting plasmid was designated pC1a. Additionally, a 6026 bp large SalI/ClaI fragment was released from the phage clone and subcloned into a Bluescript plasmid and designated pC1b. The promoter for the gene cab12 is present on a PstI fragment that was also subcloned into a plasmid vector and completely sequenced. The fragment is 4002 bp long and the harboring plasmid was designated pC2.
From both genomic clones 1148 and 3049 base pairs containing most of the upstream region were deposited in GenBank (Acc. Nr. AX449166 and AX449167). The 1148 bp promoter fragment is designated cab11 promoter and the 3049 bp fragment cab12 promoter.
Promoter reporter gene constructs
For transient gene expression assays, promoter fragments were linked as translational fusions to the luciferase reporter gene from Photinus pyralis in the reporter gene vector pGEM-luc (Promega, Mannheim). To introduce a plant polyA addition signal into pGEM-luc the respective fragment was isolated from pBI101.3 (Clontech, Heidelberg) by EcoRI digestion, followed by a Klenow fill in reaction and by redigestion with SacI. This released a 260 bp DNA fragment from the nopaline synthase (nos) gene containing the polyA addition signal. To directionally clone this fragment into pGEM-luc, this plasmid was first linearised with SfiI, treated with T4-polymerase to generate blunt ends and subsequently redigested with SacI. After inserting the nos fragment the resulting plasmid was designated pLuc-nos2. To insert the cab11 promoter fragment, a SalI(fill in)-AviII fragment was cloned into the ApaI linearised and T4-polymerase treated plasmid pLuc-nos2. This plasmid harbors 1145 bp from the cab11 promoter including the coding sequence for the first 16 amino acids of the cab11 gene. This plasmid was designated pC1L-1097. In this plasmid the luciferase gene is translationally fused with the first 16 amino acids from the cab11 gene. A second plasmid was generated which harbors additional upstream sequences. Towards these ends a 6099 bp KpnI fragment was released from the plasmid pC1b (see above) and the ends treated with T4-polymerase. The fragment was redigested with NotI and the desired fragment was directionally cloned as a KpnI(blunt end)-NotI fragment upstream of the cab11 fragment in pC1L-1097. To generate compatible ends pC1L-1097 was digested with HindIII treated with T4-polymerase and redigested with NotI. The resulting plasmid was designated pC1L-7126.
To clone the promoter for gene cab12 upstream to the luciferase coding region, the promoter fragment from pC2 was released by NotI/EcoRI digestion and subsequently subjected to a partial digestion with AviII. A 3100 bp long NotI/AviII fragment was purified and subcloned into pLuc-nos2. The plasmid pLuc-nos2 was digested with ApaI, the ends treated with T4-polymerase and redigested with NotI. After ligation the resulting plasmid was designated pC2L-2998. In this plasmid the luciferase gene is translationally fused with the first 16 amino acids from the cab12 gene. To generate 5' promoter deletions pC2L-2998 was (1) digested with KpnI/NotI, T4-polymerase treated, and religated to yield pC2L-1827, (2) digested with SmaI and religated to yield pC2L-989, and (3) digested with NotI and SalI (partial), Klenow polymerase treated, and religated to yield pC2L-342.
For stable transformation the cab11 and cab12 promoters were cloned 5' to the β-glucuronidase gene (uidA). Towards these ends a 1.17 kb HindIII/BamHI fragment was released from pC1L-1097 and cloned into the binary vector pBI101.3 (Clontech, Heidelberg). The resulting plasmid pC1G-1097 harbors a translational fusion between the first 16 amino acids of the cab11 gene and the uidA gene. Similarly, the cab12 promoter was released as a PstI fragment from plamid pC2L-2998, treated with T4-polymerase then digested with BamHI and subcloned into pBI101.3 which was linearised with SalI, ends filled in with Klenow and redigested with BamHI. The resulting plasmid was named pC2G-2998.
Transient and stable gene expression assays
The luciferase expression from plasmids pC1L-1097, pC1L-7126, pC2L-2998, pC2L-1827, pC2L-989, and pC2L-342 were measured in sugar beet leaves after biolistic transformation [37]. For biolistic transformation the PDS-1000/He Particle Delivery System (BioRad, München, Germany) was used. Microcarrier was gold powder type 200-03 (Heraeus, Hanau, Germany) with a diameter of 1.09–2.04 micrometer. The transformation protocol supplied by the manufacturer of the particle delivery system was followed. Equimolar amounts of plamids pC1L-1097 and pC1L-7126 were used. Similarly, equimolar amounts of plamids pC2L-2998, pC2L-1827, pC2L-989, and pC2L-342 were used. To quantify gene expression the transformation control plasmid p70Sruc harboring the luciferase gene from Renilla reniformis under the control of the doupled CaMV 35S promoter was employed as a second reporter gene [38]. For each reporter gene construct three (pC1L-series) or four (pC2L-series) bombardments were made, gene expression strength of both luciferases measured and normalised relative to the luciferase expression of p70Sruc (see below). For each bombardment 13 leaf discs of equal diameter were cut out of sugar beet leaves and preincubated for 6 hours in petri dishes on MS-media containing 0.4 M mannitol at 25°C. The particle delivery conditions were 1550 psi, 9 cm distance and 27 Hg low pressure. After bombardment the petri dishes with the leaf discs were incubated for 16 h at 25°C under constant light. The Photinus and Renilla luciferase activity were measured with the dual-luciferase reporter assay system (Promega, Mannheim, Germany) in a Lumat 9501 luminometer (PE Biosystem) according to the protocol of the supplier.
For the generation of transgenic plants pC1G-1097 and pC2G-2998 were directly transformed into Agrobacterium tumefaciens strain GV2260 [39]. Agrobacterium tumefaciens mediated transformation techniques were performed with the binary T-DNA plasmids on sugar beet (Beta vulgaris, var. VRB) according to [40]. Selection of the transgenic plants was carried out on kanamycin. β-Glucuronidase (GUS) activity in crude leaf extracts was determined as described by Jefferson et al. [41] using 4-methylumbelliferone beta-glucuronide as a substrate. The concentration of the product 4-methylumbelliferone (Mu) was determined with a multiwell fluorescence plate reader (Millipore CytoFluor 2350). Protein content was measured by the method of Bradford (BioRad protein assay kit). Enzyme activity was calculated as pmol Mu × min-1 × mg-1.
Authors' contributions
DS isolated the cDNA and genomic clones, analysed the transcription during different developmental stages and tissues, generated promoter reporter gene constructs, transformed sugar beet, and performed the quantitative reporter gene assays. DUK performed the suppression subtractive hybridization and analysed the homology and tissue specificity of the cDNA fragments. RH identified cis-regulatory elements. RH and DS conceived of the study, and participated in its design and coordination.
Acknowledgments
Acknowledgements
We thank Maike Baumeister, Iris Grocholl, Jeanette Kurrasch, Dorothea Pralle and Corinna Rohlf for excellent technical assistance, Klaus Schmidt for providing the plasmid p70Sruc, and Frank Breuer for helpful suggestions on the manuscript.
Contributor Information
Dietmar J Stahl, Email: d.stahl@kws.de.
Dorothee U Kloos, Email: Dorothee.Kloos@web.de.
Reinhard Hehl, Email: r.hehl@tu-bs.de.
References
- Elliott MC, Weston GD. Biology and physiology of the sugar-beet plant. In: Cooke DA, Scott RK, editor. The sugar beet crop: science into practice. London: Chapman and Hall; 1993. pp. 37–66. [Google Scholar]
- Graham MW, Craig S, Waterhouse PM. Expression patterns of vascular-specific promoters RolC and Sh in transgenic potatoes and their use in engineering PLRV-resistant plants. Plant Mol Biol. 1997;33:729–735. doi: 10.1023/A:1005726918110. [DOI] [PubMed] [Google Scholar]
- Zhang J, Van Toai T, Huynh L, Preiszner J. Development of flooding-tolerant Arabidopsis thaliana by autoregulated cytokinin production. Mol Breed. 2000;6:135–144. doi: 10.1023/A:1009694029297. [DOI] [Google Scholar]
- Outchkourov NS, Peters J, De Jong J, Rademakers W, Jongsma MA. The promoter-terminator of chrysanthemum rbcS1 directs very high expression levels in plants. Planta. 2003;216:1003–1012. doi: 10.1007/s00425-002-0953-8. [DOI] [PubMed] [Google Scholar]
- Jaeger GD, Scheffer S, Jacobs A, Zambre M, Zobell O, Goossens A, Depicker A, Angenon G. Boosting heterologous protein production in transgenic dicotyledonous seeds using Phaseolus vulgaris regulatory sequences. Nat Biotechnol. 2002;20:1265–1268. doi: 10.1038/nbt755. [DOI] [PubMed] [Google Scholar]
- Herwig R, Schulz B, Weisshaar B, Hennig S, Steinfath M, Drungowski M, Stahl D, Wruck W, Menze A, O'Brien J, Lehrach H, Radelof U. Construction of a 'unigene' cDNA clone set by oligonucleotide fingerprinting allows access to 25 000 potential sugar beet genes. Plant J. 2002;32:845–857. doi: 10.1046/j.1365-313X.2002.01457.x. [DOI] [PubMed] [Google Scholar]
- Diatchenko L, Lukyanov S, Lau YF, Siebert PD. Suppression subtractive hybridization: a versatile method for identifying differentially expressed genes. Methods Enzymol. 1999;303:349–380. doi: 10.1016/s0076-6879(99)03022-0. [DOI] [PubMed] [Google Scholar]
- Kloos DU, Oltmanns H, Dock C, Stahl D, Hehl R. Isolation and molecular analysis of six taproot expressed genes from sugar beet. J Exp Bot. 2002;53:1533–1534. doi: 10.1093/jexbot/53.373.1533. [DOI] [PubMed] [Google Scholar]
- Schwartz E, Shen D, Aebersold R, McGrath JM, Pichersky E, Green BR. Nucleotide sequence and chromosomal location of Cab11 and Cab12, the genes for the fourth polypeptide of the photosystem I light-harvesting antenna (LHCI) FEBS Lett. 1991;280:229–234. doi: 10.1016/0014-5793(91)80299-I. [DOI] [PubMed] [Google Scholar]
- Hehl R, Wingender E. Database-assisted promoter analysis. Trends Plant Sci. 2001;6:251–255. doi: 10.1016/S1360-1385(01)01954-9. [DOI] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel A E, Kel-Margoulis OV, Kloos DU, Land S, Lewicki-Potapov B, Michael H, Münch R, Reuter I, Rotert S, Saxel H, Scheer M, Thiele S, Wingender E. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y, Suzuki A, Wu CY, Washida H, Takaiwa F. A rice functional transcriptional activator, RISBZ1, responsible for endosperm-specific expression of storage protein genes through GCN4 motif. J Biol Chem. 2001;276:14139–14152. doi: 10.1074/jbc.M007405200. [DOI] [PubMed] [Google Scholar]
- de Pater S, Katagiri F, Kijne J, Chua NH. bZIP proteins bind to a palindromic sequence without an ACGT core located in a seed-specific element of the pea lectin promoter. Plant J. 1994;6:133–140. doi: 10.1046/j.1365-313X.1994.6020133.x. [DOI] [PubMed] [Google Scholar]
- Chen W, Chao G, Singh KB. The promoter of a H2O2-inducible, Arabidopsis glutathione S-transferase gene contains closely linked OBF- and OBP1-binding sites. Plant J. 1996;10:955–966. doi: 10.1046/j.1365-313X.1996.10060955.x. [DOI] [PubMed] [Google Scholar]
- Hobo T, Asada M, Kowyama Y, Hattori T. ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J. 1999;19:679–689. doi: 10.1046/j.1365-313x.1999.00565.x. [DOI] [PubMed] [Google Scholar]
- Hobo T, Kowyama Y, Hattori T. A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc Natl Acad Sci U S A. 1999;96:15348–15353. doi: 10.1073/pnas.96.26.15348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa H, Ohmiya K, Hattori T. A rice bZIP protein, designated OSBZ8, is rapidly induced by abscisic acid. Plant J. 1996;9:217–227. doi: 10.1046/j.1365-313X.1996.09020217.x. [DOI] [PubMed] [Google Scholar]
- Katagiri F, Lam E, Chua NH. Two tobacco DNA-binding proteins with homology to the nuclear factor CREB. Nature. 1989;340:727–730. doi: 10.1038/340727a0. [DOI] [PubMed] [Google Scholar]
- Niggeweg R, Thurow C, Kegler C, Gatz C. Tobacco transcription factor TGA2.2 is the main component of as-1-binding factor ASF-1 and is involved in salicylic acid- and auxin-inducible expression of as-1-containing target promoters. J Biol Chem. 2000;275:19897–19905. doi: 10.1074/jbc.M909267199. [DOI] [PubMed] [Google Scholar]
- Mikami K, Sakamoto A, Takase H, Tabata T, Iwabuchi M. Wheat nuclear protein HBP-1 binds to the hexameric sequence in the promoter of various plant genes. Nucleic Acids Res. 1989;17:9707–9717. doi: 10.1093/nar/17.23.9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green BR, Pichersky E, Kloppstech K. Chlorophyll a/b-binding proteins: an extended family. Trends Biochem Sci. 1991;16:181–186. doi: 10.1016/0968-0004(91)90072-4. [DOI] [PubMed] [Google Scholar]
- Knoetzel J, Svendsen I, Simpson DJ. Identification of the photosystem I antenna polypeptides in barley. Isolation of three pigment-binding antenna complexes. Eur J Biochem. 1992;206:209–215. doi: 10.1111/j.1432-1033.1992.tb16918.x. [DOI] [PubMed] [Google Scholar]
- Schmid VH, Cammarata KV, Bruns BU, Schmidt GW. In vitro reconstitution of the photosystem I light-harvesting complex LHCI-730: Heterodimerization is required for antenna pigment organization. Proc Natl Acad Sci U S A. 1997;94:7667–7672. doi: 10.1073/pnas.94.14.7667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotor C, Romero LC, Inouye K, Lam E. Analysis of three tissue-specific elements from the wheat Cab-1 enhancer. Plant J. 1993;3:509–518. doi: 10.1046/j.1365-313X.1993.03040509.x. [DOI] [PubMed] [Google Scholar]
- Lübberstedt T, Bolle CEH, Sopory S, Flieger K, Herrmann RG, Oelmüller R. Promoters from genes for plastid proteins possess regions with different sensitivities toward red and blue light. Plant Physiol. 1994;104:997–1006. doi: 10.1104/pp.104.3.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S, Bogorad L. A rice cab gene promoter contains separate cis-acting elements that regulate expression in dicot and monocot plants. Plant Cell. 1992;4:971–981. doi: 10.1105/tpc.4.8.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffus JE, Ruppel EG. Diseases. In: Cooke DA, Scott RK, editor. The sugar beet crop: science into practice. London: Chapman and Hall; 1993. pp. 347–427. [Google Scholar]
- Benfey PN, Ren L, Chua NH. The CaMV 35S enhancer contains at least two domains which can confer different developmental and tissue-specific expression patterns. EMBO J. 1989;8:2195–2202. doi: 10.1002/j.1460-2075.1989.tb08342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger M, Daubert S, Goodman RM. Characteristics of a strong promoter from figwort mosaic virus: comparison with the analogous 35S promoter from cauliflower mosaic virus and the regulated mannopine synthase promoter. Plant Mol Biol. 1990;14:433–443. doi: 10.1007/BF00028779. [DOI] [PubMed] [Google Scholar]
- Gatz C. Chemical control of gene expression. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:89–108. doi: 10.1146/annurev.arplant.48.1.89. [DOI] [PubMed] [Google Scholar]
- Logemann J, Schell J, Willmitzer L. Improved method for the isolation of RNA from plant tissue. Anal Biochem. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York, Greene and Wiley Interscience; 1988. [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci U S A. 1984;81:8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marck C. 'DNA Strider': a 'C' program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1988;16:1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JC. The biolistic process. Trends Biotechnol. 1988;6:299–302. doi: 10.1016/0167-7799(88)90023-6. [DOI] [Google Scholar]
- Schmidt K. Identifizierung eines Cercospora beticola responsiven Bereichs innerhalb des Kernpromotors des pal-Gens der Zuckerrübe sowie eines daran bindenden, neuen Transkriptionsfaktors. Dissertation, Technische Universität Braunschweig, Germany; 2003. [Google Scholar]
- An G. Binary Ti vectors for plant transformation and promoter analysis. Methods Enzymol. 1987;153:292–305. [Google Scholar]
- Lindsey K, Gallois P, Eady C. Plant Tissue Culture Manual B7. Kluwer Academic Publishers; 1991. Regeneration and transformation of sugar beet by Agrobacterium tumefaciens; pp. 1–13. [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopriva S, Bauwe H. Cloning and sequencing of two isoforms of serine hydroxymethyltransferase from Flaveria pringlei (accession nos. Z25859 and Z25860) Plant Physiol. 1998;116:1603. doi: 10.1104/pp.116.4.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steppuhn J, Hermans J, Nechushtai R, Herrmann GS, Herrmann RG. Nucleotide sequences of cDNA clones encoding the entire precursor polypeptide for subunit VI and of the plastome-encoded gene for subunit VII of the photosystem I reaction center from spinach. Curr Genet. 1989;16:99–108. doi: 10.1007/BF00393402. [DOI] [PubMed] [Google Scholar]
- Pelzer-Reith B, Penger A, Schnarrenberger C. Plant aldolase: cDNA and deduced amino-acid sequences of the chloroplast and cytosol enzyme from spinach. Plant Mol Biol. 1993;21:331–340. doi: 10.1007/BF00019948. [DOI] [PubMed] [Google Scholar]
- Schneider K, Borchardt DC, Schäfer-Pregl R, Nagl N, Glass C, Jeppsson A, Gebhardt C, Salamini F. PCR-based cloning and segregation analysis of functional gene homologues in Beta vulgaris. Mol Gen Genet. 1999;262:515–524. doi: 10.1007/s004380051113. [DOI] [PubMed] [Google Scholar]
- Falkenstein E, von Schaewen A, Scheibe R. Full-length cDNA sequences for both ferredoxin-thioredoxin reductase subunits from spinach (Spinacia oleracea L.) Biochim Biophys Acta. 1994;1185:252–254. doi: 10.1016/0005-2728(94)90218-6. [DOI] [PubMed] [Google Scholar]
- Werneke JM, Zielinski RE, Ogren WL. Structure and expression of spinach leaf cDNA encoding ribulosebisphosphate carboxylase/oxygenase activase. Proc Natl Acad Sci U S A. 1988;85:787–791. doi: 10.1073/pnas.85.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flieger K, Oelmüller R, Herrmann RG. Isolation and characterization of cDNA clones encoding a 18.8 kDa polypeptide, the product of the gene psaL, associated with photosystem I reaction center from spinach. Plant Mol Biol. 1993;22:703–709. doi: 10.1007/BF00047411. [DOI] [PubMed] [Google Scholar]


